Abstract
Background
Tripartite motif-containing protein 11 (TRIM11), encoded by the TRIM11 gene, has been studied in some human malignant tumors. MicroRNA-5193 (miRNA-5193) was predicted to target TRIM11, according to bioinformatics data from TargetScan. However, the roles of TRIM11 and miRNA-5193 in prostate cancer remain unknown. This study aimed to investigate the regulatory effects of miRNA-5193 on the expression of TRIM11 in prostate cancer tissues compared with adjacent normal prostate, and in human prostate cancer cell lines, PC3 and DU145 in vitro.
Material/Methods
Prostate tumor tissue and adjacent normal tissue from 137 patients with stage T1c (n=66), stage T2 (n=48), and stage T3 (n=23) prostate cancer were studied. Expression levels of the TRIM 11 protein and the TRIM11 gene in prostate cancer, normal prostate tissue, and human prostate cancer cell lines, PC3 and DU145, were measured by Western blot and quantitative real-time polymerase chain reaction (qRT-PCR), respectively. Transfection with TRIM11 small interfering RNA (siRNA) resulted in gene knockdown. Transfection with a miR-5193 mimic resulted in overexpression of miR-5193. Proliferation and invasion assays were performed for PC3 and DU145 cells in vitro.
Results
TRIM11 expression was upregulated in prostate cancer specimens compared with normal prostate tissue and was significantly correlated with reduced outcome. In human prostate cancer cell lines, PC3 and DU145, TRIM11 overexpression promoted cell proliferation. Upregulation of miR-5193 downregulated the expression of TRIM11.
Conclusions
TRIM11 was upregulated in prostate cancer tissue and was associated with reduced prognosis. TRIM11 expression increased cell proliferation in vitro and was downregulated by miR-5193.
MeSH Keywords: Cell Proliferation, Prognosis, Prostatic Neoplasms
Background
Worldwide, prostate cancer is the second most common malignancy in men, and is one of the leading causes of male cancer-related deaths, especially in economically developed countries [1]. According to the World Cancer Research Fund (WCRF) prostate cancer statistics, the global incidence of prostate cancer is expected to double by 2030, but despite the increasing prevalence, mortality from prostate cancer has not increased at the same rate, which may be partly attributed to the use of serum prostate-specific antigen (PSA) as a screening method [2]. PSA has had a key role in the early detection of prostate cancer, but the use of the PSA biomarker has some limitations in discriminating between benign prostate disease, such as prostatitis, and between benign and malignant prostate tumors, and is currently not used as a prognostic biomarker [3,4]. Further studies are still needed to identify both diagnostic and prognostic biomarkers and potential molecular markers that may lead to the development of new targeted therapy for prostate cancer.
Tripartite motif-containing protein 11 (TRIM11), encoded by the TRIM11 gene, is a member of a large family of TRIM proteins. The TRIM family of proteins is characterized by their shared three conserved structural motifs, including RING-finger, B-Box, and coiled-coil domains, which are also known collectively as the tripartite RBCC (RING, B-box, coiled-coil) or TRIM proteins [5–7]. As with all the TRIM family members, TRIM11 functions as an E3 ubiquitin ligase in humans [8]. Members of the TRIM family have roles as regulators in diverse cell processes, and early published studies have identified roles for TRIM11 in neurodegenerative disorders and viral infections [9]. TRIM11 has been shown to have a role in the regulation of Alzheimer’s disease by destabilizing intracellular humanin, which is a newly identified neuroprotective peptide [10]. TRIM11 has been shown to function as a ubiquitin ligase, controlling the degradation of Pax6, a conserved transcription factor that contains two DNA-binding domains [11]. TRIM11 has also been shown to inhibit HIV infection by reducing levels of the TRIM5α protein [12]. Recently, TRIM11 expression was reported to be increased in high-grade gliomas and to have an oncogenic role and to be associated with the progression of glioma [13]. Chen et al. showed that TRIM11 was upregulated in ovarian cancer tissue, and that knockdown of the TRIM11 gene suppressed proliferation and induced apoptosis of cancer cells [14]. Also, the overexpression of TRIM11 has been shown to promote cell proliferation, migration, and invasion of lung cancer cells [8]. However, the role of TRIM11 in prostate cancer remains poorly understood.
MicroRNAs (miRNAs) are composed of approximately non-protein coding RNAs, which are 22-nucleotides-long, and which regulate post-transcriptional gene expression by targeting the 3′ untranslated regions (3′ UTR) and coding mRNA regions for degradation or translational repression [15,16]. Recently, miRNAs have been shown to provide diagnostic, prognostic, and therapeutic targets for several diseases, including cancer [17]. In this study, miR-5193 was predicted to target TRIM11, according to bioinformatics data from TargetScan.
Therefore, this study aimed to investigate the regulatory effects of miRNA-5193 on the expression of TRIM11 in prostate cancer tissues compared with adjacent normal prostate tissue, and in human prostate cancer cell lines, PC3 and DU145 in vitro. The expression levels of TRIM11 were measured in prostate cancer tissue and correlated with patient outcome. The in vitro study on the effects of TRIM11 on cell proliferation allowed for investigation of the possible regulatory effects of miRNA-5193.
Material and Methods
Patients and specimen collection
Between October 2009 to October 2016, there were 206 men diagnosed with prostate cancer in our hospital. The patients were initially considered for inclusion in the study as they satisfied the following inclusion criteria: all men had available tissue samples from prostatectomy, and medical records; they were clinically followed-up for more than 12 months after surgery; there were no severe perioperative complications, and no other tumors were identified; none of the patients had chemotherapy or radiotherapy before surgical resection. The 95% confidence interval (CI) level was used with a confidence interval set at 5, which showed that at least 134 patients were required for the study population.
Therefore, we randomly selected 137 patients from the initial population of 206. The cohort of 137 men with prostate cancer included 66 patients with stage T1c, 48 patients with stage T2, and 23 patients with stage T3. The diagnosis of prostate cancer was histologically confirmed in all cases. Prostate cancer samples and the matched adjacent normal tissue specimens were obtained from the Shandong Provincial Hospital Affiliated to Shandong University (Shandong, China), and were sampled by pathologists. This study was approved by the Ethical Committee for Clinical Research, and all study participants signed informed consent forms.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissue samples containing prostate cancer and adjacent normal prostate tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). A qRT-PCR kit (Takara Bio Inc., Kusatsu, Shiga, Japan) was used to prepare cDNA according to the manufacturer’s instructions. The gene expression of TRIM11 was detected using PCR with SYBR Premix Ex Taq II (Takara Bio Inc., Kusatsu, Shiga, Japan). The 2−ΔΔCt method was used to calculate the expression of TRIM11 relative to β-actin. Experiments were performed in triplicate for each data point. The primers included in this study (Genepharma, Shanghai, China) were as follows:
TRIM11 forward: GAGAACGTGAACAGGAAGGAG;
TRIM11 reverse: CCATCGGTGGCACTGTAGAA;
β-actin forward: GACCTCTATGCCAACACAGT;
β-actin reverse: AGTACTTGCGCTCAGGAGGA.
Cell culture and transfection conditions
Human prostate carcinoma cell lines, PC3 and DU145, were obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The cell lines were cultured with Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Hyclone, South Logan, UT, USA) in a humidified atmosphere of 5% CO2 at 37°C. The small interfering RNA (siRNA) of TRIM11 used in this study were purchased from Genechem (Shanghai, China). The transfection of TRIM11 siRNA was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
MicroRNA-5193 (miRNA-5193) was predicted to target TRIM11 according to bioinformatics data from TargetScan (http://www.targetscan.org). The miRNA-5193 mimic and negative control were synthesized by RiboBio (Guangzhou, China), and RiboFect™ chemically pure (CP) transfection reagent (RiboBio, Guangzhou, China) was used to transfect miRNA mimics, according to the manufacturer’s recommendations.
Western blot
Total cell extracts were prepared in RIPA buffer (RiboBio, Guangzhou, China) supplemented with a complete protease inhibitor cocktail (Invitrogen, Carlsbad, CA, USA) at 4°C for an hour. The lysates were cleared by centrifugation at 12000 rpm for 15 min, and the protein concentrations of the samples were determined using a Pierce™ BCA protein assay kit (Thermo Fisher Scientific, Pittsburgh, PA, USA). The lysates were isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride (PVDF) membrane filters, and probed with anti-TRIM11 polyclonal antibody and anti-β-actin mouse antibody (Sigma-Aldrich, St Louis, MO, USA), following by incubating with horseradish peroxidase (HRP)-conjugated secondary antibodies (Santa Cruz Biotech., Santa Cruz, CA, USA). Densitometry of obtained signals was semi-quantified with the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Cell proliferation and viability using the MTT assay
The PC3 and DU145 cell lines with transient knockdown of TRIM11 were reseeded in 96-well plates at 3,000 cells per well. The effect of TRIM11 on prostate cancer cell proliferation was determined by the MTT assay performed daily for five days. After adding 20 μL of 5 mg/ml MTT (Abcam, Cambridge, UK) to each well and incubating for another 4 hours, the supernatant was discarded, and 150 μL of dimethyl sulfoxide (DMSO) was added to dissolve the precipitated formazan. Absorbance was measured at 450 nm.
Statistical analysis
All statistical analysis was performed using SPSS version 22.0 Software (SPSS, Inc., Chicago, IL, USA). Student’s t-test (two-tailed) was used to analyze the differences between the two groups. Statistical analysis of the groups was conducted using the chi-squared (χ2) test. Survival curves were performed using Kaplan-Meier curves and the log-rank test was used to compare the curves. The Cox regression model was performed to modify potential prognostic variables. A P-value <0.05 was considered as statistically significant.
Results
Upregulation of the tripartite motif-containing protein 11 (TRIM11) gene correlated with the clinicopathological features in patients with prostate cancer
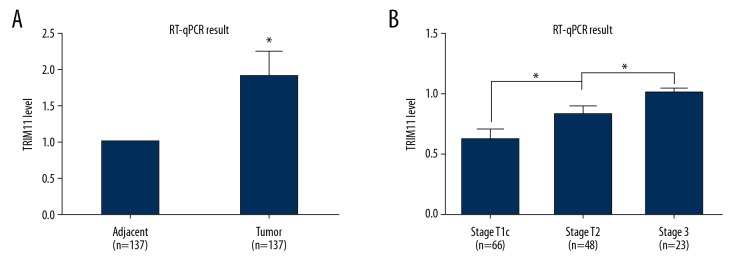
To investigate the role of tripartite motif-containing protein 11 (TRIM11) in cases of prostate cancer, prostate cancer tissue from 137 patients, and the matched adjacent normal prostate tissue, were analyzed for TRIM11 expression was detected by quantitative real-time polymerase chain reaction (qRT-PCR), which showed that TRIM11 was significantly upregulated in prostate cancer compared with normal prostate tissue (Figure 1A). The mRNA expression of TRIM11 was profiled in three stages of prostate cancer, stage T1c, stage T2, and stage T3, and as shown in Figure 1B, patients with a more advanced stage of prostate cancer has significantly increased expression levels of TRIM11. The 137 patients included in the study were divided into low-expression group and a high-expression group, according to the expression level of TRIM11. As summarized in Table 1, TRIM11 over-expression was significantly correlated with an increased serum level of prostate-specific antigen (PSA), advanced tumor stage (TNM stage), and a significantly increased association with seminal vesicle invasion (SVI).
Figure 1.
The tripartite motif-containing protein 11 (TRIM11) gene was upregulated in prostate cancer tissues. (A) TRIM11 mRNA levels in prostate cancer tissues, measured by quantitative real-time polymerase chain reaction (qRT-PCR), were significantly increased when compared with the matched adjacent normal prostate tissue (n=137). * p<0.05. (B) The gene expression profiles of TRIM11 gene in 137 patients with three stages of prostate cancer, stage T1c (n=66), stage T2 (n=48), and stage T3 (n=23) show a positive association with the advanced stage prostate cancer when compared with lower stage prostate cancer. * p<0.05
Table 1.
Clinical correlation between the expression levels of tripartite motif-containing protein 11 (TRIM11) and clinicopathological characteristics of prostate cancer patients.
| Variables | Cases (n) | TRIM11 level | P-value | |
|---|---|---|---|---|
| High (n=73) | Low (n=64) | |||
| Age (years) | 0.332 | |||
| <61 | 66 | 38 | 28 | |
| ≥61 | 71 | 35 | 36 | |
| PSA level (ng/100 mL) | 0.011* | |||
| <10 | 69 | 28 | 41 | |
| 10–20 | 33 | 22 | 11 | |
| >20 | 35 | 23 | 12 | |
| Gleason score | 0.412 | |||
| <7 | 38 | 20 | 18 | |
| 7 | 62 | 30 | 32 | |
| >7 | 37 | 23 | 14 | |
| T stage | <0.001* | |||
| T1c | 66 | 25 | 41 | |
| T2–T3 | 71 | 48 | 23 | |
| Lymph node metastasis | 0.107# | |||
| Negative | 121 | 61 | 60 | |
| Positive | 16 | 12 | 4 | |
| SVI | 0.028* | |||
| Negative | 53 | 22 | 31 | |
| Positive | 84 | 51 | 33 | |
| Surgical margin | 0.154 | |||
| Negative | 106 | 53 | 53 | |
| Positive | 31 | 20 | 11 | |
indicates P <0.05 by the chi-squared test;
P-value analyzed by two-tail Fisher exact test, due to limited case numbers.
PSA – prostate-specific antigen; SVI – seminal vesicle invasion.
TRIM11 expression and prognosis in patients with prostate cancer
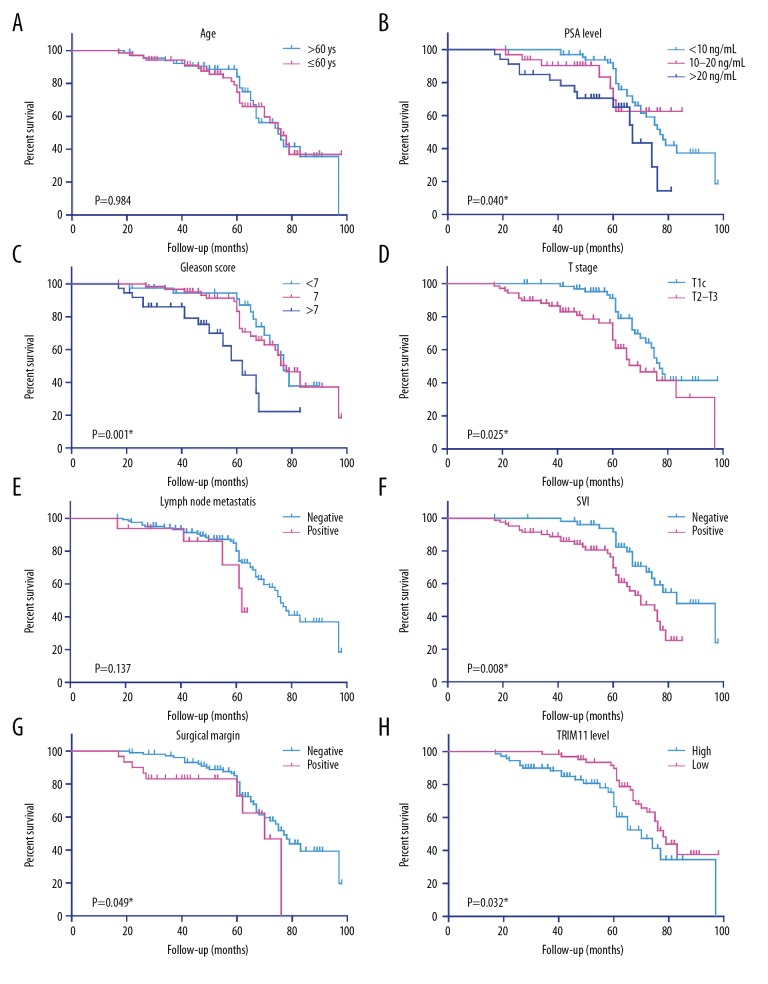
To determine whether TRIM11 expression was a potential prognostic biomarker for prostate cancer, the TRIM11 low-expression group, and the TRIM11 high-expression group were analyzed using Kaplan-Meier plots to determine the associated progression-free survival (PFS). As shown in Figure 2H, men with prostate cancer with high TRIM11 expression levels had a significantly shorter PFS compared with those with a low level of TRIM11 expression (P=0.032). The relationship between the clinicopathological features and PFS were also analyzed. According to the results of analysis using the log-rank test, prostate-specific antigen (PSA) levels (P=0.040), Gleason scores for tumor grade (P=0.001), T stage (in the TNM staging system) (P=0.025), seminal vesicle invasion (SVI) (P=0.008), and measurement of the surgical tumor-free resection margin (P=0.049) were statistically significant prognostic factors (Figures 2B–2D, 2F, 2G) (Table 2). Univariate regression analysis showed that the PSA level (HR=2.111; 95% CI, 1.182–3.767; P=0.012), Gleason score (HR=3.562; 95% CI, 1.783–7.116; P=0.006), seminal vesicle invasion (HR=2.096; 95% CI, 1.100–3.993; P=0.025) and high (compared with low) TRIM11 expression (HR=0.593; 95% CI, 0.325–0.897; P=0.039) were significantly associated with poor outcome (Table 3). In contrast, neither age nor lymph node metastasis affect patients’ PFS according to our data (Figure 2A, 2E). These findings showed that, in this study, TRIM11 was an independent prognostic biomarker in prostate cancer.
Figure 2.
Kaplan-Meier survival analyses of progression-free survival (PFS) in patients with prostate cancer. Kaplan-Meier analysis of the progression-free survival (PFS) in 137 patients with prostate cancer in relation to age (A), prostate-specific antigen (PSA) level (B), Gleason score (C), T stage (D), lymph node metastasis (E), seminal vesicle invasion (SVI) (F), surgical margin (G), and TRIM11 expression level (H).
Table 2.
The progression-free survival (PFS) was assessed by Kaplan-Meier univariate analysis.
| Variables | Cases (n) | 5-year PFS rate (%) | Mean PFS survival (months) | P-value |
|---|---|---|---|---|
| Age (years) | 0.984 | |||
| <61 | 66 | 84.1% | 75.1±3.1 | |
| ≥61 | 71 | 74.6% | 74.9±3.3 | |
| PSA level (ng/100 mL) | 0.040* | |||
| <10 | 69 | 88.5% | 78.6±2.4 | |
| 10–20 | 33 | 69.5% | 72.2±4.0 | |
| >20 | 35 | 65.1% | 60.6±3.8 | |
| Gleason score | 0.001* | |||
| <7 | 38 | 90.8% | 76.0±3.0 | |
| 7 | 62 | 83.3% | 77.7±2.9 | |
| >7 | 37 | 53.4% | 58.7±4.0 | |
| T stage | 0.025* | |||
| T1c | 66 | 91.1% | 80.2±2.5 | |
| T2–T3 | 71 | 66.0% | 69.8±3.7 | |
| Lymph node metastasis | 0.137 | |||
| Negative | 121 | 80.0% | 75.7±2.3 | |
| Positive | 16 | 71.6% | 57.3±3.2 | |
| SVI | 0.008* | |||
| Negative | 53 | 91.5% | 82.0±2.8 | |
| Positive | 84 | 69.8% | 66.2±2.4 | |
| Surgical margin | 0.049* | |||
| Negative | 106 | 81.4% | 77.0±2.3 | |
| Positive | 31 | 72.8% | 62.9±4.1 | |
| TRIM11 level | 0.032* | |||
| High | 48 | 66.6% | 70.3±3.9 | |
| Low | 80 | 89.8% | 79.3±2.6 |
The P-value was analyzed by log-rank test.
Indicates P<0.05 with statistical significance.
PSA – prostate-specific antigen; SVI – seminal vesicle invasion; TRIM11 – tripartite motif-containing protein 11.
Table 3.
Multivariate analysis of progression-free survival (PFS).
| Variable | HR | 95% CI | P-value |
|---|---|---|---|
| PSA (≥10 vs. <10 ng/100 mL) | 2.111 | 1.182±3.767 | 0.012* |
| Gleason score (>7 vs. ≤7) | 3.562 | 1.783±7.116 | <0.001* |
| T stage (T2–T3 vs. T1c) | 1.493 | 0.815±2.737 | 0.194 |
| SVI (positive vs. negative) | 2.096 | 1.100±3.993 | 0.025* |
| Surgical margin (positive vs. negative) | 1.481 | 0.683±3.211 | 0.320 |
| TRIM11 level (low vs. high) | 0.593 | 0.325±0.897 | 0.039* |
Indicates P<0.05 with statistical significance.
CI – confidence interval; HR – hazard ratio; PSA – prostate-specific antigen; SVI – seminal vesicle invasion; TRIM11 – tripartite motif-containing protein 11.
MTT assays with TRIM11 gene silencing increased cell proliferation and cell growth of PC3 and DU145 prostate cancer cells in vitro
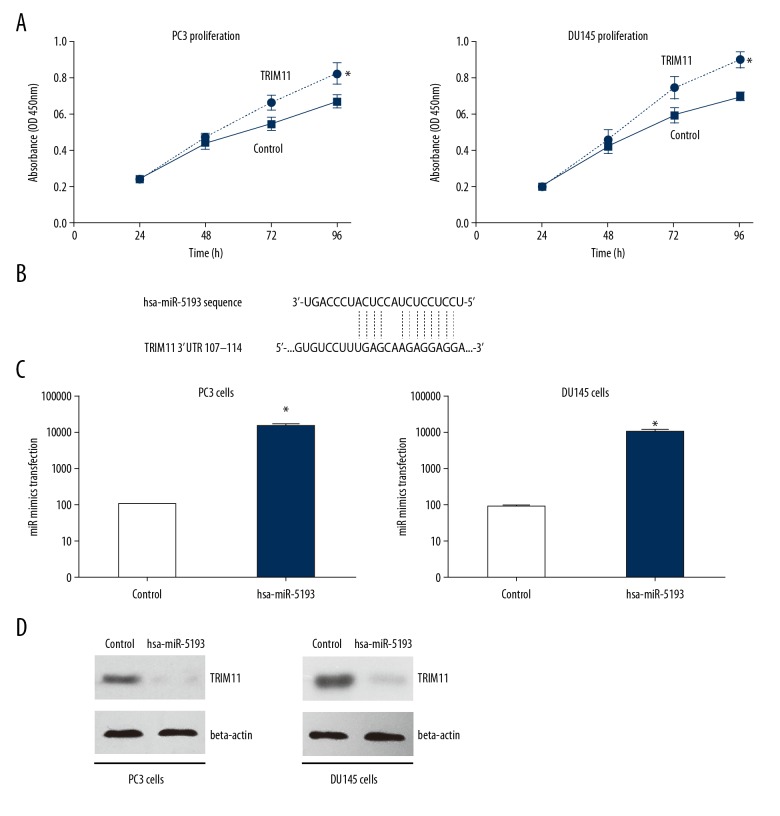
To determine whether TRIM11 was required for prostate cancer cell growth, we depleted TRIM11 was depleted in PC3 and DU145 cell lines with small interfering RNA (siRNA). The MTT assay was used to detect cell proliferation in vitro. As shown in Figure 3A, there was a significant reduction in cell viability of TRIM11-knocked down cells when compared with the control group (Figure 3A). These data showed that TRIM11 had a role in the proliferation of prostate cancer cells in vitro.
Figure 3.
Tripartite motif-containing protein 11 (TRIM11) inhibited prostate cancer growth and was downregulated by miR-5193 in PC3 and DU143 prostate cancer cells in vitro. (A) MTT assays of PC3 and DU143 cells with tripartite motif-containing protein 11 (TRIM11) gene silencing shows the effect of TRIM11 on increasing cell proliferation when compared with the negative control. * P<0.05. (B) The microRNA-5193 (miRNA-5193) binding sites in the 3′UTR of TRIM11 was predicted by TargetScan analysis. (C) The level of miR-5193 in the miR-5193 mimic group was significantly increased compared with the negative control group. * P<0.01. (D) TRIM11 expression was downregulated by overexpression of miR-5193 in both PC3 and DU143 prostate cancer cell lines in vitro.
Upregulation of microRNA-5193 (miRNA-5193) resulted in downregulation of TRIM11 in PC3 and DU145 prostate cancer cells in vitro
The upstream miRNA regulators of TRIM11 were investigated, as microRNA-5193 (miRNA-5193) was predicted to specifically target TRIM11 according to bioinformatics data from TargetScan (http://www.targetscan.org). The targeting sites on 3′-UTR of TRIM11 by miR-5193 are listed in Figure 3B. The miR-5193 mimic was successfully transfected in both PC3 and DU145 cell lines (Figure 3C). The endogenous protein level of TRIM11 was measured by Western blot, which showed that the overexpression of the miR-5193 mimic significantly reduced TRIM11 expression. These findings demonstrated a potential novel signaling axis for miR-5193 and TRIM11 in the development of prostate cancer.
Discussion
Worldwide, prostate cancer is regarded as a serious threat to men’s health and still has a high mortality rate, due to late presentation and lack of sensitive and specific methods of detection of early prostate cancer [18]. Because of the increasing number of aging people in the world, the number of expected cases of prostate cancer is expected to increase in the future [19]. Currently, prostate biopsy and histology remains the definitive method for the diagnosis of prostate cancer but is invasive. The prostate-specific antigen (PSA) test is recommended as a screening method for prostate cancer, but has significant problems with specificity and has limited capacity as a prognostic biomarker [20]. The identification of more sensitive and specific diagnostic and prognostic biomarkers for prostate cancer and a better understanding of the mechanisms underlying the progression of prostate cancer will improve early clinical diagnosis and treatment.
Posttranslational modification of protein affects the activation of protein and the regulation of cell function, including ubiquitination and phosphorylation. E3 ubiquitin ligases belong to a large family of proteins that mediate protein ubiquitination, typically by recruiting a catalytic reaction resulting in protein degradation [21]. Currently, E3 ligases are known to have important roles in cell proliferation and apoptosis and have been considered as the potential novel targets for the treatment of certain diseases [22]. Tripartite motif-containing protein 11 (TRIM11), encoded by the TRIM11 gene, is an E3 ubiquitin ligase. Recent studies have demonstrated that the expression levels of TRIM11 are increased in gliomas [13], ovarian cancer [14], and lung cancers [8]. These studies have shown that TRIM11 has a role in regulating the development of human malignancies.
In the present study, the level of TRIM11 expression in prostate cancer tissue was significantly upregulated when compared with patient-matched, adjacent, non-neoplastic tissues, and an increased expression level of TRIM11 was an independent risk factor for poor prognosis in patients with prostate cancer. This finding indicated that TRIM11 should be studied further as a potential diagnostic and prognostic biomarker for prostate cancer. For the first time, the findings of this study showed, that TRIM11 has a role in the progression of prostate cancer, but also provided results that showed that TRIM11 functions as an oncogenic protein via promoting prostate cancer cell proliferation. The findings of this study also showed that miR-5193 could downregulate TRIM11, as overexpression of miR-5193 significantly decreased the level of TRIM11 expression in both PC3 and DU145 cells in vitro. Yan et al. previously showed that TRIM11 was specifically targeted by miR-24-3p in colon cancer, promoting cell proliferation and inhibiting apoptosis [23]. The findings of the present study, that TRIM11 was a target of miR-5193, might be due to the tissue and cell specificity of miRNAs, which is a finding that has been previously reported [24,25].
This study had several limitations. The study sample population was small and was collected from a single medical center, and may not be representative of prostate cancer in global populations. Also, the signaling pathway remains to be verified in further prostate carcinoma cell lines and animal models. The functional mechanisms of TRIM11 and its other upstream regulators require further investigation in different tumor types.
Conclusions
In this study, in patients with prostate cancer, the tripartite motif-containing protein 11 (TRIM11) gene was significantly upregulated in prostate cancer tissue compared with normal prostate tissue, and increased expression levels were associated with reduced prognosis. In prostate cancer cells in vitro, TRIM11 expression increased cell proliferation and was downregulated by microRNA-5193 (miRNA-5193). The results of this study support that further studies on the miR-5193 and TRIM11 axis may provide future prognostic, diagnostic or therapeutic biomarkers to improve the early diagnosis, treatment, and prognosis for patients with prostate cancer.
Footnotes
Source of support: This study was supported by a grant from the City Technology Bureau (Yue Pan, No. Y20170441)
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Johnson IR, Parkinson-Lawrence EJ, Keegan H, et al. Endosomal gene expression: A new indicator for prostate cancer patient prognosis? Oncotarget. 2015;6:37919–29. doi: 10.18632/oncotarget.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Nunzio C, Kramer G, Marberger M, et al. The controversial relationship between benign prostatic hyperplasia and prostate cancer: The role of inflammation. Eur Urol. 2011;60:106–17. doi: 10.1016/j.eururo.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 4.Tarhan F, Orcun A, Kucukercan I, et al. Effect of prostatic massage on serum complexed prostate-specific antigen levels. Urology. 2005;66:1234–38. doi: 10.1016/j.urology.2005.06.077. [DOI] [PubMed] [Google Scholar]
- 5.Borden KL. RING fingers and B-boxes: Zinc-binding protein-protein interaction domains. Biochem Cell Biol. 1998;76:351–58. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 6.Reymond A, Meroni G, Fantozzi A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–14. [PubMed] [Google Scholar]
- 8.Wang X, Shi W, Shi H, et al. TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. J Exp Clin Cancer Res. 2016;35:100. doi: 10.1186/s13046-016-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Lee Y, Song B, Park C, Kwon KS. TRIM11 negatively regulates IFNbeta production and antiviral activity by targeting TBK1. PLoS One. 2013;8:e63255. doi: 10.1371/journal.pone.0063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niikura T, Hashimoto Y, Tajima H, et al. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer’s disease-relevant insults. Eur J Neurosci. 2003;17:1150–58. doi: 10.1046/j.1460-9568.2003.02553.x. [DOI] [PubMed] [Google Scholar]
- 11.Tuoc TC, Stoykova A. Trim11 modulates the function of neurogenic transcription factor Pax6 through ubiquitin-proteosome system. Genes Dev. 2008;22:1972–86. doi: 10.1101/gad.471708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchil PD, Quinlan BD, Chan WT, et al. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008;4:e16. doi: 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di K, Linskey ME, Bota DA. TRIM11 is overexpressed in high-grade gliomas and promotes proliferation, invasion, migration and glial tumor growth. Oncogene. 2013;32:5038–47. doi: 10.1038/onc.2012.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Sun J, Ma J. Proliferation and invasion of ovarian cancer cells are suppressed by knockdown of TRIM11. Oncol Lett. 2017;14:2125–30. doi: 10.3892/ol.2017.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan ZY, Cai GY, Bu R, et al. Selection of urinary sediment miRNAs as specific biomarkers of IgA nephropathy. Sci Rep. 2016;6:23498. doi: 10.1038/srep23498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian J, Hu L, Li X, et al. MicroRNA-130b promotes lung cancer progression via PPARgamma/VEGF-A/BCL-2-mediated suppression of apoptosis. J Exp Clin Cancer Res. 2016;35:105. doi: 10.1186/s13046-016-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Sethi S, Sethi S, Bluth MH. Clinical implication of microRNAs in molecular pathology: An update for 2018. Clin Lab Med. 2018;38:237–51. doi: 10.1016/j.cll.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 19.Morlando M, Pelullo CP, Di Giuseppe G. Prostate cancer screening: Knowledge, attitudes and practices in a sample of men in Italy. A survey. PLoS One. 2017;12:e0186332. doi: 10.1371/journal.pone.0186332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–28. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 21.Gossage L, Eisen T, Maher ER. VHL, the story of a tumor suppressor gene. Nat Rev Cancer. 2015;15:55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 22.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 23.Yin Y, Zhong J, Li SW, et al. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7:86755–65. doi: 10.18632/oncotarget.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Fan H, Zou Q, et al. TEAD4 exerts pro-metastatic effects and is negatively regulated by miR6839-3p in lung adenocarcinoma progression. J Cell Mol Med. 2018;22:3560–71. doi: 10.1111/jcmm.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiaoli Z, Yawei W, Lianna L, et al. Screening of target genes and regulatory function of miRNAs as prognostic indicators for prostate cancer. Med Sci Monit. 2015;21:3748–59. doi: 10.12659/MSM.894670. [DOI] [PMC free article] [PubMed] [Google Scholar]