Abstract
Translin-associated protein X (TSNAX), also called trax, was first identified as a protein that interacts with translin. Subsequent studies demonstrated that these proteins form a heteromeric RNase complex that mediates degradation of microRNAs, a pivotal finding that has stimulated interest in understanding the role of translin and trax in cell signaling. Recent studies addressing this question have revealed that trax plays key roles in both synaptic plasticity and DNA repair signaling pathways. In the context of synaptic plasticity, trax works together with its partner protein, translin, to degrade a subset of microRNAs. Activation of the translin/trax RNase complex reverses microRNA-mediated translational silencing to trigger dendritic protein synthesis critical for synaptic plasticity. In the context of DNA repair, trax binds to and activates ATM, a central component of the double-stranded DNA repair process. Thus, these studies focus attention on trax as a critical signaling protein that interacts with multiple partners to impact diverse signaling pathways. To stimulate interest in deciphering the multifaceted role of trax in cell signaling, we summarize the current understanding of trax biology and highlight gaps in our knowledge about this protean protein.
Keywords: translin, microRNA system, synaptic tagging, ATM, adenosine A2A receptors
Introduction
Since the initial discovery of trax and its partner protein, translin, about 20 years ago [1,2], there has been substantial progress in deciphering their interaction and function. However, figuring out how they operate in the context of cellular signaling pathways has posed a formidable challenge. Recent studies have succeeded in positioning trax in two major cell signaling pathways, important breakthroughs that will stimulate broader interest in its function. In one line of research, Park et al. [3] implicate trax, working in combination with translin, in mediating an important form of synaptic plasticity in the hippocampus, referred to as synaptic tagging and capture [4]. In a completely separate area of investigation, Chern and co-workers [5,6] have demonstrated in fibroblasts and neurons that trax, acting without translin, plays a key role in a major signaling pathway that repairs double-stranded DNA breaks. Thus, these findings suggest that trax plays critical roles in multiple signaling pathways and that further studies are warranted to elucidate how these ostensibly disparate functions may be related. Furthermore, these lines of research have unexpectedly revealed that trax exerts its actions on DNA repair and synaptic plasticity in fundamentally different ways, prompting a re-appraisal of the mode of action of trax. Previous models postulated that trax interacts exclusively with translin because trax protein is unstable in cells or organisms lacking translin [7]. However, these new findings indicate that trax is a versatile protein that is able to participate in diverse signaling pathways via its interaction with multiple partner proteins.
To highlight the significance of these findings, we will first provide a brief overview of these recent studies. Then, we will summarize the initial discovery and characterization of translin and trax, focusing on evidence that indicated that trax interacts exclusively with translin. Lastly, we will highlight how recent findings have prompted an expanded model of trax action that incorporates its interaction with translin as well as other partner proteins.
Translin/ trax RNase complex mediates synaptic plasticity
As rapid changes in translation in response to environmental cues play a key role in mediating neuronal plasticity, there is considerable interest in defining the cellular mechanisms capable of triggering rapid translational responses [8–10]. Given the prominent role that the microRNA system plays in translational silencing, the possibility that rapid reversal of microRNA-mediated translational silencing could elicit translational responses underlying plasticity has attracted attention [11]. Accordingly, this line of thinking has heightened interest in identifying microRNA degradation pathways that are rapidly activated by cell surface receptor stimulation and can trigger this translational response. The demonstration that Lin-28a responds quickly to growth factor stimulation and mediates rapid transcription of plasticity transcripts in hippocampal cultures has provided critical support for this mechanism [12]. However, since Lin-28a acts selectively on let-7 family members, these findings have prompted a search for other microRNA degradation pathways capable of targeting other subsets of microRNAs. From this perspective, the observation that the translin/trax RNAs complex targets a different subset of microRNAs for degradation [11] raised the possibility that it might mediate rapid translational responses underlying synaptic plasticity. The recent study by Park et al. (2017) [3] directly tests this hypothesis and provides compelling evidence that activation of translin/trax RNase by synaptic stimulation in the mammalian hippocampus, triggers the rapid reversal of translational silencing and, thereby, plays a key role in mediating long-lasting synaptic plasticity and long-term memory. Thus, these findings establish the translin/trax RNase complex as a key component of the signaling pathway linking synaptic stimulation to translation of “plasticity” proteins (Figure 1).
Figure 1: Rapid reversal of translational silencing by synaptic stimulation: role of translin/ trax.
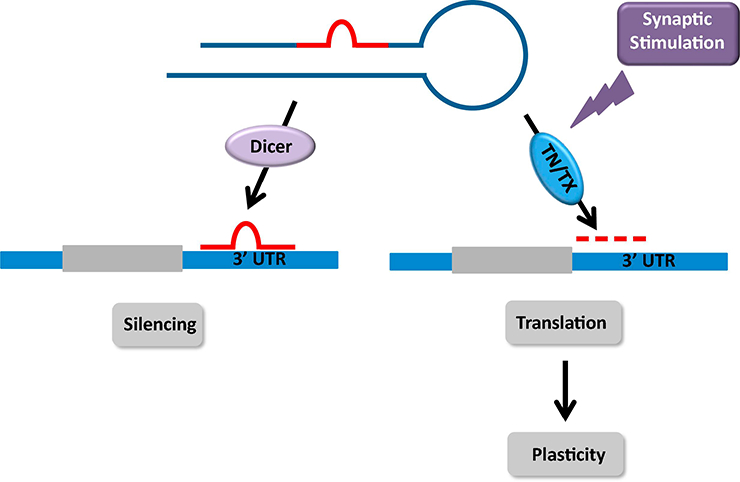
Dicer mediated processing of pre-miRNAs produces mature miRNAs that bind to the 3’UTR of dendritic mRNAs to silence their translation. Synaptic stimulation can reverse silencing of these transcripts via activation of translin/ trax, a microRNA-degrading enzyme. Rapid degradation of microRNAs targeted by translin/trax triggers de novo translation of proteins underlying long-term synaptic plasticity.
Trax mediates DNA repair
In a separate line of research, Chern and co-workers [5,6] have uncovered a key role for trax in DNA repair. The initial studies that led to the identification of translin and its partner protein, trax sought to identify DNA binding proteins that mediate chromosomal translocations associated with malignant transformations, such as those involving Bcl-2 [1,2]. In addition, subsequent studies identified trax as a binding partner of C1D, a protein that binds to and activates DNA- dependent protein kinase (DNA-PK), a key enzyme responsible for the repair of double-stranded breaks (DSBs) [13,14]. Furthermore, γ - irradiation increases expression of C1D and its interaction with trax [14], suggesting that trax may contribute to the maintenance of genome integrity. In following up on these earlier findings, Wang et al. [6] found that trax-null cells are more susceptible to DNA damage produced by UV- or gamma-irradiation. In addition, studies aimed at deciphering how trax participates in the cellular response to DNA damage revealed that tiax binds to and promotes auto-phosphorylation of ATM, which plays a major role in promoting repair of double-strand breaks (DSBs).
As it was well established that tiax forms a complex with translin, these investigators checked whether tianslin was also present in the trax/ATM complex. Unexpectedly, they found that it was not, indicating that the interaction of trax with ATM occurs independently of translin, generally thought to be an obligatory partner of tiax. However, this observation fits well with the previous report that binding of trax to C1D and translin are mutually exclusive [14]. Furthermore, a mutant form of trax that abolishes the RNase activity of the TN/TX complex retains the ability to promote ATM phosphorylation, providing additional evidence that the effect of trax on DNA repair is not mediated via its interaction with tianslin.
Thus, taken together, these studies indicate that trax is a versatile signaling protein that acts in fundamentally different ways in distinct contexts: together with translin it can form an RNase complex that mediates microRNA degradation; working independently of translin it binds to ATM to promote DNA repair. To help explain how this expanded view of trax action contrasts with earlier notions that trax interacts exclusively with translin, we will present a brief overview of the classical findings that formed the basis for the initial, limited view of trax action.
Initial identification of trax as a partner for translin
Translin was initially identified in the search for DNA binding factors that might mediate chromosomal translocations [2]. To this end, Kasai and coworkers looked for proteins that bind to a ssDNA fragment from the region flanking Bcl-2, a well-known site of chromosomal translocation linked to lymphoma. This approach led to the isolation of translin, named for its putative role in mediating chromosomal translocations. Yeast two-hybrid studies looking for proteins that interact with translin yielded translin-interacting protein X, or trax [1] which shares a high degree of homology with translin. Subsequent studies revealed that the translin/trax complex is an octomer composed of two translin homodimers and two translin/trax heterodimers [15,16]. Thus, the native, octameric translin/trax complex contains 2 trax subunits and 6 translin subunits. Of note, trax, unlike translin, is unable to form a homomeric complex, suggesting that its stability may depend on its interaction with translin. This concept was reinforced by the striking observation in multiple species, including yeast, Drosophila, and mice, that deletion of translin leads to complete or near complete loss of trax protein [17–20], even though trax mRNA levels remain at wild type levels. Conversely, deletion of trax in Drosophila or mouse does not elicit a similar loss of translin protein [6,18]. Accordingly, the observation that trax protein is unstable in the absence of translin led to the conclusion that trax stability is dependent on its interaction with translin, but not vice versa.
Following these initial advances in identifying and characterizing the translin/trax complex, efforts to confirm the initial hypothesis that this complex might be involved in chromosomal translocation or DNA repair met with negative results. Deletion of translin from yeast, Drosophila or mice did not affect the frequency of chromosomal translocations or susceptibility to DNA damage [17–19]. On the other hand, following exposure to gamma radiation, trax was found to bind to C1D, an activator of a DNA-dependent protein kinase implicated in DNA repair [14], leaving open the possibility that trax might play a role in the DNA repair process. Attempts to gain clues to the function of the translin/trax complex by defining a consensus binding motif were also unhelpful, as the complex binds to single stranded DNA or RNA with little apparent sequence specificity [21]. Fortunately, pivotal new insights into the function of the translin/trax complex came from a surprising direction, attempts to elucidate the cellular machinery mediating RNA-induced silencing in Drosophila.
Discovery of Translin/Trax RNase activity: Role in translational silencing
In a series of elegant studies aimed at identifying cellular factors that could reconstitute efficient RNA-inducing silencing activity in Drosophila extracts, Liu et al. (2009) [22] identified the translin/trax complex as a key factor in this process. Addition of the complex to a mixture of recombinant components of the silencing pathway, including Dicer and Ago, hastened loading of the guide strand of siRNAs onto the RISC complex. Tracking down how the translin/trax complex exerts this effect revealed that it possesses RNase activity enabling it to degrade the passenger strand. In retrospect, this critical insight was missed in prior studies characterizing the nucleic acid binding properties of the translin/trax complex for two reasons. First, sequence analysis did not reveal any domains in translin or trax that would suggest the presence of nuclease activity. Second, earlier nucleic acid binding studies were routinely performed in the presence of divalent ion chelators to minimize degradation of the radiolabeled probe by nucleases inevitably present in tissue or cell extracts. As the nuclease activity of translin/trax is magnesium-dependent, inclusion of these chelators inhibited the nuclease activity of the complex. Thus, these studies elucidating the role of translin/trax in processing siRNA in Drosophila were pivotal in advancing our understanding of the function of the translin/trax complex by uncovering its RNase activity and implicating it in RNA-induced silencing.
As there are marked differences in the machinery mediating siRNA-mediating silencing in Drosophila and microRNA-mediated silencing in mammalian cells [23,24], one could not assume that the translin/trax complex exerts a similar effect on mammalian microRNA signaling pathways. In fact, subsequent studies examining this question revealed that translin/ trax has the opposite effect, i.e. it suppresses miRNA-mediating silencing [25]. In vitro studies with recombinant translin/trax have demonstrated that it is able to cleave selected pre-microRNAs that have mismatches in their stems. In contrast, the complex is unable to cleave the corresponding “siRNA” version of these microRNAs generated by deleting the mismatches present in the stem (Figure 2). Thus, these findings indicate that the translin/ trax complex competes with Dicer for pre-microRNA substrates and suggest that activation of translin/ trax RNase activity could provide a mechanism for reversing microRNA-induced silencing. Direct support for this hypothesis has emerged from recent studies, summarized below, demonstrating that the translin/trax complex mediates de novo translation underlying synaptic plasticity via this mechanism.
Figure 2: Selective cleavage of pre-microRNAs by recombinant translin/ trax in vitro.
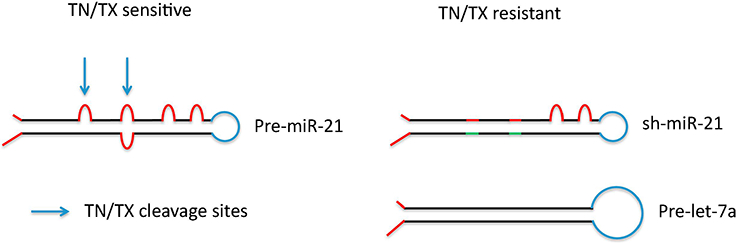
Incubation of recombinant translin/trax with a prototypical pre-miRNA, such as pre-miR-21, indicates that it is cleaved initially at mismatches in the stem, indicated by arrows. Furthermore, these mismatches are required for cleavage to occur as their removal makes the corresponding stem-loop structure (sh-miR-21) resistant to cleavage Similarly, pre-let-7a, which does not contain mismatches in its stem is not cleaved by translin/trax (TN/TX) (adapted from [25]).
Role of Translin/ Trax in Synaptic Tagging
One of the most fascinating challenges facing neuroscience is deciphering the molecular mechanisms mediating learning and memory. Compelling evidence indicates that these phenomena are mediated by the ability of brief bouts of neuronal stimulation that encode external stimuli to elicit long-lasting changes in synaptic strength. Furthermore, it is now clear that certain salient forms of experience or neuronal stimulation have the ability to empower routine forms of synaptic stimuli, that are normally unable to produce long-lasting potentiation, to do so. This type of mechanism is thought to underlie the ability of a salient or novel experience to enhance recall of events that occur during a window of about 1 hour prior to or following this salient experience. At a molecular level, this ability of a “strong” experience to influence synaptic plasticity both retroactively and prospectively is thought to reflect the coordination of two distinct molecular events: 1) de novo translation of plasticity products that underlie long-lasting changes in synaptic efficacy, and 2) setting of a synaptic “tag” that can capture these plasticity products [10]. In this model, routine experiences are insufficient to activate translation but are able to set “tags” marking synaptic inputs activated by these routine stimuli. In contrast, intense experiences are able to activate translation of plasticity products that are then captured by the “tagged” synapses (Figure 3A).
Figure 3:
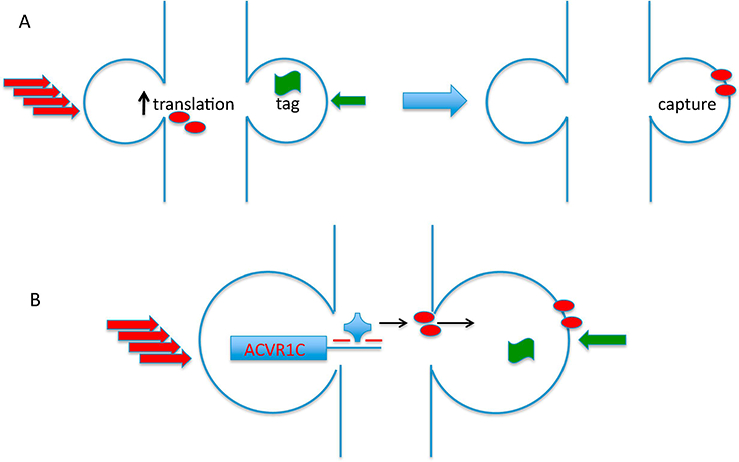
(A) Overview of synaptic tagging and capture paradigm. In this form of associative plasticity, strong stimulation of one input can prolong potentiation elicited by weak stimulation of a second input onto a common target neuron, even if these stimuli are delivered at different times within a window of approximately one hour. The stimuli can be delivered in either order. The current model explaining synaptic tagging and capture posits that the strong stimulus (depicted by a stack of red arrows) is sufficient to trigger de novo translation of dendritic proteins that are needed to mediate long-lasting potentiation (red ovals). Even though the weak stimulus is unable to drive translation, it is able to set a “tag” (depicted by green banner) to mark the activated synapses and “capture” plasticity proteins induced by strong stimulation at other synapses. If the weak stimulus is delivered first, then the longevity of the tag set by the weak stimulus determines the duration of time that these activated synapses can “capture” plasticity proteins induced by strong stimulation. On the other hand, if the strong stimulus is delivered first, then the longevity of the plasticity proteins defines the window during which weak stimulation can capture these plasticity proteins. Figure adapted from [10]. (B) ACVR1C, a member of the TGF-β receptor family, is induced by strong stimulation and mediates enhanced potentiation of the weak input. In the model proposed by Park et al. [3], strong synaptic stimulation triggers activation of the translin/trax RNase complex (blue knob) which reverses translational silencing of ACVR1C, which encodes a type II member of the TGF-β receptor family.
To explore the possibility that the translin/trax RNase might mediate the translational response elicited by strong neuronal stimuli, Park et al (2017) [3] examined whether translin deletion blocked the ability of a strong input to enhance potentiation of a weak input activated 45 minutes later in mouse hippocampal slices. Although the input receiving strong stimulation displayed normal potentiation in translin KO mice, this intense activity was not able to enhance potentiation elicited by the “weak” input activated 45 minutes later. Thus, this observation indicates that activation of the translin/ trax complex by strong stimulation is required to drive de novo translation of plasticity-related proteins that are normally captured by the weak input.
The hypothesis is supported by several findings. First, miRNA profiling of WT and TN KO mice after exposure to a novel environment, thought to be equivalent to a “strong” stimulus, revealed that translin KO mice displayed higher levels of a small subset of microRNAs. Using computational approaches to identify transcripts targeted by these microRNAs led to the identification of ALK7 (also called ACVR1C), a member of the TGF-beta receptor family, as a candidate target. Furthermore, treatment of hippocampal slices with an inhibitor of type I TGF-beta receptor family members, including ALK7, phenocopied the defect in synaptic tagging displayed by translin KO mice. Similarly, intraventricular infusion of this drug also phenocopied the memory deficit displayed by translin KO mice in the novel object location paradigm. Thus, these findings strongly support the following scenario:synaptic activation of translin/ trax RNase activity by strong stimuli triggers degradation of a subset of microRNAs, which in turn reverses translational silencing of plasticity proteins, such as ALK7. These products are then captured by synapses that have been “tagged” in response to “weak” stimulation and enable them to display prolonged potentiation (Figure 3B).
Role of trax in the nucleus: DNA repair
The findings described above imply that trax, which is essential for the catalytic activity of the translin/trax RNase, plays a key role in driving translation underlying synaptic plasticity. Parallel studies focusing on the role of trax in DNA repair have also made progress in understanding how cell signaling pathways regulate this aspect of trax function. A key starting point for this line of research was the observation that trax had been identified in a yeast two-hybrid screen looking for proteins that interact with the C-terminal segment of the A2A receptor [26]. Activation of A2AR is known to protect human striatal medium spiny neurons from oxidative DNA damage and subsequent cell death [27]. As trax had been shown to promote ATM activation, which helps prevent p53-mediated apoptosis triggered by DNA damage, these findings motivated a series of studies demonstrating that trax mediates the protective effects of A2AR stimulation. Furthermore, this line of research has revealed that A2AR stimulation triggers dissociation of trax from this cell surface receptor allowing it to translocate to the nucleus where it can associate with ATM. In fact, deletion of the nuclear localization signal present in trax blocks its translocation and protective effects [6].
As A2AR is Gsα-coupled receptor that activates the cAMP/PKA pathway[28], Chien et al. [5] asked if dissociation of trax from this receptor is mediated by this pathway. Furthermore, A2AR stimulation has been shown to trigger phosphorylation of GSK3β, which suppresses its activity. Using inhibitors of PKA or GSK3beta to test the role of these signaling pathways in mediating dissociation of trax from A2AR, they found that: 1) inhibition of PKA blocks the ability of A2AR stimulation to phosphorylate GSK3β, and 2) GSK3β inhibition triggers dissociation of trax from the A2AR. Further examination of the interaction between GSK3β and trax revealed that they co-precipitate along with Disrupted-in-Schizophrenia 1 (DISCI), a protein known to interact with GSK3P [5]{Chien, 2018 #6}. Detection of a physical interaction between DISCI and trax is of particular interest because these two proteins are encoded by two genes (namely DISCI and TSNAX) that are located adjacent to each other on human chromosome 1 in a region that has been implicated in major mental disorders (such as schizophrenia, autism, depression, and bipolar disorder) [29–34].
Taken together, these findings indicate that under basal conditions trax binds directly to the C-terminal segment of A2AR and participates in a complex with DISCI and GSK3β, which is constitutively active. Stimulation of A2AR activates PKA, which inhibits GSK3P by phosphorylation at Ser9. GSK3β inhibition triggers disruption of the trax/DISCI/GSK3P complex, as well as dissociation of trax from A2AR. Following its liberation from A2AR, trax translocates to the nucleus where it associates with ATM to promote DNA repair (Figure 4).
Figure 4: Regulation of DNA repair by A2A adenosine receptor (A2AR) in a trax-dependent pathway.
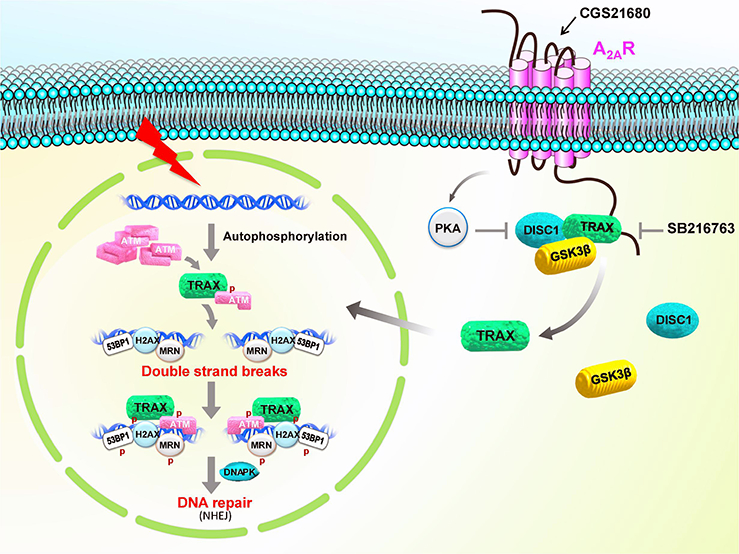
The A2AR interacts with TRAX, which forms a complex with GSK3β and DISCI. Inhibition of GSK3P by A2AR activation or GSK3β inhibition releases TRAX from this complex allowing it to translocate to the nucleus where it facilitates ATM-mediated DNA repair.
Concluding remarks
Although trax was first identified over 20 years ago [1], it has eluded attempts to decipher its role in cell signaling pathways. Accordingly, recent studies demonstrating that it plays key roles in both DNA repair and synaptic plasticity represent welcome advances in this area of research. However, paradoxically, these studies show that trax acts in very different ways in these paradigms suggesting that we have only begun to uncover how it operates. In the context of synaptic plasticity, trax acts in concert with translin to form an RNase that degrades microRNAs; in the context of DNA repair, it acts without translin to bind to and activate ATM. Thus, these studies suggest that trax is a versatile signaling protein with multiple partners and modes of action. As a multi-tasking protein, it should be included in the growing list of “moonlighting” proteins, such as GAPDH and aconitase that have both enzymatic and non-enzymatic modes of action [35,36].
From this perspective, it will be interesting to explore whether the role of trax in DNA repair, via its interaction with ATM, and its role in microRNA degradation, via its interaction with translin, may be part of a coordinated cellular response. Recent studies indicate that double-stranded DNA breaks and subsequent DNA repair are not only pathological events but also occur during physiological transcription of immediate early genes, key components of cellular plasticity [37,38]. Thus, trax may help coordinate changes in both transcription and translation that underlie a plasticity response.
The observation that trax is able to interact with multiple partner proteins is particularly puzzling because, as noted above, it has been documented in multiple species that deletion of translin triggers degradation of trax protein [17– 20]. This finding suggested that the stability of trax protein was dependent on its physical interaction with translin and, therefore, taken to imply that it interacted exclusively with translin. However, this inference needs to be re-evaluated in light of the ability of trax to form complexes with A2aR and ATM that do not contain translin [6]. These new findings suggest that the presence of translin protein may block degradation of trax via an indirect mechanism rather than via their physical interaction.
Recent studies examining the role of trax in DNA repair and synaptic plasticity suggest that this protein can mediate responses to extracellular cues. In the DNA repair paradigm, stimulation of A2aR induces phosphorylation and inhibition of GSK3β, which triggers dissociation of trax from A2aR. Accordingly, it will be of interest to determine if trax may also bind to other cell surface receptors that can regulate its availability. In a similar vein, PLCβ has been implicated in regulating the activity of the translin/trax RNase complex [39,40]. In this scenario, PLCβ binds to the translin/trax complex and inhibits its activity. Activation of Gq-linked receptors favors association of PLCβ with Gq and its dissociation from translin/ trax, thereby stimulating translin/ trax RNase activity.Further studies are needed to assess if this mechanism may mediate activation of translin/trax following synaptic stimulation.
Recent studies indicate that receptor stimulation can reverse silencing and trigger translation in neurons by stimulating microRNA degradation. BDNF elicits activation of the Lin28a microRNA degradation pathway to drive expression of plasticity proteins in cultured hippocampal neurons [12]. In the synaptic tagging and capture paradigm, synaptic stimulation triggers expression of activin receptors via activation of the translin/trax RNase complex. Thus, these findings indicate that Lin28a and translin/trax microRNA degradation pathways act in a similar fashion to trigger translation of plasticity proteins. Accordingly, it will be interesting in future studies to compare and contrast their roles in this process. The discovery that the substrate specificity of the Lin28a pathway is determined by the presence of a short sequence motif in the loop of pre-microRNAs, commonly found in let-7 family members, has been especially helpful in identifying candidate target mRNAs regulated by this pathway. Accordingly, studies aimed at defining the substrate specificity of the translin/trax RNase, which is poorly understood, would be welcome.
In summary, recent studies have revealed that trax is a multi-functional protein that plays key roles in disparate cellular signaling pathways. Thus, we anticipate that these seminal findings will stimulate interest in elucidating the regulation and function of this versatile protein.
Highlights.
Trax is a versatile signaling protein that interacts with multiple partner proteins.
Together with translin, it forms a microRNA-degrading enzyme.
This RNase mediates rapid reversal of translational silencing underlying plasticity.
Trax also plays a key role in DNA repair by binding to and activating ATM.
Acknowledgments
This work was supported by grants from the National Institutes of Health (MH087463 to T Abel; DA00266 to JM Baraban) and the Institute of Biomedical Sciences of Academia Sinica (103-Academia Sinica Investigation Award-06 to Y Chern).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aoki K, Ishida R, Kasai M: Isolation and characterization of a cDNA encoding a Translin-like protein TRAX., FEBS Lett 1997, 401:109–112. [DOI] [PubMed] [Google Scholar]
- 2.Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M: A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet 1995,10:167–174. [DOI] [PubMed] [Google Scholar]
- 3.Park AJ, Havekes R, Fu X, Hansen R, Tudor JC, Peixoto L, Li Z, Wu YC, Poplawski SG, Baraban JM, et al. : Learning induces the translin/trax RNase complex to express activin receptors for persistent memory. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frey U, Morris RG: Synaptic tagging and long-term potentiation. Nature 1997, 385:533–536. [DOI] [PubMed] [Google Scholar]
- 5.Chien T, Weng YT, Chang SY, Lai HL, Chiu FL, Kuo HC, Chuang DM, Chern Y: GSK3beta negatively regulates TRAX, a scaffold protein implicated in mental disorders, for NHEJ-mediated DNA repair in neurons. Mol Psychiatry 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JY, Chen SY, Sun CN, Chien T, Chern Y: A central role of TRAX in the ATM-mediated DNA repair. Oncogene 2016, 35:1657–1670. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Wu Y, Baraban JM: The Translin/Trax RNA binding complex: clues to function in the nervous system. Biochim Biophys Acta 2008,1779:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle M, Kiebler MA: Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 2011, 30:3540–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayford M, Siegelbaum SA, Kandel ER: Synapses and memory storage. Cold Spring Harb Perspect Biol 2012,4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redondo RL, Morris RG: Making memories last: the synaptic tagging and capture hypothesis. Nat Rev Neurosci 2011,12:17–30. [DOI] [PubMed] [Google Scholar]
- 11.Fu X, Shah A, Baraban JM: Rapid reversal of translational silencing: Emerging role of micro RNA degradation pathways in neuronal plasticity. Neurobiol Learn Mem 2016,133:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang YW, Ruiz CR, Eyler EC, Lin K, Meffert MK: Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell 2012,148:933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erdemir T, Bilican B, Cagatay T, Goding CR, Yavuzer U: Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol Microbiol 2002, 46:947–957. [DOI] [PubMed] [Google Scholar]
- 14.Erdemir T, Bilican B, Oncel D, Goding CR, Yavuzer U: DNA damage-dependent interaction of the nuclear matrix protein C1D with Translin-associated factor X (TRAX). J Cell Sci 2002, 115:207–216. [DOI] [PubMed] [Google Scholar]
- 15.Tian Y, Simanshu DK, Ascano M, Diaz-Avalos R, Park AY, Juranek SA, Rice WJ, Yin Q, Robinson CV, Tuschl T, et al. : Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat Struct Mol Biol 2011, 18:658–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H: Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol 2011, 18:650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chennathukuzhi V, Stein JM, Abel T, Donlon S, Yang S, Miller JP, Allman DM, Simmons RA, Hecht NB: Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol Cell Biol 2003, 23:6419–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claussen M, Koch R, Jin ZY, Suter B: Functional characterization of Drosophila Translin and Trax. Genetics 2006, 174:1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaendling A, Ramayah S, Pryce DW, McFarlane RJ: Functional characterization of the Schizosaccharomyces pombe homologue of the leukaemia- associated translocation breakpoint binding protein translin and its binding partner, TRAX. Biochim Biophys Acta 2008, 1783:203–213. [DOI] [PubMed] [Google Scholar]
- 20.Yang S, Cho YS, Chennathukuzhi VM, Underkoffler LA, Loomes K, Hecht NB: Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J Biol Chem 2004, 279:12605–12614. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Baraban JM: High affinity binding of the Translin/Trax complex to RNA does not require the presence of Y or H elements. Brain Res Mol Brain Res 2004, 120:123–129. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q: C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 2009, 325:750–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, Hannon GJ: MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004, 5:522–531. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q, Paroo Z: Biochemical principles of small RNA pathways. Annu Rev Biochem 2010, 79:295–319. [DOI] [PubMed] [Google Scholar]
- 25.Asada K, Canestrari E, Fu X, Li Z, Makowski E, Wu YC, Mito JK, Kirsch DG, Baraban J, Paroo Z: Rescuing dicer defects via inhibition of an anti-dicing nuclease. Cell Rep 2014, 9:1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun CN, Cheng HC, Chou JL, Lee SY, Lin YW, Lai HL, Chen HM, Chern Y: Rescue of p53 blockage by the A(2A) adenosine receptor via a novel interacting protein, translin-associated protein X. Mol Pharmacol 2006, 70:454–466. [DOI] [PubMed] [Google Scholar]
- 27.Chiu FL, Lin JT, Chuang CY, Chien T, Chen CM, Chen KH, Hsiao HY, Lin YS, Chern Y, Kuo HC: Elucidating the role of the A2A adenosine receptor in neurodegeneration using neurons derived from Huntington’s disease iPSCs. Hum Mol Genet 2015, 24:6066–6079. [DOI] [PubMed] [Google Scholar]
- 28.Chen JF, Lee CF, Chern Y: Adenosine receptor neurobiology: overview. Int Rev Neurobiol 2014, 119:1–49. [DOI] [PubMed] [Google Scholar]
- 29.Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, Nieminen-von Wendt T, von Wendt L, Paunio T, Peltonen L: Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry 2008, 13:187–196. [DOI] [PubMed] [Google Scholar]
- 30.Meng G, Aoki K, Tokura K, Nakahara K, Inazawa J, Kasai M: Genomic structure and chromosomal localization of the gene encoding TRAX, a Translin- associated factor X. J Hum Genet 2000, 45:305–308. [DOI] [PubMed] [Google Scholar]
- 31.Millar JK, Christie S, Semple CA, Porteous DJ: Chromosomal location and genomic structure of the human translin-associated factor X gene (TRAX; TSNAX) revealed by intergenic splicing to DISC1, a gene disrupted by a translocation segregating with schizophrenia. Genomics 2000, 67:69–77. [DOI] [PubMed] [Google Scholar]
- 32.Schosser A, Gaysina D, Cohen-Woods S, Chow PC, Martucci L, Craddock N, Farmer A, Korszun A, Gunasinghe C, Gray J, et al. : Association of DISC1 and TSNAX genes and affective disorders in the depression case-control (DeCC) and bipolar affective case-control (BACCS) studies. Mol Psychiatry 2010, 15:844–849. [DOI] [PubMed] [Google Scholar]
- 33.Thomson PA, Wray NR, Millar JK, Evans KL, Hellard SL, Condie A, Muir WJ, Blackwood DH, Porteous DJ: Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry 2005, 10:657–668, 616. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Tochigi M, Ohashi J, Maeda K, Kato T, Okazaki Y, Kato N, Tokunaga K, Sawa A, Sasaki T: Association study of the DISC1/TRAX locus with schizophrenia in a Japanese population. Schizophr Res 2005, 79:175–180. [DOI] [PubMed] [Google Scholar]
- 35.Huberts DH, van der Klei IJ: Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta 2010, 1803:520–525. [DOI] [PubMed] [Google Scholar]
- 36.Kim JW, Dang CV: Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 2005, 30:142–150. [DOI] [PubMed] [Google Scholar]
- 37.Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, Mucke L: Physiologic brain activity causes DNA doublestrand breaks in neurons, with exacerbation by amyloid-beta. Nat Neurosci 2013, 16:613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson LA, Tsai LH: In the loop: how chromatin topology links genome structure to function in mechanisms underlying learning and memory.Curr Opin Neurobiol 2017, 43:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aisiku OR, Runnels LW, Scarlata S: Identification of a novel binding partner of phospholipase cbeta1: translin-associated factor X. PLoS One 2010, 5:e15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garwain O, Scarlata S: Phospholipase Cbeta-TRAX Association Is Required for PC12 Cell Differentiation. J Biol Chem 2016, 291:22970–22976. [DOI] [PMC free article] [PubMed] [Google Scholar]


