Abstract
Growing evidence indicates that phosphoserine phosphatase (PSPH) is up-regulated and correlates with prognosis in multiple types of cancer. However, little is known about the roles of PSPH in NSCLC. Thus, the aim of the present study was to demonstrate the expression of PSPH in human NSCLC and reveal its biological functions and the underlying mechanisms. qRT-PCR, western blot and immunohistochemistry were used to assess the expression of NSCLC patient specimens and NSCLC cell lines. The functions of PSPH in migration and invasion were determined using trans-well and wound-healing assays. Cell proliferation capacity was performed by cell counting kit-8 (CCK-8), colony formation assays and cell cycle analysis. We demonstrated that PSPH was overexpressed in NSCLC specimens compared with the adjacent non-tumorous specimens, and high expression of PSPH was associated with clinical stage, metastasis and gender in NSCLC. Decreased expression of PSPH inhibited NSCLC cells migration, invasion and proliferation. Most importantly, further experiments demonstrated that PSPH might regulate NSCLC progress through MAPK signaling pathways. Lastly, immunohistochemistry (IHC) revealed that the PSPH expression level was positively correlated with the clinical stage in NSCLC patients. These results suggest that PSPH may act as a putative oncogene and a potential therapeutic target in NSCLC.
Keywords: PSPH, NSCLC, metastasis, proliferation, MAPK
Introduction
Lung cancer is the most frequently-occurring tumor and the leading cause of cancer-related death worldwide 1. As the predominant type of lung cancer, non-small cell lung cancer (NSCLC) accounts for about 85% of all lung cancer cases 2. According to the histopathology, NSCLC can be divided into adenocarcinoma, squamous cell carcinoma and large cell carcinoma. Among these types, adenocarcinoma cases account for the largest number of NSCLC cases, especially in China. Despite great clinical improvements in the systematic treatment were performed for lung cancer patients, the 5-year survival rate remains less than 15% after initial diagnosis 3. More than 90% of NSCLC patients are triggered by tumor progression, especially distant metastasis, and metastasis has been a critical contributor to mortality rate 4. Furthermore, though conventional tumor biomarker examination techniques are wildly used, there is no effective and predictable biomarker for the early NSCLC metastasis. Therefore, exploring potential therapeutic targets of NSCLC metastasis is required for more effective clinical therapies.
Phosphoserine phosphatase (PSPH) is a member of haloacid dehalogenase (HAD) superfamily and contains the conserved N-terminal DXDXT(T/V) motif sequence, which is utilized in the L-serine biosynthesis process and converts phospho-L-serine to L-serine 5. Several reports indicate that PSPH mainly participates in multiple fundamental aspects of cell behavior such as proliferation and differentiation by producing a precursor for the biosynthesis of diverse compounds including neurotransmitters, phospholipids, glycolipids, purines, and thymidine. For example, PSPH deficiency induces congenital neurological abnormalities such as Williams syndrome, and neural tube defects by inhibiting precursor compounds biosynthesis 6. On the other hand, augmented PSPH expression level is associated with a number of cancers including cutaneous squamous cell carcinoma, breast cancer, colorectal cancer, gastric cancer and hepatocellular carcinoma 7-11. Interesting, PSPH is located on the short arm of chromosome 7p11.2, a frequent chromosome amplification region has been observed in NSCLC 12. Although various studies of PSPH are being conducted in other tumors, very little is known about the functional significance of PSPH dysregulation in NSCLC. In this study, we aimed to identify the potential role and mechanism of PSPH in NSCLC.
Results
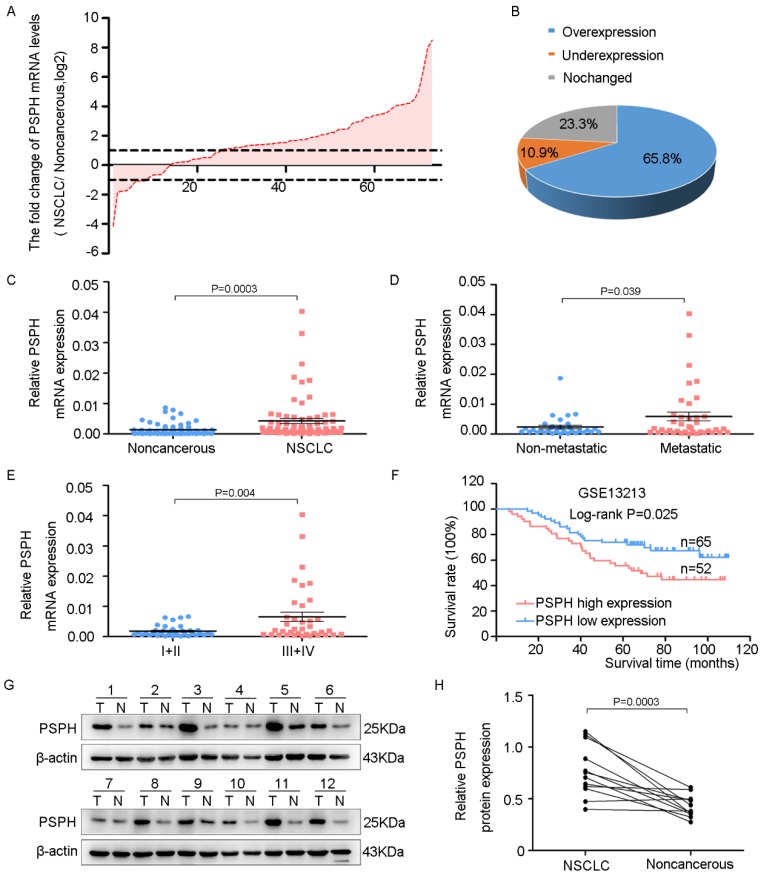
PSPH is overexpressed in NSCLC and associated with NSCLC metastasis and progression
To assess the potential clinical significance of PSPH expression in NSCLC progression, we analyzed PSPH mRNA expression in 73 pairs of human NSCLC tissues and their corresponding nontumorous tissues by qRT-PCR. The relationship between the PSPH expression levels and the clinicopathological characteristics of the NSCLC patients is summarized in Table 1. The results showed no statistically significant correlations between PSPH mRNA expression and age, pathological type, differentiation and tumor size. However, we observed that PSPH expression was almost up-regulated threefold in NSCLC tissues compared with corresponding nontumorous tissues (Figure 1C). In addition, PSPH expression was up-regulated in 65.8% of NSCLC cases (Figure 1A and 1B). Moreover, its expression was positively correlated with advanced TNM stages (III and IV), lymph node and distal metastasis, and gender (Figure 1D and 1E). Kaplan-Meier survival was performed to analyze the correlation between the PSPH expression levels and overall survival. Importantly, high levels of PSPH mRNA were remarkably associated with poor outcomes in the 117 NSCLC patient cohort downloaded from GEO (Figure 1F). Furthermore, compared with corresponding nontumorous tissues. PSPH protein was overexpressed in NSCLC patient tissues using western blot analysis (Figure 1G and 1H). Taken together, our data show that PSPH is highly expressed in lung cancer and related to clinical severity and prognosis. These results indicated that PSPH potentially has a pivotal role in the aggressiveness of NSCLC.
Table 1.
Relationship between PSPH mRNA expression and their clinicopathologic parameters in 73 NSCLC patients by qRT-PCR.
| Characteristics | Number of cases (%) | PSPH expression | |
|---|---|---|---|
| Mean± SD | P value | ||
| Age | |||
| < 60 | 47 (64.4%) | 0.00299±0.00473 | 0.280 |
 60 60 |
26 (35.6%) | 0.00489±0.00814 | |
| Gender | * | ||
| Female | 29 (39.7%) | 0.00201±0.00178 | 0.033 |
| Male | 44 (60.3%) | 0.00567±0.00883 | |
| Tumor size(cm) | |||
| < 3 | 28 (38.4%) | 0.00382±0.00709 | 0.720 |
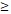 3 3 |
45 (61.6%) | 0.00446±0.00721 | |
| Tissue | *** | ||
| NSCLC | 73 | 0.00426±0.00716 | 0.000 |
| Noncancerous | 73 | 0.00139±0.00190 | |
| Pathological type | |||
| Adenocarcinoma | 57 (78.1%) | 0.00336±0.00593 | 0.057 |
| Squamous cell carcinoma | 16 (21.9%) | 0.00725±0.00988 | |
| Degree of differentiation | |||
| Well and moderately | 26 (35.6%) | 0.00326±0.00523 | 0.403 |
| Poorly | 47 (64.4%) | 0.00474±0.00800 | |
| Clinical stage | ** | ||
| Stage I+II | 36 (49.3%) | 0.00182±0.00181 | 0.004 |
| Stage III+IV | 37 (50.7%) | 0.00655±0.00934 | |
| Metastasis¹ | * | ||
| No | 34 (46.6%) | 0.00236±0.00334 | 0.039 |
| Yes | 39 (53.4%) | 0.00584±0.00899 | |
¹The patients were classified into tumor metastasis negative and positive groups (lymph node metastasis and/ or distal metastasis).
P-value represents the probability from a Student's t-test for PSPH expression between variable subgroups. *P<0.05, **P<0.01 and ***P<0.001, which was considered to have a significant difference.
Figure 1.
PSPH is overexpressed in NSCLC and correlated with advanced clinical stage and tumor invasion and metastasis. A. The expression levels of PSPH in 73 paired NSCLC and corresponding noncancerous tissues were measured by qRT-PCR analysis and β-Actin was used as the internal reference gene. B. Overexpression of PSPH mRNA was observed in 65.8% of the NSCLC compared with the corresponding non-cancerous tissues. C. PSPH mRNA was overexpressed in NSCLC tissues compared with the noncancerous tissues (P value =0.0003). D. The up-regulation of PSPH mRNA in NSCLC was associated with tumor metastasis (P value =0.039). The patients were classified into tumor metastasis negative and positive groups (lymph node metastasis and/ or distal metastasis). E. PSPH mRNA expression was detected in different clinical stages of NSCLC (P value =0.004). F. Kaplan-Meier analyses of the correlation between PSPH mRNA levels and overall survival in the 117-patient-GEO cohort (P value =0.025). G. The expression of PSPH protein was detected in paired NSCLC and corresponding noncancerous tissues (T is the abbreviation tumor tissue, N is the abbreviation for noncancerous tissue). H. Overexpression of PSPH protein was observed in NSCLC tissues (P value =0.0003). Error bars represent SD (standard deviation). The statistical analysis was performed using paired t-test for C and H, and Student's t-test for D and E.
PSPH enhances the migration and invasion ability of NSCLC cells in vitro
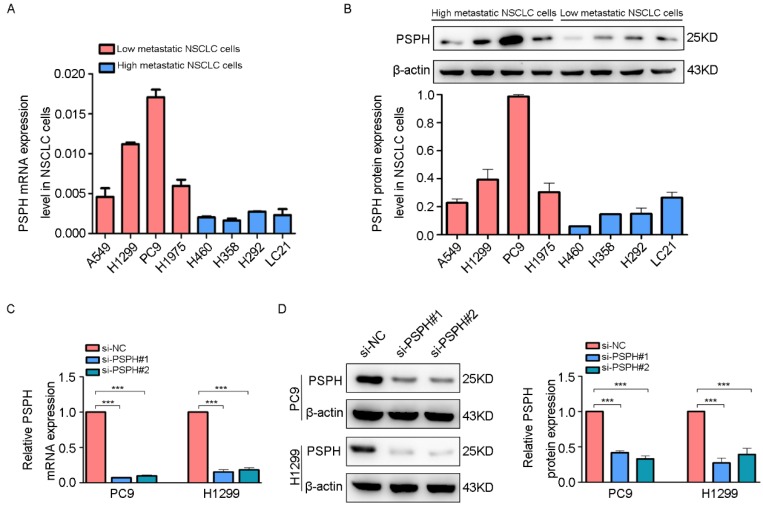
To clarify the significance of PSPH in human NSCLC metastasis, we investigated the endogenous expression level of PSPH in eight human NSCLC cell lines (A549, H1299, PC9, H1975, H460, H358, H293 and LC-21) through qRT-PCR and western blot analysis. Analysis of the PSPH mRNA and protein expression levels in these cell lines showed that PSPH was markedly up-regulated in A549, H1299, PC9, and H1975 cell lines compared with H460, H358, H293 and LC-21cell lines (Figure 2A and 2B). In our previous study, we found that the migration and invasion ability of A549, H1299, PC9, and H1975 cells was higher than that of H460, H358, H293 and LC-21 cells using trans-well assays 13. In general, these results showed that PSPH was significantly overexpressed in highly metastatic lung cancer cells compared with weakly metastatic lung cancer cells.
Figure 2.
PSPH is overexpressed in highly metastatic cancer cells. A and B. qRT-PCR and western blot analysis to quantify the endogenous levels of PSPH in NSCLC cell lines. β-actin was used as the internal reference gene. C and D. PSPH expression was determined by qRT-PCR and western blot analysis after PC9 and H1299 cells were transfected with the si-control or the si-PSPH. β-actin served as an internal control.
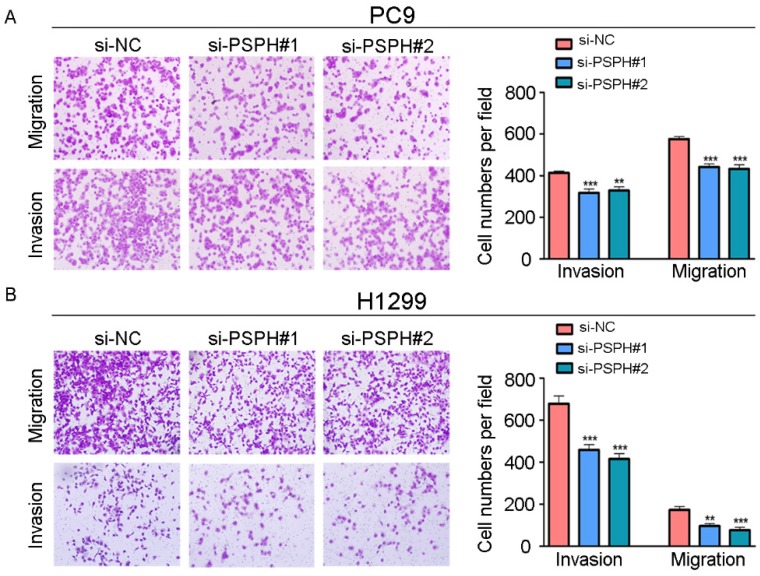
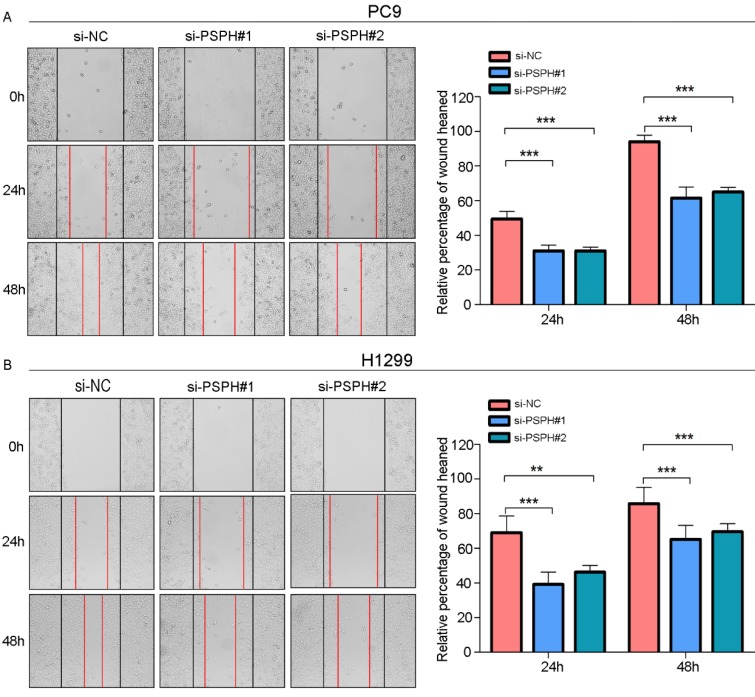
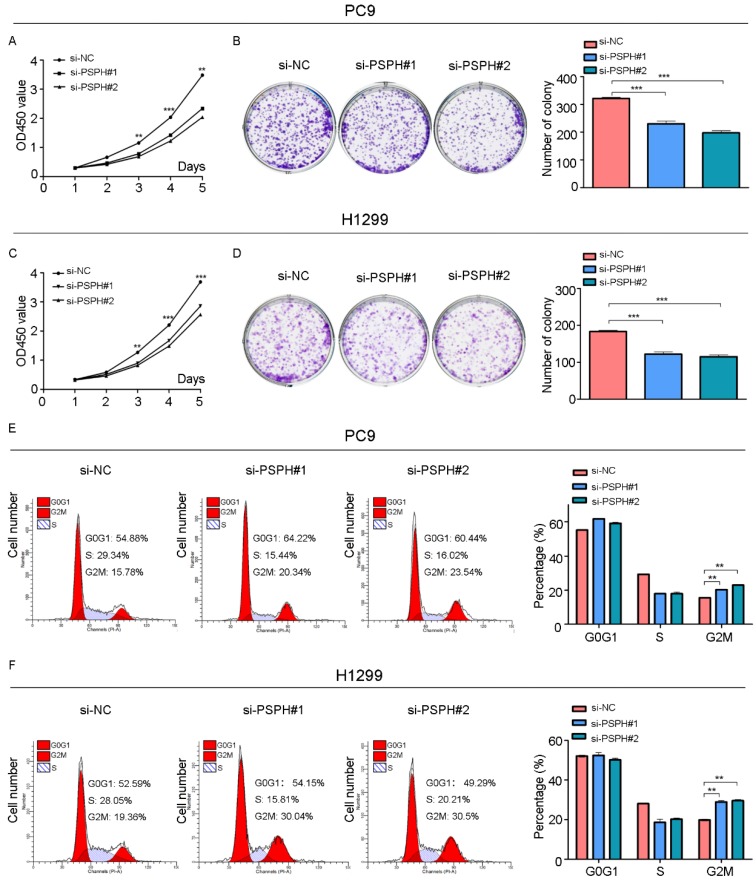
On the basis of the results showing the facilitation of PSPH expression in highly metastatic NSCLC cells, H1299 and PC9 cells were transfected using a transient transfection strategy by targeting at PSPH to downregulate PSPH expression. PSPH mRNA and protein expression were determined by qRT-PCR and western blot respectively after H1299 and PC9 cells were transfected with the siRNA-control or siRNA-PSPH. qRT-PCR and western blot analysis showed more than 80% efficiency in the reduction of endogenous PSPH expression when β-actin used as the internal standard (Figure 2C and 2D). Compared with the control cells, the invasion and migration of H1299 and PC9 cells, which were infected with si-PSPH, were significantly suppressed (Figure 3A and 3B). Subsequently, wound-healing assays were applied to determine the migratory abilities of PSPH, and we observed that migration rates were suppressed in H1299 and PC9 cells transfected with si-PSPH compared to si-NC (Figure 4A and 4B). These results showed that downregulation of PSPH decreased the invasion and migration of human NSCLC cells.
Figure 3.
PSPH mediates NSCLC cells migration and invasion. A and B. Trans-well migration and invasion assays for PC9 and H1299 were determined after transduction with the si-control or the si-PSPH. Data are expressed as the mean ± SEM. **P<0.01 and ***P<0.001 by Student's t test.
Figure 4.
PSPH mediates NSCLC cells motility. A and B. Wound-healing assay for PC9 and H1299 after transduction with the si-control or the si-PSPH. Error bars represent SEM. **P<0.01 and ***P<0.001 by Student's t test.
PSPH enhances proliferation of NSCLC cells during G2/M phase in vitro
We then examined the effect of PSPH on cell growth in H1299 and PC9 cells. Cell proliferation was evaluated using a CCK-8 assay, which showed that the inhibition of PSPH suppressed the proliferation of H1299 and PC9 cells (Figure 5A and 5C). A colony forming assay was also used as a direct method to detect the proliferation ability of cells. We found that the number of colony forming units was significantly reduced after transfected si-PSPH in H1299 and PC9 cell lines (Figure 5B and 5D). These results suggested that the expression of PSPH may enhance proliferation of NSCLC cells.
Figure 5.
Knockdown of PSPH inhibits cells proliferation through influencing G2/M cell cycle of NSCLC cells. A and C. Diagrams showing results of the CCK-8 assays. PC9 and H1299 cells proliferation was inhibited by downregulation of PSPH expression. **P<0.01 and ***P<0.001 by Student's t test. B and D. Decrease in PSPH impairs NSCLC cells colony formation capacity. Representative photomicrographs of PC9 and H1299 cell colonies in culture plates and significant reduction in the colony formation efficacy in PC9 and H1299 cells following PSPH knockdown. Data are expressed as the mean ± SEM. ***P<0.001 by Student's t test. E and F. Cell cycle profiles determined by propidium iodide (PI) staining and flow cytometry assays of PC9 and H1299 cells transfected with si-NC or si-PSPH. Data are expressed as the mean ± SEM. **P<0.01 by Student's t-test.
To gain deeper insight into the effect of PSPH in the aspect of cell growth, we examined the function of PSPH on cell cycle progression by flow cytometry. The cell cycle of H1299 and PC9 cell lines was compared before and after PSPH knockdown. Compared with si-NC, si-PSPH resulted in an arrest in G2/M phase in H1299 and PC9 cell lines (Figure 5E and 5F). Consistent with these findings, we can conclude that PSPH promoted cell growth during the G2/M phase.
PSPH regulates the ERK MAPK and P38 MAPK signaling pathways in NSCLC cells
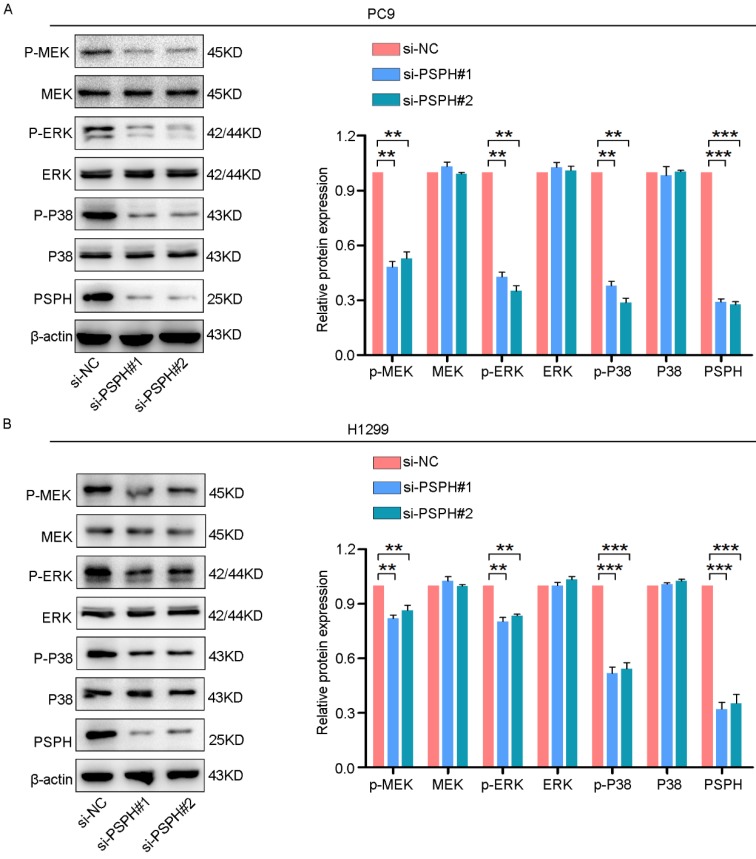
On the basis of previous studies showing that MAPK signaling pathways could regulate cancer cell proliferation, invasion and migration in multiple ways. Phosphorylated proteins are the form in which they function. Therefore, we examined whether PSPH could regulate phosphorylated protein levels downstream of these pathways. As shown in Figure 6A and 6B, si-PSPH suppressed the phosphorylation of ERK, MEK and P38 compared with the si-NC cohort in H1299 and PC9 cell lines. However, total protein levels of ERK, MEK and P38 were not changed. These data indicate that PSPH potentially facilitate the migration, invasion and proliferation of NSCLC cells by regulating the MAPK signaling pathways.
Figure 6.
PSPH regulates the ERK MAPK and P38 MAPK signaling pathways. Western blot analysis of p-MEK, MEK, p-ERK, ERK, p-P38 and P38 in PC9 and H1299 cells transfected with si-NC and si-PSPH. Data are expressed as the mean ± SEM. **P<0.01 and ***P<0.001 by Student's t-test.
PSPH protein levels in NSCLC are associated with advanced clinical stage
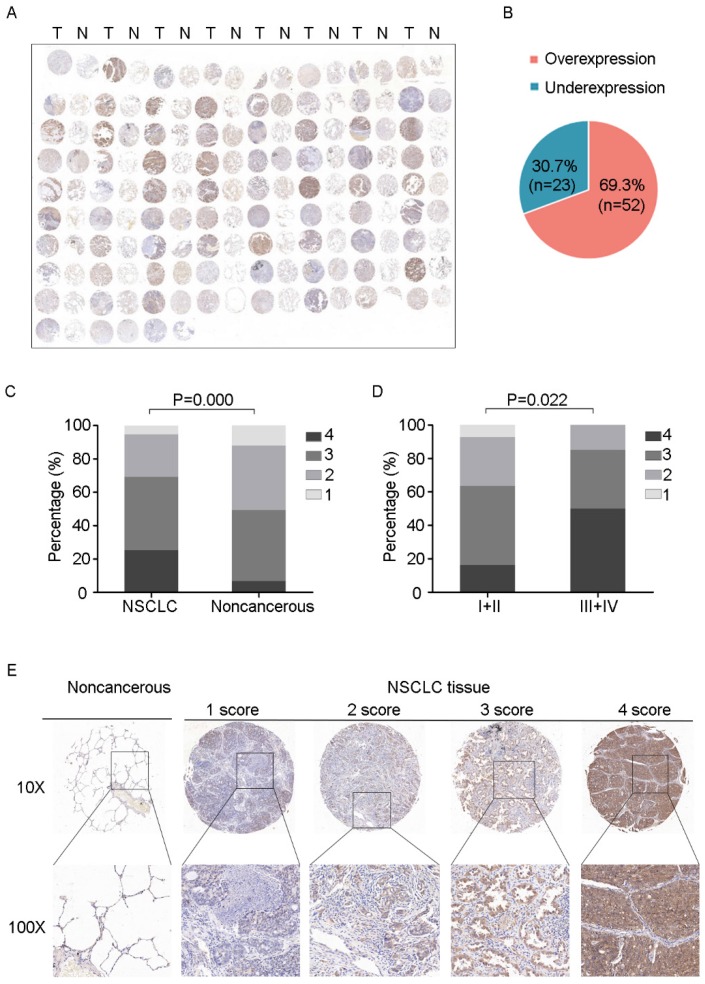
To further determine the clinical relevance of this finding, we performed immunohistochemical analysis of PSPH protein levels expression in a tissue micarray (TMA) that included an independent set of 75 paired adenocarcinomas and their adjacent noncancerous tissues. Correlations between the PSPH expression level and clinicopathologic characteristics of NSCLC are summarized in Table 2 and Figure 7. As showed in Figure 7, not only was the PSPH levels significantly higher in NSCLC tissues compared with matched adjacent controls (Figure 7A and 7C), but the proportion of NSCLC specimens with PSPH overexpression was also predominant (7B), which was consistent with our genomic and transcriptional results. Additionally, the expression level of PSPH was higher in patients with clinical advanced stage NSCLC than early stage (Figure 7D). However, the results showed no statistically significant correlations between PSPH protein expression and age, gender, differentiation and tumor size. Additionally, due to the limitation of clinical tissues with distant metastasis, we found that no significantly statistical correlations between PSPH protein expression and metastasis, but there was a trend that high PSPH level correlated positively with lymph node metastasis and/ or distal metastasis (P = 0.106). In general, we found that a high level of PSPH positively correlated with clinical stage in NSCLC patients, indicating that PSPH may play an important role in the development and progression of NSCLC.
Table 2.
Relationship between PSPH protein expression and clinicopathologic characteristics in 75 NSCLC patients by IHC.
| PSPH expression level1 | ||||||
|---|---|---|---|---|---|---|
| Characteristics | 1 score | 2 score | 3 score | 4 score | P value2 | |
| Age | ||||||
| < 60 | 3 (4.0%) | 7 (9.3%) | 11 (14.7%) | 9 (12.00%) | 0.036 | |
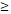 60 60 |
1 (1.3%) | 12 (16.0%) | 22 (29.3%) | 10 (13.3%) | ||
| Gender | 0.648 | |||||
| Female | 3 (4.0%) | 8 (10.7%) | 16 (21.3%) | 8 (10.7%) | ||
| Male | 1 (1.3%) | 11 (14.7%) | 17 (22.7%) | 11 (14.7%) | ||
| Tumor size(cm) | 0.347 | |||||
| < 3 | 0 (0.0%) | 6 (8.0%) | 5 (6.7%) | 5 (6.7%) | ||
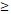 3 3 |
4 (5.3%) | 13 (17.3%) | 28 (37.3%) | 14 (18.7%) | ||
| Degree of differentiation | ||||||
| Well and moderately | 2 (2.7%) | 12 (16.0%) | 24 (32.0%) | 15 (20.0%) | 0.565 | |
| Poorly | 2 (2.7%) | 7 (9.3%) | 9 (12.0%) | 4 (5.3%) | ||
| Clinical stage | * | |||||
| Stage I+II | 4 (5.3%) | 16 (21.3%) | 26 (34.7%) | 9 (12.0%) | 0.022 | |
| Stage III+IV | 0 (0.0%) | 3 (4.0%) | 7 (9.3%) | 10 (13.3%) | ||
| Metastasis3 | ||||||
| No | 4 (5.3%) | 12 (16.0%) | 22 (29.3%) | 8 (10.7%) | 0.106 | |
| Yes | 0 (0.0%) | 7 (9.3%) | 11 (14.7%) | 11 (14.7%) | ||
| Carcinoma | *** | |||||
| NSCLC | 4 (5.3%) | 19 (25.3%) | 33 (44.0%) | 19 (25.3%) | 0.000 | |
| Adjacent lung | 9(12.0%) | 29 (38.7%) | 32 (42.7%) | 5 (6.7%) | ||
1Scoring was measured by the percentage of positive cells with the following staining intensities: less than 25% scored “1”; 25-49.0% scored “2”; 50-74.9% scored “3”; and more than 75% scored “4”. 2The Chi-square (X2) test was used to evaluate the association between the PSPH protein expression and clinicopathologic parameters. P < 0.05 was considered significant. 3The patients were classified into tumor metastasis negative and positive groups (lymph node metastasis and/ or distal metastasis).
Figure 7.
IHC images with differential expression levels of PSPH between lung cancer and adjacent noncancerous tissue. A. Tissue microarray images reflect PSPH protein expression level in 75 paired lung adenocarcinoma tissues (T) and adjacent noncancerous tissues (N). B. The pie chart represents the proportions of NSCLC samples showing PSPH overexpression and underexpression. C. PSPH was overexpressed in NSCLC tissues compared with the noncancerous tissues (P value =0.000). D. Association between PSPH expression patterns and clinical stages of NSCLC (P value =0.022). The patients were classified into tumor metastasis negative and positive groups (lymph node metastasis and/ or distal metastasis). E. Representative IHC images of the express of PSPH between lung cancer tissues and their adjacent noncancerous tissues. Score was measured by the percentage of positive cells with the following staining intensities: less than 25% scored “1”; 25-49.9% scored “2”; 50- 74.9% scored “3”; and more than 75% scored “4”.
Discussion
Lung cancer has become one of the most frequently-occurring cancer and the leading cause of cancer-related death. Median survival for patients with NSCLC is 13 months 14. Metastasis, one of the most critical hallmarks of cancer, is the leading cause of cancer-related death worldwide, particularly in NSCLC. For example, approximately 10-25% of patients with lung cancer have brain metastases when the cancer is first diagnosed, and 40-50% of these eventually develop brain metastases during the course of their disease 15. The median survival of these patients decreases to 2 months 16. Therefore, there is an urgent need for the identification of metastatic factors and understanding the underlying molecular mechanisms of NSCLC.
Previous studies on PSPH have yielded various results in different malignant cancers. Clinical studies have shown that PSPH overexpression is a promising prognostic and predictive biomarker for advanced colorectal cancer, hepatocellular carcinoma, breast cancer, endometrial cancer and thyroid cancer patients 11, 17-20. Another report demonstrated that suppression of PSPH could increase the sensitivity to 5-fluorouracil in colorectal cancer 21. The proposed mechanism for improved colorectal cancer efficacy involves the depletion of PSPH-mediated serine synthesis and glutathione reservoirs, resulting in the increased accumulation of 5-fluorouracil induced reactive oxygen species. However, PSPH overexpression may play a role in determining sensitivity to erlotinib 22. In addition, PSPH inhibits apoptosis in hepatocellular carcinoma 11, promotes cell proliferation in colorectal cancer [20]and promotes osteoclastogenesis in bone metastatic breast cancer 23. These findings suggest that PSPH may have important functions in human carcinogenesis and may serve as a potential and predictive therapy target for human cancer.
Of note, PSPH gene is located on very close to EGFR on chromosome 7 (positions 7p11.2 and 7p12, respectively). Previous study has indicated that the EGFR region on chromosome 7 is frequently amplified in lung adenocarcinoma 12. However, little is known regarding its precise biological role in NSCLC process.
In this study, we focused on the effect of PSPH on NSCLC process and showed that PSPH might act as an oncogene during NSCLC metastasis. We observed that PSPH was up-regulated in NSCLC tissue samples compared with the corresponding non-cancerous tissues. Furthermore, the expression was highly associated with lymph node or distant metastasis and clinical stage. In addition, PSPH were significantly overexpressed in highly metastatic cells. Furthermore, our results showed that PSPH could promote the migration, invasion and proliferation of NSCLC cells.
To date, PSPH-mediated serine synthesis is known to be up-regulated in cancer cells as a mechanism contributing to enhanced nucleotide and amino acid synthesis metabolism 24. L-serine synthesis may serve as a metabolic gatekeeper in cell cycle progression by MAPK activation 25. Ross et al. reported that the MAPK cascade is activated inducing serine biosynthesis in metastatic melanoma cells 26. Based on previous reports, we speculated that PSPH mediated the deterioration of NSCLC through MAPK signaling pathways. In the present study, our results demonstrated that PSPH overexpression could promote the phosphorylation of MEK, ERK and P38 through western blot analysis. On the basis of these data, we speculated that PSPH may play an important role in NSCLC malignant process through ERK MAPK and P38 MAPK signaling pathways.
In conclusion, we demonstrated that PSPH was up-regulated in NSCLC tissues, and that PSPH overexpression was correlated with clinical stage and metastasis of human NSCLC in this study. PSPH Knockdown inhibited NSCLC cell migration, invasion and proliferation of NSCLC cells through MAPK signaling pathways. Importantly, these findings provide an insight into a novel therapeutic target for NSCLC.
Materials and methods
Cells culture
Eight NSCLC cell lines were used in this study: PC9, A549, H1299, H1975, H460, H358, H292 and LC-21. The human NSCLC cell lines A549, H1299, H1975, H460, H358 and H292 cells used in this study were obtained from American Type Culture Collection (ATCC). PC9 and LC-21 cell lines were gifted from Professor Yao from Shanghai center institute (Shanghai, China). All of them were cultured in Dulbecco's Modified Eagle Media (DMEM) (Gibco, Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Biowest, South America origin) and 1% Pen/Strep Cells (Sigma-Aldrich) and were kept at 37 °C in a humidified air atmosphere incubator containing 5% carbon dioxide. These cell lines were regularly certified as free of mycoplasma contamination, and cell growth and survival quality were assessed by Trypan blue exclusion assay.
Clinical human NSCLC specimens
The freshly snap-frozen human NSCLC and their matched adjacent normal adjacent tissues for quantitative real-time PCR (n=73) and western blot (n=12) were collected from Huashan hospital, Fudan University (Shanghai, China) between 2014 and 2017. Paired normal adjacent tissues were taken at a distance of at least 3cm from the tumors. All human surgical specimens were pathologically diagnosed at Huashan hospital. Approval from the Ethical Review Committee of Huashan hospital and written informed consent was obtained from all patients.
RNA extraction and qRT-PCR analysis
Total RNA samples from the paired NSCLC, adjacent noncancerous NSCLC tissue specimens and cell lines in this study were isolated and purified with TRIzol reagent (Invitrogen, CA) according to the manufacturer's protocol, and quantified with Nanodrop 2000 (Thermo, Japan). The reverse transcription was performed with the PrimeScript™ RT Reagent Kit (TaKaRa, Shiga, Japan) according to the manufacturer's instruction. Reverse transcription conditions were as follows: 37°C for 45 min followed by 85°C for 5 s and 4°C forever. The quantitative real-time polymerase chain reaction (qRT-PCR) was subsequently performed with SYBR Green Premix Ex Taq kit (TaKaRa, Japan). RNA expression levels were determined by 7500 software systems, version 2.0.5 (Applied Biosystems, Foster City, CA). The PCR reaction conditions were as follows: 95°C for 10 s followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The information of the primer sequences used was as follows: PSPH (forward primer: 5'-CACGGTCATCAGAGAAGAAG-3' and reverse primer: 5'-GGTTGCTCTGCTATGAGTCT-3'); β-Actin (forward primer: 5'- TGTGGCCGAGGACTTTGATT -3' and reverse primer: 5'- CCTGTGTGGACTTGGGAGAG -3') was used as the internal reference gene for quantifying mRNA levels. Three independent assays were performed. The relative expression levels were calculated by the following formula: 2^- ∆CT (∆Ct=Ct target gene - Ct β-actin). When analyzing the mRNA expression level of PSPH in clinical NSCLC samples (n=73), the log2(fold change, FC≤-1) was defined as low expression, >1 as high expression, and-1~1 as no change.
Protein extraction
All protein extraction procedures were performed on ice. The cells protein was extracted using a mixture of T-PER Protein Extraction Reagent lysates (Thermo Fisher Scientific, USA) plus protease (Yeasen, China) and phosphatase inhibitors (Yeasen, China) according to the manufacturer's instructions, and then incubated on ice for 30 min. For tissue samples protein extraction, tissue samples were thawed on ice and ground in liquid nitrogen, then dissolved on ice for 45 min. Both lysates were centrifuged with a speed of 12000 rpm for 15 min at 4°C, and the supernatant was collected and frozen at -80 °C. A bicinchoninic acid (BCA) protein assay kit (Pierce, USA) was used to determine the protein concentrations according to the manufacturer's instructions. PBS was used as the standard.
Western blot analysis
Proteins samples were separated by 8% and 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After blocking in 5% non-fat dried milk for 1 h at room temperature, the membranes were incubated with mouse anti-PSPH (1:200) form Santa Cruz Biotechnology, rabbit anti-p-ERK (1:1000), rabbit anti-ERK (1:1000), rabbit anti-p-P38 (1:1000), rabbit anti-P38 (1:1000), rabbit anti-p-MEK (1:1000) and rabbit anti-MEK (1:1000) from Cell Signaling overnight at 4 °C. Horseradish peroxidase (HRP) conjugated anti-mouse IgG1 (1:5000) from Sigma-Aldrich and HRP conjugated anti-rabbit IgG (1:5000) from Sigma-Aldrich were incubated as the secondary antibodies for 2 h at room temperature. Then the membranes were washed in PBS-T for three times between each antibody incubation step. After washing, the bands were detected using LumiBest ECL reagent solution kit (Share-bio, China). Band intensities were quantified SuperSignal West Femto Maximun Sensitivity Substrate (Thermo Fisher Scientific, USA) with β-actin levels used as the loading control. The experiments were repeated twice.
Cell migration assays
Cell migration assays were performed in 8-µm pore size Transwell filter chamber inserts (Corning, USA). Cells were diluted 1:1 in 0.2% trypan blue and counted in duplicate using an Olympus microscope (Tokyo, Japan). For migration assays, 5×10^4 cells in 200 µl DMEM without FBS medium were suspended into the upper chamber and 800 µl DMEM supplemented with 10% FBS was added in the lower chamber per well. After incubation for 14 h at 37°C and 5% CO2, cells moved to the bottom surface of the membrane were fixed with 100% methanol for 20 min and stained with 0.1% crystal violet for 10 min and washed with water for three times. Then, the invading cells were counted and imaged in at least 10 random fields under a light microscope (Olympus, Japan). The assays were performed three times independently.
Cell invasion Assays
Before invasion assays, Matrigel (BD Biosciences, CA) was diluted to 1 mg/mL with DMEM without FBS medium and immediately applied to the upper chamber per trans-well. After hydrated for 3 h or 4 h, 1×10^5 cells in 200 µl DMEM without FBS medium were placed into the upper chamber and 800 µl DMEM supplemented with 10% FBS was added in the lower chamber per well. After incubation for 24 h at 37°C and 5% CO2, cells moved to the bottom surface of the membrane were fixed with 100% methanol for 30 min and stained with 0.1% crystal violet for 10 min and washed with water for three times. Then, the invading cells were counted and imaged in at least 10 random fields under a light microscope. The assays were performed three times independently.
Wound-healing assays
For the wound-healing assay, cells were seeded in six-well plates. After the cells reached 80% confluence, a linear wound was carefully made by a 200 ml sterile pipette tip across the confluent cell monolayer, and the cell debris was washed and removed with phosphate-buffered saline (PBS) and then incubated with DMEM supplemented with 1% FBS. Five selected fields at the lesion border were then photographed at 0, 24 and 48 h after wounding, and the migrated areas were measured. All experiments were performed three times.
Clone formation assays
After transfected with siRNA for 48 h, about 1 × 10^3 of H1299 and PC9 cells were seeded in 6-well plates. The plates were incubated at 37°C with 5% CO2 for 7 days until cells had formed sufficiently large clones. Then, the cells were fixed with 100% methanol for 30 min and stained with 0.1% crystal violet for 10 min. The crystal violet was removed carefully and rinsed with tap water. The number of visible colonies was counted. The assays were performed three times independently.
Cell proliferation
In vitro proliferation of NSCLC was determined using WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl) -5-(2,4-disulfophenyl) -2H-tetrazolium) assay kit (CCK-8 assay kit; Dojindo, Japan) according to the manufacturer's instructions. NSCLC cells were seeded in triplicate wells of 96-well plates at 1.5 × 10^3 cells per well in a final volume of 200 µl. Then 10 µl of CCK-8 solution was added into 100 µl fresh DMEM each well and incubated for 2 h at 37°C. The absorbance of each well was measured at 450 nm to calculate the number of viable cells. The experiments were repeated twice.
Cell cycle analysis
The distribution of cell cycle stages was analyzed using flow cytometry. Cells were cultured in six-well plates, harvested and washed twice with ice-cold PBS. Subsequently, cells were and fixed with 70% ethanol diluted in PBS at -20 °C overnight. Following PBS washing, the cells were then collected by centrifugeation at 1000 rpm for 5 min, resuspended and stained with 500 µl propidium iodide (PI) (Beyotime, China) in the dark for 30 min according to the manufacturer's instructions and analyzed by a FACSCalibur flow cytometer (BD Biosciences, USA). The percentage of cells in G0-G1, S, and G2-M phase was counted and compared. The assays were performed three times independently.
RNA interference using siRNA
Cells were transfected with the indicated small interfering RNA (siRNA). Two siRNA oligonucleotides targeted at PSPH were designed and synthesized by RiboBio (Guangzhou, China). The target sequences were as follows: si-PSPH#1: 3′-GGAGCGAAATGTTCAGGTT-5′; si-PSPH#2: 3′-GGCAACAAGTCAAGGATAA-5′; si-NC was used as the control. PSPH was knocked down by transfecting cells using Lipofectamine 2000 Reagents in 6-well plates (Invitrogen, CA) according to the manufacturer's protocols. After transfection for 48 hours, the cells were collected, assessed the specific silencing of PSPH expression using qRT-PCR and WB, and used for migration and invasion assays and so on.
Microarray and expression data analysis
We performed online-available data sets downloaded from NCBI to screen the relationship between the expression level of PSPH and NSCLC patient clinical features. RNA-seq data of NSCLC tumor tissues and/or adjacent non-cancerous tissues were obtained and downloaded from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo). Overall survival data of 117 NSCLC patients from GEO (GSE13213) were analyzed using a Kaplan-Meier survival plot.
Immunohistochemistry (IHC)
Patient samples in this study were obtained following informed consent, according to an established protocol approved by the Ethics Committee of the Huashan Hospital, Fudan University. Matched pairs (n= 75) of lung adenocarcinoma tissues and adjacent noncancerous tissues were used for the construction of a tissue microarray (Shanghai Biochip Co., Ltd. Shanghai, China). Immunohistochemical staining was performed to detect the expression of PSPH protein in NSCLC tissues and matched noncancerous tissues. The primary antibody against PSPH was obtained from Proteintech (1:100). The slides were examined and scored by a pathologist who had no prior knowledge of the clinical origins of the specimens.
Statistical analysis
The results were presented as mean ± standard error of the mean (SEM) from one representative experiment out of three independent experiments unless stated otherwise and imaged by using GraphPad Prism 5 software (GraphPad Software, USA). The comparisons of quantitative data between two groups or between more than two groups were analyzed by Student's t test between two groups or one-way analysis of variance (ANOVA) respectively using SPSS. P < 0.05 was considered statistically significant.
Acknowledgments
This work was financially supported by the Shanghai Municipal Committee of Health and Family planning (201440584) and Baoshan District Committee of Science and Technology (14-E-28).
Abbreviations
- CCK-8
cell counting kit-8
- PSPH
Phosphoserine phosphatase
- NSCLC
non-small cell lung cancer
- IHC
immunohistochemistry
- HAD
haloacid dehalogenase
- TMA
tissue micarray
- ATCC
American Type Culture Collection
- DMEM
Dulbecco's Modified Eagle Media
- FBS
fetal bovine serum
- qRT-PCR
quantitative real-time polymerase chain reaction
- BCA
bicinchoninic acid
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PVDF
polyvinylidene difluoride
- HRP
horseradish peroxidase
- PBS
phosphate-buffered saline
- WST-8
2-(2-methoxy-4-nitrophenyl) -3-(4-nitrophenyl)-5-(2,4-disulfophenyl) -2H-tetrazolium
- siRNA
small interfering RNA
- GEO
Gene Expression Omnibus
- PI
propidium iodide.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Hubbs JL, Boyd JA, Hollis D, Chino JP, Saynak M, Kelsey CR. Factors associated with the development of brain metastases: Analysis of 975 Patients with Early Stage Nonsmall Cell Lung Cancer. Cancer. 2010;116(21):5038–5046. doi: 10.1002/cncr.25254. [DOI] [PubMed] [Google Scholar]
- 3.Field JK, Oudkerk M, Pedersen JH, Duffy SW. Prospects for population screening and diagnosis of lung cancer. The Lancet. 2013;382(9893):732–741. doi: 10.1016/S0140-6736(13)61614-1. [DOI] [PubMed] [Google Scholar]
- 4.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Strunck E, Frank K, Tan MI, Expression of l-3-Phosphoserine Phosphatase Is Regulated by Reconstituted Basement Membrane. 2001; 281(3): 747 - 753. [DOI] [PubMed]
- 6.Tabatabaie L, Klomp LW, Berger R, de Koning TJ. l-Serine synthesis in the central nervous system: A review on serine deficiency disorders. Molecular Genetics and Metabolism. 2010;99(3):256–262. doi: 10.1016/j.ymgme.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Gylfe AE, Katainen R, Kondelin J. et al. Eleven candidate susceptibility genes for common familial colorectal cancer. PLoS Genet. 2013;9(10):e1003876. doi: 10.1371/journal.pgen.1003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachelor MA, Lu Y, Owens DM. L-3-Phosphoserine phosphatase (PSPH) regulates cutaneous squamous cell carcinoma proliferation independent of L-serine biosynthesis. J Dermatol Sci. 2011;63(3):164–72. doi: 10.1016/j.jdermsci.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimomura K, Sakakura C, Takemura M. et al. Combination of L-3-phosphoserine phosphatase and CEA using real-time RT-PCR improves accuracy in detection of peritoneal micrometastasis of gastric cancer. Anticancer Res. 2004;24(2C):1113–20. [PubMed] [Google Scholar]
- 10.Wu Q, Li J, Sun S. et al. YAP/TAZ-mediated activation of serine metabolism and methylation regulation is critical for LKB1-deficient breast cancer progression. Bioscience Reports. 2017;37(5):BSR20171072. doi: 10.1042/BSR20171072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Song L, Wan Q. et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25(4):429–444. doi: 10.1038/cr.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weir BA, Woo MS, Getz G, Perner S, Li D. Characterizing the cancer genome in lung adenocarcinoma. nature. 2007;450(10):893–901. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu T, Li J, Yan M. et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34(4):413–23. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- 14.Riihimäki M, Hemminki A, Fallah M. et al. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86(1):78–84. doi: 10.1016/j.lungcan.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 15.Steeg PS, Camphausen KA, Smith QR. Brain metastases as preventive and therapeutic targets. Nat Rev Cancer. 2011;11(5):352–63. doi: 10.1038/nrc3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sajama C, Lorenzoni J, Tagle P. Diagnosis and treatment of brain metastasis. Rev Med Chil. 2008;136(10):1321–6. [PubMed] [Google Scholar]
- 17.Sun WY, Kim HM, Jung W, Koo JS. Expression of serine/glycine metabolism-related proteins is different according to the thyroid cancer subtype. Journal of Translational Medicine. 2016;14(1):168–180. doi: 10.1186/s12967-016-0915-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Arcy M, Fleming J, Robinson WR, Kirk EL, Perou CM, Troester MA. Race-associated biological differences among Luminal A breast tumors. Breast Cancer Res Treat. 2015;152(2):437–448. doi: 10.1007/s10549-015-3474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allard JE, Chandramouli GV, Stagliano K. et al. Analysis of PSPHL as a Candidate Gene Influencing the Racial Disparity in Endometrial Cancer. Front Oncol. 2012;2:65. doi: 10.3389/fonc.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato K, Masuda T, Hu Q. et al. Phosphoserine Phosphatase Is a Novel Prognostic Biomarker on Chromosome 7 in Colorectal Cancer. Anticancer Res. 2017;37(5):2365–2371. doi: 10.21873/anticanres.11574. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Xun Z, Yang Y. Inhibition of phosphoserine phosphatase enhances the anticancer efficacy of 5-fluorouracil in colorectal cancer. Biochem Biophys Res Commun. 2016;477(4):633–639. doi: 10.1016/j.bbrc.2016.06.112. [DOI] [PubMed] [Google Scholar]
- 22.Tan EH, Ramlau R, Pluzanska A. et al. A multicentre phase II gene expression profiling study of putative relationships between tumour biomarkers and clinical response with erlotinib in non-small-cell lung cancer. Annals of Oncology. 2010;21(2):217–222. doi: 10.1093/annonc/mdp520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollari S, Nen SKK, Edgren H, Wolf M, Kohonen P. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125(2011):421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 24.Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57(1):87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayano T, Kawano Y, Kusada W. et al. Adaptive response to l-serine deficiency is mediated by p38 MAPK activation via 1-deoxysphinganine in normal fibroblasts. FEBS Open Bio. 2016;6(4):303–16. doi: 10.1002/2211-5463.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross KC, Andrews AJ, Marion CD, Yen TJ, Bhattacharjee V. Identification of the Serine Biosynthesis Pathway as a Critical Component of BRAF Inhibitor Resistance of Melanoma, Pancreatic, and Non-Small Cell Lung Cancer Cells. Molecular Cancer Therapeutics. 2017;16(8):1596–1609. doi: 10.1158/1535-7163.MCT-16-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]