Abstract
Mucolipidosis II α/β, mucolipidosis III α/β, and mucolipidosis III γ are autosomal recessive disorders belonging to the family of lysosomal storage disorders caused by deficiency of the UDP-N-acetylglucosamine, a lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc-phosphotransferase) localized in the Golgi apparatus, which is essential for normal processing and packaging of soluble lysosomal enzymes with initiating the first step of tagging lysosomal enzymes with mannose-6-phosphate (M6P). Mucolipidosis II and III are caused by mutations in the GNPTAB and GNPTG genes, and patients with these diseases are characterized by short stature, skeletal abnormalities, and developmental delay. In this study we report 38 patients with mucolipidosis II and III enrolled in Eastern China during the past 8 years. The diagnosis was made based on clinical characteristics and measurement of plasma lysosomal enzyme activity. Sanger sequencing of GNPTAB and/or GNPTG for all patients and real-time quantitative PCR were performed to confirm the diagnosis. In addition, 11 cases of prenatal mucolipidosis II were diagnosed based on measurement of the enzyme activity in amniotic fluid supernatant and genetic testing of cultured amniotic cells. Based on molecular genetic tests, 30 patients were diagnosed with mucolipidosis II α/β, 6 were diagnosed with III α/β and 2 were diagnosed with III γ. Thirty-seven different GNPTAB gene mutations were identified in 29 patients with mucolipidosis II α/β and six patients with III α/β. These mutations included 22 new mutations (p.W44X, p.E279X, p.W416X, p.W463X, p.Q802X, p.Q882X, p.A34P, p.R334P, p.D408N, p.D534N, p.Y997C, p.D1018V, p.L1025S, p.L1033P, c.88_89delAC, c.890_891insT, c.1150_1151insTTA, c.1523delG, c.2473_2474insA, c.2980_2983delGCCT, c.3094delA, and deletion of exon 9). Four new GNPTG gene mutations were identified (c.13delC, p.Y81X, p.G126R and c.609+1delG) in two mucolipidosis III γ patients. Among the 11 cases of prenatal diagnosis, four were mucolipidosis II fetuses, three were heterozygous, and the remaining four were normal fetuses. This study expands the mutation spectrum of the GNPTAB and GNPTG genes and contributes to specific knowledge of mucolipidosis II/III in a population from Eastern China.
Keywords: mucolipidosis, lysosomal enzymes, genotype, GNPTAB, GNPTG, gene mutation, phenotype, prenatal diagnosis
Introduction
Mucolipidosis II α/β (ML II originally; MIM 252500, which was later renamed ML II α/β by Cathey et al.) [1], mucolipidosis III α/β (ML III α/β; MIM 252600) and mucolipidosis III γ (ML III γ; MIM 252605) are autosomal recessive disorders belonging to the family of lysosomal storage disorders caused by deficiency of the uridine diphosphate (UDP)-N-acetylglucosamine: lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc-phosphotransferase; EC 2.7.8.17), which is localized in the Golgi apparatus and is essential for normal processing and packaging of soluble lysosomal enzymes by initiating the first step of tagging lysosomal enzymes with mannose-6-phosphate (M6P). GlcNAc-phosphotransferase comprises three different subunits, α, β, and γ, in a hexameric α2β2γ2 subunit complex [2, 3]. These three subunits are the products of two genes, GNPTAB (MIM:607840), which encodes the α- and β-subunits [4–6], and GNPTG (MIM:607838), which encodes the γ-subunit [7, 8]. ML II α/β and ML III α/β are caused by a homozygous or compound heterozygous mutation in the GNPTAB gene [9–13], whereas ML III γ results from mutations in the GNPTG gene [14–17].
ML II α/β and III are both rare diseases, and patients share a number of physical characteristics, such as coarse face, short stature, progressive joint stiffness, skeletal abnormalities, cardiomegaly, and developmental delay [18]. In addition, these two disorders have unique features. ML II is more severe than ML III, with onset in infancy or early childhood leading to death during the first decade of life [19–21]. ML III has a relatively mild phenotype, a later onset of clinical symptoms, which occur at approximately 3 years of age, and a more slowly progressing and prolonged course, and thus permits survival into adulthood [22–24].
α-Mannosidase and β-hexosaminidase A+B (Hex A+B) are transported to lysosomes in an M6P-dependent pathway. Their activities are deficient in lysosomes; however, they are dramatically elevated in plasma and other extracellular fluids from patients with ML II or III when measured by artificial fluorescent substrates [13]. In the past 8 years, 38 patients with ML II or III were diagnosed by clinical characteristics and elevated α-mannosidase and Hex A+B activities in the plasma in our hospital, which is a diagnostic center in Eastern China. Here, we report the clinical, biochemical, and molecular characteristics of the 38 probands and those of 11 cases of prenatal diagnosis from Eastern China. Our results expand the mutation spectrum of the GNPTAB and GNPTG genes and contribute to specific knowledge of mucolipidosis II/III in a population from Eastern China.
Materials and methods
Subjects
All ML II/III patients were from unrelated families in Eastern China, and their parents had no perspicuous consanguinity. Among them, 11 ML II families underwent prenatal diagnosis. We collected blood samples from all patients after obtaining informed consent from their guardians, and all the high-risk pregnancy patients with prenatal diagnosis signed a written informed consent. All procedures were approved by the local committee on human experimentation in Shanghai Xinhua Hospital (approval no. XHEC-D-2014-006) and were in accordance with the Helsinki Declaration of 1975, as revised in 2000. The clinical diagnosis of ML II/III was based on typical clinical, radiographic, and biochemical tests [18]. Biochemical criteria included an at least tenfold increase in plasma lysosomal enzyme levels (α-mannosidase and Hex A+B activities).
Lysosomal enzyme analysis for the probands
Plasma was isolated from fresh peripheral blood and stored at −80 °C until analysis. The α-mannosidase activity was measured by incubating plasma with 2 mM 4-methylumbelliferyl-α-D-mannopyranoside (Sigma) substrate solution, pH 4.25, at 37 °C for 1 h, as described previously with minor modification [25, 26]. Hex A+B determination was also based on a method described in the literature [27, 28]. Plasma was tenfold diluted with McIlvaine buffer, pH 4.4, containing 0.6% bovine serum albumin and then incubated with 3 mM 4-methylumbelliferyl-N-acetyl-β-D-glucosaminide (Glycosynth) for 30 min at 37 °C. Both reactions were stopped by 0.17 M glycine/NaOH buffer, pH 10.5. A 4-methylumbelliferyl standard was used. Fluorescence signals were measured with a fluorimeter at an excitation wavelength of 365 nm and an emission wavelength of 450 nm.
Lysosomal enzyme analysis for prenatal diagnosis
Prenatal diagnoses of ML II/III disease were achieved by examining enzyme activity in the amniotic supernatant fluid [29]. Amniotic fluid was obtained by amniocentesis at approximately 16 weeks gestation. The collected amniotic fluid was centrifuged at 2000 × g to isolate the supernatant, and the enzyme activities of the supernatant were detected immediately. The α-mannosidase activity detection method for amniotic fluid was the same as that used for plasma.
Sanger sequencing of genomic DNA
For the ML II/III probands, elevation of the levels of both α-mannosidase and Hex A+B activities was a prerequisite for molecular characterization of the GNPTAB or GNPTG genes. Genomic DNA in peripheral blood from patients was extracted using a Lab-Aid blood DNA isolation Kit (Zhishan Biotech) according to the manufacturer’s protocol. The 21 exons and flanking regions of GNPTAB and the 11 exons and flanking regions of GNPTG were amplified via polymerase chain reaction and Sanger sequencing. The obtained sequences were compared with the reference genes GNPTAB (NM_024312.4) and GNPTG (NM_032520.4) to identify nucleotide variations.
For the prenatal diagnoses, genomic DNA was extracted from cultured amniotic cells using a QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s protocol, and then, Sanger sequencing was used to detect the mutation sites of the probands.
Real-time quantitative PCR
One patient (P8) was suspected of absence of exon 9 in the GNPTAB gene because of the presence of a seeming homogenous variation with only a uniparental origin based on Sanger sequencing. The verification was conducted through real-time quantitative PCR with ALB (MIM: 103600) as the reference gene and mixed normal human DNA as the normal reference. Real-time PCR samples with SYBR® Premix Ex Taq™ (TaKaRa, Dalian, China) were prepared according to the manufacturer’s protocol and were run on an ABI7500 instrument.
Results
Enzyme activity determination of probands
Plasma from 473 healthy individuals was analyzed for α-mannosidase and Hex A+B enzyme activities to determine the reference range according to the 99% confidence interval: α-mannosidase activity reference range, <36.5 nmol/h · mL; Hex A+B activity reference range, <1970 nmol/h · mL. The α-mannosidase and Hex A+B activities were remarkably elevated in the available plasma from the 38 ML II/III patients (Table 1). The mean α-mannosidase activity of the ML patients was 423.9 nmol/h · mL, which reflected an ≥29-fold increase compared with the reference group (mean, 14.2 nmol/h · mL); the mean Hex A+B activity was 14,420 nmol/h · mL, an increase of ≥11-fold compared with the reference group (mean, 1242 nmol/h · mL).
Table 1.
Phenotypes and genotypes of 38 Eastern Chinese individuals with mucolipidosis II/III
| P | ML type | Gender | Age of onset | Age at diagnosis | First symptom | Defected gene | Genotypes | Amino acid changes | Location | α-mannosidase activity | Hex A+B activity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | II α/β | M | 6 m | 7 m | The lower-limb exercise less | GNPTAB | c.1090C>T+ | p.R364X+ | E9+E15 | 475 | 16,107 |
| c.2990A>G* | p.Y997C* | ||||||||||
| 2 | II α/β | F | NK | 20 m | ND | GNPTAB | c.2473_2474insA* | p.T825NfsX37* | E13+E13 | 503.5 | 17,116 |
| (Homozygous) | +p.T825NfsX37* | ||||||||||
| 3 | II α/β | M | 1 y | 4 y 11 m | Short stature, mental retardation, walking instability | GNPTAB | c.88_89delAC*+c.835G>T* | p.T30HfsX24*+p.E279X* | E1+E8 | 466.8 | 13,782 |
| 4 | II α/β | M | 1 y 6 m | 3 y | The finger joint stiff | GNPTAB | c.100G>C*+ | p.A34P*+ | E1+E2 | 422 | 14,945 |
| c.136C>T | p.R46X | ||||||||||
| 5 | II α/β | F | 3 m | 6 m | Knee joint stiffness | GNPTAB | c.1090C>T+ | p.R364X+ | E9+E15 | 499.6 | 18,153 |
| c.3091C>T | p.R1031X | ||||||||||
| 6 | II α/β | M | NK | 9 m | Coarse facial features | GNPTAB | c.99delC+ | p.A34PfsX49+ | E1+E13 | 390.4 | 13,735 |
| c.2533C>T | p.Q845X | ||||||||||
| 7 | II α/β | F | After birth | 7 m | Growth retardation, mental retardation | GNPTAB | c.136C>T+ | p.R46X+ | E2+E9 | 405.9 | 12,366 |
| c.1090C>T | p.R364X | ||||||||||
| 8 | II α/β | M | 8 m | 16 m | Pneumonia | GNPTAB | c.1000C>T+ | p.R334X+ | E9+E9 | 418.2 | 11,280 |
| del E9* | del E9* | ||||||||||
| 9 | II α/β | M | After birth | 4 y 2 m | Skeletal malformation | GNPTAB | c.1120T>C+ | p.F374L+ | E10+E12 | 248.9 | 15,513 |
| c.1600G>A* | p.D534N* | ||||||||||
| 10 | II α/β | F | NK | 1 y | ND | GNPTAB | c.3136-2A>G+ | IVS15-2A>G+ | IVS15+IVS15 | 541.2 | 13,273 |
| (Homozygous) | IVS15-2A>G | ||||||||||
| 11 | II α/β | M | 6 m | 3 y 5 m | Developmental retardation | GNPTAB | c.3094delA*+ | p.T1032HfsX11*+ | E15+E15 | 464 | 17,215 |
| c.3098T>C* | p.L1033P* | ||||||||||
| 12 | II α/β | F | NK | 15 m | Bone disease | GNPTAB | c.1090C>T | p.R364X+ | E9+E9 | 416.3 | 11,754 |
| (Homozygous) | p.R364X | ||||||||||
| 13 | II α/β | F | After birth | 14 m | Weak cry, eating less | GNPTAB | c.2404C>T*+ | p.Q802X*+ | E13+E13 | 284.2 | 13,772 |
| c.2455G>T | p.E819X | ||||||||||
| 14 | II α/β | M | NK | 3 y | Backward development | GNPTAB | c.2404C>T*+ | p.Q802X*+ | E13+E13 | 428.1 | 13,049 |
| c.2644C>T* | p.Q882X* | ||||||||||
| 15 | II α/β | F | 4 m | 4 y | Coarse facial features | GNPTAB | c.1001G>C*+ | p.R334P*+ | E9+E19 | 520.7 | 18,701 |
| c.3565C>T | p.R1189X | ||||||||||
| 16 | II α/β | M | NK | 9 m | Developmental retardation | GNPTAB | c.2455G>T+ | p.E819X+ | E13+E20 | 310.8 | 12,410 |
| c.3613C>T | p.R1205X | ||||||||||
| 17 | II α/β | M | NK | 20 m | Slow growth, mental retardation | GNPTAB | c.1090C>T+ | p.R364X+ | E9+E13 | 484.6 | 13,706 |
| c.2533C>T | p.Q845X | ||||||||||
| 18 | II α/β | F | NK | 4 y | Short stature, skeletal deformity | GNPTAB | c.835G>T*+ | p.E279X*+ | E8+E9 | 511 | 18,082 |
| c.1090C>T | p.R364X | ||||||||||
| 19 | II α/β | M | NK | 6 m | Developmental retardation, scoliosis | GNPTAB | c.1090C>T+ | p.R364X+ | E9+E19 | 419 | 17,370 |
| c.3565C>T | p.R1189X | ||||||||||
| 20 | II α/β | M | 8 m | 3 y | Coarse facial features, skeletal deformity | GNPTAB | c.1090C>T+ | p.R364X+ | E9+E10 | 512.7 | 11,143 |
| c.1222G>A* | p.D408N* | ||||||||||
| 21 | II α/β | F | After birth | 9 m | Coarse facial features, mental retardation | GNPTAB | c.1090C>T | p.R364X+ | E9+E9 | 432 | ND |
| (Homozygous) | p.R364X | ||||||||||
| 22 | II α/β | M | 3 m | 8 m | Developmental retardation | GNPTAB | c.1150_1151insTTA*+ | p.F383_S384insI*+ | E10+E11 | 404.5 | 15,558 |
| c.1389G>A* | p.W463×* | ||||||||||
| 23 | II α/β | M | NK | 5 m | Developmental retardation | GNPTAB | c.1090C>T+ | p.R364X+ | E9+E15 | 399.4 | 18,749 |
| c.3094delA* | p.T1032HfsX11* | ||||||||||
| 24 | II α/β | F | NK | 10 m | Suspected mucopolysaccharide disease | GNPTAB | c.132G>A*+ | p.W44×*+ | E2+E13 | ND | ND |
| c.2693dupA | p.Y899VfsX21 | ||||||||||
| 25 | II α/β | M | NK | 5 m | Long-term neonatal recurrent pneumonia | GNPTAB | c.132G>A*+ | p.W44×*+ | E2+E15 | 317.9 | ND |
| c.3094delA* | p.T1032HfsX11* | ||||||||||
| 26 | II α/β | M | NK | 2 y 6 m | Developmental retardation | GNPTAB | c.1001G>C*+ | p.R334P *+ | E9+E10 | 157.9 | ND |
| c.1247G>A* | W416X* | ||||||||||
| 27 | II α/β | F | NK | 5 y | Coarse facial features, Skeletal dysplasia | GNPTAB | c.648_651delAGAA+ | p. E217Sfs*4+ | E7+IVS15 | ND | ND |
| c.3136-2A>G | IVS15-2A>G | ||||||||||
| 28 | II α/β | F | 6 m | 15 m | Kyphosis, cyclic vomiting | GNPTAB | c.1523delG*+NK | p.G508DfsX39*+NK | E12+NK | 395.1 | 15,252 |
| 29 | II α/β | M | NK | 2 y | Coarse facial features, developmental retardation | GNPTAB | c.1001G>A+NK | p. R334Q+NK | E9+NK | 531.1 | 12,544 |
| 30 | II α/β | F | 3 m | 5 m | Unable to raise head | NK | NK | NK | NK | 359.8 | 13,778 |
| 31 | III α/β | F | NK | 6 y 9 m | ND | GNPTAB | c.2715+1G>A+ | IVS13+1G>A+p. | IVS13+E15 | 486.4 | 16,909 |
| c.2980_2983delGCCT* | A994SfsX8* | ||||||||||
| 32 | III α/β | M | 2 y | 7 y | Back prominent | GNPTAB | c.1090C>T+ | p.R364X+ | E9+IVS13 | 460.9 | 15,414 |
| c.2715+1G>A | IVS13+1G>A | ||||||||||
| 33 | III α/β | F | 4 y | 8y2m | Short, prominent forehead | GNPTAB | c.890_891insT*+ | p.298KfsX16*+ | E8+E15 | 489.5 | 14,478 |
| c.3053A>T* | p.D1018V* | ||||||||||
| 34 | III α/β | M | 6 y | 9y6m | Finger flexion deformity, abnormal heart | GNPTAB | c.2715+1G>A+ | IVS13+1G>A+ | IVS13+E15 | 213.9 | 10,921 |
| c.3094delA* | p.T1032HfsX11* | ||||||||||
| 35 | III α/β | M | NK | 17 y | Skeletal dysplasia | GNPTAB | c.2715+1G>A+ | IVS13+1G>A+ | IVS13+E15 | 471.2 | ND |
| c.3074T>C* | p.L1025S* | ||||||||||
| 36 | III α/β | M | NK | 14 y | Pectus carinatum, finger stiffness | GNPTAB | c.99delC+ND | p. A34PfsX49+NK | E1+NK | 382.6 | 11,346 |
| 37 | III γ | M | NK | 10 y | Abnormal flexion of wrist | GNPTG | c.13delC*+ | p.L5WfsX23*+ | E1+IVS8 | 479.5 | 9622 |
| c. 609+1delG* | IVS8+1deG* | ||||||||||
| 38 | III γ | M | 8 y | 10 y | Scoliosis | GNPTG | c.243T>A*+ | p.Y81X*+ | E5+E6 | 467.1 | 7826 |
| c.376G>C* | G126R* |
Data summary of 38 mucolipidosis II/III Chinese patients
Note: P indicated patient number, NK indicated unknown; ND indicated not done,* indicated unreported mutation, E indicated exon, the unit of enzyme activity is nmol/h · mL. Reference range: α-mannosidase activity <36.5 nmol/h· mL, Hex A+B activity <1970 nmol/h · mL
The enzyme activity values of the 30 ML II patients and 8 ML III patients were analyzed with SPSS 17.0 statistical software. An independent sample t-test was utilized, and a P value of less than 0.05 was considered statistically significant. There was no significant difference in plasma α-mannosidase activity between ML II and ML III patients (the average activity was 421.8 nmol/h · mL and 431.4 nmol/h·mL, respectively; P = 0.796), but the Hex A+B activity in the two groups was significantly different (the average activity was 14,975 nmol/h · mL and 12,359 nmol/h·mL, respectively; P = 0.031). The plasma Hex A+B activity of ML II patients was significantly higher than that of ML III patients, which supported our understanding that ML II has a more severe disease course than ML III.
Enzyme activity determination for prenatal diagnosis
We detected the α-mannosidase enzyme activity in the amniotic fluid supernatant from 32 cases of high-risk pregnancy with organic acidemias or amino acid metabolic diseases to determine the reference range according to the 99% confidence interval, which was <9.3 nmol/h · mL.
Prenatal diagnosis was performed for 11 ML II high-risk pregnant women. The α-mannosidase activity in the amniotic supernatant fluid from four cases was significantly higher than the reference range, which we concluded to indicate an affected fetus. The activity in the amniotic fluid from the seven remaining cases was within the normal range, and these women were considered to be carrying unaffected fetuses (Table 2).
Table 2.
Prenatal diagnosis of 11 families with mucolipidosis II
| Case no | ML type | Genotypes | Amino acid changes | Location | α-mannosidase activity |
|---|---|---|---|---|---|
| P3 | II α/β | c.88_89 del AC+ c.835G>T | T30Hfs*24+ E279X | E1+E8 | 72.5 |
| P5 | II α/β | c.1090C>T | R364X | E9 | 6.4 |
| P7 | II α/β | N | N | 7.4 | |
| P8 | II α/β | c.1000C>T+ del E9 | R334X+ del E9 | E9+E9 | 72.3 |
| P12 | II α/β | c.1090C>T (Homozygous) | R364X+ R364X | E9+E9 | 127.2 |
| P13 | II α/β | c.2455G>T | E819X | E13 | 6.6 |
| P18 | II α/β | N | N | 8.3 | |
| P20 | II α/β | N | N | 7.3 | |
| P26 | II α/β | c.1001G>C | R334P | E9 | 7.4 |
| P27 | II α/β | c.648_651delAGAA+ c.3136-2A>G | p. E217Sfs*4+ IVS15-2A>G | E7+IVS15 | 83.8 |
| P29 | II α/β | N | N | 7.9 |
Data summary of 11 mucolipidosis II prenatal diagnosis
Note: P indicated patient number, N indicated no mutations were detected, E indicated exon, the unit of enzyme activity is nmol/h · mL. Reference range: α-mannosidase activity <9.3 nmol/h · mL
Patients with ML II α/β
The ML II α/β group included 30 patients with an average age at onset of 5 months old. Two patients had a positive family history, with an elder brother or sister having similar symptoms. However, only ten patients had been diagnosed before the age of 1 year. The average age at diagnosis was 1 year and 9 months old. All these patients had unanimous typical characteristics, e.g., coarse facial features, gingival hypertrophy, short stature, mental retardation, skeletal deformity, joint stiffness, and hepatomegaly. Four of them had documented hearing loss at diagnosis. It should be mentioned that one patient (P28) initially presented with cyclic vomiting from the age of 9 months with interval periods of 13–29 days, which had not been reported to be associated with ML II α/β. She had been diagnosed with ML II α/β at the age of 15 months.
GNPTAB gene in ML II α/β
Biallelic nucleotide variations of the GNPTAB gene were identified in 27 of 30 patients (P1–P27) with Sanger sequencing (Table 1). A total of 14 reported mutations and 18 novel variations were detected. In two patients (P28 and P29), only one mutant allele could be identified. For patient 30, who had typical clinical manifestations and a positive family history, with an elder sister having similar symptoms and death at 3 years old, ML II α/β was diagnosed based on elevated plasma α-mannosidase and Hex A+B activities. However, no mutation was detected in the GNPTAB gene or the GNPTG gene by Sanger sequencing.
Patients with ML III
The clinical manifestations of eight patients with ML III were relatively mild, with an average age of onset at 4 years old and an average age of diagnosis at 10 years and 3 months old. As reported, ML III α/β and ML III γ are clinically indistinguishable [6]. The intelligence of the patients was normal or slightly lagged, and their stature was short compared with the mid-height of their parents. We first conducted GNPTAB gene testing. If no mutation was identified in the GNPTAB gene, then the GNPTG gene was further tested.
GNPTAB gene in ML III α/β
Six patients were categorized as ML III α/β because variations were identified in the GNPTAB gene. Both mutant alleles had been identified in five patients (P31–P35), but only one mutant allele was detected in patient 36. In total, eight different variations were identified, with three reported and five novel mutations. Notably, four patients carried the same reported mutation: c.2715+1G>A.
GNPTG gene in ML III γ
Two patients (P37 and P38) possessed compound heterozygous mutations of the GNPTG gene, with all of the mutant alleles being novel, and thus were categorized as ML III γ.
Prenatal gene diagnosis
Among the 11 cases of prenatal gene testing, four fetuses with remarkably elevated α-mannosidase activity in the amniotic supernatant fluid carried the same GNPTAB mutations as the respective probands. Additionally, three fetuses were heterozygous, and no mutations were detected in the remaining four fetuses (Table 2).
Characterization of mutations
In summary, we detected 37 different GNPTAB gene variations in 67 mutant alleles out of 72 alleles in 36 ML II and III α/β patients. The distribution of all the detected mutations is shown in Fig. 1a. Fifteen mutations were listed in the HGMD database or reported in the literature, c.99delC [30], p.R46X, c.648_651del, p.R334X, p.R334Q, p.R364X, p.F374L, p.E819X [30], p.Q845X, c.2693dupA, p.R1031X, p.R1189X, p.R1205X, c.2715+1G>A, c.3136-2A>G [30].
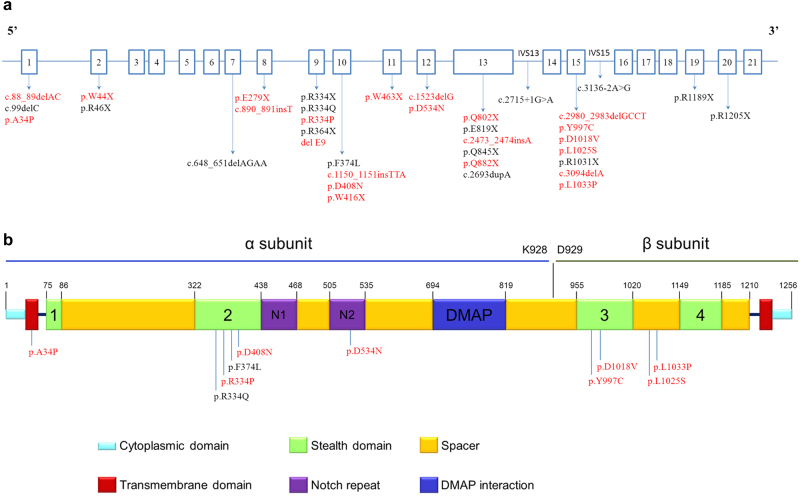
Fig. 1.
a Distribution of GNPTAB gene mutations in 21 exons from 36 Eastern Chinese patients with ML II/III. In total, 37 different mutations were identified in the GNPTAB gene. The 15 previously reported mutations are in black, and the 22 novel mutations are marked in red. b Distribution of 10 missense mutations in the α/β-subunit precursor. The two previously reported mutations are in black, and the eight novel mutations are marked in red
Twenty-two mutations are novel according to the HGMD mutation database (http://www.hgmd.cf.ac.uk). These included six new termination mutations: p.W44X, p.E279X, p.W416X, p.W463X, p.Q802X, and p.Q882X, which were predicted as “disease causing” by Mutationtaster (http://www.mutationtaster.org/).
For the eight new missense mutations, p.A34P, p.R334P, p.D408N, p.D534N, p.Y997C, p.D1018V, p.L1025S, and p.L1033P, PROVEAN, SIFT (http://sift.jcvi.org), and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) were applied to evaluate their pathogenicity potential. The majority of the prediction results were “Deleterious” in PROVEAN, “Damaging” in SIFT, and “probably damaging” in PolyPhen-2, supporting the notion that these variations were the causative mutations. The only exception was p.A34P, which was predicted to be “Neutral” by PROVEAN but was “Damaging” according to SIFT and “probably damaging” according to PolyPhen-2. None of these eight missense mutations were found in ExAC nor in 1000G except the p.D534N mutation, which was present twice in ExAC.
Seven new exonic small deletions/duplications, c.88_89delAC, c.890_891insT, c.1150_1151insTTA, c.1523delG, c.2473_2474insA, c.2980_2983delGCCT, and c.3094delA, were predicted as “disease causing” by Mutationtaster. Additionally, the absence of the entirety of exon 9 had not been reported.
Figure 1a shows that there are 24 mutations in four exons (E9, E10, E13, and E15) or in their exon–intron junctions of the GNPTAB gene. The mutations in these regions account for the majority of 37 mutations (24/37 = 64.9%). The allele frequency of these 24 mutations was 68% (49/72) in the analyzed alleles of these ML II α/β and III α/β patients. The most frequent GNPTAB mutation was the nonsense mutation c.1090C>T (p.R364X), accounting for 18% (13/72) of the analyzed alleles detected. Eleven patients carried this mutation, with two of them harboring it homozygously (P12 and P21).
Figure 1b shows the distribution of the ten missense mutations identified, including the eight novel and two reported mutations, in the α/β-subunit precursor. The α/β-subunit precursor contains two transmembrane domains and three identifiable domains separated by spacer regions: the Stealth domain, two Notch repeat modules, and a DNA methyltransferase-associated protein (DMAP) interaction domain [13, 31]. The ten mutations in this study were distributed as follows: one mutation in the transmembrane domain of the N terminus, six mutations in the Stealth region, one mutation in the second Notch module, and two mutations in the spacer region. Consistent with a previous report, most of the missense mutations were located in the Stealth region [31].
GNPTG gene analysis showed four novel mutations, c.13delC, p.Y81X, p.G126R, and c.609+1delG. All the prediction results for these four mutations supported their pathogenicity.
Discussion
Patients with mucolipidosis II/III have clinical features similar to those of patients with mucopolysaccharidosis (MPS), which is caused by a deficiency in the metabolism of one or more types of mucopolysaccharides or glycosaminoglycans. Both ML and MPS are lysosomal storage disorders and show a number of similar physical characteristics, such as coarse facial features, short stature, skeletal deformity, hepatosplenomegaly, and a reduced life expectancy [32]. Lysosomal enzyme activities of leucocytes or plasma provide a differential diagnosis for ML II/III and MPS, while gene testing helps provide a definitive diagnosis.
In our cohort of ML II α/β and ML III α/β patients, p.R364X was the most frequent mutation. Unlike our results, Liu et al. found that the most frequent mutation among their patient cohort was the splicing mutation c.2715+1G>A (allele frequency 9/32 = 28%), which was identified in both ML II α/β (three patients) and ML III α/β (six patients), with the secondary frequent mutations being p.R364X (allele frequency 4/32 = 13%) in their cohort of patients that represented the northern populations of China [30]. The c.2715+1G>A mutation was detected in only four patients with ML III α/β (P31, P32, P34, and P35) in our cohort that represented a population from Eastern China. Compared with their results, we found that the frequent mutations in populations from Northern and Eastern China differ. All the reported mutations of the GNPTAB gene in 54 Chinese patients are summarized in Table 3, including 18 cases from the literature [30, 33] and our 36 cases. p.R364X is still a frequent mutation throughout the Chinese population. The nonsense mutation c.3565C>T (p.R1189X) was reported to be the most frequent mutation in 40 Japanese patients (allele frequency 33/80 = 41%) [34] and in 13 Korean patients (allele frequency 5/26 = 19.2%) [9, 20, 35]. This mutation was detected twice in our patients (P15 and P19). However, it was not detected among the northern Chinese population.
Table 3.
Review of GNPTAB mutations in 54 Chinese patients with ML II/III
| Mutation type | Exon | Nucleotide change | Amino acid change | No. of alleles | Allele frequency | Reference | Geographic location |
|---|---|---|---|---|---|---|---|
| Missense | E1 | c.100G>C | p.A34P | 1 | 0.93% (1/108) | This study | East China |
| Missense | E9 | c.1001G>A | p.R334Q | 1 | 0.93% (1/108) | [18] | East China |
| Missense | E9 | c.1001G>C | p.R334P | 2 | 1.85% (2/108) | This study | East China |
| Missense | E10 | c.1120T>C | p.F374L | 1 | 0.93% (1/108) | [34] | East China |
| Missense | E10 | c.1222G>A | p.D408N | 1 | 0.93% (1/108) | This study | East China |
| Missense | E12 | c.1600G>A | p.D534N | 1 | 0.93% (1/108) | This study | East China |
| Missense | E13 | c.2345C>T | p.S782F | 3 | 2.78% (3/108) | [30] | North China |
| Missense | E15 | c.2990A>G | p.Y997C | 1 | 0.93% (1/108) | This study | East China |
| Missense | E15 | c.3053A>T | p.D1018V | 1 | 0.93% (1/108) | This study | East China |
| Missense | E15 | c.3074T>C | p.L1025S | 1 | 0.93% (1/108) | This study | East China |
| Missense | E15 | c.3098T>C | p.L1033P | 1 | 0.93% (1/108) | This study | East China |
| Nonsense | E2 | c.132G>A | p.W44× | 2 | 1.85% (2/108) | This study | East China |
| Nonsense | E2 | c.136C>T | p.R46X | 2 | 1.85% (2/108) | [18] | East China |
| Nonsense | E8 | c.835G>T | p.E279X | 2 | 1.85% (2/108) | This study | East China |
| Nonsense | E9 | c.1000C>T | p.R334X | 1 | 0.93% (1/108) | [18] | East China |
| Nonsense | E9 | c.1071G>A | p.W357× | 3 | 2.78% (3/108) | [33] | North China |
| Nonsense | E9 | c.1090C>T | p.R364X | 19 | 17.59% (19/108) | [18] | East and North China |
| Nonsense | E10 | c.1212C>G | p.Y404X | 2 | 1.85% (2/108) | [30] | North China |
| Nonsense | E10 | c.1247G>A | p.W416× | 1 | 0.93% (1/108) | This study | East China |
| Nonsense | E11 | c.1389G>A | p.W463× | 1 | 0.93% (1/108) | This study | East China |
| Nonsense | E13 | c.2213C>A | p.S740X | 1 | 0.93% (1/108) | [30] | North China |
| Nonsense | E13 | c.2356C>T | p.Q786X | 1 | 0.93% (1/108) | [30] | North China |
| Nonsense | E13 | c.2404C>T | p.Q802X | 2 | 1.85% (2/108) | This study | East China |
| Nonsense | E13 | c.2455G>T | p.E819X | 4 | 3.70% (4/108) | [30] | East and North China |
| Nonsense | E13 | c.2533C>T | p.Q845X | 2 | 1.85% (2/108) | [11] | East China |
| Nonsense | E13 | c.2644C>T | p.Q882X | 1 | 0.93% (1/108) | This study | East China |
| Nonsense | E15 | c.3091C>T | p.R1031X | 1 | 0.93% (1/108) | [18] | East China |
| Nonsense | E19 | c.3565C>T | p.R1189X | 2 | 1.85% (2/108) | [9] | East China |
| Nonsense | E20 | c.3613C>T | p.R1205X | 2 | 1.85% (2/108) | [11] | East and North China |
| Frameshift | E1 | c.88_89delAC | p.T30Hfs*24 | 1 | 0.93% (1/108) | This study | East China |
| Frameshift | E1 | c.99delC | p.A34Pfs*49 | 3 | 2.78% (3/108) | [30] | East and North China |
| Frameshift | E5 | c.523_524delAAinsG | p.N175Afs38X | 1 | 0.93% (1/108) | [30] | North China |
| Frameshift | E7 | c.648_651delAGAA | p.E217Sfs*4 | 1 | 0.93% (1/108) | [18] | East China |
| Frameshift | E8 | c.890_891insT | p.S298Kfs*16 | 1 | 0.93% (1/108) | This study | East China |
| Frameshift | E10 | c.1150_1151insTTA | p.-384I | 1 | 0.93% (1/108) | This study | East China |
| Frameshift | E12 | c.1523delG | p.G508Dfs*39 | 1 | 0.93% (1/108) | This study | East China |
| Frameshift | E13 | c.2473_2474insA | p.T825Nfs*37 | 2 | 1.85% (2/108) | This study | East China |
| Frameshift | E13 | c.2550_2554delGAAAA | p.K850Nfs10X | 1 | 0.93% (1/108) | [13] | North China |
| Frameshift | E13 | c.2693dupA | p.Y899Vfs*21 | 1 | 0.93% (1/108) | [18] | East China |
| Frameshift | E14 | c.2821dupA | p.I941Nfs4X | 1 | 0.93% (1/108) | [30] | North China |
| Frameshift | E15 | c.2980_2983delGCCT | p.A994Sfs*8 | 1 | 0.93% (1/108) | This study | East China |
| Frameshift | E15 | c.3094delA | p.T1032Hfs*11 | 4 | 3.70% (4/108) | This study | East China |
| Splicing | IVS1 | c.118-1G>A | Splicing | 1 | 0.93% (1/108) | [30] | North China |
| Splicing | IVS13 | c.2715+1G>A | Splicing | 13 | 12.04% (13/108) | [9] | East and North China |
| Splicing | IVS15 | c.3136-2A>G | Splicing | 4 | 3.70% (4/108) | [30] | East and North China |
| Deletion | E9 | delE9 | 1 | 0.93% (1/108) | This study | East China |
Note: Bold letters indicate number of alleles >1
The mutation c.3503_3504delTC is a reported high frequency mutation. Tappino et al. reported that its allele frequency is 51% (47/92) in their patients from four countries (Italy, Argentina, Bangladesh, and Hungary) [13]. The frequency of this allele in American patients is 22% (27/122) [18] and is 45% (9/20) in Portuguese patients [23]. However, it was not detected in Chinese patients. Thus, patients in different ethnic backgrounds carry different frequent mutations.
The reported p.R334X mutation of the GNPTAB gene was found to be homozygous in patient 8. However, it was identified only in the paternal allele. His mother did not carry this allele. We argue that a genomic deletion in this region could explain this phenomenon. The real-time quantitative PCR results confirmed that this patient and his mother had only one copy of exon 9. This patient inherited p.R334X from his father and the deletion of exon 9 from his mother. Thus, it is essential to determine the genotype of parents to identify the pathogenic mutation of a patient.
ML II α/β is defined by two severe mutations, whereas ML III α/β is defined by at least one attenuated mutation that can produce a small amount of active GlcNAc-phosphotransferase [5]. The p.R334X mutant was reported to be associated with ML III α/β in the HGMD database, but this ML III α/β patient also carried another missense mutation, p.D190V [18]. Kudo et al. found that the substitution of valine for aspartic acid at position 190 produced 86% of GlcNAc-phosphotransferase activity and did not significantly affect the enzyme activity [5]. Therefore, it was presumed that p.D190V was an attenuated mutation. In our study, patient 8 with p.R334X had the clinical characteristics of ML II α/β, with early onset at 8 months old. Thus, both p.R334X and deletion of exon 9 were presumed to be severe mutations.
Cathey et al. reported previously that the mutation p.R364X was found in the homozygous state in an ML II α/β patient [18]. Here, we also had two homozygous patients (P12 and P21) with ML II α/β, supporting the notion that p.R364X is associated with a clinically severe phenotype in ML patients. Additionally, we detected p.R364X in a heterozygous form in an ML III α/β patient (P32), in combination with the splice mutant c.2715+1G>A. Thus, the heterozygous splicing mutant c.2715+1G>A was presumed to retain some of the enzymatic functions of GNPTAB-encoded proteins and be associated with an attenuated phenotype, which was consistent with previous results [9, 34].
In the current study, 34 patients were confirmed to possess two mutant alleles. Only one mutant allele was detected in three patients, and none was detected in one patient. These four patients were biochemically confirmed to have ML II α/β and III α/β via plasma lysosomal enzyme analysis. We presumed that large genomic deletions or rearrangements, which could not be identified by conventional Sanger sequencing, or an unknown mutation in the region we did not test (e.g., the promoter and intronic regions) could not be excluded. Another possibility is that there is polymorphism or mutation in the region of the primers, which caused the observed allele-specific dropout [36]. For these four patients, further molecular studies are warranted to identify the types of mutations present.
In our study, there were two patients, P9 and P24, with a positive family history but without definite diagnosis. Thus, they were born without prenatal diagnosis. Unfortunately, they had the same disease as their older siblings.
The Stealth domain mediates the catalytic function of GlcNAc-1-phosphotransferase. Thus, mutations in this domain may cause severe enzyme deficiency [31]. The Notch and DMAP interaction domain is hypothesized to function in the specific recognition of protein determinants on lysosomal acid hydrolases. van Meel et al. found that the Notch 2 module mediated a function that could not be carried out by Notch 1. The Notch 2 module might be a critical site of the transferase [37]. In our study, patient 9 carried the mutation F374L at the Stealth domain and D534N at the Notch 2 module on different alleles, which may explain his severe ML II phenotype. The R334P, D408N, and Y997C mutation on the Stealth domain of one allele and the nonsense mutation on the other allele may explain why patients 15, 20, and 1, respectively, manifested with a severe ML II phenotype. The combination of A34P mutation in the N-terminal transmembrane domain and a nonsense mutation may explain the severe phenotype of patient 4. More functional studies should be performed for L1025S and L1033P in the spacer region. Since there is no effective treatment for ML II and III, recognition of typical clinical characteristics, establishment of definite diagnosis, and identification of causative mutations are very important for genetic counseling, prenatal diagnosis, and prevention of these diseases. Among our 30 ML II families with probands, 11 chose prenatal diagnosis when the mothers were pregnant again. We carried out a lysosomal enzyme activity assay along with gene testing. The enzyme activities in the amniotic fluid supernatant of four fetuses were markedly higher than the reference range, and genetic testing verified the same mutations as the probands, both of which indicated ML II fetuses. The enzyme activities in the remaining seven cases were within the normal range, and genetic tests identified three of them as heterozygous fetuses, while four did not carry any of the mutations present in the proband. ML II fetuses were diagnosed by enzyme activity tests, but enzymatic detection could not further distinguish between heterozygotes and normal fetuses. Therefore, identification of mutations is important for prenatal diagnosis and genetic counseling. By combining the lysosomal enzyme assay and gene testing, prenatal diagnosis for pregnant women at high risk of ML II can be achieved and would contribute to the prevention of ML II occurrence.
Conclusion
In conclusion, ML II α/β has a higher incidence than ML III in a population from Eastern China. We identified 22 novel mutations in the GNPTAB gene and four novel mutations in the GNPTG gene. In this study, 11 prenatal tests with ML II α/β were successfully performed. Our study expands the GNPTAB gene and GNPTG gene mutation spectrum, which is helpful for prenatal testing, and established genetic targets for further structure-based drug design to rescue the defective GlcNAc-phosphotransferase activity in ML II/III disease.
Acknowledgements
The authors thank Shi-chao Zhao for her assistance with gene molecular analysis. This project was supported by Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152520), Shanghai Science and Technology Committee (16JC1404600), National Natural Science Foundation of China (81570516 and 81270936), and the National Key Research and Development Program (2016YFC0905100 and 2016YFC0901505).
Author contributions
Hui-wen Zhang and Xue-fan Gu conceived of the study; Yu Wang carried out enzymatic tests and molecular testing and drafted the manuscript; X-fG carried out the enzymatic tests; Hui-wen Zhang, Jun Ye, Lian-shu Han, Wen-juan Qiu, Li-li Liang, and Xiao-lan Gao participated in patient history collection and physical examination; Hui-wen Zhang revised the final manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xue-fan Gu, Email: guxuefan@xinhuamed.com.cn.
Hui-wen Zhang, Email: zhanghuiwen@xinhuamed.com.cn.
References
- 1.Cathey SS, Kudo M, Tiede S, Raas-Rothschild A, Braulke T, Beck M, et al. Molecular order in mucolipidosis II and III nomenclature. Am J Med Genet A. 2008;146A:512–3. doi: 10.1002/ajmg.a.32193. [DOI] [PubMed] [Google Scholar]
- 2.Bao M, Booth JL, Elmendorf BJ, Canfield WM. Bovine UDP-N-acetylglucosamine:lysosomal-enzyme N-acetylglucosamine-1-phosphotransferase. I. Purification and subunit structure. J Biol Chem. 1996;271:31437–45. doi: 10.1074/jbc.271.49.31437. [DOI] [PubMed] [Google Scholar]
- 3.Bao M, Elmendorf BJ, Booth JL, Drake RR, Canfield WM. Bovine UDP-N-acetylglucosamine:lysosomal-enzyme N-acetylglucosamine-1-phosphotransferase. II. Enzymatic characterization and identification of the catalytic subunit. J Biol Chem. 1996;271:31446–51. doi: 10.1074/jbc.271.49.31446. [DOI] [PubMed] [Google Scholar]
- 4.Kudo M, Bao M, D’Souza A, Ying F, Pan H, Roe BA, et al. The α- and β-subunits of the human UDP-N-acetylglucosamine:lysosomal enzyme phosphotransferase are encoded by a single cDNA. J Biol Chem. 2005;280:36141–9. doi: 10.1074/jbc.M509008200. [DOI] [PubMed] [Google Scholar]
- 5.Kudo M, Brem MS, Canfield WM. Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase α/β–subunits precursor gene. Am J Hum Genet. 2006;78:451–63. doi: 10.1086/500849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarghooni M, Dittakavi SS. Molecular analysis of cell lines from patients with mucolipidosis II and mucolipidosis III. Am J Med Genet A. 2009;149A:2753–61. doi: 10.1002/ajmg.a.33134. [DOI] [PubMed] [Google Scholar]
- 7.Raas-Rothschild A, Cormier-Daire V, Bao M, Genin E, Salomon R, Brewer K, et al. Molecular basis of variant pseudo-hurler polydystrophy (mucolipidosis IIIC) J Clin Invest. 2000;105:673–81. doi: 10.1172/JCI5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raas-Rothschild A, Bargal R, Goldman O, Ben-Asher E, Groener JE, Toutain A, et al. Genomic organisation of the UDP-N-acetylglucosamine-1-phosphotransferase gamma subunit (GNPTAG) and its mutations in mucolipidosis III. J Med Genet. 2004;41:e52. doi: 10.1136/jmg.2003.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik KH, Song SM, Ki CS, Yu HW, Kim JS, Min KH, et al. Identification of mutations in the GNPTA (MGC4170) gene coding for GlcNAc-phosphotransferase α/β subunits in Korean patients with mucolipidosis type II or type IIIA. Hum Mutat. 2005;26:308–14. doi: 10.1002/humu.20205. [DOI] [PubMed] [Google Scholar]
- 10.Tiede S, Muschol N, Reutter G, Cantz M, Ullrich K, Braulke T. Missense mutations in N-acetylglucosamine-1-phosphotransferase α/β subunit gene in a patient with mucolipidosis III and a mild clinical phenotype. Am J Med Genet A. 2005;137A:235–40. doi: 10.1002/ajmg.a.30868. [DOI] [PubMed] [Google Scholar]
- 11.Tiede S, Storch S, Lübke T, Henrissat B, Bargal R, Raas-Rothschild A, et al. Mucolipidosis II is caused by mutations in GNPTA encoding the α/β GlcNAc-1-phosphotransferase. Nat Med. 2005;11:1109–12. doi: 10.1038/nm1305. [DOI] [PubMed] [Google Scholar]
- 12.Bargal R, Zeigler M, Abu-Libdeh B, Zuri V, Mandel H, Ben Neriah Z, et al. When mucolipidosis III meets mucolipidosis II: GNPTA gene mutations in 24 patients. Mol Genet Metab. 2006;88:359–63. doi: 10.1016/j.ymgme.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Tappino B, Chuzhanova NA, Regis S, Dardis A, Corsolini F, Stroppiano M, et al. Molecular characterization of 22 novel UDP-N-acetylglucosamine-1-phosphate transferase α- and β-subunit (GNPTAB) gene mutations causing mucolipidosis types IIα/β and IIIα/β in 46 patients. Hum Mutat. 2009;30:E956–73. doi: 10.1002/humu.21099. [DOI] [PubMed] [Google Scholar]
- 14.Pohl S, Tiede S, Castrichini M, Cantz M, Gieselmann V, Braulke T. Compensatory expression of human N-acetylglucosaminyl-1-phosphotransferase subunits in mucolipidosis type III gamma. Biochim Biophys Acta. 2009;1792:221–5. doi: 10.1016/j.bbadis.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Pohl S, Encarnacão M, Castrichini M, Müller-Loennies S, Muschol N, Braulke T. Loss of N-acetylglucosamine-1-phosphotransferase gamma subunit due to intronic mutation in GNPTG causes mucolipidosis type III gamma: implications for molecular and cellular diagnostics. Am J Med Genet A. 2010;152A:124–32. doi: 10.1002/ajmg.a.33170. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Zhang W, Shi H, Meng Y, Qiu Z. Three novel homozygous mutations in the GNPTG gene that cause mucolipidosis type III gamma. Gene. 2014;535:294–8. doi: 10.1016/j.gene.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 17.van Meel E, Kornfeld S. Mucolipidosis III GNPTG missense mutations cause misfolding of the γ subunit of GlcNAc-1-phosphotransferase. Hum Mutat. 2016;37:623–6. doi: 10.1002/humu.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cathey SS, Leroy JG, Wood T, Eaves K, Simensen RJ, Kudo M, et al. Phenotype and genotype in mucolipidoses II and III alpha/beta: a study of 61 probands. J Med Genet. 2010;47:38–48. doi: 10.1136/jmg.2009.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unger S, Paul DA, Nino MC, McKay CP, Miller S, Sochett E, et al. Mucolipidosis II presenting as severe neonatal hyperparathyroidism. Eur J Pediatr. 2004;164:236–43. doi: 10.1007/s00431-004-1591-x. [DOI] [PubMed] [Google Scholar]
- 20.Heo JS, Choi KY, Sohn SH, Kim C, Kim YJ, Shin SH, et al. A case of mucolipidosis II presenting with prenatal skeletal dysplasia and severe secondary hyperparathyroidism at birth. Korean J Pediatr. 2012;55:438–44. doi: 10.3345/kjp.2012.55.11.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggarwal S, Coutinho MF, Dalal AB, Mohamed Nurul Jain SJ, Prata MJ, Alves S. Prenatal skeletal dysplasia phenotype in severe MLII alpha/beta with novel GNPTAB mutation. Gene. 2014;542:266–8. doi: 10.1016/j.gene.2014.03.053. [DOI] [PubMed] [Google Scholar]
- 22.Tylki-Szymańska A, Czartoryska B, Groener JE, Ługowska A. Clinical variability in mucolipidosis III (pseudo-Hurler polydystrophy) Am J Med Genet. 2002;108:214–8. doi: 10.1002/ajmg.10224. [DOI] [PubMed] [Google Scholar]
- 23.Encarnação M, Lacerda L, Costa R, Prata MJ, Coutinho MF, Ribeiro H, et al. Molecular analysis of the GNPTAB and GNPTG genes in 13 patients with mucolipidosis type II or type III - identification of eight novel mutations. Clin Genet. 2009;76:76–84. doi: 10.1111/j.1399-0004.2009.01185.x. [DOI] [PubMed] [Google Scholar]
- 24.Persichetti E, Chuzhanova NA, Dardis A, Tappino B, Pohl S, Thomas NS, et al. Identification and molecular characterization of six novel mutations in the UDP-N-acetylglucosamine-1-phosphotransferase gamma subunit (GNPTG) gene in patients with mucolipidosis III gamma. Hum Mutat. 2009;30:978–84. doi: 10.1002/humu.20959. [DOI] [PubMed] [Google Scholar]
- 25.Shanske S, Miranda AF, Penn AS, DiMauro S. Mucolipidosis II (I-cell disease): studies of muscle biopsy and muscle cultures. Pediatr Res. 1981;15:1334–9. doi: 10.1203/00006450-198110000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Prence EM, Natowicz MR. Diagnosis of alpha-mannosidosis by measuring alpha-mannosidase in plasma. Clin Chem. 1992;38:501–3. [PubMed] [Google Scholar]
- 27.Brown CA, Mahuran DJ. beta-Hexosaminidase isozymes from cells cotransfected with alpha and beta cDNA constructs: analysis of the alpha-subunit missense mutation associated with the adult form of Tay-Sachs disease. Am J Hum Genet. 1993;53:497–508. [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma R, Bukovac S, Callahan J, Mahuran D. A single site in human β-hexosaminidase A binds both 6-sulfate-groups on hexosamines and the sialic acid moiety of GM2 ganglioside. Biochim Biophys Acta. 2003;1637:113–8. doi: 10.1016/S0925-4439(02)00221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owada M, Nishiya O, Sakiyama T, Kitagawa T. Prenatal diagnosis of I-cell disease by measuring altered alpha-mannosidase activity in amniotic fluid. J Inherit Metab Dis. 1980;3:117–21. doi: 10.1007/BF02312545. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Zhang W, Shi H, Yao F, Wei M, Qiu Z. Mutation analysis of 16 mucolipidosis II and III alpha/beta Chinese children revealed genotype-phenotype correlations. PLoS ONE. 2016;11:e0163204. doi: 10.1371/journal.pone.0163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian Y, van Meel E, Flanagan-Steet H, Yox A, Steet R, Kornfeld S. Analysis of mucolipidosis II/III GNPTAB missense mutations identifies domains of UDP-GlcNAc: lysosomal enzyme GlcNAc-1-phosphotransferase involved in catalytic function and lysosomal enzyme recognition. J Biol Chem. 2015;290:3045–56. doi: 10.1074/jbc.M114.612507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Hoof F. Mucopolysaccharidoses and mucolipidoses. J Clin Pathol Suppl. 1974;8:64–93. doi: 10.1136/jcp.27.Suppl_8.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Wu J, Liu H, Chen X, Wang Y, Zhao M, et al. Two homozygous nonsense mutations of GNPTAB gene in two Chinese families with mucolipidosis II alpha/beta using targeted next-generation sequencing. Genomics. 2013;102:169–73. doi: 10.1016/j.ygeno.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Otomo T, Muramatsu T, Yorifuji T, Okuyama T, Nakabayashi H, Fukao T, et al. Mucolipidosis II and III alpha/beta: mutation analysis of 40 Japanese patients showed genotype–phenotype correlation. J Hum Genet. 2009;54:145–51. doi: 10.1038/jhg.2009.3. [DOI] [PubMed] [Google Scholar]
- 35.Yang M, Cho SY, Park HD, Choi R, Kim YE, Kim J, et al. Clinical, biochemical and molecular characterization of Korean patients with mucolipidosis II/III and successful prenatal diagnosis. Orphanet J Rare Dis. 2017;12:11. doi: 10.1186/s13023-016-0556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coutinho MF, Encarnação M, Laranjeira F, Lacerda L, Prata MJ, Alves S. Solving a case of allelic dropout in the GNPTAB gene: implications in the molecular diagnosis of mucolipidosis type III alpha/beta. J Pediatr Endocrinol Metab. 2016;29:1225–8. doi: 10.1515/jpem-2016-0173. [DOI] [PubMed] [Google Scholar]
- 37.van Meel E, Lee WS, Liu L, Qian Y, Doray B, Kornfeld S. Multiple domains of GlcNAc-1-phosphotransferase mediate recognition of lysosomal enzymes. J Biol Chem. 2016;291:8295–307. doi: 10.1074/jbc.M116.714568. [DOI] [PMC free article] [PubMed] [Google Scholar]