Abstract
DNA methylation has emerged as promising target linking environmental exposures and cancer. The World Trade Center (WTC) responders sustained exposures to potential carcinogens, resulting in increased risk of cancer. Previous studies of cancer risk in WTC exposed responders were limited by the deficiency in quantitative and individual information on exposure to carcinogens. The current study introduces a new exposure ranking index (ERI) for estimating cancer related acute and chronic exposures, which aimed to improve the ability of future analyses to estimate the cancer risk. An epigenome-wide association study (EWAS) based on DNA methylation and a weighted gene co-expression network analysis (WGCNA) were conducted to identify CpG sites, modules of correlated CpG sites and biological pathways associated with the new ERI. Methylation was profiled on blood samples using Illumina 450K Beadchip. No significant epigenome-wide association was found for ERI at an FDR 0.05. Several cancer related pathways emerged in pathway analyses for the top ranking genes from EWAS as well as enriched module from WGCNA. The current study was the first DNA methylation study which aimed to identify methylation signature for cancer related exposure in the WTC population. No CpG sites survived multiple testings adjustment. However, enriched gene sets involved in cancer, were identified in both acute and chronic ERIs, supporting the view that multiple genes play a role in this complex exposure.
Keywords: Environmental Exposure, EWAS, Methylation, Cancer, World Trade Center, Pathway analysis
Introduction
Concern has arisen about the potential for increased risk of cancer among World Trade Center (WTC) responders, who sustained exposures to a complex mix of toxic chemicals that included multiple known and suspected human carcinogens (1). The combustion of jet fuel at high temperatures released soot, metals, benzene and other volatile organic compounds, and strong inorganic acids. The burning and subsequent collapse of the towers resulted in the release of particulate matter comprising asbestos, silica, cement dust, glass fibers, heavy metals like arsenic, beryllium, cadmium, chromium VI, nickel, polycyclic aromatic hydrocarbons, polychlorinated biphenyls, and polychlorinated dibenzofurans and dioxins (1–6). Evidence on increased cancer risk are emerging, in which the three cohort studies including WTC responders showed modest elevations in the risk of all cancers combined, with standardized incidence ratio (SIR) ranging from 1.06 to 1.14 across studies, and substantial overlaps in the 95% confidence intervals (7–10).
The lack of quantitative, individual information on exposure to potential carcinogens is an important limitation of previous analyses of cancer risk in WTC exposed populations. Two approaches can be considered, to overcome this limitation: an individualized, high resolution assessment of circumstances of exposure experienced by WTC responders, and the use of biomarkers of exposure. The current study combines these two approaches by introducing a new exposure ranking index for estimating cancer related exposure, in an effort to improve the ability of future analyses to elucidate the cancer risk experienced by this population.
The epigenome acts as an interface between the genome and the environment. It is plastic, changing with environmental exposures (11) thereby, regulating transcription. It has been suggested that perturbation in the epigenome in response to the environment is more stable than changes in the transcriptome (12) making epigenetic changes a potentially valuable tool for exposure assessment. One of the most studied epigenetic mechanisms is DNA methylation, a heritable epigenetic modification that does not change the underlying DNA sequence, and is involved in the regulation of gene expression (13). The most common methylation site in mammals is a cytosine located next to a guanosine (CpG). CpG islands are found mainly in the 5′ regulatory and promoter regions of genes; most are unmethylated in normal cells (14).
Epigenetic changes in tumor tissues have been linked to specific environmental exposures. For instance, a genes specific methylation in lung cancer was associated with tobacco smoking compared to alcohol consumption (15). Similarly, DNA methylation patterns were able to distinguish various environmental risk factors associated with hepatocellular carcinoma (16). Recently, specific epigenetic profiles have been identified in the methylome of peripheral blood DNA in patients exposed to a wide variety of environmental exposures including tobacco smoking (17), benzene (18), air pollution (19) and arsenic (20). This holds the tremendous promise that these epigenetic changes in the blood may be exploited as biomarkers of exposure. In this study, we identify DNA methylation patterns associated with WTC exposure. The identified DNA methylation signature could serve not only as a biomarker of exposure, but may give us insight into the susceptibility to develop a variety of diseases, including cancer.
Methods
Development of WTC “Exposure Ranking Indices” (WTC-ERIs)
WTC “Exposure Ranking Indices” (WTC-ERIs) were developed for ranking potential exposures to hazardous substances related to post-9/11 activities of responders/workers. The calculation procedures and associated factors for estimating WTC-ERIs were adapted for the present work specifically to utilize information available for responders registered with the WTC General Responder Cohort (WTCGRC) (21). The exposure-related information in the WTCGRC dataset, collected through the Exposure Activities Questionnaire (EAQ) (see (8, 22)) was combined with information developed through various studies that characterized exposure-relevant attributes and factors related to post-WTC activities ((1, 23–27). The WTC-ERI is an ordinal metric (see, e.g., (28, 29)) WTC-ERI values represent numerical scores derived via a Multi-Criteria Decision Analysis (MCDA) procedure (see, e.g. (30)) involving factors related to date, duration, location, type of activity, microenvironment, type and usage of personal protective equipment associated with each exposure event experienced by post-9/11 responders and workers. The index for a specific subject/worker is calculated by summing over the time-sequence of all WTC-related “exposure events” for that subject/worker (where a typical exposure event is a work shift, though other types of events are possible). The MCDA score calculation procedure for the ERI was implemented primarily in Python 2.7 (www.python.org) for retrieving and analyzing information from the EAQ data available through the WTC-HPDC, complemented with a set of Matlab codes (www.themathworks.com), and is summarized in the following equation,:
where
| ERIw | is the Exposure Ranking Index for a specific subject (responder or worker) identified by index w (who may be have been involved in different activities during different exposure events) |
| Fcliud, w | is an exposure-related factor accounting for direct contact of the specific responder or worker with the dust cloud on September 11, 2001 |
| fact | is an exposure-related factor accounting for each type of WTC-related activity (search and rescue, cleanup, etc.) |
| i = 1, … M | is a counting index for outdoor exposure events during day i where i=1 on 9/11/2001 |
| j = 1, … N | is a counting index for indoor (i.e. confined space) exposure events during day j where j=1 on 9/11/2001 |
| f*time | is an “adjusted” exposure-related factor accounting for the time aspects of each exposure event [f*time=function(fdate, i) where fdate, i is a factor associated with date relative to 9/11] |
| floc, ϕloc | are exposure-related factors accounting for the locations where subject spend the majority of the exposure event (typically, but not exclusively, work shift) |
| μj | is a microenvironmental adjustment factor reflecting any information, if available, specific to the confined space settings of the exposure event |
| p | is a personal protective equipment (PPE) factor, reflecting the type and usage of PPE during the exposure event |
| Δti, Δτj | is a factor reflecting the duration of exposure event during day i or j |
It should be noted that ERI for each subject is explicitly calculated as the sum
where ERI_a accounts for a subject’s acute exposure (exposed during 9/11/2001 to 9/13/2001, i, j ≤ 3); ERI_c accounts for a subject’s chronic exposure (exposed during 9/14/2001 to 6/30/2002, i, j > 3).
Time/Location-Related Factors
The values of the exposure factors related to the location, and the time period(s) spent at that location, for each worker/responder are summarized in Supplementary Table S1.
Dust cloud factor
A special “location” of particular concern is the area that was covered by the dust cloud on September 11, 2001. So, a worker/responder-specific exposure-related factor (Fcliud, w) accounting for direct contact with the dust cloud on September 11, 2001, is used, and a numerical value of 0 to 500 is assigned for each of six different exposure scenarios (corresponding to situations ranging from no contact with the cloud to “full immersion” without PPE).
Time-Related Factors
For the exposure-related factor (f*time;) that accounts for the date, time and the duration of each exposure event, the following equation is used:
where
| tDay | is total number of working days during each time period |
| tPosD | is total number of possible working days during each time period derived from first and last day (if first day or last day is missing it is assumed that first day is 9/11/2001 and last day is 6/30/2002) |
| tshift | is the number of work shifts in a day (typically one work shift is 8 hours) |
| fdate | is a day-specific exposure-related factor accounting for severity of environmental and microenvironmental conditions on the date of each exposure event |
Location-Related Factors
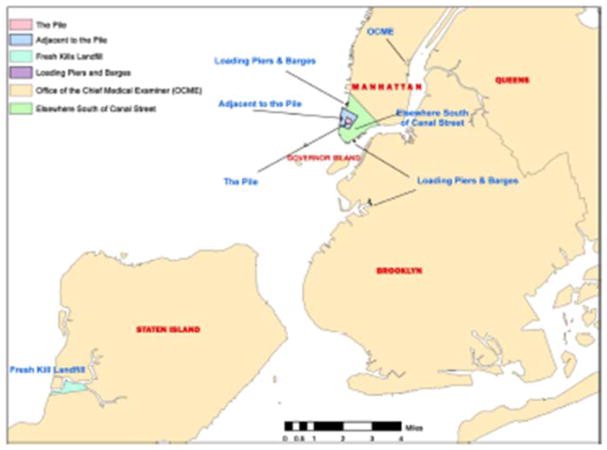
Exposure-related factors (floc, ϕloc) accounting for the location of each exposure event consider differences in environmental and microenvironmental conditions in the general location areas identified on the map of Figure 1 (22). These factors are assigned numerical values in the range of 1 to 5 for different WTC work areas identified in the WTCGRC dataset.
Figure 1.
World Trade Center response and cleanup workforce locations (22)
Activity-Related Factors
Exposure-related factors (fact) that account for different types of WTC-related activities during an exposure event, are assigned numerical values in the range of 1 to 10 for the activity categories identified in the WTC General Responder Cohort (WTCGRC). These categories include barge workers, carpenters, dock builders, electricians, glaziers, insulation workers, laborers, mechanics, roofers, truck drivers, military, canteen service, EMT, fire fighters, morgue workers, police officers, etc. It is important to note here that, in cases where an activity type different from that corresponding to the job title is reported in the questionnaire (e.g. “search and rescue” by a police officer), that activity is used to determine the appropriate factors for each of the activity categories listed in Supplementary Table 2.
Personal Protective Equipment (PPE) Related Formulas and Factors
Exposure-related factors accounting for PPE type and usage are specific to (1) respirators and (2) dust masks. A total PPE factor is calculated during each exposure event by averaging of the factors for the PPE types used during the event:
where
| pi | is the total personal protective equipment (PPE) factor, reflecting the type and usage of all types of PPE used during an exposure event i (range from 1 to 0.1) |
| pi, q | is the personal protective equipment (PPE) factor, reflecting the type and usage of a specific PPE type q used during the exposure event i |
| q = 1, … n | is an index for different types of PPE |
The only information available for the usage of “surgical/disposable mask” is the start date, so a constant PPE factor (0.55) for wearing a mask is assigned to the subject after and including their first day of wearing a mask.
For respirators the following general relation is used:
where
| fresp_type | is the type of respirator used |
| fFreq_G | is the frequency of respirator use |
| fMaint_R | is the frequency of respirator replacement |
| fMaint_C | is the frequency of respirator maintenance (cartridge replacement) |
| fMaint_Clean | is the frequency of respirator cleaning |
| fseal | is the frequency of respirator seal cleaning |
A numerical value is assigned for each aspect of PPE (these values are listed in Supplementary Table S3), with the highest value indicating best protection, most frequent usage, and correct usage of PPE. The lowest value indicates worst or no protection, most infrequent usage, and the incorrect usage of the respirator. Information on the type of respirator (full face or half face) is only available for the first week (9/11–18/2001); an average between full and half face respirator factors is used for days after 9/18/2001.
It should be noted that the quality of exposure relevant information in the WTCGRC database varied considerably. Therefore, based on evaluation of data gaps, uncertainties and resolution for the records of each subject, these subjects were classified in Groups A to E, with Group A including the subjects with the most complete and unambiguous exposure data and Group E including those with the highest uncertainties (see details in Supplementary Methods and in Supplementary Table S4 on the classification criteria and the numbers of subjects assigned to each of the groups).
Participants
Participants were recruited through the Stony Brook WTC-Health Program, part of a consortium of Clinical Centers of Excellence in the New York metropolitan area established in 2002 to monitor and treat WTC-related conditions in responders to the WTC disaster (31). Enrollees with documented WTC experience were enlisted from extensive outreach efforts involving partnerships with volunteer organizations, labor unions, and public outlets. The current study was approved annually by the Committees on Research Involving Human Subjects at Stony Brook University (IRB number: 604113). Written informed consent was obtained.
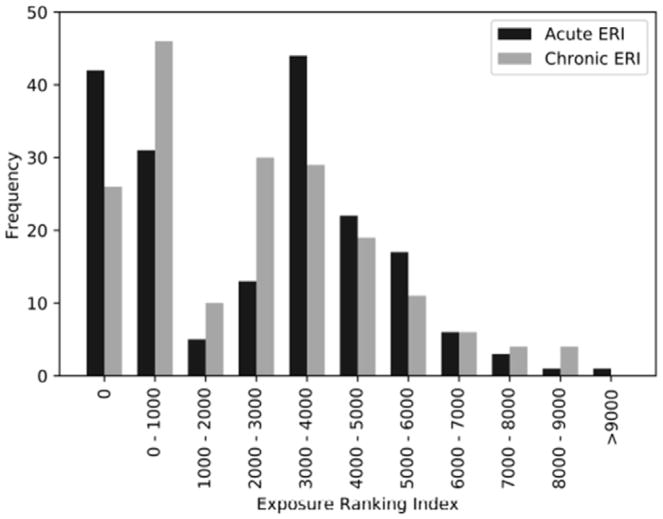
The EAQ data for 6,110 subjects in the Stony Brook database, who are part of the larger WTCGRC, were processed using the MCDA classification and scoring system developed for WTC-ERI. Sufficient and consistent information for developing “unambiguous” estimates of ERI values was available for 2,625 of these subjects that were assigned to quality Group A, while another 1,390 subjects with less complete information - though sufficient to develop rankings - were assigned to Groups B and C (see Supplementary Methods. The distribution of acute and chronic ERI values for all Stony Brook subjects in Group A is shown in Figure 2 (not including zero acute ERI values for 632 subjects). The ERI reflecting overall exposure of responders from post-WTC activities was calculated as the sum of the acute and chronic ERI values. High and low ERI were defined as ERI ≤ 356 and ERI ≥ 5599, where the values 356 and 5599 were the top and bottom 10 percentile, respectively (8).
Figure 2.
Distribution of ERI_a (acute ERI) and ERI_c (chronic ERI) values for 503 Stony Brook subjects with complete and consistent exposure-relevant information in the EAQ component of the WTC-HPDC dataset.
A subset of 185 responders with ERI values in groups A–C, assessed between February, 2012 and March, 2014 were included in DNA methylation profiling. All participants provided blood samples for the epigenetics assays. Inclusion criteria were signed consent, sufficient English language skills to participate in a diagnostic interview, and being male. We included only males because females show notably different methylation patterns from males, and very few responders were females. Participants were 51.3 years of age on average at the time of blood drawing, predominantly Caucasian (83.2%) and non-smokers (95.7%) (Table 1).
Table 1.
Clinical characteristics. The p-values were computed from t-test (for age) and chi-squared test (for race and smoking status) comparing high to low ERI.
| ERI | Low n = 69 |
High n = 116 |
P-value |
|---|---|---|---|
| Age (Mean (SD)) | 54.3 (8.1) | 49.5 (6.3) | < 0.001 |
| Race (N (%)) | |||
| Caucasian | 61 (0.88) | 93 (0.80) | 0.213 |
| Other | 8 (0.12) | 23 (0.20) | |
| Smoker (N (%)) | |||
| Yes | 3 (0.04) | 5 (0.04) | 0.999 |
| No | 66 (0.96) | 111 (0.96) | |
Illumina Infinium Human Methylation450K Beadchip
Blood samples were obtained from each participant via venipuncture and sent to Roswell Park Cancer Institute for DNA extraction. Genomic DNA was isolated from 0.3 ml of whole blood using the Qiagen BioRobot Universal System and the QIAamp DNA blood BioRobot MDx Kit (Qiagen, Valencia, CA) following the manufacturer’s recommended protocol. DNA methylation profiling was performed by Roswell Park Cancer Institute using the Human Methylation 450K BeadChip (Illumina Inc., San Diego, CA). DNA extraction and methylation profiling were done blinded to group assignment. 500ng of high quality genomic DNA measured by picogreen quantitation (Life technologies, Grand Island, NY) was bisulphite converted, amplified, fragmented and hybridized to the Illumina Infinium Human Methylation450K Beadchip using standard Illumina protocol. Data was processed using Illumina’s GenomeStudio methylation module (v1.9.0).
Data Preprocessing and Normalization
The 450K BeadChip methylation data from the GenomeStudio were imported into R (http://cran.r-project.org). Preprocessing of methylation data at the 485,557 CpG sites were performed as follows. CpG sites with detection p-value > 0.001 are set to missing and CpG sites with more than 20% missing were filtered. Beta mixture quantile (BMIQ) normalization (32) was applied to the beta values for correction of bias due to the type I and type II probes. Non-specific, cross-hybridized CpG sites (33, 34), CpG sites overlapping with a SNP and CpG sites mapping to repeat regions were filtered. The final data consisted of 375,223 CpG sites.
Estimation of Blood Cell Type Proportions
Cell type proportions have been implicated in DNA methylation analysis of whole blood samples (35). The proportions of CD8T, CD4T, natural killer (NK), B-cell, monocytes (Mono) and granulocytes (Gran) were estimated using the R packages minfi and FlowSorted.Blood.450 based on the procedures described in (36). We normalized the sum of the proportions per sample to one, and include five out of six estimated cell types as adjustment factor in our epigenomewide association analysis. In addition, two-sample t-tests were used to assess the association between each cell type and ERI.
Statistical Method for Epigenomewide Association Analysis (EWAS)
To identify CpG sites associated with each phenotype, separate multple linear regression for each CpG site was first fitted on logit transformed beta values (log(β/(1 − β))) as response, and ERI, adjusting for age, race, smoking status and cell types. Statistical significance for CpG association with ERI was assessed via the Wald test. A false discovery rate (FDR) (37) was used to account for multiple testings.
Pathway and Gene Ontology Analysis
Pathway and gene ontology analysis were carried out using gometh function in Bioconductor package missMethyl (38). Since the number of CpG sites mapping to each gene varied in the Methylation 450K BeadChip, pathway and gene ontology analysis would be biased and inaccurate (39). gometh accounted for the varying number of CpG sites per gene by providing a prior probability for each gene based on gene length, followed by a modified hypergeometric test for over-representation of a gene set (40). We tested for over-representation among the top 500 CpG sites from EWAS, against the background list of 375,223 CpG sites. 290 KEGG pathways (minimum and maximum number of genes for each gene set were 15 and 500, respectively) were tested. Gene sets significant at FDR < 0.05 were reported.
Weighted Gene Co-expression Network Analysis
The weighted gene co-expression network analysis (WGCNA) (41) was used to identify modules of correlated CpG sites on the logit transformed beta values. The Pearson correlation matrix was raised to the tenth power to achieve scale free topology. The minimum module size was set as 30, and the cut-offs for splitting and merging modules were 2 and 0.25, respectively. The methylation profiles for each module were represented by the eigenCpG. The association between module eigenCpG and ERI was performed using two-sample t-test, and the p-values were corrected via FDR. Over-representation analysis described above was also conducted on the CpG sites within the identified modules from WGCNA.
Results
Cell Type Proportions
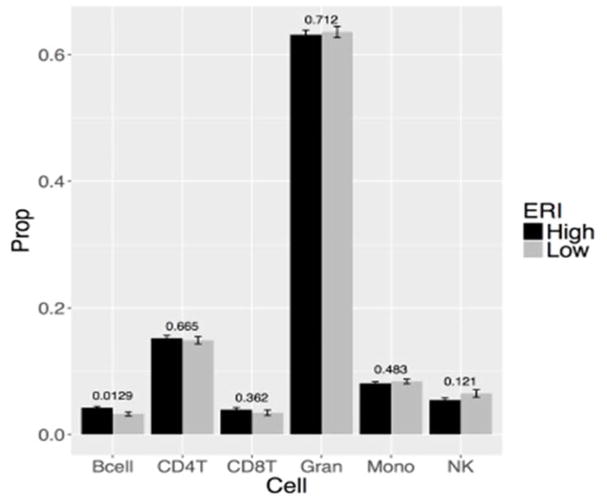
Figure 3 displayed the association between the estimated cell type proportions and ERI. At nominal p-value 0.05, Bcell was higher in high ERI compared to low ERI group.
Figure 3.
Barplots of estimated cell type proportions. Each barplot depicts the mean value (y-axis) and 1 standard error. The p-values on each cell type were computed from two-sample t-tests.
Epigenome-wide Association Analysis
The volcano plots in the top panel of Supplementary Figure S1 showed an approximately equal amount of hyper- and hypo-methylation pattern comparing high to low ERI. EWAS with ERI did not identify statistically significant CpG sites at FDR 0.05. 2 CpG sites were significant at nominal p-value 0.0001. Both CpG sites were hyper-methylated and mapped to 3′UTR of GABRA4 and gene body of TUBB, respectively. Top 10 CpG sites were provided in Supplementary Table S5.
Pathway and Gene Ontology Analysis
Table 2 displayed the enriched KEGG pathways at FDR 0.05 for the top 500 CpG sites from EWAS with ERI. 21 KEGG pathways were found to be enriched among the top 500 CpG sites associated with ERI, including several cancer related pathways, i.e., PPAR, MAPK, Ras and PI3K-Akt signaling pathways.
Table 2.
List of significant KEGG pathways (FDR < 0.05) for ERI among the top 500 CpG sites. N is the size of the pathway, DE is the number of genes in our list overlapping with the pathway, Gene is the list of overlapping genes.
| ERI
| ||||
|---|---|---|---|---|
| Pathway | N | DE | FDR | Gene |
| Neuroactive ligand-receptor interaction | 270 | 10 | 4.76E-05 | CHRND;ADRA1D;GABRA2;GABRA4;GALR1;MCHR1;GRIN2A;GRM7;HTR5A;PTGFR |
| Human papillomavirus infection | 309 | 10 | 0.00172 | COL1A1;IFNAR2;NOTCH4;HES6;CCND1;MPP5;THBS1;TSC1;WNT3A;CREB3L1 |
| Thiamine metabolism | 15 | 3 | 0.00305 | AK4;AK5;TPK1 |
| Calcium signaling pathway | 180 | 7 | 0.00438 | ADRA1D;GRIN2A;HTR5A;PTGFR;CACNA1H;CACNA1G;MCU |
| PPAR signaling pathway | 72 | 4 | 0.00753 | FABP5;ACSL3;SLC27A6;ANGPTL4 |
| Amyotrophic lateral sclerosis (ALS) | 51 | 4 | 0.00753 | GRIN2A;NEFM;NEFH;SOD1 |
| Phagosome | 143 | 5 | 0.0104 | ATP6V0E2;TUBB;TAP2;THBS1;COLEC11 |
| Synaptic vesicle cycle | 63 | 4 | 0.0104 | CPLX2;CPLX1;ATP6V0E2;STX1A |
| PI3K-Akt signaling pathway | 325 | 8 | 0.0108 | COL1A1;ANGPT1;IFNAR2;CCND1;THBS1;TSC1;CREB3L1;FGF19 |
| Proteoglycans in cancer | 203 | 6 | 0.032 | GPC1;IHH;MIR21;CCND1;THBS1;WNT3A |
| Ras signaling pathway | 224 | 6 | 0.0346 | RGL1;ANGPT1;GRIN2A;MAPK10;RASAL2;FGF19 |
| Nicotine addiction | 40 | 3 | 0.0377 | GABRA2;GABRA4;GRIN2A |
| Pathways in cancer | 394 | 8 | 0.038 | CTBP2;CTNNA2;MAPK10;RARA;RARB;CCND1;WNT3A;FGF19 |
| Choline metabolism in cancer | 98 | 4 | 0.0389 | DGKG;MAPK10;SLC22A5;TSC1 |
| MAPK signaling pathway | 254 | 6 | 0.0486 | MAPT;ECSIT;MAPK10;CACNA1H;CACNA1G;FGF19 |
| Hedgehog signaling pathway | 46 | 3 | 0.0486 | ADRBK2;IHH;CCND1 |
| Hippo signaling pathway | 153 | 5 | 0.0486 | YAP1;CTNNA2;CCND1;MPP5;WNT3A |
| Circadian entrainment | 96 | 4 | 0.0486 | GRIN2A;PRKG1;CACNA1H;CACNA1G |
| Ubiquitin mediated proteolysis | 136 | 4 | 0.0492 | UBE4B;ERCC8;PARK2;UBE2B |
| Platelet activation | 123 | 4 | 0.0492 | COL1A1;PRKG1;P2RY12;FERMT3 |
| Taste transduction | 79 | 3 | 0.0492 | HCN4;GABRA2;GABRA4 |
Weighted Gene Co-expression Network Analysis
109 modules were identified from the WGCNA analysis. The eigenCpG of two modules (M1 and M2) were associated with ERI at nominal p-value < 0.05, but did not survive FDR control. M1 and M2 contained 388 and 59 CpG sites, respectively. 22 KEGG pathways were identified to be enriched among the 388 CpGs sites from M1 (Table 3), including B cell receptor signaling pathway, hematopoietic cell lineage, primary immunodeficiency and cytokine-cytokine receptor interaction, chemokine signaling pathway and several other cancer related pathways. No statistically significant pathway was identified for M2 module.
Table 3.
List of significant KEGG pathways (FDR < 0.05) for M1 module. N is the size of the pathway, DE is the number of genes in our list overlapping with the pathway, Gene is the list of overlapping genes.
| M1
| ||||
|---|---|---|---|---|
| Pathway | N | DE | FDR | Gene |
| B cell receptor signaling pathway | 70 | 9 | 0 | BLNK;INPP5D;NFATC1;NFKBIE;VAV2;CD19;CD22;CD79B;CD81 |
| Hematopoietic cell lineage | 90 | 6 | 1.29E-05 | FCER2;ITGB3;KIT;IL1R2;CD19;CD22 |
| Primary immunodeficiency | 36 | 5 | 1.29E-05 | TNFRSF13B;BLNK;JAK3;RAG1;CD19 |
| Cytokine-cytokine receptor interaction | 246 | 7 | 0.000149 | CCR6;TNFRSF13B;IL10RA;KIT;CXCR5;TGFB1;IL1R2 |
| Chemokine signaling pathway | 179 | 7 | 0.000243 | CCR6;ADRBK1;GNG3;GRK6;JAK3;CXCR5;VAV2 |
| Epstein-Barr virus infection | 196 | 7 | 0.000425 | FCER2;IL10RA;JAK3;NFKBIE;USP7;CD19;HDAC4 |
| Regulation of actin cytoskeleton | 211 | 7 | 0.00158 | DIAPH1;CYFIP1;CYFIP2;ITGB3;SSH3;VAV2;ARHGEF7 |
| mTOR signaling pathway | 150 | 6 | 0.00179 | FLCN;LRP5;TBC1D7;RPTOR;RPS6KA2;SLC7A5 |
| Osteoclast differentiation | 121 | 5 | 0.00179 | LILRA2;BLNK;ITGB3;NFATC1;TGFB1 |
| Transcriptional misregulation in cancer | 179 | 6 | 0.00521 | PTCRA;PAX5;CDK14;TCF3;IL1R2;LDB1 |
| Cholinergic synapse | 112 | 5 | 0.00708 | PIK3R5;GNG3;KCNQ1;CAMK2A;KCNQ4 |
| PI3K-Akt signaling pathway | 325 | 7 | 0.0128 | PIK3R5;GNG3;ITGB3;JAK3;KIT;RPTOR;CD19 |
| Th17 cell differentiation | 105 | 4 | 0.0155 | JAK3;NFATC1;NFKBIE;TGFB1 |
| cAMP signaling pathway | 198 | 5 | 0.0291 | ADORA2A;NFATC1;VAV2;CAMK2A;ACOX3 |
| Morphine addiction | 91 | 4 | 0.0291 | ADRBK1;GABRD;GNG3;GRK6 |
| FoxO signaling pathway | 129 | 4 | 0.0341 | PRKAG2;RAG1;TGFB1;USP7 |
| Tuberculosis | 163 | 4 | 0.0346 | IL10RA;TGFB1;CAMK2A;KSR1 |
| Apelin signaling pathway | 136 | 4 | 0.0426 | PIK3R5;GNG3;PRKAG2;HDAC4 |
| Proteoglycans in cancer | 203 | 5 | 0.0426 | ANK1;ITGB3;TGFB1;VAV2;CAMK2A |
| Inositol phosphate metabolism | 73 | 3 | 0.0448 | INPP5J;INPP5D;ITPKB |
| SNARE interactions in vesicular transport | 33 | 2 | 0.0457 | VAMP1;STX7 |
| MicroRNAs in cancer | 281 | 5 | 0.0457 | FOXP1;ITGB3;RPTOR;ST14;HDAC4 |
Discussion
While it might still be too early to see the full effect of WTC exposure on cancer incidence (10) it is important to assess the presence in this population of biomarkers which may be related to cancer risk. The current study introduced a new exposure index for estimating cancer related exposure in the WTC cohort. The ERI allowed ranking quantitatively WTCGRC members in terms of their acute and chronic WTC exposure, and provided a refinement of previous approaches based on qualitative information (8, 42). A limitation of the present application of the ERI framework is that the available exposure data were not sufficient to attribute separate exposure to specific agents that contributed to mixture of WTC contaminants. Although certain measurements for specific agents at various locations are available, the resolution of exposure-relevant time/location data for the study subjects was too “coarse” to allow morespecific assessments and in any case strong correlations between the agents could have hampered any such effort.
This was the first DNA methylation study which aimed to identify methylation signature for cancer related exposure in the WTC population. Although no CpG site was significant after adjustment for multiple testings, pathway analysis revealed enriched gene sets involved in cancer. Specifically, several cancer related KEGG pathways were identified among the top ranking CpG sites associated with ERI and CpG sites within enriched module from WGCNA, including PPAR, Ras, MAPK, PI3K-Akt, mTOR and chemokine signaling, as well as pathways, proteoglycans, choline metabolism and microRNAs in cancer; although different overlapping genes were implicated. Ras proteins are involved in cellular signal transduction and the Ras genes (HRas, KRas and NRas) are the most common oncogenes in cancer (43). On the other hand, MAPK pathways are involved in stress signaling, and abberant MAPK pathways leads to uncontrolled growth and tumerigenesis (44). Akt has been shown to be the hub of signaling pathway implicated in tumorigenesis (45). These pathways are involved in the major hallmarks of cancer, including cell cycle, survival, motility and genomic instability (46). The identified cancer related enriched gene sets support the importance of pathway analysis compared to single-gene approach, i.e., the search for the combined effect of multiple genes acting in concert in this complex exposure.
Limitations
The current study had several strengths, including the use of an enhanced exposure assessment approach and the first and largest EWAS sample to date in environmental exposure study. Nonetheless, our findings must be considered in the context of several limitations. First, the methylation data was profiled on whole blood collected more than 10 years post September 11 attack. Our analysis was conducted under the assumption that DNA methylation changes from high exposure is persistent over time, as methylation is known to be a stable marker (47). Second, unexposed control group was not available in this study. We sought to address this issue by selecting extreme exposure groups within our populations, i.e., top 10% versus bottom 10%. It is important to note that a comparison with an external population might be affected by selection bias and confounding, which are less likely to occur in internal comparisons within a relatively homogeneous population. Third, the sample size of our study is relatively small for EWAS. Fourth, our methylation analysis was performed in DNA samples derived from whole blood cells and were thus a mix of cell types. We controlled for the cell types heterogeneity using state-of-the-art statistical method, but future work needs to isolate and examine each cell type individually.
Conclusions
The current study aimed to provide a better understanding of the relationship between epigenetic alteration and WTC-related exposure, with potential relevance to cancer risk. Enriched gene sets were involved in several biological pathways associated with cancer, including Ras, MAPK and PI3K-Akt signaling pathways. Taken together, this provides biological evidence supporting a possible association between exposure and risk of cancer among the WTC responders.
Supplementary Material
Acknowledgments
Financial Support
The analysis was supported by grants # 200-2011 41815 (PI: Paolo Boffetta), U01 OH010987 (PI: Paolo Boffetta) and U01 OH010416-01 (PI: Benjamin J. Luft) from the National Institute for Occupational Safety and Health (NIOSH)/Centers for Disease Control and Prevention (CDC).
We gratefully acknowledge the support of the first responders and rescue/recovery workers for participating in this study. We also thank the staff of the Stony Brook World Trade Center Health Program and the World Trade Center Health Program Data Monitoring Center for ongoing support. We are also grateful to Sean Glenn at the Roswell Park Cancer Institute for his contributions to the methylation assessment. The findings and conclusions in this article are those of the authors and do not represent the official position of NIOSH, the CDC or the US Public Health Service.
Footnotes
Conflict of Interest
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors reported no biomedical financial interests or other potential conflicts of interest.
References
- 1.Lioy PJ, Gochfeld M. Lessons learned on environmental, occupational, and residential exposures from the attack on the World Trade Center. American journal of industrial medicine. 2002;42(6):560–5. doi: 10.1002/ajim.10160. [DOI] [PubMed] [Google Scholar]
- 2.Clark RN, Green RO, Swayze GA, Meeker G, Sutley S, Hoefen TM, et al. Environmental studies of the World Trade Center area after the September 11, 2001 attack. 2001:1258. [Google Scholar]
- 3.Edelman P, Osterloh J, Pirkle J, Caudill SP, Grainger J, Jones R, et al. Biomonitoring of chemical exposure among New York City firefighters responding to the World Trade Center fire and collapse. Environmental health perspectives. 2003;111(16):1906. doi: 10.1289/ehp.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litten S, McChesney DJ, Hamilton MC, Fowler B. Destruction of the World Trade Center and PCBs, PBDEs, PCDD/Fs, PBDD/Fs, and chlorinated biphenylenes in water, sediment, and sewage sludge. Environmental science & technology. 2003;37(24):5502–10. doi: 10.1021/es034480g. [DOI] [PubMed] [Google Scholar]
- 5.McGee JK, Chen LC, Cohen MD, Chee GR, Prophete CM, Haykal-Coates N, et al. Chemical analysis of World Trade Center fine particulate matter for use in toxicologic assessment. Environmental health perspectives. 2003;111(7):972. doi: 10.1289/ehp.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Offenberg J, Eisenreich S, Chen L, Cohen M, Chee G, Prophete C, et al. Persistent organic pollutants in the dusts that settled across lower Manhattan after September 11, 2001. Environmental science & technology. 2003;37(3):502–8. doi: 10.1021/es025730g. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Cone JE, Kahn AR, Brackbill RM, Farfel MR, Greene CM, et al. Association between World Trade Center exposure and excess cancer risk. Jama. 2012;308(23):2479–88. doi: 10.1001/jama.2012.110980. [DOI] [PubMed] [Google Scholar]
- 8.Solan S, Wallenstein S, Shapiro M, Teitelbaum SL, Stevenson L, Kochman A, et al. Cancer incidence in world trade center rescue and recovery workers, 2001–2008. Environ Health Perspect. 2013;121(6):699–704. doi: 10.1289/ehp.1205894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeig-Owens R, Webber MP, Hall CB, Schwartz T, Jaber N, Weakley J, et al. Early assessment of cancer outcomes in New York City firefighters after the 9/11 attacks: an observational cohort study. The lancet. 2011;378(9794):898–905. doi: 10.1016/S0140-6736(11)60989-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boffetta P, Zeig-Owens R, Wallenstein S, Li J, Brackbill R, Cone J, et al. Cancer in World Trade Center responders: Findings from multiple cohorts and options for future study. Am J Ind Med. 2016;59(2):96–105. doi: 10.1002/ajim.22555. [DOI] [PubMed] [Google Scholar]
- 11.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 12.Barouki R, Gluckman PD, Grandjean P, Hanson M, Heindel JJ. Developmental origins of non-communicable disease: implications for research and public health. Environmental Health. 2012;11(1):42. doi: 10.1186/1476-069X-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plass C, Soloway PD. DNA methylation, imprinting and cancer. European journal of human genetics: EJHG. 2002;10(1):6. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 14.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40(1):91–9. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 15.Vaissiere T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69(1):243–52. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez-Vargas H, Lambert M-P, Le Calvez-Kelm F, Gouysse G, McKay-Chopin S, Tavtigian SV, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PloS one. 2010;5(3):e9749. doi: 10.1371/journal.pone.0009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan ES, Qiu W, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Human molecular genetics. 2012;21(13):3073–82. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji Z, Zhang L, Peng V, Ren X, McHale C, Smith M. A comparison of the cytogenetic alterations and global DNA hypomethylation induced by the benzene metabolite, hydroquinone, with those induced by melphalan and etoposide. Leukemia. 2010;24(5):986–91. doi: 10.1038/leu.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environmental health perspectives. 2009;117(2):217. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, García-Vargas G, et al. Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. Journal of biochemical and molecular toxicology. 2013;27(2):106–15. doi: 10.1002/jbt.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasaro CR, Holden WL, Berman KD, Crane MA, Kaplan JR, Lucchini RG, et al. Cohort Profile: World Trade Center Health Program General Responder Cohort. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woskie SR, Kim H, Freund A, Stevenson L, Park BY, Baron S, et al. World Trade Center disaster: assessment of responder occupations, work locations, and job tasks. American journal of industrial medicine. 2011;54(9):681–95. doi: 10.1002/ajim.20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber A, Georgopoulos P, Gilliam R, Stenchikov G, Wang S-W, Kelly B, et al. Modeling Air Pollution from the Collapse of the World Trade Center and Assessing the Potential Impacts on Human Exposures. Environmental Manager. 2004 Feb;:35–40. [Google Scholar]
- 24.Landrigan PJ, Lioy PJ, Thurston G, Berkowitz G, Chen LC, Chillrud SN, et al. Health and environmental consequences of the world trade center disaster. Environ Health Perspect. 2004;112(6):731–9. doi: 10.1289/ehp.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lioy PJ, Weisel CP, Millette JR, Eisenreich S, Vallero D, Offenberg J, et al. Characterization of the dust/smoke aerosol that settled east of the World Trade Center (WTC) in lower Manhattan after the collapse of the WTC 11 September 2001. Environmental health perspectives. 2002;110(7):703. doi: 10.1289/ehp.02110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stenchikov G, Lahoti N, Diner DJ, Kahn R, Lioy PJ, Georgopoulos PG. Multiscale plume transport from the collapse of the World Trade Center on September 11, 2001. Environmental Fluid Mechanics. 2006;6(5):425–50. [Google Scholar]
- 27.Yiin L-M, Millette JR, Vette A, Ilacqua V, Quan C, Gorczynski J, et al. Comparisons of the dust/smoke particulate that settled inside the surrounding buildings and outside on the streets of southern New York City after the collapse of the World Trade Center, September 11, 2001. Journal of the Air & Waste Management Association. 2004;54(5):515–28. doi: 10.1080/10473289.2004.10470935. [DOI] [PubMed] [Google Scholar]
- 28.Brüggemann R, Patil GP. Ranking and prioritization for multi-indicator systems: Introduction to partial order applications. Springer Science & Business Media; 2011. [Google Scholar]
- 29.Myatt GJ. Making sense of data: a practical guide to exploratory data analysis and data mining. John Wiley & Sons; 2007. [Google Scholar]
- 30.Linkov I, Moberg E. Multi-criteria decision analysis: environmental applications and case studies. CRC Press; 2011. [Google Scholar]
- 31.Herbert R, Moline J, Skloot G, Metzger K, Baron S, Luft B, et al. The World Trade Center disaster and the health of workers: five-year assessment of a unique medical screening program. Environmental health perspectives. 2006:1853–8. doi: 10.1289/ehp.9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–96. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y-a, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price EM, Cotton AM, Lam LL, Farré P, Emberly E, Brown CJ, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics & chromatin. 2013;6(1):1. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houseman EA, Kim S, Kelsey KT, Wiencke JK. DNA methylation in whole blood: uses and challenges. Current environmental health reports. 2015;2(2):145–54. doi: 10.1007/s40572-015-0050-3. [DOI] [PubMed] [Google Scholar]
- 36.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13(1):1. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995:289–300. [Google Scholar]
- 38.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32(2):286–8. doi: 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- 39.Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C. Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics. 2013;29(15):1851–7. doi: 10.1093/bioinformatics/btt311. [DOI] [PubMed] [Google Scholar]
- 40.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wisnivesky JP, Teitelbaum SL, Todd AC, Boffetta P, Crane M, Crowley L, et al. Persistence of multiple illnesses in World Trade Center rescue and recovery workers: a cohort study. Lancet. 2011;378(9794):888–97. doi: 10.1016/S0140-6736(11)61180-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2(3):344–58. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26(22):3279–90. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 45.Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 47.Riggs AD. DNA methylation and cell memory. Cell Biochemistry and Biophysics. 1989;15(1):1–13. doi: 10.1007/BF02991574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.