Abstract
Interferons (IFNs) mediate cellular defence against viral pathogens by upregulation of interferon-stimulated genes (ISGs) whose products interact with viral components or alter cellular physiology to suppress viral replication (1–3). Among the ISGs that can inhibit influenza A virus (IAV) (4) are the myxovirus resistance 1 (MX1) GTPase (5) and IFN-induced transmembrane protein 3 (IFITM3) (6, 7). Here we use ectopic expression and gene knock-out to demonstrate that the IFN-inducible 219 amino acid short isoform of human nuclear receptor coactivator 7 (NCOA7) is an inhibitor of IAV as well as other viruses that enter the cell by endocytosis, including hepatitis C virus (HCV). NCOA7 interacts with the vacuolar H+-ATPase (V-ATPase) and its expression promotes cytoplasmic vesicle acidification, lysosomal protease activity and the degradation of endocytosed antigen. Step-wise dissection of the IAV entry pathway demonstrates that NCOA7 inhibits fusion of the viral and endosomal membranes and subsequent nuclear translocation of viral ribonucleoproteins (vRNPs). NCOA7, therefore, provides a mechanism for immune regulation of endo-lysosomal physiology that not only suppresses viral entry into the cytosol from this compartment but may also regulate other V-ATPase-associated cellular processes such as physiological adjustments to nutritional status, or the maturation and function of antigen presenting cells.
Previously we characterized a panel of primary and immortalized cells for IFNα-inducible resistance to human immunodeficiency virus type-1 (HIV-1) infection as the basis for the transcriptomic-led identification of ISGs that suppress virus infection (8, 9). Among the cDNAs of interest, the IFN-inducible short isoform (isoform 4) of NCOA7 (also termed NCOA7-Alternative-Start (AS) (10)) was cloned into pEasiLV-MCS, a doxycycline-inducible, E2 crimson expressing lentiviral vector (9), and evaluated for anti-retroviral activity.
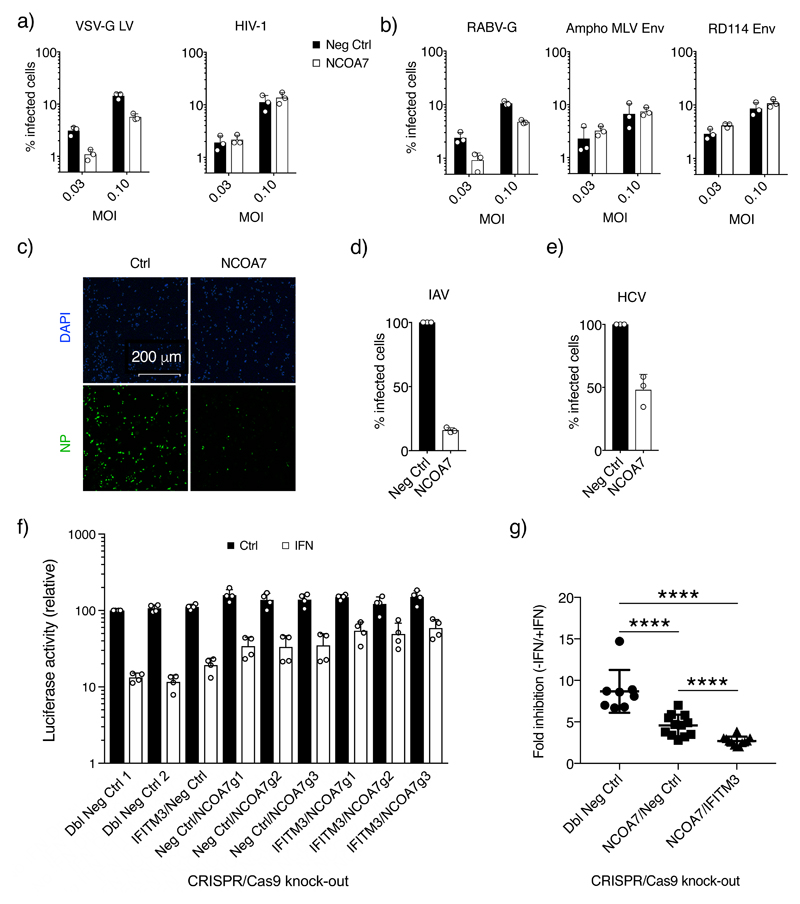
HIV-1 susceptible U87MG CD4+ CXCR4+ cultures were transduced with NCOA7 expressing EasiLV or a negative control vector. Cells were challenged with a vesicular stomatitis virus G-glycoprotein (VSV-G)-pseudotyped HIV-1 derived lentiviral vector that expresses GFP (11). 48 hr later, E2 crimson-positive cells were gated and the percentage of infected cells enumerated by flow cytometry. At multiple viral doses, NCOA7 expression decreased virus infectivity (Fig. 1a), with similar effects seen with VSV-G pseudotyped vectors derived from diverse retroviruses, including simian immunodeficiency virus (SIV), equine infectious anaemia virus (EIAV) and feline immunodeficiency virus (FIV) (Supplementary Fig. 1a). In contrast, in challenges with HIV-1/Nef-internal ribosome entry signal (IRES)-GFP carrying its natural envelope glycoprotein (Env), NCOA7 displayed no discernable anti-viral activity (Fig. 1a). We surmised that the differences in NCOA7 sensitivity between HIV-1 particles carrying HIV-1 Env or VSV-G could be dictated by the viral entry pathway: HIV-1 is believed to mediate pH independent membrane fusion (12) whereas VSV-G undergoes pH dependent fusion in early endosomes (13). We therefore extended this pseudotyping analysis to additional viral envelope glycoproteins; the rabies virus glycoprotein (RABV-G) mediates entry via endosomes whereas amphotropic murine leukemia virus (MLV) and the feline endogenous retrovirus, RD114, Env are thought to facilitate pH independent membrane fusion. NCOA7 inhibited infection by pseudotypes carrying RABV-G but not amphotropic MLV or RD114 Env (Fig. 1b), consistent with suppressed entry from the endocytic pathway.
Figure 1. NCOA7 inhibits infection by viruses entering through the endocytic pathway.
U87MG CD4+ CXCR4+ cells were transduced with EasiLV expressing NCOA7 or a control (CD8 or luciferase) and treated with doxycycline for 48 hr. Cells were challenged with a) VSV-G-pseudotyped HIV-1 based, GFP-expressing lentiviral vector (VSV-G LV) or HIV-1 (NL4.3/Nef-IRES-GFP) or b) lentiviral vectors pseudotyped with: rabies virus GP (RABV-G), or amphotropic MLV Env or RD114 Env. 48 hr later, E2 crimson-positive cells were gated and the percentage of infected, GFP-positive cells determined by flow cytometry. c) A549 cells expressing NCOA7 or CD8 were infected with A/Eng/195/2009, fixed at 5 hr, stained with anti-NP antibody (green) and visualized by confocal microscopy, and d) shows the percentage of infected cells in c) as quantitated by ImageJ. e) Huh-7.5 cells were transduced with lentiviral vectors expressing NCOA7 or GFP, and infected with HCV. After 48 hr, cells were fixed, stained with anti-NS5A antibody and the percentage of infected cells was determined by flow cytometry. f) NCOA7 requirement for effective IFNα-induced inhibition of IAV. A549-Cas9 cells were transduced with lentiviral vectors expressing CRISPR non-targeting guide RNAs (g1/g2-CTRL) or guide RNAs targeting IFITM3 or NCOA7, singly or in combination. Cells were antibiotic selected for at least 15 days, seeded and treated for 24 hr with IFNα prior to challenge with 5 x 104 PFU/well of IAV NanoLuc. The cells were lysed 7 hr post infection and luciferase activity measured. Relative luminescence results for IFNα-treated and untreated conditions are shown. g) Statistical analysis of infectivity in the absence (-) or presence (+) of IFNα from (f) combining different guides from control (n=8), NCOA7 knock-out (n=12) and NCOA7/IFITM3 double knock-out (n=12) cell populations, ****p-value (95% Confidence Interval) was 6.351e-05 (2.2-5.3), 1.588e-05 (4.3-6.3) and 4.955e-05 (0.8-2.9) for Dbl Neg Ctrl vs NCOA7/Neg Ctrl, Dbl Neg Ctrl vs NCOA7/IFITM3 and NCOA7/Neg Ctrl vs NCOA7/IFITM3 respectively. Mann Whitney test, unadjusted, two-sided. Charts show the mean of 3 independent experiments for a), b), d) and e) and 4 for f). Error bars indicate one standard deviation from the mean.
We next tested the effect of NCOA7 on two replication competent viruses from diverse families that naturally enter by endocytic uptake, namely IAV (an orthomyxovirus) and HCV (a flavivirus). A549 cells transduced with a constitutively expressing NCOA7 lentiviral vector, or control cells, were challenged with IAV, fixed at 5 hr and stained with an antibody specific for viral NP, a viral protein that accumulates to high levels in the nucleus during productive viral replication following de novo expression. NCOA7 expression reduced the number of cells with nuclear NP staining by ~6-fold (Fig. 1, c and d); similarly, an engineered IAV expressing NanoLuc luciferase was also inhibited across a range of viral doses (Supplementary Fig. 1b). The suppressive effects of NCOA7 were most evident at earlier time-points, as illustrated by IAV infection of control versus NCOA7 expressing 293T cultures and time-lapse microscopy (Supplementary Movie 1). For HCV infection, Huh-7.5 cell cultures were stably transduced with an NCOA7 expressing (or control) lentiviral vector, challenged with HCV and harvested and stained with an anti-NS5A antibody at 48 hr: NCOA7 expression resulted in a ~50% decrease in HCV infection (Fig. 1e).
Having demonstrated that provision of NCOA7 was sufficient to inhibit IAV infection, we complemented this approach by assessing the contribution of endogenous NCOA7 to the IFNα induced suppression of IAV infection. We used CRISPR/Cas9-mediated genome editing and guide RNAs targeting the unique region of the short isoform to generate A549 cell populations (Fig. 1f and 1g, Supplementary Fig. 2a) and clones (Supplementary Fig. 2b, 2c, 2d and 2e) lacking this isoform. All cultures in which the short isoform had been inactivated displayed diminished IFNα-induced suppression of IAV relative to control cells. We additionally created matching cell populations in which the integral membrane protein IFITM3 (6) was knocked-out alone or in combination with NCOA7 (Fig. 1f and 1g). Reductions in IFNα-mediated inhibition were seen in cells that expressed or lacked NCOA7 in the context of IFITM3 knock-out (~6-fold inhibition in single IFITM3 knock-out versus 2.7-fold inhibition in IFITM3/NCOA7 double knock-out), indicating that the anti-viral effects of NCOA7 and IFITM3 are cumulative as well as independent.
To verify further that NCOA7 and IFITM3 inhibit IAV independently of each other, we showed (using knock-out clonal lines) that NCOA7 mediated inhibition of infection is unaffected by IFITM3 knock-out and vice versa (Supplementary Fig 3a, 3b, and 3c). Reintroduction of codon-optimized NCOA7 isoform 4, which can’t be targeted by our guide RNAs, into NCOA7 knock-out cell lines reconstituted the full IFNα-mediated inhibition of IAV (Supplementary Fig. 3d). Together, these findings confirm that NCOA7 is a significant component of IFNα-induced protection against IAV infection.
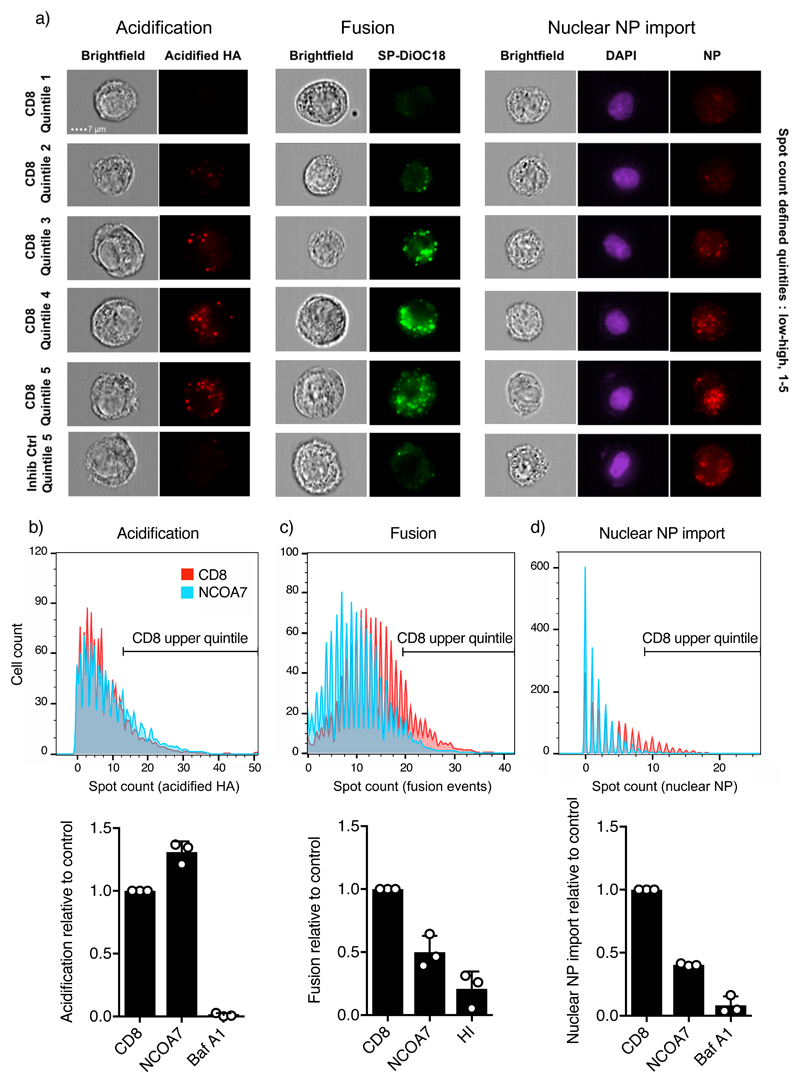
We next employed a series of sequential imaging flow cytometry-based assays to dissect the step(s) in IAV infection that is inhibited by NCOA7 (Fig. 2a) (14, 15). First, using A549 cells expressing NCOA7 or a negative control (CD8), we quantified low pH-induced conformational changes in HA at 1 hr post-infection using monoclonal antibody A1, which specifically recognizes acidified HA (14, 16). HA acidification was not compromised in the presence of NCOA7 (Fig. 2b). Second, to study viral membrane fusion we labelled IAV particles with the self-quenching dye SP-DiOC18. Fluorescence dequenching of SP-DiOC18 occurs upon viral-endosome membrane fusion (reported to occur at pH<5 for A/Victoria/3/75 and ~pH 5.5 for A/Eng/195/2009 (17, 18)) and there was ~50% decrease in the proportion of cells exhibiting high numbers of fusion events in the presence of NCOA7 (defined by the upper quintile of the control population) (Fig. 2c) (14). NCOA7’s ability to inhibit viral-endosome membrane fusion was independently demonstrated by challenging U87MG CD4+ CXCR4+ cells expressing NCOA7 (or a control) with VSV-G pseudotyped HIV-1 containing a β-lactamase/Vpr fusion protein that induces the cleavage of CCF2-AM dye upon viral entry into the cytosol. After loading cells with CCF2-AM dye, cleavage was analyzed by flow cytometry, showing that cytosolic entry was consistently reduced in NCOA7 expressing cells (Supplementary Fig 4(a and b)). Third, to examine the nuclear import of incoming virion RNPs, we infected A549 cells with IAV at high MOI in the presence of cycloheximide (to prevent de novo protein synthesis), and quantified nuclear NP staining at 5 hr. Consistent with the fusion assays, NCOA7 expression resulted in ~60% decrease in the number of cells exhibiting a high level of NP import (defined by the control upper quintile) (Fig. 2d). Supplementary Fig. 4c confirms this result by confocal microscopy. In sum, our mapping strategy indicated that NCOA7 inhibits viral membrane fusion, whereas neither HA acidification nor viral uptake into the endocytic pathway (Supplementary Fig. 4d) were compromised.
Figure 2. NCOA7 function inhibits IAV membrane fusion.
a) Images from HA-acidification, IAV membrane fusion and nuclear vRNP import assays performed by imaging flow cytometry are shown. For the acidification assay, A549 cells expressing NCOA7 or CD8, or treated for 1 hr with bafilomycin A1, were challenged with A/Victoria/3/75 in serum free medium. At 1 hr, cells were fixed, and stained with the A1 antibody that recognizes the acidified form of HA. For the fusion assay, cells were challenged with SP-DiOC18-labelled A/Victoria/3/75 and fixed at 1.5 hr post infection. The control virus was heat-inactivated (HI) prior to labelling and infection. For the nuclear vRNP import assay, cells were challenged with A/Eng/195/2009, the medium changed at 1 and 3 hr while maintained in 1 mM cycloheximide. Cells were fixed at 5 hr and stained with anti-NP antibody; nuclei were demarcated with DAPI. Acidified HA staining, fluorescent SP-DiOC18, and nuclear NP spot counts were quantitated by imaging flow cytometry. Representative images (from 3 independent experiments) from spot count defined quintiles for the CD8 control population are shown in addition to the highest quintile for the bafilomycin A1 and heat inactivation (inhibition) controls. b-d) Histograms show spot count distributions across the CD8 control and NCOA7 populations (equal numbers of cells displayed, >1000 per condition) from a representative experiment for each assay (n=3 independent experiments). Populations are gated on the upper quintile of the CD8 control, with the bar graphs indicating the mean relative proportion of the gated populations from 3 independent experiments. Error bars represent one standard deviation from the mean.
NCOA7 was first identified as an oestrogen receptor-associated protein that localizes to the nucleus upon oestradiol treatment (19, 20). It is thought to have arisen from gene duplication of its homologue, OXR1, with which it shares oxidation resistance properties (20). The short isoform of NCOA7, studied herein, has a unique amino-terminal region of 25 amino acids and otherwise contains the carboxy-terminal five exons of the longest isoform (10). The carboxy-terminal region of OXR/NCOA7 comprises a TLDc (TBC/LysM Domain containing) domain, the function of which is currently not understood (20). In contrast to the longer isoforms, the short isoform displays cytoplasmic staining upon stable expression (Supplementary Fig. 5a) with no observable co-localization or accumulation with markers of early/late endosomes (Supplementary Fig. 5 a), and is the only isoform that is IFN-inducible (and LPS-inducible) (10), a property presumably conferred by the canonical IFN-sensitive response element sequence located upstream of its unique amino-terminal exon (Supplementary Fig. 5b) (10).
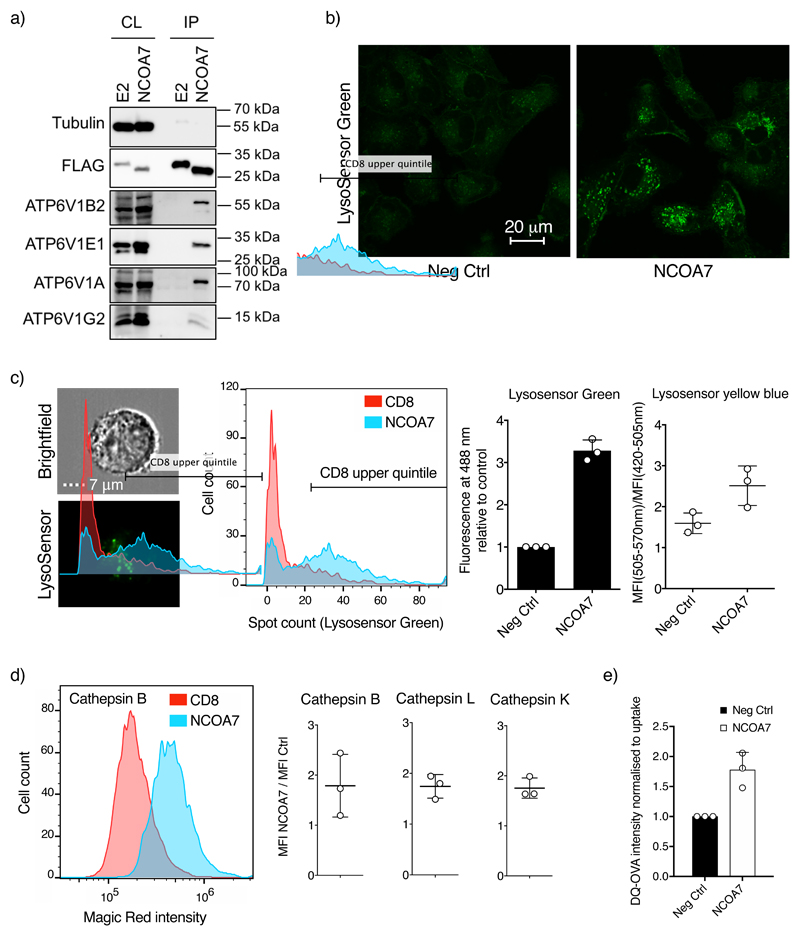
Prior proteomic analyses of the V-ATPase have reported interactions between the ATP6V1B1 subunit (a component of the catalytic motor) and both NCOA7 and OXR1 (21, 22), consistent with activity as potential V-ATPase regulators (23). ATP6V1B1 is the B-subunit that is expressed primarily in kidney cells and in cells of the inner ear, whereas ATP6V1B2 is expressed in non-renal tissues. Because NCOA7 disrupts endosome-mediated virus entry, we used co-immunoprecipitation to assess whether NCOA7 associates with ATP6V1B2. Flag-tagged NCOA7 (or E2-crimson) was expressed in U87MG cells, and whole cell lysates subjected to anti-Flag immunoprecipitation, followed by immunoblot analysis using an ATP6V1B2-specific antibody. As shown in Fig. 3a, interactions between the short isoform of NCOA7 and ATP6V1B2, as well as multiple other subunits of the V-ATPase, were readily detected.
Figure 3. NCOA7 interacts with the V-ATPase and promotes cytoplasmic vesicle acidification and lysosomal protease activation.
a) U87MG cells were transduced with lentiviral vectors constitutively expressing Flag-tagged E2-crimson (E2) or NCOA7. Cells were lysed, and the Flag-tagged proteins recovered by immunoprecipitation and analyzed by immunoblotting using anti-tubulin (loading control), anti-Flag, anti-ATP6V1B2, anti-ATP6V1A, anti-ATP6V1E or anti-ATP6V1G2 antibodies. CL, whole cell lysate; IP, immunoprecipitate. One representative immunoblot from 3 independent experiments is shown. b) A549 cells expressing NCOA7 or CD8 were plated on glass-bottomed dishes and treated for 1 hr with 1 mM LysoSensor Green. Cells were subsequently washed, incubated in clear medium and were visualized (live) by confocal microscopy. Representative images from 3 independent experiments are shown. c) A549 cells (as in panel b) were seeded, treated with 0.1 μM LysoSensor Green DND-189 for 1 hr, or with 10 μM Lysosensor Yellow/Blue DND-160 for 30 min, trypsinized and washed. For LysoSensor Green, fluorescent spot counts were determined by imaging flow cytometry and the data are represented as in Fig 2. For Lysosensor Yellow/Blue the ratio of the mean fluorescence intensity of the cells in channels 420-505 nm and 505-570 nm was measured for the analysis (>2,000 cells per condition) d) Arranged as in c) but the cells were trypsinized prior to treatment with Magic Red cathepsin B, L or K reagents for 1.5 hr. Cells were fixed and analyzed by imaging flow cytometry (>3,000 cells per condition are displayed). e) Arranged as in c) but cells were incubated in serum-free media containing 250 μg/ml of DQ Ovalbumin (Ova-DQ) or Alexa Fluor 647 Ovalbumin (Ova-647) for 1 hr followed by washing. Ova-647 containing cells were subsequently fixed whereas Ova-DQ containing samples were incubated for a further 4 hr before washing and fixation. Ova-DQ fluorescence was enumerated by flow cytometry and Ova-647 was used to normalize for uptake. The mean of 3 independent experiments is shown with error bars representing one standard deviation from the mean.
We next used cell-based assays of V-ATPase function to seek evidence of NCOA7-mediated up-regulation. The effect of NCOA7 on cytoplasmic vesicle acidification was examined using LysoSensor Green DND-189 dye, the fluorescence dequenching of which is pH dependent (pKa 5.2). Living cells were treated for 1 hr prior to visualization, with NCOA7 expression markedly increasing the number of LysoSensor Green-positive punctae (Fig. 3c), as well as the fluorescence intensity in individual vesicles (Fig. 3b). Ratiometric pH analysis using Lysosensor Yellow/Blue demonstrated that this was at least partly due to true decreases in vesicular pH (Fig 3c). These observations are consistent with NCOA7 promoting increased vesicular acidification through a regulatory interaction with the V-ATPase.
Regulation of the V-ATPase has been well described in the context of LPS-induced maturation of dendritic cells, where increased acidification results in increased antigen degradation in the lysosome and increased antigen presentation on MHC Class II (24, 25). To examine the effects of NCOA7 on V-ATPase dependent lysosomal activation we quantified the degradative activity of three different lysosomal cysteine proteases, cathepsins B, L and K, using Magic Red assays which employ cathepsin-specific target peptide sequences coupled to a fluorophore that is activated upon peptide cleavage (thought to occur primarily within endolysosomes (26)). As shown in Fig. 3d, the degradative activity of each cathepsin was substantially increased by NCOA7 expression. To verify that NCOA7 increases the degradation of endocytosed antigen (as opposed to membrane permeable substrates), we measured the degradation rate of BODIPY dye conjugated ovalbumin (DQ-Ovalbumin) by flow cytometry in NCOA7 expressing and control cells (controlling for any difference in endocytic uptake with Alexa-647 conjugated ovalbumin). Fig. 3e shows that NCOA7 promotes the degradation of Ova DQ demonstrating that the increase in acid-dependent lysosomal protease activity enhances the degradation of material entering cells by endocytosis, further supporting the assertion that NCOA7 promotes V-ATPase activity.
Here we identify the short isoform of human NCOA7 (isoform 4) as an IFNα-inducible inhibitor of viruses entering the cell by endocytosis. NCOA7 interacts with the V-ATPase and stimulates vesicle acidification, lysosomal protease activity and the degradation of endocytosed material. These perturbations compromise the fusion capabilities of viruses that are adapted to the dynamics of the endo-lysosomal pathway, thereby inhibiting infection. The endocytic pathway is also important for the entry of many bacterial pathogens and toxins and a role for OXR1/NCOA7 homologues has been suggested in protection from Vibrio cholera infection in Drosophila models (27). These findings add innate immunity to the growing number of biological contexts for which regulation of the V-ATPase is a focus of investigation: for instance, cellular nutrition, the pathology of neurodegeneration, adaptive immunity and the growth of transformed cells (24, 28, 29). Finally, in addition to providing important insights into the antiviral action of IFN, the identification of this cytokine-responsive mechanism for regulating the endo-lysosomal system may facilitate therapeutic manipulation of disease processes that stem from V-ATPase-associated endo-lysosomal dysfunction.
Materials and Methods
Plasmids and constructs
The EasiLV system including pEasiLV-MCS and pEasiLV-CD8-Flag (negative control) has been described, as has the lentiviral vector system employing pRRL.sin.cPPT.CMV/IRES-puro.WPRE used for the generation of cell lines stably expressing genes of interest (9). The CMV promoter from the latter system was replaced with SFFV promoter (amplified from HR SIN CSGW, a gift from Paul Lehner, and cloned using ClaI/BamHI) to yield pRRL.sin.cPPT.SFFV/IRES-puro.WPRE. NCOA7 variant 6 (NM_001199622.1) (encoding isoform 4 NP_001186551.1) was amplified using the SuperScript® III One-Step RT-PCR System with Platinum® Taq (Invitrogen) from 100 ng RNA obtained from IFNα-treated monocyte-derived macrophages (MDMs) using primers 5’- aaatttggatccATGAGAGGCCAAAGATTACCCTTGG-3’ and 5’-aaatttCTCGAGTCAATCAAATGCCCACACCTCCAG-3 (or the Flag-tagged 5’-aaatttggatccATGGACTACAAAGACGACGACAAAAGAGGCCAAAGATTACCCTTGG-3’). A codon-optimized version of NCOA7 (coNCOA7) CDS with the 25 first codons modified was synthesized (Eurofins; 5’-atgcgcggtcagcgtctgccactagatattcaaatcttttactgcgcgcgtcccgatgaggaaccgttcgtaaaa) and amplified by PCR. Firefly luciferase cDNA was amplified from pGL4 (Promega) and E2-crimson cDNA from pEasiLV-MCS. The cDNAs were inserted into BamHI-XhoI-digested pRRL.sin.cPPT.CMV/IRES-puro.WPRE, pRRL.sin.cPPT.SFFV/IRES-puro.WPRE or pEasiLV-MCS. NL4-3/Nef-IRES-GFP and NL4-3/Nef-IRES-Renilla were obtained from the NIH AIDS Reagent and Reference Program (pBR43IeG, cat no. 11349) and from Sumit Chanda, respectively. pBlaM-Vpr and pAdVAntage have been described (30).
HIV-1, FIV, SIV, EIAV based vector plasmids which express GFP upon infection have previously been described (11, 31–34) as have the Env-encoding constructs phCMV-MLV-A (35), phCMV-RD114/TR (35) and pRABIES Env (36).
In order to modify the A/Victoria/3/75 polymerase PA gene for co-expression with NanoLuc luciferase, a PA-NanoLuc encoding fragment (37) in which PA is fused with the porcine teschovirus P2A and the NanoLuc luciferase gene was inserted into the PasI site of PolI-RT-Victoria PA plasmid. The 12-plasmid system used to rescue IAV Victoria-NanoLuc reporter virus was provided by Wendy Barclay. The firefly luciferase gene from pHSP1-Firefly (9) was replaced by the eGFP coding sequence to generate pHSP1-eGFP.
HCV particles were generated using the cell culture-adapted clone J6/JFH-1 (38).
The LentiCas9-Blast and LentiGuide-Puro vectors were a gift from Feng Zhang (39) (Addgene). The LentiGuide-Neo vector was generated by replacing the puromycin resistance gene in LentiGuide-Puro with the neomycin resistance gene (amplified using PCR from pcDNA3.1+) and using BsiWI and MluI sites. Guide RNA coding oligonucleotides were annealed and ligated into BsmBI-digested LentiGuide-Puro or LentiGuide-Neo vectors, as described (Addgene). The gRNA coding sequences used were as follow: g1-GFP 5’-gagctggacggcgacgtaaa, g1-CTRL 5’-agcacgtaatgtccgtggat; g2-CTRL 5’-caatcggcgacgttttaaat; g-IFITM3 5’-taggcctggaagatcagcac; g1-NCOA7 5’-aaaaggctcttcgtcaggtc; g2-NCOA7 5’-ttcacaaaaggctcttcgtc; g3-NCOA7 5’-ggatgtccaagggtaatctt. The 3 NCOA7 guides were designed to target the first coding exon of NCOA7 variant 6 (NM_001199622.1), which is unique to the short IFN-inducible isoform 4 (NP_001186551.1) studied herein.
Cell lines
Human 293T, U87MG CD4+ CXCR4+(9), A549, MDCK and Huh-7.5 cells were maintained in complete Dulbecco's modified Eagle medium (DMEM) (Gibco) and THP-1 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco), both supplemented with 10% foetal bovine serum and penicillin/streptomycin. Non-essential amino acids were added for Huh-7.5 cells. 293T, A549, MDCK and THP-1 cells were obtained from American Type Culture Collection (ATCC). Huh-7.5 were originally obtained from the Rice laboratory (40). U87MG cells were originally obtained from the NIH AIDS Reagent Program and modified to express CD4 and CXCR4 (9). A549, 293T, U87MG CD4+ CXCR4+, and Huh-7.5 cells stably expressing NCOA7, CD8 or E2-crimson were generated by transduction with either RRL.sin.cPPT.CMV/IRES-puro.WPRE or RRL.sin.cPPT.SFFV/IRES-puro.WPRE containing-vectors (cDNA as indicated) and were maintained under 1 μg/ml puromycin selection.
For CRISPR-Cas9-mediated gene disruption, A549 cells stably expressing Cas9 were first generated by transduction with the LentiCas9-Blast vector followed by blasticidin selection at 10 μg/ml. Cas9-expressing A549s were then transduced with guide RNA expressing LentiGuide-Puro and Lentiguide-Neo vectors and cells selected with antibiotics for at least 15 days. Single CRISPR/Cas9 knock-out cellular clones were isolated using a FACSAria IIu cell sorter (Beckton Dickinson).
When indicated, IFNα (INTRON® A; Merck Sharp & Dohme Corp.) was added at 1,000 U/ml for 16-24 hr prior to virus infection. Cells were treated with 50 nM bafilomycin A1 (an inhibitor of the V-ATPase) (Sigma-Aldrich) for 1 hr prior to infection and throughout the assay where indicated; cycloheximide (Sigma-Aldrich) was used at 1 mM to prevent de novo protein synthesis and the media replenished every 2 hr if required.
Lentiviral production and infection
Lentiviral vector stocks were obtained by polyethylenimine (PEI), TransIT-2020 (Mirus Bio LLC) or Lipofectamine 3000 (Thermo Scientific)-mediated multiple transfection of 293T cells in 6-well plates with vectors expressing Gag-Pol, the miniviral genome, the Env glycoprotein, and when required (i.e. for EasiLV production), a TetR-KRAB expression plasmid (41) (pptTRKrab,) at a ratio of 1:1:0.25:0.5. The culture medium was changed 6 hr post-transfection, and vector containing supernatants harvested 36 hr later, filtered and stored at -80°C. Wild type HIV-1 particles (NL4-3/Nef-IRES-GFP, NL4-3/Nef-IRES-Renilla) were generated by polyethylenimine (PEI)-based transfection of 293T cells. β-lactamase-Vpr (BlaM-Vpr)-carrying viruses, VSV-G-pseudotyped or bearing the wild type Env, were produced by co-transfection of 293T cells with pMD.G (or not), the NL4-3/Nef-IRES-Renilla provirus expression vector, pBlaM-Vpr and pAdVAntage at a ratio of (1.33:)4:1:0.5, as previously described (30).
For EasiLV-based experiments, U87MG CD4+ CXCR4+ cells were transduced with EasiLV stocks encoding genes of interest and the medium changed at 6 hr. Doxycycline (0.4-1 μg/ml) was added for 24-48 hr, the cells plated at 2.5 x 104 per well in 96-well plates, and then challenged with GFP-expressing virus stocks or NL4-3. The percentage of infected, GFP-expressing cells was enumerated by flow cytometry 48 hr after infection.
HIV-1NL4-3 containing, filtered supernatants were routinely purified by ultracentrifugation through a sucrose cushion (20% w/v; 75 min; 4°C, 28,000 rpm using a Beckman Coulter SW 32 rotor), resuspended in DMEM medium without serum and stored in small aliquots at -80°C. Viral particles were normalized by p24Gag ELISA or p24Gag AlphaLisa (Perkin-Elmer) and/or by determining their infectious titers on U87MG cells (for GFP expressing LVs, RVs and HIV-1). The multiplicity of infection (MOI) for LV stocks was determined by infecting a known number of cells with standardized amounts of viral particles and evaluating by flow cytometry the percent of infected cells 2-3 days later. For instance, an MOI of 0.1 would theoretically correspond to the volume of virus necessary to obtain 10% of GFP-expressing cells.
IAV production and infection
A/Eng/195/2009 and A/Victoria/3/75 particles were provided by Wendy Barclay and amplified in MDCK cells using serum-free DMEM containing 0.5 μg/ml TPCK-treated trypsin (Sigma-Aldrich). Stocks were titred by plaque assays on MDCK cells, or on A549 cells to determine multiplicities of infection (MOI) defined by the proportion of NP-positive nuclei at 12 hr post infection. A 12-plasmid system was used to rescue A/Victoria/3/75 and the IAV Victoria-NanoLuc reporter virus. The 8 PolI plasmids (0.5 μg each) and the 4 rescue plasmids (PB1, PB2, PA: 0.32 μg each, NP: 0.64 μg) were co-transfected into 293T cells in 6-well plates using Lipofectamine3000 (Thermo Scientific). After 24 hr, the cells were removed and co-cultured with MDCK cells in 25 ml flasks. The first 8 hr of co-culture was carried out in 10% serum, after which the medium was replaced with serum-free medium containing 0.5 μg/ml of TPCK-treated trypsin. Supernatants from day 5 post-transfection were used for virus amplification on MDCK cells.
All IAV challenges were performed in serum-free DMEM for 1 hr (unless otherwise specified) and the medium was subsequently replaced with DMEM containing 10% FBS (where the experiment lasted more than 1.5 hr). For microscopy-based infectivity assays, A549 cells stably expressing CD8 or NCOA7 were plated on coverslips in 12-well plates at a density of 1 x 105 cells per well. The following day, cells were infected with A/Eng/195/2009 (MOI of 10). Fixation was performed at 5 hr post-infection using 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 10 min, and the cells were permeabilized using 0.2% Triton X-100 for 10 min. After quenching and blocking in buffer NGB (50 mM NH4Cl, 2% goat serum, 2% bovine serum albumin in PBS) for 1 hr, cells were incubated with anti-NP antibody (Hb65; provided by Yohei Yamauchi) (14), diluted in buffer NGB for 1 hr and subsequently incubated with an Alexa-488 conjugated anti-mouse secondary antibody. Cells were then stained with 1 μg/ml 4',6-diamidino-2-phenylindole (DAPI), washed, mounted on coverslips and examined using a Nikon A1 confocal microscope (10X objective) with captured images analyzed using FIJI (42) to determine the percentage of GFP-expressing cells. For time-lapse microscopy, 293T cells stably expressing CD8 or NCOA7 were seeded (5 x 104 per well) in a 48-well plate and transfected with 0.1 μg pHSP1-eGFP using Lipofectamine 2000 (Thermo Scientific). pHSP1-eGFP is a minigenome-like reporter construct that expresses GFP and when transfected into target cells is transcribed by host RNA polymerase to produce a negative sense, viral genomic like segment (with 5’-triphosphate and 3’-hydroxyl). Upon IAV infection, this segment is recognized by the viral RNA polymerase to generate GFP-expressing mRNA. After ~24 hr, the cells were infected with A/Eng/195/2009 (MOI of 1) and then monitored by time-lapse microscopy with images acquired at 20 min intervals from 2 to ~18 hr post-infection using a Nikon Ti-Eclipse wide-field inverted microscope (40X objective) and controlled by NIS Elements software. Movies were created using FIJI (42). The Victoria-Nanoluc infection experiments were performed in duplicate or triplicate in 96-well plates with cultures maintained for 7 hr post-challenge. NanoLuc activity was measured with the Nano-Glo assay system (Promega), and luminescence was detected using a plate reader (Infinite® 200 PRO, Tecan).
HCV production and infection
Cell culture-derived HCV particle stocks were produced by electroporation of in vitro-transcribed viral RNA into Huh-7.5 cells using a Bio-Rad Gene Pulser Xcell (Bio-Rad) (43). A cell culture-adapted J6/JFH-1 derivative virus, named Clone 2, was used for this study. Virus-containing supernatants were collected 48, 72 and 96 hours post-electroporation and titrated by limiting dilution assay, as described (44). Transduced Huh-7.5 cells were seeded in 24-well plates, challenged with virus (MOI 0.5) and incubated for 48 hr. Cells were harvested using AccuMax (eBioscience), fixed with 4% PFA, permeabilized with PBS-saponin (0.2%) and stained for 1 hr at room temperature with an Alexa-647-conjugated antibody (9E10 clone) specific for the HCV non-structural protein NS5A in PBS-saponin (0.2%). The percentage of infected cells determined by flow cytometry.
Quantification of mRNA expression
3-5 x 105 cells with or without 24hr treatment with IFNα were harvested and total RNA was extracted using the RNeasy kit (Qiagen) employing on-column DNase treatment. 500 ng RNA was used to generate cDNA, which was analyzed by qPCR using TaqMan gene expression assays (Applied Biosystems) for ACTB (Hs99999903_m1) or GAPDH (Hs99999905_m1). For the short isoform of NCOA7, the primers used were 5’-GATTACCCTTGGACATCCAGATTTTCTATT-3’ and 5’-CACCTCTTCGTCCTCGTCTTCATAGT-3’ and the probe sequence was 5’-FAM-AGGCAAAGCGCAGGAAGAGCACATGC-TAMRA-3’. Triplicate reactions were run according to the manufacturer’s instructions using an ABI Prism model 7900HT sequence detection platform, and data was analyzed using RQ software (Applied Biosystems). GAPDH and ACTB mRNA expression was used to normalize samples.
T7-endonuclease assay
To assess CRISPR/Cas9 mediated editing of the NCOA7 and IFITM3 loci, genomic DNA was extracted from cell populations (DNeasy, Qiagen) and an amplicon spanning the targeted genomic region was generated on 50 ng DNA by touch-down PCR using the primers 5’-tgctggtagaaatgtcagcacaatcctttc-3’ and 5’-aaagttcattttactctaaaatccatttttggccc-3’, 5’-tccctgatctcagggcgggg and 5’-aggcccgaaaccagaaggc, respectively. Amplicons were then subject to denaturation, reannealing and T7-endonuclease (NEB) digestion.
Indirect imunofluorescence
For visualization of NCOA7 localization by confocal microscopy, U87MG CD4+ CXCR4+ cultures were transduced with lentiviral vectors containing Flag-tagged NCOA7 as described above and were subsequently plated on coverslips treated with poly-L-lysin. Fixation, staining with anti-Flag antibody (Sigma-Aldrich F3165 or F7425, or Miltenyi 130-101-576), anti-EEA1 (BD Biosciences 610457), anti-CD63 (Santa-Cruz sc5275) and anti-Lamp1 (Sigma-Aldrich L1418), and Alexa-488 and Alexa-546 conjugated anti-mouse or anti-rabbit secondary, and and image acquisition was performed as in previous sections with either a Zeiss LSM880 confocal microscope.
Influenza A virus uptake assay
Stably expressing E2-crimson- or NCOA7-A549 cells were seeded at 105 cells and 2 x 105 cells per well, respectively, in 12-well plates. The following day, medium was removed and cells were incubated in serum-free DMEM with 3 x 105 and 1.5 x 106 PFU of A/Victoria/3/75 for 20 min at 37°C. A heat-inactivated virus was used at the highest viral input as a negative control. Cells were then trypsinized, washed and fixed in 2% PFA. BD Perm/wash™ (BD Biosciences) was used to permeabilized and stained the cells with anti-NP antibody (Hb65) and Alexa 488 secondary antibody according to the manufacturer’s instructions. Cells were analyzed using a NovoCyte™ (Ozyme) cytometer.
HA acidification assay
A549 cells, parental or stably expressing CD8 or NCOA7, were seeded in 6-well plates. Unmodified cells were treated with 50 nM bafilomycin A1 for 1 hr prior to infection (inhibition control). Medium was removed and cells were infected in serum-free DMEM with A/Victoria/3/75 at MOI of 30 and incubated for 1 hr at 37°C. Cells were then trypsinized, and both fixed and stained using IntraStain (Dako) with monoclonal antibody A1 (14), which is specific for HA that has undergone low pH induced conformational change (provided by Yohei Yamauchi). Alexa-647 conjugated anti-mouse total IgG (Life Technologies) was used as secondary antibody, and cells were analyzed by imaging flow cytometry (see below) using the 647 nm laser for excitation.
Membrane fusion assay
A/Victoria/3/75 was labeled with the self-quenching dye SP-DiOC18(3) (Molecular Probes). SP-DiOC18(3) was added to 1 ml of virus stock in serum-free DMEM (containing approximately 2.5 x 107 infectious units per ml) at a final concentration of 0.2 μM in a screw-capped eppendorf tube (45). The tube was protected from light, incubated while rolling for 1 hr at room temperature and subsequently stored at -80°C or used as required. For the heat inactivation control, virus was kept at 75°C for 30 min and returned to room temperature prior to labelling. Labeled virus preparations were passed through a 0.45μm filter. Labeled virus was diluted in serum free DMEM for infection of CD8 or NCOA7 expressing A549 monolayers at MOI of 10. Cells were incubated for 1.5 hr after infection, harvested with trypsin, fixed with 4% PFA for 10 min and analyzed by imaging flow cytometry using the 488 nm laser for excitation.
BlaM-VPR assay for lentiviral entry
The BlaM-VPR assay has previously been described (30). Briefly, 2 x 105 E2-crimson or NCOA7-expressing U87MG CD4+ CXCR4+ cells were plated in 24 well plates the day prior to infection. Cells were incubated with increasing amounts of VSV-G pseudotyped or wild-type Envelope, BlaM-VPR containing NL4-3/Nef-IRES-Renilla viruses (31.2, 62.5 or 125 ng p24) or mock infected for 3 hr. Cells were then washed once in CO2-independent media and loaded with CCF2-AM substrate (Invitrogen) containing development media (CO2-independent media containing 1.6 mM probenecid) for 2 hr at room temperature before 2 washes and incubation at room temperature for 16 hr in development media. Finally, cells were harvested, washed and fixed in 2% PFA, before analysis with using a FACSCanto II (BD Biosciences).
Nuclear virion RNP (vRNP) import assay
A549 cells were plated and unmodified cells treated with bafilomycin A1 as above. 1 mM cycloheximide was maintained in the medium throughout the experiment. Cells were infected with A/Eng/195/2009 at MOI of 10. The medium was changed at 1 hr (as above) and again at 3 hr to ensure optimal cycloheximide activity. Cells were fixed in 4% PFA at 5 hr, stained using Permwash (BD Biosciences) according to the manufacturer’s instructions with anti-NP (Hb65) antibody and secondary as above. DAPI staining was used to demarcate the nucleus (0.1 μg/ml for 5 min). Cells were analyzed by imaging flow cytometry using a 642 nm laser for excitation. For the parallel visualization of nuclear vRNP import by confocal microscopy, cells were plated on coverslips as described above, infections conditions were identical and staining and image acquisition was performed as described.
Vesicle acidification and cathepsin activity assays
Transduced A549 cells were plated in 6-well plates and treated with 0.1 μM LysoSensor Green DND-189 (Molecular Probes) for 1 hr, or with 10 μM Lysosensor Yellow/Blue DND-160 for 30 min at 37°C. Cells were subsequently trypsinized, washed and resuspended for immediate analysis by imaging flow cytometry. A 488 nm laser was used for Lysosensor Green. For Lysosensor Yellow/Blue a 405 nm laser was used for excitation and the ratio of the mean fluorescence intensity of the cells in channels 420-505 nm and 505-570 nm was measured for the analysis. For analysis by confocal microscopy, A549 cells were plated in glass-bottomed dishes and, the following day, treated with 1 μM LysoSensor Green for 1 hr. Living cells were subsequently washed and visualized using a Nikon A1R confocal microscope and a 488 nm laser. To analyze cathepsin B, L and K activity, the Magic Red reagents (Immunochemistry Technologies) specific for each of these proteases were reconstituted in DMSO and subsequently diluted in distilled H20 to form a 26X solution as per the manufacturer’s instructions. Approximately 5 x 105 transduced A549 cells were trypsinized from a 6-well plate and resuspended in 150 μl medium, to which the reagent was added. Cells were incubated for 1.5 hr at 37°C, washed, fixed in 2% PFA and analyzed by imaging flow cytometry using a 561 nm laser for excitation.
Ovalbumin degradation assay
CD8 or NCOA7 expressing cells were plated in 48 well plates. Cells were incubated in serum-free media containing 250μg/ml of DQ Ovalbumin (Ova-DQ), which emits fluorescence upon degradation, or Alexa Fluor 647 Ovalbumin (Ova-647) for 1 hr at 37°C followed by washing. Ova-647 containing cells were subsequently fixed whereas Ova-DQ containing samples were incubated for a further 4 hr at 37°C before washing and fixation. Ova-DQ and Ova-647 fluorescence was enumerated by flow cytometry. Ova-DQ fluorescence at 4 hr was normalized by Ova-647 at 1 hr to allow for any differences in uptake of reagent.
Imaging flow cytometry and flow cytometry
Cells were processed using the Amnis ImageStreamX at 60X magnification with the indicated excitation laser, in addition to the brightfield system, according to the manufacturer’s instructions. Amnis IDEAS software was used to analyze the data. The IDEAs automated spot counting wizard, performed on the CD8 expressing population, was used to determine the optimal spot counting algorithm for individual experiments. For the nuclear vRNP import assay, spot counting was performed on the “mask” (region of interest) demarcated by DAPI staining using a threshold of 40% of the maximum DAPI staining intensity. Standard flow cytometry was performed using the FACSCanto II (BD Biosciences) or the NovoCyte™ (Ozyme).
Co-Immunoprecipitation and immunoblot analysis
U87MG cells were stably transduced with vectors carrying RRL.sin.cPPT.CMV/Flag-NCOA7.WPRE or RRL.sin.cPPT.CMV/Flag-E2-crimson-.WPRE and selected with 1 μg/ml puromycin. 15-22 x 106 cells were washed twice in cold PBS and resuspended in lysis buffer (20mM Hepes-NaOH pH 7.5, 150 mM NaCl, 1% NP-40, protease inhibitor cocktail), the lysates clarified by centrifugation at 1000g, for 10 min at 4°C and then incubated with Flag-magnetic beads (Life Technologies) for 3 hr at 4°C. The beads were washed 5 times and the immunoprecipitated proteins eluted using 3x Flag peptide (150 μg/ml, Sigma-Aldrich) for 1.5-2 hr. Cell lysates and immunoprecipitation eluates were supplemented with sample buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 5% glycerol, 100 mM DTT, 0.02% bromphenol blue), resolved by SDS-PAGE and analyzed by immunoblotting using primary antibodies specific for the Flag epitope (Sigma-Aldrich F3165), tubulin (mouse monoclonal DM1A, Sigma-Aldrich T9026), ATP6V1B2 (Proteintech 15097-1-AP), ATP6V1A (Proteintech 17115-1-AP), ATP6V1E1 (Abcam Ab111733), ATP6V1G2 (Proteintech 25316-1-AP), followed by secondary horseradish peroxidase-conjugated anti-mouse, or anti-rabbit immunoglobulin antibodies and chemiluminescence Clarity or Clarity max substrate (Bio-Rad). A Bio-Rad ChemiDoc imager was used.
Statistical analysis
Mann Whitney test (two-tailed) was used for statistical analysis in Fig 1g and Supplementary Fig 2d.
Supplementary Material
Acknowledgements
We wish to thank, Wendy Barclay, Fabien Blanchet, Matteo Bonazzi, Lucile Espert, Moona Huttunen, Jason Long, Eric Martinez, Jason Mercer, Delphine Muriaux, Paula Rocha, Reiner Schulz and Yohei Yamauchi, for the generous provision of reagents and for helpful discussions. We are grateful to Aisling Vaughan for instruction in IAV amplification, PJ Chana from Biomedical Research Centre (BRC) Flow core for training and advice on imaging flow cytometry, Isma Ali from the King’s College London Nikon Imaging Centre for advice with confocal microscopy and image acquisition, Ian Parham for help with the time-lapse microscopy, Myriam Boyer and Baptiste Monterroso from the imaging and flow cytometry facility MRI, for cell sorting and for advice with flow cytometry and confocal microscopy, respectively. This work was supported by the U.K. Medical Research Council (G1000196) (to MM), the Wellcome Trust (106223/Z/14/Z) (to MM), The National Institutes of Health (DA033773), a Wellcome Trust Research Training Fellowship and a National Institute for Health Research (NIHR) BRC King’s Prize Fellowship (to TD), the Institut National de la Santé et de la Recherche Médicale (INSERM) (to CG), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement n°759226) (to CG), the ATIP-Avenir programme (to CG) and institutional funds from the Centre National de la Recherche Scientifique (CNRS) and Montpellier University (to CG), France REcherche Nord&Sud Sida/HIV et Hépatites (ANRS) (to OM), a PhD studentship from the Ministry of Higher Education and Research (to BB), King’s College London Departmental start-up funds (to MTC), and the Department of Health via a NIHR BRC award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust. We acknowledge the imaging facility MRI, member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04).
Footnotes
Requests for materials:
Requests for material should be addressed to Michael Malim or Caroline Goujon at the corresponding addresses above.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Michael H. Malim: 000-0002-7699-2064
Caroline Goujon: 0000-0001-8571-1108
Conflicts of interest statement:
The authors have no conflicts of interest to declare in relation to this manuscript.
Author contribution statement:
T.D., O.M., M.-T.C., C.G. and M.H.M. conceived and designed the experiments; T.D., O.M., B.B., D.P., M.L., M.T., L.A. and C.G. performed the experiments; all authors analyzed the data; and T.D., C.G. and M.H.M. wrote the manuscript with input from all authors.
References
- 1.Doyle T, Goujon C, Malim MH. HIV-1 and interferons: who's interfering with whom? Nature reviews Microbiology. 2015;13(7):403–13. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nature reviews Immunology. 2015;15(2):87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. The Journal of general virology. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nature reviews Immunology. 2014;14(5):315–28. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31(1):79–87. doi: 10.1089/jir.2010.0076. [DOI] [PubMed] [Google Scholar]
- 6.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–54. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519–23. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goujon C, Malim MH. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. Journal of virology. 2010;84(18):9254–66. doi: 10.1128/JVI.00854-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502(7472):559–62. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu L, Croze E, Yamaguchi KD, Tran T, Reder AT, Litvak V, et al. Induction of a unique isoform of the NCOA7 oxidation resistance gene by interferon beta-1b. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2015;35(3):186–99. doi: 10.1089/jir.2014.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 12.Herold N, Anders-Osswein M, Glass B, Eckhardt M, Muller B, Krausslich HG. HIV-1 entry in SupT1-R5, CEM-ss, and primary CD4+ T cells occurs at the plasma membrane and does not require endocytosis. Journal of virology. 2014;88(24):13956–70. doi: 10.1128/JVI.01543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Annual review of biochemistry. 2010;79:803–33. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee I, Yamauchi Y, Helenius A, Horvath P. High-content analysis of sequential events during the early phase of influenza A virus infection. PloS one. 2013;8(7):e68450. doi: 10.1371/journal.pone.0068450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee I, Miyake Y, Nobs SP, Schneider C, Horvath P, Kopf M, et al. Influenza A virus uses the aggresome processing machinery for host cell entry. Science. 2014;346(6208):473–7. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 16.Webster RG, Brown LE, Jackson DC. Changes in the antigenicity of the hemagglutinin molecule of H3 influenza virus at acidic pH. Virology. 1983;126(2):587–99. doi: 10.1016/s0042-6822(83)80015-4. [DOI] [PubMed] [Google Scholar]
- 17.Shelton H, Roberts KL, Molesti E, Temperton N, Barclay WS. Mutations in haemagglutinin that affect receptor binding and pH stability increase replication of a PR8 influenza virus with H5 HA in the upper respiratory tract of ferrets and may contribute to transmissibility. The Journal of general virology. 2013;94(Pt 6):1220–9. doi: 10.1099/vir.0.050526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russier M, Yang G, Rehg JE, Wong SS, Mostafa HH, Fabrizio TP, et al. Molecular requirements for a pandemic influenza virus: An acid-stable hemagglutinin protein. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(6):1636–41. doi: 10.1073/pnas.1524384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao W, Halachmi S, Brown M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Molecular and cellular biology. 2002;22(10):3358–72. doi: 10.1128/MCB.22.10.3358-3372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durand M, Kolpak A, Farrell T, Elliott NA, Shao W, Brown M, et al. The OXR domain defines a conserved family of eukaryotic oxidation resistance proteins. BMC cell biology. 2007;8:13. doi: 10.1186/1471-2121-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkulova M, Paunescu TG, Azroyan A, Marshansky V, Breton S, Brown D. Mapping the H(+) (V)-ATPase interactome: identification of proteins involved in trafficking, folding, assembly and phosphorylation. Scientific reports. 2015;5 doi: 10.1038/srep14827. 14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162(2):425–40. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merkulova M, Paunescu TG, Nair AV, Wang CY, Capen DE, Oliver PL, et al. Targeted deletion of the Ncoa7 gene results in incomplete distal renal tubular acidosis in mice. American journal of physiology Renal physiology. 2018;315(1):F173–F85. doi: 10.1152/ajprenal.00407.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299(5611):1400–3. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 25.Cotter K, Stransky L, McGuire C, Forgac M. Recent Insights into the Structure, Regulation, and Function of the V-ATPases. Trends in biochemical sciences. 2015;40(10):611–22. doi: 10.1016/j.tibs.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bright NA, Davis LJ, Luzio JP. Endolysosomes Are the Principal Intracellular Sites of Acid Hydrolase Activity. Curr Biol. 2016;26(17):2233–45. doi: 10.1016/j.cub.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Berkey CD, Watnick PI. The Drosophila protein mustard tailors the innate immune response activated by the immune deficiency pathway. Journal of immunology. 2012;188(8):3993–4000. doi: 10.4049/jimmunol.1103301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colacurcio DJ, Nixon RA. Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing research reviews. 2016 doi: 10.1016/j.arr.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sennoune SR, Martinez-Zaguilan R. Vacuolar H+-ATPase Signaling Pathway in Cancer. Curr Protein Pept Sc. 2012;13(2):152–63. doi: 10.2174/138920312800493197. [DOI] [PubMed] [Google Scholar]
- 30.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nature biotechnology. 2002;20(11):1151–4. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 31.Mangeot PE, Duperrier K, Negre D, Boson B, Rigal D, Cosset FL, et al. High levels of transduction of human dendritic cells with optimized SIV vectors. Molecular therapy : the journal of the American Society of Gene Therapy. 2002;5(3):283–90. doi: 10.1006/mthe.2002.0541. [DOI] [PubMed] [Google Scholar]
- 32.Saenz DT, Teo W, Olsen JC, Poeschla EM. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5alpha proteins. Journal of virology. 2005;79(24):15175–88. doi: 10.1128/JVI.79.24.15175-15188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Rourke JP, Newbound GC, Kohn DB, Olsen JC, Bunnell BA. Comparison of gene transfer efficiencies and gene expression levels achieved with equine infectious anemia virus- and human immunodeficiency virus type 1-derived lentivirus vectors. Journal of virology. 2002;76(3):1510–5. doi: 10.1128/JVI.76.3.1510-1515.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarrosson-Wuilleme L, Goujon C, Bernaud J, Rigal D, Darlix JL, Cimarelli A. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. Journal of virology. 2006;80(3):1152–9. doi: 10.1128/JVI.80.3.1152-1159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandrin V, Boson B, Salmon P, Gay W, Negre D, Le Grand R, et al. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100(3):823–32. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 36.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature methods. 2007;4(1):47–9. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran V, Moser LA, Poole DS, Mehle A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. Journal of virology. 2013;87(24):13321–9. doi: 10.1128/JVI.02381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, et al. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS pathogens. 2009;5(1):e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11(8):783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. Journal of virology. 2002;76(24):13001–14. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangeot PE, Dollet S, Girard M, Ciancia C, Joly S, Peschanski M, et al. Protein transfer into human cells by VSV-G-induced nanovesicles. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19(9):1656–66. doi: 10.1038/mt.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9(7):676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–6. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 44.Catanese MT, Loureiro J, Jones CT, Dorner M, von Hahn T, Rice CM. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. Journal of virology. 2013;87(15):8282–93. doi: 10.1128/JVI.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai T, Ohuchi M, Imai M, Mizuno T, Kawasaki K, Kuroda K, et al. Dual wavelength imaging allows analysis of membrane fusion of influenza virus inside cells. Journal of virology. 2006;80(4):2013–8. doi: 10.1128/JVI.80.4.2013-2018.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.