Abstract
Mouse PUMILIO1 (PUM1) and PUMILIO2 (PUM2) belong to the PUF (Pumilio/FBF) family, a highly conserved RNA binding protein family whose homologues play critical roles in embryonic development and germ line stem cell maintenance in invertebrates. However, their roles in mammalian embryonic development and stem cell maintenance remained largely uncharacterized. Here we report an essential requirement of the Pum gene family in early embryonic development. A loss of both Pum1 and Pum2 genes led to gastrulation failure, resulting in embryo lethality at E8.5. Pum-deficient blastocysts, however, appeared morphologically normal, from which embryonic stem cells (ESCs) could be established. Both mutant ESCs and embryos exhibited reduced growth and increased expression of endoderm markers Gata6 and Lama1, making defects in growth and differentiation the likely causes of gastrulation failure. Furthermore, ESC Gata6 transcripts could be pulled down via PUM1 immunoprecipitation and mutation of conserved PUM-binding element on 3′UTR (untranslated region) of Gata6 enhanced the expression of luciferase reporter, implicating PUM-mediated posttranscriptional regulation of Gata6 expression in stem cell development and cell lineage determination. Hence, like its invertebrate homologues, mouse PUM proteins are conserved posttranscriptional regulators essential for embryonic and stem cell development.
INTRODUCTION
Posttranscriptional regulation represents a fundamental mode of gene expression critical for key cellular decisions as well as human physiology and diseases (Day and Tuite, 1998; Wickens et al., 2002; Keene, 2007; Agami, 2010; Ye and Blelloch, 2014). RNA binding proteins are the critical players in posttranscriptional regulation and often bind to tens or hundreds of RNA targets with related function to achieve coordinated biological control (Keene and Tenenbaum, 2002; Prasad et al., 2016). Understanding the roles of RNA binding proteins could provide insights into the mechanism and impact of posttranscriptional regulation during development and growth.
One of the highly conserved eukaryotic RNA binding proteins is the PUF (PUMILIO/FBF) family, identified initially for its requirement in Drosophila embryonic development but later found to be important in diverse cellular and developmental processes (Lehmann and Nusslein-Volhard, 1987; Wickens et al., 2002; Quenault et al., 2011). A common theme of PUF family protein function among invertebrate homologues appeared to regulate stem cell maintenance/proliferation (Parisi and Lin, 2000; Crittenden et al., 2002; Wickens et al., 2002). PUF proteins bind the consensus PUM-binding element (PBE) on the 3′UTR (untranslated region) of their target mRNAs to regulate the stability, localization, and translation of the target mRNAs (Zhang et al., 1997; Quenault et al., 2011). Functions of the mammalian PUF homologues were recently reported to be important in the nervous and reproductive systems (Xu et al., 2007; Chen et al., 2012; Gennarino et al., 2015; Zhang et al., 2015; Mak et al., 2016; Lin et al., 2017; Zhang et al., 2017). Functional overlap and redundancies have been found for many paralogues within subfamilies of PUF proteins (Wickens et al., 2002). In mice there are two Pumilio genes with canonical eight PUF repeats in the genome, Pum1 and Pum2 (Spassov and Jurecic, 2003). Based on sequence similarity and phylogeny, Pum1 and Pum2 are mammalian orthologues of Drosophila pumilio required for embryonic patterning and germ line stem cell maintenance (Lehmann and Nusslein-Volhard, 1987; Lin and Spradling, 1997; Forbes and Lehmann, 1998; Kuo et al., 2009). It is not known whether such embryonic and stem cell function is also conserved in mammals.
Mice with single-knockout mutation of Pum1 or Pum2 are viable and fertile (Xu et al., 2007; Chen et al., 2012; Gennarino et al., 2015; Lin et al., 2017). A role of Pum in mouse stem cell was reported in haploid Pum1 deletion embryonic stem cells (ESCs) derived from parthenogenetic embryos of unfertilized eggs; neural-specific removal of both Pum genes also affects neurogenesis (Leeb et al., 2014; Zhang et al., 2017). It remained unknown whether mouse Pum1 and Pum2 could compensate for the loss of each other and function redundantly to regulate stem cell development during embryogenesis. Hence we set out to determine the primary requirement of both Pum genes in mice and what effect loss of the entire Pum family has on embryonic and stem cell development.
RESULTS AND DISCUSSION
Absence of Pum double-knockout mutant mice from interbreeding of double heterozygotes
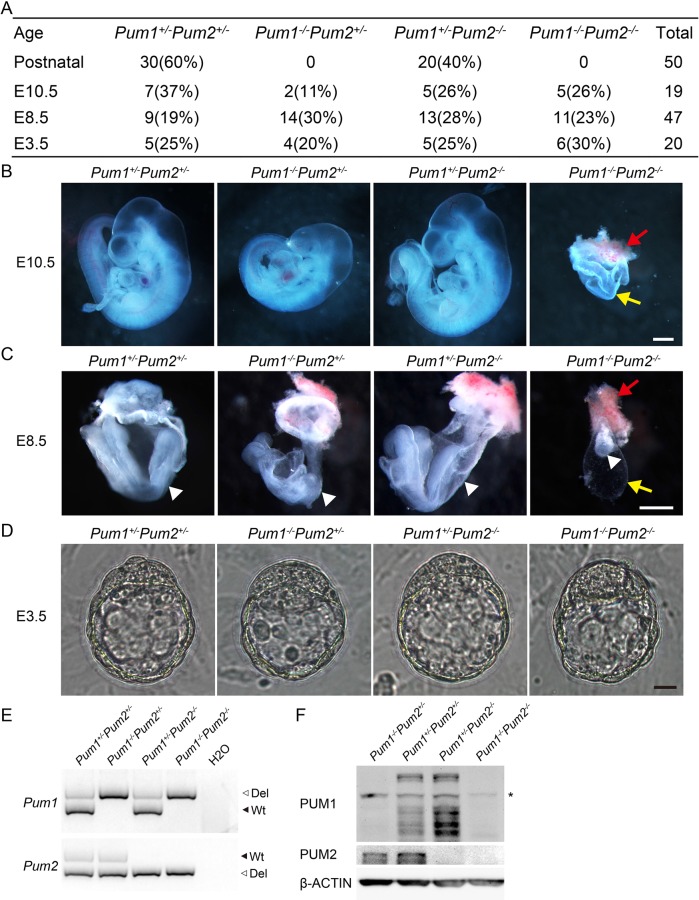
Mouse Pum1 and Pum2 mutations have been reported to affect spermatogenesis but mutants are otherwise viable and fertile (Xu et al., 2007; Chen et al., 2012; Lin et al., 2017). A gene-trap Pum1 mutation failed to produce any homozygous pups or blastocysts, implicating a role of Pum1 in early embryonic development or fertilization (Zhang et al., 2015). Such roles in embryonic development may merely indicate a gain-of-function nature of this Pum1 allele since mice with Pum1 loss-of-function mutation could be recovered and are viable like their homologue Pum2 mutants (Xu et al., 2007; Chen et al., 2012; Lin et al., 2017). To determine the requirement of mammalian PUM family, we hence interbred mice heterozygous for both Pum1 and Pum2 loss-of-function mutations. From 974 progeny, we failed to recover any pups with double-knockout genotype (Pum1−/−; Pum2−/−), indicating mice with a loss of both Pum1 and Pum2 died before birth (Table 1). Hence, PUM family is essential for mammalian embryonic development.
TABLE 1:
Absence of double-knockout offspring from Pum1 and Pum2 double-heterozygous interbreeding.
| Genotype | Males | Females | Total (***) | Expected |
|---|---|---|---|---|
| Pum1+/+;Pum2+/+ | 36 | 41 | 77 | 60.875 |
| Pum1−/−;Pum2−/− | 0 | 0 | 0 | 60.875 |
| Pum1+/+;Pum2−/− | 39 | 37 | 76 | 60.875 |
| Pum1−/−;Pum2+/+ | 26 | 25 | 51 | 60.875 |
| Pum1+/−;Pum2−/− | 60 | 57 | 117 | 121.75 |
| Pum1−/−;Pum2+/− | 16 | 5 | 21 | 121.75 |
| Pum1+/+;Pum2+/− | 77 | 80 | 157 | 121.75 |
| Pum1+/−;Pum2+/− | 157 | 162 | 319 | 243.5 |
| Pum1+/−;Pum2+/+ | 77 | 79 | 156 | 121.75 |
| Total | 488 | 486 | 974 | 974 |
Data were analyzed by Chi-square test, ***p < 0.001.
We also noticed a significant reduction in the number of Pum1−/−; Pum2+/−. Similarly, Pum1 null mice were fewer than expected in Table 1, which was consistent with the results of Pum1 heterozygotes interbreeding (unpublished data) and previous reports (Xu et al., 2007; Chen et al., 2012; Gennarino et al., 2015). The different recovery rates of Pum1 and Pum2 homozygotes in single and double mutants suggest that Pum1 loss has a bigger impact on embryonic development than Pum2 loss. Such a difference may be attributed to different mRNAs targets, which PUM1 and PUM2 bind, besides their common targets. Indeed, Zhang et al reported that PUM1 binds to many more unique targets than PUM2 in the brain (Zhang et al., 2017).
Double-knockout mutant embryos die by E8.5
To further determine the stage when double-knockout embryos die, we set up a cross utilizing a maternally expressed Cre, Ddx4-cre, by mating Ddx4-cre; Pum1+/−; Pum2+/− females and Pum1F/F; Pum2−/− males. The presence of maternal Cre in embryos from Ddx4-cre mother will generate knockout alleles from all Pum1 Flox alleles. This cross scheme is expected to produce double-knockout embryos at one quarter of the total number of embryos, a significant increase from double-heterozygote interbreeding, reducing the number of embryos needed for analysis. Indeed, no double-knockout pups were born at birth, consistent with the results from our interbreeding of double heterozygotes (Figure 1A). We then collected embryos from different time points. Double-knockout embryos were recovered at E10.5 (embryonic day 10.5). The number of E10.5 double-knockout decidua appeared smaller, and the embryos inside were degenerated and almost absent (Figure 1B). Development of placenta and yolk sac in the double-knockout embryos were also severely retarded (Figure 1B). At E8.5, double-knockout embryos were very small and degenerating. Placenta and yolk sac were severely retarded but present, suggesting E8.5 double-knockout embryos were highly retarded and already dead (Figure 1C). To determine whether mutant embryos were affected before implantation, we collected blastocysts at E3.5 day for examination. We not only were able to collect the expected number of embryos of all the genotypes including double knockout but also observed that the mutant blastocysts were morphologically indistinguishable from those of control (Figure 1D). The genotypes of the embryos produced were determined by PCR and later confirmed by Western blot, validating the loss-of-function nature of Pum1 and Pum2 alleles (Figure 1, E and F). Hence mouse embryos without the entire Pum family were able to form inner cell mass and develop all the way to the blastocyst stage. Embryonic stem cell lines were successfully established from those double-knockout blastocysts, suggesting that Pum1 and Pum2 genes are not required for the establishment of pluripotent embryos or stem cells. However, it remains to be determined whether PUM 1 and PUM2 proteins are dispensable for earlier embryonic development up to blastocyst stage and for establishing the stemness, as maternal PUM contribution has not been excluded in those double-knockout mutants.
FIGURE 1:
Pum1 and Pum2 double-knockout embryos died at E8.5. (A) Genotypic results of progeny and embryos produced from male Pum1F/F; Pum2−/− and female Ddx4-cre; Pum1+/−; Pum2+/− intercross. (B) Representative images of dissected E10.5 embryos from four genotypes, except that yolk sac and placenta were not removed from Pum1−/−Pum2−/− embryos. Red arrow, placenta; yellow arrow, yolk sac; scale bar represents 500 μm. (C) Representative images of dissected E8.5 embryos of all four genotypes except that Pum1−/−Pum2−/− embryos were within the yolk sac. Arrowhead, embryo; red arrow, placenta; yellow arrow, yolk sac; Scale bar represents 500 μm. (D) E3.5 blastocysts from Pum1+/−Pum2+/−, Pum1−/−Pum2+/−, Pum1+/−Pum2−/−, Pum1−/−Pum2−/− were recovered and appeared similar in morphology. Scale bar represents 20 μm. (E, F) Genotypes of embryos for Pum1 and Pum2 were determined by PCR (E) and further confirmed by Western blot (F). Asterisk represents nonspecific band. Both PUM1 and PUM2 proteins are completely absent in the double knockout, validating the loss of function nature for both mutations.
Embryo proper exhibited strong expression of Pum1 and Pum2 during embryogenesis
Given the essential requirement of Pum genes during postimplantation embryonic development, we asked how Pum genes are expressed during these stages of early embryogenesis. We first determine the expression pattern of Pum2 during early embryonic development using gene-trap line in Pum2 locus, Pum2XE772 (Xu et al., 2007). We found that embryos as early as blastocysts were already positive for LacZ (Supplemental Figure S1A). At embryonic stage E3.5 blastocyst, we could see LacZ-positive cells in some of the inner cell mass and trophectoderm cells. At E6.5, Pum2 expression was high in almost the entire epiblast cell populations and extraembryonic ectoderm, weak in visceral endoderm, and not detectable in parietal endoderm and ectoplacental cone (Supplemental Figure S1B). At E7.5, embryonic ectoderm, allantois, and visceral endoderm were positive for LacZ staining while ectoplacental cone remained negative for LacZ staining (Supplemental Figure S1C). At E8.5, strongest expression was detected in head fold, somites, primitive streak, and placenta (Supplemental Figure S1D). Overall by E8.5, most parts of embryos were positive for LacZ staining, suggesting that Pum2 is expressed in most embryonic tissues at E8.5.
Pum1-LacZ activities were previously detected in the inner cell mass and trophectoderm of blastocysts, suggesting a potential role of Pum1 in preimplantation stage (Zhang et al., 2015). We further asked when PUM1 protein is expressed via immunohistochemistry. We found that PUM1 protein is expressed extensively during early embryonic development. Strong expression was seen in epiblast, extraembryonic ectoderm, and ectoplacental cone at E6.5 and E7.5, with strongest signal at epiblast (Supplemental Figure S1, E and F). The overlapping expression of Pum1 and Pum2 from blastocyst stage through embryonic development supports the hypothesis that Pum1 and Pum2 may function redundantly during mammalian embryonic development and ESC maintenance.
Mouse embryos lacking both Pum1 and Pum2 could go through early embryonic development and reach blastocysts at the expected ratio, suggesting that the establishment of embryonic pluripotent cells do not require Pum family genes. Nor are Pum genes essential for the development to blastocysts. It should be noted, however, these double-knockout embryos could still have maternal PUM proteins from mother. Indeed, single-cell RNA sequencing data showed that Pum1 is barely detectable at the one-cell and two-cell stages, but Pum2 is expressed at a significant level, suggesting potential maternal PUM2 contribution (Supplemental Figure S2A) (Xue et al., 2013). Further experiments are needed to determine whether embryos with neither zygotic nor maternal PUM proteins could be formed or develop into blastocyst stage.
Gastrulation failure in double-knockout embryos
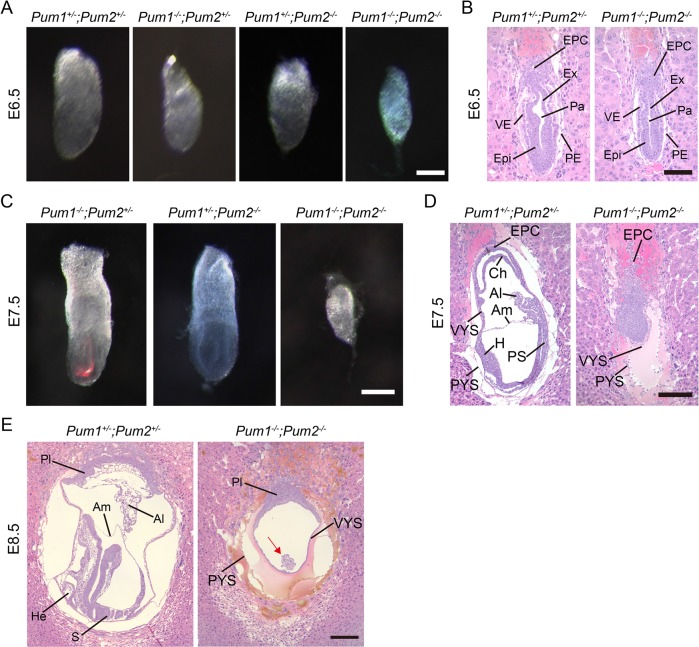
To determine the developmental defects in double-knockout embryos, we collected embryos for analysis prior to E8.5. At E6.5 and E7.5, we recovered the expected ratio of double-knockout embryos, respectively. The double-mutant embryos were smaller at E6.5 and became very small at E7.5 (Figure 2, A and C). The significantly reduced size of E6.5 mutant embryos exhibited grossly normal histology (Figure 2B), suggesting reduced growth in absence of PUM family. However, Pum1−/−; Pum2−/− embryos exhibited severe anatomic defect with sign of degeneration by E7.5. At E7.5, Pum1+/−; Pum2+/− embryos undergo normal gastrulation, resulting in the formation of primitive streak and amniotic cavitation, while Pum1−/−; Pum2−/− embryos remained at the beginning of gastrulation with no signs of primitive streak (Figure 2D). In addition, those Pum1−/−; Pum2−/− embryos also lack allantois, amnion, and chorion and consist of largely a lump of embryonic cells (Figure 2D). By E8.5, normal embryos underwent normal gastrulation and turning; in contrast, the double-knockout embryos undergo further degeneration with only a small number of cells left (Figures 1C and 2E). Our analysis showed that double-knockout embryos were reduced in size at E6.5 with grossly normal structure; however, by E7.5 development was completely disrupted, resulting in a developmental arrest, embryonic degeneration, and, eventually, lethality at E8.5.
FIGURE 2:
Developmental defects in postimplantation embryos lacking both Pum1 and Pum2 genes at E6.5 and E7.5. (A, B) Representative images of intact embryos and HE sections from E6.5 embryos of various genotypes. Scale bars represent 100 μm. (C, D) Representative images of intact embryos and HE-stained sections from E7.5 embryos of various genotypes. E7.5 double-knockout embryos were very small (C), and HE staining shows that embryonic development in tiny embryos was retarded or disrupted with clear signs of degeneration (D). Scale bars represent 200 μm. (E) HE staining of E8.5 Pum1+/−; Pum2+/− and Pum1−/−; Pum2−/− embryo section. The double-knockout embryo was degenerated and retained few embryonic cells and little extraembryonic structure. Scale bars represent 200 μm. Al, allantois; Am, amnion; PYS, parietal yolk sac; VYS, visceral yolk sac; EPC, ectoplacental cone; H, head folds; Ch, chorion; PE, parietal endoderm; PS, primitive streak; Ex, extraembryonic ectoderm; Epi, epiblast; Pa, proamniotic cavity; VE, visceral endoderm.
A major developmental event during this period is gastrulation, in which epiblast cells proliferate and differentiate into three germ layers. This period represents an extraordinary period of proliferation and differentiation. The epiblast cell number from E6.5 to E7.5 increased 22-fold within 1 d, while epiblast undergo gastrulation and produced three germ layers and other extraembryonic structures (Snow, 1977). Coincidentally Pum genes are highly expressed in epiblast during this period (Supplemental Figure S1), supporting important roles leading to or during gastrulation. Since double-knockout mutant embryos lack any embryonic structures such as primitive streak or extraembryonic chorion and amnion, we suspected that the double-knockout mutant embryos either experienced a complete disruption of gastrulation or failed to initiate gastrulation. Hence the loss of Pum genes led to major defects right before or during gastrulation, resulting in gastrulation failure. Drosophila Pumilio regulates anterior–posterior patterning by repressing the translation of hunchback gene during syncytium embryo (Murata and Wharton, 1995). One of Pumilio-like Caenorhabditis elegans PUF family members, Puf-9 is also involved in regulation of development patterning by regulating C. elegans hunchback expression to control developmental timing (Nolde et al., 2007). Like in Drosophila, mammalian Pum family is also required for embryonic development, supporting a conserved function of PUM proteins in embryonic development. Interestingly mouse embryonic patterning also occurs prior to gastrulation (Takaoka and Hamada, 2012). The absence of any gastrulation in E7.5 double mutants could potentially result from a patterning defect or differentiation defect. Hence, we set out to determine gene expression of signature germ layers and pluripotent markers in mutant embryos and in the established double-knockout ESCs.
Loss of PUM proteins led to reduced cell proliferation and increased apoptosis in E6.5 embryo
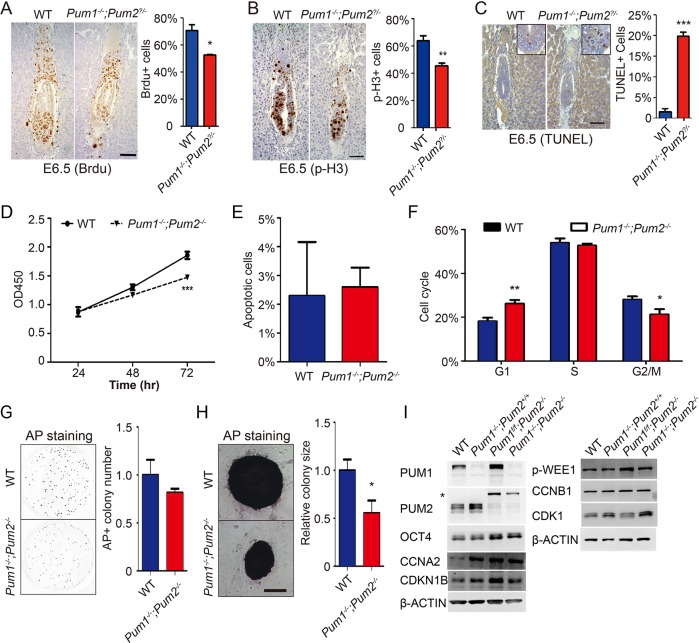
To explore the causes of embryonic lethality of double knockouts, we first study the cause of size reduction in the E6.5 double-knockout embryo. We performed BrdU labeling and phosphorylated-Histone3 (p-H3) immunohistochemistry staining to compare cell proliferation levels in all embryos by E6.5. Embryos from the interbreeding of heterozygotes were collected and stained with PUM1 and PUM2 antibodies to establish genotypes of used embryos. PUM1 negative (Pum1−/−; Pum2−/− or Pum1−/−; Pum2+/−) embryo and wild-type embryo were analyzed, and we found that loss of Pum1 and one or two alleles of Pum2 significantly decreased BrdU positive cells percentage and p-H3-positive cells percentage (Figure 3, A and B), indicating that loss of PUM1 and PUM2 led to significantly reduced cell proliferation in E6.5 embryo. And we also detected increased cell apoptosis (Figure 3C). These data indicate that reduced cell proliferation and increased cell apoptosis may contribute to the smaller embryo size of double knockout. Such reduction in cell proliferation may be small in the mutant cells, as the double-knockout mutant embryos were largely normal at blastocyst stage.
FIGURE 3:
Effects of cell proliferation and apoptosis after depletion of PUMs in E6.5 embryos and ESCs. (A, B) Cell proliferation in wild-type (WT) and Pum1−/−; Pum2–/− embryo by E6.5 was detected by BrdU labeling (A) and p-H3 staining (B). Scale bars represent 100 μm in C and D. (C) Apoptotic cells were detected by TUNEL staining in E6.5 embryo. The data represented percentages of positive cells in each embryo from at least three comparable sections of representative embryos structure (n = 3). Scale bars represent 200 μm. (D) Cell growth was examined by CCK8 assay in wild-type and Pum1−/−; Pum2−/− ESCs. (E) Cell apoptosis were assessed by Annexin V/PI staining. (F) Cell cycle of mutant cells was analyzed using flow cytometry after propidium iodide staining. (G, H) Colony formation assay of wild-type and Pum1−/−; Pum2−/− ESCs when cultured in feeder-coated plates following alkaline phosphatase staining. Scale bars represent 50 μm. (I) Potential Pum targets in cell-cycle regulators were examined using Western blot on single- or double-knockout ESCs. Results are presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
To dissect functions of PUM family in pluripotent stem cells, we isolated ESCs from E3.5 blastocyst, and those double-knockout ESCs could grow in our standard 2i stem cell medium. However, the loss of both Pum1 and Pum2 led to significantly decreased cell growth (Figure 3D) and little change in apoptosis (Figure 3E), indicating that PUMs were important for pluripotent cell growth. In addition, knockout Pum1 and Pum2 in ESCs led to accumulation of G1 phase and reduced G2/M phase (Figure 3F), suggesting potential cell-cycle defect in mutant ESCs.
We also performed a colony formation assay and found that loss of both Pum1 and Pum2 does not affect colony number significantly (Figure 3G), but colony size of Pum1−/−; Pum2−/− was significantly reduced compared with wild-type ESCs (Figure 3H). To determine the potential targets of PUM in cell-cycle regulation, we examined protein expression of CCNA2, CDKN1B, and CDK1 genes in Pum1 and Pum2 single-knockout ESCs as well as double-knockout ESCs, which contain PBEs in their 3′UTR (Kedde et al., 2010). We found that only CDK1 were significantly increased in Pum1−/−; Pum2−/− ESCs relative to wild-type and single-knockout cells (Figure 3I). Increased level of CDK1 may promote cell-cycle progression and is incompatible with the reduced cell proliferation of the mutant cells (Katsuno et al., 2009). Further investigation of other regulators involved in cell proliferation is needed. Taken together, our data suggest that the PUM family plays a significant role in ESC growth and embryo growth.
Double-knockout ESCs and E6.5 embryo display up-regulated endoderm marker- Gata6 expression
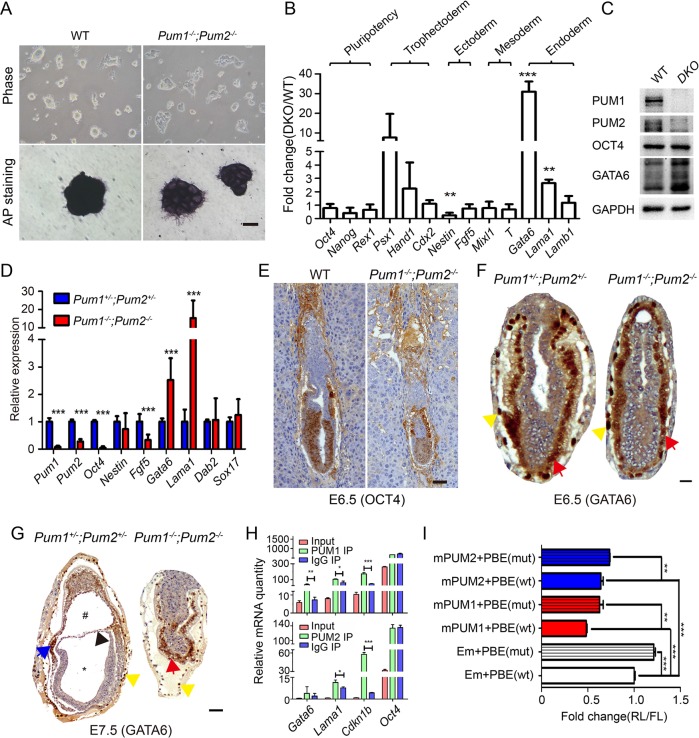
The clue that mammalian PUM proteins might retain a conserved stem cell maintenance function came from a study on special haploid Pum ESCs. Pum1 haploid mutant ESCs failed to exit self-renew (Leeb et al., 2014). Disrupted gastrulation in double-knockout embryos may suggest differentiation defect. Our results show that knockout of both Pum1 and Pum2 do not affect establishment of ESCs; however, double-knockout ESCs displayed spontaneous differentiation when cultured without feeder cells in 2i medium (Figure 4A). After alkaline phosphatase staining, wild-type ESC colonies showed strong AP-positive staining while double-knockout ESC colonies exhibited reduced AP signal, suggesting that double-knockout ESCs remained pluripotent but their full pluripotency may be compromised. To examine the extent of spontaneous differentiation for double-knockout ESCs, we performed quantitative reverse transcription PCR (qRT-PCR) on molecular markers of pluripotency and three germ layers. Although pluripotent factors such as Oct4, Nanog, and Rex1 were not affected in double-knockout ESCs, ectoderm marker-Nestin was significantly reduced while endoderm markers-Gata6 and Lama1 were significantly increased with Gata6 transcript level increasing as much as 30-fold and increased GATA6 protein level as well (Figure 4, B and C).
FIGURE 4:
Developmental defects in postimplantation embryos lacking both Pum1 and Pum2 genes at E6.5 and E7.5. (A) Representative images of wild-type and Pum1−/−; Pum2−/− ESC colonies and AP staining. Spontaneous differentiation occurred in double-knockout ESCs when cultured under feeder-free condition. Scale bar represents 20 μm. (B) Relative expression of pluripotency and three germ layers marker genes was compared by qRT-PCR between wild-type and double-knockout ESCs, and results were normalized to wild type and are shown as mean ± SD. Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001. (C) Western blot result shows that GATA6 protein level was increased in double-knockout (DKO) ESCs compared with wild type (WT). (D) Relative expression of ectoderm and endoderm markers were detected by qRT-PCR in double-heterozygote and DKO whole E6.5 embryos, and results were normalized to double heterozygote and are shown as mean ± SD. (E, F) OCT4 (E) and GATA6 (F) levels in E6.5 embryos were detected by immunohistochemistry staining. Scale bars represent 100 μm in E and 50 μm in F. (G) GATA6 protein expression was detected by immunohistochemistry staining in E7.5 embryos. Scale bars represent 50 μm. Yellow arrowhead represents parietal endoderm, red arrow represents visceral endoderm, blue arrow represents precardiac mesoderm, black arrow represents amnion, * and # represent amniotic cavity and exocoelomic cavity, respectively. (H) qRT-PCR results demonstrate the association of Gata6 and Lama1 mRNA with PUM1 and PUM2 proteins for ESCs in comparison to IgG precipitates. Results are representing as mean ± SD. Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001. (I) Bar graph results from dual-luciferase assay on HEK-293T cells expressing the reporter constructs containing either wild-type or PBE mutated Gata6 3′UTR. The cells were also cotransfected with mouse Pum1, Pum2 and empty vector. Results from A to I are presented as mean ± SD. Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001.
Next we investigate molecular changes in mutant embryos prior to gastrulation stage. Remarkably, expression of Gata6 and Lama1 mRNAs were also significantly increased in double-knockout embryo by E6.5, similarly to that in double-knockout ESCs (Figure 4D). Fgf5 and Oct4 were significantly decreased in E6.5 embryos (Figure 4D). These results indicated that pluripotent cells or epiblast stem cells in absence of both Pum1 and Pum2 might prematurely differentiate toward endoderm germ layer. To further examine to what extent stemness and differentiation potential were affected in mutant embryos, we performed immunohistochemistry staining and found that overall OCT4 signal intensity and Oct4+ cells were reduced in E6.5 double-knockout embryo (wild type [WT] 64.1% and mutant 46.9%, p < 0.001) (Figure 4G). Hence the pluripotency state of epiblast cells in double-knockout embryo may be compromised.
Given that Gata6 RNA and protein were both increased in double-knockout ESCs, we asked whether such molecular change is also present in vivo. There was no obvious difference of GATA6 protein expression between the double-heterozygote and double-knockout embryos by E6.5 (Figure 4F). However, the E7.5 double-knockout embryo exhibited broad GATA6 expression in most parts of embryo; in contrast, GATA6 expression in wild-type or double-heterozygote embryos was restricted only in parietal endoderm cells at E7.5. Taken together, loss of both Pum1 and Pum2 in both ESC and E6.5 embryos led to selective increase of gene expression in endoderm markers and possibly premature and disproportionate differentiation toward endoderm. When such lineage differentiation bias was acquired, extreme high level of GATA6 in embryonic visceral endoderm may disrupt gastrulation related genes’ expression pattern and morphogenesis, as evidenced by altered expression of components of Wnt signal pathway important for gastrulation in mutant embryos (Supplemental Figure S2B) (Schrode et al., 2014; Wamaitha et al., 2015; Rodriguez and Downs, 2017).
Gata6 expression is regulated by PUM1 at posttranscriptional level
We next asked how loss of PUM proteins led to increased expression of Gata6. Given that PUM are posttranscriptional regulators, which regulate the stability of target transcripts (Quenault et al., 2011), we performed RNA immunoprecipitation using PUM1 and PUM2 antibodies on ESC lysate. Both PUM1 and PUM2 proteins could be precipitated efficiently from ESCs by specific antibodies (Supplemental Figure S2C). Cdkn1B has been previously reported to be a target of PUM and Oct4 is a nontarget as a negative control (Kedde et al., 2010; Leeb et al., 2014). We found that Gata6 transcript is significantly enriched by PUM1 antibody but not by PUM2 antibody while Lama1 was slightly enriched by either antibody (Figure 4H). Given the association of Gata6 with PUM1 protein, we searched the 3′UTR of Gata6 and identified one PBE site while none on the 3′UTR of Lama1 (Supplemental Figure S2D). To determine whether PUM regulates expression of Gata6 via its 3′UTR, we construct a dual luciferase reporter construct containing wild-type Gata6 3′UTR and mutant 3′UTR with the PBE site disrupted (Supplemental Figure S2D). Mutation of PBE consistently up-regulated the expression of luciferase reporter in absence or presence of PUM1 or PUM2 (Figure 4I). The increased expression of luciferase in absence of Pum transgenes likely resulted from repression of endogenous PUM1 and PUM2 proteins. Overexpression of PUM1 and PUM2 led to repression of luciferase reporter containing wild-type or mutant Gata6 3′UTR, but reporters from mutant Gata6 3′UTR consistently exhibited higher expression. Although Gata6 does not appear to be associated with PUM2 protein, overexpression of PUM2 still exhibited an effect similar to that of PUM1, supporting similar binding ability of PUM1 and PUM2 when overexpressed. We hence proposed that PUM proteins, in particular PUM1, regulate Gata6 expression at posttranscriptional level and such regulation is important for stem cell and embryonic development. Gata6 is not only an endoderm marker but also a key regulator of primitive endoderm differentiation (Morgani and Brickman, 2015); PUM-mediated Gata6 repression could be important to keep ESCs and epiblast cells from premature differentiation and to ensure proper cell lineage determination and differentiation leading to gastrulation.
In conclusion, we report an essential embryonic requirement of mouse Pum gene family through the analysis of mouse double-knockout mutations of both Pum1 and Pum2, supporting a conserved embryonic function of Pumilio genes in mice. Mouse embryos lacking both Pum1 and Pum2 died by E8.5, resulting from severe defect in cell growth and gastrulation. Furthermore, double-knockout ESCs and E6.5 embryos display a significantly increased level of endoderm marker-Gata6, suggesting differentiation and cell lineage defect prior to gastrulation. Furthermore, PUM1 may regulate Gata6 expression directly at the posttranscriptional level to ensure stemness of ESCs and proper cell lineage determination during early embryo development. Hence zygotic Pum1 and Pum2 function redundantly during postimplantation embryonic development but are dispensable for establishment of embryonic pluripotent stem cells or for preimplantation embryonic development. Identification of other targets PUM binds to during embryo development could provide a full picture of PUM-mediated posttranscriptional regulation in stem cell and embryonic development. In addition, characterization of Pum mutants in pure inbred background, currently underway, should also provide greater insight into PUM’s requirement and roles in stem cell maintenance and differentiation.
MATERIALS AND METHODS
Generation of Pum1 and Pum2 double-knockout animals
We have constructed Pum1 and Pum2 mutant mice previously (Xu et al., 2007; unpublished data). Pum1flox/flox; Pum2−/− male mice and Ddx4-Cre; Pum1+/−; Pum2+/− female mice were crossbred to obtain double-knockout mice (Gallardo et al., 2007). Mouse strains were on the mixed genetic background of FVB/NJ, C57B6, and 129svj. Animals were raised under standard conditions in the animal facilities of Nanjing Medical University, Nanjing, China. All mouse procedures and protocols were approved by the Animal Care and Use Committee of Nanjing Medical University and conducted in accordance with institution guidelines for the care and use of animals.
Genotyping
Newborn pups of each litter were collected and genotyped by PCR using genomic DNA extracted from tissues. Blastocysts (E3.5 embryos) were flushed from uterus of plugged female mice. Embryos (6.5, 7.5, 8.5, and 10.5 dpc [days postcoitum]) were dissected from pregnant mice, and yolk sac or embryos were used for genotyping. Primers for Pum1 genotyping are KOF1-P1 (5′-CAT GAG TTT GGG AGG CAT TT-3′), KOR1-P3 (5′-GTG GCT AAC AAC TGC TGC AA-3′), and LOXGTR-P2 (5′-GTC TTG TGG CAA CTA GGG TA-3′). PCR products include the following: 361-base-pair fragment for Pum1-WT allele, a 453-base-pair fragment for Pum1-floxed allele, and a 557-base-pair fragment for Pum1-mutant allele. Genotyping for Pum2 transgenic mice was performed as described previously (Xu et al., 2007).
Western blot
Whole-cell lysates were prepared from yolk sac and embryo at E10.5. Aliquots of the lysates were electrophoresed in 8.0% or 12% polyacrylamide gel and transferred to PVDF (polyvinylidene difluoride) membrane (Bio-Rad). The membrane was blocked with 1% skim-milk/TBST (Tris-buffered saline with 0.1% Tween-20) and probed with various antibodies: anti-Actin (Sigma, A1978), anti-Pumilio1 (PUM1) (Abcam, ab92545), anti-PUM2 (Abcam, ab92390), anti-CDKN1B (Abcam, ab92741), anti-CDK1 (Cell Signal Technology] [CST], 9116), anti-Cyclin B1 (CST, 12231), anti-p-Wee1 (CST, 4910), anti-Cyclin A2 (CST, 4656), in 1% bovine serum albumin/TBST. Signals were detected with HRP (horseradish peroxidase)-conjugated secondary antibody (CST) and chemiluminescence detection reagents (ECL Plus Kit; Amersham).
Histology, immunohistochemistry, and TUNEL assay
Embryos in decidua were fixed overnight in Hartman’s fixative (Sigma). Fixed embryos were embedded in paraffin and cut into 5-µm-thick serial sections. Each embryo was sectioned in its entirety, and comparable sections between control and mutant embryos were used for analysis. In case of reduced mutant embryos or degenerated embryos, sections with most tissues are shown. All the comparison analysis on embryos was done on the embryos collected from the same litter. Hematoxylin and eosin (H&E) staining and immunohistochemistry followed standard protocols (VanGompel and Xu, 2010). Antibody are as follows: PUM1 (Abcam, ab92545), PUM2 (Abcam, ab92390), anti-BrdU (Invitrogen, 03-3940), anti-phospho-Histone H3 (Ser10) (CST, 3377), OCT4 (Santa Cruz, sc-5279). and GATA6 (R&D, AF1700). TUNEL analysis was performed using the In Situ Cell Death Detection Kit from Roche according to the manufacturer’s instructions.
X-gal staining
Whole-mount X-gal staining in embryo were performed as previously reported (Xu et al., 2007).
Generation of ESCs
The blastocysts were placed in a four-well plate precoated with mouse feeder cells and cultured in 2i medium as described previously for cell-line derivation (Li et al., 2012). The 2i medium consists of N2B27 medium supplemented with 1 mM MEK inhibitor PD0325901 (Stemgent), 3 mM GSK3b inhibitor CHIR99021 (Stemgent), mouse recombinant LIF (Millipore), and 5% knockout serum replacement (GIBCO). The outgrowths were digested with 0.25% trypsin after being cultured for 5–7 d. ESC colonies usually appeared after 2–3 d. For feeder-free culture, cells were maintained in a dish coated with gelatin 0.1% solution (Sigma-Aldrich). ESCs were cultured in 2i medium and passaged with a split ratio of 1:3 every 2–3 d.
CCK8 assay
ESCs were cultured in 2i with N2B27 medium. ESCs were plated in 96-well plates at 2000 cells per well and cultured for 24, 48, and 72 h. The absorbance at 450 nm was measured after incubation with 10 μl of Cell Counting Kit 8 (CCK8, Yeasen) for 2 h. Results were combined from three independent experiments.
EdU incorporation
For EdU incorporation experiments, ESCs were treated with 10 μM EdU for 30 min. EdU incorporation was determined using Click-iTEdU Flow Cytometry Assay Kits (Invitrogen) following the manufacturer’s instructions.
Cell cycle and apoptosis analysis
ESCs were seeded in 12-well plates at a density of 1 × 105 cells per well. For cell-cycle analysis, cells were harvested 20 h after seeding and fixed in 70% ethanol overnight at 4°C. Fixed cells were washed twice with phosphate-buffered saline (PBS) and stained in PI (propidium iodide)/RNase Staining Buffer (BD Biosciences) for 30 min at room temperature. At least 10,000 cells were counted for each sample, and data were analyzed with MODfit L.T. 4.0 software (Verity Software House). For cell apoptosis analysis, 1 × 105 cells were collected and washed twice with ice-cold PBS, suspended in binding buffer (100 μl), treated with Annexin V-FITC (fluorescein isothiocyanate) and PI (BD Biosciences) and incubated in the dark for 15 min. Another 100 μl of binding buffer was then added, and flow cytometry analysis was performed within 1 h to measure Annexin V-FITC-positive cells for apoptosis (BD FACVerse).
Colony formation assay
ESCs were grown on feeder-coated plates in N2B27 supplemented with 2i/LIF. For colony formation assay, 300 cells were plated on a 12-well plate with irradiated mouse embryonic fibroblasts as feeders in 2i/LIF media. Figure 3H shows the alkaline phosphatase staining in the colony-forming assay: nearly every colony stains positive. Colonies were stained and counted 5–8 d after plating (Gu et al., 2016). Relative colony number and colony size were analyzed using ImageJ software.
Alkaline phosphatase staining
Pluripotency of established ES cell lines was determined by alkaline phosphatase (AP) staining. Cultured confluent ESCs were fixed with Formalin for 15 min for AP staining according to the manufacturer’s instructions using the Alkaline Phosphatase Detection Kit (Sidansai Biotechnology Company).
RNA immunoprecipitation
RNA immunoprecipitation (RIP) was performed as described previously (Keene et al., 2006). Wild-type and double-knockout ESCs were cultured in feeder-free condition and grown to 80% confluence for collection. About 1.0 × 107 ESCs were lysed in polysome lysis buffer. Goat anti-PUM1 antibody (Bethyl Lab) and Rabbit anti-Pum2 antibody (Bethyl Lab) were coupled to Protein A Agarose (Invitrogen), respectively. ESC lysates were added to antibody-coupled beads to immunoprecipitate PUM1 or PUM2 proteins. RNA associated PUM proteins were extracted with Trizol Reagent (Invitrogen) for qPCR analysis of candidate target RNAs.
RNA isolation and real-time PCR
RNA from embryos or cell pellets was extracted with an RNeasy micro/mini kit (QIAGEN). Reverse transcription was performed using a PrimeScript RT Master Mix (Takara). Real-time PCR was performed with Taq Polymerase (Takara) according to the product manual and gene-specific primers are listed below. The number of PCR cycles ranged from 22 to 35 depending on the linearity of the reaction. All gene expression analyses were performed with samples from at least three independent experiments.
PCR primers listed are as follows (5′ to 3′):
Oct4 forward (ATGGCATACTGTGGACCTCA),
Oct4 reverse (AGCAGCTTGGCAAACTGTTC);
Nanog forward (CTCATCAATGCCTGCAGTTTTTCA),
Nanog reverse (CTCCTCAGGGCCCTTGTCAGC);
Rex1 forward (ACGAGGTGAGTTTTCCGAAC),
Rex1 reverse (CCTCTGTCTTCTCTTGCTTC);
Psx1 forward (GAATTGGTTTCGGATGAGGA),
Psx1 reverse (GTGGCTCAGAAGAAGCCATC);
Hand1 forward (GCCAAGGATGCACAAGCA),
Hand1 reverse (GGGCTGCTGAGGCAACTC);
Cdx2 forward (CGAGCCCTTGAGTCCTGTGA),
Cdx2 reverse (AACCCCAGGGACAGAACCA);
Nestin forward (TGAGGGTCAGGTGGTTCTG),
Nestin reverse (AGAGCAGGGAGGGACATTC);
Fgf5 forward (TGCGTCCGCGATCCA),
Fgf5 reverse (TCAGGGCCACGTACCACTCT);
Mixl1 forward (ACTTTCCAGCTCTTTCAAGAGCC),
Mixl1 reverse (ATTGTGTACTCCCCAACTTTCCC);
T forward (ATCACCAGCCACTGCTTTC),
T reverse (CCATTACATCTTTGTGGTCGTTTC);
Gata6 forward (CTTGCGGGCTCTATATGAAACTCCAT),
Gata6 reverse (TAGAAGAAGAGGAAGTAGGAGTCATAGGGACA);
Lama1 forward (CAGAGCCCCAGTATTCAGAATG),
Lama1 reverse (CGCTCCAAAATCCAGTTTCC);
Lamb1 forward (CCCCAATCTCTGTGAACCATG),
Lamb1 reverse (GCAATTTGCACCGACACTGA);
Dab2 forward (TCTCAGCCTGCATCTTCTGA),
Dab2 reverse (GAGCGAGGACAGAGGTCAAC);
Sox17 forward (GAGGGCCAGAAGCAGTGTTA),
Sox17 reverse (AGTGATTGTGGGGAGCAAGT);
β-Actin forward (TGACCCAGATCATGTTTGAG),
β-Actin reverse (GAGTCCATCACAATGCCTG).
Dual-luciferase report system assay
The region of the mouse Gata6 3′UTR (NM_010258.3) containing PBE was subcloned into psiCHECK-2 vector (Promega) using a ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech). Wild-type and mutant PBE sequences were as follows: PBE wt, 5′-TGTAAATA-3′; PBE mutant, 5′-acaAAATA-3′ (Weidmann and Goldstrohm, 2012). 293T cells in 24-well plates were transfected with 100 ng of psi-CHECK-2 construct carrying either wild-type or mutant Cdkn1b 3′UTR plus 500 ng of EM46-mPum1/CMV-Pum2 or control pCMV vector using FUGENE HD (Promega). After 36 h, Firefly expression and Renilla luciferase expression were measured by using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Wenan Qiang and Yanmei Chen for initial characterization of PUM mutant mice, Min Zang for technical assistance, and Alec Wang, Zhongzhou Yang, and Jun Yan for discussion and/or comments on our manuscript. We are grateful for Haixin Li’s advice and help with RIP experiments and for two anonymous reviewers’ helpful comments. This work was supported by the National Basic Research Program of China (973 program, 2015CB943002 and 2013CB945201), the National Science Foundation of China (81270737, 31771652, and 81401256), the Natural Science Foundation of Jiangsu Province (BK2012838), and a Provincial Innovation and Entrepreneurship Grant. Funding for open access charge: Provincial Shuangchuang Program.
Abbreviations used:
- dpc
days postcoitum
- ESC
embryonic stem cell
- PBE
PUM-binding element
- PUF
pumilio/FBF
- PUM
PUMILIO
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-06-0369) on September 26, 2018.
REFERENCES
- Agami R. (2010). microRNAs, RNA binding proteins and cancer. Eur J Clin Invest , 370–374. [DOI] [PubMed] [Google Scholar]
- Chen D, Zheng W, Lin A, Uyhazi K, Zhao H, Lin H. (2012). Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr Biol , 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. (2002). A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature , 660–663. [DOI] [PubMed] [Google Scholar]
- Day DA, Tuite MF. (1998). Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol , 361–371. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. (1998). Nanos and Pumilio have critical roles in the development and function of germline stem cells. Development , 679–690. [DOI] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. (2007). Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis , 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim JY, Jafar-Nejad P, di Ronza A, Kang H, Sayegh LS, et al. (2015). Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type ataxin1 levels. Cell , 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu KL, Zhang Q, Yan Y, Li TT, Duan FF, Hao J, Wang XW, Shi M, Wu DR, Guo WT, Wang Y. (2016). Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res , 350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. (2009). Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci USA , 3184––3189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. (2010). A Pumilio-induced RNA structure switch in p27-3’UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol , 1014–1020. [DOI] [PubMed] [Google Scholar]
- Keene JD. (2007). RNA regulons: coordination of post-transcriptional events. Nat Rev Genet , 533-543. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. (2006). RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc , 302–307. [DOI] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA. (2002). Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell , 1161––1167.. [DOI] [PubMed] [Google Scholar]
- Kuo MW, Wang SH, Chang JC, Chang CH, Huang LJ, Lin HH, Yu AL, Li WH, Yu J. (2009). A novel puf-A gene predicted from evolutionary analysis is involved in the development of eyes and primordial germ-cells. PLoS One , e4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. (2014). Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell , 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. (1987). Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature , 167. [Google Scholar]
- Li VC, Ballabeni A, Kirschner MW. (2012). Gap 1 phase length and mouse embryonic stem cell self-renewal. Proc Natl Acad Sci USA , 12550–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development , 2463–2476. [DOI] [PubMed] [Google Scholar]
- Lin KB, Zhang SK, Chen JL, Yang D, Zhu MY, Yujun Xu E. (2017). Generation and functional characterization of a conditional Pumilio2 null allele. J Biomed Res, DOI: 10.7555/JBR.32.20170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak W, Fang C, Holden T, Dratver MB, Lin H. (2016). An important role of pumilio 1 in regulating the development of the mammalian female germline. Biol Reprod , 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani SM, Brickman JM. (2015). LIF supports primitive endoderm expansion during pre-implantation development. Development , 3488–3499. [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. (1995). Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell , 747–756. [DOI] [PubMed] [Google Scholar]
- Nolde MJ, Saka N, Reinert KL, Slack FJ. (2007). The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3’UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol , 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Lin H. (2000). Translational repression: a duet of Nanos and Pumilio. Curr Biol , R81–R83. [DOI] [PubMed] [Google Scholar]
- Prasad A, Porter DF, Kroll-Conner PL, Mohanty I, Ryan AR, Crittenden SL, Wickens M, Kimble J. (2016). The PUF binding landscape in metazoan germ cells. RNA , 1026–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A. (2011). PUF proteins: repression, activation and mRNA localization. Trends Cell Biol , 104–112. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Downs KM. (2017). Visceral endoderm and the primitive streak interact to build the fetal-placental interface of the mouse gastrula. Dev Biol , 98–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N, Saiz N, Di Talia S, Hadjantonakis AK. (2014). GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev Cell , 454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow MHL. (1977). Gastrulation in the mouse: Growth and regionalization of the epiblast. J Embryol Exp Morphol , 293–303. [Google Scholar]
- Spassov DS, Jurecic R. (2003). Mouse Pum1 and Pum2 genes, members of the Pumilio family of RNA-binding proteins, show differential expression in fetal and adult hematopoietic stem cells and progenitors. Blood Cells Molec Dis , 55–69. [DOI] [PubMed] [Google Scholar]
- Takaoka K, Hamada H. (2012). Cell fate decisions and axis determination in the early mouse embryo. Development , 3–14. [DOI] [PubMed] [Google Scholar]
- VanGompel MJ, Xu EY. (2010). A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum Mol Genet , 2360–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamaitha SE, del Valle I, Cho LT, Wei Y, Fogarty NM, Blakeley P, Sherwood RI, Ji H, Niakan KK. (2015). Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev , 1239–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann CA, Goldstrohm AC. (2012). Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol Cell Biol , 527––540.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. (2002). A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet , 150–157. [DOI] [PubMed] [Google Scholar]
- Xu EY, Chang R, Salmon NA, Pera RAR. (2007). A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol Reprod Dev , 912–921. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature , 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Blelloch R. (2014). Regulation of pluripotency by RNA binding proteins. Cell Stem Cell , 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. (1997). A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature , 477–484. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhu T, Chen Y, Xu EY. (2015). Loss of preimplantation embryo resulting from a Pum1 gene trap mutation. Biochem Biophys Res Commun , 8–13. [DOI] [PubMed] [Google Scholar]
- Zhang M, Chen D, Xia J, Han W, Cui X, Neuenkirchen N, Hermes G, Sestan N, Lin H. (2017). Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev , 1354–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.