Abstract
Hepatic ischemia/reperfusion injury (IRI) is a common cause of morbidity and mortality in liver transplantation settings and involves severe cell death and inflammatory responses. MicroRNA-191 has recently been reported to be abnormally expressed in hepatocellular carcinoma and other liver diseases in the regulation of important cellular processes. However, little is known about its function and molecular mechanism in IRI. Here, we demonstrate that miR-191 is significantly upregulated in a cultured cell line during hypoxia/reperfusion (H/R) and in liver tissue during IRI in mice. The activation of miR-191 under hypoxic conditions is mediated by hypoxia-inducible factor-1α (HIF1α) binding to its promoter region. Global miR-191 KO mice were constructed by CRISPR/Cas9 system, and we found that miR-191 deficiency markedly reduces liver tissue damage, cell inflammatory responses and cell death in a mouse hepatic IRI model. Under the H/R condition, miR-191 overexpression promotes G0/G1 cell cycle arrest and cell apoptosis, but inhibition of miR-191 facilitates cell cycle progression and decreases cell death. Mechanistically, upon induction by hypoxia or ischemia, miR-191 suppresses expression of ZO-1-associated Y-box factor (ZONAB) and its downstream factor Cyclin D1, consequently resulting in cell death and tissue injury. Moreover, the effects of miR-191 on cell cycle arrest and cell apoptosis are abrogated by ZONAB overexpression, and vice versa. Taken together, our results indicate an important role of the HIF1α/miR-191/ZONAB signaling pathway in hepatic IRI and suggest miR-191 as a novel therapeutic target for the treatment of liver IRI.
Subject terms: RNA, Experimental models of disease, Acute inflammation
Introduction
Liver transplantation (LTx) is an effective therapeutic modality for the treatment of end-stage liver disease [1]. However, ischemia/reperfusion injury (IRI) leads to serious liver graft damage after LTx. Liver IRI contributes to a higher incidence of acute primary liver graft non-function or late chronic graft rejection, especially in marginal livers with greater susceptibility to ischemic insults [2]. Some therapeutic strategies for liver IRI have been reported, including pharmacological agents, ischemia preconditioning, surgical interventions and gene therapy. Unfortunately, they are limited by poor validity and scientific controversy [3–5].
MicroRNAs (miRNAs) are a class of small noncoding RNAs (18–22 nucleotides) that play important regulatory roles in plants and animals by repressing the translation of proteins from mRNAs (messenger RNAs) or by increasing the degradation of mRNAs by binding to their 3′-untranslated regions (3′-UTRs) [6–8]. miRNAs play critical roles in a broad range of biological processes including proliferation, differentiation, apoptosis and stress responses [9]. Recently, the altered expression of several miRNAs has been described in liver diseases, including hepatocellular carcinoma (HCC) [10–12], non-alcoholic fatty liver disease [13] and hepatitis C [14]. A class of specific targets of miRNAs that modulate liver diseases has been identified and includes miR-221 (highly expressed in HCC), which targets the cell cycle [11], miR-122 (low expression in HCC), which controls Cyclin G1 in genomic instability [12], and miR-296 (low expression in NAFLD), which targets the p53-upregulated mediator of apoptosis that induces hepatocyte lipoapoptosis [13]. These findings provide strong evidence that miRNAs have important regulatory roles in liver diseases.
miR-191 is a recently discovered miRNA that plays an important regulatory role in several human tumors, such as colorectal cancer [15], HCC [16], thyroid follicular tumors [17] and acute myeloid leukemia [18], and may be associated with clinical stage, patient survival and disease prognosis. Previous reports have characterized miR-191 as a stress-response gene that can be regulated by hypoxia and nutrient deprivation [19, 20], estrogen [20] or other chemicals such as 5-Fu [15] or dioxin [16]. Therefore, miR-191 may likely emerge as a regulator in stress-induced tissue injury or other diseases, such as IRI. Li et al. demonstrated the inhibitory effect of miR-191 on neointima formation after carotid injury and uncovered the interaction between miR-191 and EGR-1 [21]. However, the underlying function and mechanism of miR-191 in other injuries or diseases remain largely unknown.
In this study, we demonstrated that miR-191 expression was upregulated in hepatocytes under hypoxia or ischemia conditions. Hypoxia-inducible factor-1α (HIF1α) could bind to the promoter region of miR-191 and elevated its expression. miR-191-deficient mice were protected against liver IRI, whereas miR-191 overexpression mediated hepatocyte death. ZO-1-associated Y-box factor (ZONAB) was identified as a direct target of miR-191 during IRI and hypoxia/reperfusion (H/R) stresses. We further revealed that ZONAB inhibition was crucial for miR-191-mediated cell apoptosis and cell cycle arrest. Our data suggest that miR-191 may serve as a novel therapeutic target for liver IRI interventions.
Results
miR-191 is transcriptionally activated by HIF1α
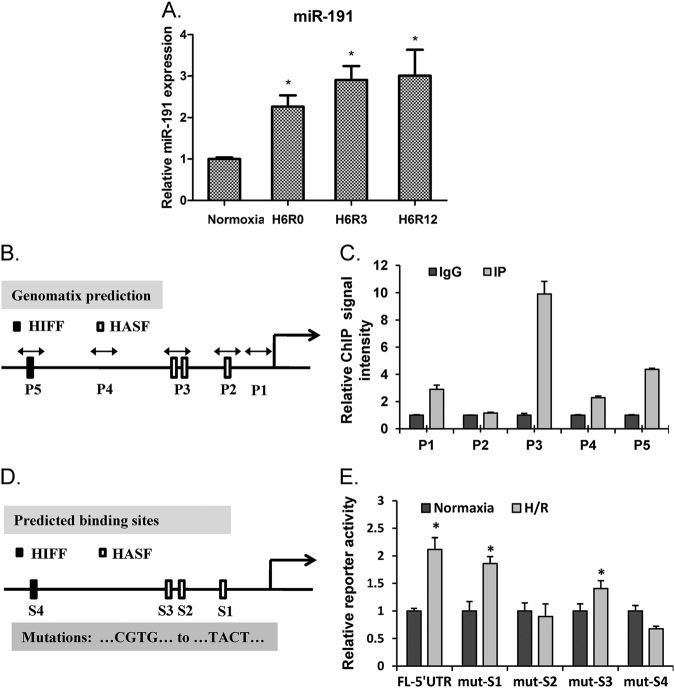
Previous reports have characterized miR-191 as a stress-response gene that can be regulated by hypoxia in breast cancer [19, 20]. Hypoxia is one of the key microenvironment in ischemic tissues. In order to investigate the possible role of miR-191 in IRI, we first tested the regulation of hypoxia on the transcription activity of miR-191 by subjecting a human liver cell line (LO2) to 6 h of hypoxia followed by indicated time periods of reoxygenation. As expected, we observed that miR-191 levels increased dramatically following 6 h of hypoxia and reoxygenation for 0, 3 and 12 h (Fig. 1a). To address the role of HIFs in the regulation of miR-191, we first analyzed the sequence of the miR-191 gene promoter region by Genomatix software tools. The results revealed a putative hypoxia responsive element (HRE), or HIF-binding site, in the miR-191 promoter region, suggesting that miR-191 may be a direct transcriptional target of HIF (Fig. 1b). HIF1a is the inducible subunit of HIF, which is crucial for regulating a variety of cellular activities in response to oxygen deprivation. To verify whether miR-191 induction depends on HIF1a, a chromatin immunoprecipitation (ChIP) assay was performed with LO2 cells. The possible binding sites in the miR-191 promoter region were designated as P1 to P5 (Fig. 1b). The results showed that HIF1a ChIPs were mainly enriched in the P3 promoter region and secondarily accumulated in the P5 promoter region, but not in other regions (P1, P2 and P4), suggesting that the P3 region contains the primary site for HIF1a binding (Fig. 1c). The miR-191 promoter region of approximately 1.5 kb and its HRE deletion mutant were cloned into luciferase promoter reporter vectors (Fig. 1d), which were then transfected into HEK 293T cells to determine their promoter activity. As shown in Fig. 1e, hypoxia induced a twofold increase in the activity of the full-length miR-191 promoter. However, the promotion effect of HIF1a on miR-191 promoter activity was abrogated in cells transfected with mutated S2 and S4 binding sequences (Fig. 1e). Together, these results indicate that miR-191 is mainly induced via HIF1α under hypoxic conditions, and suggest an involvement of miR-191 in IRI.
Fig. 1.
HIF1α induces miR-191 expression during hypoxia. a Quantification of miR-191 levels in liver cells (LO2) subjected to H/R. (N = 6). b Schematic diagram of the miR-191 promoter region and possible HIF1α-binding sites. HIFF HIF-binding sequence, HASF HIF auxiliary sequences. P1, P2, P3, P4 and P5 represent primer locations. c ChIP analysis of the binding of HIF1α to the miR-191 promoter region. d The mutation (“CGTG” to “TACT”) of predicted HIF1α-binding sites on the miR-191 promoter region. e Luciferase assay quantifying the effect of hypoxia on miR-191 promoter activity in LO2 cells. *p < 0.05 vs Normoxia. The data are expressed as the mean ± SD
miR-191 participates in liver injury in mice
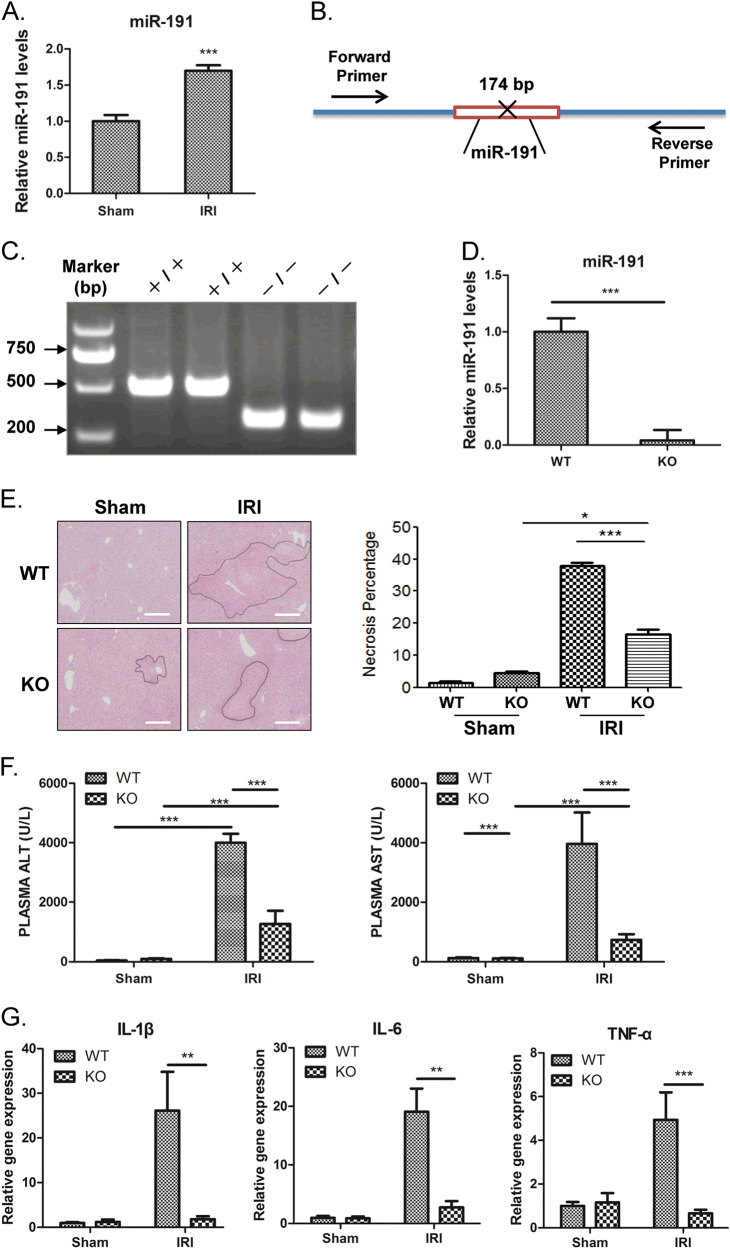
Hepatic IRI triggers a multifaceted cascade of physiological and biochemical events. To determine whether miR-191 was involved in hepatic IRI, the expression levels of hepatic miR-191 after IR injury were evaluated by quantitative PCR. The results showed that liver miR-191 levels were significantly increased after 6 h of reperfusion following 1 h of ischemia compared with sham-operated liver tissues (Fig. 2a). To investigate the function of miR-191 in vivo, global miR-191-knockout (KO) mice were established using the CRISPR-Cas9 genome-editing system (Fig. 2b). The genotyping of KO mice was performed using quantitative PCR (Fig. 2c). The deletion was also confirmed by Sanger sequencing (supplementary Fig 1) and quantification of miR-191 by real-time PCR (Fig. 2d).
Fig. 2.
The effect of miR-191 deficiency on I/R-induced liver damage. a Quantitative PCR indicates that miR-191 expression levels were significantly increased in the liver tissues of mice after 1 h of ischemia and 6 h of reperfusion. (N = 6, ***p < 0.001 vs Sham). b Schematic representation of the CRISPR-Cas9 knockout system. c WT and KO mice were genotyped by tail DNA amplification. d miR-191 deletion was confirmed using quantitative PCR in the liver tissues of WT and KO mice (right). (N = 6). e Representative images of liver I/R damage and quantification of necrotic areas in miR-191 KO and WT mice after IRI. (N = 6, scale = 100 μm). f Quantification of serum ALT and AST levels in miR-191 KO and WT mice after IRI. (N = 6). g Quantification of the mRNA levels of TNF-α, IL-1β and IL-6 in miR-191 KO and WT mice after IRI. (N = 6). *p < 0.05, **p < 0.01, ***p < 0.001. The data are expressed as the mean ± SD
Loss of miR-191 protects the liver from IRI injury
miR-191 KO and wild-type (WT) mice were subjected to liver IRI and damaged liver tissues were harvested for histopathologic analysis. As shown in Fig. 2e, IR induced significant liver tissue necrosis in both miR-191 KO and WT mice. miR-191 deficiency remarkably reduced liver damage induced by IRI (Fig. 2e). It was noteworthy that miR-191 deletion led to more necrosis of liver tissues after sham surgery (Fig. 2e). Hepatic ischemia is accompanied by elevated levels of plasma alanine aminotransferase (ALT) and aspartate transaminase (AST). We found that miR-191 deletion dramatically orchestrated the induction of serum ALT and AST after IRI (Fig. 2f). After sham operation, the levels of AST in livers from miR-191 KO mice was significantly higher than that in livers from WT mice (Fig. 2f), which is consistent with the results of hematoxylin/eosin (H/E) staining. The mRNA levels of proinflammatory factors, including tumor necrosis factor (TNF)-a, interleukin-6 (IL-6) and IL-1β, were measured by real-time PCR after 1 h of ischemia and 6-h reperfusion. The results indicated that miR-191 deletion significantly reduced the levels of these factors (Fig. 2g). These data indicated that miR-191 loss-of-function protects the liver from IRI-induced tissue damage and inflammatory insult.
miR-191 deficiency alleviates cell death after hepatic I/R
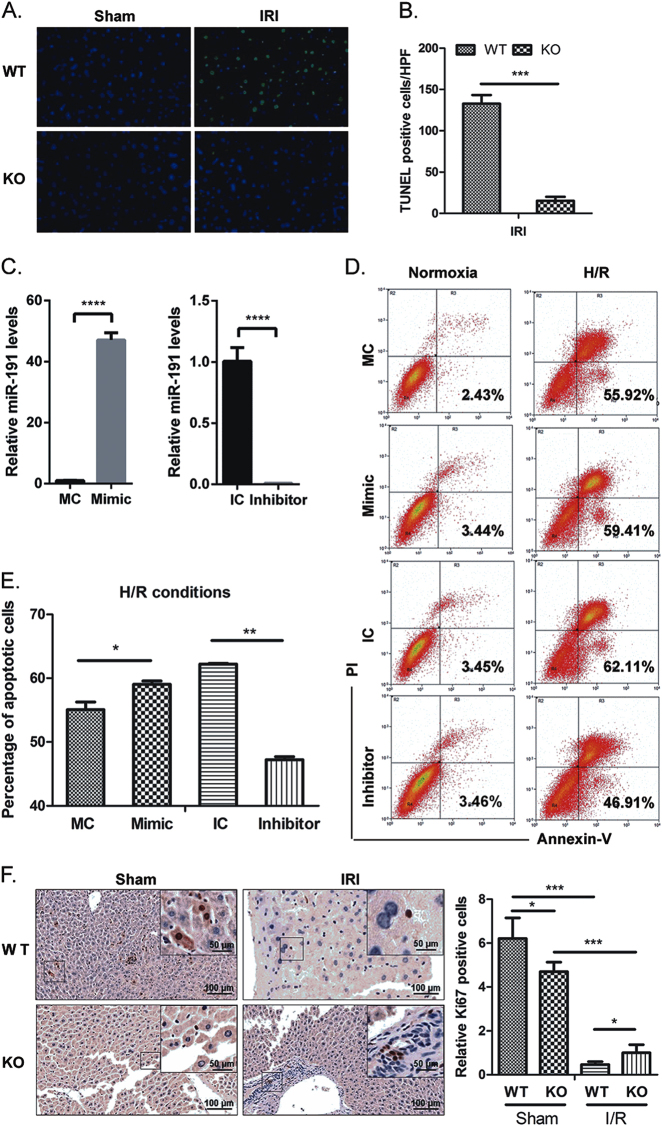
To further confirm the functional influence of miR-191 on cell death during IRI, cell apoptosis and cell cycle progression were evaluated by terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) staining and fluorescence activated cell sorting (FACS) assay. As shown in Figs. 3a, b, the number of TUNEL-positive cells in the livers of WT mice was greater than that in the livers of miR-191 KO mice after IRI (Figs. 3a, b). miR-191 mimics/inhibitors and corresponding control oligos were utilized to modulate endogenous miR-191 levels in LO2 (Fig. 3c). Annexin V-FITC cell apoptosis analysis demonstrated that H/R treatment triggered significant liver cell apoptosis. miR-191 overexpression increased the apoptotic rates of LO2 cells under the H/R condition, whereas miR-191 inhibition clearly decreased cell death from 62.11 to 46.91% (Figs. 3d, e). The data were consistent with the TUNEL assay.
Fig. 3.
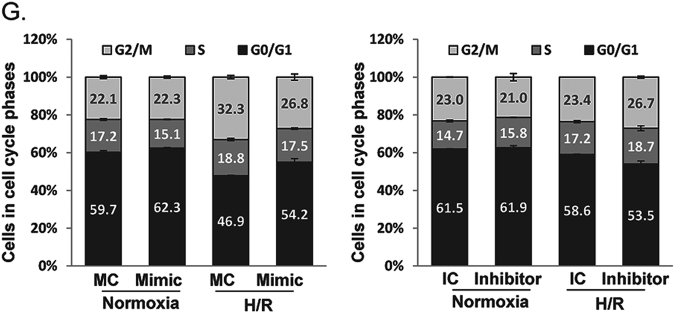
The effect of miR-191 deficiency on I/R- or H/R-induced cell death. a Representative images of liver TUNEL staining and quantification of TUNEL-positive cells in miR-191 KO and WT mice after sham or IR surgery. (N = 6). b Quantitative analysis of TUNEL-positive cells. c Quantitative analysis of endogenous miR-191 levels in LO2 cells transfected with miR-191 mimics, inhibitors or corresponding control oligos. (MC: mimic control oligos; Mimic: miR-191 mimic oligos; IC: inhibitor control oligos; Inhibitor: miR-191 inhibitor oligos). d Cell apoptosis analysis of LO2 cells transfected with miR-191 mimics, inhibitors or corresponding control oligos under normoxia and H/R conditions. e Quantitative analysis of apoptotic cells. f Immunohistochemical analysis of Ki67 in liver tissues from miR-191 KO and WT mice after sham or IR surgery. g Cell cycle analysis of LO2 cells transfected with miR-191 mimics, inhibitors or corresponding control oligos under normoxia and H/R conditions. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The data are expressed as the mean ± SD
Hepatocyte proliferation can alleviate tissue damage after liver I/R, and the capacity of liver regeneration determine the function of postischemic liver. Therefore, we next investigated the effects of miR-191 on hepatocyte proliferation by immunohistochemical analysis and FACS analysis. miR-191 KO mice exhibited a higher percentage of Ki67-positive cells than WT mice after liver IRI, although Ki67-positive cells in miR-191 KO mice were significantly lower than that in WT mice after sham operation (Fig. 3f). Furthermore, we observed an increase in the G0/G1-phase cell population (54.2 vs. 46.9%) in miR-191 mimic-transfected cells compared with corresponding control cells under H/R. In contrast, miR-191 suppression decreased the G0/G1-phase cell population (Fig. 3g). Taken together, these results showed that miR-191 directly regulates cell survival in IR- or H/R-induced tissue damage.
miR-191 directly suppresses ZONAB
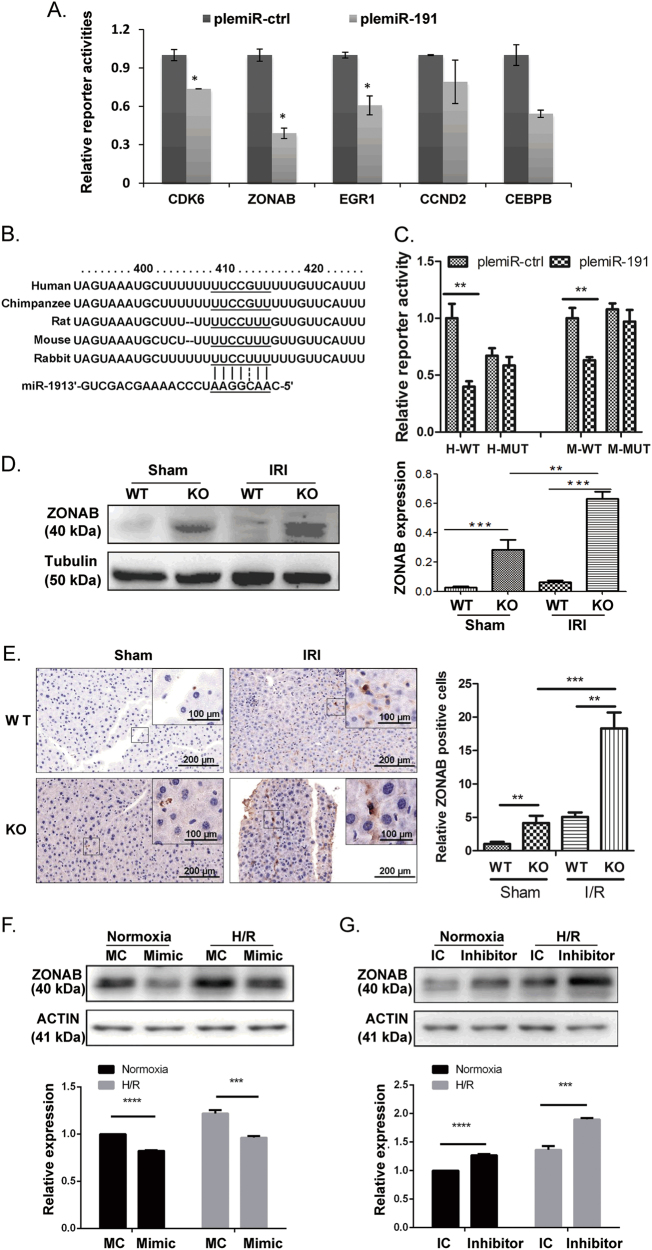
Next, we explored the downstream targets of miR-191. Target prediction databases, including TargetScan (http://www.targetscan.org/vert_71/) and miRanda (http://www.microrna.org/microrna/home.do), were utilized. Several candidate targets of miR-191 were selected, including CDK6, ZONAB, EGR1, CCND2 and CEBPB. The 3′-UTR reporter activities of these genes were detected by luciferase assay. The results showed that miR-191 inhibited the reporter activities of different targets in various degree, among which the downtrend of ZONAB was most obvious (Fig. 4a). Given the known function of CDK6 and ZONAB in regulating cell cycle progression and cell apoptosis [22, 23], we chose them for further study. The mRNA and protein levels of CDK6 were measured. However, we found an obvious reduction of CDK6 in miR-191 KO mice, which was not as expected (Supplementary Fig 3). We observed that miR-191 seed region perfectly paired with the human ZONAB 3′-UTR (UUCCGUU), but imperfectly paired with the mouse/rat ZONAB 3′-UTR (UUCCUUU) (Fig. 4b). However, the results from the luciferase assay indicated that miR-191 could repress the reporter activities of both human and mouse WT ZONAB 3′-UTRs, but not the mutant ZONAB 3′-UTRs, indicating the direct binding of miR-191 with the ZONAB 3′-UTR regardless of species (Fig. 4c). Then, in vivo protein levels of ZONAB were determined and we observed apparent ZONAB upregulation in miR-191 KO mice. Interestingly, IR injury could further induce the expression of ZONAB (Fig. 4d). Accordingly, further immunohistochemistry staining showed that ZONAB expression was increased after IR surgery, and miR-191 KO further induced the expression of ZONAB (Fig. 4e). The results from the in vitro gain- and loss-of-function experiments showed that miR-191 overexpression inhibited ZONAB expression. miR-191 inhibitor transfection further enhanced the hypoxia-induced increase in ZONAB expression (Figs. 4f, g). To validate the correlation of miR-191 and ZONAB, we analyzed the expression of miR-191 and ZONAB in liver tissues following IRI treatment by RT-PCR. As shown in Fig. 4h, miR-191 level was inversely correlated with ZONAB expression (Fig. 4h). These data suggested that miR-191 directly targeted the 3′-UTR of ZONAB to repress ZONAB expression.
Fig. 4.
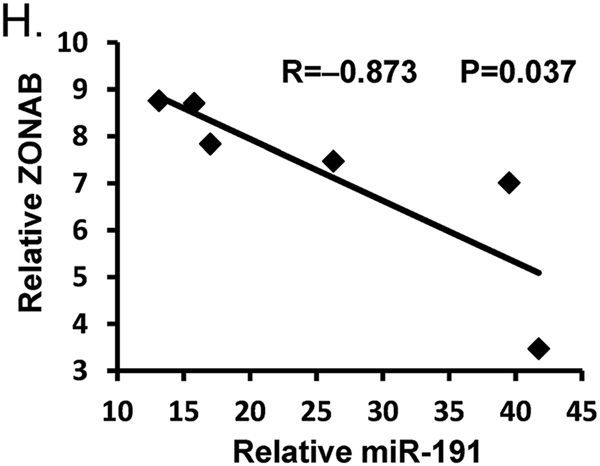
MiR-191 targets ZONAB. a Luciferase analysis detecting the reporter activities of CDK6, ZONAB, EGR1, CCND2 and CEBPB. (N=3, *p < 0.05 vs plemiR-ctrl) b The predicted binding sites of miR-191 in the 3′-UTRs of ZONAB. c Luciferase analysis detecting the reporter activities of the ZONAB 3′-UTR with wild-type or mutated seed regions for miR-191 binding. (**p < 0.01 vs plemiR-ctrl, H human, M mouse). d Western blot analysis of ZONAB levels in liver tissues from WT/miR-191-KO mice after sham surgery or IRI. β-Tubulin served as the loading control. (N = 6). e Immunohistochemical analysis of ZONAB in liver tissues from miR-191 KO and WT mice after sham or IR surgery. f, g Western blot analysis of ZONAB levels in LO2 cells transfected with the indicated oligos under normoxia or the H/R condition. β-ACTIN served as the loading control. h Inverse correlation between miR-191 and ZONAB in liver tissues following IRI. miR-191 level was normalized to the U6, and ZONAB level was normalized to the GAPDH. Statistical analysis was performed using Person’s correlation coefficient analysis. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The data are expressed as the mean ± SD
The interaction of miR-191 with ZONAB is crucial for cell death under hypoxic conditions
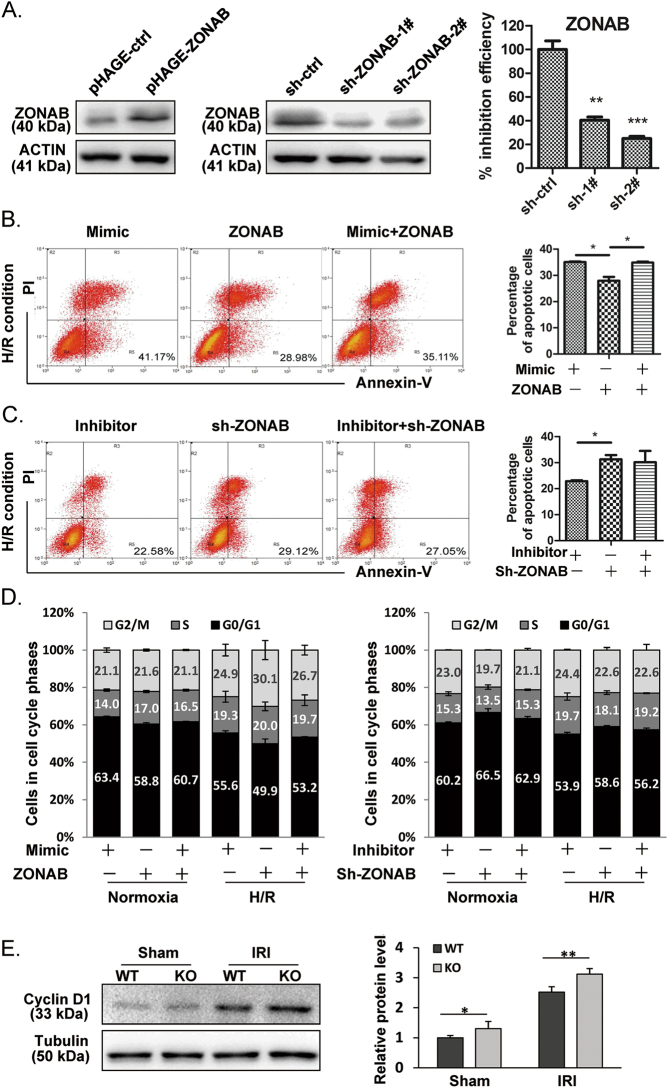
To further confirm that ZONAB is the major downstream target of miR-191, we constructed a ZONAB overexpression plasmid without the 3′-UTR (pHAGE-ZONAB), which did not interact with miR-191, and a knockdown plasmid (pLKO.1-PTK6 sh-ZONAB#2) that could effectively decrease the protein level of endogenous ZONAB with a >70% inhibition rate. (Fig. 5a). In vitro co-transfection studies were performed to investigate the relationship between miR-191 and ZONAB. The results from the cell apoptosis and cell cycle analyses showed that co-transfection of ZONAB without the 3′-UTR rescued the effect of miR-191 overexpression on cell apoptosis and G0/G1 phase arrest, indicating that the function of miR-191 during hypoxia was at least partly due to the downregulation of ZONAB expression (Figs. 5b, d, left panel). In contrast, after H/R, more apoptotic cells were observed in the sh-ZONAB group (29.12%) compared with the miR-191 inhibition group (22.58%), and co-transfection of sh-ZONAB impaired the inhibitory effect of miR-191 inhibitor on cell apoptosis. (Fig. 5c) The cell cycle analysis also demonstrated that sh-ZONAB could induce G0/G1 phase arrest and disturb the function of miR-191 inhibition in H/R damage. (Fig. 5d, right panel)
Fig. 5.
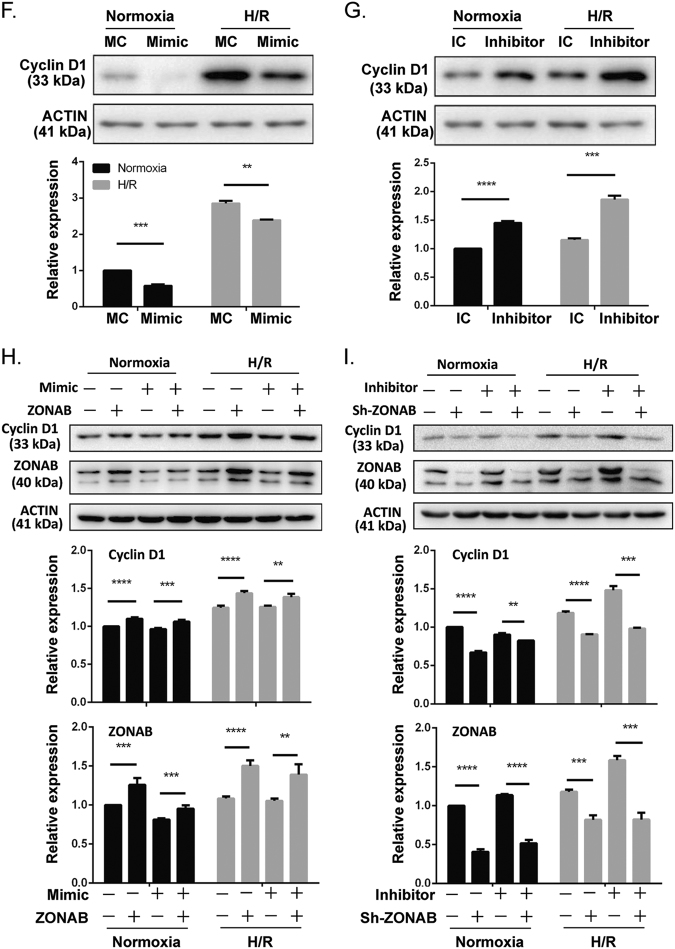
The effect of the miR-191-ZONAB interaction on H/R-induced cell death. a Western blot analysis of ZONAB levels in LO2 cells following pHAGE-ctrl/pHAGE-ZONAB or pLKO.1-sh-ctrl/pLKO.1-sh-ZONAB-1# and 2# transfection. **p < 0.01, ***p < 0.001 vs pLKO.1-sh-ctrl. b, c Cell apoptosis analysis of LO2 cells co-transfected with the indicated oligos and plasmids after H/R treatment. d Cell cycle analysis of LO2 cells co-transfected with the indicated oligos and plasmids under normoxia or the H/R condition. e Western blot analysis of Cyclin D1 levels in hepatic tissues from WT/miR-191-KO mice after sham surgery or IRI. β-Tubulin served as the loading control. (N = 6). f, g Western blot analysis of Cyclin D1 levels in LO2 cells transfected with the indicated oligos under normoxia or the H/R condition. β-ACTIN served as the loading control. h, i Western blot analysis of the indicated proteins in LO2 cells transfected with the indicated oligos or plasmids under normoxia or the H/R condition. β-ACTIN served as the loading control. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The data are expressed as the mean ± SD
Previous studies have reported that ZONAB is an activator of Cyclin D1 [23–26]. To verify Cyclin D1 expression and the effect of miR-191/ZONAB signaling on Cyclin D1 during hypoxia, we first determined Cyclin D1 levels in the liver tissues of miR-191 KO mice following IR injury. As shown in Fig. 5e, Cyclin D1 expression was increased following IRI; miR-191 deficiency enhanced Cyclin D1 expression after ischemic injury (Fig. 5e). An in vitro study indicated that miR-191 overexpression eliminated Cyclin D1 protein levels, whereas miR-191 inhibition increased Cyclin D1 expression under both normoxia and hypoxia conditions (Figs. 5f, g). To further explore the regulatory relationship between miR-191, ZONAB and Cyclin D1, we performed a western blot on cells co-transfected with mimic/inhibitor oligos and ZONAB overexpression or knockdown plasmids. A lower Cyclin D1 level was observed in miR-191-transfected cells than in ZONAB-overexpressing cells. In contrast, the expression of Cyclin D1 was increased following miR-191 inhibitor transfection. In addition, co-transfection of ZONAB could restore the decrease of Cyclin D1 induced by miR-191 mimic, and co-transfection of sh-ZONAB diminished the increase of Cyclin D1 following miR-191 inhibitor transfection (Figs. 5h, i). These data suggested that Cyclin D1 is a downstream factor of ZONAB in the mediation of miR-191-induced cell death under hypoxic or ischemic conditions. Taken together, we showed that miR-191 enhanced IRI or H/R stress-induced cell death by inhibiting the ZONAB/Cyclin D1 pathway.
Discussion
IRI is a condition in which the restoration of blood flow in a previously ischemic organ or tissue leads to exacerbated damage, further insulting functional and anatomical integrity [27]. In the current study, we described a novel and highly significant function of miR-191 in liver IRI. We found that increased liver miR-191 expression at a transcriptional level under hypoxia or ischemia conditions contributes to liver damage and cell death, whereas miR-191 deficiency provides resistance to I/R injury. miR-191, activated by HIF1α, targeted and repressed ZONAB to induce cell cycle arrest and cell apoptosis during liver injury.
miR-191 is ubiquitously expressed in various tissues and is highly conserved in humans and other organisms [28, 29]. It is a hepatocyte-abundant miRNA and is detectable in serum during hepatic injury [30]. Dysregulated miR-191 expression was correlated with liver cancer initiation and development. For example, miR-191 is increased in liver cancer tissues and functions in the regulation of cell proliferation, apoptosis and epithelial mesenchymal transition by targeting TIMP metallopeptidase inhibitor 3 or various cancer-related pathways [16, 31]. In the present study, the mice with global KO of miR-191 exhibited significantly lower aminotransferase levels, less tissue necrosis and obviously reduced inflammation, with lower levels of proinflammatory cytokines, including IL-1β, IL-6 and TNFα. Therefore, the effect of miR-191 on liver damage appears to be due to its potential regulatory role in cell death and the immune response. It is interesting that miR-191 KO caused more necrotic areas at baseline but less after IRI, indicated by H/E staining. Correspondingly, we observed an significantly increased serum AST and decreased cell proliferation (Ki67 staining) in livers at our following studies. Our further results showed that there were no damage in lung, kidney and heart tissues from WT and miR-191 KO mice after sham operation (Supplementary Table 1 and Fig. 2). Multiple studies have illustrated that ischemic preconditioning protects livers against ischemic/reperfusion damage and induces hepatic regeneration, which involves multiple molecular inducers and signaling pathways [4, 32–35]. An elegant study has reported that miR-199a inhibition can recapitulates hypoxia preconditioning in cardiac myocytes against hypoxic damage [36]. We speculated that miR-191 KO can bring forth an ischemic preconditioning effect on liver tissue. That, in turn, decreased hepatocyte susceptibility to IR injury. In our future study, we will try to uncover the molecular mechanism of this interesting issue.
Extensive studies have shown that miR-191 has important roles in cell proliferation and apoptosis by regulating proteins involved in related signaling pathways [37, 38]. In our previous study, we found that miR-191 targeted CEBPB and promoted the G0/G1 to S phase transition and inhibited 5-Fu-induced cell apoptosis in colorectal cancer [15]. In the present study, miR-191 deficiency resulted in fewer apoptotic cells after IRI. The results from in vitro loss- or gain-of-function studies indicated an inhibitory effect of miR-191 on cell cycle progression and cell survival during I/R- or H/R-induced cell damage. Our data revealed miR-191 as a novel proapoptotic mediator in IR injury; however, its function and mechanism in the inflammatory response remain to be determined.
HIF1 is the essential regulator of gene expression in the cellular response to ischemia or hypoxia. miRNAs can be regulated by HIF1 under hypoxic conditions and HIF1 also can exert some of its effect through various miRNAs. However, the role of HIF1α in cell survival and inflammation most likely depends on cell type and context. In our study, miR-191 was found to be upregulated in response to IRI or H/R stress, which is partially consistent with previous reports [19, 20]. Further experiments using ChIP and luciferase assays indicated that HIF1α could directly bind to the promoter region of miR-191 and activate its expression. These data verify the role of miR-191 during IRI.
ZONAB is a Y-box transcription factor. ZONAB exerts diverse biological effects that are dependent on cell type and context [23]. Previous studies have demonstrated that ZONAB regulates CDK4, proliferating cell nuclear antigen (PCNA) and Cyclin D1 and consequently influences G1/S phase progression and cell proliferation in mammary epithelial cells [23, 24]. In this study, we found that miR-191 targeted ZONAB by directly binding to the 3′-UTR region of ZONAB mRNA. Additionally, ZONAB inhibited hypoxia-induced cell apoptosis and cell cycle arrest, which is partially consistent with previous reports [23, 26]. Disruption of the miR-191/ZONAB interaction impaired the effect of miR-191, suggesting the important function of miR-191-ZONAB interaction in IRI. We also excluded the regulation of miR-191 on CDK6 expression in IRI. Moreover, the levels of ZONAB and Cyclin D1 were increased under H/R stress. ZONAB overexpression could increase the level of Cyclin D1 and sh-ZONAB reduced the level of Cyclin D1, indicating a possible ZONAB/Cyclin D1 interaction in liver cells under hypoxia conditions. Notably, the levels of miR-191, ZONAB and Cyclin D1 were all increased following hypoxia or ischemia. However, the negative regulation of miR-191 on the expression of ZONAB and Cyclin D1 was obvious, suggesting a regulatory balance between gene expression and the injury microenvironment.
In conclusion, the current study highlighted miR-191 as an important miRNA in the pathogenesis of liver IRI (Fig. 6). Under IRI or H/R stress, miR-191 is markedly upregulated via HIF1α. miR-191 represses ZONAB to facilitate cell cycle arrest and induce cell death. Our findings suggest a novel and important mechanism for miR-191 regulation during IRI. The deletion of miR-191 or the suppression of the miR-191/ZONAB interaction in hepatocytes may provoke potentially therapeutic effects on hepatic IRI.
Fig. 6.
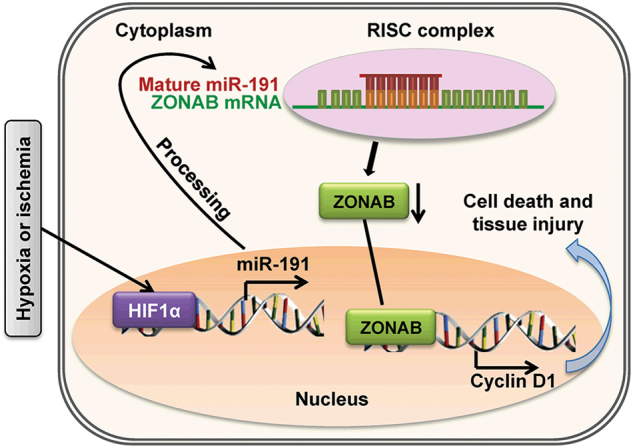
Schematic of the underlying mechanism of miR-191 in hepatic IRI. Upon hypoxic or ischemic stimulation, hypoxia-inducible factor-1α (HIF1α) was activated, triggering the transcription of miR-191 by directly binding to its promoter region. After transport into the cytoplasm and further processing, miR-191 matured, incorporated into the RISC complex to pair to the ZONAB 3′-UTR, and suppressed the levels of ZONAB, further impairing ZONAB-mediated transcription of Cyclin D1 and ultimately inducing cell death and liver damage
Materials and methods
Animals
The experimental protocols were approved by the Institutional Animal Care and Use Committee of the Tongji Medical College (affiliated with the Huazhong University of Science and Technology). C57BL/6 WT mice were purchased from the Animal Experiment Center of Wuhan university. The CRISPR-Cas9 system of genome editing was employed to generate miR-191 KO mice. mRNA was microinjected into fertilized embryos (C57BL/6J background) to produce global KO mice and four independent KO lines were established. Genomic DNA was extracted from the tail for genotyping using PCR analysis with the primers 5′-CGGATCAGGACCTCGTCCCGGG-3′ (forward) and 5′-GCCATCAGCCCGAGCTTTTTTC-3′ (reverse). All mice were 8–12 weeks old and were housed in an environment with controlled light (12-h light/dark cycle), humidity and temperature, with food and water available ad libitum. All experimental procedures involving animals were performed in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publications nos. 80–23, revised 1996) and were approved by the Animal Care and Use Committee of Wuhan Union Hospital.
Hepatic I/R model
Nonlethal segmental (70%) liver ischemia was induced as previously described [39]. Male C57BL/6 mice were anesthetized with ketamine (80 mg/kg, ip) and xylazine (10 mg/kg, ip). A midline laparotomy was performed and ligaments were dissected carefully. The portal vein, hepatic artery and the bile duct supplying the median and left lateral lobes of the liver were clamped with an artery clip (Cat. 18055-03, FST, USA). The temperature during ischemia was maintained at 32–33 °C using a warming incubator chamber. After 60 min of segmental liver ischemia, the clip was removed to initiate liver reperfusion. The mice were sacrificed after 6 h of reperfusion and then plasma and liver tissues were collected for analysis. The sham group received identical treatment without micro-vascular clamp placement.
Liver damage assessment
Plasma ALT and AST levels were measured using a spectrophotometer (ELx800, BioTek, USA). For histopathological analysis, formalin-fixed liver samples were embedded in paraffin and evaluated using H/E staining as previously described [40]. All images were captured on an Olympus Provis microscope. Necrotic areas were assessed quantitatively using Image J software (National Institutes of Health, USA) as previously described [40]. The necrotic area/whole area ratio was normalized by a pathologist blinded to our experimental groups.
TUNEL assay
Cell death in liver paraffin sections was detected using the TUNEL method (ApopTag® Plus In Situ Apoptosis Fluorescein Detection Kit, EMD Millipore Corporation, S7111) according to the manufacturer’s instructions. The numbers of labeled cells in the liver sections were blindly evaluated in 10 high-power fields/sections under light microscopy (40 × ).
Cell lines and hypoxia treatment
The human liver cell line (LO2) was obtained from ATCC and grown in Dulbecco’s modified Eagle’s medium culture medium supplemented with 10% fetal bovine serum. For hypoxia treatment, cells cultured overnight were transferred into a hypoxic chamber with 1% oxygen and incubated in pre-equilibrated medium for indicated time periods.
Flow cytometry analysis
Flow cytometry was performed to assess cell cycle and cell apoptosis. In brief, LO2 cells transfected with the indicated oligos or plasmids were subjected to hypoxia treatment. After 6 h of hypoxia and 12 h of reperfusion, the cells were collected and washed. For cell cycle analysis, after fixing with 70% ethanol overnight at −20 °C, the cells were suspended in phosphate-buffered saline (PBS) and incubated with 100 μg/ml of RNaseA (Sigma) for 30 min and 50 μg/ml of propidium iodide (PI) (Sigma, St Louis, MO) for another 10 min. Then, the cells were subjected to a DNA content analysis using a CyAN analyzer (Beckman Coulter, Fullerton, CA). Apoptosis was determined using Annexin V-FITC staining (B32117, Bimake, Beijing, China) according to the manufacturer’s protocol. In brief, LO2 cells (1 × 105) were prepared in a single-cell suspension and incubated with Annexin V-FITC and PI for 15 min at RT in the dark, and an additional 400 μl of binding buffer was added to the reaction prior to analysis. The results were analyzed with Summit v4.3 software.
ChIP assay
ChIP analysis of HIF1a binding to the miR-191 promoter was conducted using an assay kit from R&D Systems (Minneapolis, MN) following the manufacturer’s instructions. Briefly, stable HIF1α-transfected LO2 cells were cultured and fixed with 1% formaldehyde, washed with ice-cold PBS, and sonicated. The soluble chromatin was then immunoprecipitated with anti-HIF1a antibody (#14179, Cell Signaling Technology) or normal rabbit IgG (#2729 S, Cell Signaling Technology) followed by protein G-magnetic beads (10004D, Invitrogen). The resultant immunoprecipitates were washed and harvested. DNA was purified by phenol/chloroform extraction and subjected to PCR analysis using specific primers designed according to the gene sequence of amplification. The PCR primers for the ChIP assays were as follows: P1: forward primer 5′-TCCAAGTCAAGGGCCTACTG-3′ and reverse primer 5′-AAAGCGACAATGTTCCCCTG-3′; P2: forward primer 5′-GCTTCTCGGGTTCTCTAGGG-3′ and reverse primer 5′-ACACTATCACAGCTTGCCCT-3′; P3: forward primer 5′-GCTCAAGTTTCTCCGGCTAG-3′ and reverse primer 5′-AGAGAACCCGAGAAGCCAAT-3′; P4: forward primer 5′-CTCAGTCTTAGGAGGCCGAG-3′ and reverse primer 5′-CGGGACTATGGGCAAGGG-3′; P5: forward primer 5′-GAAAGCCAAGTTTGCAGGGA-3′ and reverse primer 5′-CTCGGTGGTGCAGTGGAA-3′.
Luciferase reporter assay
The ZONAB 3′-UTR fragment was amplified from human or mouse genomic DNA and subcloned into a psiCHECK2 vector (Promega, Madison, WI). The sequences of the primers used for PCR amplification were as follows: human 3′-UTR forward primer: 5′-TTAGGAGATAGGGTGCAGCC-3′ and reverse primer 5′-ACAAACACACGCTAAGGTCG-3′; mouse 3′-UTR forward primer: 5′-ACTACAAAGGGAACGAGCCA-3′ and reverse primer 5′-AGCCTGTAAACCTCATGCCT-3′. The primer sequences for the mutagenesis of seed sequences were as follows: human forward primer: 5′-AATGCTTTTTTTCTGCACTTTGTTCA-3′ and reverse primer 5′-AAATGAACAAAGTGCAGAAAAAAAGC-3′; mouse forward primer: 5′-AAATGCTCTTTTCTGCACGTTGTTCA-3′ and reverse primer 5′-AAATGAACAACGTGCAGAAAAGAGCA-3′. The WT binding sequence “TTCCGTT” in the 3′-UTR was mutated into “TCTGCAC”. For the luciferase assay, LO2 cells were seeded in 24-well plates. After transfection with 100 ng of psiCHECK2-wt/mutant 3′-UTR plasmids (100 ng) and plemiR-ctrl/ plemiR-191 (200 ng) for 48 h, the cells were lysed and reporter activities were measured using the Dual Luciferase kit (Promega, Madison, WI).
Plasmid constructs
Has-miR-191-5p mimic, has-miR-191-5p inhibitor and the corresponding mimic/inhibitor control oligos were purchased from Guangzhou RiboBio Co., LTD (Guangzhou, China). The human miRNA expression vectors plemiR and plemiR-191 were kindly provided by Dr. Yendamuri, Sai (Department of Thoracic Surgery, Roswell Park Cancer Institute, Buffalo, USA) [41]. Human ZONAB complementary DNA (cDNA) was amplified using the following primers: 5′-CGGGATCCATGAGTGAGGCGGGCGAGGC-3′ (forward) and 5′-CCGCTCGAGTTACTCAGCACTGCTCTGCTGG-3′ (reverse). The fragments digested by BamHI and XhoI were ligated into a pHAGE vector. The shRNA sequences against ZONAB were as follows: sequence 1, 5′-GGTTCATCGAAATCCAACTT-3′; sequence 2, 5′-CAAAGAAACATCCAAGCAAT-3′; and the scramble shRNA sequence was 5′-AATTCTCCGAACGTGTCACGT-3′. ShRNAs were ligated into the pLKO.1 puro vector (Addgene, #8453). All plasmids were sequenced to verify the inserted targets. LO2 cells were transfected with oligos (100 nmol/L) or indicated plasmids using Lipofectamine2000TM (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The OptiMEM media (Invitrogen, Carlsbad, CA, USA) were replaced with cell growth media after 8 h of transfection. Stable cell lines expressing the indicated genes were selected with 1–2 μg/ml puromycin and verified through detection of the indicated miRs and genes by quantitative PCR or western blotting.
Quantitative real-time PCR
Total mRNA was extracted from liver or cells using TRIZOL reagent (Cat. 15596-026, Invitrogen, USA) and cDNA was generated from 1 µg of RNA using an RT reagent kit (Cat. k1622, Thermo Fisher Scientific, USA). Real-time PCR was performed using a Bio-Rad CFX96 Detection System (Bio-Rad Laboratories, USA) with SYBR-green (Cat.1725121, Bio-Rad, USA). All reactions were run in triplicate and relative gene expression was calculated using the comparative threshold cycle (Ct) method (relative gene expression = 2−(ΔCtsample-ΔCtcontrol)). All reagents for miR-191 detections were obtained from RiboBio (Guangzhou, China). The U6 snRNA was used as an internal control. The gene-specific primers included the following: TNFα: forward: 5′-ACTGAACTTCGGGGTGATCG-3′, reverse: 5′-GGCTACAGGCTTGTCACTCG-3′; IL-6: forward: 5′-AGTTGCCTTCTTGGGACTGA-3′, reverse: 5′-TCCACGATTTCCCAGAGAAC-3′; IL-1β: forward: 5´-GGATGAGGACATGAGCACCT-3´, reverse: 5´-GGAGCTGTAGTGCAGTTGT-3´;ZONAB: forward: 5′-CCCGTAATGCTGGTGAGATT-3′, reverse: 5′GGGTTCTCAGTTGGTGCTTC-3′; β-ACTIN: forward: 5′-AGCCATGTACGTAGCCATCC-3′, reverse: 5′-CTCTCAGCTGTGGTGGTGAA-3′.
Western blotting
Total protein lysates were extracted from liver tissues or cells using lysis buffer (Cell Signaling Technology, Beverly, MA) containing complete protease inhibitors (Roche Applied Sciences) and western blotting as previously described [42]. The antibodies used in this study were as follows: rabbit monoclonal anti-Cyclin D (#2978 s) purchased from Cell Signaling Technology (Beverly, MA); mouse monoclonal anti-β-ACTIN (#AC004) purchased from Abclonal Biotechnology (Cambridge, MA, USA); rabbit polyclonal anti-β-Tubulin (10068-1-AP) purchased from Proteintech (Wuhan, Hubei, P.R. China); rabbit polyclonal anti-ZONAB (AV100917) purchased from Sigma Aldrich (St. Louis, MO); peroxidase-conjugated goat anti-mouse IgG (H + L) (115-035-003) and peroxidase-conjugated goat anti-rabbit IgG (H + L) (111-035-003) purchased from Jackson ImmunoResearch (Jackson, West Baltimore Pike West Grove, PA, USA).
Immunohistochemistry
For immunohistochemical analysis, formalin-fixed liver samples were embedded in paraffin, routinely deparaffinised in xylene and an ethanol gradient, incubated in 3% hydrogen peroxide for 10 min, treated with 10 mM Na citrate buffer, pH 6.0, in a pressure cooker at 120 °C for 10 min and cooled to room temperature. Then sections were incubated with the following primary antibodies: anti-ZONAB (rabbit polyclonal, Sigma) and anti-Ki67 (rabbit polyclonal, Abcam), and secondary antibodies at 37 °C for 30 min. The sections were counterstained with hematoxylin, dehydrated and cover-slipped. Positive cells on 10 random fields at ×400 magnification in each slide were counted by two pathologists blinded to our experimental groups.
Statistical analysis
The data are expressed as the means ± S.D. and were analyzed with the statistical analysis software SPSS 15.0 software (SPSS, Inc., Chicago, IL) and the GraphPad Prism Software (version 6, GraphPad Software, San Diego, CA, USA). Significant differences were evaluated using a one-way analysis of variance (ANOVA) analysis. Statistical significance was considered when p < 0.05.
Electronic supplementary material
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (nos. 81070355 to JXZ, 81570570 to HW, 81470375 to X-DZ and 31501148 to R-LD). We also appreciate Jinhui Zhang (Laboratory of general surgery, Union Hospital, Wuhan, China) for providing assistance with RT-PCR.
Author contributions
W-MP, LW, X-FZ and H-JZ performed and analyzed experiments. HW, J-XZ, W-MP, X-FZ and H-JZ performed statistical analyses. X-DZ and R-LD provided plasmids and reagents. G-LW and PX helped to construct plasmids and stable cell lines. Y-WZ and PH performed histopathologic analysis. HW, R-LD, J-XZ and X-DZ designed the overall research and directed the work. HW, R-LD and X-FZ wrote the manuscript. All authors discussed the results, reviewed and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Wenming Pan, Lin Wang, Xiao-Fei Zhang, Hongji Zhang.
Edited by R. De Maria.
Contributor Information
Run-Lei Du, Phone: +86 27 68752099, Email: runleidu@whu.edu.cn.
Hui Wang, Phone: +86 18995591866, Email: wanghuipitt@hust.edu.cn.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0120-9) contains supplementary material, which is available to authorized users.
References
- 1.Zhai Y, Petrowsky H, Hong JC, Busuttil RW, Kupiec-Weglinski JW. Ischaemia-reperfusion injury in liver transplantation--from bench to bedside. Nat Rev Gastroenterol Hepatol. 2013;10:79–89. doi: 10.1038/nrgastro.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaked A, Ghobrial RM, Merion RM, Shearon TH, Emond JC, Fair JH, et al. Incidence and severity of acute cellular rejection in recipients undergoing adult living donor or deceased donor liver transplantation. Am J Transplant. 2009;9:301–8. doi: 10.1111/j.1600-6143.2008.02487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peralta C, Jimenez-Castro MB, Gracia-Sancho J. Hepatic ischemia and reperfusion injury: effects on the liver sinusoidal milieu. J Hepatol. 2013;59:1094–106. doi: 10.1016/j.jhep.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Shin JK, Kang JW, Lee SM. Enhanced nitric oxide-mediated autophagy contributes to the hepatoprotective effects of ischemic preconditioning during ischemia and reperfusion. Nitric Oxide. 2016;58:10–19. doi: 10.1016/j.niox.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Suyavaran A, Thirunavukkarasu C. Preconditioning methods in the management of hepatic ischemia reperfusion- induced injury: update on molecular and future perspectives. Hepatol Res. 2017;47:31–48. doi: 10.1111/hepr.12706. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897–901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 9.Weiss JB, Eisenhardt SU, Stark GB, Bode C, Moser M, Grundmann S. MicroRNAs in ischemia-reperfusion injury. Am J Cardiovasc Dis. 2012;2:237–47. [PMC free article] [PubMed] [Google Scholar]
- 10.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 11.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 12.Scisciani C, Vossio S, Guerrieri F, Schinzari V, De Iaco R, D’Onorio de Meo P, et al. Transcriptional regulation of miR-224 upregulated in human HCCs by NFkappaB inflammatory pathways. J Hepatol. 2012;56:855–61. doi: 10.1016/j.jhep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Cazanave SC, Mott JL, Elmi NA, Bronk SF, Masuoka HC, Charlton MR, et al. A role for miR-296 in the regulation of lipoapoptosis by targeting PUMA. J Lipid Res. 2011;52:1517–25. doi: 10.1194/jlr.M014654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA. 2012;109:941–6. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang XF, Li KK, Gao L, Li SZ, Chen K, Zhang JB, et al. miR-191 promotes tumorigenesis of human colorectal cancer through targeting C/EBPbeta. Oncotarget. 2015;6:4144–58. doi: 10.18632/oncotarget.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, et al. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 2010;70:8077–87. doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 17.Colamaio M, Borbone E, Russo L, Bianco M, Federico A, Califano D, et al. miR-191 down-regulation plays a role in thyroid follicular tumors through CDK6 targeting. J Clin Endocrinol Metab. 2011;96:E1915–24. doi: 10.1210/jc.2011-0408. [DOI] [PubMed] [Google Scholar]
- 18.Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–9. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagpal N, Ahmad HM, Chameettachal S, Sundar D, Ghosh S, Kulshreshtha R. HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFbeta-signaling in hypoxic microenvironment. Sci Rep. 2015;5:9650. doi: 10.1038/srep09650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagpal N, Ahmad HM, Molparia B, Kulshreshtha R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis. 2013;34:1889–99. doi: 10.1093/carcin/bgt107. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, McRobb LS, Khachigian LM. MicroRNA miR-191 targets the zinc finger transcription factor Egr-1 and suppresses intimal thickening after carotid injury. Int J Cardiol. 2016;212:299–302. doi: 10.1016/j.ijcard.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–6. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 23.Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, et al. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol. 2006;26:2387–98. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol. 2003;160:423–32. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balda MS, Matter K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000;19:2024–33. doi: 10.1093/emboj/19.9.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie M, Balda MS, Matter K. Stress- and Rho-activated ZO-1-associated nucleic acid binding protein binding to p21 mRNA mediates stabilization, translation, and cell survival. Proc Natl Acad Sci USA. 2012;109:10897–902. doi: 10.1073/pnas.1118822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galaris D, Barbouti A, Korantzopoulos P. Oxidative stress in hepatic ischemia-reperfusion injury: the role of antioxidants and iron chelating compounds. Curr Pharm Des. 2006;12:2875–90. doi: 10.2174/138161206777947614. [DOI] [PubMed] [Google Scholar]
- 28.Nagpal N, Kulshreshtha R. miR-191: an emerging player in disease biology. Front Genet. 2014;5:99. doi: 10.3389/fgene.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiezun A, Artzi S, Modai S, Volk N, Isakov O, Shomron N. miRviewer: a multispecies microRNA homologous viewer. BMC Res Notes. 2012;5:92. doi: 10.1186/1756-0500-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farid WR, Pan Q, van der Meer AJ, de Ruiter PE, Ramakrishnaiah V, de Jonge J, et al. Hepatocyte-derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl. 2012;18:290–7. doi: 10.1002/lt.22438. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, et al. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia. 2011;13:841–53. doi: 10.1593/neo.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Degli Esposti D, Sebagh M, Pham P, Reffas M, Pous C, Brenner C, et al. Ischemic preconditioning induces autophagy and limits necrosis in human recipients of fatty liver grafts, decreasing the incidence of rejection episodes. Cell Death Dis. 2011;2:e111. doi: 10.1038/cddis.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yadav SS, Sindram D, Perry DK, Clavien PA. Ischemic preconditioning protects the mouse liver by inhibition of apoptosis through a caspase-dependent pathway. Hepatology. 1999;30:1223–31. doi: 10.1002/hep.510300513. [DOI] [PubMed] [Google Scholar]
- 34.Selzner N, Selzner M, Jochum W, Clavien PA. Ischemic preconditioning protects the steatotic mouse liver against reperfusion injury: an ATP dependent mechanism. J Hepatol. 2003;39:55–61. doi: 10.1016/S0168-8278(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 35.Franco-Gou R, Rosello-Catafau J, Casillas-Ramirez A, Massip-Salcedo M, Rimola A, Calvo N, et al. How ischaemic preconditioning protects small liver grafts. J Pathol. 2006;208:62–73. doi: 10.1002/path.1859. [DOI] [PubMed] [Google Scholar]
- 36.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9:e1003311. doi: 10.1371/journal.pgen.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, et al. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641–9. doi: 10.1158/0008-5472.CAN-10-0527. [DOI] [PubMed] [Google Scholar]
- 39.Huang H, Evankovich J, Yan W, Nace G, Zhang L, Ross M, et al. Endogenous histones function as alarmins in sterile inflammatory liver injury through Toll-like receptor 9 in mice. Hepatology. 2011;54:999–1008. doi: 10.1002/hep.24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Wang G, Zhang L, Zhang J, Zhang J, Wang Q, et al. ADAR1 suppresses the activation of cytosolic RNA-sensing signaling pathways to protect the liver from ischemia/reperfusion injury. Sci Rep. 2016;6:20248. doi: 10.1038/srep20248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patnaik SK, Kannisto E, Yendamuri S. Overexpression of microRNA miR-30a or miR-191 in A549 lung cancer or BEAS-2B normal lung cell lines does not alter phenotype. PLoS ONE. 2010;5:e9219. doi: 10.1371/journal.pone.0009219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H, Nace GW, McDonald KA, Tai S, Klune JR, Rosborough BR, et al. Hepatocyte-specific high-mobility group box 1 deletion worsens the injury in liver ischemia/reperfusion: a role for intracellular high-mobility group box 1 in cellular protection. Hepatology. 2014;59:1984–97. doi: 10.1002/hep.26976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.