Abstract
Hematopoietic stem and progenitor cell (HSPC) transplantation represents a treatment option for patients with malignant and nonmalignant hematological diseases. Initial steps in transplantation involve the bone marrow homing and engraftment of peripheral blood–injected HSPCs. In recent work, we identified the tetraspanin CD82 as a potential regulator of HSPC homing to the bone marrow, although its mechanism remains unclear. In the present study, using a CD82 knockout (CD82KO) mouse model, we determined that CD82 modulates HSPC bone marrow maintenance, homing, and engraftment. Bone marrow characterization identified a significant decrease in the number of long-term hematopoietic stem cells in the CD82KO mice, which we linked to cell cycle activation and reduced stem cell quiescence. Additionally, we demonstrate that CD82 deficiency disrupts bone marrow homing and engraftment, with in vitro analysis identifying further defects in migration and cell spreading. Moreover, we find that the CD82KO HSPC homing defect is due at least in part to the hyperactivation of Rac1, as Rac1 inhibition rescues homing capacity. Together, these data provide evidence that CD82 is an important regulator of HSPC bone marrow maintenance, homing, and engraftment and suggest exploiting the CD82 scaffold as a therapeutic target for improved efficacy of stem cell transplants.
INTRODUCTION
Hematopoietic stem and progenitor cells (HSPCs) provide the cellular reservoir that gives rise to the highly varied blood and immune cells required to support the lifespan of an organism. Thus, it is necessary that HSPCs maintain a finely tuned balance between quiescence, self-renewal, proliferation, and differentiation. While key signaling pathways intrinsic to HSPCs are involved in regulating this delicate balance, HSPCs are also regulated by a variety of signals they receive from their microenvironment or niche. The bone marrow microenvironment is the primary residence for HSPCs, where they are regulated by both secreted signals and cell–cell interactions (Morrison and Spradling, 2008; Morrison and Scadden, 2014; Mendelson and Frenette, 2014). Under physiological conditions, HSPCs are maintained in the bone marrow, but also circulate within the blood at low levels (Mazo and von Andrian, 1999; Sahin and Buitenhuis, 2012). Then, from the peripheral blood, the HSPCs can migrate back to the bone marrow, using a process called homing, which is the critical first step in the repopulation of the bone marrow after stem cell transplantation. Currently, allogeneic hematopoietic stem cell (HSC) transplantation is a standard treatment option for patients suffering from a variety of malignant and nonmalignant hematological diseases (Gyurkocza et al., 2010). Effective treatment requires the successful homing of donor HSPCs back to the bone marrow microenvironment, where they can engraft and repopulate the blood and immune cell lineages. Notably, only a small percentage of transplanted HSPCs have the capacity to engraft, and while graft failure is rare, it remains a significant contributor to patient morbidity and mortality (Ratajczak and Suszynska, 2016). With the ultimate goal of improving transplantation therapies, there is significant interest in understanding the molecular mechanisms that regulate the repopulation potential or fitness of HSPCs, which requires productive bone marrow homing and engraftment.
HSPC bone marrow homing is a dynamic process, which includes various adhesion and signaling molecules such as chemokines and integrins. The chemokine CXCL12 is an important chemoattractant and regulator of HSPCs. The expression and secretion of CXCL12 is abundant in the bone marrow microenvironment (expressed by osteoblasts and endothelial cells) and promotes the homing and maintenance of HSPCs within the bone marrow. The receptor for CXCL12 is the C-X-C Chemokine receptor type 4 (CXCR4), which is highly expressed on HSPCs and controls HSPC homing, mobilization, and niche localization (Prosper and Verfaillie, 2001; Nie et al., 2008; Sahin and Buitenhuis, 2012). The loss of CXCR4 on HSPCs results in a significant decrease in HSPC homing to the bone marrow (Nie et al., 2008). Interestingly, the reexpression of CXCR4 restored hematopoiesis upon bone marrow engraftment. In addition, integrins such as α4, α6, and β1 facilitate adhesion in the bone marrow, but also mediate tethering of HSPCs within blood vessels to enable transendothelial HSPC migration (Papayannopoulou et al., 1995, 2001; Mazo and von Andrian, 1999). The treatment of bone marrow HSPCs with an antibody to α4β1 resulted in decreased bone marrow homing (Papayannopoulou et al., 1995, 2001). In addition, bone marrow cells from a conditional α4 KO mouse showed a delay in bone marrow homing and defect in short-term engraftment (Scott et al., 2003). Similarly, HSPCs deficient in integrin β1 (Hirsch et al., 1996; Potocnik et al., 2000) and treatment of bone marrow cells with an α6 antibody (Qian et al., 2006) led to a decrease in bone marrow homing and engraftment. Together, these data highlight the critical role of integrins and the CXCR4/CXCL12 signaling axis in HSPC homing and engraftment. Thus, understanding how these signaling and adhesion pathways are regulated in HSPCs is critical for improved transplantation therapies.
Tetraspanins are a family of scaffold proteins that are known to regulate adhesion and signaling molecules at the plasma membrane (Boucheix and Rubinstein, 2001; Hemler, 2005). Tetraspanins have an evolutionary conserved structure that spans the plasma membrane four times and interact with other tetraspanins and signaling and adhesion molecules to form tetraspanin-enriched microdomains (TEMs) that are important for modulating cell migration and adhesion (Charrin et al., 2009; van Deventer et al., 2017). Previous work from our laboratory identified the tetraspanin CD82 as a potential regulator of HSPC adhesion and migration, demonstrating that human CD34+ HSPC bone marrow homing was diminished when CD82 was neutralized with a monoclonal antibody (Larochelle et al., 2012). CD82 was first described as a tumor metastasis suppressor in solid tumors (Bienstock and Barrett, 2001) and is expressed in both normal and malignant hematopoietic cells (Burchert et al., 1999). Using an acute myeloid leukemia (AML) cell line model, our laboratory also identified decreased bone marrow homing upon CD82 knockdown (Marjon et al., 2016) and went on to show that CD82 regulates the density of the α4 integrin at the plasma membrane, which contributes significantly to HSPC adhesive potential (Termini et al., 2014). More recently, CD82 was shown to be highly expressed in the long-term hematopoietic stem cell population (LT-HSCs) with a potential role in the regulation of HSPC quiescence (Hur et al., 2016). In the current study, we set out to determine how CD82 impacts the homing and engraftment of HSPCs.
Using a global CD82 knockout (CD82KO) mouse model, we identify a reduction in the LT-HSCs localized within the bone marrow compartment, which we find results from LT-HSC activation. Moreover, measurements of HSPC fitness identified both engraftment and homing defects resulting from CD82 deficiency. Isolated HSPCs analyzed by confocal imaging demonstrated additional defects in migration and cell spreading. Recognizing the critical role for the Rho GTPase Rac1 in cell migration and spreading, we analyzed the expression and activity of Rac1, finding Rac1 hyperactivation in CD82KO HSPCs. Inhibition of Rac1 hyperactivation using pharmacological inhibitors restored the bone marrow homing capacity of the CD82KO HSPCs, suggesting that CD82-mediated regulation of HSPC homing and engraftment involves the modulation of Rac1 activity.
RESULTS
Diminished LT-HSCs within the bone marrow of CD82KO mice
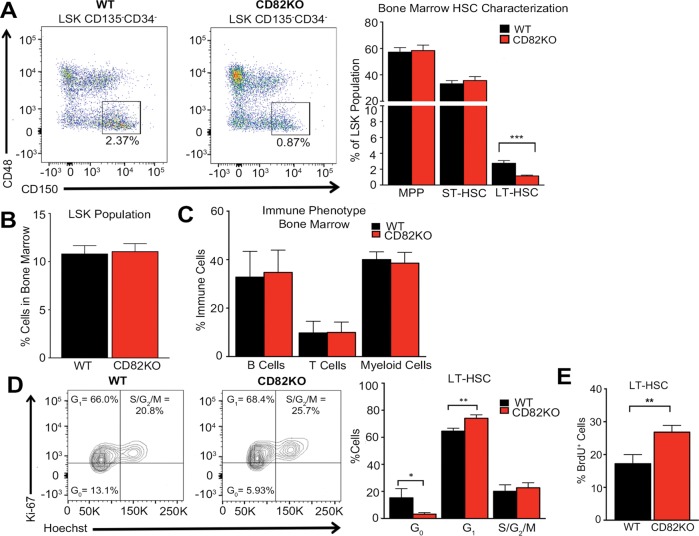
To understand the mechanism by which the CD82 scaffold impacts HSPC regulation, we took advantage of the CD82KO mice previously described (Wei et al., 2014; Jones et al., 2016). To address the consequences of CD82KO for HSPC homeostasis, we first compared the bone marrow frequencies of HSPCs in wild-type (WT) and congenic CD82KO mice. Using flow cytometry, we observed a reduction in the frequency of long-term HSCs (LT-HSCs), defined as Lin–Sca1+Kit+CD34–CD135–CD48–CD150+, in CD82KO, whereas short-term HSCs (ST-HSCs: Lin–Sca1+Kit+CD34+CD135–CD48–CD150+), multipotent progenitors (MPP: Lin–Sca1+Kit+CD34+CD135+CD48–CD150–), and LSK (Lin–Sca1+Kit+) populations showed no significant change (Figure 1, A and B). Further characterization of the immune phenotype of isolated bone marrow cells identified similar percentages of B-cells, T-cells, and myeloid cells in the WT and CD82KO mice (Figure 1C). Together, these data suggest that CD82 functions in the maintenance of the LT-HSC population.
FIGURE 1:
CD82 expression maintains LT-HSCs within the bone marrow. (A) Representative flow cytometry plots of LT-HSCs gated on the LSK CD135-CD34-CD48-CD150+ population. Flow cytometry analysis of the percentage of MPP, ST-HSC, and LT-HSCs from the bone marrow LSK population of WT and CD82KO mice. Error bars, SEM: n = 8–9 mice per strain (***p < 0.001). (B) Flow cytometry analysis of the percentage of the LSK population from WT and CD82KO mice. n = 8 mice per strain. (C) Flow cytometry analysis of the percentage of immune cells (B-cells [B220], T-cells [CD3], and myeloid cells [Gr1/Mac1]) within the bone marrow of WT and CD82KO mice. n = 15 mice per strain. (D) Flow cytometry plots of DNA (Hoechst) and the proliferative nuclear antigen (Ki-67) expression of the bone marrow to measure the cell cycle status of LT-HSC population from WT and CD82KO mice. Error bars, SEM; n = 3 independent experiments (*p < 0.05 and **p < 0.01). (E) Flow cytometry analysis of BrdU expression in the LT-HSC population after 3 d of BrdU incorporation in vivo. Error bars, SEM; n = 3 independent experiments (**p < 0.01).
To address the cause of the reduction in LT-HSCs in the CD82KO bone marrow, we first analyzed extramedullary tissues and identified no increase in the number of LT-HSCs in CD82KO mice (unpublished data). Therefore, extramedullary hematopoiesis does not appear to contribute to the observed reduction in bone marrow LT-HSCs. Next, we analyzed the proliferation and cell cycle status of CD82KO LT-HSCs. Combining the Ki67 marker with DNA content analysis, we find that CD82KO LT-HSCs increase cell cycle entry (Figure 1D). We also completed bromodeoxyuridine (BrdU) incorporation assays to assess proliferation changes in vivo, identifying a significant increase in BrdU+ LT-HSCs within the bone marrow of CD82KO mice (Figure 1E). These data suggest that the cell cycle activation of the CD82KO LT-HSCs ultimately results in reduction of the quiescent LT-HSC population localized to the bone marrow. Collectively, these data are consistent with a previous study using an alternative CD82KO mouse model, which described a similar reduction in the LT-HSCs resulting from cell cycle entry (Hur et al., 2016).
Reduced competitive repopulation capacity of CD82KO HSPCs
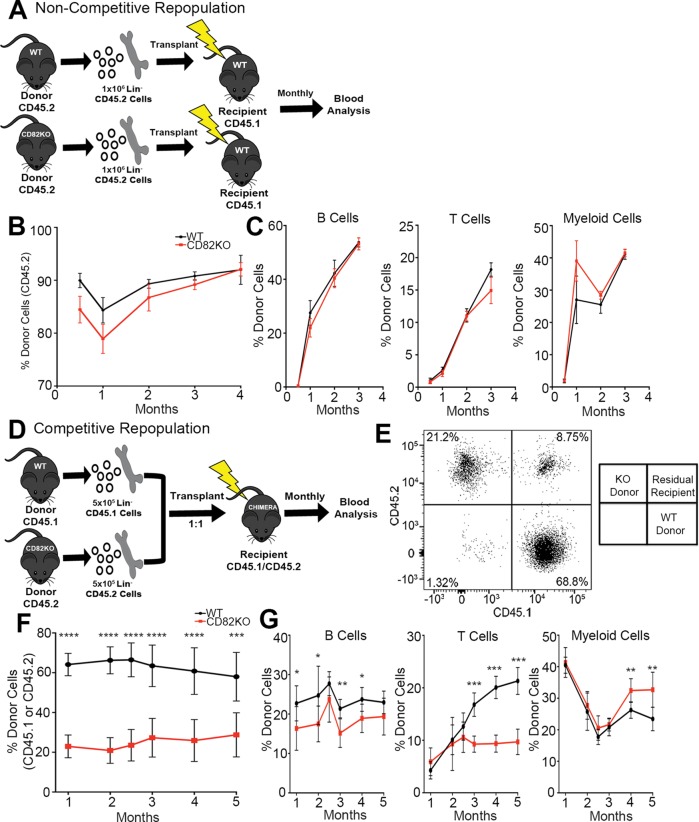
To assess how CD82 deficiency impacts stem cell repopulation, we carried out long-term engraftment assays where we analyzed the reconstitution ability of WT and CD82KO HSPCs. We transplanted donor WT or CD82KO Lin– HSPCs into lethally irradiated recipients (Figure 2A). The B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) mouse strain were used as recipients because they carry the differential panleukocyte marker CD45.1, which can be distinguished from the WT and CD82KO donor cell populations that express the CD45.2 allele. Monthly peripheral blood analysis confirmed a similar engraftment of both CD82KO and WT donor-derived CD45.2 cells (Figure 2B). Additionally, analysis of the immune cell phenotype of the recipient mice identified no significant changes in the production of B, T, or myeloid cells (Figure 2C). Therefore, CD82KO HSPCs have the capacity to repopulate a recipient and generate similar percentages of differentiated immune cells.
FIGURE 2:
CD82KO HSPCs display decreased repopulation in a competitive environment. (A) Experimental scheme for the noncompetitive repopulation experiment. (B) The percentage of donor cell repopulation of peripheral blood collected monthly from tail bleeds. Donor cell chimerism (CD45.2) status measured via flow cytometry. n = 6 mice per strain, (C) Flow cytometry analysis of the percentage of donor immune cells (B-cells [B220], T-cells [CD3], and myeloid cells [Gr1/Mac1]) from donor (CD45.2) population in B. (D) Experimental scheme for the competitive repopulation experiment. (E) Representative flow cytometry plot for the competitive repopulation assay gated for donor cells (WT, CD45.1 and CD82KO, CD45.2) in the peripheral blood recipient mice. (F) The percentage of donor cell repopulation of peripheral blood collected monthly from tail bleeds. Donor cell chimerism (WT, CD45.1 and CD82KO, CD45.2) status measured via flow cytometry. Error bars, SEM; n = 7 mice per strain (***p < 0.001 and ****p < 0.0001). (G) Flow cytometry analysis of the percentage of donor immune cells (B-cells [B220], T-cells [CD3], and myeloid cells [Gr1/Mac1]) from donor population in F (*p < 0.05 and **p < 0.01).
Next, we went on to assess the long-term engraftment potential of CD82KO HSPCs when transplanted into a competitive environment. Isolated Lin– HSPCs were harvested from WT (CD45.1) and CD82KO (CD45.2) donors and transplanted into lethally irradiated chimeric mice (CD45.1/CD45.2) at a ratio of 1:1 (Figure 2D). The use of chimeric mice enables us to distinguish the WT (CD45.1) and CD82KO (CD45.2) donor cells from the CD45.1/CD45.2 recipient cells by flow cytometry (Figure 2E). After transplant, blood cell chimerism was analyzed monthly using flow cytometry to measure repopulation. Under these competitive conditions, we identified a significant decrease in the repopulation capacity of CD82KO derived cells (CD45.2) when compared with WT cells (CD45.1) (Figure 2E). These data suggest that while CD82KO HSPCs have the capacity to successfully engraft, when cotransplanted with WT HSPCs, CD82KO HSPCs display a decreased efficiency to reconstitute a recipient, indicating an overall reduction in HSPC fitness (Figure 2F). Additionally, we analyzed the immune cell differentiation potential of competitively engrafted HSPCs, identifying a significant decrease in B- and T-cells derived from CD82KO HSPCs in comparison with WT HSPCs (Figure 2G). Interestingly, we detect a significant increase in the number of CD82KO-derived myeloid cells in comparison with WT cells (Figure 2G), which suggests that CD82KO HSPCs present a myeloid skewing phenotype following competitive repopulation.
CD82KO HSPCs displayed reduced bone marrow homing
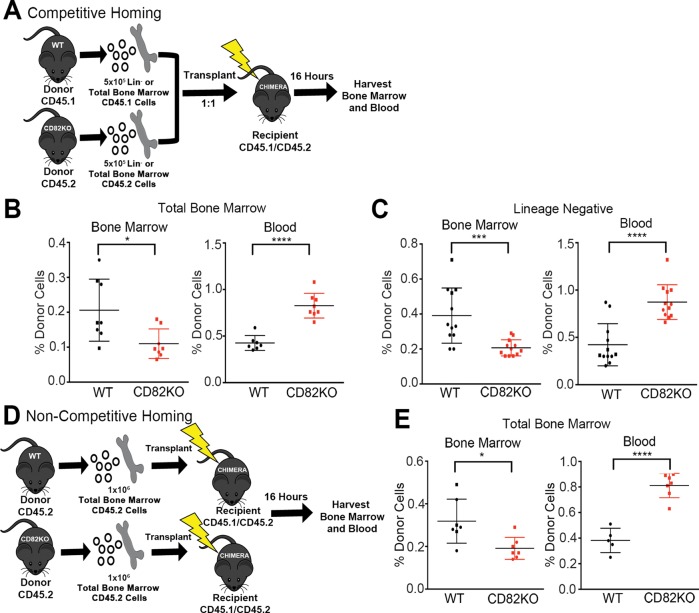
Successful competitive repopulation requires the initial migration or homing of HSPCs to the bone marrow. Therefore, we next evaluated how CD82KO contributes to the early steps of HSPC repopulation by performing a competitive homing experiment (Figure 3A). Total bone marrow (Figure 3B) or Lin– cells (Figure 3C) were harvested from CD82KO (CD45.2) and WT (CD45.1) mice and transplanted into lethally irradiated chimeric mice (CD45.1/CD45.2) at a ratio of 1:1. The blood and bone marrow of recipient mice were harvested 16 h postinjection to assess the chimerism status using flow cytometry. Analysis of the bone marrow indicates a significant decrease in the number of CD82KO cells that migrate to the bone marrow in comparison with WT cells. Consistent with the decrease in bone marrow homing, we measure a significant increase in the number of CD82KO cells detected within the blood in comparison with WT cells. Additionally, we completed homing experiments with total bone marrow injected into a noncompetitive environment (Figure 3D), identifying a homing defect of the CD82KO cells similar to that observed in the competitive environment (Figure 3E). Together, these data suggest that the reduced competitive repopulation capacity of the CD82KO HSPCs is likely due to the reduced bone marrow homing potential of these cells.
FIGURE 3:
CD82KO HSPCs demonstrate decreased bone marrow homing. (A) Experimental scheme for the competitive homing experiment. (B) The percentage of total bone marrow and (C) of lineage-negative donor cells homed within the blood and bone marrow 16 h postinjection. Donor cell chimerism (WT, CD45.1 and CD82KO, CD45.2) status measured via flow cytometry. Error bars, SEM; n = 8–12 mice per strain (*p < 0.05, ***p < 0.001, and ****p < 0.0001). (D) Experimental scheme for the noncompetitive homing experiment. (E) The percentage of total bone marrow homed within the blood and bone marrow 16 h postinjection. Donor cell chimerism (CD45.2) status measured via flow cytometry. Error bars, SEM; n = 5–7 mice per strain (*p < 0.05 and ****p < 0.0001).
CD82KO does not impact CXCR4 expression or activity
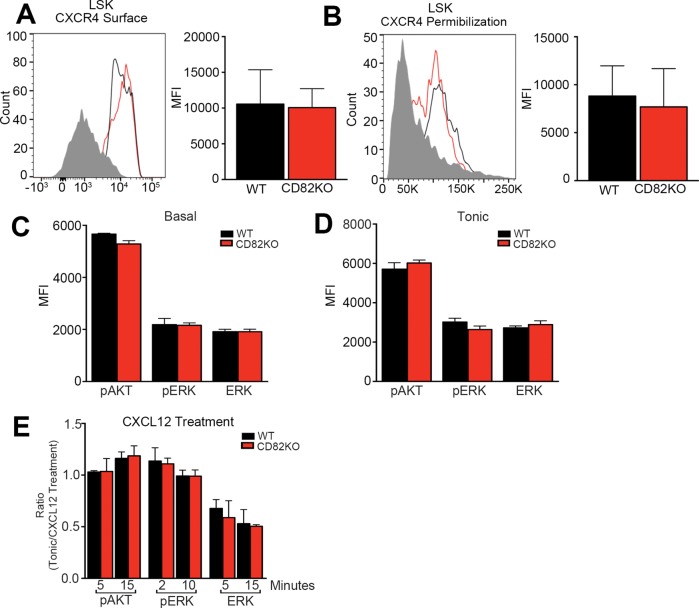
The CXCR4–CXCL12 signaling axis is a critical regulator of HSPC homing to and maintenance within the bone marrow. CXCR4 is highly expressed on HSPCs and serves as the chemokine receptor for CXCL12, which is produced by bone marrow stromal cells and controls HSPC homing, mobilization, and localization (Nie et al., 2008; Prosper and Verfaillie, 2001; Sahin and Buitenhuis, 2012). To determine whether the reduced homing behavior observed with the CD82KO HSPCs is due to changes in the expression of CXCR4, we measured the surface expression of CXCR4 by flow cytometry. In Figure 4A we detect no difference in the CXCR4 mean fluorescence intensity between CD82KO and WT LSKs. Recognizing that CXCR4 is internalized following activation, we also fixed and permeabilized WT and CD82KO LSKs to measure total CXCR4 expression. Similarly to the surface expression, we find no difference in mean fluorescence intensity between CD82KO and WT LSK HSPCs, indicating no changes in overall CXCR4 expression (Figure 4B).
FIGURE 4:
CD82KO does not impact CXCR4 expression or activity. Representative histograms of (A) surface and (B) total (permeablized) CXCR4 expression of bone marrow HSPCs from WT and CD82KO mice. Flow cytometry analysis measured the mean fluorescence intensity (MFI) of CXCR4 on the LSK population. Phosphoflow cytometry analysis of (C) basal and (D) tonic (1 h serum starvation) conditions to assess the mean fluorescence intensity (MFI) of pAKT, pERK, and total ERK signaling of the LSK population. n = 3 mice per strain. (E) Phosphoflow cytometry analysis of SDF-1 treatment at various time points post 1 h of serum starvation to assess pAKT, pERK, and total ERK signaling of the LSK population. Quantification ratio calculated by dividing tonic signaling by SDF-1 treatment conditions. n = 3 mice per strain.
In addition to modulating the surface expression of membrane proteins, tetraspanins also have the capacity to cluster membrane-associated proteins, promoting their activation (Marjon et al., 2016; Termini et al., 2016). Thus, we went on to evaluate CXCR4 signal transduction downstream of CXCL12 activation with a focus on AKT and ERK signaling. Phosphoflow cytometry of phosphorylated AKT and ERK revealed no change in the basal or tonic signaling levels between WT or CD82KO LSKs (Figure 4, C and D). Moreover, following CXCL12 stimulation, we detected similar AKT and ERK phosphorylation, indicating no difference between WT and CD82KO LSKs in CXCR4 activity (Figure 4E). Taken together, the CD82KO homing defect does not result from altered CXCR4 expression or activation.
CD82KO HSPCs display a disruption in migratory behavior
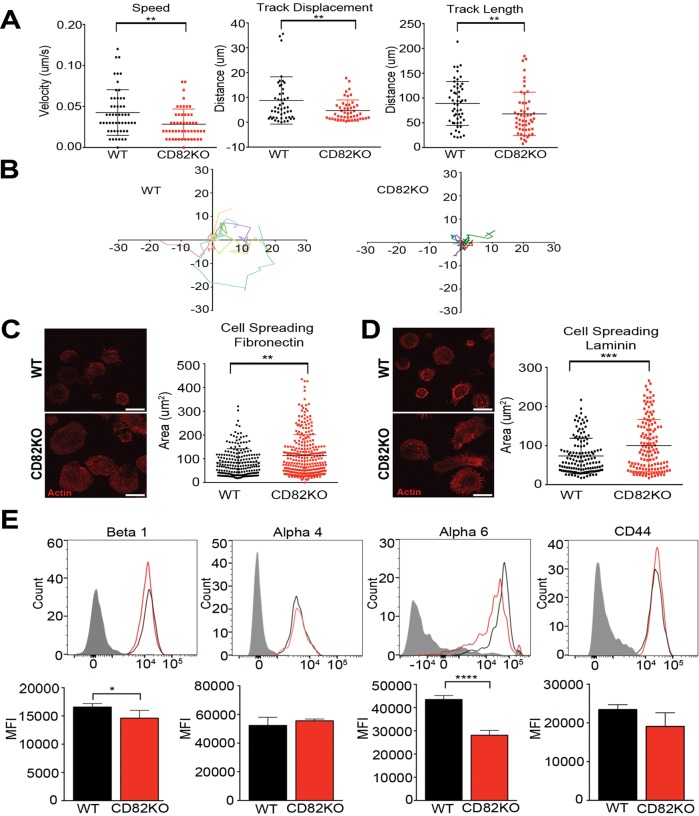
To further evaluate the mechanism by which the capacity of CD82KO HSPCs to home and engraft is reduced, HSPC migration was assessed in vitro. Lin– HSPCs were isolated from CD82KO and WT mice and imaged by live cell confocal microscopy. The time-lapse images were analyzed using Imaris tracking software to obtain measurements of track speed, displacement, and length for individual HSPCs. Figure 5A illustrates that isolated CD82KO HSPCs have a significant decrease in track speed, track displacement, and track length when compared with WT HSPCs. Moreover, single-cell trajectory rose plots indicate that CD82KO HSPC track movements are short and consolidated at the point of origin and lack directional movement, in comparison with WT HSPC tracks (Figure 5B). These data demonstrate a two-dimensional (2D) migratory defect for CD82KO HSPCs, which is consistent with the observed disruption in homing behavior, and further implies an important role for CD82 in HSPC migration.
FIGURE 5:
CD82KO HSPCs display decreased migration and increased cell spreading. (A) Live-cell confocal imaging analysis of HSPC migration from WT and CD82KO bone marrow. Imaris imaging software was used to assess track speed, track displacement, and track length. Error bars, SEM; n = 2 independent experiments (**p < 0.01). (B) Rose plots were generated using the WT and CD82KO HSPC track length coordinates. Cell spreading potential of isolated WT and CD82KO HSPCs plated on (C) fibronectin and (D) laminin. Representative images show actin staining used to quantify HSPC area using ImageJ software. Error bars, SEM; n = 2 independent experiments (**p < 0.01 and ***p < 0.001). (E) Representative histograms of surface adhesion molecule expression of bone marrow HSPCs from WT and CD82KO mice. Flow cytometry analysis measuring the mean fluorescence intensity (MFI) of each adhesion molecule on the LSK population. Error bars, SEM; n = 3 mice per strain (*p < 0.05 and ****p < 0.0001).
Adhesive strength and cell spreading play key roles in generating the required traction for cell migration. Therefore, we next measured the cell spreading capacity of WT and CD82KO cells plated on specific extracellular matrices. Isolated Lin– HSPCs were plated on either fibronectin or laminin for 4 h before being fixed and fluorescently labeled with the cytoskeletal marker phalloidin. Confocal microscopy images were analyzed using ImageJ software to quantify the area of cell spreading. In Figure 5, C and D, we measure a significant increase in the area of CD82KO Lin– HSPCs plated on fibronectin or laminin from that of WT HSPCs. To further assess the role of CD82 deficiency in HSPC adhesion, we quantified the surface expression of specific adhesion molecules, including integrins α4, α6, β1, and CD44. Flow cytometry analysis of the CD82KO LSK HSPCs measured a significant decrease in mean fluorescence intensity of α6 and a modest decrease in β1, with no mean fluorescence intensity changes detected for α4 and CD44 from WT HSPCs (Figure 5E). Collectively, the decreased surface expression of integrins α6 and β1 and the enhanced cell spreading likely contribute to the decrease in homing and engraftment potential seen with the CD82KO HSPCs.
CD82KO HSPCs have increased Rac1-GTPase activity
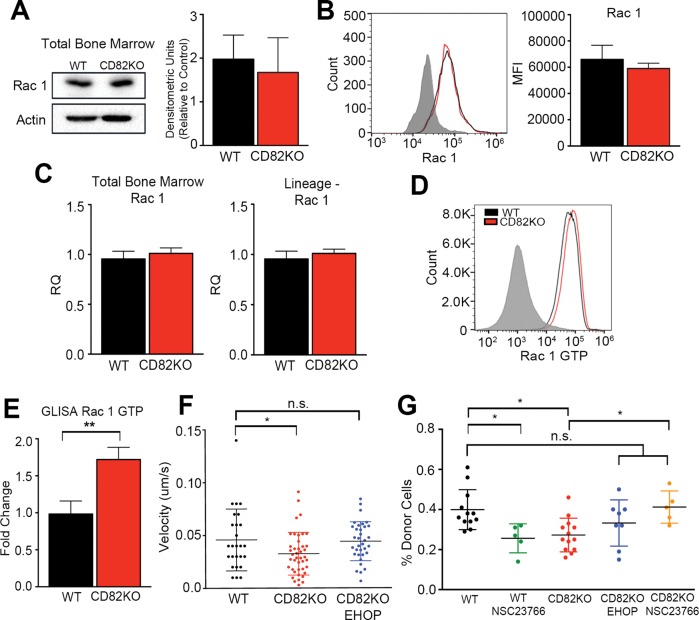
Rho GTPases play an essential role in cell spreading and cell migration. Moreover, previous studies have demonstrated that the specific RhoGTPase, Rac1, can control hematopoietic stem cell activities such as marrow homing and retention (Yang et al., 2001; Gu et al., 2003; Cancelas et al., 2005; Williams et al., 2008; Liu et al., 2011; Shang et al., 2011; Dorrance et al., 2013). On the basis of the increased cell spreading observed in Figure 5, C and D, by the CD82KO cells, we set out to measure Rac1 expression and activity. Western blot analysis of total bone marrow suggests no change in Rac1 expression when WT and CD82KO cells are compared (Figure 6A). Additional flow cytometry analysis of the LSK cells also indicates no difference in Rac1 expression when WT and CD82KO cells are compared (Figure 6B). Similarly, we were unable to detect any changes in Rac1 gene expression between WT and CD82KO mice when either the total bone marrow or the Lin– fraction of HSPCs was analyzed (Figure 6C). Recognizing that Rac activity is a key contributor to cell spreading, we went on to measure changes in Rac1 activity by flow cytometry using an active Rac1-specific antibody. Figure 6D indicates that the CD82KO LSK HSPCs have increased Rac1 activity in comparison with WT cells. A similar result was measured using an active Rac1 ELISA, where we identified a significant increase in the active form of Rac1 in the CD82KO Lin– bone marrow lysates in comparison with WT (Figure 6E). Collectively, these data suggest that while CD82 deficiency does not impact overall Rac1 expression, Rac1 hyperactivation is detected in the HSPCs upon CD82KO. Last, we wanted to determine whether the measured increase in Rac1 activity contributes to the disruption in cell migration and bone marrow homing observed with the CD82KO HSPCs. Thus, we completed cell migration studies of the CD82KO Lin– HSPCs that were treated with and without the Rac1-specific inhibitor EHOP-016. Analysis of track speed indicates that inhibition of Rac1 hyperactivation restores the velocity of the CD82KO HSPCs to WT HSPC speeds (Figure 6F). Moreover, we completed the bone marrow homing experiments with WT and CD82KO total bone marrow, where we also pretreated cells with EHOP-016 or another Rac1 inhibitor, NSC23766, for 1 h prior to injection. Bone marrow was isolated 16 h after injection and analyzed as previously described. Again, we measure a decrease in bone marrow homing of CD82KO cells in comparison with WT cells. Moreover, we find that WT cells pretreated with the Rac1 inhibitor NSC23766 display a significant reduction in bone marrow homing, consistent with previous reports establishing the importance of Rac1 in HSC migration (Yang et al., 2001; Gu et al., 2003; Cancelas et al., 2005; Liu et al., 2011; Dorrance et al., 2013). In contrast, when Rac1 hyperactive CD82KO cells were pretreated with Rac1 inhibitors, bone marrow homing capacity was restored to WT levels (Figure 6G). Taken together, the evidence suggests that CD82 deficiency inhibits HSPC migration to the bone marrow at least in part by promoting a shift in the balance of Rac1 activity to a hyperactivated state.
FIGURE 6:
Rac1 hyperactivity in the CD82KO HSPCs contributes to diminished HSPC homing. (A) Western blot analysis of WT and CD82KO total bone marrow measuring Rac1 expression. Densitometry was used to quantify Rac1 expression relative to an actin control. n = 3 independent experiments. (B) Representative histogram of surface Rac1 expression. Flow cytometry analysis measured the MFI of Rac1 on the LSK population. n = 3 mice per strain. (C) Rac1 mRNA expression of total bone marrow and FACS-sorted Lineage-negative cells using real-time qPCR. Rac1 expression was normalized to GAPDH to obtain relative quantification (RQ) values. n = 3 independent experiments. (D) Representative histogram of Rac GTP expression. Flow cytometry analysis measured Rac GTP on WT and CD82KO lineage-negative bone marrow cells. (E) Rac1 activity of WT and CD82KO lineage-negative cells measured using a small G-protein enzyme-linked immunosorbent assay (GLISA). Quantification represents fold change relative to WT. Error bars, SEM; n = 3 independent experiments (**p < 0.01). (F) Live cell confocal imaging analysis of HSPC migration from WT and CD82KO bone marrow. CD82KO HSPCs were treated with EHOP-016 1 h prior to migration. Imaris software was used to assess track speed. Error bars, SEM; n = 2 independent experiments. One-way ANOVA (*p < 0.05 and n.s. nonsignificant). (G) The percentage of total bone marrow cells homed within bone marrow 16 h postinjection. Total bone marrow were treated with EHOP-016 or NSC23766 1 h prior to injection. Donor cell chimerism (CD45.2) status was measured via flow cytometry. Error bars, SEM; n = 5–13 mice per strain, one-way ANOVA (*p < 0.05 and n.s. nonsignificant).
DISCUSSION
Successful clinical outcomes from transplantation depend on the efficient bone marrow homing and engraftment of HSPCs. The current study analyzing HSPCs from CD82KO mice provides strong evidence that the tetraspanin CD82 regulates the maintenance of LT-HSCs within the bone marrow as well as the processes of homing and engraftment. Moreover, if we inhibit the activation of the Rac1 GTPase in the CD82KO HSPCs, we recover the homing defect observed upon loss of CD82. These results have led us to propose a model where the CD82 scaffold functions to 1) promote bone marrow niche interactions that maintain cell cycle quiescence and 2) enhance the bone marrow homing required for HSPC engraftment through the modulation of Rac1 activation.
Regulation of HSPC activation has been described for a number of tetraspanins. For example, tetraspanin CD81 was shown to be important for the return of HSCs to quiescence through the inhibition of the AKT signaling pathway (Lin et al., 2011). The polarized organization of CD81 on the surfaces of murine HSCs led to the deactivation of AKT and the nuclear translocation of FoxO1a, which was important for the return of HSCs into quiescence from a highly proliferative state. This study also demonstrated that CD81KO HSCs have a marked engraftment defect. Previous work by Rossi et al. (2015) also demonstrated a role for tetraspanin CD63 in the regulation of HSPC proliferation through its interaction with the tissue inhibitor metalloproteinase-1 (TIMP1) and the activation of the PI3K/AKT signaling pathway. Furthermore, they found that upon TIMP1 treatment of HSPCs, cyclin D1 gene expression was increased downstream of AKT phosphorylation. Previous work from our group identified a correlation between CD82 membrane organization and cell cycle progression (Larochelle et al., 2012), where CD34+ HSPCs sorted based on the G0 cell cycle stage showed a polarized membrane distribution of CD82, suggesting that CD82 organization may also impact HSPC quiescence. More recently, a CD82KO mouse model was used to identify CD82 as an important regulator of LT-HSC quiescence (Hur et al., 2016). In this study, a cell line system was predominantly used to describe a mechanism where CD82 binds to the Duffy antigen receptor complex on bone marrow macrophages downstream of TGFβ activation to modulate LT-HSC quiescence. In our own study, we use an alternative CD82KO mouse model that was previously described (Wei et al., 2014; Jones et al., 2016) and identify a similar LT-HSC defect with the loss of CD82. Our cell cycle activation studies demonstrating a loss of LT-HSC quiescence nicely parallel the findings of the Hur et al. study. However, we then went on to characterize the fitness of the CD82KO HSPCs, further identifying homing and engraftment defects.
Once introduced into the bloodstream, HSPCs have the capacity to migrate back to the bone marrow in a process that involves intercellular signaling and adhesive interactions (Vermeulen et al., 1998; Caocci et al., 2017). The major chemokine signal responsible for this homing process is the CXCR4/CXCL12 signaling axis. As mentioned previously, CXCR4 on the surface of HSPCs migrates toward its ligand, CXCL12, produced within the bone marrow. Recognizing the critical importance of this receptor/ligand interaction in homing, we thoroughly investigated the potential influence of CD82KO on receptor expression and activation. However, we demonstrate that CD82 deficiency results in no significant impact on CXCR4 expression or activation. Therefore, the observed CD82-mediated homing defect appears to occur through an alternative mechanism, which led us to analyze in vitro adhesion and migration behavior.
Tetraspanin–tetraspanin and tetraspanin–integrin interactions are known to regulate adhesion and migration in a variety of different biological systems. In HSPCs specifically, the tetraspanin CD63 in a complex with TIMP1 and the β1 integrin were shown to modulate adhesion and migration of human CD34+ HSPCs (Wilk et al., 2013). Moreover, the tetraspanin CD9 was shown to regulate HSPC migration and adhesion, although its role in homing remains a bit unclear. In the initial study described by Leung et al. (2011), CD9 expression on human cord blood CD34+ HSPCs was shown to be modulated by SDF-1 and CXCR4 activity, resulting in increased HSPC migration and adhesion. Additionally, this group went on to show that enhancing CD9 expression on the surface of CD34+ HSPCs by treatment with a protein kinase C agonist ε, ingenol 3,20-dibenzoate (IDB), increases homing to the bone marrow. However, in a follow-up study, Desmond et al. (2011) found that while IDB increases CD9 expression on CD34+ HSPCs, it does not increase HSPC homing in comparison with that in control treated CD34+ HSPCs intravenously injected into NSG mice. Our own work with CD82 demonstrated that human CD34+ HSPC pretreatment with a CD82-specific neutralizing antibody significantly reduced bone marrow homing of these cells in an animal model (Larochelle et al., 2012). In the current study, we identify a significant defect in the bone marrow homing and engraftment capacity of CD82KO HSPCs, demonstrating a clear role for CD82 in these two processes. We also detect a myeloid skewing phenotype, which is consistent with a HSPC aging phenotype (Liang et al., 2005; Rossi et al., 2005). Interestingly, aged HSPCs are also known to have reduced bone marrow homing and engraftment capabilities (Morrison et al., 1996; Liang et al., 2005), which suggests that CD82 may also impact HSPC aging.
Once HSPCs are injected into the peripheral blood, homing is the initial process that enables them to traffic to the bone marrow, where the cells will ultimately engraft and repopulate the blood and immune cell lineages. Despite the clear homing defect we observe with the CD82KO HSPCs, it is interesting to note that noncompetitive engraftment assays illustrate no detectable impact of CD82 expression. Only in a competitive environment do we identify a significant disruption in repopulation. These data suggest that perhaps CD82KO HSPCs have a reduced rate of migration and homing, but when given unlimited time, as in noncompetitive engraftment, the CD82KO cells will eventually make their way to the bone marrow, where they have the capacity to repopulate. Conversely, in a competitive environment, the WT HSPCs home more efficiently to the bone marrow, where they perhaps fill many of the available niche sites, reducing the engraftment capabilities of the CD82KO HSPCs with delayed bone marrow homing. These data are further supported by the in vitro migration data in Figure 5, where CD82KO cells display reduced 2D migration. Additionally, we find that CD82KO promotes increased cell spreading, which likely contributes to the reduction in cell migration. Previous work focusing on bone marrow–derived dendritic cells from CD82KO mice identified a similar cell spreading defect when cells were plated on fibronectin (Jones et al., 2016). Moreover, they went on to show altered GTPase activities in the CD82KO cells, illustrating an important role for tetraspanins in the regulation of GTPase activity.
Tetraspanins can interact with Rho GTPases at the plasma membrane and mediate downstream signaling (Termini and Gillette, 2017). In fact, evidence in the literature suggests that tetraspanins are important for the regulation of both Rac1 expression and activity. For example, the overexpression of CD82 was shown to inhibit Rac1 activity, resulting in actin disorganization (Liu et al., 2012), whereas the interaction of Rac1 with the c-terminal tail of CD81 led to an overall decrease in Rac1 expression (Tejera et al., 2013). Similarly, tetraspanins CD9 and CD151 are both implicated in the activation of Rac1 (Arnaud et al., 2015; Hong et al., 2012). The Rho family GTPases function as molecular switches that can coordinate cytoskeletal rearrangements, which ultimately impact a range of cellular behaviors including migration and adhesion. Among the Rho family GTPases, Rac is known to play a clear role in HSPCs migration and homing (Ridley, 2001; Yang et al., 2001; Gu et al., 2003; Cancelas et al., 2005; Williams et al., 2008; Liu et al., 2011; Shang et al., 2011; Dorrance et al., 2013). Conditional knockout mouse studies for Rac1 demonstrated both impaired HSPC engraftment and reduced adhesion to fibronectin (Gu et al., 2003). Similarly, knockout of the HSPC-specific Rac2 identified defects in actin cytoskeleton remodeling and α4β1-mediated adhesion (Yang et al., 2001). Interestingly, in the case of Rac2 deficiency, HSPCs showed increased migration toward a CXCL12 gradient, which the authors suggest may be the result of compensatory up-regulation of Rac1 and Cdc42 activities (Yang et al., 2001). Collectively, these studies illustrate the significant impact of Rac knockout on HSPCs, but much less is known about how Rac hyperactivation alters phenotype. In a recent study, c-Kit+ HSPCs overexpressing a constitutively active form of Rac1 GTPase (Rac1 V12) displayed increased cell migration and adhesion (Chen et al., 2016). In contrast, a study by Shang et al. modulated endogenous Rac1 activation through manipulation of R-Ras expression and identified diminished bone marrow homing of HSPCs upon Rac1 hyperactivation (Shang et al., 2011). Our studies measuring changes in endogenous Rac1 activation identify a similar defect in bone marrow homing of HSPCs when Rac1 is hyperactive. These data suggest that once Rac1 activity goes beyond a certain threshold, cell migration is diminished. Previously, when Rac inhibitors were used, decreased cell migration and adhesion were observed (Montalvo-Ortiz et al., 2012; Chen et al., 2016). Even in our own studies, we find that WT HSPC homing is decreased upon treatment with the Rac1 inhibitor NSC23766. In contrast, the use of two different Rac1 inhibitors restored the bone marrow homing deficit of the CD82KO HSPCs. We speculate that upon Rac1 hyperactivation, the inhibitor reduces the Rac1 activity threshold, resulting in baseline migration and adhesion. However, when inhibitors are used to disrupt basal Rac1 activity, an overall reduction in migration and adhesion is observed (Montalvo-Ortiz et al., 2012; Chen et al., 2016). Together, these data highlight the importance of the tight regulation of a Rac1 activity threshold to maintain HSPC fitness. At this time, how the CD82 scaffold functions to regulate Rac1 activity remains unclear. While there is precedence from the literature that direct interactions can occur between tetraspanins and GTPases (Tejera et al., 2013), we speculate that CD82 is more likely to modulate a Rac1 regulator, and thus affect Rac1 activity indirectly. For example, our own work has established a role for CD82 in the regulation of adhesion molecules α4β1 and N-cadherin (Termini et al., 2014; Marjon et al., 2016), which can both modulate Rac activation (Arthur et al., 2002; Rose, 2006; Rose et al., 2007). Additionally, the tetraspanin CD151 was shown to facilitate interactions between α3β1 and α6β1 and several small GTPases (Hong et al., 2012). Alternatively, the modulation of specific guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) can also have a significant impact on small GTPase activity (Lawson and Burridge, 2014). Therefore, future studies will be directed at understanding how CD82 potentially modulates the expression and/or activity of key regulators of Rac1 activation.
The goal of the current study was to identify the mechanism by which CD82 regulates bone marrow homing and engraftment of HSPCs. Our data suggest that CD82 can modulate not only the activation of LT-HSCs, but also the overall fitness of HSPCs. Activation of LT-HSCs is likely related to how tightly HSPCs interact with specific components of the bone marrow microenvironment, although at this point it is unclear how CD82KO impacts cellular localization within the niche. HSPC fitness is characterized in part by successful bone marrow homing and engraftment, for which our data demonstrate a key role for CD82. Thus, we propose a model where CD82 serves to modulate the activation of Rac1, which significantly impacts the migration, adhesion, and bone marrow homing behavior of HSPCs. Finally, our detailed insight into how CD82 contributes to the homing and engraftment of HSPCs implicates CD82 as an attractive therapeutic target to enhance the efficacy of HSPC transplantation therapies.
MATERIALS AND METHODS
Mice
C57BL/6 WT and B6.SJL-Ptprca Pepcb/BoyJ mice were obtained from the Jackson Laboratory. CD45.1/CD45.2 chimeric mice were generated by mating C57BL/6 and B6.SJL-Ptprca Pepcb/BoyJ mice. CD82KO mice were generated from cre-loxP recombination (Wei et al., 2014). All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of New Mexico Health Science Center. Mice were housed under pathogen-free conditions in the UNM Animal Facility. The age and sex of mice were matched for each experiment.
Isolation and analysis of bone marrow cells
Bone marrow cells were isolated from the front and back limb bones using a mortar and pestle. Isolated bone marrow cells were passed through a 40-µm strainer to remove bone fragments. Red blood cells were lysed using ACK lysis Buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). To assess HSC populations, terminally differentiated cells were removed using a lineage cell depletion kit (Miltenyi Biotec). Isolated Lin– cells were treated with Fc block (2.4G2; BD Pharmingen) prior to surface marker staining. Isolated Lin– cells were stained with antibodies against surface markers using the following antibodies: mouse APC lineage cocktail (BD Pharmingen), BV605 CD117 (2B8; BD Pharmingen), Pe-Cy7 Sca-1 (D7; BD Bioscience), FITC CD34 (RAM34; BD Pharmingen), BV421 CD135 (A2F10.1; BD Pharmingen), BV510 CD48 (HM48-1; BD Pharmingen), and PE CD150 (Q38-480; BD Pharmingen). Labeled bone marrow samples were analyzed using the LSR Fortessa (BD Bioscience). For the sorting of the HSPC population, the mouse HSC isolation kit (BD Pharmingen) was used. Labeled cells were fluorescence activated–cell sorted (FACS) using the iCyt Sony sy32000 Sorter to obtain the Lin–Sca-1+c-kit+ (LSK) population. In addition, bone marrow immune cells were assessed using the following antibodies: PerCP-Cy5.5 CD3 (145-2C11; BD Pharmingen), BV421 B220 (RA3-6B2; Biolegend), Pe-Cy7 Ly6G (IA8; BD Pharmingen), and Pe-Cy7 CD11b (MI/70; BD Pharmingen). Labeled bone marrow samples were analyzed using the LSR Fortessa (BD Bioscience).
Noncompetitive and competitive repopulation assay
For noncompetitive repopulation assay, 1 × 106 donor Lin– bone marrow cells from CD82KO or WT (CD45.2) were retroorbitally injected into recipient BoyJ mice (CD45.1). For a competitive repopulation assay, 1 × 106 donor Lin– bone marrow cells from CD82KO (CD45.2) were mixed at a ratio of 1:1 with Lin– BoyJ cells (CD45.1) and were retroorbitally injected into recipient chimeric mice (CD45.1/CD45.2). Recipient mice underwent total body irradiation, which was administered as a single dose of 10 Gy. Each month, blood was taken from tail snips to assess chimerism and immune cell differentiation. Cells were treated with Fc block prior to staining with the following directly conjugated fluorescent antibodies: FITC CD45.1 (A20; BD Pharmingen), APC CD45.2 (104; BD Pharmingen), PerCP-Cy5.5 CD3 (145-2C11; BD Pharmingen), BV421 B220 (RA3-6B2; Biolegend), Pe-Cy7 Ly6G (IA8; BD Pharmingen), and Pe-Cy7 CD11b (MI/70; BD Pharmingen). Labeled blood samples were analyzed using the LSR Fortessa (BD Bioscience).
Cell cycle analysis
Lineage-depleted bone marrow cells were incubated with Hoechst 33342 (Sigma-Aldrich) and then labeled with PE Ki67 (16A8; BD Bioscience) in addition to LT-HSC surface markers: lineage, Sca-1, CD117, CD135, CD34, CD48, and CD150. Cells were analyzed on the LSR Fortessa (BD Bioscience). For in vivo BrdU incorporation studies, one BrdU (5-bromo-2-deoxyuridine; Sigma-Aldrich) dose of 1 mg was intraperitoneally injected into mice and bone marrow was collected 3 d later. Bone marrow samples were processed using the FITC BrdU Flow kit (BD Bioscience) per the manufacturer’s instructions. Processed bone marrow cells were labeled with surface markers lineage, Sca-1, CD117, CD34, CD135, and CD150 to assess BrdU incorporation in the LT-HSC population. Cells were analyzed on the LSR Fortessa (BD Bioscience).
Noncompetitive and competitive homing
For a noncompetitive homing experiment, 1 × 106 donor bone marrow cells from CD82KO or WT (CD45.2) were retroorbitally injected into recipient chimeric mice (CD45.1/CD45.2). For a competitive homing experiment, donor bone marrow cells from CD82KO were mixed at a ratio of 1:1 with BoyJ competition bone marrow cells and injected into chimeric recipient mice. Competitive homing experiments were also performed using isolated HSPCs from the bone marrow as described above. Recipient mice underwent total body irradiation, which was administered as a single dose of 10 Gy. Mice were killed 16 h postinjection to assess chimerism of the bone marrow and peripheral blood. Blood and bone marrow samples were treated with Fc block prior to labeling with directly conjugated fluorescent antibodies, FITC CD45.1 (A20; BD Pharmingen), and APC CD45.2 (104; BD Pharmingen) to assess chimerism. Samples were analyzed on the LSR Fortessa (BD Bioscience).
Flow cytometry
Bone marrow cells were analyzed for the surface expression of PE CXCR4 (L276F12; Biolegend), PE β1 integrin (HM B1-1; BD Pharmingen), PE α4 integrin (9C10; BD Pharmingen), PE-α6 integrin (GoH3; BD Pharmingen), and PE-CD44 (IM7; BD Pharmingen) on the LSK population. In addition, we also assessed Rac-1 (Cytoskeleton) surface expression. Isolated bone marrow cells were treated with Fc block prior to staining with the LSK markers. All samples were labeled in MACs buffer (phosphate-buffered saline [PBS], 0.5% bovine serum albumin [BSA], 2 mM EDTA, pH 7.2; Miltenyi Biotec) for 30 min on ice. Samples were washed three times with MACs buffer after staining and analyzed using the LSR Fortessa (BD Bioscience). Histograms were created using FlowJo software.
Phosphoflow cytometry
Basal signaling activity was assessed by fixing isolated bone marrow cells with 4% paraformaldehyde and permeabilizing with 100% methanol prior to antibody staining. Tonic signaling activity was assessed by serum-starving bone marrow cells in SFEM for 1 h at 37°C. Starved cells were then fixed and permeabilized. After 1 h of serum starvation, bone marrow cells were also treated with 100 ng/ml SDF-1 for 2, 5, 10, and 15 min. After each time point, samples were fixed and permeabilized. Permeabilized samples were stained for LSK markers and the following signaling molecules: p-AKT Pacific Blue (BD Pharmingen), p-ERK FITC (Biolegend), and MEK2 PE (BD Pharmingen). Samples were analyzed on the LSR Fortessa (BD Bioscience). The ratio of SDF-1 treatment was calculated by dividing tonic mean fluorescence intensity (MFI) by SDF-1 treatment MFI.
HSPC migration
Four-chamber coverslips (Thermo Scientific) were coated with fibronectin (50 μg/ml in PBS; Millipore). Isolated HSPCs were plated at 250,000 cells/well on fibronectin in StemSpan SFEM (StemCell Technologies) media supplemented with murine cytokines IL-3, SCF, and FLT-3 (20 ng/ml; Peprotech) overnight at 37°C. For EHOP-016 treatment, isolated HSPCs were treated with 5 μM EHOP-016 for 1 h at 37°C prior to plating. Each well was washed twice with PBS to remove nonadherent cells. CO2-independent media supplemented with 0.5% fetal bovine serum (FBS) were added to each well for 1 h and migration was measured using a Zeiss Axiovert confocal microscope. Images were taken every 10 s for 50 min and were analyzed using Imaris tracking software (Bitplane Oxford Instruments) to measure track length, track displacement, and speed. The single cell–trajectory rose plots were produced from track position parameter data generated by Imaris tracking software. The beginning X, Y position and ending X, Y position of each HSPC were taken into account. Using the position data of each HSPC, each track was normalized to shift each HSPC to an origin of (0,0). A total of 15 positions between the beginning and end positions were randomly selected to produce a track. For each condition, a total of 10 HSPCs were assessed.
Cell spreading
Isolated Lin– bone marrow cells were plated on fibronectin- or laminin-coated coverslips (Neuvitro) and placed in IMDM media supplemented with 10% FBS for 1 h at 37°C. Adherent cells were fixed with 4% PFA for 15 min at room temperature. Each well was washed three times and then blocked with 5% BSA in PBS for 30 min at room temperature. Cells were stained with rhodamine phalloidin F-actin stain (Invitrogen) for 1 h at room temperature. Each well was washed out three times and replaced with PBS to image. Cells were imaged using an LSM510 confocal microscope. Cell spreading was analyzed with ImageJ software to measure the area of each cell.
Western blot analysis
Total bone marrow was isolated from WT and CD82KO mice. Cells were lysed in RIPA buffer containing 150 mM NaCl, 0.1% SDS, 0.5% DoC, 50 mM Tris pH 8.0, and 1% IGEPAL-NP40. Cell lysates were run on SDS–polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were blotted with a mouse monoclonal antibody against mouse Rac1. Blots were developed using ECL (Thermo Scientific).
Quantitative RT-PCR
Total RNA was extracted from total bone marrow and FACS-isolated LSKs using the RNeasy Micro Kit (Qiagen). RNA samples were quantified using a Nanodrop 2000 (Thermo Scientific). cDNA was made using a qScript cDNA synthesis kit (Quanta Bioscience) and amplified using a MyCycle Thermocycler (Bio Rad). A real-time polymerase chain reaction was done using a SYBR Green PCR Master Mix (Applied Biosystems). Primers for target genes were designed using Primer-BLAST (NCBI). Primer sets for each gene: Rac1, Forward: 5′-AGA GTA CAT CCC CAC CGT CT-3′ and Reverse: 5′-CAT GTG TCT CCA ACT GTC TGC-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Forward: 5′-AAC TTT GGC ATT GTG GAA GG-3′ and Reverse: 5′-ACA CAT TGG GGG TAG GAA CA-3′ (Scoumanne et al., 2011). Target genes were amplified using the AB75000 Fast Real Time PCR System (Applied Biosystems). The endogenous control gene GAPDH was used to normalize each sample. The relative change in gene expression was calculated using the 2–∆∆CT algorithm.
Rac1 activity
WT and CD82KO lineage-negative bone marrow HSPCs were lysed for a G protein–linked immunosorbent assay. A G-LISA Rac-1 Activation Assay Biochem Kit (catalogue no. BK128, Cytoskeleton) was performed per the manufacturer’s instructions. In addition, Rac1 GTP was assessed via flow cytometry. Lineage-depleted bone marrow cells were fixed, permeabilized, and treated with Fc block prior to staining with Rac1 GTP antibody (New East Biosciences). All samples were labeled in MACs buffer for 30 min on ice. Samples were washed three times with MACs buffer after staining and analyzed using the Accuri C6 (BD Bioscience). Histograms were created using FlowJo software.
Rac1 inhibition and homing
Prior to injection, 1 × 106 donor bone marrow cells from CD82KO (CD45.2) or WT (CD45.2) were treated with 5 μM EHop-016 (Selleckchem) or 50 μM NSC23766 (Cayman Chemical) in SFEM for 1 h at 37°C. Treated cells were then retroorbitally injected into BoyJ recipient mice (CD45.1). Recipient mice underwent total body irradiation 24 h prior to injection, which was administered as a single dose of 10 Gy. Mice were killed 16 h postinjection to assess chimerism of the bone marrow and peripheral blood. Blood and bone marrow samples were treated with Fc block prior to labeling with directly conjugated fluorescent antibodies FITC CD45.1 (A20; BD Pharmingen) and APC CD45.2 (104; BD Pharmingen) to assess chimerism. Samples were analyzed on the LSR Fortessa (BD Bioscience).
Statistical analysis
Statistical significance was calculated using Student’s t test, for which significance was labeled as *p < 0.05, **p < 0.01,***p < 0.001, and ****p < 0.0001. One-way analysis of variance (ANOVA) was used to calculate significance of EHOP migration and Rac-1 inhibitor (EHOP and NSC23766) homing studies, for which significance was labeled as *p < 0.05. The annotation n.s. is for nonsignificance. All statistical analyses were performed using GraphPad Prism 6 Software.
ACKNOWLEDGMENTS
We thank Mark Wright (Monash University, Australia) and Günter J. Hämmerling (German Cancer Research Center, Heidelberg, Germany) for the CD82KO mice. Additionally, we thank C.M. Termini, K.L. Karlen, and R.J. Dodd for their experimental advice and assistance. This study was supported by the following grants: National Institutes of Health (NIH) R01-HL12248301A1 (to J.M.G.), American Heart Association SDG14630080 (to J.M.G.), NIH-T32-HL07736IH (to Tom C. Resta for C.A.S.-R. and K.D.M.), NIH R25-GM075149 (to Kelly Miller for E.M.P.), NIH F31-CA232480 (to M.F.), and NIH P50-GM085273 (to Bridget S. Wilson for J.M.G.). Data were generated using the UNM Flow Cytometry Shared Resource and the Animal Models Shared Resource supported by the University of New Mexico Comprehensive Cancer Center, NCI 2P30 CA118100-11 (to C.L.W.).
Abbreviations used:
- AKT
protein kinase B
- BrdU
bromodeoxyuridine
- CXCL12
C-X-C motif chemokine ligand 12
- CXCR4
C-X-C motif chemokine receptor 4
- ERK
extracellular signal-regulated kinase
- FN
fibronectin
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GLISA
small GTPase activation assay
- HSPCs
hematopoietic stem and progenitor cells
- Lin-
lineage-negative
- LN
laminin
- LSK
markers used to identify HSPC population
- LT-HSCs
long-term HSCs
- MFI
mean fluorescence intensity
- MPP
multipotent progenitors
- PI3K
phosphatidylinositol-3-kinase
- Rac1
Ras-related C3 botulinum toxin substrate 1
- ST-HSCs
short-term HSCs
- TEMs
tetraspanin-enriched microdomains.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-05-0305) on August 22, 2018.
REFERENCES
- Arnaud MP, Vallee A, Robert G, Bonneau J, Leroy C, Varin-Blank N, Rio AG, Troadec MB, Galibert MD, Gandemer V. (2015). CD9, a key actor in the dissemination of lymphoblastic leukemia, modulating CXCR4-mediated migration via RAC1 signaling. Blood , 1802–1812. [DOI] [PubMed] [Google Scholar]
- Arthur WT, Noren NK, Burridge K. (2002). Regulation of Rho family GTPases by cell–cell and cell–matrix adhesion. Biol Res , 239–246. [DOI] [PubMed] [Google Scholar]
- Bienstock RJ, Barrett JC. (2001). KAI1, a prostate metastasis suppressor: prediction of solvated structure and interactions with binding partners; integrins, cadherins, and cell-surface receptor proteins. Mol Carcinog , 139–153. [DOI] [PubMed] [Google Scholar]
- Boucheix C, Rubinstein E. (2001). Tetraspanins. Cell Mol Life Sci , 1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchert A, Notter M, Dietrich Menssen H, Schwartz S, Knauf W, Neubauer A, Thiel E. (1999). CD82 (KAI1), a member of the tetraspan family, is expressed on early haemopoietic progenitor cells and up-regulated in distinct human leukaemias. Br J Haematol , 494–504. [DOI] [PubMed] [Google Scholar]
- Cancelas JA, Lee AW, Prabhakar R, Stringer KF, Zheng Y, Williams DA. (2005). Rac GTPases differentially integrate signals regulating hematopoietic stem cell localization. Nat Med , 886–891. [DOI] [PubMed] [Google Scholar]
- Caocci G, Greco M, La Nasa G. (2017). Bone marrow homing and engraftment defects of human hematopoietic stem and progenitor cells. Mediterr J Hematol Infect Dis , e2017032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. (2009). Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J , 133–154. [DOI] [PubMed] [Google Scholar]
- Chen S, Li H, Li S, Yu J, Wang M, Xing H, Tang K, Tian Z, Rao Q, Wang J. (2016). Rac1 GTPase promotes interaction of hematopoietic stem/progenitor cell with niche and participates in leukemia initiation and maintenance in mouse. Stem Cells , 1730–1741. [DOI] [PubMed] [Google Scholar]
- Desmond R, Dunfee A, Racke F, Dunbar CE, Larochelle A. (2011). CD9 up-regulation on CD34+ cells with ingenol 3,20-dibenzoate does not improve homing in NSG mice. Blood , 5774–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrance AM, De Vita S, Radu M, Reddy PN, McGuinness MK, Harris CE, Mathieu R, Lane SW, Kosoff R, Milsom MD, et al. (2013). The Rac GTPase effector p21-activated kinase is essential for hematopoietic stem/progenitor cell migration and engraftment. Blood , 2474–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Filippi MD, Cancelas JA, Siefring JE, Williams EP, Jasti AC, Harris CE, Lee AW, Prabhakar R, Atkinson SJ, et al. (2003). Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science , 445–449. [DOI] [PubMed] [Google Scholar]
- Gyurkocza B, Rezvani A, Storb RF. (2010). Allogeneic hematopoietic cell transplantation: the state of the art. Expert Rev Hematol , 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler ME. (2005). Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol , 801–811. [DOI] [PubMed] [Google Scholar]
- Hirsch E, Iglesias A, Potocnik AJ, Hartmann U, Fassler R. (1996). Impaired migration but not differentiation of haematopoietic stem cells in the absence of beta1 integrins. Nature , 171–175. [DOI] [PubMed] [Google Scholar]
- Hong IK, Jeoung DI, Ha KS, Kim YM, Lee H. (2012). Tetraspanin CD151 stimulates adhesion-dependent activation of Ras, Rac, and Cdc42 by facilitating molecular association between beta1 integrins and small GTPases. J Biol Chem , 32027–32039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J, Choi JI, Lee H, Nham P, Kim TW, Chae CW, Yun JY, Kang JA, Kang J, Lee SE, et al. (2016). CD82/KAI1 maintains the dormancy of long-term hematopoietic stem cells through interaction with DARC-expressing macrophages. Cell Stem Cell , 508–521. [DOI] [PubMed] [Google Scholar]
- Jones EL, Wee JL, Demaria MC, Blakeley J, Ho PK, Vega-Ramos J, Villadangos JA, van Spriel AB, Hickey MJ, Hammerling GJ, et al. (2016). Dendritic cell migration and antigen presentation are coordinated by the opposing functions of the tetraspanins CD82 and CD37. J Immunol , 978–987. [DOI] [PubMed] [Google Scholar]
- Larochelle A, Gillette JM, Desmond R, Ichwan B, Cantilena A, Cerf A, Barrett AJ, Wayne AS, Lippincott-Schwartz J, Dunbar CE. (2012). Bone marrow homing and engraftment of human hematopoietic stem and progenitor cells is mediated by a polarized membrane domain. Blood , 1848–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CD, Burridge K. (2014). The on–off relationship of Rho and Rac during integrin-mediated adhesion and cell migration. Small GTPases , e27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KT, Chan KY, Ng PC, Lau TK, Chiu WM, Tsang KS, Li CK, Kong CK, Li K. (2011). The tetraspanin CD9 regulates migration, adhesion, and homing of human cord blood CD34+ hematopoietic stem and progenitor cells. Blood , 1840–1850. [DOI] [PubMed] [Google Scholar]
- Liang Y, Van Zant G, Szilvassy SJ. (2005). Effects of aging on the homing and engraftment of murine hematopoietic stem and progenitor cells. Blood , 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KK, Rossi L, Boles NC, Hall BE, George TC, Goodell MA. (2011). CD81 is essential for the re-entry of hematopoietic stem cells to quiescence following stress-induced proliferation via deactivation of the Akt pathway. PLoS Biol , e1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Feng Y, Shang X, Zheng Y. (2011). Rho GTPases in hematopoietic stem/progenitor cell migration. Methods Mol Biol , 307–319. [DOI] [PubMed] [Google Scholar]
- Liu WM, Zhang F, Moshiach S, Zhou B, Huang C, Srinivasan K, Khurana S, Zheng Y, Lahti JM, Zhang XA. (2012). Tetraspanin CD82 inhibits protrusion and retraction in cell movement by attenuating the plasma membrane-dependent actin organization. PLoS One , e51797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjon KD, Termini CM, Karlen KL, Saito-Reis C, Soria CE, Lidke KA, Gillette JM. (2016). Tetraspanin CD82 regulates bone marrow homing of acute myeloid leukemia by modulating the molecular organization of N-cadherin. Oncogene , 4132–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo IB, von Andrian UH. (1999). Adhesion and homing of blood-borne cells in bone marrow microvessels. J Leukoc Biol , 25–32. [DOI] [PubMed] [Google Scholar]
- Mendelson A, Frenette PS. (2014). Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med , 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Ortiz BL, Castillo-Pichardo L, Hernandez E, Humphries-Bickley T, De la Mota-Peynado A, Cubano LA, Vlaar CP, Dharmawardhane S. (2012). Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J Biol Chem , 13228–13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. (2014). The bone marrow niche for haematopoietic stem cells. Nature , 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell , 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. (1996). The aging of hematopoietic stem cells. Nat Med , 1011–1016. [DOI] [PubMed] [Google Scholar]
- Nie Y, Han YC, Zou YR. (2008). CXCR4 is required for the quiescence of primitive hematopoietic cells. J Exp Med , 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulou T, Craddock C, Nakamoto B, Priestley GV, Wolf NS. (1995). The VLA4/VCAM-1 adhesion pathway defines contrasting mechanisms of lodgement of transplanted murine hemopoietic progenitors between bone marrow and spleen. Proc Natl Acad Sci USA , 9647–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulou T, Priestley GV, Nakamoto B, Zafiropoulos V, Scott LM. (2001). Molecular pathways in bone marrow homing: dominant role of alpha(4)beta(1) over beta(2)-integrins and selectins. Blood , 2403–2411. [DOI] [PubMed] [Google Scholar]
- Potocnik AJ, Brakebusch C, Fassler R. (2000). Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity , 653–663. [DOI] [PubMed] [Google Scholar]
- Prosper F, Verfaillie CM. (2001). Regulation of hematopoiesis through adhesion receptors. J Leukoc Biol , 307–316. [PubMed] [Google Scholar]
- Qian H, Tryggvason K, Jacobsen SE, Ekblom M. (2006). Contribution of alpha6 integrins to hematopoietic stem and progenitor cell homing to bone marrow and collaboration with alpha4 integrins. Blood , 3503–3510. [DOI] [PubMed] [Google Scholar]
- Ratajczak MZ, Suszynska M. (2016). Emerging strategies to enhance homing and engraftment of hematopoietic stem cells. Stem Cell Rev , 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. (2001). Rho GTPases and cell migration. J Cell Sci , 2713–2722. [DOI] [PubMed] [Google Scholar]
- Rose DM. (2006). The role of the alpha4 integrin-paxillin interaction in regulating leukocyte trafficking. Exp Mol Med , 191–195. [DOI] [PubMed] [Google Scholar]
- Rose DM, Alon R, Ginsberg MH. (2007). Integrin modulation and signaling in leukocyte adhesion and migration. Immunol Rev , 126–134. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, Weissman IL. (2005). Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA , 9194–9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L, Forte D, Migliardi G, Salvestrini V, Buzzi M, Ricciardi MR, Licchetta R, Tafuri A, Bicciato S, Cavo M, et al. (2015). The tissue inhibitor of metalloproteinases 1 increases the clonogenic efficiency of human hematopoietic progenitor cells through CD63/PI3K/Akt signaling. Exp Hematol , 974–985.e1. [DOI] [PubMed] [Google Scholar]
- Sahin AO, Buitenhuis M. (2012). Molecular mechanisms underlying adhesion and migration of hematopoietic stem cells. Cell Adh Migr , 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LM, Priestley GV, Papayannopoulou T. (2003). Deletion of alpha4 integrins from adult hematopoietic cells reveals roles in homeostasis, regeneration, and homing. Mol Cell Biol , 9349–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoumanne A, Cho SJ, Zhang J, Chen X. (2011). The cyclin-dependent kinase inhibitor p21 is r egulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res , 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X, Cancelas JA, Li L, Guo F, Liu W, Johnson JF, Ficker A, Daria D, Geiger H, Ratner N, et al. (2011). R-Ras and Rac GTPase cross-talk regulates hematopoietic progenitor cell migration, homing, and mobilization. J Biol Chem , 24068–24078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera E, Rocha-Perugini V, Lopez-Martin S, Perez-Hernandez D, Bachir AI, Horwitz AR, Vazquez J, Sanchez-Madrid F, Yanez-Mo M. (2013). CD81 regulates cell migration through its association with Rac GTPase. Mol Biol Cell , 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termini CM, Cotter ML, Marjon KD, Buranda T, Lidke KA, Gillette JM. (2014). The membrane scaffold CD82 regulates cell adhesion by altering alpha4 integrin stability and molecular density. Mol Biol Cell , 1560–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termini CM, Gillette JM. (2017). Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol , 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termini CM, Lidke KA, Gillette JM. (2016). Tetraspanin CD82 regulates the spatiotemporal dynamics of PKCalpha in acute myeloid leukemia. Sci Rep , 29859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deventer SJ, Dunlock VE, van Spriel AB. (2017). Molecular interactions shaping the tetraspanin web. Biochem Soc Trans , 741–750. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Le Pesteur F, Gagnerault MC, Mary JY, Sainteny F, Lepault F. (1998). Role of adhesion molecules in the homing and mobilization of murine hematopoietic stem and progenitor cells. Blood , 894–900. [PubMed] [Google Scholar]
- Wei Q, Zhang F, Richardson MM, Roy NH, Rodgers W, Liu Y, Zhao W, Fu C, Ding Y, Huang C, et al. (2014). CD82 restrains pathological angiogenesis by altering lipid raft clustering and CD44 trafficking in endothelial cells. Circulation , 1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk CM, Schildberg FA, Lauterbach MA, Cadeddu RP, Frobel J, Westphal V, Tolba RH, Hell SW, Czibere A, Bruns I, et al. (2013). The tissue inhibitor of metalloproteinases-1 improves migration and adhesion of hematopoietic stem and progenitor cells. Exp Hematol , 823–831.e2. [DOI] [PubMed] [Google Scholar]
- Williams DA, Zheng Y, Cancelas JA. (2008). Rho GTPases and regulation of hematopoietic stem cell localization. Methods Enzymol , 365–393. [DOI] [PubMed] [Google Scholar]
- Yang FC, Atkinson SJ, Gu Y, Borneo JB, Roberts AW, Zheng Y, Pennington J, Williams DA. (2001). Rac and Cdc42 GTPases control hematopoietic stem cell shape, adhesion, migration, and mobilization. Proc Natl Acad Sci USA , 5614–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]