Abstract
The aim of the study was to determine the effect of carboxylesterase 1(CES1) genetic variation on the activation of angiotensin-converting enzyme inhibitor (ACEI) prodrugs. In vitro incubation study of human liver, intestine, kidney s9 fractions demonstrated that the ACEI prodrugs enalapril, ramipril, perindopril, moexipril, and fosinopril are selectively activated by CES1 in the liver. The impact of CES1/CES1VAR and CES1P1/CES1P1VAR genotypes and diplotypes on CES1 expression and activity on enalapril activation was investigated in 102 normal human liver samples. Neither the genotypes nor diplotypes affected hepatic CES1 expression and activity. Moreover, among several CES1 nonsynonymous variants studied in transfected cell lines, the G143E (rs71647871) was a loss-of-function variant for the activation of all ACEIs tested. The CES1 activity on enalapril activation in human livers with the 143G/E genotype was approximately 1/3 of that carrying the 143G/G. Thus, some functional CES1 genetic variants (e.g. G143E) may impair ACEI activation, and consequently affect therapeutic outcomes of ACEI prodrugs.
Introduction
Angiotensin-converting enzyme inhibitors (ACEIs) are among the most prescribed medications in the world, and are the cornerstone for the treatment of patients with hypertension, heart failure, and chronic kidney diseases. The responses to ACEIs therapy vary significantly within individual patients. A meta-analysis concluded that relative to other antihypertensives, the use of ACEIs was associated with wider interindividual variations in systolic blood pressure (BP) 1. In fact, target BP was not achieved in approximate 50% of the intent-to-treat patients receiving ACEIs 2–7. Additionally, unacceptable side effects are commonly reported in patients treated with ACEIs 8. The current clinical management of ACEI pharmacotherapy is largely based on an empirical “trial and error” approach due to the lack of reliable predictors of drug response. Therefore, there is a pressing need to identify the factors contributing to interindividual variability in responses to ACEI therapy.
With the exception of lisinopril and captopril, all ACEIs are formed as ester prodrugs to improve otherwise poor bioavailability. The activation of ACEI prodrugs is fundamental for successful ACEI pharmacotherapy since the active metabolites are 10–1000 times more potent on ACE inhibition compared to their respective parent compounds. Carboxylesterase 1 (CES1) is the major hydrolase in humans, contributing to 80%- 95% of total hepatic hydrolytic activity 9, while carboxylesterase 2 (CES2), another primary hydrolase involved in drug metabolism in humans, attributes to the residual 5%−20% activity in the liver10. CES1 and CES2 exhibit distinct substrate specificity, i.e. CES1 is highly efficient for hydrolyzing the substrates with small alcohol group and large carboxyl group whereas CES2 prefers to hydrolyze the esters with bulky alcohol group11. CES1-mediated hydrolysis is involved in the deactivation of many medications such as methylphenidate and clopidogrel12, 13. Additionally, CES1 is critical for the activation of a number of prodrugs such as oseltamivir and several ACEIs including trandolapril, benazepril, quinaparil, temocapril, cilazapril, delapril and imidapril 14–16. CES2 is the enzyme responsible for the metabolism of several ester medications, such as aspirin and irinotecan, but is not involved in the activation of several ACEI prodrugs studied previously 15, 17, 18.
CES1 is encoded by CES1 gene. CES1 consists of 14 exons located on chromosome 16q13-q22.1. The CES1P1 gene is a nonfunctional pseudogene located in proximity with the CES1 gene. The CES1P1VAR is a functional variant of CES1P1, which is identical to the CES1 gene except the differences of 5 nucleotides in the exon 1. The CES1P1VAR encodes for the same protein as the CES1 gene. Interestingly, the exon 1 of CES1 gene can be converted to that of CES1P1VAR resulting in the CES1 variant CES1VAR 19. The CES1 and CES1P1 gene structures and its nomenclature were illustrated in the Supplementary Figure S1. It has been speculated that the CES1/CES1VAR and CES1P1/CES1P1VAR genotypes could affect expression levels of CES1, and consequently affect the metabolism of CES1 substrate drugs. However, the findings from the published studies were inconsistent or even contradictive 20,21.
Besides the CES1/CES1VAR and CES1P1/CES1P1VAR genotypes, over 1000 single nucleotide polymorphisms (SNPs) have been identified in coding and non-coding regions of CES1 gene. Some of these variants, such as the G143E (rs71647871) originally discovered in our laboratory, markedly affected CES1 activity 13, 15, 22, 23, and significantly altered pharmacokinetics (PK) and/or pharmacodynamics (PD) of several drugs metabolized by CES1, including methylphenidate, clopidogrel, and oseltamivir 23–26. Therefore, these functional CES1 SNPs may have the potential to impair the activation of ACEI prodrugs, and lead to therapeutic failure. In the present study, we demonstrated that hepatic CES1 is the enzyme responsible for the activation of the ACEI prodrugs enalapril, ramipril, perindopril, moexipril, and fosinopril. We then assessed the effect of the CES1/CES1VAR and CES1P1/CES1P1VAR genotypes, diplotypes, and several selected CES1 nonsynonymous SNPs on the activation of ACEI prodrugs utilizing transfected cell lines and a large set of individual human liver samples.
Materials and Methods
Materials
Enalapril maleate, moexipril hydrochloride, perindopril erbumine, and 5-hydroxyomeprazole were purchased from Sigma-Aldrich (St. Louis, MO). Fosinopril sodium salt and ramipril were products from Toronto Research Chemicals Inc. (Toronto, Canada). The enalapril hydrolytic metabolite enalaprilat dehydrate was purchased from Sellechchem (Houston, TX). The hydrolytic metabolites of other tested ACEI prodrugs including ramipril, perindopril, moexipril and fosinopril were obtained via incubation of the parent compounds (100 μM, 30 μM, 40 μM, 40 μM, respectively) with the cell s9 fractions (10 mg protein/ml) prepared from the transfected cells stably expressing wild type (WT) CES1 23. The hydrolytic reactions for ramipril, perindopril, moexipril and fosinopril were completed at 37°C in 20min, 1h, 24h, and 24h, respectively. The completion of the CES1-mediated biotransformation of those ACEI prodrugs was confirmed by monitoring the reduction of the parent compounds via an HPLC-MS/MS analysis. Pooled human liver s9 fractions (HLS9), human intestine s9 fractions (HIS9), and human kidney s9 fractions (HKS9) were obtained from XenoTech LLC (Lenexa, KS). CES1 antibody was the product from Abcam Inc. (Cambridge, MA). Taq polymerase and MseI restriction endonuclease were obtained from New England Biolabs Inc. (Ipswich, MA). All other chemicals and agents were of the highest analytical grade commercially available.
A total of 102 individual normal human liver samples were obtained from the XenoTech LLC (Lenexa, KS) and the Cooperative Human Tissue Network (CHTN, Columbus, OH). The liver samples consist of 46 males and 56 females with ages ranging from 22 to 81 years. The donors included 92 Caucasians, 6 African-Americans, 2 Hispanics, and 2 classified as ‘others’.
Preparation of human liver S9 fractions
To prepare individual HLS9 samples from 102 human liver tissues, approximate 200 mg frozen tissues were sliced into 1×1×1 mm pieces, and homogenized in 0.5mL ice-cold phosphate buffered saline (PBS) in 1.5 ml microcentrifuge tubes using microcentrifuge pestle (VWR International LLC, Chicago, IL). The homogenate was centrifuged for 20 min at 9000 ×g at 4 °C, followed by the removal of fats on the top layer. The S9 fractions were then transferred to new centrifuge tubes for centrifugation at 9000 ×g at 4 °C for another 20 min 27. The remaining fats were carefully removed, and the S9 fractions were collected and diluted to 2 mg/ml with PBS. Pooled HLS9 samples from these 102 liver tissues were prepared by mixing an aliquot (20 μl) from each of HLS9 (2 mg/ml) samples. The HLS9 samples were stored at −80 °C until use.
CES1 genotyping
Total genomic DNA was extracted from 102 human liver tissues with Pure Link™ Genomic DNA Mini Kit (Life technology, Austin, TX) according to the manufacture’s instruction. The CES1 and CES1VAR genes differ in a set of variants located in the exon 1 of the genes 21.To determine CES1/CES1VAR genotypes, PCR was carried out to amplify the exon 1 of the CES1 and CES1VAR genes using the PCR conditions and primers outlined in the Table S1 and Table S2. The PCR products were purified with Pure Link™ Quick PCR Purification Kit (Life technologies, Austin, TX), then subjected to Sanger sequencing. The first batch of 10 samples were bidirectionally sequenced using the same primers for the PCR reactions (Supplementary Table S2). The data form the two sequencing directions were in full agreement with each other. Thereafter, the samples were sequenced using the forward primers.
It is challenging to determine the CES1P1/CES1P1VAR genotypes due to high similarity of DNA sequences between the CES1 and CES1P1 genes. A restriction fragment length polymorphism (RFLP) method was developed and utilized for the determination of CES1P1/CES1P1VAR genotypes in two previously published studies 21, 28. This assay was based upon the differences of several nucleotides in the exon 5 between the CES1P1 and CES1P1VAR (Figure S2). However, the CES1P1 genotype is defined by the presence of a stop codon in exon 3 of the gene, and the stop codon cannot be directly detected by this RFLP assay. Though the authors claimed that the nucleotides for differentiating CES1P1 from CES1P1VAR are in complete linkage with the CES1P1 stop codon 21, we feel that the assay needs to be validated independently. Accordingly, we developed a novel RFLP CES1P1/CES1P1VAR genotyping method by taking advantage of the differences of exon 3 between the CES1P1 and CES1P1VAR. In brief, a set of primers were designed based on the shared sequences flanking the exon 3 of the CES1, CES1VAR, CES1P1, and CES1P1VAR genes for the amplification of the exon 3 where the stop codon of CES1P1 resides. Thus, the PCR products contain the exon 3 of all 4 genes if present. The primers sequences and PCR conditions were summarized in the Supplementary Table S1 and S2. The digestion of the PCR products with the restriction endonuclease MseI at 37ºC for 1h yielded 359 and 130 bp fragments for the CES1, CES1VAR and CES1P1VAR genes, and 246 and 243 bp fragments for the CES1P1 gene. The purified digestions (10 μl) were electrophoresed on 2% agarose gel and visualized by ethidium bromide staining. The 246 bp and 243 bp fragments were observed as a single band under the present experimental conditions. The ratios of the band intensities of the 246/243 bp to the 359 bp fragments were quantified and normalized for the fragment lengths for the determination of CES1P1/CES1P1VAR genotypes. The ratios of 246/243 bp to 359 bp bands are 1:0.7 and 1:2.2 for CES1P1/CES1P1 and CES1P1/CES1P1VAR, respectively, while the 246/243 band is absent for the CES1P1VAR/CES1P1VAR genotype. The RFLP assay is illustrated in details in the Supplementary Figure S3. Additionally, all liver samples were genotyped for the CES1P1/CES1P1VAR genotypes using the previous exon 5-based RFLP assay for cross validation of the two genotyping methods 21, 28. Briefly, the exon 5 was amplified with the primers (CES1/1P1-int4F and CES1/1P1-int5R) under the thermocycling conditions summarized in Supplementary Table S1 (C) and Table S2. The PCR products were incubated with Pvu II restriction endonuclease at 37ºC for 8h for complete digestion of CES1P1 PCR products. The CES1P1/CES1P1VAR genotypes were discriminated based on the different ratios of the intensities of the 407bp to 248bp bands (Supplementary Figure S2). According to the combination of CES1/CES1VAR and CES1P1/CES1P1VAR genotypes, 4 CES1 and CES1P1 haplotypes were designated as haplotypes A to D (Supplementary Figure S1), which form 10 possible diplotypes (Table 1).
Table 1.
Inferred diplotypes based on CES1/CES1VAR and CES1P1/CES1P1VAR genotypes
| Diplotypes | A/A | A/B | A/C | A/D | B/B | B/C | B/D | C/C | C/D | D/D |
|---|---|---|---|---|---|---|---|---|---|---|
| CES1/CES1VAR genotypes | 1/1 | 1/1 | 1/1VAR | 1/1VAR | 1/1 | 1/1 VAR | 1/1VAR | 1VAR/1VAR | 1VAR/1VAR | 1VAR/1VAR |
| CES1P1/CES1P1 VAR genotypes | 1P1/1P1 | 1P1/1P1VAR | 1P1/1P1 | 1P1/1P1VAR | 1P1VAR/1P1VAR | 1P1/1P1VAR | 1P1VAR/1P1VAR | 1P1/1P1 | 1P1/1P1VAR | 1P1VAR/1P1VAR |
| Number of functional Genes | 2 | 3 | 2 | 3 | 4 | 3 | 4 | 2 | 3 | 4 |
Note: The diplotypes A/D and B/C share the same genotypes CES1/CES1VAR and CES1P1/CES1P1VAR and cannot be differentiated under the present experimental conditions.
To determine the G143E genotypes, the exon4 of CES1 were amplified with the experimental conditions and primers described in the Supplementary Table S1 and Table S2. The PCR products were purified with Pure Link™ Quick PCR Purification Kit (Life technologies, Austin, TX), then subjected to Sanger sequencing to determine the presence of the variant utilizing the same PCR primers indicated in the Supplementary Table S2.
Western blot studies
Western blot studies were conducted to determine CES1 expression levels in individual human liver tissues. Protein concentrations of HLS9 samples were measured using a Pierce BCA assay kit (Rockford, IL). The expression of CES1 in individual human liver S9 fraction samples was determined using a western blot assay similar to that we published previously 15, with the exception that, instead of β-action expression, total protein staining (Pierce reversible protein stain kits, Thermo Scientific, Rockford, IL) was performed to ensure that equal amounts of proteins were loaded for analysis as our preliminary study demonstrated significant interindividual variability in β-action expression in some human liver samples. Pooled HLS9 samples with total protein amounts ranging from 0.2 to 3.75 μg were included in each run for semi-quantification of CES1 expression to control for inter-blot variations.
Hydrolysis of enalapril, ramipril, perindopril, moexipril, and fosinopril by human liver, intestine and kidney s9 fractions
An incubation study was conducted to assess enzymatic activity of HLS9, HIS9 and HKS9 on the hydrolysis (activation) of enalapril, ramipril, perindopril, moexipril, and fosinopril. Preliminary studies revealed significant nonspecific bindings of enalapril and enalaprilat to plastic test tubes. Therefore, the enalapril incubation experiment was carried out in 4 ml silanized glass vials, while the other 4 ACEIs were incubated in 1.5 ml Eppendorf tubes. The parent compound solutions were freshly prepared in phosphate buffered saline (PBS, pH 7.4) at a concentration of 200 μM, and s9 fraction samples were diluted in PBS as well. The reaction was initiated by mixing 100 μl of substrates with 100 μl of different concentrations (0.2, 0.4, 1.0 mg/ml) of HLS9, HIS9, or HKS9.
To determine kinetic parameters of the HLS9-catalyzed hydrolysis of the ACEI prodrugs, a series concentrations of enalapril (2, 10, 20, 50, 100, 200, 500, 1000 μM), ramipril (10, 20, 50, 100, 200, 500, 1000, 2000 μM), perindopril (10, 20, 50, 100, 200, 500, 1000, 2000, 4000 μM), moexipril (10, 50, 100, 200, 500, 1000, 2000, 4000 μM), and fosinopril (20, 100, 500, 1000, 2000 μM) were incubated with HLS9 in PBS buffer (pH 7.4). The final concentrations of HLS9 were 0.5, 0.1, 0.1, 0.2, and 0.2 mg/ml for enalapril, ramipril, perindopril, moexipril, and fosinopril, respectively. After incubation at 37ºC for 10 min (enalapril, ramipril and moexipril) or 5 min (perindopril and fosinopril), the reactions were terminated by adding a 4-fold volume of methanol containing analytical internal standards. The internal standards for the analysis of enalaprilat, moexiprilat, and fosinoprilat were 5-hydroxyomeprazole (10 ng/ml), fosinoprilat (50 ng/ml), and moexiprilat (50 ng/ml), respectively. Enalaprilat (35 ng/ml) was used as the internal standard for the analysis of both ramiprilat and perindoprilat. A preliminary experiment confirmed that the formation of active metabolites was linear with the concentrations of HLS9 (0.1 – 1.0 mg/ml) and the incubation times (5 – 10 min). Following the addition of methanol, the samples were vortexed briefly, and then centrifuged at 13,200 rpm at 4ºC for 20 min to remove precipitated proteins. The supernatant was collected and analyzed for the concentrations of the active metabolites enalaprilat, ramiprilat, perindoprilat, moexiprilat, and fosinoprilat utilizing the HPLC-MS/MS methods described below.
To determine CES1 activity in 102 individual human liver samples, the s9 fractions prepared from those samples were incubated with enalapril at 37ºC for 10 min. The final concentrations of the HLS9 and enalapril were 0.5 mg/ml and 500 μM, respectively. Following incubation, the reactions were terminated, and the concentrations of the metabolite enalaprilat were determined using the aforementioned methods.
Hydrolysis of enalapril, ramipril, perindopril, moexipril, and fosinopril by wild type CES1 and the variants G19V, S83L, G143E, and A270S
The established cell lines stably expressing WT CES1 as well as the variants G19V, S83L, G143E, and A270S were used to investigate the effect of those CES1 nonsynonymous variants on ACEIs activation 23. Cells transfected with vector plasmid were included as negative control. The s9 fractions of the transfected cells were prepared according to a previously described method 13, 15. To demonstrate the effect of the variant G143E on the hydrolysis of enalapril, ramipril, perindopril, moexipril and fosinopril, the in vitro hydrolysis experiments were performed utilizing the methods similar to that of the HLS9 incubation study described above except that the incubation time for ramipril and moexipril was adjusted to 5 min, and the s9 concentrations for the kinetic studies of ramipril, moexipril and fosinopril were 0.05, 0.1, and 0.1 mg/ml, respectively. The influence of other three CES1 SNPs, G19V, S83L and A270S, on enalapril activation was evaluated by comparing their catalytic activities on the hydrolysis of enalapril (500 μM) to that of the WT CES1. CES1 specific activity was calculated by subtracting non-enzymatic hydrolysis of the prodrugs observed after incubation with the vector cell s9 fractions.
HPLC-MS/MS assay
HPLC-MS/MS assays were developed to determine the concentrations of the ACEI active metabolites enalaprilat, ramiprilat, perindoprilat, moexiprilat, and fosinoprilat based on previously published methods with some modifications 29–33. The HPLC-MS/MS system consists of a Shimadzu HPLC system and an Applied Biosystems API 4000 QTRAP® mass spectrometer. The analytes were separated on a reverse phase column (Restek, Ultra II® C18, 2.0 × 150 mm, 5 micron). A constant flow rate (0.3 ml/min for enalapril, ramipril, moexipril and fosinopril and 0.4 ml/min for perindopril) was applied using the gradient conditions summarized in the Supplementary Table S3. Column temperature was set at 50ºC for enalaprilat and ramiprilat, 65ºC for perindoprilat, and 55ºC for moexiprilat and fosinoprilat. An injection volume of 10 μl was used for all analytes.
Ionization was achieved via positive electrospray ionization (ESI) mode, and ions were monitored by multiple reaction monitoring using the following m/z transitions: enalapril (377.0>234.0), ramipril (417.4>234.3), perindopril (369.4>172.4), moexipril (499. 4>234.2), fosinopril (564.3>346.2), enalaprilat (349.0>206.0), ramiprilat (389.3>206.2), perindoprilat (341.0>170.2), moexiprilat (471.3>238.2), fosinoprila (436.2>390.2), and 5-hydroxyomeprazole (362.3>213.9). The collision energy was 26 eV for enalapril and ramipril, and 20 eV for perindopril, moexipril, and fosinopril. Ionspray voltage, source temperature, and curtain gas was 3500 V, 350ºC and 25 psi, respectively, for all analytes.
The methods were validated for precision and accuracy by analyzing blank samples spiked with the analytes at three concentrations; The precision (CV%) was <11%, and the accuracy was 89.8% to 103.2% for all analytes.
Data analysis
Data are presented as mean ± SD of triplicated independent experiments. Data were fit to the Michaelis-Menten equation for the analysis of the kinetic parameters Km and Vmax via nonlinear regression analysis. Apparent intrinsic clearance (Clint) was calculated as the ratio of Vmax to Km. Mann-Whitney test, one way ANOVA, and linear regression analysis were utilized to analyze the differences of CES1 expression and activity between genotypes, among diplotypes, and the correlation between CES1 expressions and activities, respectively (Graphpad Prism software version 6.0; Graphpad Software, Inc., San Diego, CA). The western blot results were semiquantified with VisionWorks®LS program.
Results
Enalapril, ramipril, perindopril, moexipril, and fosinopril were hydrolyzed to their respective metabolites by human liver but not intestine and kidney s9 fractions
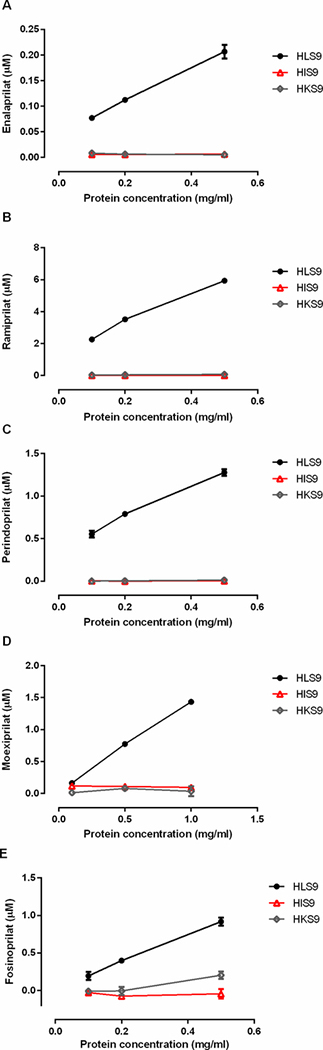
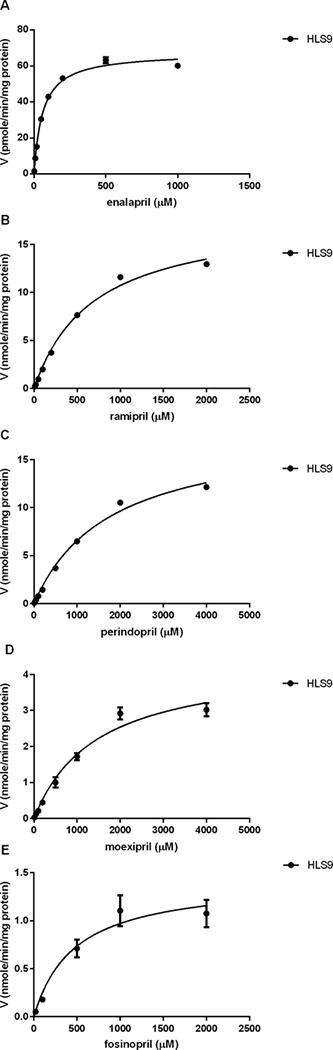
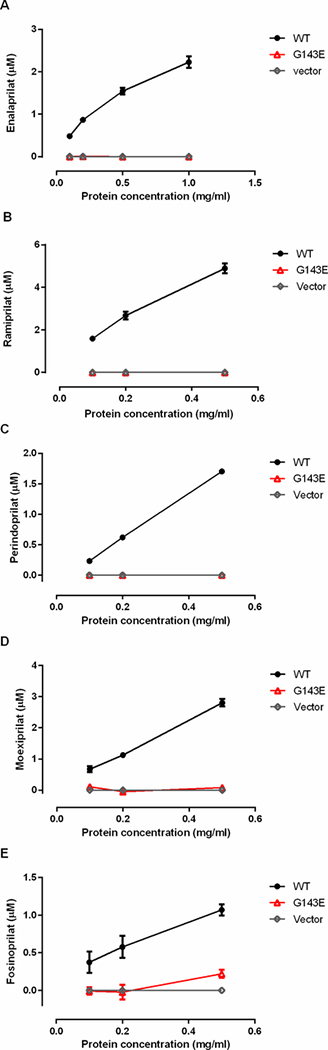
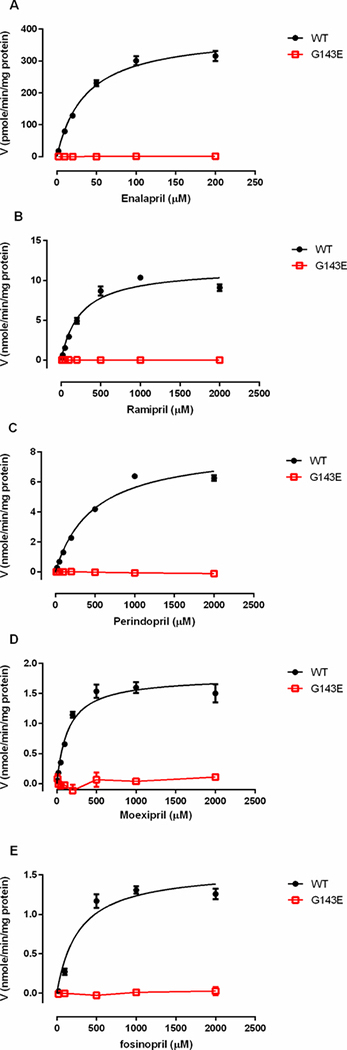
The incubation of the tested ACEI prodrugs with different concentrations of human liver, intestine, and kidney s9 fractions demonstrated that enalapril, ramipril, perindopril, moexipril and fosinopril were efficiently hydrolyzed to their respective active metabolites by human liver but not intestine or kidney s9 fractions (Figure 1). The formation of the active metabolites increased in proportion to the increase of the concentrations of HLS9. No appreciable activation of the tested ACEI prodrugs was observed after incubation with various concentrations of HIS9 and HKS9. To evaluate hydrolytic activity in the HIS9 and HKS9 samples, p-nitrophenyl acetate (PNPA) was included as the positive substrate in the incubation study under the same conditions for ACEI prodrugs. As expected, PNPA was readily hydrolyzed to its metabolite p-nitrophenol (PNP) following the incubation (Supplementary Figure S4). In humans, CES1 is the most abundant hydrolase expressed in the liver 9, but is absent in the intestine and kidney where CES2 is the predominant hydrolytic enzyme34. Thus, the data suggest that CES1 but not CES2 is the enzyme responsible for the activation of the ACEI prodrugs tested. The kinetic data of hydrolysis of the ACEI prodrugs catalyzed by HLS9 fit well to the Michaelis-Menten kinetic equation (Figure 2). The kinetic parameters including Vmax, Km, and Clint (Vmax/Km) of enalapril, ramipril, perindopril, moexipril and fosinopril were summarized in Table 2.
Figure 1.
Hydrolysis (activation) of enalapril (A), ramipril (B), perindopril (C), moexipril (D) and fosinopril (E) by human liver, intestine, and kidney s9 fractions. The ACEI prodrugs (100 μM) were incubated with 3 different concentrations of HLS9, HIS9 and HKS9. Catalytic activity was determined based on the formation of respective hydrolytic products. Data are expressed as mean ±S.D. from 3 independent experiments.
Figure 2.
Kinetics of the hydrolysis of enalapril (A), ramipril (B), perindopril (C), moexipril (D) and fosinopril (E) by human liver s9 fractions. Various concentrations of the ACEI prodrugs were incubated with HLS9 samples in order to determine kinetic parameters of hydrolysis of the ACEI prodrugs. The incubation time was 10 min for enalapril, moexipril and fosinopril, and 5 min for ramipril and perindopril. Data are mean ±S.D. (n=3).
Table 2.
Kinetic parameters of enzymatic hydrolysis of enalapril, ramipril, perindopril, moexipril and fosinopril
| Substrates | Enzymes | Vmax | Km | Clint (Vmax/Km) |
|---|---|---|---|---|
| Enalapril | HLS9 | 67.5 ± 1.3 | 60.1 ± 4.2 | 1.1 |
| WT CES1 | 389.0 ± 16.1 | 36.6 ± 4.5 | 10.6 | |
| Ramipril | HLS9 | 18.1 ± 0.7 | 690.4 ± 61.8 | 26.2 |
| WT CES1 | 11.6 ± 0.6 | 245.6 ± 39.9 | 47.1 | |
| Perindopril | HLS9 | 18.1 ± 0.8 | 1767.0 ± 165.0 | 23.3 |
| WT CES1 | 8.3 ± 0.4 | 477.5 ± 60.9 | 17.4 | |
| Moexipril | HLS9 | 4.4 ± 0.3 | 1457.0 ± 265.5 | 12.7 |
| WT CES1 | 1.8 ± 0.1 | 143.6 ± 24.5 | 12.4 | |
| Fosinopril | HLS9 | 1.4 ± 0.2 | 471.3 ± 211.9 | 3.0 |
| WT CES1 | 1.6 ± 0.1 | 276.1 ± 83.2 | 5.7 | |
The Km and Vmax values were calculated using nonlinear regression analysis with Graphpad Prism software. Data represent the mean ± S.D. from 3 independent experiments. The Vmax values are expressed in pmole/min/mg protein for enalapril, and nmole/min/mg protein for other ACEIs. The Km and Vmax/Km values are expressed in μM and μl/min/mg protein, respectively, for all substrates.
CES1 protein expression and activity in human livers did not differ among CES1/CES1VAR and CES1P1/CES1P1VAR genotypes and diplotypes
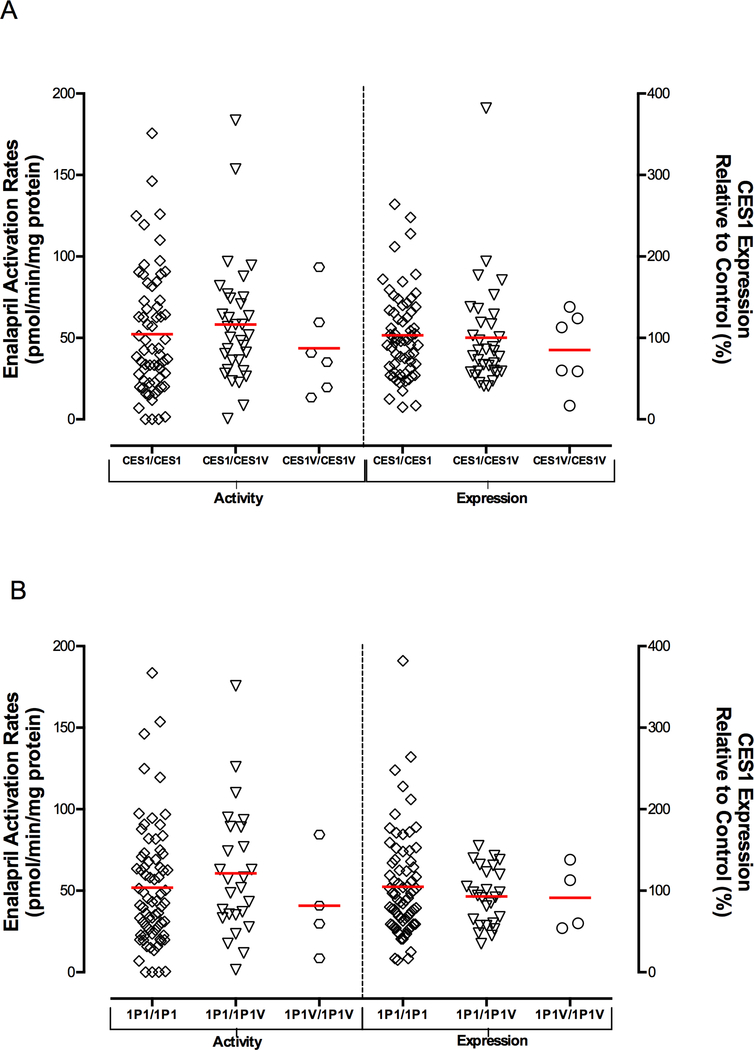
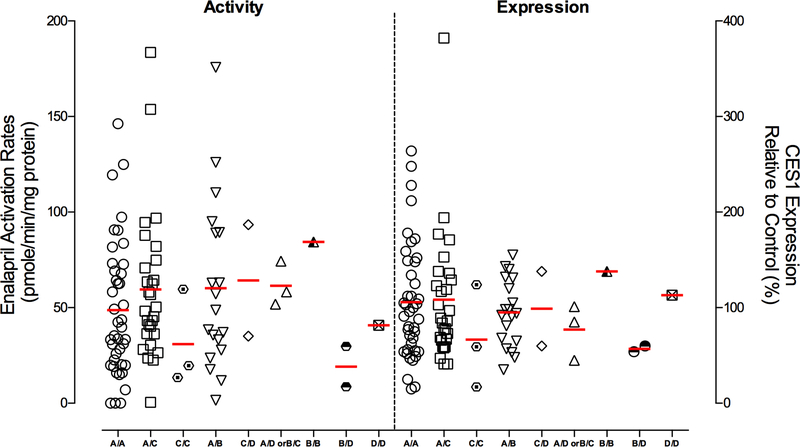
A total of 102 human liver samples were genotyped for the CES1P1/CES1P1VAR genotypes using both the previously published exon 5-based RFLP genotyping assay 21, 28 and our novel exon 3-based RFLP method. The genotyping results from the two methods were in agreement with each other except for 2 samples which might be due to random experimental errors. Thus, both RFLP methods are valid for genotyping CES1P1/CES1P1VAR. The two samples with inconclusive CES1P1/CES1P1VAR genotypes were excluded from the final data analysis of the studies involving the CES1P1/CES1P1VAR genotypes. The frequencies of CES1/CES1VAR and CES1P1/CES1P1VAR genotypes as well as the diplotypes in 92 Caucasian human liver donors were summarized in Table 3. The frequencies in other populations were not determined due to the small sample size (8 in total). We first assessed potential function of the CES1/CES1VAR genotypes using the 102 normal human liver samples. Marked interindividual variability of CES1 protein expression and activity on enalapril hydrolysis was observed among the individuals tested in the present study (Figure 3 and 4). CES1 expression and activity were found to be comparable among the CES1/CES1, CES1/CES1VAR, and CES1VAR/CES1VAR genotypes (Figure 3A). To avoid potential confounding effect of CES1P1/CES1P1VAR genotypes, we further analyzed the expression and activity data obtained from CES1P1 homozygotes (i.e. CES1P1/CES1P1). Again, the CES1/CES1VAR genotypes were not associated with the variability in CES1 expression and activity in the human livers tested (data not shown). Secondly, we evaluated if the conversion of the pseudogene CES1P1 to the functional CES1P1VAR gene affects CES1 expression and/or activity in human livers. Interestingly, no significant differences in CES1 expression and activity on enalapril activation were observed between the CES1P1/CES1P1, CES1P1/CES1P1VAR, and CES1P1VAR/CES1P1VAR genotypes (Figure 3B). Moreover, to study the impact of CES1 and CES1P1 diplotypes on the CES1 function, the samples were classified into nine diplotypes for the analysis of the expression and activity of CES1. There were no significant differences in CES1 expression and activity within the diplotypes as analyzed by one way ANOVA (expression: P=0.726; activity: P=0.549, Figure 4).
Table 3.
Frequency of CES1/CES1VAR and CES1P1/CES1P1VAR genotypes and diplotypes in 92 Caucasian subjects
| Genotypes | Number of subjects | Frequency |
|---|---|---|
| MAF of CES1/CES1VAR: 0.217 | ||
| CES1/CES1 | 58 | 63% |
| CES1/CES1VAR | 28 | 30% |
| CES1VAR/CES1VAR | 6 | 7% |
| MAF of CES1P1/CES1P1VAR: 0.168 | ||
| CES1P1/CES1P1 | 66 | 72% |
| CES1P1/CES1P1VAR | 22 | 24% |
| CES1P1VAR/CES1P1VAR | 4 | 4% |
| Diplotypes | Number of subjects | Frequency |
|---|---|---|
| AA | 39 | 42% |
| A/B | 17 | 18% |
| A/C | 25 | 27% |
| A/D or B/C | 2 | 2% |
| B/B | 1 | 1% |
| B/D | 2 | 2% |
| C/C | 3 | 3% |
| C/D | 2 | 2% |
| D/D | 1 | 1% |
Figure 3.
Effect of CES1/CES1VAR (A) and CES1P1/CES1P1VAR (B) genotypes on enalapril activation rates (left y axis) and CES1 expressions (right y axis) in 100 human liver samples. Total protein staining was used as the loading control for semi-quantification of CES1 protein expression. Horizontal bars represent means values in each group.
Figure 4.
Influence of CES1 diplotypes on CES1 activity on enalapril activation (left) and CES1 expression (right) in human liver samples (n=100). Divided by the middle dotted line, the activity data were plotted with the left y axis, while the expression levels were plotted with the y axis on the right. CES1 protein expression was semi-quantified using total protein staining as the loading control. Horizontal bars represent mean values in each group.
Enzymatic activity of CES1 and its variants G19V, S83L, G143E and A270S on the activation of ACEIs
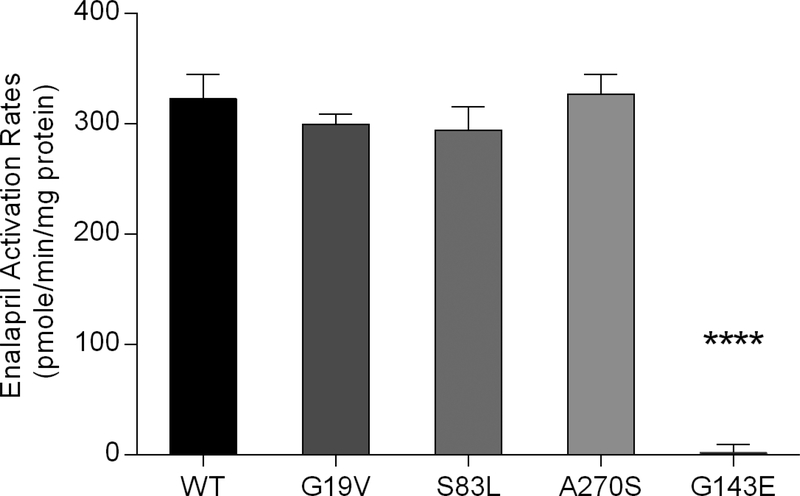
Enalapril, ramipril, perindopril, moexipril, and fosinopril were readily hydrolyzed to their respective active metabolites by WT CES1 in an enzyme concentration-dependent manner. However, active metabolites of all tested ACEI prodrugs were not detected after the incubation with the CES1 variant G143E (Figure 5). The calculated Clint (Vmax/Km) values were higher for the s9 fractions prepared from the WT CES1 expressing cells relative to that observed in the HLS9 incubation studies (Table 2). This might be due to different CES1 expression levels between these two enzyme preparations. The kinetic study confirmed that the G143E was a lossof-function variant for catalyzing the hydrolysis of those ACEI prodrugs under the experimental conditions (Figure 6). Additionally, consistent with our previous in vitro clopidogrel and methylphenidate hydrolysis studies 13, the activity of the variants G18V, S82L, and A269S on enalapril hydrolysis was comparable to WT CES1 (Figure 7).
Figure 5.
Enzymatic activity of wild type CES1 and the variant G143E on the hydrolysis of enalapril (A), ramipril (B), perindopril (C), moexipril (D) and fosinopril (E). The ACEI prodrugs (100 μM) were incubated with 3 different concentrations of the s9 fractions prepared from the WT CES1 and G143E cells. The data are mean ±S.D. from three independent experiments.
Figure 6.
Kinetics of the hydrolysis of enalapril (A), ramipril (B), perindopril (C), moexipril (D) and fosinopril (E) catalyzed by WT CES1 and the variant G143E. Hydrolytic (active) metabolites of the ACEI prodrugs were determined after incubation of various concentrations of the prodrugs with the cell s9 fractions. The incubation time was 10 min for enalapril and fosinopril, and 5 min for the rest three ACEI prodrugs. Data are means ±S.D. (n=3).
Figure 7.
Activation of enalapril by CES1 and its variants G19V, S83L, A270s and G143E. Enalapril (500 μM) was incubated with the s9 fractions prepared from the WT CES1 and the variants (G143E, G19V, S83L and A270s) transfected cells. Activation rate was determined by measuring the formation of the hydrolytic metabolite enalaprilat. Data are expressed as mean± S.D. (n=3). **** P<0.0001 (G143E vs wild type CES1).
Activation of enalapril was impaired in human livers carrying the CES1 SNP G143E
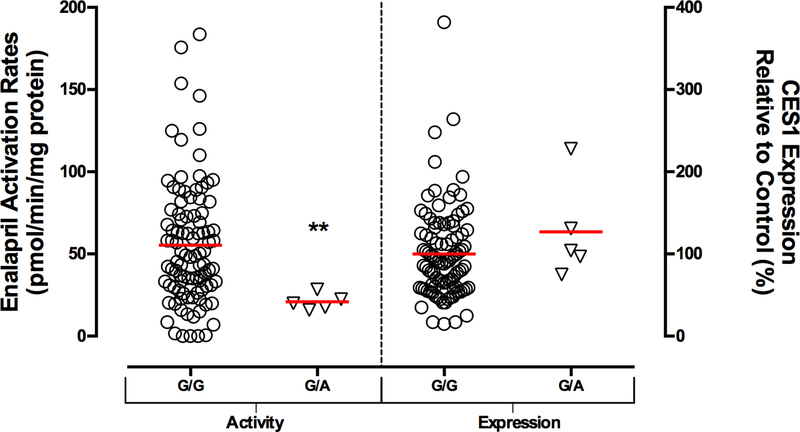
To further examine the influence of the CES1 variant G143E on the activation of ACEI prodrugs in human, we sequenced the CES1 exon4 region of 102 individual human liver samples for the G143E genotypes, and measured CES1 expression and activity on enalapril hydrolysis. Five subjects were determined to be 143G/E genotype while the others were non-carriers (i.e. 143G/G), which gives a minor allele frequency (MAF) of 2.5% for this variant, an observation consistent with our previous study 23. The CES1 SNP G143E was associated with significantly decreased enalapril activation rates (55.4±37.3 (G/G) vs 20.8±4.9 (G/E) pmole/min/mg protein, P = 0.0074), while CES1 protein expression levels were comparable between the 2 genotypes (Figure 8).
Figure 8.
Comparison of hepatic CES1 activity and expression between the G143E heterozygotes and non-carriers. CES1 activity on enalapril activation was impaired in the subjects with 143G/E genotype while the expressions were comparable between the 2 genotypes in 102 human livers. Total protein staining was used as the loading control for the CES1 protein expression assay. ** P<0.01, G143E heterozygotes (G/E) vs non-carriers (G/G).
Discussion
CES1 and CES2, the primary carboxylesterases (CESs) in humans, are the major hydrolases catalyzing the metabolism of many clinically important medications. CES1 is the predominant carboxylesterase expressed in the liver while absent in the intestine in humans. In contrast, human CES2 is highly expressed in the intestine, but only accounts for approximate 10% hepatic hydrolytic activity 9, 35. In addition, human kidney contributes to hydrolytic metabolism of a number of ester-containing medications to a less extent than the liver and intestine 36. CES1 expression in human kidney was considered extremely low, and 95% renal hydrolytic activity was due to CES2 according to a recently published in vitro investigation 34. Because of these distinct expression patterns of CES1 and CES2 in human liver, intestine, and kidney, the microsomes or s9 fraction samples prepared from those tissues can serve as a valid in vitro model to determine substrate specificity of CES1 and CES2 15.
In this study, hydrolytic activation of enalapril, ramipril, perindopril, moexipril, and fosinopril was determined after incubation with the s9 fractions of human liver, intestine, and kidney. All five tested ACEI prodrugs were readily hydrolyzed to their respective active metabolites by HLS9. However, no appreciable active metabolites were formed after incubation with HIS9 or HKS9, indicating that human liver but not the intestine or kidney is the site where ACEI prodrugs activation occurs. Thus, enalapril, ramipril, perindopril, moexipril, and fosinopril are the substrates of CES1 instead of CES2, a finding in line with previous reports from our laboratory and others showing that other ACEI prodrugs including trandolapril, benazepril, quinaparil, temocapril, cilazapril, delapril, imidapril were efficiently activated by CES1 14, 15. Moreover, the data are in agreement with the substrate selectivity of CES1, i.e. CES1 prefers substrates with a small alcohol group and a large acyl group (Supplementary Figure S6) 37. Based upon the calculated Clint values, the order of the efficacy of CES1-mediated hydrolysis is: ramipril > perindopril > enalapril, moexipril, and fosinopril. During the process of preparing this report, Thomsen and colleagues reported that the activation of ramipril and enalapril were catalyzed by CES1 in an incubation study with purified CES1 and CES2 enzymes 17. Consistent with our findings, the study suggested that ramipril was a more efficient CES1 substrate relative to enalapril.
Significant interindividual variability of CES1 expression and activity has been consistently reported by independent investigators 16, 28, 38. Varied CES1 function may lead to marked interindividual variability in pharmacokinetics of many drugs metabolized by CES1, and consequently affect therapeutic outcomes of those medications. Genetic variation is increasingly recognized as a major contributing factor to CES1 variability. In the present study, we determined the effects of CES1/CES1VAR and CES1P1/CES1P1VAR genotypes and diplotypes on CES1 expression and activity on ACEI prodrug activation in 100 human liver tissues. No significant differences in CES1 expression and activity have been observed among the CES1/CES1, CES1/CES1VAR, CES1VAR/CES1VAR genotypes. To avoid potential interference of CES1P1/CES1P1VAR with CES1/CES1VAR genotypes on CES1 function, we reanalyzed the data only from subjects carrying homozygous CES1P1. Again, the expression and activity of CES1 were comparable between the CES1/CES1VAR genotypes. The data are in agreement with the fact that the CES1 and CES1VAR genes encode for identical protein product and share same promoter structure. Our results were also consistent with the previous clinical observation that the metabolism of the CES1 substrate oseltamivir were not significantly different in healthy subjects with different CES1/CES1VAR genotypes 20. Thus, CES1/CES1VAR genotypes are unlikely a factor contributing to varied CES1 function in human liver.
The CES1P1VAR is a variant of the pseudogene of CES1P1, and encodes for the same protein product of the CES1 gene. It has been speculated that the presence of the CES1P1VAR may enhance the expression of CES1 as a result of extra functional gene copies in addition to two copies from CES1/CES1VAR genes (i.e. total function gene copies for CES1 expression: CES1P1/CES1P1: 2, CES1P1/CES1P1VAR: 3; CES1P1VAR/CES1P1VAR: 4). However, our study showed that the CES1P1/CES1P1VAR genotypes were not associated with CES1 expression and activity on enalapril activation in 100 human liver samples. The lack of contribution of the CES1P1VAR to total CES1 expression is likely due to that the transcription efficiency of the CES1P1VAR is only 2% of the CES1 gene in the liver 28. Consistent with our findings, a clinical pharmacokinetic study indicated that the conversion of the pseudogene CES1P1 to the functional CES1P1VAR gene did not affect the pharmacokinetics of oseltamivir in healthy volunteers 20. In a study in Japanese cancer patients receiving irinotecan therapy, the median AUC ratios of the metabolites SN-38 and SN-38G to the parent compound were 24% higher in CES1P1VAR heterozygous and homozygous patients relative to those with CES1P1/CES1P1 genotype 21. However, it should be noted that CES2 is believed to play a greater role in the metabolism of irinotecan than CES1 39. Thus, it is unclear whether the ratio of (SN-38+SN38G)/irinotecan is a reliable indicator of CES1 activity in vivo. Furthermore, we demonstrated that the expression and activity of CES1 did not differ significantly among various diplotypes of the CES1 and CES1P1 genes in 100 human livers, providing additional evidence suggesting the CES1/CES1VAR and CES1P1/CES1P1VAR genotypes are not associated with interindividual variability in CES1 function.
Beyond the CES1/CES1VAR and CES1P1/CES1P1VAR genotypes and diplotypes, several CES1 nonsynonymous SNPs including the G19V, S83L, G143E, and A270S were studied for their potential impact on ACEI prodrug activation in vitro. The SNP G143E is the first clinically significant CES1 variant, originally identified in a methylphenidate poor metabolizer during a healthy volunteer methylphenidate pharmacokinetic study 23. The MAFs of the G143E was determined to be 3.7%, 4.3%, 2.0%, and 0% in White, Black, Hispanic, and Asian populations, respectively 23. In vitro function study demonstrated that the G143E is a loss-of-function variant for the metabolism of several CES1 substrates including methylphenidate, trandolapril, and clopidogrel, while retaining approximate 25% of catalytic activity towards the hydrolysis (activation) of oseltamivir relative to the WT enzyme 13, 15, 22, 23. Accordingly, the magnitude of the impact of the G143E on drug metabolism may vary depending on different substrates. For instance, the mean Cmax value of oseltamivir carboxylate in 143E heterozygotes was 17.5% lower than that in non-carriers 26, whereas the concentrations of clopidogrel active metabolite were approximately 60% higher in patients carrying heterozygous 143E relative to patients with 143GG genotype 24. It is noted that, instead of acting as a prodrug-activating enzyme, as seen in the cases of ACEIs, dabigatran, and oseltamivir, CES1 is the enzyme responsible for the deactivation of clopidogrel and its active metabolite 13. Therefore, loss-of-function CES1 variants (e.g. G143E) and CES1 inhibition could lead to the increase of the formation of clopidogrel active metabolite 13, 24. The data from the cell lines stably expressing the G143E demonstrate that the G143E is a loss-of-function variant for the activation of all five tested ACEI prodrugs. Additionally, the CES1 activity on enalapril activation in human liver samples with 143G/E genotype was approximately 1/3 of that in subjects carrying the 143G/G, while the expression of CES1 was similar between these 2 genotypes. Therefore, the patients carrying the 143E allele may experience difficulty of converting ACEI prodrugs to active metabolites. The correlation between CES1 expression and activity was analyzed in the 102 human liver samples. In general, the enalapril activation rates were significantly correlated with CES1 protein expression levels (R2 = 0.40, P < 0.0001) (Supplementary Figure S5). However, some G143E non-carriers exhibited null CES1 activity towards enalapril activation even though CES1 expressions in these samples were comparable to pooled HLS9 (Figure 8). Besides enalapril, trandolapril, another ACEI prodrug selectively activated by CES1, was also tested for its activation in the 102 human liver samples. The trandolapril data were highly consistent with that from the enalapril study (Supplementary Figure S7). The null CES1 samples identified from the enalapril study were also inactive on trandolapril hydrolysis. On the other hand, the samples with complete loss of CES1 activity were found to exhibit normal catalytic activity of other drug-metabolizing enzymes including CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4, suggesting that the null CES1 activity is not related to sample integrity (data not shown). The data strongly indicate that other unknown nonsynonymous functional CES1 variants may exist.
The common CES1 variants G19V, S83L, and A270S are predicted by the in silico programs SIFT and Polyphen2 to have potential damaging effect on CES1 enzymatic function. However, the catalyzing activity of the G19V, S83L, and A270S on the activation of enalapril was comparable to the WT CES1 indicating that variants G19V, S83L, and A270S are nonfunctional SNPs for enalapril activation, which is in consistent with our previously study of clopidogrel and methylphenidate metabolism by those three CES1 variants 13.
Beyond CES1 nonsynonymous variants, many CES1 SNPs have been discovered in the promoter and other regulatory regions of the gene. Geshi and co-workers reported that the −816A>C, a SNP located in the promoter region of CES1P1 gene, was associated with significantly greater response to the ACEI prodrug imidapril 40. Additionally, the −816C allele was found to be associated with significantly lower antiplatelet activity of clopidogrel in patients with coronary heart diseases 41. Both studies suggest that −816C carriers may exhibit higher expression of CES1, and thus represent the phenotype of rapid metabolizers of CES1 substrates. However, a recently published clinical investigation showed that the −816A>C was associated with greater response to the antiplatelet activity of clopidogrel in patients undergoing percutaneous coronary intervention 42, indicating that CES1 expression is impaired in the −816C carriers, which is contrary to the previous imidapril and clopidogrel studies. Of note, the CES1P1 gene, in which the −816A>C resides, appears unlikely to significantly contribute to hepatic CES1 expression in humans 28, given that the CES1P1 is a pseudogene and the transcription efficiency of CES1P1VAR is only 2% of that of CES1 in the liver. Importantly, our present in vitro human liver studies have revealed that CES1P1/CES1P1VAR genotypes did not significantly affect CES1 expression and activity on enalapril hydrolysis. Thus, the function and clinical significance of the −816A>C variant warrant further investigation.
Two recently published studies examined clinical significance of the variant −75 T>G (rs3815583) located in the 5’-untranslated region of CES1 43, 44. Though preliminary, the studies support that the G allele is associated with hepatotoxicity of isoniazid in patients with latent tuberculosis infection 44, and the increased side effects on appetite reduction in attention deficiency hyperactive disorder (ADHD) patients treated with methylphenidate 43. However, the effect of the −75T>G variant on CES1 expression and/or activity remains unexplored.
In summary, we have demonstrated that the ACEI prodrugs enalapril, ramipril, perindopril, moexipril, and fosinopril are activated in the liver by CES1. For the first time, we revealed that CES1/CES1VAR and CES1P1/CES1P1VAR genotypes and diplotypes have no significant effect on CES1 expression and ACEI activation in human livers. Additionally, the CES1 SNP G143E is a loss-of-function variant for the activation of the five tested ACEIs, while the G18V, S82L, and A269S are non-functional variants for enalapril activation. Thus, the G143E and other functional CES1 variants could impair ACEI prodrug activation, resulting in therapeutic failure of ACEI prodrug treatment. Furthermore, patients with functional CES1 variants may experience unexpected adverse effects as a result of excessive accumulation of parent compounds of ACEI prodrugs secondary to impaired CES1 function. To date, more than 1000 CES1 genetic variants have been identified and registered in various databases, however, the majority of these variants have not been studied for their function yet. A comprehensive study of CES1 pharmacogenetics will allow us to better understand individual variability of CES1 expression and activity, and eventually develop CES1 pharmacogenetics-based personalized medicine to improve therapeutic outcomes of ACEI prodrugs and many other CES1 substrate medications.
Supplementary Material
Acknowledgment
Research reported in this publication was supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number 2UL1TR000433 (Hao-Jie Zhu) and the American Association of Colleges of Pharmacy (AACP) 2015 New Investigator Award (Hao-Jie Zhu). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interests
No conflict of interests was declared.
Supplementary information is available at The Pharmacogenomics Journal’s website
References
- 1.Webb AJ, Fischer U, Mehta Z, Rothwell PM. Effects of antihypertensive-drug class on interindividual variation in blood pressure and risk of stroke: a systematic review and meta-analysis. Lancet 2010; 375(9718): 906–915. [DOI] [PubMed] [Google Scholar]
- 2.Tsoukas G, Anand S, Yang K. Dose-dependent antihypertensive efficacy and tolerability of perindopril in a large, observational, 12-week, general practice-based study. Am J Cardiovasc Drugs 2011; 11(1): 45–55. [DOI] [PubMed] [Google Scholar]
- 3.Malacco E, Omboni S, Volpe M, Auteri A, Zanchetti A. Antihypertensive efficacy and safety of olmesartan medoxomil and ramipril in elderly patients with mild to moderate essential hypertension: the ESPORT study. J Hypertens 2010; 28(11): 2342–2350. [DOI] [PubMed] [Google Scholar]
- 4.Ionescu DD. Antihypertensive efficacy of perindopril 5–10 mg/day in primary health care: an open-label, prospective, observational study. Clin Drug Investig 2009; 29(12): 767–776. [DOI] [PubMed] [Google Scholar]
- 5.Mugellini A, Dobovisek J, Planinc D, Cremonesi G, Fogari R. Efficacy and safety of delapril plus manidipine compared with enalapril plus hydrochlorothiazide in mild to moderate essential hypertension: results of a randomized trial. Clin Ther 2004; 26(9): 1419–1426. [DOI] [PubMed] [Google Scholar]
- 6.Kostis JB, Packer M, Black HR, Schmieder R, Henry D, Levy E. Omapatrilat and enalapril in patients with hypertension: the Omapatrilat Cardiovascular Treatment vs. Enalapril (OCTAVE) trial. Am J Hypertens 2004; 17(2): 103–111. [DOI] [PubMed] [Google Scholar]
- 7.Poirier L, Bourgeois J, Lefebvre J, Archambault F, Lacourciere Y. ACE Inhibitors as First-Line Treatment Agents: A Comparative Study of Trandolapril and Enalapril on Casual and Ambulatory Blood Pressures. Am J Ther 1995; 2(3): 159–164. [PubMed] [Google Scholar]
- 8.Mahmoudpour SH, Leusink M, van der Putten L, Terreehorst I, Asselbergs FW, de Boer A, et al. Pharmacogenetics of ACE inhibitor-induced angioedema and cough: a systematic review and meta-analysis. Pharmacogenomics 2013; 14(3): 249–260. [DOI] [PubMed] [Google Scholar]
- 9.Imai T Human carboxylesterase isozymes: catalytic properties and rational drug design. Drug Metab Pharmacokinet 2006; 21(3): 173–185. [DOI] [PubMed] [Google Scholar]
- 10.Satoh T, Hosokawa M. Structure, function and regulation of carboxylesterases. Chem Biol Interact 2006; 162(3): 195–211. [DOI] [PubMed] [Google Scholar]
- 11.Imai T, Taketani M Fau - Shii M, Shii M Fau - Hosokawa M, Hosokawa M Fau - Chiba K, Chiba K. Substrate specificity of carboxylesterase isozymes and their contribution to hydrolase activity in human liver and small intestine. (0090–9556 (Print)). [DOI] [PubMed]
- 12.Sun Z, Murry DJ, Sanghani SP, Davis WI, Kedishvili NY, Zou Q, et al. Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1. J Pharmacol Exp Ther 2004; 310(2): 469–476. [DOI] [PubMed] [Google Scholar]
- 13.Zhu HJ, Wang X, Gawronski BE, Brinda BJ, Angiolillo DJ, Markowitz JS. Carboxylesterase 1 as a determinant of clopidogrel metabolism and activation. The Journal of pharmacology and experimental therapeutics 2013; 344(3): 665–672. [DOI] [PubMed] [Google Scholar]
- 14.Takai S, Matsuda A, Usami Y, Adachi T, Sugiyama T, Katagiri Y, et al. Hydrolytic profile for ester- or amide-linkage by carboxylesterases pI 5.3 and 4.5 from human liver. Biol Pharm Bull 1997; 20(8): 869–873. [DOI] [PubMed] [Google Scholar]
- 15.Zhu HJ, Appel DI, Johnson JA, Chavin KD, Markowitz JS. Role of carboxylesterase 1 and impact of natural genetic variants on the hydrolysis of trandolapril. Biochem Pharmacol 2009; 77(7): 1266–1272. [DOI] [PubMed] [Google Scholar]
- 16.Yang D, Pearce RE, Wang X, Gaedigk R, Wan YJ, Yan B. Human carboxylesterases HCE1 and HCE2: ontogenic expression, inter-individual variability and differential hydrolysis of oseltamivir, aspirin, deltamethrin and permethrin. Biochem Pharmacol 2009; 77(2): 238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomsen R, Rasmussen HB, Linnet K. In Vitro Drug Metabolism by Human Carboxylesterase 1: Focus on Angiotensin-converting Enzyme Inhibitors. Drug Metab Dispos 2013. [DOI] [PubMed] [Google Scholar]
- 18.Vistoli G, Pedretti A Fau - Mazzolari A, Mazzolari A Fau - Testa B, Testa B. Homology modeling and metabolism prediction of human carboxylesterase-2 using docking analyses by GriDock: a parallelized tool based on AutoDock 4.0. (1573–4951 (Electronic)). [DOI] [PubMed]
- 19.Tanimoto K, Kaneyasu M, Shimokuni T, Hiyama K, Nishiyama M. Human carboxylesterase 1A2 expressed from carboxylesterase 1A1 and 1A2 genes is a potent predictor of CPT-11 cytotoxicity in vitro. Pharmacogenet Genomics 2007; 17(1): 1–10. [DOI] [PubMed] [Google Scholar]
- 20.Suzaki Y, Uemura N, Takada M, Ohyama T, Itohda A, Morimoto T, et al. The effect of carboxylesterase 1 (CES1) polymorphisms on the pharmacokinetics of oseltamivir in humans. Eur J Clin Pharmacol 2013; 69(1): 21–30. [DOI] [PubMed] [Google Scholar]
- 21.Sai K, Saito Y, Tatewaki N, Hosokawa M, Kaniwa N, Nishimaki-Mogami T, et al. Association of carboxylesterase 1A genotypes with irinotecan pharmacokinetics in Japanese cancer patients. Br J Clin Pharmacol 2010; 70(2): 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu HJ, Markowitz JS. Activation of the antiviral prodrug oseltamivir is impaired by two newly identified carboxylesterase 1 variants. Drug Metab Dispos 2009; 37(2): 264–267. [DOI] [PubMed] [Google Scholar]
- 23.Zhu HJ, Patrick KS, Yuan HJ, Wang JS, Donovan JL, DeVane CL, et al. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am J Hum Genet 2008; 82(6): 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis JP, Horenstein RB, Ryan K, O’Connell JR, Gibson Q, Mitchell BD, et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet Genomics 2013; 23(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nemoda Z, Angyal N, Tarnok Z, Gadoros J, Sasvari-Szekely M. Carboxylesterase 1 gene polymorphism and methylphenidate response in ADHD. Neuropharmacology 2009; 57(7–8): 731–733. [DOI] [PubMed] [Google Scholar]
- 26.Tarkiainen EK, Backman JT, Neuvonen M, Neuvonen PJ, Schwab M, Niemi M. Carboxylesterase 1 polymorphism impairs oseltamivir bioactivation in humans. Clin Pharmacol Ther 2012; 92(1): 68–71. [DOI] [PubMed] [Google Scholar]
- 27.al-Dirbashi O, Kuroda N, Menichini F, Noda S, Minemoto M, Nakashima K. Enantioselective high-performance liquid chromatography with fluorescence detection of methamphetamine and its metabolites in human urine. Analyst 1998; 123(11): 2333–2337. [DOI] [PubMed] [Google Scholar]
- 28.Fukami T, Nakajima M, Maruichi T, Takahashi S, Takamiya M, Aoki Y, et al. Structure and characterization of human carboxylesterase 1A1, 1A2, and 1A3 genes. Pharmacogenet Genomics 2008; 18(10): 911–920. [DOI] [PubMed] [Google Scholar]
- 29.Gu Q, Chen X, Zhong D, Wang Y. Simultaneous determination of enalapril and enalaprilat in human plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2004; 813(1–2): 337–342. [DOI] [PubMed] [Google Scholar]
- 30.Yuan B, Wang X, Zhang F, Jia J, Tang F. Simultaneous Determination of Ramipril and Its Active Metabolite Ramiprilat in Human Plasma by LC–MS–MS. Chromatographia 2008; 68(7–8): 533–539. [Google Scholar]
- 31.Jain DS, Subbaiah G, Sanyal M, Pande UC, Shrivastav P. First LC-MS/MS electrospray ionization validated method for the quantification of perindopril and its metabolite perindoprilat in human plasma and its application to bioequivalence study. J Chromatogr B Analyt Technol Biomed Life Sci 2006; 837(1–2): 92–100. [DOI] [PubMed] [Google Scholar]
- 32.Karra VK, Mullangi R, Pilli NR, Inamadugu JK, Ravi VB, Seshagiri Rao JV. A rapid and sensitive liquid chromatography-tandem mass spectrometric assay for moexipril, an angiotensin-converting enzyme inhibitor in human plasma. Biomedical chromatography : BMC 2012; 26(12): 1552–1558. [DOI] [PubMed] [Google Scholar]
- 33.Cui S, Feng F, Ma M, Liu H, Chen Y. Development and validation of liquid chromatography-tandem mass spectrometric method for simultaneous determination of fosinopril and its active metabolite fosinoprilat in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci 2007; 854(1–2): 143–151. [DOI] [PubMed] [Google Scholar]
- 34.Hatfield MJ, Tsurkan L, Garrett M, Shaver TM, Hyatt JL, Edwards CC, et al. Organspecific carboxylesterase profiling identifies the small intestine and kidney as major contributors of activation of the anticancer prodrug CPT-11. Biochemical pharmacology 2011; 81(1): 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taketani M, Shii M, Ohura K, Ninomiya S, Imai T. Carboxylesterase in the liver and small intestine of experimental animals and human. Life Sci 2007; 81(11): 924–932. [DOI] [PubMed] [Google Scholar]
- 36.Hosokawa M Structure and catalytic properties of carboxylesterase isozymes involved in metabolic activation of prodrugs. Molecules 2008; 13(2): 412–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosokawa M Structure and catalytic properties of carboxylesterase isozymes involved in metabolic activation of prodrugs. (1420–3049 (Electronic)). [DOI] [PMC free article] [PubMed]
- 38.Hosokawa M, Endo T, Fujisawa M, Hara S, Iwata N, Sato Y, et al. Interindividual variation in carboxylesterase levels in human liver microsomes. Drug Metab Dispos 1995; 23(10): 1022–1027. [PubMed] [Google Scholar]
- 39.Humerickhouse R, Lohrbach K, Li L, Bosron WF, Dolan ME. Characterization of CPT11 hydrolysis by human liver carboxylesterase isoforms hCE-1 and hCE-2. Cancer Res 2000; 60(5): 1189–1192. [PubMed] [Google Scholar]
- 40.Geshi E, Kimura T, Yoshimura M, Suzuki H, Koba S, Sakai T, et al. A single nucleotide polymorphism in the carboxylesterase gene is associated with the responsiveness to imidapril medication and the promoter activity. Hypertens Res 2005; 28(9): 719–725. [DOI] [PubMed] [Google Scholar]
- 41.Xie C, Ding X, Gao J, Wang H, Hang Y, Zhang H, et al. The effects of CES1A2 A(816)C and CYP2C19 loss-of-function polymorphisms on clopidogrel response variability among Chinese patients with coronary heart disease. Pharmacogenet Genomics 2014. [DOI] [PubMed] [Google Scholar]
- 42.Zou J-J, Chen S-L, Fan H-W, Tan J, He B-S, Xie H-G. The CES1A −816C as a genetic marker to predict greater platelet clopidogrel response in patients with percutaneous coronary intervention. Journal of Cardiovascular Pharmacology 2014; 63(2): 178–183. [DOI] [PubMed] [Google Scholar]
- 43.Bruxel EM, Salatino-Oliveira A, Genro JP, Zeni CP, Polanczyk GV, Chazan R, et al. Association of a carboxylesterase 1 polymorphism with appetite reduction in children and adolescents with attention-deficit/hyperactivity disorder treated with methylphenidate. Pharmacogenomics J 2012. [DOI] [PubMed] [Google Scholar]
- 44.Yamada S, Richardson K, Tang M, Halaschek-Wiener J, Cook VJ, Fitzgerald JM, et al. Genetic variation in carboxylesterase genes and susceptibility to isoniazid-induced hepatotoxicity. Pharmacogenomics J 2010; 10(6): 524–536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.