Abstract
Background and Aims: This study was aim to investigate the relationship between the four intron SNPs (rs3087404, rs2029167, rs2029166 and rs7296239) of SMUG1 and the susceptibility of cervical squamous cell carcinoma.
Methods: Four SMUG1 intron SNPs (rs3087404, rs2029167, rs2029166 and rs7296239) were genotyped by MA-PCR in 400 CSCCs, 400 CIN III and 1200 controls. qRT-PCR and Western blot were used to detect the SMUG1 mRNA and protein expression.
Results: Interestingly, we found that the homozygous GG of rs3087404 had a significantly increased risk of CIN III [OR=1.78(1.27-2.51), P= 0.001] and CSCCs [OR=4.04(2.94-5.55), P=0.000]. The individuals with G allele or G carrier (AG +GG) at rs3087404 were at higher risk for CSCCs [OR=1.34 (1.04-1.71), P= 0.022]. Similarly, the homozygous GG of rs2029167 also had an increased risk of CIN III [OR=2.56 (1.91-3.43), P= 0.000] and CSCCs [OR=4.05(3.02-5.44), P=0.000]. The individuals with G allele or G carrier (AG +GG) at rs2029167 were at higher risk for CINIII [OR=1.41(1.10-1.80), P= 0.006] and CSCCs [OR=1.91 (1.48-2.47), P= 0.000]. In HR-HPV positive group, both the homozygous GG of rs3087404 and the homozygous GG of rs2029167 had an increased risk to CIN III and CSCC. Stratified analysis of the number of sexual partners and the age of first sexual intercourse found that the rs3087404 (A/G) had a particularly high level of enrichment in the CIN III or CSCCs groups. About the rs2029167 (A/G), we only found a particularly high level of enrichment grouping by the number of sexual partners in the CIN III and CSCCs groups. Meanwhile, we also found that there is a correlation between the SNPs of SMUG1 rs3087404 (A/G) and rs2029167 (A/G) with tumor cell differentiation and family heredity. But we didn't find that there was an association between the deferent genotypes of SMUG1 rs2029166 and rs7296239 with SMUG1 gene mRNA or protein expression. During the linkage disequilibrium analysis between rs3087404 (A/G) and rs2029167 (A/G), the genotype with AA-GG [OR=3.14(1.95-5.05)], AG-GG [OR=2.45(1.58-3.89)], GG-AA [OR=2.24(1.28-3.90)] and GG-AG [OR=2.58(1.54-4.32)] significantly increased the risk of CIN III. More notably, this risk is much greater in CSCCs: AA-GG [OR=7.13(4.03-12.61)], AG-GG [OR=7.22(4.21-12.38)], GG-AA [OR=8.60(4.73-15.63)], GG-AG [OR=9.64(5.43-17.13)]. Additionally, most GG (rs3087404) genotypes were linkage GG-AG (44/77, 80/140) in the CIN III and CSCCs, while most GG (rs2029167) genotypes were linkage genotype AG-GG (79/145, 112/184) in the CIN III and CSCCs, respectively.
Conclusions: These findings suggested that there was association between the two genetic polymorphisms of SMUG1 rs3087404(A/G) and rs2029167(A/G) with the susceptibility of CIN III and CSCCs, and there was a linkage disequilibrium between the rs3087404 with the rs2029167 in CIN III and CSCCs. This particular linkage disequilibrium can be used as predictive biomarkers of CIN III and CSCC.
Keywords: SMUG1, intron, genetic variant, linkage disequilibrium, cervical squamous cell carcinoma, CIN III
Introduction
Around the world, cervical cancer (CC) is the fourth most common cancer among women, accounting for an estimated 529,572 diagnosed new cases and 274,967 deaths per year 1. This cancer is the 3rd-leading cause of death in women' neoplasis worldwide and the morbidity of cervical cancer has increased recently 2. In China, the cervical cancer has become the first major female cancer (98.9 per 100000) in addition to breast cancer. The mortality rate is up to 30.5 per 100000. And the incidence rate has the increasing trends 3.
Although several factors that contribute to cervical cancer development have been identified—mainly intrinsic factors (genetic), and extrinsic factors belonging to the high risk Human Papillomavirus (HR-HPV)—genetic factors show great potential as susceptibility or prognosis indicators4,5. Only a small fraction (~1%) of cervical HR-HPV infection outcomes to cervical neoplasia, and the factors determining risk of progression are not entirely understood 6. Many genetic variants were associated with the risk of cervical cancer as supported by the epidemiological evidence 7.
Genomic instability due to DNA damage by carcinogens has been implicated in the initiation and development of cancer. DNA damage response and repair counteract the threats to genomic integrity, and variations in DNA repair capacity resulting from genetic variants could correlate with cancer predisposition 8-11. The base excision repair (BER) pathway is the major DNA genetic damage repair pathway involved in genomic instability and tumorigenesis. Previous candidate gene studies showed that selected functional single nucleotide polymorphisms (SNP) in BER genes are associated with higher risk of several solid cancers 12-17.
Single-strand selective mono-functional uracil-DNA glycosylase (SMUG1) is one of the BER genes which remove uracil from double-stranded and single-stranded DNA to maintain genomic stability following oxidative attacks 18. SNPs in this gene could have an effect on its enzyme capability of repairing DNA damage.
Xie et al evaluated the associations of 167 SNPs from 19 genes of the BER pathway with the risk of bladder cancer. 13 SNPs in 10 BER pathway genes were significantly associated with bladder cancer risk. The most significant SNP was rs2029167 in the SMUG1 gene 19. Similar studies also found a correlation between SNP of SMUG1 with breast cancer 20 and colorectal cancer (CRC) 21. Until now, there is no report of SNP of SMUG1 in cervical cancer.
In this large scale case-control study, the aim was to investigate the relationship between the four intron SNPs (rs3087404, rs2029167, rs2029166 and rs7296239) of SMUG1 and the susceptibility of cervical squamous cell carcinoma (CSCC). Genotyping analyses of the four SMUG1 SNPs were performed in 400 CSCCs, 400 precursor lesion CIN III and 1200 normal controls.
Materials and methods
Study samples selection
400 CSCC cases, 400 CIN III cases and 1200 normal controls were recruited from Zhejiang Province, China. The diagnosis was determined by two pathologists. All subjects were unrelated ethnic Chinese women and recruited between 2004 Jun to 2008 Dec. Normal controls were randomly selected from healthy women volunteers during gynecologic examinations. The inclusion criteria for healthy volunteers were without gynecological neoplasm, cytological findings, endometriosis, other solid cancer and immune disorders.
Patients with pathological diagnosis of CINIII and cervical squamous cell carcinoma (CSCC) were included in the study. Considering that patients with CINI and CINII have unstable disease progression, we excluded these patients and only selected patients with CINIII. All cases of CSCC were FIGO stage Ia-IIb, histologically confirmed primary cervical carcinoma, treated radical hysterectomy with pelvic lymph node dissection, and did not receive any anticancer therapy prior to their surgery. Patients who are eligible for any of the following criteria are excluded: over 70 years of age, with other serious complications, or previous malignant disease.
Of these, 201 CSCC patients, 357 CIN III patients and 609 normal controls agreed to provide cervical brush-off samples for detecting HR-HPV. This study was approved by the Medical Ethical Committee of Women's Hospital, School of Medicine, Zhejiang University (No.2004002). All patients signed informed consent.
DNA Extraction and Genotyping
Genomic DNA was extracted from anticoagulant peripheral blood using a DNA extraction kit according to the manufactor's guideline (Sangon Bioengineering Co., Shanghai, China). All DNA samples were dissolved in water and hypothermic preservation ready to use.
The four intron SNPs (rs3087404[A/G], rs2029167 [A/G], rs2029166 [C/T] and rs7296239 [C/T]) of SMUG1was detected by Modified polymerase chain reaction-mismatch amplification (MA-PCR) (As described in detail previously 22). The PCR forward and reverse primers and product length were showed in following table 1.
Table 1.
The PCR forward and reverse primers
| SNP No. | Forward primer | Reverse primer | Product length | |
|---|---|---|---|---|
| rs3087404 [A/G] | For “A” | 5'-CTCATCAAGAGACTGCTGGA-3' | 5'-ACTTTCATTGTTCCATAACT-3' | 240bp |
| For “G” | 5'-CTCATCAAGAGACTGCTGGG-3' | |||
| rs2029167 [A/G] | For “A” | 5'-GGGTGGTCCTCAGCTTGGCA-3' | 5'-GCAGTGACTGGCAGGAGGCG-3' | 184bp |
| For “G” | 5'-GGGTGGTCCTCAGCTTGGCG-3' | |||
| rs2029166 [C/T] | For “C” | 5'-GCCATCTCTCATGGATTAAC-3' | 5'-TTATGAGATAGCAGTGACTG-3' | 228bp |
| For “T” | 5'-GCCATCTCTCATGGATTAAT-3' | |||
| rs7296239 [C/T] | For “C” | 5'-CAGCCTCAACCCCAAAAGAC-3' | 5'-TGGCTAATGTTGAGCGAAAT-3' | 128bp |
| For “T” | 5'-CAGCCTCAACCCCAAAAGAT-3' | |||
The PCR was performed in a 25ul reaction mixture, containing 50 ng of genomic DNA, 5.0 pmol of each primer, 0.2 mM of each dNTP and 1.0U of Taq DNA polymerase (TAKARA, Dalian, China). PCR undertook the following conditions: an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30s, 57°C for 30s, and 72°C for 1min, and a final step of 72°C for 10min. The PCR products were developed by 1.5% agarose gel electrophoresis, stained with ethidium bromide and visualized with a TyphoonTM 9410 Imaging System (GE Healthcare, USA). All samples were tested twice in double blind by two different technicians, and the reproducibility of the experiment was 100%.
HR-HPV detection
HR-HPV infection was identified using the Hybrid Capture II(HC II) assay (Digene Diagnostics Inc., Gaitherburg, MD, USA) using probe B, which includes a pool of RNA probes for HR-HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68. Cervical sampling for HR-HPV DNA was performed with the Digene Cervical Sampler.
SMUG1 mRNA expression by qRT-PCR
Freshly frozen tumor tissues of eighty seven CSCCs were selected. Total RNA was extracted using Trizol reagent according to the manufacturer's protocol (Invitrogen, USA). Total RNA was treated with RNase-free DNase I. cDNA was reversed transcription and used as a template for qPCR detection. The following PCR primer pairs were used for quantitative amplification; 95°C 30s, 40 cycles at 95°C 5s followed with 60°C 30s. The primers of SMUG1 (mRNA: NM_001243787.1) were 5'-CGCAACTACGTGACTCGCTA-3'; 5'-GTCCCAGCACTGGTCGTTTA- 3'. GAPDH was used as internal control. The primers of GAPDH (mRNA: NM_001256799.2) were 5'-GAGAAGGCTGGGGCTCATTT-3'; 5'-AGTGATGGCATGGACTGTGG-3'. The PCR product length of SMUG1 and GAPDH were 190bp and 231 bp, respectively. All reactions were performed with a ViiA 7 Dx System (ABI). The cutoff point (Ct) was defined as the value when the fluorescent signal increased above the background threshold. The ΔCt for gene-specific mRNA expression was calculated relative to the Ct of GAPDH. Relative mRNA expression was calculated with the formula: 2-ΔCt.
SMUG1 protein expression by Western blotting
Eighty seven freshly CSCCs tissues were used to detect SMUG1 protein expression. Briefly, the tissue samples were minced on ice, and then the tissue was homogenized in the RIPA protein lysis buffer. The homogenized mixtures were rotated in the tubes at 4℃ for 1 h, and after centrifugation at 12,000 rpm at 4℃, the supernatant was collected and the protein concentrations were quantified.
10μl protein lysates were loaded into an 8% PAGE gel. Subsequently, electrophoretic separated proteins were transferred onto a 0.45µm PVDF membrane. After blocking with 5% non-fat milk for 1h, PVDF membrane was incubated with primary mouse monoclonal antibodies: SMUG1 (1:2000) purchased from NOVUS Biologicals (Cat No. H00023583-M07) and GAPDH (1:5000) purchased from Proteintech(Cat No. Cat.60004-1-Ig) for 4°C overnight, then were washed with TBS containing 0.05%Tween-20 for three times, followed by a 1h incubation with an HRP-conjugated secondary antibody (1:5000). After washing with TBS, the membranes were imaged with ECL using an Image Quant LAS 4000 mini (GE Healthcare).
Statistical Analysis
For the association between the genotypes and risk of cervical carcinoma, the odds ratio (OR), 95% confidence intervals (CIs) and P-values were obtained by binary logistic regression analysis. The control was set as the reference group for analysis. Stratified analysis of life style habits and genotype frequencies were evaluated with Kruskal-Wallis H test. The differences of quantitative mRNA and protein expression were calculated by ANOVA with a post hoc analysis (Fisher least significant difference). All reported values are two-tailed. The level of statistically significant difference was set at P≤0.05. All statistical analysis was done with SPSS software 18.0 ver for Windows.
Results
Clinical Features of Cases and Controls
40 years old/40 years old individuals were 602/598, 258/ 142 and 160/240 in the control, CIN III and carcinoma respectively. The carcinoma group had significantly more individuals >40 years old, but the CIN III group had more <40 years old individuals (P<0.001) compared to the control. There was no significant difference beside the increase of the proportion of individuals with number of parities more than 3 in the CIN III and carcinoma groups. The HR- HPV infection rate was 31.4%, 86.8% and 88.6% in the control, the CIN III and the carcinoma, respectively. HR-HPV infection in CIN III and cervical carcinoma cases were more than in controls. The Table 2 and data are quoted from our previously published work 23.
Table 2.
Frequency distribution of select features by case control status
| Variable | Control | CIN III | χ2* | P | Carcinoma | χ2* | P | |
|---|---|---|---|---|---|---|---|---|
| N=1200, N(%) | N=400, N(%) | N=400, N(%) | ||||||
| Age | ≤40 | 602 (50.2) | 258(64.5) | 24.793 | <0.001 | 160(40.0) | 12.431 | <0.001 |
| >40 | 598(49.8) | 142(35.5) | 240(60.0) | |||||
| Age at the first intercourse | ≤20 years | 359(29.9) | 130(32.5) | 0.943 | 0.331 | 125(31.3) | 0.253 | 0.615 |
| >20 years | 841(70.1) | 270(67.5) | 275(68.8) | |||||
| Number of sexual partners | ≤ 1 | 963(80.3) | 316(79.0) | 0.292 | 0.589 | 309(77.3) | 1.657 | 0.198 |
| >1 | 237(19.8) | 84(21.0) | 91(22.8) | |||||
| Age at the first birth | ≤22years | 235(19.6) | 91(22.8) | 1.854 | 0.173 | 89(22.3) | 1.321 | 0.25 |
| >22 years | 965(80.4) | 309(77.3) | 311(77.8) | |||||
| Number of parities** | ≤ 3 | 548(45.7) | 158(39.5) | 4.627 | 0.031 | 131(32.8) | 20.49 | <0.001 |
| >3 | 652(54.3) | 242(60.5) | 269(67.3) | |||||
| Smoking status | smoker | 4(0.3) | 2(0.5) | 0.223 | 0.637 | 2(0.5) | 0.223 | 0.637 |
| nonsmoker | 1196(99.7) | 398(99.5) | 398(99.5) | |||||
| HR-HPV infection | Positive | 191(31.4) | 310(86.8) | 277.107 | <0.001 | 178(88.6) | 199.315 | <0.001 |
| Negative | 418(68.6) | 47(13.2) | 23(11.4) | |||||
| total | 609 | 357 | 201 | |||||
Bold values show statistical data with significant difference.* Two-sided χ2 test. ** Parities including full-term pregnancy and abortion at or after 28 weeks
Correlation analysis of SMUG1 SNPs Genotypes with risk of CSCC
Table 3 represents the genotypic and allelic frequencies of SMUG1 rs3087404, rs2029166, rs2029167 and rs7296239. Genotype distributions were in Hardy-Weinberg equilibrium. The CC, CT, and TT frequency of SMUG1 rs2029166 was 42.1%, 46.1% and 11.6% in the controls, 40.8%, 45.3% and 14.0% in the CIN III, 43.5%, 39.8% and 16.8% in CSCCs. The TT, TC, and CC frequency of SMUG1 rs7296239 was 33.0%, 51.9% and 15.1% in the controls, 36.0%, 51.8% and 12.3% in the CIN III, 34.8%, 49.3% and 16.0% in CSCCs, respectively. These results indicated that the SMUG1 rs2029166 and rs7296239 polymorphism were not associated with the risk for CIN III or CSCCs.
Table 3.
Association between SMUG1 polymorphisms and the risk of CIN III and cervical carcinoma
| SMUG1 Genotypes | All patients and controls | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control N=1200 |
CIN III N=400 |
adjusted OR* (95% CI) | P | Carcinoma N=400 |
adjusted OR* (95% CI) | P | |||||||
| N | % | N | % | N | % | ||||||||
| rs3087404 | |||||||||||||
| AA | 414 | 34.5 | 141 | 35.3 | 1.00(ref) | 113 | 28.3 | 1.00(ref) | |||||
| AG | 659 | 54.9 | 182 | 45.5 | 0.81(0.63-1.04) | 0.103 | 147 | 36.8 | 0.82(0.62-1.08) | 0.149 | |||
| GG | 127 | 10.6 | 77 | 19.3 | 1.78(1.27-2.51) | 0.001 | 140 | 35.0 | 4.04(2.94-5.55) | 0.000 | |||
| AG+GG | 786 | 65.5 | 259 | 64.8 | 0.97(0.76-1.23) | 0.785 | 287 | 71.8 | 1.34(1.04-1.71) | 0.022 | |||
| Allelic frequency | |||||||||||||
| Allele A | 1487 | 62.0 | 464 | 58.0 | 1.00(ref) | 373 | 46.6 | 1.00(ref) | |||||
| Allele G | 913 | 38.0 | 336 | 42.0 | 1.78(1.00-1.39) | 0.047 | 427 | 53.4 | 1.86(1.59-2.19) | 0.000 | |||
| rs2029166 | |||||||||||||
| CC | 504 | 42.0 | 163 | 40.8 | 1.00(ref) | 174 | 43.5 | 1.00(ref) | |||||
| CT | 557 | 46.4 | 181 | 45.3 | 1.01(0.79-1.28) | 0.969 | 159 | 39.8 | 0.83(0.65-1.06) | 0.131 | |||
| TT | 139 | 11.6 | 56 | 14.0 | 1.25(0.87-1.78) | 0.228 | 67 | 16.8 | 1.40(1.00-1.96) | 0.053 | |||
| CT+TT | 696 | 58.0 | 237 | 59.3 | 1.05(0.84-1.33) | 0.661 | 226 | 56.5 | 0.94(0.75-1.18) | 0.599 | |||
| Allelic frequency | |||||||||||||
| Allele C | 1565 | 65.2 | 507 | 63.4 | 1.00(ref) | 507 | 63.4 | 1.00(ref) | |||||
| Allele T | 835 | 34.8 | 293 | 36.6 | 1.08(0.92-1.28) | 0.347 | 293 | 36.6 | 1.08(0.92-1.28) | 0.347 | |||
| rs2029167 | |||||||||||||
| AA | 455 | 37.9 | 121 | 30.3 | 1.00(ref) | 97 | 24.3 | 1.00(ref) | |||||
| AG | 532 | 44.3 | 134 | 33.5 | 0.95(0.72-1.25) | 0.700 | 119 | 29.8 | 1.05(0.78-1.41) | 0.750 | |||
| GG | 213 | 17.8 | 145 | 36.3 | 2.56(1.91-3.43) | 0.000 | 184 | 46.0 | 4.05(3.02-5.44) | 0.000 | |||
| AG+GG | 745 | 62.1 | 279 | 69.8 | 1.41(1.10-1.80) | 0.006 | 303 | 75.8 | 1.91(1.48-2.47) | 0.000 | |||
| Allelic frequency | |||||||||||||
| Allele A | 1442 | 60.1 | 376 | 47.0 | 1.00(ref) | 313 | 39.1 | 1.00(ref) | |||||
| Allele G | 958 | 39.9 | 424 | 53.0 | 1.70(1.45-1.99) | 0.000 | 487 | 60.9 | 2.34(1.99-2.76) | 0.000 | |||
| rs7296239 | |||||||||||||
| TT | 396 | 33.0 | 144 | 36.0 | 1.00(ref) | 139 | 34.8 | 1.00(ref) | |||||
| TC | 623 | 51.9 | 207 | 51.8 | 0.91(0.71-1.17) | 0.474 | 197 | 49.3 | 0.90(0.70-1.16) | 0.415 | |||
| CC | 181 | 15.1 | 49 | 12.3 | 0.74(0.52-1.08) | 0.117 | 64 | 16.0 | 1.00(0.71-1.42) | 0.967 | |||
| TC+CC | 804 | 67.0 | 256 | 64.0 | 0.88(0.69-1.11) | 0.272 | 261 | 65.3 | 0.93(0.73-1.17) | 0.521 | |||
| Allelic frequency | |||||||||||||
| Allele T | 1415 | 59.0 | 495 | 61.9 | 1.00(ref) | 475 | 59.4 | 1.00(ref) | |||||
| Allele C | 985 | 41.0 | 305 | 38.1 | 0.89(0.75-1.04) | 0.145 | 325 | 40.6 | 0.98(0.84-1.16) | 0.836 | |||
Bold values show statistical data with significant difference. *All P-values are adjusted for age, number of sexual partners, age at first intercourse, parities (including full-term pregnancy and abortion at or after 28 weeks) and age at first full-term pregnancy.
The AA, AG, and GG frequency of SMUG1 rs3087404 was 34.5%, 54.9% and 10.6% in the controls, 35.3%, 45.5% and 19.3% in the CIN III, 28.3%, 36.8% and 35.0% in CSCCs, respectively. These results revealed that women with the homozygous GG of rs3087404 had a significantly increased risk of CIN III [OR=1.78(1.27-2.51), P= 0.001] and CSCC [OR=4.04 (2.94-5.55), P=0.000]. We observed “A” allele is the major form at rs3087404 in controls (62.0%, 1487/ 2400), but “G” allele is the major form in CSCCs (53.4%, 427/800). The increased risk of “G” allele in CIN III and CSCCs were 1.78(1.00-1.39) and 1.86 (1.59-2.19) respectively. Data also indicated that individuals with “G” allele or “G” carrier (AG +GG) at rs3087404 were at higher risk for CSCCs [OR=1.34 (1.04-1.71), P= 0.022].
The AA, AG, and GG frequency of SMUG1 rs2029167 was 37.9%, 44.3% and 17.8% in the controls, 30.3%, 33.5% and 36.3% in the CIN III, 24.3%, 29.8% and 46.0% in CSCCs, respectively. These results revealed that women with the homozygous GG of rs2029167 had an increased risk of CIN III [OR=2.56 (1.91-3.43), P= 0.000] and CSCCs [OR=4.05(3.02-5.44), P=0.000]. “A” allele is the major form at rs2029167 in controls (60.1%, 1442/2400), but “G” allele is the major form in CIN III (53.0%, 424/800) and in CSCCs (60.9%, 487/800). “G” allele at rs2029167 was significantly higher in CIN III and CSCCs compared with normal controls. The increased risk of “G” allele in CIN III and CSCCs were OR=1.70(1.45-1.99) and OR=2.34(1.99-2.76) respectively. “G” allele or “G” carrier (AG +GG) at rs2029167 were at higher risk for CINIII [OR=1.41(1.10-1.80), P= 0.006] and CSCCs [OR=1.91 (1.48-2.47), P= 0.000].
As show in Table 4, in the HR-HPV positive group, though the homozygous GG of rs3087404 have not increased the risk of CIN III [OR=1.43(0.80-2.53, P= 0.226], it significantly increased the risk of CSCCs [OR=3.91(2.15-7.15), P=0.000], the increased risk of “G” allele in CSCCs were OR=1.94(1.45-2.60). The homozygous GG of rs2029167 had an increased risk of CIN III [OR=2.40 (1.48-3.89), P= 0.000] and CSCCs [OR=3.88(2.26-6.88), P=0.000]. Meanwhile, the increased risk of “G” allele in CIN III and CSCCs were OR=1.66(1.29-2.15) and OR=2.35(1.75-3.15).
Table 4.
Association between SMUG1 rs3087404 and rs2029167 polymorphisms with the risk of HR-HPV positive cervical carcinoma and CIN III
| SMUG1 Genotypes | HPV-positive patients and controls | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control N=191 |
CIN III N=310 |
adjusted OR*(95% CI) | P | carcinoma N=178 |
adjusted OR*(95% CI) | P | |||||||
| N | % | N | % | n | % | ||||||||
| rs3087404 | |||||||||||||
| AA | 65 | 34.0 | 111 | 35.8 | 1.00(ref) | 49 | 27.5 | 1.00(ref) | |||||
| AG | 103 | 53.9 | 143 | 46.1 | 0.81(0.55-1.21) | 0.307 | 61 | 34.3 | 0.79(0.48-1.28) | 0.332 | |||
| GG | 23 | 12.0 | 56 | 18.1 | 1.43(0.80-2.53) | 0.226 | 68 | 38.2 | 3.91(2.15-7.15) | 0.000 | |||
| AG+GG | 126 | 66.0 | 199 | 64.2 | 0.93(0.63-1.35) | 0.686 | 129 | 72.5 | 1.36(0.87-2.12) | 0.177 | |||
| Allelic frequency | |||||||||||||
| Allele A | 233 | 61.0 | 365 | 58.9 | 1.00(ref) | 159 | 44.7 | 1.00(ref) | |||||
| Allele G | 149 | 39.0 | 255 | 41.1 | 1.09(0.84-1.42) | 0.506 | 197 | 55.3 | 1.94(1.45-2.60) | 0.000 | |||
| rs2029167 | |||||||||||||
| AA | 71 | 37.2 | 92 | 29.7 | 1.00(ref) | 42 | 23.6 | 1.00(ref) | |||||
| AG | 83 | 43.5 | 103 | 33.2 | 0.96(0.63-1.46) | 0.841 | 51 | 28.7 | 1.04(0.62-1.74) | 0.885 | |||
| GG | 37 | 19.4 | 115 | 37.1 | 2.40(1.48-3.89) | 0.000 | 85 | 47.8 | 3.88(2.26-6.88) | 0.000 | |||
| AG+GG | 120 | 62.8 | 218 | 70.3 | 1.40(0.96-2.05) | 0.083 | 136 | 76.4 | 1.92(1.22-3.02) | 0.005 | |||
| Allelic frequency | |||||||||||||
| Allele A | 225 | 58.9 | 287 | 46.3 | 1.00(ref) | 135 | 37.9 | 1.00(ref) | |||||
| Allele G | 157 | 41.1 | 333 | 53.7 | 1.66(1.29-2.15) | 0.000 | 221 | 62.1 | 2.35(1.75-3.15) | 0.000 | |||
Bold values show statistical data with significant difference. *All P-values are adjusted for age, number of sexual partners, age at first intercourse, parities (including full-term pregnancy and abortion at or after 28 weeks) and age at first full-term pregnancy.
Association between SMUG1 rs3087404, rs2029167 polymorphisms and the Sexual, Reproductive History in CSCCs and CIN III
As show in Table 5, the participants were divided into two groups according to Age, age of first sexual intercourse, number of sexual partners, age at first parity, number of parities and HR-HPV infection, then stratified analysis was done with the SMUG1 rs3087404 (A/G) and rs2029167 (A/G) genotype. Stratified analysis of age, number of parities, and age at first parity showed no correlation with rs3087404 (A/G) polymorphisms. However, we find a particularly high level of enrichment between groups with stratified analysis of the number of sexual partners in the CIN III (χ2=15.610, P=0.000) and CSCCs (χ2=13.468, P=0.000), and the age of first sexual intercourse in the CIN III (χ2=18.453, P=0.000) and CSCCs (χ2=15.528, P=0.000). We also did not find a high level of enrichment between HR-HPV positive and negative group of CIN III (χ2=0.176, P=0.675) and CSCCs (χ2=0.017, P=0.895).
Table 5.
Association between SMUG1 rs3087404 polymorphisms and the risk for CIN III and cervical carcinoma stratified by the sexual, reproductive history
| High risk exposure | Controls | χ2 | P | CIN III | χ2 | P | Carcinoma | χ2 | P | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | AA | AG | GG | |||||||||||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | ||||||||
| Age | |||||||||||||||||||||||||
| ≤ 40 | 212 | 35.2 | 323 | 53.7 | 67 | 11.1 | 0.036 | 8.490 | 93 | 36.0 | 120 | 46.5 | 45 | 17.4 | 0.837 | 0.360 | 44 | 27.5 | 61 | 38.1 | 55 | 34.4 | 0.000 | 0.991 | |
| >40 | 202 | 33.8 | 336 | 56.2 | 60 | 10.0 | 48 | 33.8 | 62 | 43.7 | 32 | 22.5 | 69 | 28.8 | 86 | 35.8 | 85 | 35.4 | |||||||
| Number of sexual partners | |||||||||||||||||||||||||
| ≤ 1 | 321 | 33.3 | 538 | 55.9 | 104 | 10.8 | 2.590 | 0.108 | 120 | 38.0 | 151 | 47.8 | 45 | 14.2 | 15.610 | 0.000 | 93 | 30.1 | 126 | 40.8 | 90 | 29.1 | 13.468 | 0.000 | |
| >1 | 93 | 39.2 | 121 | 51.1 | 23 | 9.7 | 21 | 25.0 | 31 | 36.9 | 32 | 38.1 | 20 | 22.0 | 21 | 23.1 | 50 | 54.9 | |||||||
| Age at the first intercourse | |||||||||||||||||||||||||
| ≤20 | 122 | 34.0 | 187 | 52.1 | 50 | 13.9 | 1.364 | 0.243 | 32 | 24.6 | 57 | 43.8 | 41 | 31.5 | 18.453 | 0.000 | 26 | 20.8 | 36 | 28.8 | 63 | 50.4 | 15.528 | 0.000 | |
| >20 | 292 | 34.7 | 472 | 56.1 | 77 | 9.2 | 109 | 40.4 | 125 | 46.3 | 36 | 13.3 | 87 | 31.6 | 111 | 40.4 | 77 | 28.0 | |||||||
| Number of parities | |||||||||||||||||||||||||
| ≤ 3 | 184 | 33.6 | 299 | 54.6 | 65 | 11.9 | 1.055 | 0.304 | 51 | 32.3 | 74 | 46.8 | 33 | 20.9 | 1.094 | 0.296 | 34 | 26.0 | 52 | 39.7 | 45 | 34.4 | 0.069 | 0.792 | |
| >3 | 230 | 35.3 | 360 | 55.2 | 62 | 9.5 | 90 | 37.2 | 108 | 44.6 | 44 | 18.2 | 79 | 29.4 | 95 | 35.3 | 95 | 35.3 | |||||||
| Age at the first parity | |||||||||||||||||||||||||
| ≤22 | 84 | 35.7 | 131 | 55.7 | 20 | 8.5 | 0.675 | 0.411 | 31 | 34.1 | 43 | 47.3 | 17 | 18.7 | 0.014 | 0.905 | 22 | 24.7 | 36 | 40.4 | 31 | 34.8 | 0.183 | 0.669 | |
| >22 | 330 | 34.2 | 528 | 54.7 | 107 | 11.1 | 110 | 35.6 | 139 | 45.0 | 60 | 19.4 | 91 | 29.3 | 111 | 35.7 | 109 | 35.0 | |||||||
| HR-HPV infection status | |||||||||||||||||||||||||
| Positive | 65 | 34.0 | 103 | 53.9 | 23 | 12.0 | 0.595 | 0.440 | 111 | 35.8 | 143 | 46.1 | 56 | 18.1 | 0.176 | 0.675 | 49 | 27.5 | 61 | 34.3 | 68 | 38.2 | 0.017 | 0.895 | |
| Negative | 151 | 36.1 | 227 | 54.3 | 40 | 9.6 | 16 | 34.0 | 21 | 44.7 | 10 | 21.3 | 6 | 26.1 | 9 | 39.1 | 8 | 34.8 | |||||||
Bold values show statistical data with significant difference. Stratified analysis were applied by the Kruskal-Wallis H. A p value less than 0.05 was considered significant.
Data display of rs2029167 (A/G) as show in Table 6, we did not find a particularly high level of enrichment between groups, except for the number of sexual partners in the CIN III (χ2=10.214, P=0.001) and CSCCs (χ2=12.366, P=0.000), there was a particularly high level of enrichment.
Table 6.
Association between SMUG1 rs2029167 polymorphisms and the risk for CIN and cervical carcinoma stratified by the sexual, reproductive history
| High risk exposure | Controls | χ2 | P | CIN III | χ2 | P | Carcinoma | χ2 | P | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | AA | AG | GG | |||||||||||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | ||||||||
| Age | |||||||||||||||||||||||||
| ≤ 40 | 234 | 38.9 | 278 | 46.2 | 90 | 15.0 | 2.702 | 0.100 | 75 | 29.1 | 88 | 34.1 | 95 | 36.8 | 0.325 | 0.569 | 38 | 23.8 | 44 | 27.5 | 78 | 48.8 | 0.513 | 0.474 | |
| >40 | 221 | 37.0 | 254 | 42.5 | 123 | 20.6 | 46 | 32.4 | 46 | 32.4 | 50 | 35.2 | 59 | 24.6 | 75 | 31.3 | 106 | 44.2 | |||||||
| Number of sexual partners | |||||||||||||||||||||||||
| ≤ 1 | 362 | 37.6 | 431 | 44.8 | 170 | 17.7 | 0.074 | 0.786 | 103 | 32.6 | 112 | 35.4 | 101 | 32.0 | 10.214 | 0.001 | 83 | 26.9 | 99 | 32.0 | 127 | 41.1 | 12.366 | 0.000 | |
| >1 | 93 | 39.2 | 101 | 42.6 | 43 | 18.1 | 18 | 21.4 | 22 | 26.2 | 44 | 52.4 | 14 | 15.4 | 20 | 22.0 | 57 | 62.6 | |||||||
| Age at the first intercourse | |||||||||||||||||||||||||
| ≤20 | 132 | 36.8 | 164 | 45.7 | 63 | 17.5 | 0.121 | 0.728 | 38 | 29.2 | 42 | 32.3 | 50 | 38.5 | 0.312 | 0.576 | 29 | 23.2 | 36 | 28.8 | 60 | 48.0 | 0.274 | 0.601 | |
| >20 | 323 | 38.4 | 368 | 43.8 | 150 | 17.8 | 83 | 30.7 | 92 | 34.1 | 95 | 35.2 | 68 | 24.7 | 83 | 30.2 | 124 | 45.1 | |||||||
| Number of parities | |||||||||||||||||||||||||
| ≤ 3 | 202 | 36.9 | 238 | 43.4 | 108 | 19.7 | 1.539 | 0.215 | 46 | 29.1 | 51 | 32.3 | 61 | 38.6 | 0.494 | 0.482 | 31 | 23.7 | 36 | 27.5 | 64 | 48.9 | 0.416 | 0.519 | |
| >3 | 253 | 38.8 | 294 | 45.1 | 105 | 16.1 | 75 | 31.0 | 83 | 34.3 | 84 | 34.7 | 66 | 24.5 | 83 | 30.9 | 120 | 44.6 | |||||||
| Age at the first parity | |||||||||||||||||||||||||
| ≤22 | 88 | 37.4 | 101 | 43.0 | 46 | 19.6 | 0.237 | 0.626 | 25 | 27.5 | 29 | 31.9 | 37 | 40.7 | 0.933 | 0.334 | 19 | 21.3 | 23 | 25.8 | 47 | 52.8 | 1.797 | 0.180 | |
| >22 | 367 | 38.0 | 431 | 44.7 | 167 | 17.3 | 96 | 31.1 | 105 | 34.0 | 108 | 35.0 | 78 | 25.1 | 96 | 30.9 | 137 | 44.1 | |||||||
| HR-HPV infection status | |||||||||||||||||||||||||
| Positive | 71 | 37.2 | 83 | 43.5 | 37 | 19.4 | 0.028 | 0.867 | 92 | 29.7 | 103 | 33.2 | 115 | 37.1 | 0.330 | 0.566 | 42 | 23.6 | 51 | 28.7 | 85 | 47.8 | 0.135 | 0.713 | |
| Negative | 149 | 35.6 | 191 | 45.7 | 78 | 18.7 | 16 | 34.0 | 15 | 31.9 | 16 | 34.0 | 5 | 21.7 | 6 | 26.1 | 12 | 52.2 | |||||||
Bold values show statistical data with significant difference. Stratified analysis were applied by the Kruskal-Wallis H. A p value less than 0.05 was considered significant.
Association between SMUG1 rs3087404, rs2029167 polymorphisms and the Clinical pathological characteristics in CSCCs
The correlation of SMUG1 rs3087404 and rs2029167 polymorphisms with CSCCs clinicopathological characteristics is shown in Table 7.The CSCCs were divided into two groups according to age, tumor family history, FIGO stage, tumor size, differentiation grade, lymph node metastasis, vascular involvement, stromal invasion, vaginal wall extension, parametrial extension, and endometrial extension, then stratified analysis was done with the SMUG1 rs3087404 (A/G) and rs2029167 (A/G) genotype.
Table 7.
Association between SMUG1 rs3087404 and rs2029167 polymorphisms and the risk for cervical carcinoma stratified by clinical pathological characteristics
| Clinical pathological characteristics | SMUG1 rs3087404 | χ2 | P | SMUG1 rs2029167 | χ2 | P | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | ||||||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | ||||||
| Age | |||||||||||||||||
| ≤ 40 | 42 | 26.3 | 66 | 41.3 | 52 | 32.5 | 0.022 | 0.882 | 36 | 22.5 | 52 | 32.5 | 72 | 45.0 | 0.003 | 0.955 | |
| >40 | 71 | 29.6 | 81 | 33.8 | 88 | 36.7 | 61 | 25.4 | 67 | 27.9 | 112 | 46.7 | |||||
| Tumor family history | |||||||||||||||||
| Negative | 108 | 29.4 | 139 | 37.9 | 120 | 32.7 | 8.792 | 0.003 | 87 | 23.7 | 113 | 30.8 | 167 | 45.5 | 0.01 | 0.919 | |
| positive | 5 | 15.2 | 8 | 24.2 | 20 | 60.6 | 10 | 30.3 | 6 | 18.2 | 17 | 51.5 | |||||
| FIGO stage | |||||||||||||||||
| I | 96 | 28.4 | 123 | 36.4 | 119 | 35.2 | 0.002 | 0.966 | 84 | 24.9 | 103 | 30.5 | 151 | 44.7 | 1.337 | 0.248 | |
| II | 17 | 27.4 | 24 | 38.7 | 21 | 33.9 | 13 | 21.0 | 16 | 25.8 | 33 | 53.2 | |||||
| Tumor size | |||||||||||||||||
| <4cm | 96 | 28.8 | 117 | 35.1 | 120 | 36.0 | 0.091 | 0.763 | 81 | 24.3 | 95 | 28.5 | 157 | 47.1 | 0.474 | 0.491 | |
| ≥4cm | 17 | 25.4 | 30 | 44.8 | 20 | 29.9 | 16 | 23.9 | 24 | 35.8 | 27 | 40.3 | |||||
| Differentiation grade | |||||||||||||||||
| Grade I-II | 104 | 30.1 | 131 | 37.9 | 111 | 32.1 | 9.265 | 0.002 | 89 | 25.7 | 108 | 31.2 | 149 | 43.1 | 8.112 | 0.004 | |
| Grade III | 9 | 16.7 | 16 | 29.6 | 29 | 53.7 | 8 | 14.8 | 11 | 20.4 | 35 | 64.8 | |||||
| Lymph node metastasis | |||||||||||||||||
| Negative | 101 | 28.1 | 133 | 37.0 | 125 | 34.8 | 0.003 | 0.953 | 85 | 23.7 | 107 | 29.8 | 167 | 46.5 | 0.599 | 0.439 | |
| positive | 12 | 29.3 | 14 | 34.1 | 15 | 36.6 | 12 | 29.3 | 12 | 29.3 | 17 | 41.5 | |||||
| Vascular involvement | |||||||||||||||||
| Negative | 96 | 28.1 | 127 | 37.1 | 119 | 34.8 | 0.001 | 0.979 | 82 | 24.0 | 104 | 30.4 | 156 | 45.6 | 0.017 | 0.896 | |
| positive | 17 | 29.3 | 20 | 34.5 | 21 | 36.2 | 15 | 25.9 | 15 | 25.9 | 28 | 48.3 | |||||
| Stromal invasion | |||||||||||||||||
| <2/3 | 84 | 29.8 | 106 | 37.6 | 92 | 32.6 | 2.363 | 0.124 | 74 | 26.2 | 84 | 29.8 | 124 | 44.0 | 2.263 | 0.132 | |
| ≥2/3 | 29 | 24.6 | 41 | 34.7 | 48 | 40.7 | 23 | 19.5 | 35 | 29.7 | 60 | 50.8 | |||||
| Vaginal wall extension | |||||||||||||||||
| Negative | 92 | 29.0 | 121 | 38.2 | 104 | 32.8 | 2.224 | 0.136 | 76 | 24.0 | 92 | 29.0 | 149 | 47.0 | 0.438 | 0.508 | |
| positive | 21 | 25.3 | 26 | 31.3 | 36 | 43.4 | 21 | 25.3 | 27 | 32.5 | 35 | 42.2 | |||||
| Parametrail extension | |||||||||||||||||
| Negative | 104 | 28.8 | 135 | 37.4 | 122 | 33.8 | 1.888 | 0.169 | 89 | 24.7 | 107 | 29.6 | 165 | 45.7 | 0.249 | 0.618 | |
| positive | 9 | 23.1 | 12 | 30.8 | 18 | 46.2 | 8 | 20.5 | 12 | 30.8 | 19 | 48.7 | |||||
| Endometrial extension | |||||||||||||||||
| Negative | 106 | 28.7 | 139 | 37.7 | 124 | 33.6 | 2.767 | 0.096 | 91 | 24.7 | 111 | 30.1 | 167 | 45.3 | 1.017 | 0.313 | |
| positive | 7 | 22.6 | 8 | 25.8 | 16 | 51.6 | 6 | 19.4 | 8 | 25.8 | 17 | 54.8 | |||||
Bold values show statistical data with significant difference. Stratified analysis were applied by the Kruskal-Wallis H. A P value less than 0.05 was considered significant.
Stratified analysis of age, FIGO stage, tumor size, lymph node metastasis, vascular involvement, stromal invasion, vaginal wall extension, parametrail extension, and endometrial extension showed no correlation with rs3087404 (A/G) or rs2029167 (A/G) polymorphism. However, we found a particularly high level of enrichment of rs3087404 (χ2=9.265, P= 0.002)) and rs2029167 (χ2=8.112, P=0.004) when stratified by differentiation grade. This means that GG homozygotes of rs3087404 and rs2029167 are significantly associated with the degree of malignancy of tumor differentiation. In addition, interestingly, we found that GG homozygote of rs3087404 is also associated with a family history of the tumor (χ2=8.792, P=0.003).
mRNA and protein expression from patients with different genotypes of SMUG1 rs3087404 (A/G) or rs2029167 (A/G)
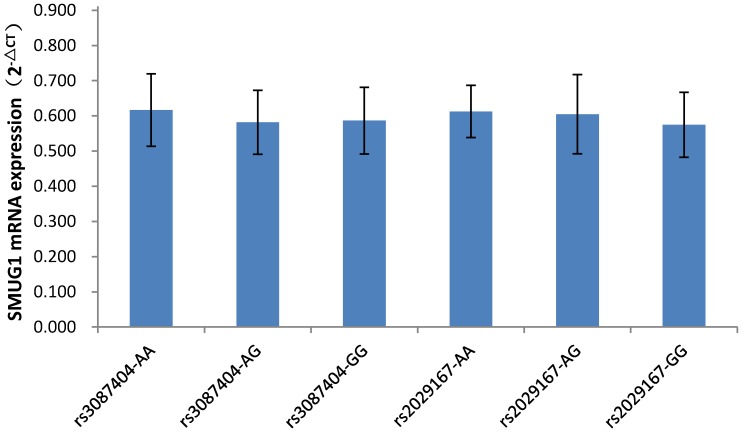
As shown in Figure 1, among the 87 CSCCs patients, the genotypes of AA, AG, and GG at rs3087404 were 25(28.7%), 32(36.8%) and 30(34.5%) cases, while the genotypes of AA, AG, and GG at rs2029167 were 22(25.3%), 26(30.0%) and 39(44.8%) cases, respectively. There was no significant difference in the expression of SMUG1 mRNA with different genotypes at rs3087404 (F=1.022, P=0.364) or at rs2029167 (F=2.067, P=0.133).
Figure 1.
SMUG1 mRNA expression in CSCCs with different genotypes of rs3087404 and rs2029167 (qPCR).
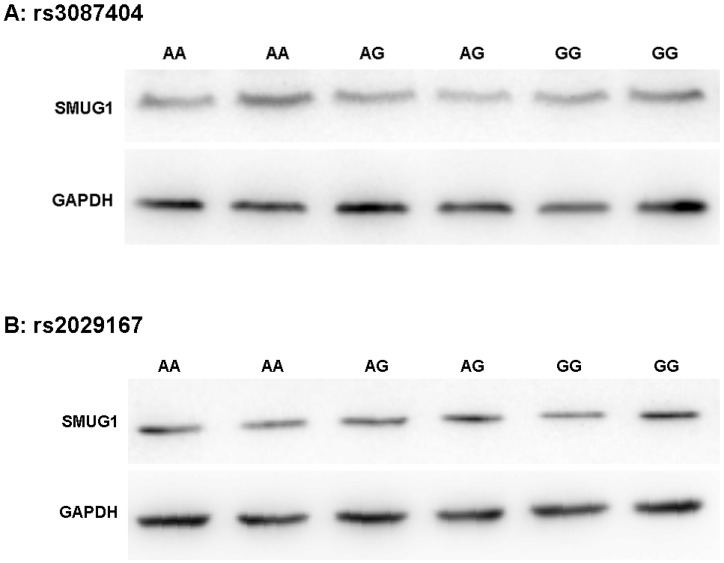
As shown in Figure 2, Western Blot experiments confirmed that the polymorphism of rs3087404 did not affect the expression of SMUG1 protein (F=0.254, P=0.781). Similarly, the polymorphisms of rs2029167 is also independent of the expression level of SMUG1 protein (F=1.346, P=0.308).
Figure 2.
SMUG1 protein expression in CSCCs with different genotypes of rs3087404 and rs2029167 (Western Blot) (A) AA: rs3087404 genotype is AA; AG: rs3087404 genotype is AG; GG: rs3087404 genotype is GG; (B) AA: rs2029167 genotype is AA; AG: rs2029167 genotype is AG; GG: rs2029167 genotype is GG
Haplotype Analysis between the Linkage Disequilibrium of the SMUG1 rs3087404 and rs2029167 Variants Genotypes and the Risk of CIN III and CSCCs
Since that the frequencies of both rs3087404 (A/G) and rs2029167 (A/G) genotypes change the risk of CIN III or CSCCs significantly, we further analyzed the linkage disequilibrium between rs3087404 (A/G) and rs2029167 (A/G). The frequencies of the nine haplotypes were shown in Table 8. GG (rs3087404)-GG (rs2029167) was not detected in normal control and CSCCs, except for 1 case detected in CIN III. Compared to AA (rs3087404)-AA (rs2029167), the genotype with AA-GG [OR=3.14 (1.95-5.05), P=0.000], AG-GG [OR=2.45(1.58-3.89), P= 0.000], GG-AA [OR=2.24(1.28-3.90), P=0.005], GG-AG [OR=2.58(1.54-4.32), P=0.027] significantly increased the risk of CIN III. More notably, this risk is much greater in CSCC: AA-GG [OR=7.13(4.03-12.61), P= 0.000], AG-GG [OR=7.22(4.21-12.38), P=0.000], GG- AA [OR=8.60(4.73-15.63), P=0.000], GG-AG [OR=9.64 (5.43-17.13, P=0.000]. This means that whether the rs3087404 or rs2029167 is GG homozygote, the linkage mode is at high risk. We also found that women with the AG-AG genotype had a decreased risk for CSCCs [OR=0.49(0.25-0.96), P=0.038].
Table 8.
Genotypes and the risk of all CIN III and cervical carcinoma subjects
| SMUG1 Genotypesa | All patients and controls | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | CIN III | adjusted ORb(95% CI) | P | Carcinoma | adjusted ORb(95% CI) | P | |||||
| 1200 | 400 | 400 | |||||||||
| N | % | N | % | N | % | ||||||
| AA-AA | 158 | 13.2 | 39 | 9.8 | 1.00(ref) | 19 | 4.8 | 1.00(ref) | |||
| AA-AG | 172 | 14.3 | 37 | 9.3 | 0.87(0.53-1.44) | 0.589 | 22 | 5.5 | 1.06(0.56-2.04) | 0.853 | |
| AA-GG | 84 | 7.0 | 65 | 16.3 | 3.14(1.95-5.05) | 0.000 | 72 | 18.0 | 7.13(4.03-12.61) | 0.000 | |
| AG-AA | 239 | 19.9 | 50 | 12.5 | 0.85(0.53-1.35) | 0.485 | 18 | 4.5 | 0.63(0.32-1.23) | 0.174 | |
| AG-AG | 291 | 24.3 | 53 | 13.3 | 0.74(0.47-1.17) | 0.192 | 17 | 4.3 | 0.49(0.25-0.96) | 0.038 | |
| AG-GG | 129 | 10.8 | 79 | 19.8 | 2.45(1.58-3.89) | 0.000 | 112 | 28.0 | 7.22(4.21-12.38) | 0.000 | |
| GG-AA | 58 | 4.8 | 32 | 8.0 | 2.24(1.28-3.90) | 0.005 | 60 | 15.0 | 8.60(4.73-15.63) | 0.000 | |
| GG-AG | 69 | 5.8 | 44 | 11.0 | 2.58(1.54-4.32) | 0.027 | 80 | 20.0 | 9.64(5.43-17.13) | 0.000 | |
| GG-GG | 0 | 0.0 | 1 | 0.3 | _ | _ | 0 | 0.0 | _ | _ | |
Bold values show statistical data with significant difference. agenotypes are composed of two polymorphic sites: rs3087404(A/G), rs2029167(A/G). bAll P-values are adjusted for age, number of sexual partners, age at first intercourse, parities (including full-term pregnancy and abortion at or after 28 weeks) and age at first full-term pregnancy.
Additionally, most GG (rs3087404) genotypes were linkage GG-AG (44/77, 80/140) in the CIN III and CSCC, while most GG (rs2029167) genotypes were linkage genotype AG-GG (79/145, 112/184) in the CIN III and CSCCs, respectively. These indicated that the majority of GG genotype distributions are caused by the linkage disequilibrium with the corresponding alleles.
Discussion
Uracil misincorporation into DNA arises spontaneously at low level as a result of cytosine deamination or misincorporation of dUMP during DNA replication 24, 25. Under normal conditions, such lesions are rapidly recovered by the BER pathway initiated by uracil-DNA glycosylases (UDG) 26, 27. In most organisms, including humans, uracil is generally an undesirable ingredient in the genome. Thus strategies are in place to remove uracil once occurring of the DNA damage. So, sophisticated mechanisms are essential for the removal of uracil from DNA and prevention of its misincorporation, and maintain genomic integrity and stability. The failure of removing misincorporated uracil from DNA will result in base abnormity during DNA replication, even lead to dsDNA breaks and chromosomal aberrations, these two events are the key genetic factors of tumorigenesis25, 28-30.
BER is a highly conserved DNA repair system from bacteria to humans 31-33. A great variety of DNA-damaging agents can cause genome instability, which would be a tremendous matter for cells if the damaged DNA is not recovered 11. The most important role of BER is to remove DNA damage caused by various carcinogens, such as reactive oxygen species (ROS), ionizing radiation and so on34. In humans, four UDGs have been identified, encoded by the UNG, SMUG1, MBD4 and TDG genes 33,35,36. Most of these critical BER genes are highly polymorphic 37. Genetic variations of these genes are likely to alter BER enzyme functional activity, and influence cancer risk 36.
The human single-strand-selective monofunctional uracil-DNA glycosylase 1(SMUG1; also named: FDG, UNG3 and HMUDG) is located in the Chromosome 12q13.11-13.3 26, 38. This gene encodes for a uracil DNA glycosylase (UDG) of the BER pathway that removes uracil, from single stranded (ssDNA) as well as double stranded DNA (dsDNA) 39. As SMUG1 removes uracil and 5-hmeU from ssDNA and dsDNA, this enzyme may take participate in the repair of deamination and oxidation damage. The SMUG1 is the major enzyme involved in the removal of 5-hmeU from damaged DNA 40.
Several researchers reported the SNPs of SMUG1were correlationship with bladder cancer, breast cancer and CRC susceptibility. In a matched study of 801 bladder cancer cases, Xie et al. found 13 SNPs in10 BER pathway genes significantly increased the risk of bladder cancer. The most significant variant was SMUG1 rs2029167 (A/G). The homozygous GG genotype increased a 1.42-fold risk of bladder cancer 19.In another 1,077 case-controls matched study of incident breast cancer, Marian et al suggested that there was increased risk of breast cancer among postmenopausal women who were heterozygous of two of SMUG1 SNPs which is thought to be the most active glycosylase in vivo, raises the possibility that subtle 'heterosis' effects on cancer risk might be produced by these SNPs 20. In a study of CRC, Broderick et al reported that genetic variations in TDG, UNG and SMUG1 may play a role in the susceptibility of CRC 21. These reports remind us to make a hypothesis which there is an association between the genetic variants of SMUG1 gene with cancer risk. We carried out the correlation study of cervical cancer and SMUG1 SNPs.
The initiation and development of cervical carcinoma involves reversible transformation in the cervical squamous cells resulting in various cellular abnormalities and ultimately to cervical tumorigenesis. The development of cervical carcinoma usually requires multiple stages, eventually developing from precursor lesion cervical intraepithelial neoplasia (CIN) to cervical malignant carcinoma 21. In our results, the two of SMUG1 rs2029166 and rs7296239 polymorphisms were not associated with the risk for CIN III or CSCC. Interestingly, the homozygous GG of rs3087404 and rs2029167 had a significantly increased risk of CIN III and CSCC. We also observed the increased risk of G allele of these two SNP in CIN III and CSCC. The individuals with G allele or G carrier (AG +GG) at rs3087404 and rs2029167 were at higher risk for CSCC. These findings indicated that the SMUG1 rs2029166 and rs7296239 polymorphisms (G allele) maybe play a role in initiation and progression of precancerous lesions (CIN) and cervical carcinoma. So far, there is no study about the correlation between cervical carcinoma and the SMUG1 rs2029166 and rs7296239 polymorphisms (G allele). The present study is the first time to discover the association between the SMUG1 rs2029166 and rs7296239 polymorphisms (G allele) and cervical carcinoma or CIN III. In general, SNP loci that affect the structure and function of genes are located in the 5' UTR promoter, coding region, or 3' UTR region. Although our two variants both are located in the intron which cannot change the amino acid, it is possible that there is linkage disequilibrium with other functional genetic variants and serves as a genetic marker of susceptibility 41. Another possibility is that the SMUG1 rs2029166 and rs7296239 genetic variants maybe influence primary mRNA splicing and regulation, and affects SMUG1 protein expression or produce alternative spliceosome. To validate the SMUG1 expression change, we detected the mRNA and protein expression in fresh tumor tissues in the different genotype groups of SMUG1 rs2029166 and rs7296239, but we discovered that there was no association between the genotype of SMUG1 rs2029166 and rs7296239 with SMUG1 gene mRNA or protein expression. These indicate that the tumor susceptibility induced by the polymorphism of this locus was not achieved by altering gene expression. Bonnet et al. speculated that the introns take participate in maintaining genetic stability at certain locations, particularly in highly expressed genes 42, and repair genes are often high expression genes.
During the linkage disequilibrium analysis between rs3087404 (A/G) and rs2029167 (A/G), we found that whether the rs3087404 or rs2029167 is GG homozygote, the linkage mode is at high risk. Additionally, most GG (rs3087404) genotypes were linkage GG-AG (44/77, 80/140) in the CIN III and CSCCs, while most GG (rs2029167) genotypes were linkage genotype AG-GG (79/145, 112/184) in the CIN III and CSCCs, respectively. These indicated that the majority of GG genotype distributions are caused by the linkage disequilibrium with the corresponding alleles. These linkage modes can be used as genetic biomarker of early prediction of cervical carcinoma, as an indicator of primary prevention.
Stratified analysis of the number of sexual partners and the age of first sexual intercourse found that the rs3087404 (A/G) had a particularly high level of enrichment in the CIN III and CSCCs. About the rs2029167 (A/G), we only found a particularly high level of enrichment grouping by the number of sexual partners in the CIN III. This suggests that there may be a certain correlation between SMUG1 rs3087404 (A/G) and rs2029167 (A/G) variants with the female sexual behavior.
Among all of clinical parameters, we found that the genetic polymorphisms of rs3087404 (A/G) and rs2029167 (A/G) are significantly associated with the degree of malignancy of tumor differentiation; homozygous GG genotype increases the risk of malignant cell differentiation grade of tumors. In addition, interestingly, we found that GG homozygote of rs3087404 is also associated with a family history of the tumor. These indicate that there maybe a correlation between the SNPs of SMUG1 rs3087404 (A/G) and rs2029167 (A/G) with tumor cell differentiation and family heredity.
In HR-HPV positive group, we found that the homozygous GG of rs3087404 and rs2029167 both significantly increased the risk of CSCCs, only “G” allele or “G” carrier (AG +GG) at rs2029167 were at higher risk for CSCCs. But, in stratified analysis, we did not find a high level of enrichment between HR-HPV positive and negative groups of CIN III and CSCCs. These indicated that the rs3087404 and rs2029167 involved in the cervical tumorigenesis, but they maybe not affect the HR-HPVs infection at early onset of disease. In the process of affecting cervical tumorigenesis, rs2029167 variant may be more effective than those of rs3087404 variant.
These findings suggested that there was association between the two of SMUG1 rs3087404 (A/G) and rs2029167(A/G) genetic variant with the susceptibility of CIN III and CSCCs, but not HR-HPVs infection. Whether the rs3087404 or rs2029167 is GG homozygote, there was linkage disequilibrium between these two of polymorphism leading to increase the risk of CIN III and CSCC. These linkage modes can be used as genetic biomarker of early prediction of cervical carcinoma, as an indicator of primary prevention.
Acknowledgments
The authors are grateful to Drs. Caiyun Zhou and Minhua Yu for their assistance in recruiting the subjects.
Grants
This work was supported by grants from the National Nature Science foundation of China (No. 30973380) and Zhejiang Provincial Natural Science Foundation of China (No. Y2110200).
Author Contributions
Conceived and designed the experiments: FY HC. Performed the experiments: FY HW QC JL XC. Analyzed the data: FY HC. Contributed reagents/ materials/analysis tools: FY HW QC. Wrote the paper: FY HC.
Abbreviations
- CC
cervical cancer
- HR-HPV
high-risk HPV
- BER
base excision repair
- SNP
single nucleotide polymorphisms
- SMUG1
Single-strand selective mono-functional uracil-DNA glycosylase
- CRC
colorectal cancer
- CSCC
cervical squamous cell carcinoma
- CIN
cervical intraepithelial neoplasia
- MA-PCR
Modified polymerase chain reaction-mismatch amplification
- HC II
Hybrid Capture II
- OR
odds ratio
- CIs
95% confidence intervals
- UDG
uracil-DNA glycosylases
- ROS
reactive oxygen species
- ssDNA
single stranded DNA
- dsDNA
double stranded DNA.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Torres-Poveda K, Burguete-García AI, Bahena-Román M. et al. Risk allelic load in Th2 and Th3 cytokines genes as biomarker of susceptibility to HPV-16 positive cervical cancer: a case control study. BMC Cancer. 2016;16:330. doi: 10.1186/s12885-016-2364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M. et al. Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PLoS One. 2016;11(4):e0153274. doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiffman M, Glass AG, Wentzensen N. et al. A long-term prospective study of type-specific human papillomavirus infection and risk of cervical neoplasia among 20,000 women in the Portland Kaiser Cohort Study. Cancer Epidemiol Biomarkers Prev. 2011;20(7):1398–1409. doi: 10.1158/1055-9965.EPI-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Zhang L, Tian C. et al. Genetic variants and risk of cervical cancer: epidemiological evidence, meta-analysis and research review. BJOG. 2014;121(6):664–674. doi: 10.1111/1471-0528.12638. [DOI] [PubMed] [Google Scholar]
- 8.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 9.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 10.Bonn D. How DNA-repair pathways may affect cancer risk. Lancet. 1998;351(9095):42. doi: 10.1016/S0140-6736(05)78079-X. [DOI] [PubMed] [Google Scholar]
- 11.Wood RD, Mitchell M, Sgouros J. et al. Human DNA repair genes. Science. 2001;291(5507):1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Newcomb PA, Egan KM. et al. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(2):353–358. doi: 10.1158/1055-9965.EPI-05-0653. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Liu Z, Wang LE. et al. Genetic variants of the ADPRT, XRCC1 and APE1 genes and risk of cutaneous melanoma. Carcinogenesis. 2006;27(9):1894–1901. doi: 10.1093/carcin/bgl042. [DOI] [PubMed] [Google Scholar]
- 14.Hao B, Wang H, Zhou K. et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64(12):4378–4384. doi: 10.1158/0008-5472.CAN-04-0372. [DOI] [PubMed] [Google Scholar]
- 15.Qian B, Zhang H, Zhang L. et al. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73(2):138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Chiang FY, Wu CW, Hsiao PJ. et al. Association between polymorphisms in DNA base excision repair genes XRCC1, APE1, and ADPRT and differentiated thyroid carcinoma. Clin Cancer Res. 2008;14(18):5919–5924. doi: 10.1158/1078-0432.CCR-08-0906. [DOI] [PubMed] [Google Scholar]
- 17.Figueroa JD, Malats N, Real FX. et al. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet. 2007;121(2):233–242. doi: 10.1007/s00439-006-0294-y. [DOI] [PubMed] [Google Scholar]
- 18.Pettersen HS, Sundheim O, Gilljam KM. et al. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35(12):3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie H, Gong Y, Dai J. et al. Genetic variations in base excision repair pathway and risk of bladder cancer: a case-control study in the United States. Mol Carcinog. 2015;54(1):50–57. doi: 10.1002/mc.22073. [DOI] [PubMed] [Google Scholar]
- 20.Marian C, Tao M, Mason JB. et al. Single nucleotide polymorphisms in uracil-processing genes, intake of one-carbon nutrients and breast cancer risk. Eur J Clin Nutr. 2011;65(6):683–689. doi: 10.1038/ejcn.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broderick P, Bagratuni T, Vijayakrishnan J. et al. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer. 2006;6:243. doi: 10.1186/1471-2407-6-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye F, Cheng Q, Hu Y. et al. PARP-1 Val762Ala polymorphism is associated with risk of cervical carcinoma. PLoS One. 2012;7(5):e37446. doi: 10.1371/journal.pone.0037446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye F, Cheng Q, Shen J. et al. Mismatch repair gene MLH3 Pro844Leu and Thr942Ile polymorphisms and the susceptibility to cervical carcinoma and HPV infection: a case-control study in a Chinese population. PLoS One. 2014;9(4):e96224. doi: 10.1371/journal.pone.0096224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kavli B, Otterlei M, Slupphaug G. et al. Uracil in DNA-general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6(4):505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Sousa MM, Krokan HE, Slupphaug G. DNA-uracil and human pathology. Mol Aspects Med. 2007;28(3-4):276–306. doi: 10.1016/j.mam.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA-occurrence, consequences and repair. Oncogene. 2002;21(58):8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 27.Visnes T, Doseth B, Pettersen HS. et al. Uracil in DNA and its processing by different DNA glycosylases. Philos Trans R Soc Lond B Biol Sci. 2009;364:563–568. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenech M. The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutat Res. 2001;475:57–67. doi: 10.1016/s0027-5107(01)00079-3. [DOI] [PubMed] [Google Scholar]
- 29.Kapiszewska M, Kalemba M, Wojciech U. et al. Uracil misincorporation into DNA of leukocytes of young women with positive folate balance depends on plasma vitamin B12 concentrations and methylenetetrahydrofolate reductase polymorphisms. A pilot study. J Nutr Biochem. 2005;16(8):467–478. doi: 10.1016/j.jnutbio.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 30.Mashiyama ST, Hansen CM, Roitman E. et al. An assay for uracil in human DNA at baseline: effect of marginal vitamin B6 deficiency. Anal Biochem. 2008;372(1):21–31. doi: 10.1016/j.ab.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974;71(9):3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krokan HE, Nilsen H, Skorpen F. et al. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476(1-2):73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 33.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 34.Sangrajrang S, Schmezer P, Burkholder I. et al. Polymorphisms in three base excision repair genes and breast cancer risk in Thai women. Breast Cancer Res Treat. 2008;111(2):279–288. doi: 10.1007/s10549-007-9773-7. [DOI] [PubMed] [Google Scholar]
- 35.Mohrenweiser HW, Jones IM. Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat Res. 1998;400(1-2):15–24. doi: 10.1016/s0027-5107(98)00059-1. [DOI] [PubMed] [Google Scholar]
- 36.Hung RJ, Hall J, Brennan P. et al. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162(10):925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 37.Izumi T, Wiederhold LR, Roy G. et al. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193(1-2):43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- 38.Krokan HE, Otterlei M, Nilsen H. et al. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- 39.Masaoka A, Matsubara M, Hasegawa R. et al. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42(17):5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 40.Boorstein RJ, Cummings A Jr, Marenstein DR. et al. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J Biol Chem. 2001;276(45):41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 41.Lo SF, Wan L, Huang CM. et al. Genetic polymorphisms of the DNA repair gene UNG are associated with the susceptibility of rheumatoid arthritis. Rheumatol Int. 2012;32(12):3723–3727. doi: 10.1007/s00296-011-2185-3. [DOI] [PubMed] [Google Scholar]
- 42.Bonnet A, Grosso AR, Elkaoutari A. et al. Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability. Mol Cell. 2017;67(4):608–621.e6. doi: 10.1016/j.molcel.2017.07.002. [DOI] [PubMed] [Google Scholar]