Abstract
Background
Estrogen is formed by the enzyme aromatase (CYP19A1) and signals via three identified receptors ERα (ESR1), ERß (ESR2), and the G protein-coupled estrogen receptor (GPER). Understanding the relative contribution of each receptor to estrogenic signaling may elucidate the disparate effects of this sex hormone across tissues, and recent developments in PCR technology allow absolute quantification and direct comparison of multiple targets. We hypothesized that this approach would reveal tissue- and sex-specific differences in estrogen receptor mRNA.
Methods
ESR1, ESR2, GPER, and CYP19A1 were measured in four cardiovascular tissues (heart, aorta, kidney, and adrenal gland), three brain areas (somatosensory cortex, hippocampus, and prefrontal cortex), and reproductive tissues (ovaries, mammary gland, uterus, testes) from six male and six female adult Sprague-Dawley rats.
Results
GPER mRNA expression was relatively stable across all tissues in both sexes, ranging from 5.49 to 113 copies/ng RNA, a 21-fold difference. In contrast, ESR1/ESR2 were variable across tissues although similar within an organ system. ESR1 ranged from 4.46 to 614 copies/ng RNA (138-fold difference) while ESR2 ranged from 0.154 to 83.1 copies/ng RNA (540-fold). Significant sex differences were broadly absent except for renal ESR1 (female 206 vs. male 614 copies/ng RNA, P < 0.0001) and GPER (62.0 vs. 30.2 copies/ng RNA, P < 0.05) as well as gonadal GPER (5.49 vs. 47.5 copies/ng RNA, P < 0.01), ESR2 (83.1 vs. 0.299 copies/ng RNA, P < 0.01), and CYP19A1 (322 vs. 7.18 copies/ng RNA, P < 0.01). Cardiovascular tissues showed a predominance of ESR1, followed by GPER. In contrast, GPER was the predominant transcript in the brain with similarly low levels of ESR1 and ESR2. CYP19A1 was detected at very low levels except for reproductive tissues and the hippocampus.
Conclusion
While the data indicates a lack of sex differences in most tissues, significant differences were found in the range of receptor gene expression across tissues as well as in the receptor profile between organ systems. The data provide a guide for future studies by establishing estrogen receptor expression across multiple tissues using absolute PCR quantification. This knowledge on tissue-specific estrogen receptor profiles will aid the development of hormonal therapies that elicit beneficial effects in specific tissues.
Keywords: Estrogen receptors, Aromatase, Droplet digital PCR (ddPCR), Sex differences, Cardiovascular
Background
The nuanced relationship that exists between normal physiology, endogenous estrogens, and menopausal hormone therapy (MHT) is highlighted by discrepancies in both basic and clinical research. The alarming publication of deleterious estrogenic effects associated with MHT in the large randomized clinical trial the Women’s Health Initiative conflict with numerous basic studies performed in animals as well as clinical observational studies, such as the Nurse’s Health Study which demonstrate beneficial and cardioprotective effects of estrogen [1, 2]. While MHT is effective for relieving menopausal symptoms, its impact on cardiovascular health and associated risks remain an area of dispute.
Many questions still surround the interaction of estrogens and their identified receptors (ERs) ERα, ERβ, and more recently the G protein-coupled estrogen receptor (GPER). The precise signaling actions of ERs and associated proteins are most likely dependent on the relative expression levels of all three ERs, or the ER expression profile, at the specific site of action. Tissue-specific and sexually dimorphic ER expression patterns may contribute to the differential effects of estrogen as well as the protective benefits observed in females but not in males. Moreover, ER profiles may aid the development of menopausal therapies that elicit only the desired effects.
Previously existing methodologies for transcript quantification, including quantitative real-time PCR (qPCR) and reverse transcription qPCR (RT-qPCR), estimate results based on an experimentally generated standard curve. This method limits accuracy when directly comparing data between discrete runs and tissue types, because standard curves are not identical across all reactions [3, 4]. Furthermore, levels of popular housekeeping genes including β-actin and GAPDH, which are used as internal controls in RT-qPCR, are not consistent among different tissues [5], leading to difficulty when comparing target gene levels in multiple tissues. In contrast, droplet digital PCR (ddPCR) is an established and validated method that allows direct comparison of multiple target sequences from discrete experiments due to absolute quantification [3, 6–8]. Utilizing ddPCR, the current study established the unique ER expression profile in 10 tissues from adult rats.
Methods
Tissue harvesting
All procedures were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved and monitored by the Tulane Institutional Animal Care and Use Committee. Sprague-Dawley rats (six female, six male) arrived from Envigo at 12–14 weeks of age (RGD Cat# 737903, RRID:RGD_737903). After 1 week of acclimation, tissues were harvested and immediately immersed in 10 μl RNAlater (QIAGEN Cat# AM7020, RRID:SCR_008539) per 1 mg of tissue to preserve RNA integrity. Larger tissues were cut into slices less than 0.5 cm thick in accordance with the manufacturer’s instructions. Tissues were stored for 24 h at 4 °C before being archived at − 20 °C.
Cell culture
Rat embryonic A7r5 aortic smooth muscle cells (ATCC Cat# CRL-1444, RRID:CVCL_0137) were cultured in 10 cm dishes with DMEM (Thermo Fisher Scientific Cat# 11330057, RRID:SCR_008452) containing 10% FBS (Sigma Cat# F6178, RRID:SCR_008988). After reaching approximately 80% confluency, media was replaced with DMEM containing 0.5% charcoal-stripped FBS (Sigma Cat# F6765) for 24 h to reduce to influence of hormones in the serum. Cells were washed, pelleted, resuspended in a small volume of PBS, then mixed with 5–10 volumes of RNAlater, and stored until analysis.
RNA extraction
RNA extraction was accomplished using the RNeasy MiniKit (QIAGEN, Cat# 74106) according to the manufacturer’s protocol. Purity and concentration of RNA was determined by a NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific) using a previously described method [9]. Samples with a 260/280 ratio greater than 1.8 were used in the study.
Droplet digital PCR
ddPCR was accomplished using a previously described method [6, 10]. Briefly, RNA was combined with One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad Laboratories, Cat# 1864021, RRID:SCR_008426) and the following PrimePCR Primers (all from Bio-Rad Laboratories): Gper, Rat (RefSeq: NM_133573, Fluorophore: FAM, Unique Assay ID: dRnoCPE5151056); Esr1, Rat (RefSeq: NM_012689, Fluorophore: HEX, Unique Assay ID: dRnoCPE5176827); Esr2, Rat (RefSeq: NM_012754, Fluorophore: FAM, Unique Assay ID: dRnoCPE5175914); and Cyp19a1, Rat (RefSeq: NM_017085, Fluorophore: HEX, Unique Assay ID: dRnoCPE5174813). Only two fluorescent signals were read for each sample (GPER-FAM/ESR1-HEX), and the same RNA sample was used to read the other two genes on a separate run (ESR2-FAM/CYP19A1-HEX). The reaction mixture was fractionated into more than 10,000 individual 1 nl droplets by oil emulsion microfluidics. Droplets were analyzed via the Bio-Rad QX200 droplet reader and QuantaSoft software and converted to copies/ng RNA based on the RNA concentration and the total volume added to the reaction. Samples were rerun or excluded if they had too many positive or negative droplets (does not satisfy Poisson statistics), Quantasoft Quality Scores below 0.85, or less than 10,000 droplets.
Statistical analysis
GraphPad Prism (RRID:SCR_002798) was utilized for statistical analysis. Sex differences in transcript expression across tissues were analyzed using 2-way ANOVA with factors of sex and tissue followed by Sidak’s multiple comparisons test. ER mRNA expression profiles within each tissue were analyzed via two-way ANOVA with factors of gene and sex followed by Sidak’s multiple comparisons test. Cohen’s d was calculated as the difference between two means divided by the average standard deviation. For tissues found in only one sex (mammary gland and uterus) as well as for cell experiments, significant differences in gene expression were determined using one-way ANOVA and Sidak’s multiple comparison test.
Results
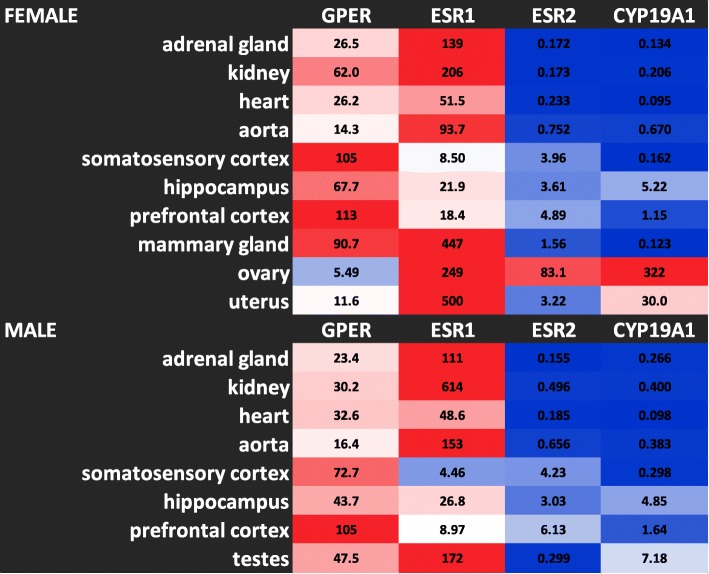
An overview of all experimental data gathered from ddPCR analysis of GPER, ESR1, ESR2, and aromatase from the isolated total RNA of 10 cardiovascular, brain, and reproductive tissues is presented as a heatmap (Fig. 1).
Fig. 1.
Heatmap of ddPCR results for GPER, ESR1, ESR2, and CYP19A1 in 10 tissue types from female and male Sprague-Dawley rats
We next analyzed each gene individually in order to determine the effect of sex on estrogen receptor as well as the enzyme aromatase across all tissues (Fig. 2). Mammary and uterine samples were excluded since they could not be paired across sex. For GPER, there was no main effect of sex but tissue and the interaction were significant (Fig. 2a, Table 1). GPER was significantly lower in the ovary (5.5 ± 1.5 copies/ng RNA) and male kidneys (30 ± 23 copies/ng RNA) in comparison with tissues of the opposite sex (testes 48 ± 16 copies/ng RNA and female kidneys 62 ± 36 copies/ng RNA). A main effect was found for sex, tissue, and the interaction when analyzing expression of ESR1, ESR2, and CYP19A1 (Fig. 2b–2d, Table 1). Opposite of the sex difference in renal GPER, renal ESR1 was significantly greater in males. Also opposite to the predominance of GPER in the testes, ESR2, and CYP19A1 expression was significantly greater in ovaries.
Fig. 2.
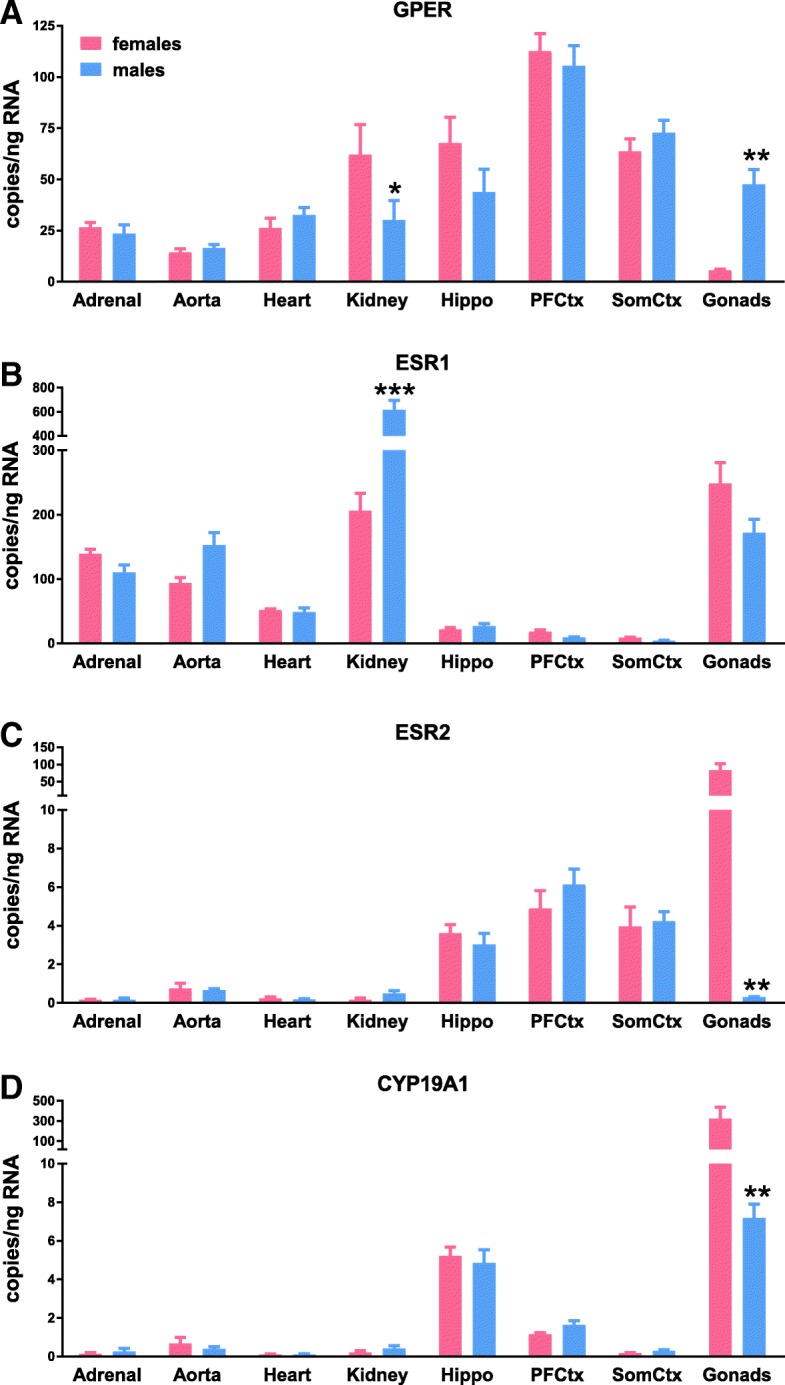
Expression of each gene was compared using two-way ANOVA (sex × tissue) with Sidak’s multiple comparisons test, *P < 0.05, **P < 0.01, and ***P < 0.001 vs. females. a GPER, b ESR1, c ESR2, and d CYP19A1
Table 1.
Statistical analysis of data presented in Fig. 2. Post hoc results are only shown for tests which reached statistical significance (P < 0.05)
| DF | eta squared Cohen’s d | F (DFn, DFd) t | P value | |
|---|---|---|---|---|
| GPER (two-way) | ||||
| Interaction | 7 | 8.56 | F (7, 77) = 4.08 | P = 0.0007 |
| Tissue | 7 | 67.1 | F (7, 77) = 32.0 | P < 0.0001 |
| Sex | 1 | 0.0137 | F (1, 77) = 0.0459 | P = 0.8310 |
| >Kidney F vs. M | 77 | 1.07 | t = 2.96 | P = 0.0327 |
| >Gonads F vs. M | 77 | − 4.75 | t = 3.72 | P = 0.0030 |
| ESR1 (two-way) | ||||
| Interaction | 7 | 20.2 | F (7, 79) = 19.2 | P < 0.0001 |
| Tissue | 7 | 65.5 | F (7, 79) = 62.3 | P < 0.0001 |
| Sex | 1 | 1.91 | F (1, 79) = 12.7 | P = 0.0006 |
| >Kidney F vs. M | 79 | − 3.08 | t = 11.8 | P < 0.0001 |
| ESR2 (two-way) | ||||
| Interaction | 7 | 37.4 | F (7, 78) = 18.6 | P < 0.0001 |
| Tissue | 7 | 34.8 | F (7, 78) = 17.3 | P < 0.0001 |
| Sex | 1 | 5.07 | F (1, 78) = 17.6 | P < 0.0001 |
| >Gonads F vs. M | 78 | 3.57 | t = 12.2 | P = 0.0030 |
| CYP19A1 (two-way) | ||||
| Interaction | 7 | 26.3 | F (7, 78) = 7.06 | P < 0.0001 |
| Tissue | 7 | 28.4 | F (7, 78) = 7.62 | P < 0.0001 |
| Sex | 1 | 3.67 | F (1, 78) = 6.89 | P = 0.0104 |
| >Gonads F vs. M | 78 | 2.19 | t = 12.2 | P = 0.0030 |
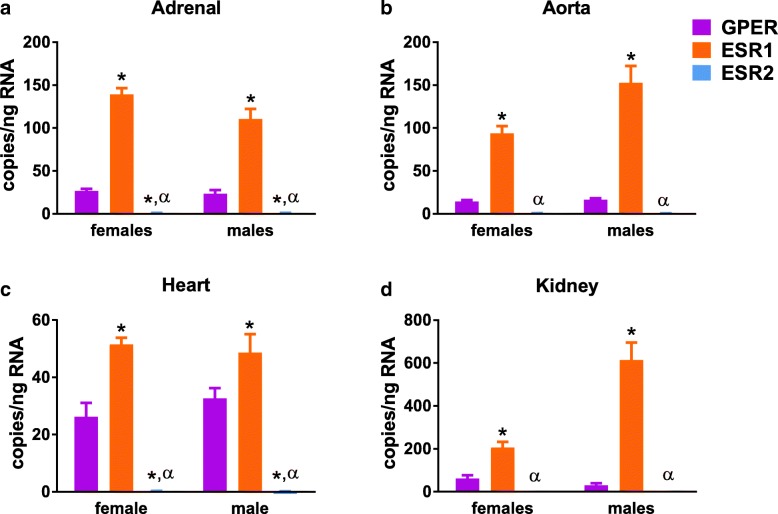
Next, we determined the estrogen receptor profile for each tissue, grouped by function. Due to nearly undetectable levels of CYP19A1 except in reproductive tissues, this gene was not included in the analysis of cardiovascular or brain ER profiles. Cardiovascular tissues (adrenal, aorta, heart, and kidney) showed a predominance of ESR1, followed by GPER (Fig. 3 and Table 2). ESR2 had near undetectable levels in all cardiovascular tissues analyzed. In the adrenal gland and heart, but not the aorta or kidney, both male and female animals had significantly more ESR1 relative to GPER and ESR2. There was a significant effect of sex on gene expression in all tissues excerpt the heart. Higher levels of ESR1 were noted in the adrenal glands of females in comparison with males, but lower ESR1 was found in the female aorta and kidney.
Fig. 3.
Estrogen receptor profiles in cardiovascular tissues. (a) Adrenal gland, (b) Aorta, (c) Heart, and (d) Kidney. Each tissue was compared using two-way ANOVA (sex × gene) with Sidak’s multiple comparisons test, *P < 0.05 vs. GPER, αP < 0.05 vs. ESR1
Table 2.
Statistical analysis of data presented in Fig. 3. Post hoc results are only shown for tests which reached statistical significance (P < 0.05)
| DF | eta squared Cohen’s d | F (DFn, DFd) t | P value | |
|---|---|---|---|---|
| Adrenal (two-way) | ||||
| Interaction | 2 | 1.309 | F (2, 29) = 3.298 | P = 0.0512 |
| Sex | 1 | 0.8823 | F (1, 29) = 4.445 | P = 0.0438 |
| >ESR1: F vs. M | 29 | 1.24 | t = 3.347 | P = 0.0068 |
| gene | 2 | 91.78 | F (2, 29) = 231.2 | P < 0.0001 |
| >F: GPER vs. ESR1 | 29 | − 9.530 | t = 13.15 | P < 0.0001 |
| >F: GPER vs. ESR2 | 29 | 8.580 | t = 3.079 | P = 0.0135 |
| >F: ESR1 vs. ESR2 | 29 | 15.81 | t = 16.23 | P < 0.0001 |
| >M: GPER vs. ESR1 | 29 | − 4.442 | t = 10.17 | P < 0.0001 |
| >M: GPER vs. ESR2 | 29 | 4.313 | t = 2.591 | P = 0.0438 |
| >M: ESR1 vs. ESR2 | 29 | 7.654 | t = 12.29 | P < 0.0001 |
| Aorta (two-way) | ||||
| Interaction | 2 | 5.10 | F (2, 29) = 6.87 | P = 0.0036 |
| Sex | 1 | 2.77 | F (1, 29) = 7.47 | P = 0.0106 |
| >ESR1: F vs. M | 29 | − 4.60 | t = 4.66 | P = 0.0002 |
| Gene | 2 | 81.0 | F (2, 29) = 109 | P < 0.0001 |
| >F: GPER vs. ESR1 | 29 | − 2.72 | t = 6.27 | P < 0.0001 |
| >F: ESR1 vs. ESR2 | 29 | 3.33 | t = 7.34 | P < 0.0001 |
| >M: GPER vs. ESR1 | 29 | − 7.31 | t = 10.8 | P < 0.0001 |
| >M: ESR1 vs. ESR2 | 29 | 8.74 | t = 11.4 | P < 0.0001 |
| Heart (two-way) | ||||
| Interaction | 2 | 0.769 | F (2, 30) = 0.804 | P = 0.457 |
| Sex | 1 | 0.0665 | F (1, 30) = 0.139 | P = 0.712 |
| Gene | 2 | 84.8 | F (2, 30) = 88.7 | P < 0.0001 |
| >F: GPER vs. ESR1 | 30 | − 2.87 | t = 4.76 | P = 0.0001 |
| >F: GPER vs. ESR2 | 30 | 4.29 | t = 4.88 | P < 0.0001 |
| >F: ESR1 vs. ESR2 | 30 | 17.2 | t = 9.64 | P < 0.0001 |
| >M: GPER vs. ESR1 | 30 | − 1.29 | t = 3.01 | P = 0.0157 |
| >M: GPER vs. ESR2 | 30 | 7.12 | t = 6.09 | P < 0.0001 |
| >M: ESR1 vs. ESR2 | 30 | 6.05 | t = 9.10 | P < 0.0001 |
| Kidney | ||||
| Interaction | 2 | 18.6 | F (2, 30) = 0.804 | P < 0.0001 |
| Sex | 1 | 7.30 | F (1, 30) = 0.139 | P = 0.0002 |
| >ESR1: F vs. M | 30 | − 3.08 | t = 8.09 | P < 0.0001 |
| Gene | 2 | 62.3 | F (2, 30) = 88.7 | P < 0.0001 |
| >F: GPER vs. ESR1 | 30 | − 2.81 | t = 4.76 | P = 0.023 |
| >F: ESR1 vs. ESR2 | 30 | 6.18 | t = 9.64 | P = 0.0009 |
| >M: GPER vs. ESR1 | 30 | − 5.25 | t = 3.01 | P < 0.0001 |
| >M: ESR1 vs. ESR2 | 30 | 6.16 | t = 9.10 | P < 0.0001 |
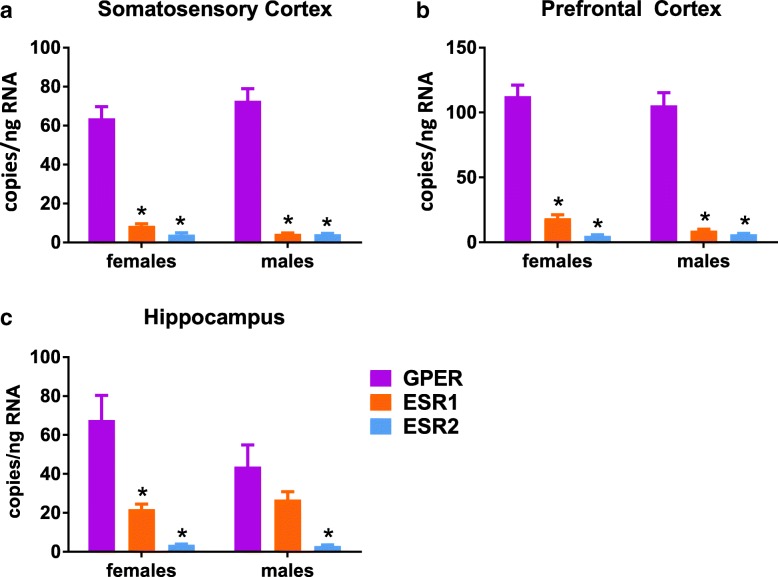
In contrast to data from cardiovascular tissues, GPER was the predominant transcript in the three selected brain regions (prefrontal cortex, somatosensory cortex, and hippocampus; Fig. 4 and Table 3). ESR1 and ESR2 displayed similarly low levels in the brain in both females and males. In addition, there was no main effect of sex in the brain. GPER was expressed significantly higher than ESR1 and ESR2 in female hippocampus, female and male prefrontal cortex, and female somatosensory cortex. In the male hippocampus and somatosensory cortex, GPER was significantly higher than only ESR2.
Fig. 4.
Estrogen receptor profiles in brain tissues. (a) Somatosensory Cortex, (b) Prefrontal Cortex, and (c) Hippocampus. Each tissue was compared using two-way ANOVA (sex × gene) with Sidak’s multiple comparisons test, *P < 0.05 vs. GPER
Table 3.
Statistical analysis of data presented in Fig. 4. Post hoc results are only shown for tests which reached statistical significance (P < 0.05)
| DF | eta squared Cohen’s d | F (DFn, DFd) t | P value | |
|---|---|---|---|---|
| Hippocampus (two-way) | ||||
| Interaction | 2 | 5.06 | F (2, 29) = 2.40 | P = 0.109 |
| Sex | 1 | 1.44 | F (1, 29) = 1.37 | P = 0.252 |
| Gene | 2 | 60.2 | F (2, 29) = 28.5 | P < 0.0001 |
| >F: GPER vs. ESR1 | 29 | 2.45 | t = 4.77 | P = 0.0001 |
| >F: GPER vs. ESR2 | 29 | 4.00 | t = 6.67 | P < 0.0001 |
| >M: GPER vs. ESR2 | 29 | 3.07 | t = 4.03 | P = 0.0011 |
| Prefrontal cortex (two-way) | ||||
| Interaction | 2 | 0.223 | F (2, 30) = 0.517 | P = 0.601 |
| Sex | 1 | 0.276 | F (1, 30) = 1.28 | P = 0.266 |
| Gene | 2 | 93.0 | F (2, 30) = 216 | P < 0.0001 |
| >F: GPER vs. ESR1 | 30 | 6.75 | t = 12.0 | P < 0.0001 |
| >F: GPER vs. ESR2 | 30 | 9.21 | t = 13.8 | P < 0.0001 |
| >M: GPER vs. ESR1 | 30 | 7.05 | t = 12.3 | P < 0.0001 |
| >M: GPER vs. ESR2 | 30 | 7.54 | t = 12.7 | P < 0.0001 |
| Somatosensory cortex (two-way) | ||||
| Interaction | 2 | 0.768 | F (2, 29) = 1.84 | P = 0.177 |
| Sex | 1 | 0.0811 | F (1, 29) = 0.389 | P = 0.538 |
| Gene | 2 | 91.4 | F (2, 29) = 219 | P < 0.0001 |
| >F: GPER vs. ESR1 | 29 | 6.84 | t = 4.767 | P = 0.0001 |
| >F: GPER vs. ESR2 | 29 | 7.52 | t = 6.668 | P < 0.0001 |
| >M: GPER vs. ESR2 | 29 | 8.30 | t = 4.031 | P = 0.0011 |
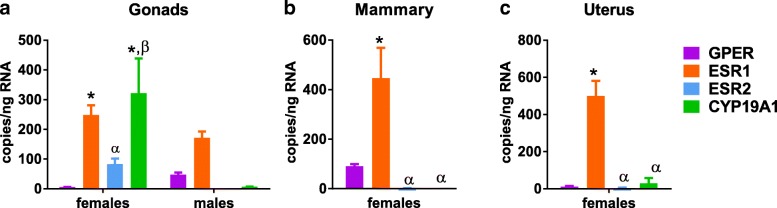
The ER profiles of reproductive tissues (gonads, mammary gland, and uterus) were dominated by significantly greater ESR1 expression relative to GPER and ESR2 (Fig. 5 and Table 4). With the exception of the ovaries and uterus, CYP19A1 expression was nearly absent. In the ovary, CYP19A1 expression was equally as high as ESR1. Data from the testes did not reach the threshold of significance, though ESR1 mRNA tended to be higher than the three other surveyed genes.
Fig. 5.
Estrogen receptor profiles in reproductive tissues. (a) Gonads, (b) Mammary gland, and (c) Uterus. Each tissue was compared using two-way ANOVA (sex × gene) with Sidak’s multiple comparisons test, *P < 0.05 vs. GPER, αP < 0.05 vs. ESR1, ßP < 0.05 vs. ESR2
Table 4.
Statistical analysis of data presented in Fig. 5. Post hoc results are only shown for tests which reached statistical significance (P < 0.05)
| DF | eta squared Cohen’s d | F (DFn, DFd) t | P value | |
|---|---|---|---|---|
| Gonads (two-way) | ||||
| Interaction | 3 | 17.8 | F (3, 38) = 5.33 | P = 0.0037 |
| Sex | 1 | 12.2 | F (1, 38) = 11.0 | P = 0.0020 |
| >CYP19A1: F vs. M | 38 | 2.19 | t = 4.95 | P < 0.0001 |
| gene | 3 | 25.4 | F (3, 38) = 7.62 | P = 0.0004 |
| >F: GPER vs. ESR1 | 38 | − 6.01 | t = 3.82 | P = 0.0029 |
| >F: GPER vs. CYP19A1 | 38 | − 2.20 | t = 4.97 | P < 0.0001 |
| >F: ESR2 vs. CYP19A1 | 38 | − 1.44 | t = 3.75 | P = 0.0035 |
| Mammary (one-way) | ||||
| Gene | 3 | 64.5 | F (3, 20) = 12.12 | P < 0.0001 |
| >F: GPER vs. ESR1 | 20 | − 2.23 | t = 4.13 | P = 0.0031 |
| >M: GPER vs. ESR1 | 20 | 2.98 | t = 5.163 | P = 0.0003 |
| >M: GPER vs. ESR2 | 20 | 3.00 | t = 5.18 | P = 0.0003 |
| Uterus (one-way) | ||||
| Gene | 3 | 92.2 | F (3, 8) = 31.7 | P < 0.0001 |
| >F: GPER vs. ESR1 | 8 | − 6.66 | t = 8.01 | P = 0.0003 |
| >M: GPER vs. ESR1 | 8 | 6.80 | t = 8.14 | P = 0.0002 |
| >M: GPER vs. ESR2 | 8 | 4.94 | t = 7.70 | P = 0.0003 |
Since all of the samples tested thus far were whole tissue homogenates, we next determined the ER profile in A7r5 cells, a rat aortic smooth muscle cell line frequently used in our lab (Table 5). Since this cell line originated from rat embryos, we assumed that the sex of the cells was a combination of male and female. GPER expression was predominant in this cell type, with similarly low levels of ESR1, ESR2, and CYP19A1 (Fig. 6).
Table 5.
Statistical analysis of data presented in Fig. 6. Post hoc results are only shown for tests which reached statistical significance (P < 0.05)
| DF | eta squared Cohen’s d | F (DFn, DFd) t | P value | |
|---|---|---|---|---|
| Rat aortic SMC (one-way) | ||||
| Gene | 3 | 88.1 | F (3, 31) = 76.52 | P < 0.0001 |
| >F: GPER vs. ESR1 | 31 | 4.52 | t = 12.09 | P < 0.0001 |
| >F: GPER vs. ESR2 | 31 | 5.90 | t = 11.51 | P < 0.0001 |
| >F: GPER vs. CYP19A1 | 31 | 6.60 | t = 12.57 | P < 0.0001 |
Fig. 6.
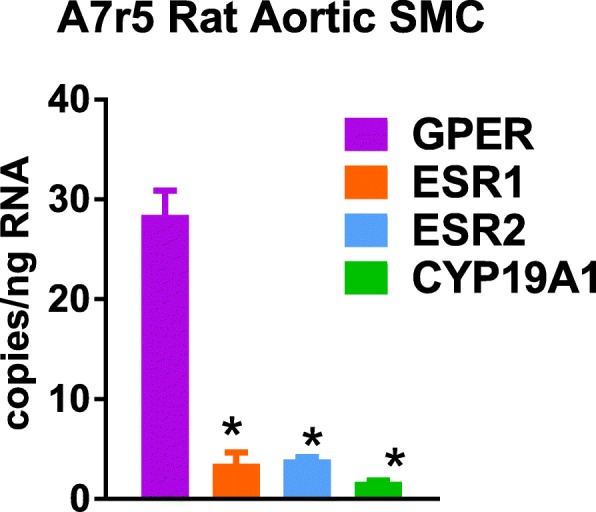
Estrogen receptor profile in rat embryonic aortic smooth muscle A7r5 cell line. One-way ANOVA with Sidak’s multiple comparisons test, *P < 0.05 vs. GPER
In order to get a more thorough picture of the ER profile across tissues, Fig. 7 presents the normalized data for the three estrogen receptors across all samples.
Fig. 7.
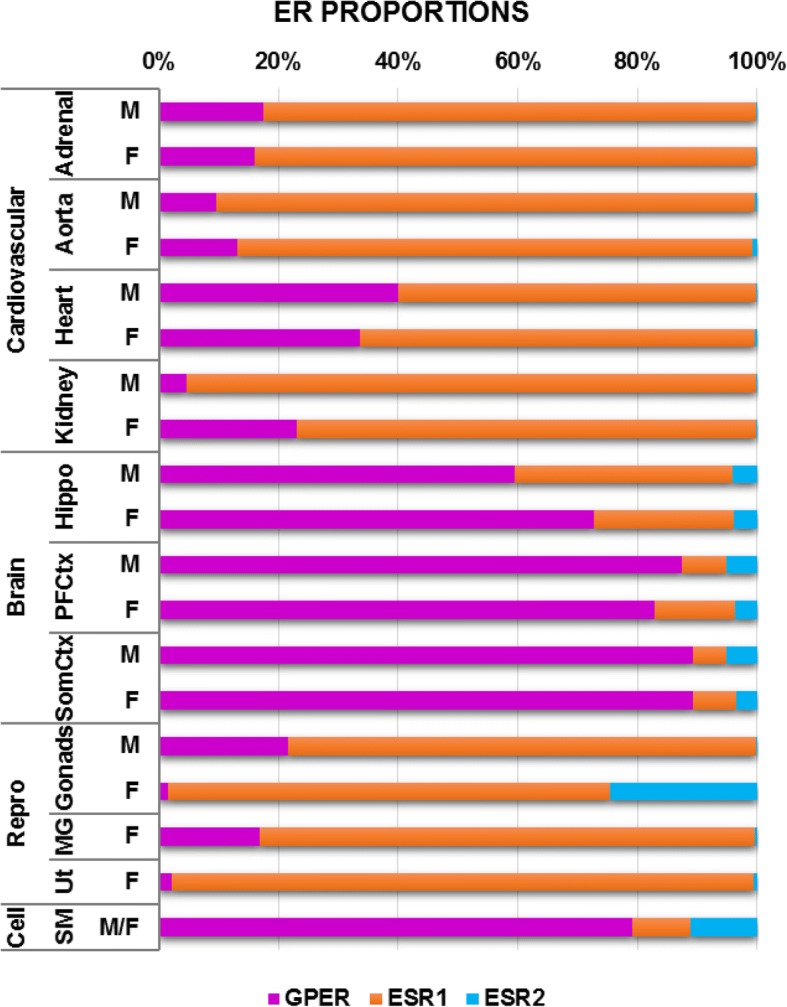
Estrogen receptor profiles for all tissues expressed as a percent of total receptor expression
Discussion
The current study quantified and compared mRNA for the three known estrogen receptors and the enzyme aromatase in 10 tissues using a previously validated PCR technique that allows direct quantification of absolute transcript number. The data indicates a surprising lack of sexual dimorphism in key players for the primary female sex hormone. However, significant differences were noted in the range of receptor expression across tissues as well as the predominant estrogen receptor in each organ system. The data provide a guide for future studies by establishing the absolute amounts of estrogen receptor mRNA in a significant number of target organs.
Sex differences were broadly absent except in the kidneys (GPER and ESR1) and gonads (GPER, ESR2, and CYP19A1). The greater expression of renal ESR1 in male versus female Sprague-Dawley rats was also detected by others using the branched DNA signal amplification assay [11]. The direction of this sexual dimorphism is surprising considering that deletion of this gene in mice increases proteinuria and glomerular damage in females but not in males [12]. This same study found that genetic deletion of ESR2 does not impact renal health and reflects the nearly undetectable levels of this transcript in kidneys from both sexes in the current study. Similarly, kidneys from adult AKR mice lack ESR2 and have greater ESR1 in males compared with females [13]. Opposite of our ESR1 results, renal GPER was greater in females than males, which may compensate for lower ESR1 since its activation protects against renal damage in female rats [14]. The gonads showed the most sexual dimorphism, with greater ESR2 and CYP19A1 but less GPER in ovaries versus testes. Higher levels of ovarian ESR2 may underlie the more prominent reproductive phenotype in females with ESR2 deletion in comparison with males [15]. The lack of a sex difference in ESR1 between the testes and ovary was surprising considering the obvious differences in anatomy as well as sex hormones. Analysis by qPCR in zebrafish found that in contrast to our findings, ESR1 is significantly lower in the testes versus the ovaries [16]. However, their findings of greater CYP19A1 in female fish gonads supports our results and is most likely related to the more important role for this enzyme in sexual differentiation of females in comparison with males [17].
GPER was relatively stable across all tissues of both sexes, ranging from 5.5 copies/ng RNA (ovary) to 113 copies/ng RNA (prefrontal cortex), an approximately 20-fold difference. This finding is consistent with current literature that describes GPER expression as ubiquitous [18, 19]. In contrast, ERα and ERβ were variable across all tissues although relatively stable within each organ system. ESR1 displayed a range of 4.5 (somatosensory cortex) to 614 (kidney) copies/ng RNA, a fold change of ~ 136 while ESR2 ranged from 0.15 (adrenal gland) to 83 (ovary) copies/ng RNA or ~ 550-fold. The stability of GPER across all tissues of a given organ system as opposed to the more variable expression of ERα and ERβ may be indicative of the rapid signaling role of GPER [20–22]. In contrast, the transcriptional role of nuclear ERs may necessitate a more tailored pattern of receptor expression [23, 24]. GPER-mediated rapid signaling is demonstrated in tissues such as blood vessels, where it induces vasodilation via nitric oxide (NO) and cAMP signaling [20] and impacts calcium mobilization [25]. GPER activation also increases mitochondrial calcium retention in the heart within minutes [26] and decreases central control of food intake within an hour [27], both of which demonstrate the importance of this non-genomic pathway. The greater variability and tissue specificity for ESR1 and ESR2 may be important for regulating the large number of genes that are important for growth and reproduction. Genomic signaling by ERα and ERβ involves direct binding to estrogen-responsive elements, transcription factors, or cofactor complexes that regulate numerous genes [28]. This transcriptional role may necessitate a more tailored pattern of receptor expression [23, 24].
In cardiovascular tissues, ESR1 predominated over both ESR2 and GPER. The magnitude of ESR1 dominance was greatest in the kidney, where ESR1 was three times higher than GPER in females and sixfold greater in males. In the heart, ESR1 and GPER copy number was the most similar with a difference of 1.4-fold and 1.9-fold in males and females, respectively. Higher expression of GPER in the heart is consistent with findings of improved function as well as reduced remodeling in response to administration of the GPER agonist G-1 [29]. Studies using cardiomyocyte-specific deletion of GPER indicate that this cell type is likely the most important target for estrogenic actions mediated by GPER [30, 31]. ESR2 was nearly undetectable in the heart as well as all cardiovascular tissues, supporting other studies showing a lack of ESR2 in cardiac tissue and isolated cardiomyocytes from both sexes [32] as well as in the adult mouse kidney [13]. Since studies implicate this in attenuating the hypertrophic response to pressure overload in females [33], cardiac ESR2 expression may be induced in response to tissue injury or disease.
In the three brain areas analyzed, prefrontal cortex, somatosensory cortex, and hippocampus, GPER was dominant whereas ESR1 was about 75% lower and similar to levels of ESR2. Previous data generated in our laboratory reveals a similar expression pattern in the somatosensory cortex of Thy1 female mice, where sensory-evoked structural plasticity positively correlates with high estrogen stages of the estrus cycle [34]. These results suggest a conserved role for rapid estrogen signaling via GPER in telencephalic brain regions. In addition, GPER reduces inflammatory markers in primary cultures of microglial cells when treated with the selective agonist G1, indicating a neuroprotective role for this receptor [35]. Although expression of ESR2 was much lower than the other receptors throughout the study, the amount found in brain was greater relative to all other tissues except the ovaries, suggesting an important role in nervous tissue. In this regard, ESR2, but not ESR1, has been recently found responsible for the modulation and promotion of synaptogenesis in cortical neurons in vitro [36] and in vivo [37], indicating a crucial role of this receptor in brain function, especially for memory and cognition. Furthermore, the relative ESR2/ESR1 expression ratio is increasingly appreciated as important in memory performance and cognitive decline with respect to age [38].
ESR1 was the dominant gene in gonadal tissues of both sexes, which is in accordance with data establishing it as a vital receptor in animal fertility [39]. Second to ESR1, males had significant levels of GPER but low ESR2, while females exhibited the inverse relationship. Higher levels of ESR2 versus GPER in the ovary and uterus supports the presence of smaller litters in ESR2 knockout animals [15, 39] and the lack of any reproductive changes in GPER knockout mice [40, 41] or rats treated with the GPER agonist [42]. Opposite to females, genetic deletion of ESR2 in males does not alter fertility [39] and correlates with our data showing low receptor levels in the testes. Despite the higher expression of GPER in males, genetic deletion of this receptor is reported to not alter fertile or function of the hypothalamic-pituitary-gonadal axis [40, 41]. Since the ovaries are the primary source of circulating estrogen, it was not surprising that CYP19A1 expression was detected at very high levels in this tissue. The low levels in most other tissues (except testes and hippocampus) are consistent with previous reports of extragonadal CYP19A1 expression only in the brain and adipose tissue, which are also major sites for estrogen production [43].
The most commonly used molecular assay in biological sciences is currently qPCR [44]. This technique utilizes nucleic acid primers for a gene of interest, a fluorescent reporter dye, putative formulae, or a standard curve of known nucleic acid concentrations in order to approximate the concentration of transcript by comparing fluorescence intensity over time [45]. A baseline value of background fluorescence is established, and by determining the cycle number at which fluorescence surpasses background levels one can establish the starting concentration of the target sequence. ddPCR was developed to more accurately and reproducibly quantify nucleic acids without the use of standard curves or reference genes [3]. Unlike qPCR, ddPCR accomplishes absolute quantification by partitioning the starting materials into thousands of nanoliter droplets suspended in an oil emulsion containing one or zero target sequences. After amplification by traditional PCR, droplets are quantified as positive or negative, and the starting concentration of target can be calculated using Poisson statistics. Because ddPCR is based on analysis of a binary end state, it is far more resistant to contamination and fluctuations in PCR efficiency, both of which drastically affect results obtained via qPCR [4]. A comparison of RT-qPCR and ddPCR in quantifying strains of influenza found that ddPCR was 30-fold more sensitive than RT-qPCR and results obtained by ddPCR were more precise [8], supporting conclusions in other investigations [3, 46]. Overall, several studies find ddPCR to be more reliable, reproducible, sensitive, and consistent when compared to traditional qPCR methods with the added benefit of not requiring standard curves or normalization to a reference gene [3, 4, 8, 47].
One limitation of utilizing tissue homogenates is a heterogeneous mixture of differentiated cell types. Analysis of mRNA from the rat aortic smooth muscle cell A7r5 cell line resulted in a completely different ER profile than that obtained using the whole rat aorta, supporting the idea that each cell or subpopulation of cells may have an individual expression profile. The cell versus tissue data shows a similar paucity of ESR2, but GPER rather than ESR1 predominates implying variable responses to estrogens within the tissue and a need for cell-specific expression profiles. Other studies also show that gene expression profiles show low correlation with cell lines [48], indicating that different environments may have a strong influence on gene expression within the same tissue type.
Additional limitations are that protein levels may not correlate with mRNA levels or patterns, and gene expression does not take into account the presence of multiple splice variants for estrogen receptors [49]. In fact, Irsik et al. found that ERß protein is expressed at similar levels in the mouse kidney and ovary, while we found that ERß mRNA was more than 400 times lower in the kidney versus ovary. These disparate findings suggest that either translation to protein does not follow the patterns seen here for mRNA or that there is a lack of specificity of currently available antibodies. Indeed, care needs to be taken to validate all antibodies against a control sample where the protein of interest has been deleted [50]. Nevertheless, the method used here is to our knowledge the best method for quantification and comparison of four genes of interest in different tissues.
Conclusions
In conclusion, this study revealed a wide range of ER expression across tissues, with surprisingly few sex differences. As illustrated in Fig. 7, which represents the data as parts of a whole, ER profiles were relatively consistent across tissues of the same organ system, with large differences in the ER profile between systems. The relative stability of GPER mRNA across all tissues as opposed to the variable expression of ESR1 and ESR2 is perhaps best explained by the differing role that GPER plays in relation to the nuclear ERs. Consistent expression of GPER allows all tissues to respond rapidly and concertedly to estrogen signaling, while the differential expression of ESR1 and ESR2 allows for tissue-specific transcriptional regulation in response to the same signal. The predominance of GPER in brain and ESR1 in cardiovascular tissues, and moreover the relative ratio between these receptors may serve to further tailor the response to estrogen in different tissues. Future studies will examine the impact of aging and disease on tissue-specific ER profiles. Overall, this work is indicative of the complex nature of ER research and the nuances that remain to be elucidated. Moreover, this study provides a contextual framework to guide future work on ER roles in specific organs.
Acknowledgements
Not applicable
Funding
NIH HL133619 (SHL).
Availability of data and materials
The datasets generated during the current study are available in the Harvard Dataverse repository, https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/N58KGA
Abbreviations
- AMP
Adenosine monophosphate
- CEE
Conjugated equine estrogen
- CVD
Cardiovascular disease
- ddPCR
Droplet digital polymerase chain reaction
- ER
Estrogen receptor(s)
- Estradiol
17β-estradiol
- GMC
Glomerular mesenchymal cell(s)
- GPER
G protein-coupled estrogen receptor
- MHT
Menopausal hormone therapy
- MPA
Medroxyprogesterone acetate
- NHS
Nurse’s Health Study
- PCR
Polymerase chain reaction
- qPCR
Quantitative polymerase chain reaction
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- VSMC
Vascular smooth muscle cell(s)
- WHI
Women’s Health Initiative
Authors’ contributions
DH conceived the experiments, collected cardiovascular and reproductive tissues, extracted RNA, ran ddPCR, analyzed and interpreted the data, and was a major contributor in writing the manuscript. RG performed RNA extraction and ddPCR. RM dissected three brain areas and contributed to the manuscript sections discussing this data. RS trained and assisted in ddPCR experiments and data analysis, and edited the manuscript. SL conceived and designed the experiments, collected the tissues, analyzed and interpreted the data, and was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Ethics approval
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dillion D. Hutson, Email: dhutson1@tulane.edu
Rakesh Gurrala, Email: rgurrala@tulane.edu.
Benard O. Ogola, Email: bogola@tulane.edu
Margaret A. Zimmerman, Email: mzimmer5@tulane.edu
Ricardo Mostany, Email: rmostany@tulane.edu.
Ryousuke Satou, Email: rsato@tulane.edu.
Sarah H. Lindsey, Email: lindsey@tulane.edu
References
- 1.Dubey RK, Imthurn B, Barton M, Jackson EK. Vascular consequences of menopause and hormone therapy: importance of timing of treatment and type of estrogen. Cardiovasc Res. 2005;66(2):295–306. doi: 10.1016/j.cardiores.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Bhupathiraju SN, Grodstein F, Rosner BA, Stampfer MJ, Hu FB, Willett WC, et al. Hormone therapy use and risk of chronic disease in the nurses’ health study: a comparative analysis with the Women's Health Initiative. Am J Epidemiol. 2017;186(6):696–708. doi: 10.1093/aje/kwx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10(10):1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor SC, Laperriere G, Germain H. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci Rep. 2017;7:2409. doi: 10.1038/s41598-017-02217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics. 2007;8:127. doi: 10.1186/1471-2164-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 2011;83(17):6474–6484. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 8.Taylor SC, Carbonneau J, Shelton DN, Boivin G. Optimization of droplet digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: clinical implications for quantification of oseltamivir-resistant subpopulations. J Virol Methods. 2015;224:58–66. doi: 10.1016/j.jviromet.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Desjardins P, Conklin D. NanoDrop microvolume quantitation of nucleic acids. J Vis Exp. 2010;45:2565. doi: 10.3791/2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Edgington-Giordano F, Dugas C, Abrams A, Katakam P, Satou R, et al. Regulation of nephron progenitor cell self-renewal by intermediary metabolism. J Am Soc Nephrol. 2017;28(11):3323–3335. doi: 10.1681/ASN.2016111246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu H, Lei X, Klaassen C. Gender differences in renal nuclear receptors and aryl hydrocarbon receptor in 5/6 nephrectomized rats. Kidney Int. 2006;70(11):1920–1928. doi: 10.1038/sj.ki.5001880. [DOI] [PubMed] [Google Scholar]
- 12.Elliot SJ, Berho M, Korach K, Doublier S, Lupia E, Striker GE, et al. Gender-specific effects of endogenous testosterone: female alpha-estrogen receptor-deficient C57Bl/6J mice develop glomerulosclerosis. Kidney Int. 2007;72(4):464–472. doi: 10.1038/sj.ki.5002328. [DOI] [PubMed] [Google Scholar]
- 13.Sharma PK, Thakur MK. Estrogen receptor alpha expression in mice kidney shows sex differences during aging. Biogerontology. 2004;5(6):375–381. doi: 10.1007/s10522-004-3191-6. [DOI] [PubMed] [Google Scholar]
- 14.Lindsey SH, Yamaleyeva LM, Brosnihan KB, Gallagher PE, Chappell MC. Estrogen receptor GPR30 reduces oxidative stress and proteinuria in the salt-sensitive female mRen2.Lewis rat. Hypertension. 2011;58(4):665–671. doi: 10.1161/HYPERTENSIONAHA.111.175174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, et al. Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci U S A. 1998;95(26):15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laing LV, Viana J, Dempster EL, Uren Webster TM, van Aerle R, Mill J, et al. Sex-specific transcription and DNA methylation profiles of reproductive and epigenetic associated genes in the gonads and livers of breeding zebrafish. Comp Biochem Physiol Mol Integr Physiol. 2018;222:16–25. doi: 10.1016/j.cbpa.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Fisher CR, Graves KH, Parlow AF, Simpson ER. Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. Proc Natl Acad Sci U S A. 1998;95(12):6965–6970. doi: 10.1073/pnas.95.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmeci C, Thompson DA, Ring HZ, Francke U, Weigel RJ. Identification of a gene (GPR30) with homology to the G-protein-coupled receptor superfamily associated with estrogen receptor expression in breast cancer. Genomics. 1997;45(3):607–617. doi: 10.1006/geno.1997.4972. [DOI] [PubMed] [Google Scholar]
- 19.Hazell GG, Yao ST, Roper JA, Prossnitz ER, O'Carroll AM, Lolait SJ. Localisation of GPR30, a novel G protein-coupled oestrogen receptor, suggests multiple functions in rodent brain and peripheral tissues. J Endocrinol. 2009;202(2):223–236. doi: 10.1677/JOE-09-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsey SH, Liu L, Chappell MC. Vasodilation by GPER in mesenteric arteries involves both endothelial nitric oxide and smooth muscle cAMP signaling. Steroids. 2014;81:99–102. doi: 10.1016/j.steroids.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmerman MA, Budish RA, Kashyap S, Lindsey SH. GPER - novel membrane oestrogen receptor. Clin Sci. 2016;130(12):1005–1016. doi: 10.1042/CS20160114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filardo EJ, Quinn JA, Frackelton AR, Jr, Bland KI. Estrogen action via the G protein-coupled receptor, GPR30: stimulation of adenylyl cyclase and cAMP-mediated attenuation of the epidermal growth factor receptor-to-MAPK signaling axis. Mol Endocrinol. 2002;16(1):70–84. doi: 10.1210/mend.16.1.0758. [DOI] [PubMed] [Google Scholar]
- 23.O'Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Mol Endocrinol. 2004;18(8):1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 24.O'Lone R, Knorr K, Jaffe IZ, Schaffer ME, Martini PG, Karas RH, et al. Estrogen receptors alpha and beta mediate distinct pathways of vascular gene expression, including genes involved in mitochondrial electron transport and generation of reactive oxygen species. Mol Endocrinol. 2007;21(6):1281–1296. doi: 10.1210/me.2006-0497. [DOI] [PubMed] [Google Scholar]
- 25.Meyer MR, Field AS, Kanagy NL, Barton M, Prossnitz ER. GPER regulates endothelin-dependent vascular tone and intracellular calcium. Life Sci. 2012;91(13–14):623–627. doi: 10.1016/j.lfs.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bopassa JC, Eghbali M, Toro L, Stefani E. A novel estrogen receptor GPER inhibits mitochondria permeability transition pore opening and protects the heart against ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2010;298(1):H16–H23. doi: 10.1152/ajpheart.00588.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler MJ, Hildebrandt RP, Eckel LA. Selective activation of estrogen receptors, ERalpha and GPER-1, rapidly decreases food intake in female rats. Horm Behav. 2018;103:54–61. doi: 10.1016/j.yhbeh.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vivar OI, Zhao X, Saunier EF, Griffin C, Mayba OS, Tagliaferri M, et al. Estrogen receptor beta binds to and regulates three distinct classes of target genes. J Biol Chem. 2010;285(29):22059–22066. doi: 10.1074/jbc.M110.114116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jessup JA, Lindsey SH, Wang H, Chappell MC, Groban L. Attenuation of salt-induced cardiac remodeling and diastolic dysfunction by the GPER agonist G-1 in female mRen2.Lewis rats. PLoS One. 2010;5(11):e15433. doi: 10.1371/journal.pone.0015433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Sun X, Chou J, Lin M, Ferrario CM, Zapata-Sudo G, et al. Cardiomyocyte-specific deletion of the G protein-coupled estrogen receptor (GPER) leads to left ventricular dysfunction and adverse remodeling: a sex-specific gene profiling analysis. Biochim Biophys Acta. 2017;1863(8):1870–1882. doi: 10.1016/j.bbadis.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Sun X, Lin MS, Ferrario CM, Van Remmen H, Groban L. G protein-coupled estrogen receptor (GPER) deficiency induces cardiac remodeling through oxidative stress. Transl Res. 2018;199:39–51. doi: 10.1016/j.trsl.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugach EK, Blenck CL, Dragavon JM, Langer SJ, Leinwand LA. Estrogen receptor profiling and activity in cardiac myocytes. Mol Cell Endocrinol. 2016;431:62–70. doi: 10.1016/j.mce.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skavdahl M, Steenbergen C, Clark J, Myers P, Demianenko T, Mao L, et al. Estrogen receptor-beta mediates male-female differences in the development of pressure overload hypertrophy. Am J Physiol Heart Circ Physiol. 2005;288(2):H469–H476. doi: 10.1152/ajpheart.00723.2004. [DOI] [PubMed] [Google Scholar]
- 34.Alexander BH, Barnes HM, Trimmer E, Davidson AM, Ogola BO, Lindsey SH, et al. Stable density and dynamics of dendritic spines of cortical neurons across the estrous cycle while expressing differential levels of sensory-evoked plasticity. Front Mol Neurosci. 2018;11:83. doi: 10.3389/fnmol.2018.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao TZ, Ding Q, Hu J, He SM, Shi F, Ma LT. GPER expressed on microglia mediates the anti-inflammatory effect of estradiol in ischemic stroke. Brain Behav. 2016;6(4):e00449. doi: 10.1002/brb3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sellers KJ, Erli F, Raval P, Watson IA, Chen D, Srivastava DP. Rapid modulation of synaptogenesis and spinogenesis by 17β-estradiol in primary cortical neurons. Front Cell Neurosci. 2015;9:137. doi: 10.3389/fncel.2015.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Zhu J, Xu T. 17β-estradiol (E2) promotes growth and stability of new dendritic spines via estrogen receptor β pathway in intact mouse cortex. Brain Res Bull. 2018;137:241–248. doi: 10.1016/j.brainresbull.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Foster TC. Role of estrogen receptor alpha and Beta expression and signaling on cognitive function during aging. Hippocampus. 2012;22(4):656–669. doi: 10.1002/hipo.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development. 2000;127(19):4277–4291. doi: 10.1242/dev.127.19.4277. [DOI] [PubMed] [Google Scholar]
- 40.Otto C, Fuchs I, Kauselmann G, Kern H, Zevnik B, Andreasen P, et al. GPR30 does not mediate estrogenic responses in reproductive organs in mice. Biol Reprod. 2009;80(1):34–41. doi: 10.1095/biolreprod.108.071175. [DOI] [PubMed] [Google Scholar]
- 41.Wang C, Dehghani B, Magrisso IJ, Rick EA, Bonhomme E, Cody DB, et al. GPR30 contributes to estrogen-induced thymic atrophy. Mol Endocrinol. 2008;22(3):636–648. doi: 10.1210/me.2007-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindsey SH, Chappell MC. GPR30 receptor activation reduces blood pressure in ovariectomized female but not intact male mRen2.Lewis rats. Hypertension. 2008;52(4):E48–E4E. [Google Scholar]
- 43.Stocco C. Tissue physiology and pathology of aromatase. Steroids. 2012;77(1):27–35. doi: 10.1016/j.steroids.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dijkstra JR, van Kempen LC, Nagtegaal ID, Bustin SA. Critical appraisal of quantitative PCR results in colorectal cancer research: can we rely on published qPCR results? Mol Oncol. 2014;8(4):813–818. doi: 10.1016/j.molonc.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tellinghuisen J, Spiess A-N. Bias and imprecision in analysis of real-time quantitative polymerase chain reaction data. Anal Chem. 2015;87(17):8925–8931. doi: 10.1021/acs.analchem.5b02057. [DOI] [PubMed] [Google Scholar]
- 46.Wang P, Jing F, Li G, Wu Z, Cheng Z, Zhang J, et al. Absolute quantification of lung cancer related microRNA by droplet digital PCR. Biosens Bioelectron. 2015;74:836–842. doi: 10.1016/j.bios.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 47.White RA, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10(1):116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emig D, Albrecht M. Tissue-specific proteins and functional implications. J Proteome Res. 2011;10(4):1893–1903. doi: 10.1021/pr101132h. [DOI] [PubMed] [Google Scholar]
- 49.Irsik DL, Carmines PK, Lane PH. Classical estrogen receptors and ERα splice variants in the mouse. PLoS One. 2013;8(8):e70926. doi: 10.1371/journal.pone.0070926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks HL, Lindsey ML. Guidelines for authors and reviewers on antibody use in physiology studies. Am J Physiol Heart Circ Physiol. 2018;314(4):H724–HH32. doi: 10.1152/ajpheart.00512.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available in the Harvard Dataverse repository, https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/N58KGA