Abstract
Apical caspases initiate and effector caspases execute apoptosis. Reagents that can distinguish between caspases, particularly apical caspases-8, 9, and 10 are scarce and generally nonspecific. Based upon a previously described large-scale screen of peptide-based caspase substrates termed HyCoSuL, we sought to develop reagents to distinguish between apical caspases in order to reveal their function in apoptotic cell death paradigms. To this end, we selected tetrapeptide-based sequences that deliver optimal substrate selectivity and converted them to inhibitors equipped with a detectable tag (activity-based probes—ABPs). We demonstrate a strong relationship between substrate kinetics and ABP kinetics. To evaluate the utility of selective substrates and ABPs, we examined distinct apoptosis pathways in Jurkat T lymphocyte and MDA-MB-231 breast cancer lines triggered to undergo cell death via extrinsic or intrinsic apoptosis. We report the first highly selective substrate appropriate for quantitation of caspase-8 activity during apoptosis. Converting substrates to ABPs promoted loss-of-activity and selectivity, thus we could not define a single ABP capable of detecting individual apical caspases in complex mixtures. To overcome this, we developed a panel strategy utilizing several caspase-selective ABPs to interrogate apoptosis, revealing the first chemistry-based approach to uncover the participation of caspase-8, but not caspase-9 or -10 in TRAIL-induced extrinsic apoptosis. We propose that using select panels of ABPs can provide information regarding caspase-8 apoptotic signaling more faithfully than can single, generally nonspecific reagents.
Subject terms: Proteases, Proteolysis
Introduction
Apoptosis is a naturally occurring, tightly regulated process through which damaged or unwanted cells are removed from an organism to maintain cellular homeostasis [1]. Apoptosis is also a desired outcome of cancer therapies including irradiation, chemo-, gene-, and immune-therapies designed to eliminate the transformed cells by cell suicide [2]. Apoptosis is orchestrated by a family of proteases called caspases, which are synthesized as inactive zymogens and activated by processing or by recruitment of molecular platforms that promote their dimerization (Fig. 1a) [3]. Generally, apoptotic caspases are classified as apical (caspase-8, -9, and -10) or effector (-3, -6, and -7) (Fig. 1a) [4]. Depending on the environment, caspase-2 can act as either, or even in non-apoptotic role [5]. Once activated, caspases cleave a set of protein substrates to trigger phenotypes modulated by altered expression and/or activity of pro-apoptotic and anti-apoptotic proteins that respectively support or block cell death [6, 7]. The biochemical hallmarks of apoptosis are DNA fragmentation and membrane blebbing, as well as cell shrinkage, resulting in the formation of apoptotic bodies engulfed by surrounding cells, a process thought to be immunologically silent [8]. Intriguingly, a second branch of the caspase family, encompassing caspases 1, 4, and 5 in humans, executes an entirely different form of cell death, a highly inflammatory activity known as pyroptosis, which triggers the release of inflammatory cytokines from phagocytic cells [9].
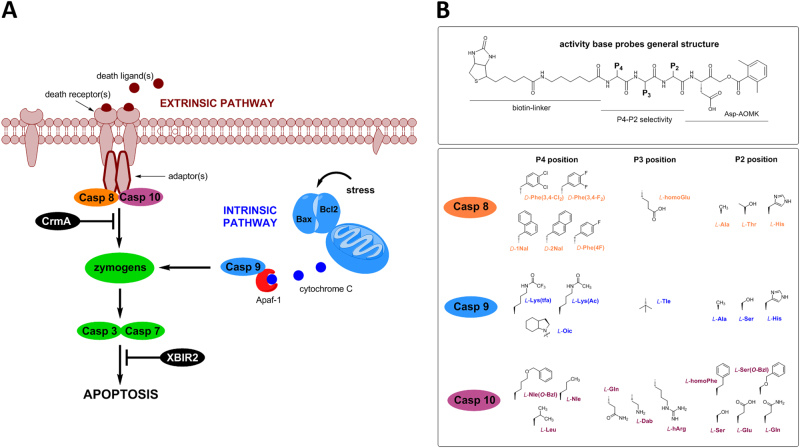
Fig. 1.
ABPs used for caspase analysis. a Two initiation pathways triggered by separate events converge at a common point to execute apoptosis in mammals. The extrinsic apoptosis pathway is triggered by extracellular ligation of death receptors, resulting in receptor clustering, adapter recruitment, and activation of the apical protease caspase-8 and possibly its close paralog caspase-10 (active enzymes are shown in orange/pink ovals—reviewed in ref. [57]). The intrinsic pathway responds primarily to cellular stress and some neurodevelopmental cues, with mitochondria acting as important integrators. Pro-apoptotic and anti-apoptotic members of the Bcl2 family (Bax and Bcl2 serve as examples) regulate the lethal stress–response threshold (reviewed in [58]). Activation of the apical caspases-8, -9, and -10 occurs when they are driven into an active conformation by their activation platforms—the DISC (death-inducing signaling complex) or the Apaf-1 apoptosome [59, 60]. Both pathways activate the effector proteases, caspases-3 and -7. Relevant to the extrinsic pathway, signals from several receptors, in response to cognate ligands, converge to activate caspases-8 and -10 (reviewed in ref. [61]). CrmA is a viral inhibitor of caspase-8 and XBIR2 is the second BIR domain of XIAP (X-linked inhibitor of apoptosis protein), highly selective for of caspases-3 and -7 [62–64]. b General structure of amino acids selected for synthesis of ABPs targeting the apoptotic apical caspases-8, -9, and -10. In each case, the P1 position was fixed as Asp
Since their active site is conserved, caspases display overlapping substrate specificity [4, 10]. Although they have distinct substrate repertoires in vivo, it has been difficult to generate diagnostic or therapeutic small molecules selective for individual caspases [11, 12]. Previously, we performed a large-scale analysis to obtain highly selective peptide-based substrates capable of distinguishing between closely related apoptotic caspases, using a process we developed called HyCoSuL (Hybrid Combinatorial Substrate Library) screening [13, 14]. The substrates we developed cannot be used to visualize specific caspase activation in complex biological material, as the product diffuses away. Accordingly, we set out to convert the caspase HyCoSuL-derived substrates into probes that would (1) remain bound to the active form of target caspases and (2) serve to inhibit individual caspases in complex samples. The general method of detecting specific active enzymes, including caspases, by activity-based profiling has been described in detail previously [15–19].
Caspases must be catalytically active to kill a cell; therefore, we reasoned that a set of selective activity-based probes (ABPs) might be helpful to better define individual caspases functioning in apoptotic processes. The most widely used reagent to inhibit caspases is cell-permeable carbobenzoxy-Val-Ala-Asp(OMe)-fluoromethyl ketone (Z-VAD(OMe)-FMK) [11, 20]. However, Z-VAD(OMe)-FMK has a broad activity spectrum and also inhibits cysteine cathepsins, and thus is not a useful tool to block individual caspases. Even the substrates and the probes synthesized based on allegedly selective caspase sequences (DEVD, LEHD, or IETD) lack specificity and can lead to erroneous conclusions [21–23]. We predicted that use of a wide set of unnatural amino acids in peptide structures, based on prior HyCoSuL screens, that would allow us to synthesize probes more selective than those containing natural amino acids only. Accordingly, we synthesized probes designed to exhibit the greatest selectivity in differentiating apical caspases-8, -9, and -10 from each other and from the primary effector caspase-3. We then used them to monitor the kinetics of apoptosis initiation in Jurkat T cells induced to undergo apoptosis by TRAIL (TNF-related apoptosis-inducing ligand) or etoposide and to define the functions of individual caspases in that activity. Specifically, we sought to differentiate the participation of apical caspases of the extrinsic apoptosis pathway, namely, caspases-8 and -10 from the activity of caspase-9, an apical caspase of the intrinsic apoptosis pathway.
Results
Conversion of substrates to activity-based probes
To determine whether we could convert substrate sequences into probes targeting individual caspases, we designed and synthesized selective ABPs to covalently and irreversibly bind (and label) only active forms of caspases, to provide tools to discriminate the functions of specific caspases during apoptosis. Since apoptotic caspases display overlapping substrate specificity and cannot be distinguished using peptides constructed of natural amino acids, we used data from our previously published HyCoSuL approach, in which we assessed the caspase preferences toward substrates comprised of unnatural amino acids [13, 14]. The approach yielded several tetrapeptide sequences displaying high selectivity for individual caspases. Here, we employed those sequences to synthesize 38 different ABPs representing five classes: (1) four broad-spectrum probes for all the 5 caspases, (2) one probe selective for the closely related caspases-3 and -7, (3) 8 caspase-9 selective probes, (4) 11 caspase-8 selective probes, and (5) 14 caspase-10 selective probes. These sequences were optimized for selectivity and thus their catalytic activity was lower than the optimal substrates for each caspase. As a scaffold for ABPs, we employed biotin-ahx-(P5)-P4-P3-P2-P1-AOMK (Fig. 1b), as biotin is bound by streptavidin, providing means to capture active caspases from lysates of apoptotic cells. To separate the biotin moiety from the caspase active site, we incorporated a 6-aminohexanoic (ahx) spacer into the ABP. As an electrophilic covalently reacting reagent (warhead), we utilized acyloxymethyl ketone (AOMK) [24]. Critical for these ABPs is the P4-P2 sequence primarily built of unnatural amino acids, a feature that significantly enhances the target selectively (Fig. 1b). For caspase-8, we selected hydrophobic, bulky D-amino acids at the P4 position, homoglutamic acid at P3, and Ala, Thr, or His at P2 (probes MP-8.01…8.11). For caspase-10 selective ABPs, we used aliphatic amino acids at P4 (Leu, hLeu—homoLeucine, Nle—norLeucine, or Nle(O-Bzl)—6-benzyloxynoreLeucine), basic amino acids Dab (aminohomoAlanine) or hArg (homoArginine) or the acidic Glu at the P3 position, and a variety of structures at P2 (hPhe—homoPhenylalanine, Ser(O-Bzl)—serine benzyl ether, Gln, Glu, or Ser) (probes MP-10.01…10.14). Finally, caspase-9 ABPs contained Oic (octahydroindole-2-carboxylic acid), Lys(tfa) (trifluoroacetylLysine), or Lys(Ac) (acetylLysine) at P4, tert-leucine at P3, and Ala, Ser, or His at P2 (probes MP-9.01…9.07) (Fig. 1b; Table S1). To increase selectivity of caspase-9 probes over caspase-3, we included an additional amino acid (proline) at P5 (Table S1). We also developed an effector caspase ABP (MP-3.01) that targeted caspases-3 and 7, and was poorly reactive with apical caspases, as well as four broad-spectrum probes (DEVD, IETD, LEHD, and LQnD) (Table S1).
Catalytic parameters of activity-based probes correlate well with substrates
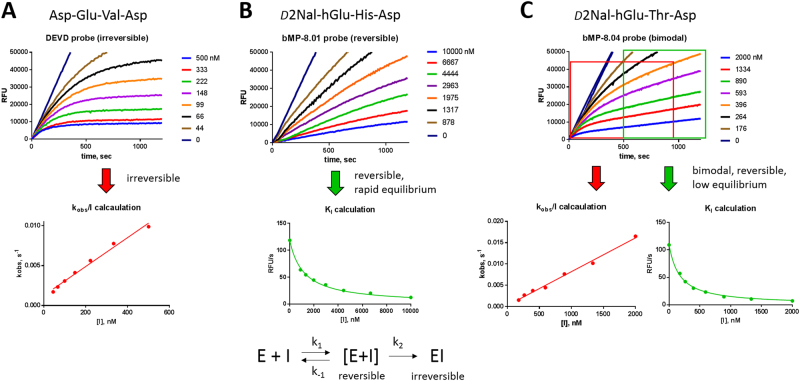
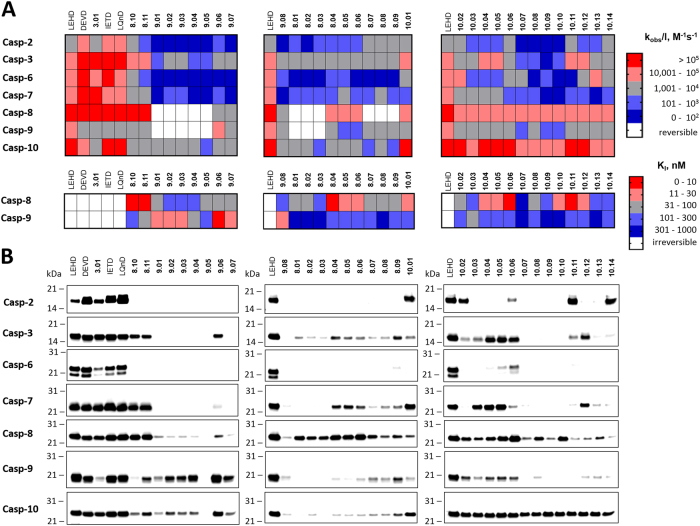
To test ABP selectivity, we measured their inhibition potency across seven recombinant apoptotic caspases: caspases-2, -3, -6, -7, -8, -9, and -10, and also two ubiquitously expressed cysteine cathepsins (L and B), which are also known to be inhibited by AOMK-based peptides (Table S2). Cathepsins were very poorly inhibited, although trace labeling was observed for some probes (Figure S1). Almost all ABPs tested displayed a classic irreversible mode of inhibition of caspase-2, -3, -6, -7, and -10, fitting a general irreversible pseudo first-order equation with no observable reversible equilibrium, enabling us to calculate second-order rates (kobs/I) (Fig. 2a; Table S2). Interestingly, however, for caspase-8 and -9, only some probes (DEVD, IETD, LEHD, and LQnD) showed fully irreversible behavior (Fig. 2a; Table S2). Other probes displayed fast-binding reversible kinetics, while others displayed bimodal inhibition kinetics. Thus, we calculated KI (equilibrium constant) for the fast-binding reversible probes, and both kobs/I (rates of inhibition) and KI parameters for the bimodal inhibitors (Fig. 2b,c; Table S2, S3). This different kinetic behavior of various ABPs is in line with the previous observation that not all of the AOMK-based peptides are irreversible inhibitors, and the mode of inhibition can also be dictated by the inhibitor peptide sequence [25]. Regardless of inhibitory mode (reversible or irreversible), our MP-8.0x, MP-9.0x, and MP-10.xx probes displayed high selectivity across caspases tested (Fig. 3a; Table S2, S3), and they are the most selective small-molecule ABPs for apical caspases reported to date [11].
Fig. 2.
Distinct mechanisms of caspase-8 inhibition by AOMK ABPs. Progress curves show that inhibition is irreversible (a), reversible (b), or bimodal (c). In b,c, the switch between reversible and bimodal inhibition occurs following exchange of one amino acid (His → Thr) in the P2 position of the AOMK-activity-based probe
Fig. 3.
ABP selectivity toward five human apoptotic caspases. a Kinetic parameters (kobs/I and KI) of ABPs for caspases displayed as heatmaps. All ABPs were irreversible for caspase-2, -3, -6, -7, and -10. Caspase-8 and -9 displayed either irreversible or reversible inhibition, depending on the ABP tested. b ABP selectivity tested by labeling of recombinant caspases. Probes (100 nM) were incubated with active site-titrated caspases (50 nM) for 30 min, followed by SDS-PAGE, membrane transfer, and detection with fluorescent streptavidin. The lane on the left in each blot is a controlled broad-spectrum caspase probe (LEHD; biotin-ahx-LEHD-AOMK) used to ensure equivalent exposure between each blot
Since we designed probes to capture and detect activity of individual caspases in apoptotic cells, we asked whether all probes, including reversible ones, formed durable adducts with target enzymes, as assessed by adduct stability under denaturing conditions. To do so, we incubated 50 nM active site-titrated caspase with 100 nM probe for 30 min, and then performed SDS-PAGE and Western blotting to detect biotin-tagged probes with fluorescent streptavidin (Fig. 3b). Using the kobs/I data, we calculated that 100 nM of broad-spectrum probe (biotin-ahx-LEHD-AOMK) is sufficient to inhibit all tested caspases over the period of 30 min (see Scheme S1 for details). As expected, the irreversible probes efficiently labeled the caspases; however, unexpectedly, reversible probes were also stable after boiling in SDS. More importantly, some probes appeared highly selective, as they robustly labeled the target caspase and either poorly labeled or did not label others (Fig. 3b). For example, caspase-9 selective probes (MP-9.01, -9.02, and 9.03) displayed minimal cross-reactivity with casapase-10, caspase-8 selective probes (MP-8.01, -8.02, and 8.03) showed some minor cross-reactivity with only caspase-3, and finally among caspase-10 ABPs series, two probes (MP-10.07 and -10.09) were very selective across all tested caspase. These observations demonstrate that out initial prediction, that caspase-selective substrate sequences can be converted to selective ABPs, is largely valid.
Since the ABPs were based on caspase substrate specificity profiling, we predicted a positive correlation between the second-order rate of inhibition (kobs/I) and second-order rate of catalytic efficiency (kcat/KM) of substrates harboring the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore, containing identical P4-P1 sequences. In other words, substrates with high turnover kinetic constants serve as good scaffolds for ageneration of potent inhibitors, by replacing the ACC fluorophore with an electrophilic warhead (i.e., AOMK), however, as shown previously, such inhibitors lose some selectivity [26]. To test this, we synthesized 19 peptidyl fluorogenic substrates of the formula of Ac-peptide-ACC and measured their kinetic parameters against caspases-2, -3, -6, -7, -8, -9, and -10 (Fig. 4; Table S4, S5). This analysis revealed that caspases-3, -6, -7, -8, -9, and -10 displayed strong positive correlations between substrate kcat/KM and ABP kobs/I values (Fig. 4b,c). Caspase-2 and -6 showed minimal catalytic activity against most of the ACC substrates, thus the correlation was not possible. We did not attempt to correlate the substrate catalytic parameters with Ki, since the dimensions are not comparable. Substrates with His at P2 were rapidly hydrolyzed, but probes or inhibitors with P2 His were less potent than those with Ala or Thr at P2, at least in terms of Ki (Fig. 4d,e). We hypothesize that His at P2 may promote substrate hydrolysis and caspase inhibition via different mechanisms. In support of this hypothesis, we found that caspase-8 and caspase-9 probes with P2 His displayed reversible inhibition, while probes with Thr or Ala at P2 demonstrated bimodal inhibition. Seeking an explanation for the differential inhibition, we note that it has previously been demonstrated (using caspase-1) that the catalytic His residue stabilizes the AOMK leaving group, thus promoting the formation of irreversible inhibitor–caspase complex [25]. However, when His is also present in inhibitor/ABP structures at the P2 position, we speculate that these two positively charged His groups (assayed at pH 7.2) may repel each other and prevent irreversible inhibition. Such His stabilization is not important for substrate hydrolysis, thus peptides with His at P2 are efficiently turned over by caspase-8 and -9 (Table S5).
Fig. 4.
Kinetic parameters of ACC substrates and AOMK-activity-based probes. a General structure of the substrate-leaving group and ABP electrophile. Correlation between kobs/I and kcat/KM for effector caspases-3 and -7 (b) and apical caspases-8, -9 and -10 (c). Red points show values obtained in the presence of broad-spectrum probes with peptide sequences DEVD, IETD, LEHD, or LQnD (where n is norleucine); green points indicate values obtained in the presence of a MP-3.01, which are selective for effector caspases-3 and -7. Pearson correlation coefficients (ρ) were calculated from linear kinetic parameters, and data were plotted as log/log to demonstrate the spread of values. In the case of caspase-8 (d) and caspase-9 (e), His at P2 differentially influences the catalytic efficiency for a substrate versus an ABP
Analysis of the extrinsic apoptosis pathway using substrates and activity-based probes
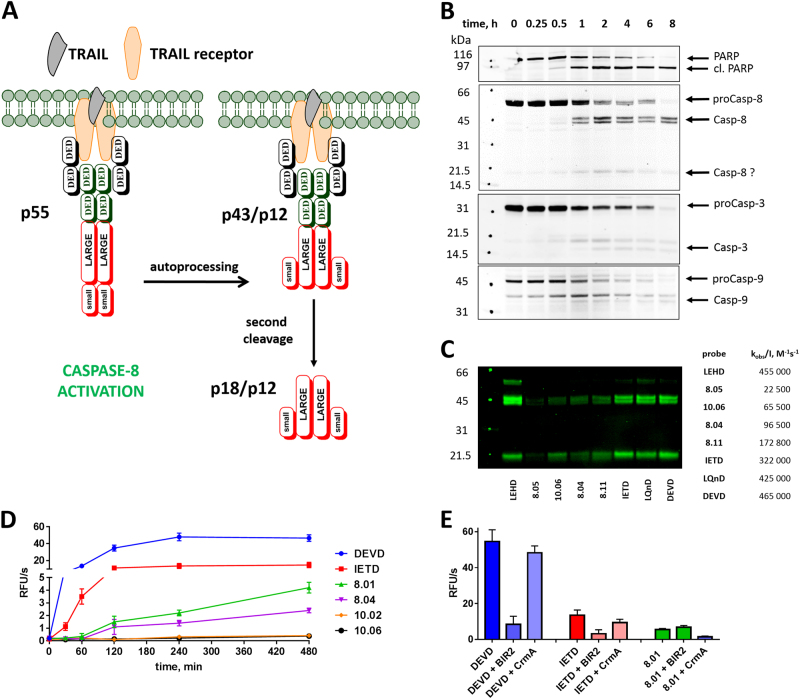
Based on our initial analysis of compounds displaying high selectivity toward individual caspases, we concluded that it would be sensible to avoid His in P2. We next asked whether a kinetically diverse set of substrates and probes can be used to dissect the role of individual caspases in apoptosis process. To do so, we focused on the highly investigated extrinsic pathway initiated by TRAIL [27, 28] and selected Jurkat T cells as a model system, as these cells readily undergo apoptosis following TRAIL stimulation (Fig. 5a; Figure S2) [29, 30]. We treated proliferating Jurkat T cells with recombinant TRAIL, and then monitored caspase and PARP cleavage at various time points (Fig. 5b). We found that within an hour of TRAIL stimulation, caspase-8 is processed from p55 into p43/p12 forms, consistent with autolytic maturation. We could not detect the large subunit devoid of the pro-domains (p18), possibly because the pro-domains had not been released during the course of our assay, or because the antibody used had poor reactivity with the large catalytic subunit in western blots. Caspase-3 was cleaved and activated, as evidenced by PARP (poly(ADP-ribose) polymerase) cleavage to 96 kDa. Caspase-9 was cleaved to its signature p35 derivative, indicative of the processing by caspase-3, an activity that does not activate caspase-9 [31, 32].
Fig. 5.
Use of small-molecule substrates and ABPs reveals caspase activation in TRAIL-stimulated Jurkat T cells. a The general scheme of caspase-8 activation upon TRAIL stimulation. Procaspase-8 (p55) is activated by dimerization at the DISC and undergoes auto-processing within the catalytic domain to generate p43/p12 fragments, which are further cleaved by the release of the DEDD recruitment domain into p18/p12 subunits [65] Jurkat T cells primarily express caspase-8 isoforms a and b, which differ by a short indel in the DED domain region, hence, the doublets seen in the 55 kDa and 43 kDa regions [66, 67] (of b,c) . b Western blots with specific antisera demonstrating the kinetics of caspase and PARP processing after TRAIL stimulation. The band around 20 kDa in the caspase-8 blot may be the p18 (large) subunit or a nonspecific band. c Cells were preincubated with indicated ABPs for 2 h and stimulated for 8 h with TRAIL. Cell lysates were then prepared and treated with streptavidin resin, the resin was washed, and the captured proteins were then eluted in boiling SDS buffer and run on SDS-PAGE for immunoblotting with caspase-8 antiserum. d Caspase activity in Jurkat cells at various time points after apoptosis induction and using indicated broad-spectrum and selective ACC substrates. e Caspase activity measured in Jurkat T cells at 8 h after apoptosis induction in the presence or absence of caspase inhibitors XBIR2 (1 µM; for caspase-3/-7) and CrmA (1 µM; for caspase-8). RFU relative fluorescence units. Western blots from b,c were performed twice, and the kinetic experiments in d,e were performed three times. Average values (with SD) are presented
To analyze the relationship between caspase-8 processing and activation, we pre-treated Jurkat cells with biotin-labeled ABPs (25 μM) for 2 h, followed by TRAIL stimulation for 8 h (Fig. 5c). We then lysed the cells, captured biotinylated probe adducts with immobilized streptavidin, and detected the probe-binding caspase-8 derivatives by immunoblotting with specific antisera. Three forms of caspase-8 (p55, p43, and p18), all of which contain the catalytic nucleophile, were captured by ABPs, indicating that each is catalytically active. Interestingly, endogenous p43 and p18 subunits showed the same specificity, since both were comparably labeled with a panel of ABPs. Moreover, the relative efficiency of ABPs in binding caspase-8 reflected their kobs/I parameters (Fig. 5c), demonstrating that various forms of endogenous caspase-8 have the same specificity as the recombinant ΔDED (death effector domain) enzyme used here for kinetic studies.
To observe the activation kinetics of caspase-8 and -3 in TRAIL-stimulated Jurkat T cells, we also utilized several caspase ACC-labeled fluorescent substrates (Fig. 5d,e). These substrates are not cell-permeable, requiring us to optimize a buffer system to conduct the assay. To do so, we induced apoptosis with TRAIL, collected cells after 8 h, permeabilized them with a panel of detergent-containing buffers, and then added Ac-DEVD-ACC substrate and monitored the hydrolysis over time. Based on permeabilization and hydrolysis outcomes, the best buffer was 0.5% Triton-X 100 in 1× caspase buffer (for components see Methods). Next, to monitor caspase activation kinetics, we stimulated Jurkat cells with TRAIL and assayed the activity at various time points using broad-spectrum caspase-8 or caspase-10 selective ACC substrates in the presence of the optimized lysis buffer (Fig. 5d). Caspase-3 was fully active between 1–2 h after TRAIL stimulation. Interestingly, we observed the same pattern of activation using the substrate Ac-IETD-ACC, which is generally used as a caspase-8 reporter. In parallel, we utilized two caspase-8 selective substrates (MP-8.01kcat/KM = 25,700 M−1 s−1 and MP-8.04kcat/KM = 10,400 M−1 s−1; Table S5), both very poorly recognized by caspases-3 and -7. Analysis of the cleavage profiles revealed that caspase-8 is active within the same time frame as caspase-3, and remains active for at least 8 h. This observation demonstrates that caspase-8 remains active for extended periods after TRAIL ligation and argues against a potential caspase-8 molecular timer mechanism similar to that proposed for caspase-9, whereby caspase-8 would be activated to cleave caspases-3 and -7, and then become inactive [33].
To confirm that the fluorescent signal from MP-8.01 cleavage is caspase-8-dependent, we added two protein caspase inhibitors, XBIR2 and the viral protein CrmA. The second BIR domain of XIAP (XBIR2) is exquisitely specific for effector caspases -3 and -7, and CrmA is selective for caspase-8 over effector caspases [34–36]. XBIR2-inhibited cleavage of DEVD and IETD substrates, but not MP-8.01, while CrmA only slightly inhibited DEVD and IETD cleavage, but substantially inhibited MP-8.01 cleavage (Fig. 5e). As a control, we stimulated FADD−/− (Fas-associated protein with death domain; unable to activate apical caspases) or Casp-8−/− Jurkat T cells with TRAIL for 8 h and were unable to detect caspase-8 activity (Figure S3). Finally, given that two caspase-10 selective substrates (MP-10.02 and MP-10.06) were not hydrolyzed (Fig. 5d), we concluded that, within the time range used here, caspase-10 does not function in TRAIL-stimulated death of Jurkat T cells. In an orthogonal experiment, we used biotincapture to investigate whether caspase-8 and caspase-10 selective ABPs could pull down the caspases from TRAIL-stimulated Jurkat T cells (as previously described for caspase-8) (Figure S4). The results demonstrated that caspase-3 was captured by almost all ABPs, except two (MP-8.04 and MP-10.07), which displayed poor kobs/I parameters. This was expected because caspase-3 is the most active and abundant enzyme in caspases cascade. Furthermore, caspase-8 was pulled down with only good and very good (high kobs/I) probes, whereas labeling of active caspase-10 was almost negligible, even with potent probes displaying excellent inactivation rates (kobs/I > 100,000 M−1s−1). Together, these data demonstrate that ABPs are useful for reflecting caspase activity, but even the most selective caspase-8 probes synthesized to date are still not able to specifically label caspase-8 because of the overwhelming activity and concentration of caspase-3, activated downstream from caspase-8.
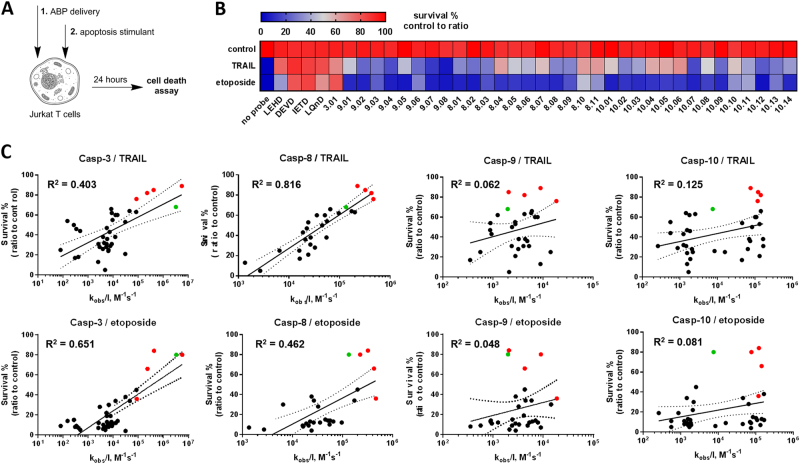
Use of a panel of activity-based probes to distinguish between apical caspases in extrinsic and intrinsic apoptosis
Given that individual probes cannot be used to dissect the role of apical caspases, we initiated a combinatorial approach and selected a panel of probe sequences showing good inhibition of apical caspases-8, -9, and -10, but poor inhibition of the effector caspases-3, -6, and -7, and treated Jurkat T cells for 24 h with TRAIL to engage the extrinsic pathway (Figure S5). Cells were pre-treated for 4 h with probes: in one set of experiments the probes were washed out before TRAIL addition, in the second set, the probes remained during the TRAIL incubation (Figure S5). We observed that only very potent and broad-spectrum probes (DEVD, LEHD, IETD) had some pro-survival effect, and other more selective, but less active probes had almost no impact on cell death.
To increase the efficiency of probe delivery, we used reversible permeabilization with digitonin, a reagent commonly used to allow molecules to penetrate the cell membranes [37]. To do so, we treated the cells with buffer containing 50% RPMI 1640 medium/50% digitonin in water (for 5 min), since the brief treatment with hypotonic buffer can substantially enhance the uptake of peptides with minimal effect on cell viability, as we observed (Figure S6). Having optimized that assay, we treated cells with ABPs in the presence of digitonin, allowed them to recover for 24 h, and then monitored the cell viability (Fig. 6a). None of the ABPs tested were toxic at a concentration of 25 μM (Fig. 6b, panel “control”). We then investigated the effects on apoptosis by treating the cells for 5 min with 25 µM of the same panel of ABPs in the presence of digitonin, replaced the medium, initiated apoptosis by TRAIL to engage the extrinsic pathway or etoposide to engage the intrinsic pathway for 24 h, and measured the cell viability by an MTS assay (Fig. 6b; Figure S7). We found that several probes (mostly broad-spectrum) blocked apoptosis with high efficiency, while others displayed moderate or no pro-survival effects (Fig. 6b). We predicted that there should be a structure–activity relationship between probe efficiencies on recombinant caspases; thus we correlated the kinetic parameters (kobs/I) of a diverse set of ABPs with their ability to rescue the cells from TRAIL-stimulated or etoposide-stimulated apoptosis (% cell survival). We performed this analysis using only probes displaying high selectivity toward apical caspases, since even if apical caspases are activated, apoptosis is blocked by inhibition downstream caspases-3 and -7, and thus the use of broad-spectrum probes may not provide accurate read-outs (Figure S5, S7, S8). In TRAIL-stimulated cells, we observed the highest correlation between ABP potency (kobs/I and also KI) and cell survival for the caspase-8 probe set (Fig. 6c; Figure S5). Interestingly, we did not observe a correlation between ABP potency toward caspase-10 and cell survival, supporting the idea that caspase-10 does not contribute to TRAIL-induced cell death in the presence of caspase-8. As expected, caspase-9 also played no role in TRAIL-induced apoptosis because the caspase-9 probe set failed to correlate with survival. Our findings confirm that caspases-3 and -7 are responsible for apoptosis execution, however, their activation was caspase-8 dependent (Fig. 6c; Figure S5B,C).
Fig. 6.
Apoptosis induction in the Jurkat T cells as analyzed using a diverse set of ABPs. a Probes were delivered to cells by reversible permeabilization, followed by apoptosis induction by TRAIL or etoposide. After 24 h, cell survival was measured relative to untreated controls using MTS assay. b Color-coded index of cell survival outcomes in the presence of a set diverse ABPs targeting caspases-8,- 9, and -10 tested in TRAIL- or etoposide-stimulated Jurkat T cells. c Correlation of probe inhibition potency (kobs/I) and cell survival (%, ratio to control). Red points show values obtained in the presence of broad-spectrum probes with peptide sequences DEVD, IETD, LEHD, or LQnD (where n is norleucine); green points indicate values obtained in the presence of a probe 3.01, which is selective for effector caspases-3 and -7. Each analysis contained the same probe panel set. Regression coefficients (R2) were calculated from semi-log plots, with dotted lines indicating 95% confidence interval
Relevant to etoposide-triggered apoptosis, we could not identify the key apical caspase(s) using our approach. We observed a good correlation between the caspases-3 and -7 probe set and cell survival, but did not observe comparable correlations for other caspases. Analysis of caspase-8 probes revealed a poor correlation dominated by broad-spectrum ABPs (DEVD, IETD, LEHD) that also target caspase-3. However, in contrast to TRAIL stimulation, we observed no correlation of ABPs with moderate kobs/I. As expected, we did not observe a correlation between etoposide-initiated cell death and caspase-10 selective probes, but unexpectedly could not correlate caspase-9 activity with etoposide-induced apoptosis based on the correlations (Fig. 6c). Since, unlike caspases-8 and -10, caspase-9 is activated within an apoptosome complex, AOMK ABPs may have had limited access to the endogenous caspase-9 active site, blunting the inhibition. To test this possibility, we used several caspase-9 ABPs, as well as some broad-spectrum probes (biotinylated DEVD, LEHD, and IETD) and zVAD-based inhibitors (-fmk, -bzl, and -AOMK) in a cell-free apoptosis system, in which we stimulated the apoptosome assembly by addition of cytochrome c (Figure S9). All probes (broad-spectrum and caspase-9-selective) were very efficient in inhibiting recombinant human ΔCARD (caspase activation and recruitment domain) caspase-9 (Figure S9A), but had only a small inhibitory effect on cytochrome c-triggered apoptosome activity, and delayed, but did not prevented caspase-3 processing and activation (Figure S9B,C). This data is consistent with previous observations that small-molecule probes are only minimally effective in inhibiting the apoptosome complex [38]. Based on this data, we conclude that AOMK probes may not be sufficient to inhibit endogenous caspase-9, and the high-cell survival ratios seen in etoposide-stimulated cells treated with select ABPs are likely attributable to the inhibition of downstream caspases-3 and -7.
Our strategy demonstrates that it is possible to dissect the initiators of apoptosis with a panel of diverse ABPs, primarily because individual probes are not selective enough due to the overpowering influence of caspases-3 and -7. To determine whether this approach has wide validity, we explored it in breast cancer cell line MDA-MB-231. As these cells overexpress inhibitors of apoptosis proteins (IAPs), the induction of apoptosis via TRAIL pathway must be supported by the addition of an IAP antagonist (birinapant) (Figure S10A) [39]. We demonstrated that the pre-incubation of MDA-MB-231 cells with a panel of ABPs (25 μM) for 4 h, followed by washing the probes out and adding TRAIL/birinapant stimulants (24 h) had only a small impact on cell survival (Figure S10B). However, when the probes were preincubated for 4 h without washout and then co-incubated with TRAIL/birinapant, their pro-survival effect was more pronounced (Figure S10B). The level of cell survival correlated well with ABP kobs/I parameters measured for caspase-8 (Pearson’s correlation coefficient = 0.775) (Figure S10C). Surprisingly, to achieve such a good anti-apoptotic effect, we did not have to use reversible permeabilization, presumably because the MDA-MB-231 cells were more permeable to ABPs than Jurkat cells. This striking difference between Jurkat T cells and MDA-MB-231 cells shows that the dynamics of ABPs uptake may be cell dependent, which should be taken into account when working with other cell types.
Discussion
Defining precise functions of caspases in apoptotic responses to distinct stimuli is critical for our understanding of mechanisms that drive degenerative and proliferative diseases, as well as for design of therapeutic agents to either block or accelerate apoptosis. An obstacle in designing these tools has been overlapping substrate specificity: chemical-based attempts to distinguish the roles of closely related enzymes in a network are hampered by activity overlap. Hence, the goal is to exploit minor differences in specificity.
Biochemical characterization of recombinant caspases and structural analysis over the last decade has provided a framework for synthesis of chemical tools now widely used in biological studies of these enzymes. Yet, as pointed out previously, the most popular reagents remain non-selective and have not yet yielded the required precision [11, 12, 40, 41]. For example, here we demonstrate that the use of LEHD and IETD-based probes, which are commonly used to analyze or even distinguish between apical caspases provide an incomplete picture, since they target effector caspases with similar efficiencies (Fig. 3) and cannot distinguish cell death mechanisms (Fig. 6, red dots on graphs). To enhance selectivity, we and others previously demonstrated that the use of unnatural amino acids significantly increases selectivity of substrates and peptide inhibitors toward individual apoptotic caspases [13, 16, 24, 42, 43]. We utilized substrate-based HyCoSuL data to design small molecule, biotin-labeled ABPs with high selectivity toward the apical caspases-8, -9, and -10. Their selectivity has been demonstrated by measuring the kinetic parameters toward individual enzymes and in affinity-labeling assays (Fig. 3). Interestingly, in this paper, we demonstrate that ABP kinetic parameters strongly correlate with cleavage efficiency of fluorogenic substrates with corresponding sequences. By this, we confirm the general rule that good substrates and be converted into good inhibitors. However, we also found that caspase-8 and caspase-9 displayed an activity switch between substrates and probes harboring His at P2 position. Although substrates with P2 His are preferred by these caspases, corresponding inhibitors display only moderate potency. We hypothesize that the His switch feature is associated with different mechanisms of substrate recognition and inhibitor binding. We also observed that some ABPs show different inhibition kinetics in interacting with caspase-8 or -9, compared to caspase-10. Differential inhibitory mechanisms associated with probes with an AOMK warhead have been reported previously [25, 44], but the reason is unknown, and why caspase-10 behaves differently than its close paralog caspase-8 awaits future analysis.
We also observed that caspase peptidyl substrates are more selective than the corresponding probes. For example, we discovered a highly selective substrate useful to detect caspase-8 activity following TRAIL-induced apoptosis (Fig. 5). Significant selectivity is lost after conversion of highly selective substrates to ABPs, suggesting that single ABPs cannot be used to label an individual apical caspase or specifically inhibit it. Nevertheless, selectivity can be assessed by calculating the second-order reaction rates, irrespective of the reaction mechanism, in order to generate structure–activity relationships for a panel of probes. Based on this approach, we conclude that caspase-8, and not caspase-10, is primarily involved in Jurkat T cell apoptosis induced by TRAIL, a finding consistent with genetic models that rule out a role for caspase-10 in extrinsic death signals [45, 46].
Finally, being unable to use individual caspase-selective ABPs in cells due to the overpowering influence of caspases-3 and -7, we designed the ABP panel approach to establish a functional assay to detect caspase activation in apoptotic cells. Many ABPs are not cell penetrant, and we found that reversible permeabilization with digitonin enhances probe delivery to Jurkat T cells, indicating that this assay could be adapted to various cancer cell lines when apoptosis (or cell death in general) is induced using different stimuli. Importantly, our set of ABPs contains several caspase-10 selective probes; therefore, it may be possible to define either cancer cell lines dependent on caspase-10 or reagents that stimulate apoptosis mediated by caspase-10. Finally, we validated our assay in breast cancer cell line MDA-MB-231. However, our experiments suggested that with this cell line, passive ABP delivery to the cytosol is sufficient to block caspase-dependent cell death. As in Jurkats, MDA-MB-231cell survival shows the strongest correlation with ABPs directed against caspase-8. Since our functional assay depends on caspase activity, it should be possible to determine whether the cells evade apoptosis and switch to caspase-independent cell death paradigms such as necrosis, autophagy, or mitotic catastrophe [47–49].
Materials and methods
Reagents
All chemicals and reagents were purchased from commercial suppliers and used without further purification, unless otherwise noted. Fmoc-protected amino acids were purchased from Sigma-Aldrich (Poznan, Poland), Iris Biotech GmbH (Marktredwitz, Germany), Creosalus (Louisville, KY, USA), Bachem (Torrance, CA, USA), and PE Biosciences Limited (Hong Kong, China). Fmoc-ACC-OH was synthesized according to a previously published method [13, 50]. Biotin, 2-chlorotrityl chloride resin (particle size 100–200 mesh, 1.59 mmol/g), Rink Amide RA resin (particle size 200–300 mesh, 0.7 mmol/g), coupling reagents (HBTU and HATU), piperidine (PIP), trifluoroacetic acid (TFA), and diisopropylcarbodiimide (DICI) were purchased from Iris Biotech GmbH. Anhydrous HOBt was purchased from Creosalus (Louisville, KY, USA). N,N-diisopropylethylamine (DIPEA), 2,4,6-trimethylpyridine (2,4,6-collidine), acetonitrile for HPLC, triisopropylsilane (TIPS), 2,2,2-trifluoroethanol (TFE), hydrobromic acid solution (30% HBr wt. in AcOH), anhydrous tetrahydrofuran (THF), 4-methylmorpholine (NMM), isobutylchloroformate (IBCF), and 2,6-dimethylbenzoic acid (2,6-DMBA) were all from Sigma-Aldrich. N,N‘-dimethylformamide (DMF), dichloromethane (DCM), methanol (MeOH), diethyl ether (Et2O), acetic acid (AcOH), and phosphorus pentoxide (P2O5) were obtained from POCh (Gliwice, Poland). Diazomethane was generated following the Aldrich Technical Bulletin (AL-180) procedure. All peptides, ACC substrates, and biotin-labeled AOMK ABPs were purified by HPLC on a Waters system (Waters M600 solvent delivery module, Waters M2489 detector system) using a semi-preparative Discovery® C8 HPLC column (particle size 10 μm) to at least 95% of purity (confirmed by analytical HPLC using an analytical Discovery® C8 column (particle size 10 μm). Solvent composition used for semi-preparative and analytical methods was as follows: phase A (water with 0.1% TFA), phase B (acetonitrile with 0.1% TFA). A linear gradient of an increasing concentration of phase B was applied. The molecular weight of each compound was confirmed using a High Resolution Mass Spectrometer WATERS LCT premier XE with electrospray ionization (ESI) and time-of-flight (TOF) module.
Cell culture, apoptosis inducers, and antibodies
Human Jurkat T cells, caspase-8-deficient Jurkat T cells, and FADD-deficient Jurkat T cells were cultured in RPMI 1640 (Corning Cellgro) supplemented with 10% fetal bovine serum (FBS, Atlas Biologicals), 2 mM l-glutamine (Omega Scientific), 100 units/mL penicillin, and 10 μg/mL streptomycin (both Gibco Life Technologies) in a humidified 5% CO2 atmosphere at 37 °C. Jurkat cell apoptosis was induced using KillerTRAILTM (Enzo Life Sciences) or etoposide (MP Biomoedicals, LLC). Birinapant, a SMAC mimetic, was purchased from Cayman Chemicals (Cat No. 19699). Caspase-3 antibody was from Cell Signaling (rabbit ab, Cat No. 9662S); caspase-9 antibody (polyclonal, rabbit), raised against recombinant catalytic domain, was as previously described; [51] monoclonal caspase-8 antibody C15 raised against an epitope of the large subunit [52] was a kind gift of Dr. Markus Peter, University of Chicago; caspase-10 antibody was from MBL Life Science (mouse, Cat No. M059-3); PARP antibody was purchased from BD-Pharmigen (mouse, Cat No. 556362). Human MDA-MB-231 breast cancer cells were cultured in Dulbecco’s Modification of Eagle’s Medium (DMEM) supplemented with 10% of fetal bovine serum, 2 mM of L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin.
ABP design and synthesis
Based on PS-SCL and HyCoSuL data, we designed and synthesized 38 ABPs targeting five human apoptotic caspases [4, 13]. The general structure of each was biotin-ahx-P4-P3-P2-P1-AOMK. These 38 probes comprised five classes: (1) 4 broad-spectrum probes for all five caspases (P4-P1 sequence: DEVD, LEHD, IETD, and LQnD), (2) 1 caspase-3/-7 selective probe (bMP-3.01), (3) 8 caspase-9 selective probes (bMP-9.01…9.08), (4) 11 caspase-8 selective probes (bMP-8.01…8.11), and (5) 14 caspase-10 selective probes (bMP-10.01…10.14). The method used to synthesize AOMK probes with P1 Asp is described elsewhere [53]. In brief, a biotin-ahx-P4-P3-P2-COOH peptide was synthesized on a solid support (2-chlorotrityl chloride resin) and used for further synthesis without purification. In a separate synthesis, Boc-Asp(O-Bzl)-AOMK was obtained through reaction of Boc-Asp(O-Bzl)-COOH with diazomethane, followed by conversion of Boc-Asp(O-Bzl)-CHN2 to bromomethyl ketone (-CH2Br) and finally to acyloxymethyl ketone (-AOMK). Next, Boc-Asp(O-Bzl)-AOMK was de-protected with TFA to obtain H2N-Asp(O-Bzl)-AOMK. The N-de-protected warhead was then coupled to biotin-ahx-P4-P3-P2-AOOH to obtain biotin-ahx-P4-P3-P2-Asp(O-Bzl)-AOMK. The protecting groups from P4-P2 amino acids were removed using TFA and Asp(O-Bzl) was de-protected via hydrogenolysis (Pd/C, H2, inert atmosphere). The final crude product was purified by RP-HPLC using a C8 column.
Recombinant caspase assay conditions
Human apoptotic caspases were expressed and purified as described [51]. Before kinetic analysis, all caspases were active site-titrated using the zVAD-fmk inhibitor (Cayman Chemical Company, Cat No. 14467), according to the protocol described by Stennicke and Salvesen [51]. Caspase assay buffer composition: 20 mM PIPES, 10 mM NaCl, 1 mM EDTA, 10% w/v sucrose, and 10 mM DTT. To allow caspase-2, -8, -9, or -10 dimerization and maintain high activity, buffers were supplemented with 0.75 M sodium citrate. Unless otherwise stated, the kinetic experiments were performed at pH 7.2.
Recombinant cathepsin L and B assay conditions
Human recombinant cathepsins L and B were a kind gift from Drs. Boris Turk and Matej Vizovisek, University of Ljubljana. Detailed description of the expression and the purification can be found elsewhere [54]. The concentration of active cathepsins were determined via titration with the irreversible inhibitor E64 (Peptide Institute).
Determination of ABP kinetic parameters
Based on the kinetic analysis, we divided ABPs into three categories: (1) irreversible—kobs/I, (2) tight-binding reversible—KI, and (3) bimodal (both kobs/I and KI). We determined kobs/I (pseudo first-order kinetic constant; Y = (V0−VS) × (1−exp(−kobs))/kobs + VS × X, where Y is enzyme activity, V0 is initial enzyme activity, and VS is enzyme activity in the equilibrium state). KI parameters were calculated using the Morrison equation [55]. For each caspase, an appropriate ACC-labeled substrate was used to monitor the reaction progress (caspase-2, Ac-VDVAD-ACC, KM = 11 μM; caspase-3, Ac-DEVD-ACC, KM = 21 μM; caspase-6, Ac-DEVD-ACC, KM = 468 μM; caspase-7, Ac-DEVD-ACC, KM = 57 μM; caspase-8, Ac-LEHD-ACC, KM = 15 μM; caspase-9, Ac-LEHD-ACC, KM = 106 μM; and caspase-10, Ac-LEHD-ACC, KM = 24 μM). For cathepsins L and B, we used the substrate Z-FR-AMC to monitor the reaction progress (cathepsin L, KM = 3.3 μM and cathepsin B, KM = 62 μM). In all assays, the lowest inhibitor concentration was at least fourfold higher than the caspase concentration. In brief, to determine inhibition parameters, the inhibitor was diluted into wells on a 96-well plate, mixed with the substrate, and the plates were preincubated at 37 °C. Separately, caspase was preincubated at 37 °C, and after 15 min, the enzyme was added and fluorescence was monitored for 30 min, starting immediately. Both kinetic parameters (kobs/I for caspases-3, -7, -8, -9, and -10, and KI for caspase-8, and -9) were determined in at least three independent experiments and presented as an average value. All assays were performed using a CLARIOstar plate reader (BMG Labtech), operating in the kinetic mode (355 nm excitation, 460 nm emission, gain 650) using 96-well plates (Corning, NY, USA).
Analysis of probe binding to recombinant caspases and cathepsins
To assess ABP selectivity, 50 nM of each zVAD-fmk active site-titrated recombinant caspases and E64 active site-titrated recombinant cathepsin L and B were incubated with 100 nM probe for 30 min at 37 °C (reaction volume 200 μL), next, the reaction products were precipitated as follows: 100 μL of ice-cold 30% trichloroacetic acid (TCA) was added to each sample, incubated on ice for 30 min, and centrifuged for 30 min (20,000 × g, 4 °C). Supernatants were gently aspirated using a vacuum device, the pellet was re-suspended in 200 μL ice-cold 10% TCA, and the samples were re-centrifuged for 15 min (20,000 × g, 4 °C). Supernatants were again removed, and the pellet was re-suspended in 200 μL of acetone. Samples were centrifuged for 10 min (20,000 × g, 4 °C) and the supernatant was removed. Samples were dried at 37 °C for 15 min, and then 80 μL of 1× SDS/DTT buffer was added to each, followed by denaturation at 95 °C for 5 min. The samples (20 μL) were subjected to SDS-PAGE (30 min, 200 V), followed by their transfer to a nitrocellulose membrane (60 min, 10 V) and blocking with 2% BSA in TBS-T (60 min, RT). The biotin-labeled probe/caspase complex was detected by probing the membrane with fluorescently labeled streptavidin (30 min, RT, 1:10,000, IRDye® 800CW, LI-COR), followed by scanning in LI-COR system (800 nm channel, Odyssey® CLx, Lincoln, NE, USA). The data were analyzed using Image Studio Lite software (Lincoln, NE, USA). To better demonstrate the discrepancy between caspases and cathepsin bands from biotin-ahx-LEHD-AOMK and biotin-ahx-DEVD-AOMK, labeling were quantified and presented in relative fluorescence units.
Determination of the substrate kinetic parameters
Substrate kinetic parameters (kcat, KM, kcat/KM) were determined as described by Poreba et al. [13, 56]. Measurements were performed using a CLARIOstar plate reader (BMG LABTECH, Cary, NC, USA), operating in the fluorescence mode (for ACC fluorescent tags 355 nm/excitation and 460 nm/emission wavelengths were used). Substrate hydrolysis was monitored for up to 30 min, and most linear segment of plots were analyzed. Statistical analysis was performed using GraphPad Prism 7 software (La Jolla, CA, USA).
Stimulation of the Jurkat T cells with TRAIL and etoposide
Jurkat T cells (5 × 105/well) were placed on a 24-well plate with fresh medium (500 μL/well), and after 1 h, apoptosis was induced by treatment with either 100 ng/mL TRAIL or 25 μM etoposide 24 h. At 0, 0.5, 1, 2, 4, 8, 12, and 24 h time points, cells were collected and centrifuged at 500 × g for 5 min. The supernatants were discarded, and the pellet was washed with 1× DPBS (Dulbecco’s Phosphate Buffered Saline). Cells were re-centrifuged at 500 × g for 5 min, and the pellet was solubilized in 100 μL 1× SDS/DTT, heated to 95 °C for 5 min, cooled to room temperature, and sonicated (Branson Sonifier 250). A 30 μL sample aliquot was loaded onto a 4–12% gel (Bis-Tris Plus) and electrophoresed for 30 min at 200 V. Proteins were transferred to a nitrocellulose membrane (0.2 μm, BIO-RAD) for 60 min at 10 V, followed by Ponceau staining to verify equal loading and transfer. The membrane was blocked with 2% BSA in TBS-T for 60 min at room temperature. Caspase activation and PARP cleavage were detected using primary antibodies (anti-caspase-3 1:1,000, anti-caspase-8 1:500, anti-caspase-9 1:10,000, and PARP 1:2,000) incubated overnight at 4 °C, followed by the incubation with fluorescent secondary antibodies (for caspase-3 and caspase-9, donkey anti-rabbit, IRDye® 680RD, 1:10,000; for caspase-8 and PARP, donkey anti-mouse, IRDye® 800CW, 1:10,000) at room temperature for 30 min. The membrane was scanned using the LI-COR system (700 nm and 800 nm channels), and the data were analyzed using Image Studio Lite software. To detect the 3 caspases plus PARP (and cleavage products) on the membranes, we performed two Western blot analyses for each apoptotic stimulant (membrane A: caspase-3, and caspase-8; and membrane B: caspase-9 and PARP). Both membranes were incubated with appropriate secondary antibodies and scanned sequentially.
Detection of caspase-3, -8, and -10 in apoptotic Jurkat T cells
To assess the specificity of endogenous caspases-3, -8, and -10, Jurkat T cells (2 × 107) were re-suspended in 2 mL fresh medium, and preincubated each for 2 h with 25 μM biotin-labeled ABP. After this time, all non-internalized probes were washed out by centrifugation (500 × g for 5 min) and the cells were re-suspended in fresh media (2 mL). Cells were then stimulated for 8 h with TRAIL (100 ng/mL), followed by centrifugation at 500 × g for 5 min. The pellet was washed with PBS and the cells re-centrifuged at 500 × g for 5 min. The resulting pellet was lysed in 200 μL RIPA buffer and centrifuged at 20,000 × g for 15 min. The supernatant was transferred to a new tube and agitated for 4 h at 4 °C with 50 μL streptavidin agarose resin (50% slurry). Samples were then centrifuged at 500 × g for 5 min, washed three times with lysis buffer (after each washing step, cells were centrifuged as above), and then 100 μL 1xSDS/DTT was added to the resin pellet and the mixture was boiled 5 min. Lysate proteins were resolved on SDS-PAGE, followed by immunoblotting, as described above (caspase-3 and caspase-8). Caspase-10 was detected with anti-caspase-10 antibody (1:500) incubated overnight at 4 °C, followed by incubation with fluorescent secondary antibody (donkey anti-mouse, IRDye® 800CW, 1:10,000) at room temperature for 30 min.
Screening of caspase-8 and -10 selective substrates
Recombinant caspases-3, -8, and -10 were pre-activated for 15 min at 37 °C and then mixed with ACC-labeled substrates (final substrate concentration, 20 μM; final caspase concentration, 50 nM). Substrate hydrolysis was monitored using a plate reader, as described above. The linear regions of the curves (10–15 min) were evaluated, and results were presented as RFU/s values.
Use of fluorescent substrates to detect caspase activity in apoptotic Jurkat T cells
Jurkat T cells (5 × 106) were seeded in 96-well white, flat bottomed plates in phenol red-free RPMI hypotonic buffer (70 μL/well) and stimulated for 8 h with TRAIL (10 μL, final conc.100 ng/mL). Then, 20 μL of 500 μM Ac-DEVD-ACC dissolved in test lysis buffers was (Fig. 5d) added to the cells, and caspase activity was measured over 30 min. Caspase buffer containing 0.5% Triton-X (which resulted in the highest fluorescence signal) was used in further analyses. Jurkat T cells (5 × 106/well) were then stimulated with TRAIL for 0, 0.5, 1, 2, 4, and 8 h, and at each time point, the caspase activity (RFU/s) was measured in the presence of 100 μM ACC-labeled caspase substrates in the indicated buffer. Substrates included: caspase-3/7 Ac-DEVD-ACC substrate, a broad-spectrum Ac-IETD-ACC substrate, caspase-8-selective MP-8.01 and MP-8.04 substrates, and caspase-10 selective MP-10.02 and MP-10.06 substrates. DEVD, IETD, and MP-8.01 substrate selectivity was confirmed in a separate experiment, in which the cells were stimulated with TRAIL for 8 h, followed by the addition of ACC substrate in the presence of the caspase inhibitors XBIR2 (1 μM; for caspase-3/-7) and CrmA (1 μM; for caspase-8). In this experiment, fluorescence was read after 15 min of inhibitor/substrate addition to allow caspase inactivation. Analysis was also performed in Casp-8−/− and FADD−/− Jurkat T cells. Kinetic experiments were performed on a CLARIOstar instrument (350 nm excitation, 460 nm emission, gain 650), and the data were analyzed using GraphPad Prism software.
Optimization of digitonin-assisted Jurkat T cells permeabilization
Five hundred micro liter Jurkat T cells (2.5 × 106) were incubated in 12-well plates with 1 μM FAM-LEHD-AOMK fluorescent probe in the presence of digitonin at various concentrations (0, 5, 10, 20, 40 μg/mL) in phenol red-free RPMI hypotonic buffer (RPMI to distilled water ratio = 1:0, 3:1, 2:2, 1:3) for 2, 5, or 10 min. Then, 1 mL RMPI was added to the wells, and the cells were collected and centrifuged at 500 × g for 5 min. Supernatants were aspirated using a vacuum device and the pellets were re-suspended in 500 μL RPMI. Cells were allowed to recover for 2 h in a 5% CO2 incubator at 37 °C. Living and dead cells were counted using Trypan blue dye, and FAM-LEHD-AOMK delivery efficiency was determined using a fluorescence microscope (FITC filter, 40×). The highest efficiency was obtained when the probes and the cells were incubated for 5 min in the presence of 10 μg/mL digitonin in 50% RPMI hypotonic buffer.
TRAIL or etoposide stimulation of Jurkat T cells in the presence of ABPs
Jurkat T cells (105) in 50% RPMI hypotonic medium were seeded into 96-well clear bottom plates (50 μL/well) and immediately incubated with a panel of biotin-labeled probes (25 μM) in the presence of 10 μg/mL of digitonin for 5 min. Then 150 μL of RPMI was added to the wells, and the plates were centrifuged at 500 × g for 5 min. The supernatants were then aspirated using an 8-channel vacuum device, and the cells were re-suspended into 100 μL fresh media and allowed to recover for 2 h. Cells were stimulated with TRAIL (100 ng/mL) or etoposide (25 μM) for 24 h, and cell viability was determined using MTS assay, according to the manufacturer’s protocol (CellTiter 96® Aqueous One Solution Cell Proliferation Assay, G3582, Promega). The number of living cells was reported as a survival ratio, calculated based on the number of unstimulated control cells.
TRAIL stimulation of Jurkat T cells in the presence of ABPs
Jurkat T cells (105) were seeded into two 96-well clear bottom plates (plate A and plate B, 80 μL/well) and incubated with a panel of biotin-labeled probes (25 μM, 20 μL) for 4 h. Then, ABPs from plate A were removed via centrifugation, and the cells were re-suspended into 100 μL fresh media. Next, cells from both plates (plate A and plate B) were stimulated with TRAIL (100 ng/mL) for 24 h, and cell viability was determined using an MTS assay, as described above.
TRAIL/birinapant stimulation of MDA-MB-231 cells in the presence of ABPs
MDA-MB-231 cells (104) were seeded into two 96-well clear bottom plates (plate A, and plate B, 80 μL/well) and incubated with a panel of biotin-labeled probes (25 μM, 20 μL) for 4 h. Then, ABPs from plate A were removed via centrifugation, and the cells were re-suspended into 100 μL fresh media. Next, cells from both plates (plate A and plate B) were stimulated with TRAIL (100 ng/mL) plus a smac mimetic, birinapant (2 μM) for 24 h, and cell viability was determined using an MTS assay, as described above.
Caspase-3 and caspase-9 inhibition profiles
Recombinant caspase-3 (10 nM) and caspase-9 (10 nM) were pre-activated in optimal caspase buffer for 15 min at 37 °C and then incubated with 13 different inhibitors (at 0.01, 0.1, 1, and 10 μM) in 96-well plates for 30 min at 37 °C. A preferred substrate (Ac-DEVD-ACC for caspase-3 or Ac-LEHD-ACC for caspase-9) was then added, and the reaction progress was monitored using CLARIOstar plate reader (BMG Labtech), operating in the fluorescence kinetic mode (355 nm excitation, 460 nm emission, gain 650) using 96-well plates (Corning, NY, USA). Percent caspase inhibition was calculated, and the results were represented as heatmaps.
Assay of the Jurkat T cell cytosolic extracts
Hypotonic lysis of the Jurkat T cells was performed and the cytosolic fraction was collected, as described [32]. The total protein concentration in the lysate was 8 mg/mL. Samples were stored at −80 °C until use, when 100 μL aliquots were thawed to room temperature and preincubated for 5 min with 10 μM of various inhibitors. The extract was then activated by the addition of dATP and horse cytochrome c in the presence of a proteasome inhibitor, as described [13]. Caspase-3 activation was monitored using 100 μM Ac-DEVD-ACC substrate. In parallel, Jurkat cell extracts were preincubated for 5 min with 10 μM of various inhibitors and activated with dATP/cytochrome c for 30 min. The samples were then resolved on SDS-PAGE, and caspase-3 and -9 were detected by immunoblotting, as described above.
Electronic supplementary material
Acknowledgements
This project received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 661187 (to MP), and was supported by the National Science Centre grant 2011/03/B/ST5/01048 in Poland (to MD) and NIH grant R01 GM099040 to (GSS).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by V Dixit
Contributor Information
Marcin Poreba, Email: marcin.poreba@pwr.edu.pl.
Marcin Drag, Phone: 0000-0001-8510-1967, Email: marcin.drag@pwr.edu.pl.
Guy S. Salvesen, Phone: 0000-0002-7933-6732, Email: gsalvesen@sbpdiscovery.org
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0110-y) contains supplementary material, which is available to authorized users.
References
- 1.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–41. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 2.Reed JC. Apoptosis-based therapies. Nat Rev Drug Discov. 2002;1:111–21. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- 3.Denault JB, Salvesen GS. Caspases: keys in the ignition of cell death. Chem Rev. 2002;102:4489–4500. doi: 10.1021/cr010183n. [DOI] [PubMed] [Google Scholar]
- 4.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–11. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 5.Bouchier-Hayes L, Green DR. Caspase-2: the orphan caspase. Cell Death Differ. 2012;19:51–57. doi: 10.1038/cdd.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhagen AM, Coulson EJ, Vaux DL. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2001;2:REVIEWS3009. doi: 10.1186/gb-2001-2-7-reviews3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/S0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 8.Saraste A, Pulkki K. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc Res. 2000;45:528–37. doi: 10.1016/S0008-6363(99)00384-3. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–5. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 10.Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, et al. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–82. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 11.Poreba M, Szalek A, Kasperkiewicz P, Rut W, Salvesen GS, Drag M. Small molecule active site directed tools for studying human caspases. Chem Rev. 2015;115:12546–629. doi: 10.1021/acs.chemrev.5b00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poreba M, Strozyk A, Salvesen GS, Drag M. Caspase substrates and inhibitors. Cold Spring Harb Perspect Biol. 2013;5:a008680. doi: 10.1101/cshperspect.a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poreba M, Kasperkiewicz P, Snipas SJ, Fasci D, Salvesen GS, Drag M. Unnatural amino acids increase sensitivity and provide for the design of highly selective caspase substrates. Cell Death Differ. 2014;21:1482–92. doi: 10.1038/cdd.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poreba M, Salvesen GS, Drag M. Synthesis of a HyCoSuL peptide substrate library to dissect protease substrate specificity. Nat Protoc. 2017;12:2189–214. doi: 10.1038/nprot.2017.091. [DOI] [PubMed] [Google Scholar]
- 15.Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, et al. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15:967–73. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers CJ, Gonzalez-Paez GE, Wolan DW. Selective detection of caspase-3 versus caspase-7 using activity-based probes with key unnatural amino acids. ACS Chem Biol. 2013;8:1558–66. doi: 10.1021/cb400209w. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sieber SA, Niessen S, Hoover HS, Cravatt BF. Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat Chem Biol. 2006;2:274–81. doi: 10.1038/nchembio781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abuelyaman AS, Jackson DS, Hudig D, Woodard SL, Powers JC. Synthesis and kinetic studies of diphenyl 1-(N-peptidylamino)alkanephosphonate esters and their biotinylated derivatives as inhibitors of serine proteases and probes for lymphocyte granzymes. Arch Biochem Biophys. 1997;344:271–80. doi: 10.1006/abbi.1997.0231. [DOI] [PubMed] [Google Scholar]
- 20.Van Noorden CJ. The history of Z-VAD-FMK, a tool for understanding the significance of caspase inhibition. Acta Histochem. 2001;103:241–51. doi: 10.1078/0065-1281-00601. [DOI] [PubMed] [Google Scholar]
- 21.McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–31. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 22.Berger AB, Sexton KB, Bogyo M. Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res. 2006;16:961–3. doi: 10.1038/sj.cr.7310112. [DOI] [PubMed] [Google Scholar]
- 23.Pereira NA, Song Z. Some commonly used caspase substrates and inhibitors lack the specificity required to monitor individual caspase activity. Biochem Biophys Res Commun. 2008;377:873–7. doi: 10.1016/j.bbrc.2008.10.101. [DOI] [PubMed] [Google Scholar]
- 24.Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, et al. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23:509–21. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 25.Brady KD, Giegel DA, Grinnell C, Lunney E, Talanian RV, Wong W, et al. A catalytic mechanism for caspase-1 and for bimodal inhibition of caspase-1 by activated aspartic ketones. Bioorg Med Chem. 1999;7:621–31. doi: 10.1016/S0968-0896(99)00009-7. [DOI] [PubMed] [Google Scholar]
- 26.Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281:12824–32. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- 27.MacFarlane M. TRAIL-induced signalling and apoptosis. Toxicol Lett. 2003;139:89–97. doi: 10.1016/S0378-4274(02)00422-8. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–33. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 29.Jang YJ, Park KS, Chung HY, Kim HI. Analysis of the phenotypes of Jurkat clones with different TRAIL-sensitivities. Cancer Lett. 2003;194:107–17. doi: 10.1016/S0304-3835(02)00680-8. [DOI] [PubMed] [Google Scholar]
- 30.Thomas WD, Hersey P. TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis in Fas ligand-resistant melanoma cells and mediates CD4 T cell killing of target cells. J Immunol. 1998;161:2195–2200. [PubMed] [Google Scholar]
- 31.Stennicke HR, Jurgensmeier JM, Shin H, Deveraux Q, Wolf BB, Yang X, et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–90. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 32.Denault JB, Eckelman BP, Shin H, Pop C, Salvesen GS. Caspase 3 attenuates XIAP (X-linked inhibitor of apoptosis protein)-mediated inhibition of caspase 9. Biochem J. 2007;405:11–19. doi: 10.1042/BJ20070288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009;28:1916–25. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645–55. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998;273:32608–13. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Q, Snipas S, Orth K, Muzio M, Dixit VM, Salvesen GS. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]
- 37.Medepalli K, Alphenaar BW, Keynton RS, Sethu P. A new technique for reversible permeabilization of live cells for intracellular delivery of quantum dots. Nanotechnology. 2013;24:205101. doi: 10.1088/0957-4484/24/20/205101. [DOI] [PubMed] [Google Scholar]
- 38.Ryan CA, Stennicke HR, Nava VE, Burch JB, Hardwick JM, Salvesen GS. Inhibitor specificity of recombinant and endogenous caspase-9. Biochem J. 2002;366:595–601. doi: 10.1042/bj20020863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finlay D, Vamos M, Gonzalez-Lopez M, Ardecky RJ, Ganji SR, Yuan H, et al. Small-molecule IAP antagonists sensitize cancer cells to TRAIL-induced apoptosis: roles of XIAP and cIAPs. Mol Cancer Ther. 2014;13:5–15. doi: 10.1158/1535-7163.MCT-13-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callus BA, Vaux DL. Caspase inhibitors: viral, cellular and chemical. Cell Death Differ. 2007;14:73–78. doi: 10.1038/sj.cdd.4402034. [DOI] [PubMed] [Google Scholar]
- 41.MacKenzie SH, Schipper JL, Clark AC. The potential for caspases in drug discovery. Curr Opin Drug Discov Devel. 2010;13:568–76. [PMC free article] [PubMed] [Google Scholar]
- 42.Vickers CJ, Gonzalez-Paez GE, Umotoy JC, Cayanan-Garrett C, Brown SJ, Wolan DW. Small-molecule procaspase activators identified using fluorescence polarization. Chembiochem. 2013;14:1419–22. doi: 10.1002/cbic.201300315. [DOI] [PubMed] [Google Scholar]
- 43.Vickers CJ, Gonzalez-Paez GE, Litwin KM, Umotoy JC, Coutsias EA, Wolan DW. Selective inhibition of initiator versus executioner caspases using small peptides containing unnatural amino acids. ACS Chem Biol. 2014;9:2194–8. doi: 10.1021/cb5004256. [DOI] [PubMed] [Google Scholar]
- 44.Brady KD. Bimodal inhibition of caspase-1 by aryloxymethyl and acyloxymethyl ketones. Biochemistry. 1998;37:8508–15. doi: 10.1021/bi9803325. [DOI] [PubMed] [Google Scholar]
- 45.Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but cannot functionally substitute caspase-8. EMBO J. 2002;21:4520–30. doi: 10.1093/emboj/cdf441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ. Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci USA. 2001;98:13884–8. doi: 10.1073/pnas.241358198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernald K, Kurokawa M. Evading apoptosis in cancer. Trends Cell Biol. 2013;23:620–33. doi: 10.1016/j.tcb.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanden Berghe T, Kaiser WJ, Bertrand MJ, Vandenabeele P. Molecular crosstalk between apoptosis, necroptosis, and survival signaling. Mol Cell Oncol. 2015;2:e975093. doi: 10.4161/23723556.2014.975093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tait SW, Ichim G, Green DR. Die another way--non-apoptotic mechanisms of cell death. J Cell Sci. 2014;127(Pt 10):2135–44. doi: 10.1242/jcs.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maly DJ, Leonetti F, Backes BJ, Dauber DS, Harris JL, Craik CS, et al. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem. 2002;67:910–5. doi: 10.1021/jo016140o. [DOI] [PubMed] [Google Scholar]
- 51.Stennicke HR, Salvesen GS. Caspases: preparation and characterization. Methods. 1999;17:313–9. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- 52.Scaffidi C, Medema JP, Krammer PH, Peter ME. FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J Biol Chem. 1997;272:26953–8. doi: 10.1074/jbc.272.43.26953. [DOI] [PubMed] [Google Scholar]
- 53.Poreba M, Solberg R, Rut W, Lunde NN, Kasperkiewicz P, Snipas SJ, et al. Counter selection substrate library strategy for developing specific protease substrates and probes. Cell Chem Biol. 2016;23:1023–35. doi: 10.1016/j.chembiol.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bromme D, Nallaseth FS, Turk B. Production and activation of recombinant papain-like cysteine proteases. Methods. 2004;32:199–206. doi: 10.1016/S1046-2023(03)00212-3. [DOI] [PubMed] [Google Scholar]
- 55.Copeland RA. A practical introduction to structure, mechanism, and data analysis. In: Enzymes. 2nd ed. Hoboken, NJ: Wiley, 2000.
- 56.Poreba M, Szalek A, Kasperkiewicz P, Drag M. Positional scanning substrate combinatorial library (PS-SCL) approach to define caspase substrate specificity. Methods Mol Biol. 2014;1133:41–59. doi: 10.1007/978-1-4939-0357-3_2. [DOI] [PubMed] [Google Scholar]
- 57.Pop C, Salvesen, GS. J Biol Chem 2009;284:21777. [DOI] [PMC free article] [PubMed]
- 58.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. Mol Cell. 2010;37:299. [DOI] [PMC free article] [PubMed]
- 59.Salvesen GS, Riedl SJ. Cell Cycle. 2009;8:2723. [DOI] [PMC free article] [PubMed]
- 60.Schafer ZT, Kornbluth S. Dev Cell. 2006;10:549. [DOI] [PubMed]
- 61.Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Cell Death Differ. 2007;14:44. [DOI] [PubMed]
- 62.Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. EMBO J. 2005;24:645. [DOI] [PMC free article] [PubMed]
- 63.Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA. J Biol Chem. 1998;273:32608. [DOI] [PubMed]
- 64.Zhou Q, Snipas S, Orth K, Muzio M, Dixit VM, Salvesen GS. J Biol Chem. 1997;272:7797. [DOI] [PubMed]
- 65.Ferreira KS, Kreutz C, Macnelly S, Neubert K, Haber A, Bogyo M, Timmer J, Borner C. Apoptosis. 2012;17:503. [DOI] [PubMed]
- 66.Fernandes-Alnemri T, Armstrong RC, Krebs J, Srinivasula SM, Wang L, Bullrich F et al. Proc Natl Acad Sci U S A. 1996;93:7464. [DOI] [PMC free article] [PubMed]
- 67.Scaffidi C, Medema JP, Krammer PH, Peter ME. J Biol Chem. 1997;272:26953. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.