Abstract
Purpose
Glaucoma, a leading cause of blindness worldwide, often remains undetected until irreversible vision loss has occurred. Treatments focus on lowering intraocular pressure (IOP), the only modifiable and readily measurable risk factor. However, IOP can vary and does not always predict disease progression. MicroRNAs (miRNAs) are promising biomarkers. They are abundant and stable in biological fluids, including plasma and aqueous humor (AqH). We aimed to identify differentially expressed miRNAs in AqH and plasma from glaucoma, exfoliation syndrome (XFS), and control subjects.
Methods
Plasma and AqH from two ethnic cohorts were harvested from glaucoma or XFS (often associated with glaucoma, n = 33) and control (n = 31) patients undergoing elective surgery. A custom miRNA array measured 372 miRNAs. Molecular target prediction and pathway analysis were performed with Ingenuity Pathway Analysis (IPA) and DIANA bioinformatical tools.
Results
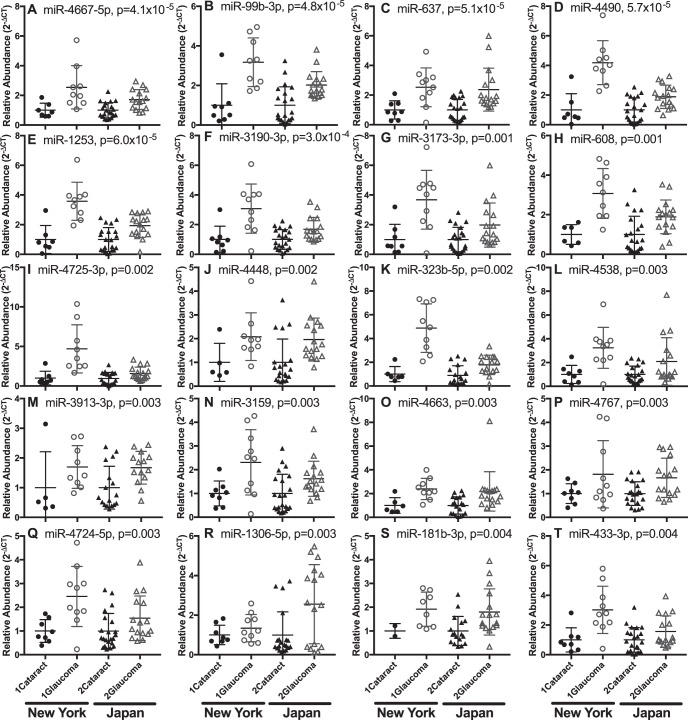
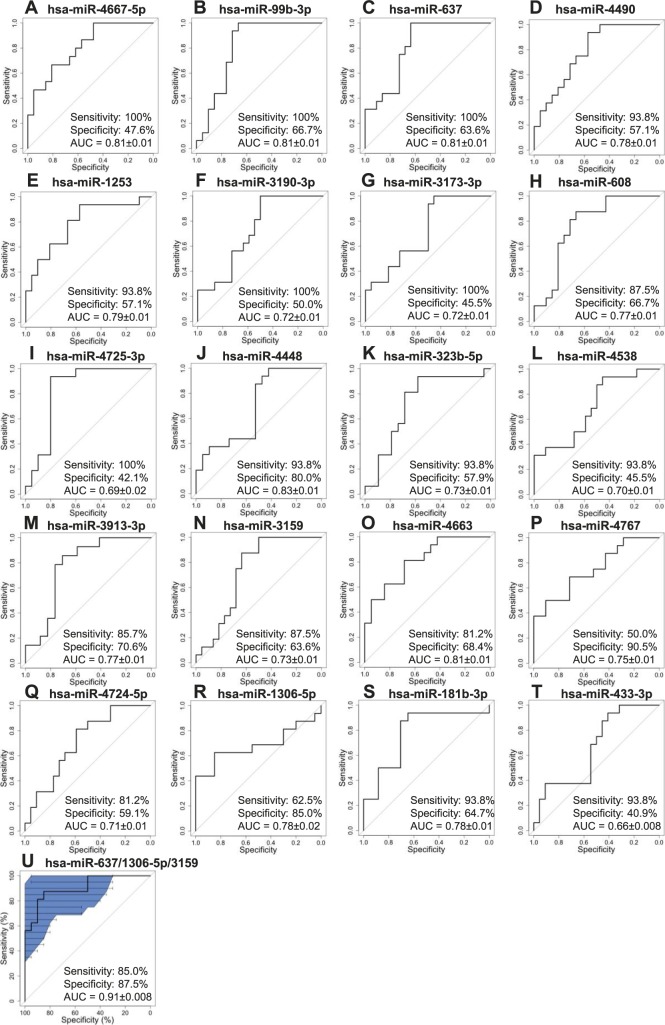
Levels of miRNAs in plasma, a readily accessible biomarker source, correlated with miRNA levels in AqH. Twenty circulating miRNAs were at least 1.5-fold higher in glaucoma or XFS patients than in controls across two ethnic cohorts: miR-4667-5p (P = 4.1 × 10−5), miR-99b-3p (P = 4.8 × 10−5), miR-637 (P = 5.1 × 10−5), miR-4490 (P = 5.7 × 10−5), miR-1253 (P = 6.0 × 10−5), miR-3190-3p (P = 3.1 × 10−4), miR-3173-3p (P = 0.001), miR-608 (P = 0.001), miR-4725-3p (P = 0.002), miR-4448 (P = 0.002), and miR-323b-5p (P = 0.002), miR-4538 (P = 0.003), miR-3913-3p (P = 0.003), miR-3159 (P = 0.003), miR-4663 (P = 0.003), miR-4767 (P = 0.003), miR-4724-5p (P = 0.003), miR-1306-5p (P = 0.003), miR-181b-3p (P = 0.004), and miR-433-3p (P = 0.004). miR-637, miR-1306-5p, and miR-3159, in combination, allowed discrimination between glaucoma patients and control subjects (AUC = 0.91 ± 0.008, sensitivity 85.0%, specificity 87.5%).
Conclusions
These results identify specific miRNAs as potential biomarkers and provide insight into the molecular processes underlying glaucoma.
Keywords: glaucoma, microRNA, aqueous humor, plasma, biomarker
Glaucoma is a progressive optic neuropathy characterized by optic nerve head damage and visual field defects that ultimately lead to irreversible blindness.1,2 Vision loss, occurring due to loss of retinal ganglion cells (RGCs) and degeneration of the optic nerve, has far-reaching effects on independent living and quality of life. In the United States, people across racial and ethnic groups state that losing eyesight has the greatest impact on their day-to-day life, more so than loss of memory.3 With increasing life expectancies and percentage of the population over 65 projected to increase over the next decade, glaucoma prevalence is forecasted to increase dramatically. The number of undetected glaucoma cases will also rise; by the year 2020, an estimated 80 million people will have glaucoma, 11 million of which will be bilaterally blind. There are no known biomarkers for early detection or to monitor disease progression and response to medication. Although several glaucoma risk factors have been identified, including elevated intraocular pressure (IOP), the molecular signaling involved in its pathogenesis remains largely unknown. Exfoliation syndrome (XFS) greatly increases the odds to develop glaucoma. Although treatment can stabilize the majority of glaucoma patients, currently available therapies to lower IOP offer incomplete protection and loss of vision may still occur. Together, these considerations highlight the acute need for strategies to identify and stratify glaucoma patients and to assess disease progression.
A class of small noncoding RNAs (>14,000 encoded in the human genome4 known as micro-RNAs [miRNAs]), regulate a wide range of cellular processes by repressing transcription or translation of their target gene.5 miRNAs are abundantly present in biological fluids6 and have shown great promise as diagnostic and predictive biomarkers, particularly in cardiovascular disease7 and cancer8,9 but also in neurological diseases.10,11 miRNAs have been implicated in a wide range of physiological processes and may contribute to disease etiology or reflect the systemic response to a pathophysiological state. Several miRNAs have previously been implicated in the pathophysiology of glaucoma,12–18 although to date these miRNAs have been assayed only in AqH, and many studies have been insufficiently powered to overcome variability and identify biomarkers that pass false discovery rate thresholds for statistical significance. We customized a high throughput PCR-based strategy to identify differentially expressed miRNAs relevant to glaucoma, then applied this tool to test AqH and plasma from glaucoma patients and cataract controls. We focused our analysis on plasma samples; in addition to its ease of collection (relative to AqH, which requires an elective procedure), at least some glaucoma endophenotypes are associated with vascular dysfunction,19,20 supporting the potential relevance for plasma biomarkers as a diagnostic tool for glaucoma.21–23 Moreover, our complete dataset included matched AqH samples for each subject, allowing comparisons between the two sample types and further investigation of ocular miRNAs in glaucoma etiology. Twenty miRNAs were differentially expressed in plasma from glaucoma or XFS patients when compared to cataract controls. We anticipate that follow-up studies building on the results presented here will lead to identification of biomarkers facilitating disease detection, stratification of patients, prediction of prognosis, and evaluation of treatment response. Additionally, identifying glaucoma-specific miRNAs and their genetic targets, followed by in-depth pathway analysis may help elucidate the complex molecular mechanisms underlying this disease.
Methods
Sample Collection
Matched plasma and AqH samples were harvested from consecutive patients as routine procedures during elective surgeries at the New York Eye and Ear Infirmary of Mount Sinai (Institutional Review Board #12.17, PI RR: Cohort 1, Caucasian) and the Ozaki Eye Hospital (Institutional Review Board #12 and #13, PI MO: Cohort 2, Japanese) conducted in accordance with the Declaration of Helsinki (see Tables 1 and 2 for demographic and clinical variables of study subjects). Nine samples from Cohort 1 were included in initial miRNA selection, with all of Cohort 2, plus additional participants from Cohort 1 included in the complete analyses (Table 1). Samples were de-identified, shipped on dry ice, and studied at the Massachusetts General Hospital under approved Institutional Review Board #2013P002175 (PI ESB). Informed consent was obtained from patients undergoing glaucoma and cataract surgery, and samples from both cohorts were collected from patients who met the following inclusion criteria: (1) age >18 years; (2) patients with open-angle glaucoma or XFS undergoing a glaucoma filtering procedure, such as a tube shunt procedure, a trabeculectomy, cataract removal, or a combined cataract removal and trabeculectomy procedure; (3) control patients undergoing a cataract removal with no diagnosis of glaucoma. Exclusion criteria included: (1) patients with closed-angle glaucoma, (2) patients who underwent a prior intraocular procedure, (3) pregnant women. Patients who underwent cataract surgery with no history of glaucoma were studied as control subjects. Patients' most recent 24-2 SITA-Standard (Carl Zeiss Meditec, Inc., Jena, Germany) visual field mean deviation were assessed and included in Table 1. Some control patients did not have visual field testing completed, and one patient had completed a 10-2 SITA-Standard visual field (Table 1). In Cohort 1, mean deviation was −7.8 ± 5.8DB and −2.1 ± 1.5DB in glaucoma patients and controls, respectively (mean ± SD, P < 0.05). In Cohort 2, mean deviation was −3.3 ± 4.8DB and −2.7 ± 5.8DB in glaucoma patients and controls, respectively (mean ± SD, P = NS).
Table 1.
Demographics and Clinical Variables
|
Analysis Group |
Study ID |
Race |
Sex |
Age |
Diagnosis |
Co-Morbidity |
Date of Diagnosis |
Medications* |
IOP (mm Hg) |
Visual Acuity |
Max IOP |
Mean Deviation |
| Cohort 1 | ||||||||||||
| Initial Selection | 101 | Caucasian | F | 66 | POAG | XFG | 12/19/2012 | 16, 19, 22 | 25 | 20/50 | 48 | −17.68 |
| Initial Selection | 105 | Caucasian | F | 77 | Control | Cataract | 12/8/2008 | 22 | 18 | 20/60 | 18 | −2.28 |
| Initial Selection | 107 | Caucasian | F | 82 | POAG | PVFL | 12/10/1992 | 8, 19 | 16 | 20/20 | 20 | −2.21 |
| Initial Selection | 108 | Caucasian | F | 80 | POAG | Peripheral vision loss | 1/29/2002 | 15, 16, 17 | 18 | 20/40 | 48 | −14 |
| Initial Selection | 109 | Caucasian | F | 71 | POAG | Narrow angle | 11/19/2012 | 18 | 13 | 20/100 | 16 | −7.33 |
| Initial Selection | 110 | Caucasian | F | 81 | POAG | PVFL | 8/12/2006 | 19 | 6 | 20/100 | 14 | −7.87 |
| Initial Selection | 111 | Caucasian | F | 77 | Control | Cataract | 6/5/2013 | 0 | 17 | 20/40 | 17 | N.D. |
| Complete Analysis | 114 | Caucasian | F | 74 | Control | Cataract | 6/24/2013 | 0 | 16 | 20/50 | 19 | −4.29 |
| Initial Selection | 115 | Caucasian | F | 75 | POAG | PVFL | 1/12/2012 | 18, 19 | 34 | 20/100 | 35 | −15.16 |
| Initial Selection | 117 | Caucasian | F | 76 | Control | Cataract | 10/14/2005 | 16 | 14 | 20/60 | 17 | −1.61 |
| Complete Analysis | 119 | Caucasian | F | 69 | POAG | PVFL | 1/28/1997 | 19 | 21 | 20/25 | 28 | −14.63 |
| Complete Analysis | 120 | Caucasian | F | 77 | Control | Cataract | 6/12/2006 | 0 | 14 | 20/20 | 17 | −2.32 |
| Complete Analysis | 122 | Caucasian | F | 71 | Control | Cataract | 2/17/2011 | 0 | 17 | 20/30 | 19 | −0.17 |
| Complete Analysis | 123 | Caucasian | F | 61 | Control | Cataract | 12/15/2011 | 0 | 18 | 20/40 | 20 | N.D. |
| Complete Analysis | 124 | Caucasian | F | 72 | POAG | PVFL | 8/4/2010 | 16, 18 | 19 | 20/25 | 34 | −2.64 |
| Complete Analysis | 128 | Caucasian | F | 71 | POAG | Peripheral vision loss | 1/3/2005 | 20, 21 | 15 | 20/20 | 20 | −4.4 |
| Complete Analysis | 136 | Caucasian | F | 86 | POAG | PVFL | 12/11/2007 | 6, 15, 19 | 13 | 20/20 | 15 | −3.35† |
| Complete Analysis | 137 | Caucasian | F | 74 | POAG | PVFL | 3/14/2012 | 6, 15, 16 | 26 | 20/50 | 35 | −6.05 |
| Complete Analysis | 140 | Caucasian | F | 69 | POAG | PVFL | 6/26/2009 | 15, 16 | 10 | 20/40 | 14 | −4.32 |
| Complete Analysis | 141 | Caucasian | F | 76 | POAG | Peripheral vision loss | 5/5/2006 | 0 | 17 | 20/40 | 17 | −6.12 |
| Complete Analysis | 142 | Caucasian | F | 82 | POAG | Peripheral vision loss | 3/10/2014 | 8, 19 | 25 | 20/400 | 32 | −18.79 |
| Complete Analysis | 143 | Caucasian | F | 63 | POAG | Peripheral vision loss | 7/23/1998 | 6, 15, 16 | 18 | 20/20 | 25 | −1.89 |
| Complete Analysis | 146 | Caucasian | F | 71 | Control | Anatomical narrow angle | 10/26/2007 | 0 | 11 | 20/20 | 14 | N.D. |
| Complete Analysis | 147 | Caucasian | F | 77 | POAG | Peripheral vision loss | 12/15/2011 | 6, 16 | 14 | 20/40 | 17 | −3.67 |
| Complete Analysis | 149 | Caucasian | F | 70 | Control | Anatomical narrow angle | 11/13/1992 | 0 | 14 | 20/20 | 27 | N.D. |
| Complete Analysis | 150 | Caucasian | F | 71 | Control | Anatomical narrow angle | 10/26/2007 | 0 | 12 | 20/20 | 13 | N.D. |
| Complete Analysis | 151 | Caucasian | F | 78 | Control | Anatomical narrow angle | 7/24/2003 | 0 | 12 | 20/25 | 21 | N.D. |
| Complete Analysis | 152 | Caucasian | F | 75 | POAG | PVFL | 5/10/2011 | 16, 18 | 14 | 20/20 | 16 | −2.47 |
| Cohort 2 | ||||||||||||
| Complete Analysis | 202 | Japanese | F | 87 | XFS | 0 | 1/18/2014 | 1 | 14 | 20/66 | 16 | N.D. |
| Complete Analysis | 203 | Japanese | F | 85 | XFS | 0 | 12/19/2013 | 2 | 16 | 20/28 | 40 | 0.41 |
| Complete Analysis | 204 | Japanese | F | 72 | XFS | 0 | 4/18/2013 | 1 | 15 | 20/22 | 19 | N.D. |
| Complete Analysis | 205 | Japanese | F | 87 | XFS | 0 | 10/6/2010 | 3, 4 | 14 | 20/28 | 15 | −3.69 |
| Complete Analysis | 206 | Japanese | F | 83 | XFS | 0 | 2/12/2014 | 1 | 19 | 20/100 | 20 | N.D. |
| Complete Analysis | 208 | Japanese | F | 84 | XFS | 0 | 3/4/2014 | 1, 5 | 14 | 20/25 | 21 | 0.66 |
| Complete Analysis | 209 | Japanese | F | 88 | XFG | 0 | 1/28/2013 | 2, 6, 7, 8 | 31 | 20/28 | 31 | −16.95 |
| Complete Analysis | 212 | Japanese | F | 86 | XFS | 0 | 9/30/2013 | 1, 8, 9 | 24 | 20/66 | 30 | −1.85 |
| Complete Analysis | 215 | Japanese | F | 77 | XFS | 0 | 10/5/2011 | 1 | 14 | 20/17 | 18 | −2.26 |
| Complete Analysis | 219 | Japanese | F | 72 | XFS | 0 | 3/3/2014 | 1 | 19 | 20/66 | 20 | −1.36 |
| Complete Analysis | 221 | Japanese | F | 92 | XFS | 0 | 11/18/2013 | 1 | 13 | 20/40 | 15 | −3.47 |
| Complete Analysis | 222 | Japanese | F | 75 | XFS | 0 | 4/12/2014 | 1 | 13 | 20/28 | 16 | N.D. |
| Complete Analysis | 224 | Japanese | F | 82 | XFG | 0 | 7/8/2014 | 2 | 13 | 20/66 | 14 | −4.54 |
| Complete Analysis | 225 | Japanese | F | 82 | XFS | 0 | 6/10/2014 | 1 | 12 | 20/130 | 16 | −1.40 |
| Complete Analysis | 226 | Japanese | F | 80 | XFS | 0 | 9/10/2013 | 10 | 10 | 20/33 | 15 | N.D. |
| Complete Analysis | 228 | Japanese | F | 82 | XFG | 0 | 5/27/2008 | 1, 9, 11 | 20 | 20/25 | 25 | −2.08 |
| Complete Analysis | 229 | Japanese | F | 75 | Control | Cataract | 3/26/2014 | 12 | 18 | 20/40 | 19 | N.D. |
| Complete Analysis | 230 | Japanese | F | 77 | Control | Cataract | 2/13/2014 | 2 | 15 | 20/50 | 17 | 0.23 |
| Complete Analysis | 231 | Japanese | F | 73 | Control | Cataract | 6/19/2014 | 1 | 18 | 20/66 | 18 | N.D. |
| Complete Analysis | 232 | Japanese | F | 72 | Control | Cataract | 12/11/2011 | 1 | 19 | 20/17 | 19 | −0.62 |
| Complete Analysis | 233 | Japanese | F | 65 | Control | Cataract | 4/24/2013 | 1 | 19 | 20/22 | 21 | N.D. |
| Complete Analysis | 234 | Japanese | M | 70 | Control | Cataract | 10/31/2014 | 1 | 16 | 20/28 | 18 | −6.11 |
| Complete Analysis | 235 | Japanese | M | 79 | Control | Cataract | 12/1/2014 | 1 | 13 | 20/50 | 14 | N.D. |
| Complete Analysis | 236 | Japanese | F | 81 | Control | Cataract | 10/28/2014 | 1 | 16 | 20/40 | 19 | −6.13 |
| Complete Analysis | 237 | Japanese | M | 67 | Control | Cataract | 12/4/2014 | 1 | 12 | 20/100 | 12 | N.D. |
| Complete Analysis | 238 | Japanese | F | 66 | Control | Cataract | 12/2/2013 | 1 | 17 | 20/20 | 22 | −0.50 |
| Complete Analysis | 239 | Japanese | F | 74 | Control | Cataract | 12/3/2014 | 1 | 13 | 20/50 | 14 | −0.83 |
| Complete Analysis | 240 | Japanese | F | 72 | Control | Cataract | 12/8/2014 | 2 | 17 | 20/50 | 17 | −1.27 |
| Complete Analysis | 241 | Japanese | F | 72 | Control | Cataract | 2/14/2013 | 1 | 19 | 20/22 | 20 | N.D. |
| Complete Analysis | 242 | Japanese | M | 82 | Control | Cataract | 7/29/2014 | 1, 13 | 15 | 20/40 | 15 | −19.06 |
| Complete Analysis | 243 | Japanese | F | 77 | Control | Cataract | 7/8/2009 | 1 | 13 | 20/17 | 17 | −2.94 |
| Complete Analysis | 244 | Japanese | M | 73 | Control | Cataract | 8/19/2014 | 1 | 16 | 20/28 | 19 | −1.92 |
| Complete Analysis | 245 | Japanese | F | 73 | Control | Cataract | 4/22/2014 | 1 | 14 | 20/66 | 16 | N.D. |
| Complete Analysis | 246 | Japanese | F | 73 | Control | Cataract | 7/7/2014 | 1 | 17 | 20/25 | 19 | N.D. |
| Complete Analysis | 247 | Japanese | M | 76 | Control | Cataract | 12/24/2014 | 1, 11 | 22 | 20/28 | 22 | 6.55 |
| Complete Analysis | 248 | Japanese | F | 80 | Control | Cataract | 9/6/2010 | 4, 14 | 15 | 20/17 | 16 | -1.05 |
| Complete Analysis | 249 | Japanese | F | 64 | Control | Cataract | 1/26/2015 | 1 | 19 | 20/66 | 23 | N.D. |
| Complete Analysis | 250 | Japanese | F | 79 | Control | Cataract | 6/5/2007 | 1 | 20 | 20/17 | 22 | -1.07 |
Demographics and clinical variables. Study ID (Cohort 1: 101-152; Cohort 2: 202-250), race (Cohort 1: Caucasian; Cohort 2: Japanese), sex (F: female; M: male), age (in years). Visual Field Mean Deviation (Carl Zeiss Meditec 24-2 SITA-Standard) and Mean Deviation were determined at the time of sampling. IOP and Visual Acuity were determined during diagnosis of the study eye. Max IOP is of the study eye. XFG, exfoliation glaucoma; XFS, exfoliation syndrome; POAG, primary open angle glaucoma; PVFL, paracentral visual field loss.
Medications at the time of sampling: 0: none; 1: Noxacin Ophthalmic Solution; 2: Bestron for Ophthalmic; 3: Colinacol; 4: Cravit ophthalmic solution; 5: Soft Santear; 6: Alphagan ophthalmic solution; 7: DuoTrav Combination Ophthalmic Solution; 8: AZOPT Ophthalmic Suspension; 9: Tafluprost; 10: Vigamox; 11: Soft Santear; 12: Gatiflo Ophthalmic Solution; 13: Proranon; 14: Hyalein; 15: Cosopt; 16: Lumigan; 17: Pilocarpine 2%; 18: Combigan; 19: Travatan; 20: Xalatan; 21: Timolol; 22: Predforte.
† 10-2 SITA-Standard, in DB.
Table 2.
Comparison of Clinical Variables in Glaucoma/XFS Patients and Control Subjects
| Cohort |
Control |
Glaucoma |
P Value |
| Cohort 1 and 2 | (N = 30) | (N = 27) | |
| Age (years) | 73 ± 5 | 79 ± 7 | 0.001 |
| Max IOP | 18 ± 3 | 22 ± 8 | 0.015 |
| Sex (female) | 24 (80%) | 27 (100%) | 0.025 |
| Cohort 1 (NY) | (N = 8) | (N = 11) | |
| Age (years) | 72 ± 5 | 74 ± 6 | 0.44 |
| Max IOP | 19 ± 4 | 23 ± 8 | 0.09 |
| Sex (female) | 8 (100%) | 11 (100%) | 1 |
| Cohort 2 (Japan) | (N = 22) | (N = 16) | |
| Age (years) | 74 ± 5 | 82 ± 6 | 0.00003 |
| Max IOP | 18 ± 3 | 21 ± 7 | 0.15 |
| Sex (female) | 16 (73%) | 16 (100%) | 0.03 |
T-tests were employed to compare age and Max IOP between glaucoma and control subjects. Gender distribution (%) was evaluated with a Fisher's exact test.
In Cohort 1, POAG eyes were defined by clinical findings consistent with glaucomatous optic neuropathy defined by vertical cup-to-disc ratio ≥0.6, asymmetry of cup-to-disc ratio ≥0.2 between eyes, and presence of localized retinal nerve fiber layer (RNFL) or neuroretinal rim defects in absence of any other abnormalities that could explain the findings on fundus examination and open anterior chamber angles. Exfoliation glaucoma (XFG) eyes had glaucomatous optic neuropathy as defined above but also had signs of exfoliation material deposition at the anterior lens capsule or pupillary margin. All glaucomatous eyes had glaucoma hemifield test (GHT) results outside normal limits on at least two consecutive reliable examinations or presence of at least three contiguous test points on pattern standard deviation (PSD) plot with P < 1%, with at least one of P < 0.5%, not including points at the edge of the field or those directly above or beneath the blind spot. All glaucoma patients underwent a comprehensive ophthalmic examination, including measurement of BCVA (Snellen), slit-lamp biomicroscopy, fundus examination with 90-D lens, IOP measurement using Goldmann applanation tonometry or rebound tonometry. In Cohort 2, the diagnosis of exfoliation syndrome (XFS), a major risk factor for glaucoma, was based on anterior segment findings of exfoliation material accompanied by an IOP of >21 mm Hg without treatment. XFG was diagnosed by the presence of the following findings: exfoliation material on the lens capsule or at the pupillary margin; an intraocular pressure of >21 mm Hg without treatment; typical glaucomatous optic nerve changes and field defects. Glaucomatous field defects were defined as a minimum presence of a cluster of three abnormal points in the same hemifield with a pattern deviation of <2% in the probability map of the Humphrey automated perimeter with at least one point of <1% or at least two adjacent points with a pattern deviation of <1%.
Blood was collected in the presence of the anticoagulant EDTA, and separated plasma was frozen in liquid nitrogen. AqH (up to 100 μl) was collected by paracentesis. A 30-gauge needle was surgically inserted to the anterior chamber at the limbus to withdraw AqH at the beginning of the elective procedures (glaucoma or cataract operation), without touching the iris or lens. Following harvest, AqH was immediately frozen in liquid nitrogen.
RNA Isolation and Initial miRNA Selection
miRNAs were evaluated from total RNA preparations using qPCR arrays. This method is simple and reliable, with very low intra-assay variability (3 samples re-run on the same array were highly correlated between runs, r = 0.95–0.99). Plasma RNA was isolated with miRNeasy Plasma/Serum Kit (217184; Qiagen) to preserve the miRNA fraction, while small-volume AqH samples were extracted directly by Qiagen. Total RNA, including miRNA, was isolated using miRNEasy Serum/Plasma Kit (217183; Qiagen) from 5 μL AqH for each sample. To normalize and monitor isolation efficiency, miRNeasy Serum/Plasma Spike-In Control (C. elegans miR-39 miRNA mimic) was added to each sample before isolation. cDNA was synthesized using miScript II RT Kit (218161; Qiagen) using HiSpec buffer chemistry, which exclusively reverse transcribed mature miRNA to cDNA. A functional quality control was performed on all cDNA samples using miScript miRNA QC PCR array, which included RTC and positive PCR control (PPC) controls to monitor reverse transcription (RT) and PCR efficiencies as well as cel-miR-39 assay. qPCR was performed on an ABI7900 HT Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). Threshold cycles (C) for PPC and RT control (miRTC) were examined to assess PCR and RT efficiencies, respectively. The expression of cel-miR-39 assay was also observed to confirm efficient RNA recovery. C values for all controls were within the manufacturer's recommended range for a successful quality control (QC). Because of low starting amount, all AqH cDNA samples underwent PCR-based amplification using miScript PreAMP PCR kit (331452; Qiagen) and miScript PreAMP primer mix (MBHS-3218Z; Qiagen). Pre-amplified cDNA was used as the template for real-time PCR analysis using miScript miRNA PCR array miRbase v18 platform (MIHS-3218Z; Qiagen), and the miScript SYBR Green Kit (218073; Qiagen). ABI 7900HT Real-time PCR instrumentation was used to perform the quantification. SDS Software 2.4 (Applied Biosystems) was used to generate raw data (Ct values). The baseline was set from cycle 2 to 10 and threshold was set to 0.2. Raw data was normalized against a global Ct mean of expressed miRNAs for each sample.
As a discovery analysis, we evaluated miRNA expression in the AqH from 9 individuals (3 control, 6 glaucoma; described in Table 1) using the commercially available Qiagen miRNA PCR Array (miRBase v18 platform, MIHS-3218Z, 1800 miRNAs screened). These data were used to inform the subsequent development of a custom miR-Finder array.
Custom miRNA Array to Evaluate Glaucoma Biomarkers
As an analysis tool to target glaucoma biomarkers, the custom miR-Finder Array (CMIHS-02263; Qiagen) included miRNAs identified in AqH as differing between cataract and glaucoma samples, as stable, or as highly expressed in the eye (Supplementary Fig. S1 includes a detailed list of inclusion criteria for the 384-well array design capable of assessing 372 selected miRNAs). Screening 372 miRNAs is more cost-effective and less statistically demanding than screening 1800 miRNAs. RNA preparation, reverse transcription and quality control were conducted in the same manner as in the commercially available MIHS-3218Z array described above.
Complete miRNA Assessment
Plasma and AqH samples (n = 57) from both cohorts were run on the CMIHS-02263 custom PCR array (Cohort 1: 11 patients and 8 controls; Cohort 2: 16 patients and 22 controls). We initially evaluated plasma data and subsequently compared our results to those for AqH. A flowchart of patient and sample distribution for initial miRNA selection and final analysis is presented in Supplementary Figure S2. All Ct data were filtered to remove background signal (all Cts >33 were discarded) and normalized by ΔCt to the mean Ct of all amplified miRNAs in each sample.
Statistical Analyses
All analyses were conducted in R.24 Plasma miRNA levels (determined using the 2ΔCT method) were analyzed using a 2-way ANOVA (factors were cohort, disease, and the interaction term) to identify miRNAs that differed between cataract control and glaucoma subjects. The P values were corrected for false discovery rate with the Benjamini-Hochberg method.25 Overlap in miRNA expression between plasma and AqH was visualized with the VennDiagrams package26 and summarized expression data from both sample types, as presented in Supplementary Table S1 Univariate logistic regression was used to assess diagnostic accuracy for each miRNA, and multivariable models were then applied to determine whether miRNA biomarkers could be combined to improve disease identification. Receiver operator characteristic (ROC) curves were plotted with the pROC package,27 which produced sensitivity and specificity metrics for each ROC curve at the optimal threshold. Plasma miRNAs identified as being associated with disease status were assessed in the AqH to determine if AqH miRNA levels also predicted disease status. Further, the degree of correlation between plasma and AqH miRNA profiles from individual-matched samples were determined using linear mixed effects models (lme4 package28), including cohort, disease state, and the interaction term as factors, adjusting for each individual subject.
Pathway Analysis
Two approaches were used to link differential miRNA expression with biological mechanisms via pathway analyses. First, mRNA targets for miRNAs of interest (20 in plasma, 6 in AqH) were identified using TargetScan.29 Enriched GO pathways were evaluated with DIANA-mirPath v330 in all high-probability mRNA targets, filtered for context + scores < −0.4.31 Second, Ingenuity Pathway Analysis (IPA) microRNA target filter (Qiagen) was used to identify mRNA targets that were either experimentally confirmed or predicted with high confidence. These mRNA targets were subsequently filtered for known associations with ophthalmic disease. A core expression analysis revealed canonical pathways that were associated with the selected miRNA ophthalmic disease mRNA targets. We predicted outcomes of miRNA expression changes on target genes based on the assumption that the prevailing action of miRNAs is to decrease target mRNA expression or translation.
Results
Our overall goal was to identify miRNAs that are differentially expressed in plasma from glaucoma or XFS patients and controls. A custom-built miRNA array detected miRNAs that may be developed as easily screenable biomarkers to diagnose glaucoma or assess treatment response (CMIHS-02263; Qiagen; Supplementary Table S1). CMIHS-02263 quantifies 372 miRNAs, selected based on screening 1800 of the most abundantly expressed and best characterized human miRNAs using a commercially available platform (MIHS-3218Z; Qiagen; Supplementary Fig. S1). We selected miRNAs that tended to be differentially expressed between patients and controls, were highly abundant in AqH, and were minimally variable.
Detecting miRNAs in the Eye
Glaucoma and XFS patients had more detectable miRNAs than in controls (Ct < 33) in AqH (P = 0.0007) and in plasma (P = 0.0014); however, the mean Ct was similar between cataract and glaucoma/XFS patients in both sample types (Fig. 1; P > 0.05). Of the 372 mature miRNAs assayed by our custom array, 153 miRNAs were detected in all AqH samples of at least one group (controls or patients in either cohort). One hundred fifty-four miRNAs were detected in all plasma samples of at least one group, 73 miRNAs were detectable in every AqH sample, and 67 miRNAs were detectable in all plasma samples. There was considerable overlap between plasma and AqH miRNAs detectable by CMIHS-02263 (Fig. 2; Supplementary Table S1).
Figure 1.
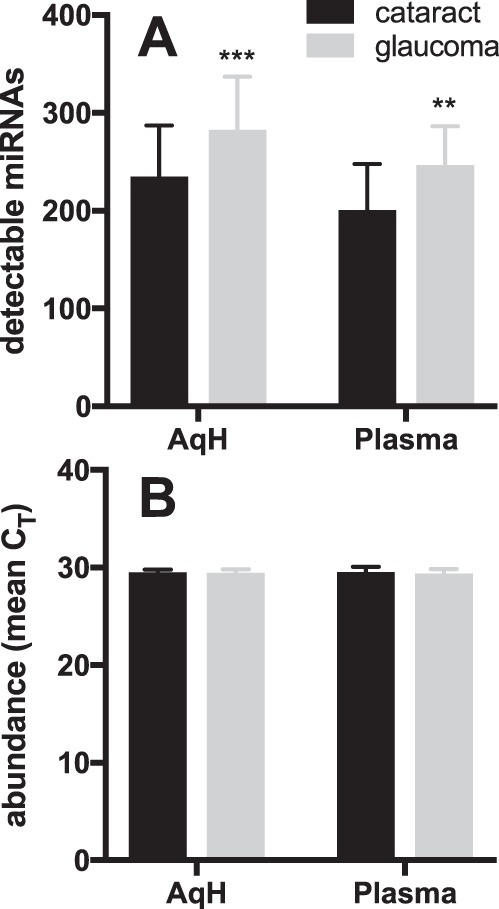
More miRNAs are detected in glaucoma/XFS patients than in controls. (A) The number of miRNAs detected with Ct <33 was higher in aqueous humor (AqH, P = 0.0007) and plasma samples (P = 0.0014) of glaucoma/XFS patients than in cataract controls. (B) Mean Ct was similar across both sample types. Cataract versus glaucoma/XFS data from both cohorts combined, were compared with 2-way ANOVA, and presented as mean ± SD.
Figure 2.
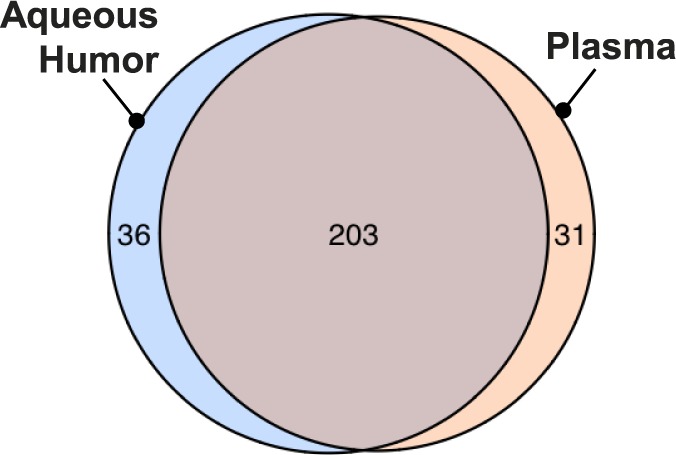
A Venn diagram demonstrates the overlap between miRNAs identified in plasma and in aqueous humor (n = 56) across both cohorts. miRNAs were considered present when they appeared in at least 75% of samples from any one group (cataract or glaucoma/XFS from either cohort). These filtered data also represent the input dataset for 2-way ANOVA comparisons.
Plasma miRNAs as Biomarkers
Expression of miRNAs was evaluated first in plasma samples, representing a more accessible and clinically useful biomarker source than AqH. Twenty plasma miRNAs were more abundant in plasma from glaucoma patients than from control subjects and achieved the threshold of significance required for the false discovery rate correction (Table 3; Fig. 3). There were no interaction effects in the 2-way ANOVA comparison between disease and cohort factors in the plasma samples for any miRNAs examined, indicating that miRNAs respond similarly to disease across the two ethnicities.
Table 3.
Results of 2-Way ANOVA in Plasma Samples
|
miR |
P |
Fold Change |
|
|
Cohort 1 |
Cohort 2 |
||
| hsa-miR-4667-5p | 4.1E-05 | 2.55 | 1.72 |
| hsa-miR-99b-3p | 4.8E-05 | 3.17 | 2.02 |
| hsa-miR-637 | 5.1E-05 | 2.53 | 2.37 |
| hsa-miR-4490 | 5.7E-05 | 4.19 | 1.88 |
| hsa-miR-1253 | 6.0E-05 | 3.58 | 1.93 |
| hsa-miR-3190-3p | 3.1E-04 | 3.08 | 1.69 |
| hsa-miR-3173-3p | 0.001 | 3.68 | 1.98 |
| hsa-miR-608 | 0.001 | 3.07 | 1.89 |
| hsa-miR-4725-3p | 0.002 | 4.69 | 1.57 |
| hsa-miR-4448 | 0.002 | 2.08 | 1.96 |
| hsa-miR-323b-5p | 0.002 | 4.88 | 1.72 |
| hsa-miR-4538 | 0.003 | 3.24 | 2.09 |
| hsa-miR-3913-3p | 0.003 | 1.70 | 1.67 |
| hsa-miR-3159 | 0.003 | 2.31 | 1.62 |
| hsa-miR-4663 | 0.003 | 2.39 | 2.19 |
| hsa-miR-4767 | 0.003 | 1.81 | 1.67 |
| hsa-miR-4724-5p | 0.003 | 2.46 | 1.54 |
| hsa-miR-1306-5p | 0.003 | 1.34 | 2.55 |
| hsa-miR-181b-3p | 0.004 | 1.92 | 1.80 |
| hsa-miR-433-3p | 0.004 | 3.02 | 1.56 |
ANOVA P values are reported for disease state (glaucoma/XFS versus cataract controls). These P values all met threshold significance after adjustment for false discovery rate using the Benjamini-Hochberg method. Fold change is 2-ΔCT of glaucoma versus cataracts for each cohort.
Figure 3.
Twenty plasma miRNAs (A–T, presented in descending order of significance) were elevated in glaucoma/XFS versus cataracts samples from both cohorts. Data are normalized to mean control abundance for each cohort. All differentially expressed miRNAs were identified by 2-way ANOVA (P values are presented). Fold changes for patient Cohort 1 and 2, along with false discovery rate corrected P values are listed in Table 3.
ROC curves were constructed for each of the 20 putative biomarkers (Figs. 4A–T). All 20 miRNAs differed significantly between patients and controls from Cohort 2 in a univariate logistic regression (Table 4). We next assessed whether detection of disease state could be improved using combinations of biomarkers. Of the 20 plasma miRNAs of interest and 190 pairwise combinations, 64 pairs were correlated (r2 > 0.70 for any simple regression between miRNA pairs), making them poor candidates for combination. A multivariate logistic regression retained a combination of three miRNAs as significant factors. miRs 637, 1306-5p, and 3159 had only limited correlation (r2 = 0.04–0.63) and in combination produced the highest AUC ± SEM of 0.91 ± 0.008 (Fig. 4U) with a specificity of 59% at a sensitivity of 95% (95% CI: 41%–97%).
Figure 4.
Area under the curve (AUC) and variance for each ROC in 20 plasma miRNAs. (A–T) ROCs for the 20 potential plasma biomarkers, presented in order of significance. (U) Of all potential miRNA combinations evaluated by multivariate logistic regression analysis, miRNAs 637, 1306-5p, and 3159 yielded the best glaucoma/XFS detection. Shaded area and error bars represent confidence interval range. All inputs showed a significant difference between cataract and glaucoma/XFS samples from Cohort 2 by univariate logistic regression. AUC is presented ± SEM.
Table 4.
Logistic Regression Statistics for Cohort 2 Data (Japan)
| miRNA |
Odds Ratio |
95% CI |
P Value |
| Univariate Analysis | |||
| hsa-miR-4667-5p | 4.14 | 1.5–11.4 | 0.006 |
| hsa-miR-99b-3p | 4.13 | 1.5–11.3 | 0.006 |
| hsa-miR-637 | 8.64 | 1.8–42.2 | 0.008 |
| hsa-miR-4490 | 3.16 | 1.4–7.3 | 0.008 |
| hsa-miR-1253 | 3.41 | 1.4–8.0 | 0.005 |
| hsa-miR-3190-3p | 2.93 | 1.2–7.1 | 0.02 |
| hsa-miR-3173-3p | 2.56 | 1.1–6.0 | 0.03 |
| hsa-miR-608 | 2.84 | 1.3–6.4 | 0.01 |
| hsa-miR-4725-3p | 2.25 | 1.0–5.1 | 0.05 |
| hsa-miR-4448 | 2.95 | 1.3–6.9 | 0.01 |
| hsa-miR-323b-5p | 2.30 | 1.1–4.8 | 0.03 |
| hsa-miR-4538 | 2.55 | 1.0–6.6 | 0.06 |
| hsa-miR-3913-3p | 2.90 | 1.2–6.8 | 0.01 |
| hsa-miR-3159 | 2.26 | 1.1–4.9 | 0.03 |
| hsa-miR-4663 | 9.80 | 1.9–50.5 | 0.006 |
| hsa-miR-4767 | 4.61 | 1.4–15.5 | 0.01 |
| hsa-miR-4724-5p | 1.91 | 1.0–3.8 | 0.07 |
| hsa-miR-1306-5p | 2.40 | 1.2–4.8 | 0.01 |
| hsa-miR-181b-3p | 3.47 | 1.2–10.2 | 0.02 |
| hsa-miR-433-3p | 1.82 | 1.0–3.6 | 0.09 |
| Multivariate Analysis* | |||
| hsa-miR-637 | 2031.9 | 2.9–1 417 266 | 0.02 |
| hsa-miR-1306-5p | 24.5 | 1.9–312 | 0.01 |
| hsa-miR-3159 | 0.002 | 5.1 × 10−06–0.9 | 0.05 |
Three of these retained significance in a multivariate analysis, suggesting that their combination may improve disease classification or detection.
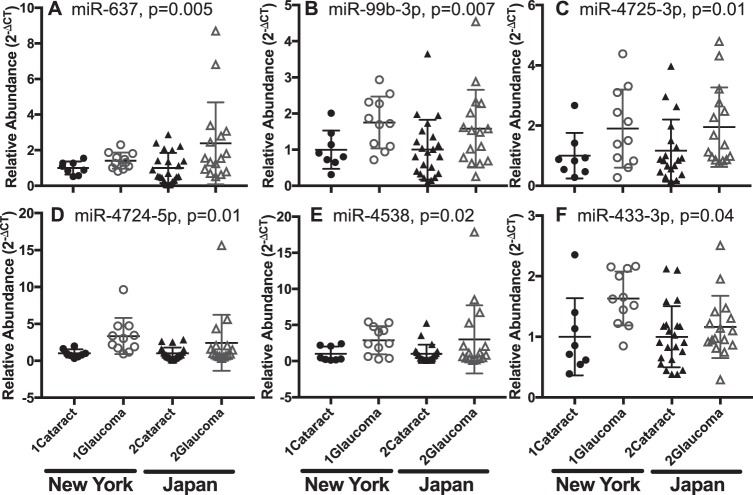
Correlation of miRNAs in AqH and Plasma
We tested the hypothesis that miRNA expression levels correlate between plasma and AqH. 6/20 miRNAs elevated in patient plasma were also significantly increased in AqH from the same subjects (miR-637, miR-99b-3p, miR-4725-3p, miR-4724-5p, miR-4358, miR-433-3p; Fig. 5). While the correlation between miRNA levels in plasma and AqH was low across the entire dataset containing 372 miRNAs (r = 0.48, P < 0.0001), it improved when only the top 20 plasma biomarkers (r = 0.72, P < 0.0001) and the top 6 AqH biomarkers (r = 0.85, P < 0.0001) were plotted. This improved correlation between the sample types when inputs are restricted to the miRNA targets identified by our analyses can be considered evidence of their usefulness as biomarkers.
Figure 5.
Relative abundance for 6/20 miRNAs (A: miR-637, B: miR-99b-3p, C: miR-4725-3p, D: miR-4724-5p, E: miR-4538, F: miR-433-3p) that differed significantly by disease state in AqH. Data are normalized to mean control abundance for each cohort. Patients in the Japan cohort include those with XFS. P values represent disease factor significance in 2-way ANOVA.
Pathway Analysis
Pathway analyses was used to assess biological pathways implicated in glaucoma by the 20 miRNA biomarkers. DIANA analysis of GO pathways linked to high-probability miRNA-mRNA interactions identified basic cellular processes (cellular nitrogen metabolism, gene expression) as well as GO terms that align with the neurological component of glaucoma (e.g., synaptic transmission; Supplementary Table S2). Identification of specific pathway enrichments (e.g., Fc-epsilon receptor and neurotrophin/tropomyosin-related kinase (TRK) receptor signaling pathways) in these subjects, which encompass multiple glaucoma subtypes, may provide generalized biological insights into the disease. There was also significant overlap between enriched GO pathways that were targeted by the 6 miRNAs differentially expressed in AqH, and the 20 miRNAs that differed in plasma samples (Supplementary Table S2), further validating the diagnostic potential of plasma.
IPA MicroRNA Target Filter produced 2829 unique potential high confidence mRNA hits that were predicted to be a target of 1 or more of the 20 miRNAs that differed in plasma samples. The ophthalmic disease filter reduced this list to 438 mRNAs. From this subset, IPA Core Expression Analysis identified canonical pathways that may be integral to the pathological processes in glaucoma (Supplementary Table S3). This analysis implicates general disease processes including neuroinflammation (Supplementary Table S4), neuronal nitric oxide synthase (nNOS) signaling (Supplementary Table S5), endothelial nitric oxide synthase (eNOS) signaling (Supplementary Table S6), and neurotrophin/TRK receptor signaling (Supplementary Table S7).
Discussion
Glaucoma is the most common cause of irreversible blindness worldwide. Subclinical disease is difficult to diagnose and can cause irreversible harm before discernable vision loss.32 There is an unmet and urgent demand for molecular biomarkers that could diagnose glaucoma early, thus making the disease more amenable to treatment and limiting disease progression. Here we present miRNAs that are more abundantly expressed in plasma (and AqH) of glaucoma patients than cataract controls from two ethnic cohorts.
The presence of relatively stable, extracellular miRNAs in plasma has generated great interest in the potential development of miRNAs to study a variety of diseases. miRNAs are easy to sample, highly stable, and simple to quantify, making them well-suited for not only the discovery of disease molecular signatures but also as disease biomarkers themselves. Some primary open angle glaucoma (POAG) endo-phenotypes, including POAG with initial parafoveal scotoma's (IPFS)33,34 and XFG,35 are associated with vascular dysfunction. Hence, circulating molecules (in plasma rather than the eye) may represent particularly useful biomarkers. Therefore, we created a customized miRNA-screen to assess expression levels of glaucoma-specific miRNAs in both plasma and AqH and used this platform to identify candidate biomarkers in clinically relevant samples from glaucoma/XFS patients and cataract controls.
In the current study, circulating levels of 20 miRNAs were higher in glaucoma/XFS patients than in cataract patients. One miRNA combination (miR-637, miR-1306-5p, miR-3159) demonstrated the best ability (AUC = 0.91), relative to single biomarkers, to classify glaucoma patients and control subjects, with high sensitivity (85%) and specificity (87.5%). Of note, miR-637 also performed well alone to identify glaucoma in AqH samples.
Despite low miRNA abundance and high sample variability observed in AqH, 6 of the 20 miRNAs more highly expressed in plasma samples from glaucoma and XFS patients than controls were also elevated in matched AqH samples from patients than controls. Detection of plasma biomarkers suggests that miRNAs related to glaucoma/XFS may not only derive from ocular tissues but may have a systemic origin. Moreover, because some miRNAs were similarly elevated with glaucoma/XFS in both plasma and AqH, there was overlap in predicted mRNA pathways systemically and in the eye (Supplementary Table S2). Whether the blood-aqueous barrier excludes circulating miRNAs from the AqH, as it does for proteins, or whether miRNAs can freely diffuse from vitreous to AqH remains a matter of ongoing debate.36 It also remains to be discovered what the source is of miRNAs detected in AqH and whether any originate from the retina or from the vasculature, possibly reflecting systemic conditions.
Few studies have so far compared miRNAs from AqH in glaucoma and control subjects.17,18,37 The first study detected 500 miRNAs using a commercially available 3D-Gene human miRNA chip-based analysis system containing miRNAs (n = 5 glaucoma, n = 10 control), identifying a subset of miRNAs that differed between groups.17 In another study,37 the potential of developing miRNAs as glaucoma biomarkers was illustrated by the differential expression of miRNAs in AqH from glaucoma patients (n = 6) and age-matched cataract controls (n = 8). Our study is the first to report glaucoma-specific biomarkers in plasma and AqH from more than 50 subjects. More recently, several POAG and XFG-associated miRNAs (querying 800 miRNAs) were identified in AqH, including miR-122-5p, miR-125b-5p, miR-302d-3p, miR-320a, miR-320e, miR-451a, miR-630, and miR-3144-3p.18 Comparison of the miRNA profile in AqH to unrelated human serum exposed potential relationships between these two fluids, but serum and AqH miRNA profiles were not significantly correlated. Interestingly, the most abundant miRNA in normal serum detected in the latter study (miR-451a) was the second-most abundant miRNA (behind miR-940) in the plasma in the current study, validating our platform. Although abundance of the top 20 miRNAs in AqH from cataract patients (n = 4) has been compared with previously published next generation sequencing reads from 10 human blood samples,38 to our knowledge, no published data identifies glaucoma-specific differential expression of miRNAs in plasma, a more readily available source of biomarkers than AqH, nor have miRNA expression levels been assessed in autologous AqH and plasma samples.
miRNAs have been identified as promising diagnostic markers for various cardiovascular diseases,7,39,40 cancer,8 and neurological disorders.41 In fact, more than 100 circulating miRNAs have been identified as biomarkers for different diseases.42 Previous studies have also identified miRNAs in cell types relevant to ocular (patho)physiology,14,16,43–45 vitreous humor,46 and AqH.47,48 Here, the combination of miR-637, miR-1306-5p, and miR-3159 displayed the strongest correlation with glaucoma/XFS. Neither miRNA had previously been described in an ocular context, nor associated with glaucoma or XFS. miR-637 was significantly elevated in both the AqH and plasma of patients compared to controls across both ethnic cohorts. miR-637 acts as a tumor suppressor in hepatocellular carcinoma49 and gliomas,50 and controls osteoblast and adipocyte differentiation,51 and C-reactive protein expression.52 In addition, miR-637 was identified as a putative predictive biomarker for long-term mortality after acute ischemic stroke.52 Similar to miR-637, miR-608 was previously determined to have anti-oncogenic properties in hepatocellular carcinoma, possibly by modulating expression of macrophage inhibitory factor,53 and in bladder cancer.54 Of all the miRNAs studied in the custom array, miR-940 was the most abundant, in both sample types examined. In contrast to a previous report that miR-940 was differentially expressed in AqH of 10 glaucoma patients and 5 control subjects,17 miR-940 expression levels did not differ between glaucoma and controls in our study. However, our results do coincide with the same prior study for miR-4725-3p: Both studies identified this miRNA as elevated in glaucoma (plasma and AqH in our study, AqH in Tanaka and colleagues17). Circulating levels of miR-4448 were also consistently higher in glaucoma and XFS patients than cataract controls; a prior next-generation sequencing study identified miR-4448 as one of the more prevalent miRNAs in AqH.38 Together, these studies validate our approach and suggest that the miRNAs identified here are putative biomarkers. Other miRNAs of interest described previously in glaucoma patients did not differ between patients and controls (including miR-143, miR-518d, miR-660, miR-3185, miR-3663-3p, and miR-4449) or were not tested on our array.17,18,37
Our study provided an opportunity to gain mechanistic insight into a complex disease process by examining putative genetic targets of the 20 identified miRNAs. Molecular target prediction and pathway analysis revealed potential mechanisms contributing to the pathophysiology of glaucoma. Enriched pathways included neuroinflammation signaling, eNOS and nNOS signaling, and neurotrophin/TRK receptor signaling. Our pathway analyses results, derived from miRNA-mRNA target predictions, are supported by other studies that have directly evaluated gene targets. For example, neurotrophin has been implicated in RGC survival, both in animal models55 and in humans,56 and neurotrophin signaling was identified as one of the most biologically relevant pathways in a recent study identifying miRNAs related to glaucoma.18 Fc-epsilon signaling has additionally been highlighted in a bioinformatics analysis of glaucoma57; enrichment of this immunoglobulin membrane receptor highlights the inflammatory component of the disease. The relevance of nitric oxide (NO) and its downstream target guanylyl cyclase (GC) to the pathophysiology of glaucoma is well-established.58–60 Both NOS and GC are expressed in both the anterior and posterior chambers of the eye. Plasma and AqH levels of NO metabolites and the secondary messenger cyclic guanosine-3′, 5′-monophosphate (cGMP), produced by GC upon activation by NO, are lower in glaucoma patients than controls.61,62 Genetic association studies have identified multiple variants in genes of the NOS signaling pathway that are associated with glaucoma.63–68 Furthermore, NO is emerging as a novel target for therapeutic lowering of IOP.69 Additional evidence for a central role of this pathway in the etiology of glaucoma may stimulate development of novel glaucoma drugs that target the NO-cGMP signaling pathway, including small molecules that synergistically increase GC enzyme activity with NO.70,71
There are limitations to the present study. Samples were collected from primarily female patients with established disease and being treated with a variety of medications, both in the glaucoma and control (cataract) group. Many of the patients included in Cohort 2 were diagnosed with XFS with no definite evidence of glaucoma, such as optic disk cupping or readily observable RNFL defects. It remains to be determined whether differences in treatment between glaucoma/XFS patients and controls confound the results, whether the predictive value of the identified biomarkers can be recapitulated when comparing glaucoma patients with healthy (non-cataract) controls, or whether these biomarkers can be predictive, maximizing their diagnostic benefit. Additional investigation will be required to assess the validity of miRNA biomarkers in men and women. The observation that plasma rather than AqH is a relevant biomarker source for glaucoma should allow for straightforward follow-up studies. Ultimately, the feasibility of miRNAs as a biomarker will depend on the ability of clinical labs to extract and detect plasma miRNAs reproducibly and reliably. Available miRNA isolation kits and highly sensitive PCR-based assays that include pre-amplification steps allowing for detection of low abundant targets and can be performed by any clinical laboratory alleviate that concern.38 Alternatively, next-generation sequencing can be a powerful tool to assess miRNA levels as it is unbiased and able to detect novel miRNAs. miRFLP assays allow for accurate quantification of miRNAs in small sample sizes.72 These techniques also obviate the historic need to pool small volume samples, allowing analysis of individual samples and maximizing statistical power and clinical utility.
Despite baseline differences between the cohorts that may affect absolute expression, identified miRNAs were differentially expressed between glaucoma patients and cataract controls in both cohorts. Statistical analysis confirmed that there are no differences in miRNA expression patterns between ethnicities related to this disease, as there was no significant interaction effect between the cohort and disease factors for any miRNAs tested. It is possible that observed cohort differences may resolve with increased sample size or they may reflect differences in sample collection methods between the two centers. The observation that the miRNA biomarkers identified in Cohort 1 could be recapitulated in Cohort 2 in patients with XFS—a syndrome that often leads to glaucomatous optic neuropathy and a population potentially enriched for subclinical glaucoma—may suggest that the identified putative biomarkers could indeed be useful for diagnosis in an earlier, subclinical stage of glaucoma. In addition, these miR biomarkers may provide valuable information for understanding the complex pathogenomics of exfoliation glaucoma. Certainly, additional screens of greater sample size and statistical power are required to validate the miRNAs we identified as potential new biomarkers, and to take the first important step toward developing improved diagnostic tools that may significantly impact the quality of care for glaucoma patients.
The data obtained throughout this study identified candidate miRNA biomarkers whose plasma levels are readily screenable and serves as a proof-of-principle, paving the way for future biomarker discovery. Detection of early, subclinical signs of glaucoma or of elevated IOP (the only modifiable risk factor for glaucoma) is essential to develop disease-modifying and potentially preventative treatments. However, diagnosis typically requires visits to ophthalmologists and often involves technically challenging, lengthy, uncomfortable, and expensive imaging protocols. Moreover, the results of these diagnostics are open to interpretation for a variety of reasons (including variability of the measures) or are not sensitive enough to detect the disease until it is too late and irreversible vision loss has occurred. A biomarker that could be readily assayed by clinical laboratories would greatly increase the potential for early detection of this disease. Blood-based biomarkers are a particularly attractive option as they can be assessed noninvasively, quickly, and inexpensively. Our investigation supports the pursuit of future studies to validate specific miRNAs as diagnostic tools for glaucoma.
Supplementary Material
Acknowledgments
Supported by the NEI R01 EY022746-01 grant from the National Institute of Health, National Eye Institute (ESB), the Wild Family Foundation, the Hassenfeld Scholar award, and the K08HL111210 grant from the National Heart, Lung, and Blood Institute (RM) and by the Matthew and Lee Sabatine Research Fund of the New York Glaucoma Research Institute, New York, NY (RR).
Disclosure: A.G. Hindle, None; R. Thoonen, None; J.V. Jasien, None; R.M.H. Grange, None; K. Amin, None; J. Wise, None; M. Ozaki, None; R. Ritch, None; R. Malhotra, None; E.S. Buys, None
References
- 1.Alward WL. The genetics of open-angle glaucoma: the story of GLC1A and myocilin. Eye. 2000;14(pt 3B):429–436. doi: 10.1038/eye.2000.127. [DOI] [PubMed] [Google Scholar]
- 2.Anderson MG, Libby RT, Mao M, et al. Genetic context determines susceptibility to intraocular pressure elevation in a mouse pigmentary glaucoma. BMC Biology. 2006;4:20. doi: 10.1186/1741-7007-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott AW, Bressler NM, Ffolkes S, Wittenborn JS, Jorkasky J. Public attitudes about eye and vision health. JAMA Ophthalmology. 2016;134:1111–1118. doi: 10.1001/jamaophthalmol.2016.2627. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 6.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 8.Jeffrey SS. Cancer biomarker profiling with microRNAs. Nat Biotechnol. 2008;26:400–401. doi: 10.1038/nbt0408-400. [DOI] [PubMed] [Google Scholar]
- 9.Larrea E, Sole C, Manterola L, et al. New concepts in cancer biomarkers: circulating miRNAs in liquid biopsies. Int J Mol Sci. 2016;17:627. doi: 10.3390/ijms17050627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum miRNA: non-invasive biomarkers for Alzheimer's disease. Exp Neurol. 2012;235:491–496. doi: 10.1016/j.expneurol.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao P, Benito E, Fischer A. MicroRNAs as biomarkers for CNS disease. Front Mol Neurosci. 2013;6:39. doi: 10.3389/fnmol.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grassmann F, Schoenberger PGA, Brandl C, et al. A circulating microRNA profile is associated with late-stage neovascular age-related macular degeneration. PLoS One. 2014;9:e107461. doi: 10.1371/journal.pone.0107461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luna C, Li G, Huang J, et al. Regulation of trabecular meshwork cell contraction and intraocular pressure by miR-200c. PLoS One. 2012;7:e51688. doi: 10.1371/journal.pone.0051688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luna C, Li G, Qiu J, Epstein DL, Gonzalez P. MicroRNA-24 regulates the processing of latent TGFbeta1 during cyclic mechanical stress in human trabecular meshwork cells through direct targeting of FURIN. J Cell Physiol. 2011;226:1407–1414. doi: 10.1002/jcp.22476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paylakhi SH, Moazzeni H, Yazdani S, et al. FOXC1 in human trabecular meshwork cells is involved in regulatory pathway that includes miR-204, MEIS2, and ITGβ1. Exp Eye Res. 2013;111:112–121. doi: 10.1016/j.exer.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Paylakhi SH, Yazdani S, April C, et al. Non-housekeeping genes expressed in human trabecular meshwork cell cultures. Mol Vis. 2012;18:241–254. [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka Y, Tsuda S, Kunikata H, et al. Profiles of extracellular miRNAs in the aqueous humor of glaucoma patients assessed with a microarray system. Sci Rep. 2014;4:5089. doi: 10.1038/srep05089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drewry MD, Challa P, Kuchtey JG, et al. Differentially expressed microRNAs in the aqueous humor of patients with exfoliation glaucoma or primary open-angle glaucoma. Hum Mol Genet. 2018;27:1263–1275. doi: 10.1093/hmg/ddy040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasquale LR, Hanyuda A, Ren A, et al. Nailfold capillary abnormalities in primary open-angle glaucoma: a multisite study. Invest Ophthalmol Vis Sci. 2015;56:7021–7028. doi: 10.1167/iovs.15-17860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cousins CC, Chou JC, Greenstein SH, et al. Resting nailfold capillary blood flow in primary open-angle glaucoma. Br J Ophthalmol. 2018] doi: 10.1136/bjophthalmol-2018-311846. [published online ahead of print April 26, [DOI] [PMC free article] [PubMed]
- 21.Bhattacharya SK, Lee RK, Grus FH;, for the Seventh ARVO/Pfizer Ophthalmics Research Institute Conference Working Group. Molecular biomarkers in glaucoma. Invest Ophthalmol Vis Sci. 2013;54:121–131. doi: 10.1167/iovs.12-11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamel K, Bourke L, O'Brien C. Clinical and laboratory biomarkers for pseudoexfoliation syndrome. J Glaucoma. 2018;2(suppl 1):S111–S113. doi: 10.1097/IJG.0000000000000924. [DOI] [PubMed] [Google Scholar]
- 23.Nath M, Halder N, Velpandian T. Circulating biomarkers in glaucoma, age-related macular degeneration, and diabetic retinopathy. Indian J Ophthalmol. 2017;65:191–197. doi: 10.4103/ijo.IJO_866_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vienna, Austria: R Foundation for Statistical Computing; 2016. R: a language and environment for statistical computing [computer program] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JSTOR, Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 26.VennDiagram: generate high-resolution Venn and Euler plots [computer program] 2016.
- 27.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:48. [Google Scholar]
- 29.Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaikh Y, Yu F, Coleman AL. Burden of undetected and untreated glaucoma in the United States. Am J Ophthalmol. 2014;158:1121–1129.e1. doi: 10.1016/j.ajo.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Feke GT, Pasquale LR. Retinal blood flow response to posture change in glaucoma patients compared with healthy subjects. Ophthalmology. 2008;115:246–252. doi: 10.1016/j.ophtha.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 34.Park SC, De Moraes CG, Teng CC, Tello C, Liebmann JM, Ritch R. Initial parafoveal versus peripheral scotomas in glaucoma: risk factors and visual field characteristics. Ophthalmology. 2011;118:1782–1789. doi: 10.1016/j.ophtha.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, He M, Zhou M, Zhang X. Ocular pseudoexfoliation syndrome and vascular disease: a systematic review and meta-analysis. PLoS One. 2014;9:e92767. doi: 10.1371/journal.pone.0092767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freddo TF. A contemporary concept of the blood-aqueous barrier. Prog Retin Eye Res. 2013;32:181–195. doi: 10.1016/j.preteyeres.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayaram H, Phillips JI, Lozano DC, et al. Comparison of microRNA expression in aqueous humor of normal and primary open-angle glaucoma patients using PCR arrays: a pilot study. Invest Ophthalmol Vis Sci. 2017;58:2884–2890. doi: 10.1167/iovs.17-21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wecker T, Hoffmeier K, Plotner A, et al. MicroRNA profiling in aqueous humor of individual human eyes by next-generation sequencing. Invest Ophthalmol Vis Sci. 2016;57:1706–1713. doi: 10.1167/iovs.15-17828. [DOI] [PubMed] [Google Scholar]
- 39.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha T-Y. MicroRNAs in human diseases: from autoimmune diseases to skin, psychiatric and neurodegenerative diseases. Immune Network. 2011;11:227–244. doi: 10.4110/in.2011.11.5.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 43.Drewry M, Helwa I, Allingham RR, Hauser MA. Liu Y. miRNA profile in three different normal human ocular tissues by miRNA-seq. Invest Ophthalmol Vis Sci. 2016;57:3731–3739. doi: 10.1167/iovs.16-19155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karali M, Persico M, Mutarelli M, et al. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016;44:1525–1540. doi: 10.1093/nar/gkw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li N, Cui J, Duan X, Chen H, Fan F. Suppression of type I collagen expression by miR-29b via PI3K, Akt, and Sp1 pathway in human Tenon's fibroblasts. Invest Ophthalmol Vis Sci. 2012;53:1670–1678. doi: 10.1167/iovs.11-8670. [DOI] [PubMed] [Google Scholar]
- 46.Ragusa M, Caltabiano R, Russo A, et al. MicroRNAs in vitreus humor from patients with ocular diseases. Mol Vis. 2013;19:430–440. [PMC free article] [PubMed] [Google Scholar]
- 47.Dunmire JJ, Lagouros E, Bouhenni RA, Jones M, Edward DP. MicroRNA in aqueous humor from patients with cataract. Exp Eye Res. 2013;108:68–71. doi: 10.1016/j.exer.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 48.Dismuke WM, Challa P, Navarro I, Stamer WD, Liu Y. Human aqueous humor exosomes. Exp Eye Res. 2015;132:73–77. doi: 10.1016/j.exer.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JF, He ML, Fu WM, et al. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54:2137–2148. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 50.Que T, Song Y, Liu Z, et al. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34:4952–4963. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 51.Zhang JF, Fu WM, He ML, et al. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell. 2011;22:3955–3961. doi: 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y, Noren Hooten N, Dluzen DF, Martindale JL, Gorospe M, Evans MK. Posttranscriptional regulation of the inflammatory marker C-reactive protein by the RNA-binding protein HuR and MicroRNA 637. Mol Biol Cell. 2015;35:4212–4221. doi: 10.1128/MCB.00645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K, Liang Q, Wei L, Zhang W, Zhu P. MicroRNA-608 acts as a prognostic marker and inhibits the cell proliferation in hepatocellular carcinoma by macrophage migration inhibitory factor. Tumour Biol. 2016;37:3823–3830. doi: 10.1007/s13277-015-4213-5. [DOI] [PubMed] [Google Scholar]
- 54.Liang Z, Wang X, Xu X, et al. MicroRNA-608 inhibits proliferation of bladder cancer via AKT/FOXO3a signaling pathway. Mol Cancer. 2017;16:96. doi: 10.1186/s12943-017-0664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson EC, Guo Y, Cepurna WO, Morrison JC. Neurotrophin roles in retinal ganglion cell survival: lessons from rat glaucoma models. Exp Eye Res. 2009;88:808–815. doi: 10.1016/j.exer.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasutto F, Matsumoto T, Mardin CY, et al. Heterozygous NTF4 mutations impairing neurotrophin-4 signaling in patients with primary open-angle glaucoma. Am J Hum Genet. 2009;85:447–456. doi: 10.1016/j.ajhg.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danford ID, Verkuil LD, Choi DJ, et al. Characterizing the “POAGome”: a bioinformatics-driven approach to primary open-angle glaucoma. Prog Retin Eye Res. 2017;58:89–114. doi: 10.1016/j.preteyeres.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wareham LK, Buys ES, Sappington RM. The nitric oxide-guanylate cyclase pathway and glaucoma. Nitric Oxide. 2018;77:75–87. doi: 10.1016/j.niox.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buys ES, Potter LR, Pasquale LR, Ksander BR. Regulation of intraocular pressure by soluble and membrane guanylate cyclases and their role in glaucoma. Front Mol Neurosci. 2014;7:38. doi: 10.3389/fnmol.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muenster S, Lieb WS, Fabry G, et al. The ability of nitric oxide to lower intraocular pressure is dependent on guanylyl cyclase. Invest Ophthalmol Vis Sci. 2017;58:4826–4835. doi: 10.1167/iovs.17-22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doganay S, Evereklioglu C, Turkoz Y, Er H. Decreased nitric oxide production in primary open-angle glaucoma. Eur J Ophthalmol. 2002;12:44–48. doi: 10.1177/112067210201200109. [DOI] [PubMed] [Google Scholar]
- 62.Galassi F, Renieri G, Sodi A, Ucci F, Vannozzi L, Masini E. Nitric oxide proxies and ocular perfusion pressure in primary open angle glaucoma. Br J Ophthalmol. 2004;88:757–760. doi: 10.1136/bjo.2003.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozel AB, Moroi SE, Reed DM, et al. Genome-wide association study and meta-analysis of intraocular pressure. Hum Genet. 2014;133:41–57. doi: 10.1007/s00439-013-1349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiggs JL, Kang JH, Yaspan BL, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011;20:4707–4713. doi: 10.1093/hmg/ddr382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang JH, Wiggs JL, Rosner BA, et al. Endothelial nitric oxide synthase gene variants and primary open-angle glaucoma: interactions with sex and postmenopausal hormone use. Invest Ophthalmol Vis Sci. 2010;51:971–979. doi: 10.1167/iovs.09-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buys ES, Ko YC, Alt C, et al. Soluble guanylate cyclase α1-deficient mice: a novel murine model for primary open angle glaucoma. PLoS One. 2013;8:e60156. doi: 10.1371/journal.pone.0060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Koolwijk LM, Ramdas WD, Ikram MK, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012;8:e1002611. doi: 10.1371/journal.pgen.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aliancy J, Stamer WD, Wirostko B. A review of nitric oxide for the treatment of glaucomatous disease. Ophthalmol Ther. 2017;6:221–232. doi: 10.1007/s40123-017-0094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evgenov OV, Pacher P, Schmidt PM, Hasko G, Schmidt HH, Stasch JP. NO-independent stimulators and activators of soluble guanylate cyclase: discovery and therapeutic potential. Nat Rev Drug Discov. 2006;5:755–768. doi: 10.1038/nrd2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buys ES, Zimmer DP, Chickering J, et al. Discovery and development of next generation sGC stimulators with diverse multidimensional pharmacology and broad therapeutic potential. Nitric Oxide. 2018;78:72–80. doi: 10.1016/j.niox.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 72.Zhu B, Zhu W, Ye S, et al. Quantification of MicroRNAs in human aqueous humor by miRFLP assay. Exp Eye Res. 2017;162:73–78. doi: 10.1016/j.exer.2017.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.