Significance
Endothelial cells (EC) subjected to laminar shear stress exhibit a specialized EC membrane architecture that promotes a nonpermeable barrier to constituents of blood. Emerging data support long-noncoding RNAs (lncRNAs) as regulators of EC phenotype. Here, levels of the lncRNA SENCR are induced with laminar shear stress in cultured EC as well as aortic EC of humanized mice. SENCR binds a new noncanonical RNA-binding protein called CKAP4. Upon knockdown of SENCR, a pool of CKAP4 binds CDH5 triggering CDH5 internalization, leading to perturbed adherens junctions, defective membrane integrity, and heightened EC permeability. These results support a function of SENCR in the maintenance of EC membrane homeostasis through CKAP4 binding and the proper localization of CDH5 at the cell membrane.
Keywords: long-noncoding RNA, endothelial cell, shear stress, adherens junction
Abstract
SENCR is a human-specific, vascular cell-enriched long-noncoding RNA (lncRNA) that regulates vascular smooth muscle cell and endothelial cell (EC) phenotypes. The underlying mechanisms of action of SENCR in these and other cell types is unknown. Here, levels of SENCR RNA are shown to be elevated in several differentiated human EC lineages subjected to laminar shear stress. Increases in SENCR RNA are also observed in the laminar shear stress region of the adult aorta of humanized SENCR-expressing mice, but not in disturbed shear stress regions. SENCR loss-of-function studies disclose perturbations in EC membrane integrity resulting in increased EC permeability. Biotinylated RNA pull-down and mass spectrometry establish an abundant SENCR-binding protein, cytoskeletal-associated protein 4 (CKAP4); this ribonucleoprotein complex was further confirmed in an RNA immunoprecipitation experiment using an antibody to CKAP4. Structure–function studies demonstrate a noncanonical RNA-binding domain in CKAP4 that binds SENCR. Upon SENCR knockdown, increasing levels of CKAP4 protein are detected in the EC surface fraction. Furthermore, an interaction between CKAP4 and CDH5 is enhanced in SENCR-depleted EC. This heightened association appears to destabilize the CDH5/CTNND1 complex and augment CDH5 internalization, resulting in impaired adherens junctions. These findings support SENCR as a flow-responsive lncRNA that promotes EC adherens junction integrity through physical association with CKAP4, thereby stabilizing cell membrane-bound CDH5.
Vascular smooth muscle cells and endothelial cells (EC), like all nucleated cells of the body plan, harbor a genome replete with millions of transcription factor binding sites and thousands of noncoding RNA genes (1). Thus, the old notion of our genome being comprised largely of “junk DNA” (2) has been debunked, particularly by the expansive class of long-noncoding RNAs (lncRNAs), defined as processed RNA transcripts greater than 200 nucleotides in length, with little or no protein-coding potential (3, 4). Numerous functions exist for lncRNAs, including the regulation of genome architecture (5), chromatin remodeling and gene expression (6), cellular differentiation (7), and a myriad of cytoplasmic activities (8). Notwithstanding these advances, the diverse spatial localization, poor nucleotide sequence conservation, unknown structures, and low cellular abundance of lncRNAs have collectively hindered efforts to elucidate their function. These challenging barriers are particularly evident in endothelial cells of the vessel wall where novel lncRNA discovery and function have only recently been reported (9–11).
EC form a tight monolayer lining all blood vessels, thereby maintaining vascular integrity and homeostasis (12, 13). An important determinant of EC homeostasis is laminar shear stress (LSS), which is the direct frictional force exerted upon EC by the flow of blood (14). LSS occurs along the long axis of blood vessels and confers protection against such vascular diseases as atherosclerosis (15). On the other hand, disturbed shear stress (DSS) occurs at branch points of the vasculature and predisposes these regions to disease (14). Studies have reported the importance of LSS in maintaining EC monolayer integrity and homeostasis through the stabilization of cell–cell junctions (14). One such junctional complex is the adherens junction, which forms cell–cell adhesive contacts through homophilic recognition of the extracellular domain of cadherin molecules (12, 16). CDH5 (also known as VE-cadherin) is an EC-restricted cadherin containing five extracellular calcium-dependent cadherin repeats, a transmembrane domain, and a conserved cytoplasmic domain (16). Several proteins have been shown to mediate CDH5 membrane localization and adherens junction integrity, including CTNND1 (catenin δ1, also known as p120-catenin), which regulates CDH5 internalization through binding of the juxtamembrane domain (JMD) of CDH5 (17). Although the mechanism of protein–protein interaction at adherens junctions has been well documented (16), the role of lncRNAs in this process is unknown.
Previous studies provided evidence for a role of SENCR in smooth muscle cell differentiation and the regulation of early EC commitment (18, 19). However, the mechanisms of action of SENCR in these or other cell types is unknown. Here, levels of SENCR are shown to be induced by LSS and SENCR knockdown disrupts EC membrane integrity and permeability. SENCR is shown to interact with a noncanonical RNA-binding domain (RBD) of cytoskeletal-associated protein 4 (CKAP4), a relatively understudied cytosolic protein in the vessel wall. SENCR knockdown enhances an association between CKAP4 and CDH5 near the EC membrane. This interaction, in turn, augments CDH5 internalization, thus weakening EC adherens junction integrity. Together, these findings illuminate a mechanism for SENCR in the maintenance of vascular EC membrane homeostasis.
Results
LSS Induces SENCR RNA in Several EC Lineages.
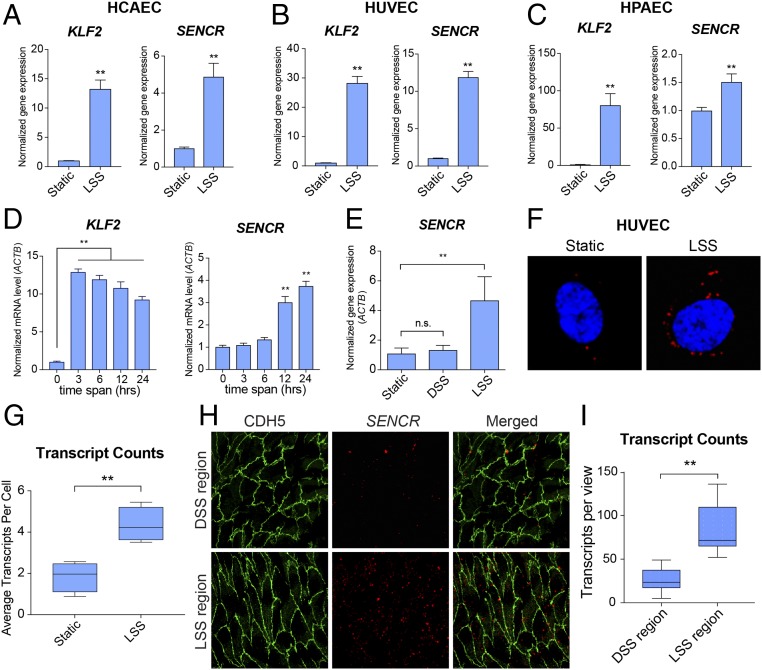
SENCR was shown initially to reside in the cytoplasm of vascular smooth muscle cells and stabilize the differentiated state of this cell type (18). To begin investigating SENCR function in differentiated EC, 10 dyne/cm2 of LSS was applied to human umbilical vein endothelial cells (HUVEC), human coronary artery endothelial cells (HCAEC), and human pulmonary artery endothelial cells (HPAEC). Real-time qRT-PCR showed that LSS treatment resulted in an elevation of SENCR in all three EC types, with HUVEC showing 10-fold induction (Fig. 1 A–C). In contrast, the EC-restricted transcription factor, FLI1, which overlaps with SENCR (18), exhibited only modest changes in mRNA expression upon LSS treatment of HUVEC (SI Appendix, Fig. S1A). Consistent with prior findings (20), KLF2 mRNA showed LSS-induced expression; however, increases were much higher than those seen with SENCR (Fig. 1 A–C). Time-course studies revealed that the increase in SENCR RNA was delayed and sustained in comparison with KLF2 mRNA (Fig. 1D). HUVEC subjected to static or DSS culture conditions exhibited little change in SENCR RNA levels (Fig. 1E). To ascertain the spatial localization of LSS-induced SENCR transcripts, RNA-FISH was performed in HUVEC under static or LSS conditions. Confocal microscopy showed more SENCR RNA transcripts in the cytoplasm of LSS-treated HUVEC (Fig. 1 F and G and SI Appendix, Fig. S1B). To extend these findings to an in vivo context, a humanized mouse line was generated by inserting a bacterial artificial chromosome (BAC) containing SENCR (SENCR-BAC) into the mouse genome using the piggyBac transposon system (SI Appendix, Fig. S1 C–E). Immuno-RNA-FISH of en face preparations of SENCR-BAC mouse aortae demonstrated more SENCR RNA transcripts in the LSS region than the DSS region (Fig. 1 H and I). These findings establish SENCR as an LSS-response gene in cultured EC and the intact vessel wall of humanized mice.
Fig. 1.
SENCR is induced by LSS. Real time-qPCR of KLF2 and SENCR RNA in static versus LSS-treated HCAEC (n = 3) (A), HUVEC (n = 5) (B), and HPAEC (n = 3) (C). (D) Real time-qPCR of time course (0–24 h) for KLF2 and SENCR RNA levels in static vs. LSS-treated HUVEC (n = 3). (E) Real-time qPCR of SENCR RNA in static, DSS-treated, and LSS-treated HUVEC (n = 3). (F) Confocal microscopy of SENCR in static vs. LSS-treated HUVEC using RNA-FISH. (Magnification: 1,000×.) (G) Quantitative analysis of SENCR transcripts in static (n = 113) vs. LSS-treated (n = 107) HUVEC. (H) Confocal microscopy of DSS region vs. LSS region in humanized SENCR-BAC mice using combined RNA-FISH and immunofluorescence staining of en face preparations of aorta (green: CDH5, red: SENCR). (Magnification: 400×.) (I) Transcript counts for combined en face staining of humanized mouse aorta. **P < 0.01; n.s., not significant.
SENCR Knockdown Impairs EC Membrane Integrity.
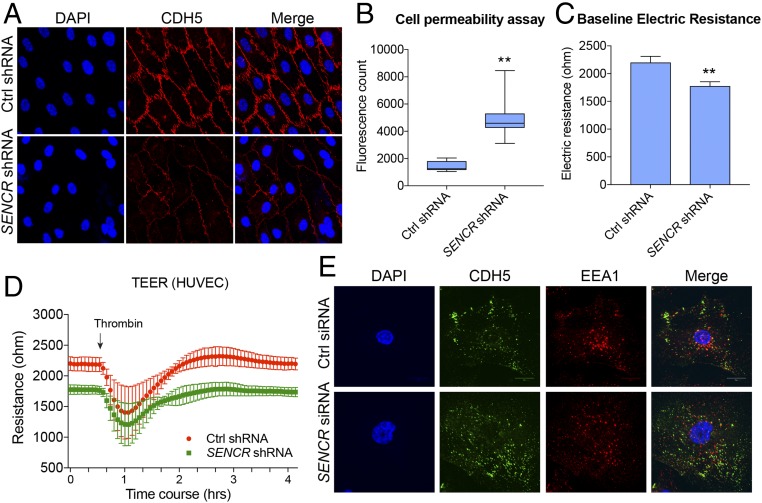
Because SENCR was induced by LSS, we tested whether reduced SENCR had any effect on normal EC morphology and function. Several small-interfering RNA (siRNA) duplexes were synthesized and tested for their efficiency in knocking down SENCR RNA (SI Appendix, Fig. S2A). siRNA-1 was most effective in knocking down SENCR in HUVEC and was selected to generate a lentiviral-based short-hairpin RNA (shRNA). Lentiviral SENCR shRNA showed >80% suppression of endogenous SENCR RNA (SI Appendix, Fig. S2B). LSS-stimulated HUVEC transduced with SENCR shRNA showed a decrease in CDH5 protein at cell–cell junctions (Fig. 2A). A similar reduction in membrane CDH5 expression was observed in static HUVEC (SI Appendix, Fig. S2C). Because CDH5 is an essential element of the EC adherens junction, necessary for membrane integrity (16), cell permeability was assessed using a FITC-Dextran/transwell assay. Results showed that EC permeability was elevated after SENCR silencing, suggesting a compromise in EC membrane integrity (Fig. 2B). To further confirm these findings, a transendothelial electrical resistance (TEER) assay was performed (21, 22). Results showed a lower electrical resistance at baseline in HUVEC with SENCR shRNA knockdown compared with control (Fig. 2 C and D). A slower recovery rate in electric resistance was observed in SENCR shRNA transduced HUVEC compared with control cells following thrombin treatment (Fig. 2D). SENCR gain-of-function in HUVECs revealed a decreasing trend in cell permeability (SI Appendix, Fig. S2D). There was little change in total CDH5 protein with SENCR shRNA knockdown (SI Appendix, Fig. S2E), suggesting a redistribution of CDH5 in HUVEC with reduced SENCR levels. To explore this possibility, we performed a CDH5 internalization assay in HUVEC ± SENCR shRNA. Results showed a significant amount of CDH5 internalized in the cytoplasm of HUVEC treated with SENCR shRNA (Fig. 2E and SI Appendix, Fig. S2 F and G). Moreover, only ∼40% of internalized CDH5 was colocalized with EEA1, an early endosome marker (Fig. 2E and SI Appendix, Fig. S2H). These results suggest that SENCR acts as a gatekeeper of EC membrane integrity and permeability by maintaining CDH5 membrane localization.
Fig. 2.
Knockdown of SENCR results in impaired EC membrane integrity through enhanced CDH5 internalization. (A) Immunofluorescence confocal microscopy of CDH5 in LSS-treated (10 dyne/cm2) HUVEC ± lentiviral SENCR shRNA treatment. (Magnification: 400×.) (B) Fluorescence count reading for cell permeability assay of control shRNA and SENCR shRNA treated HUVEC (n = 3). **P < 0.01. (C) Baseline electrical resistance of HUVEC ± SENCR shRNA by TEER (n = 4). **P < 0.01. (D) Time course of electrical resistance in static cultured HUVEC ± SENCR shRNA by TEER (4,000 Hz) (n = 4). (E) Internalization assay of CDH5 in cultured HUVEC ± SENCR shRNA using anti-CDH5 (BV9) antibody. Anti-EEA1 antibody was used to visualize early-stage endosomes. (Magnification: 600×.)
SENCR Binds CKAP4.
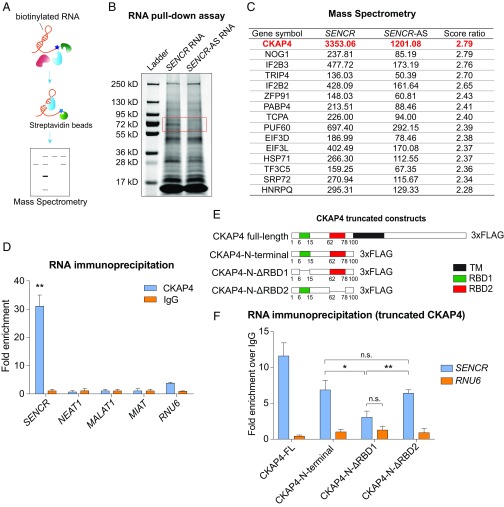
The function of SENCR in regulating EC membrane integrity may be through a change in a specific gene program via sponging of a microRNA or altered protein function, stability, or localization by way of interaction with cytosolic proteins (8). To distinguish between these possibilities, we first performed RNA sequencing (RNA-seq) in HUVEC ± SENCR shRNA. Results demonstrated mild changes in the mRNA profile of HUVEC with reduced SENCR; only 10 genes displayed >1.5-fold up-regulation and 26 genes showed <0.67-fold down-regulation (SI Appendix, Fig. S3 A–C) (Gene Expression Omnibus accession no. GSE122490). The down-regulated genes did not share a common miRNA target sequence with SENCR, suggesting that SENCR does not act as a competing endogenous RNA. We next performed an RNA pull-down assay (Fig. 3A) in static HUVEC and found a protein of molecular mass ∼70 kDa that interacted with biotinylated sense SENCR, but not antisense SENCR (Fig. 3B). Mass spectrometry of the excised ∼70-kDa band revealed a number of cytoskeletal and membrane-related proteins, with the most abundant protein CKAP4 (Fig. 3C and Dataset S1). Immunoblot following RNA pull down with sense SENCR verified enrichment of CKAP4 (SI Appendix, Fig. S3D). To validate the interaction between SENCR and CKAP4, we performed a number of complementary assays. RNA immunoprecipitation-qPCR (RIP-qPCR) demonstrated a 30-fold enrichment of SENCR RNA with anti-CKAP4 antibody; there was little change in enrichment for several other lncRNAs (Fig. 3D). RNA EMSA also showed an interaction between SENCR and CKAP4 (SI Appendix, Fig. S3E). Finally, RIP-qPCR of HUVEC transfected with a C-terminal 3xFLAG-tagged CKAP4 further demonstrated the SENCR–CKAP4 interaction (SI Appendix, Fig. S3 F and G). These findings indicate an interaction between SENCR and CKAP4.
Fig. 3.
SENCR associates with CKAP4 protein. (A) Schematic of RNA pull-down assay/mass spectrometry pipeline. (B) Coomassie blue stained SDS/PAGE gel of enriched proteins following RNA pull-down assay using full length sense SENCR RNA (SENCR RNA) or antisense SENCR RNA (SENCR-AS RNA). Red box shows position of gel slice for mass spectrometry. (C) Top hits of mass spectrometry from SENCR vs. SENCR-AS RNA pull-down assay. (D) Real-time qPCR of CKAP4 enriched RNAs following IP with anti-CKAP4 antibody (n = 3). (E) Schematic for generating FLAG-tagged CKAP4 expression constructs and truncated CKAP4 plasmids. RBD indicates a predictive RBD. (F) Real-time qPCR for FLAG-tagged CKAP4 full length (CKAP4-FL) and truncated CKAP4 proteins (CKAP4-N-terminal, CKAP4-N-ΔRBD1 and CKAP4-N-ΔRBD2) using RNA immunoprecipitation-enriched samples. *P < 0.05, **P < 0.01, n.s., not significant.
As a first step to define the SENCR-binding domain of CKAP4, we utilized Pprint, an in silico RBD prediction program (23). This analysis showed two potential RBDs in CKAP4 (SI Appendix, Fig. S3H). To test the potential function of these unconventional RBDs, structure–function studies were undertaken with a series of truncated CKAP4 expression plasmids, including CKAP4-N terminal (1–100 aa), CKAP4-N-ΔRBD1 (N terminal with 6- to 15-aa deletion), and CKAP4-N-ΔRBD2 (N-terminal with 62–78 aa deletion) (Fig. 3E). RIP-qPCR disclosed reduced SENCR enrichment with CKAP4-N-ΔRBD1. In contrast, little change in SENCR enrichment was observed in cells transfected with CKAP4-N-ΔRBD2 (Fig. 3F and SI Appendix, Fig. S3I). These results provide supportive evidence for a noncanonical RBD within CKAP4 that directly interacts with SENCR.
SENCR Mediates Protein–Protein Interactions Within Adherens Junctions.
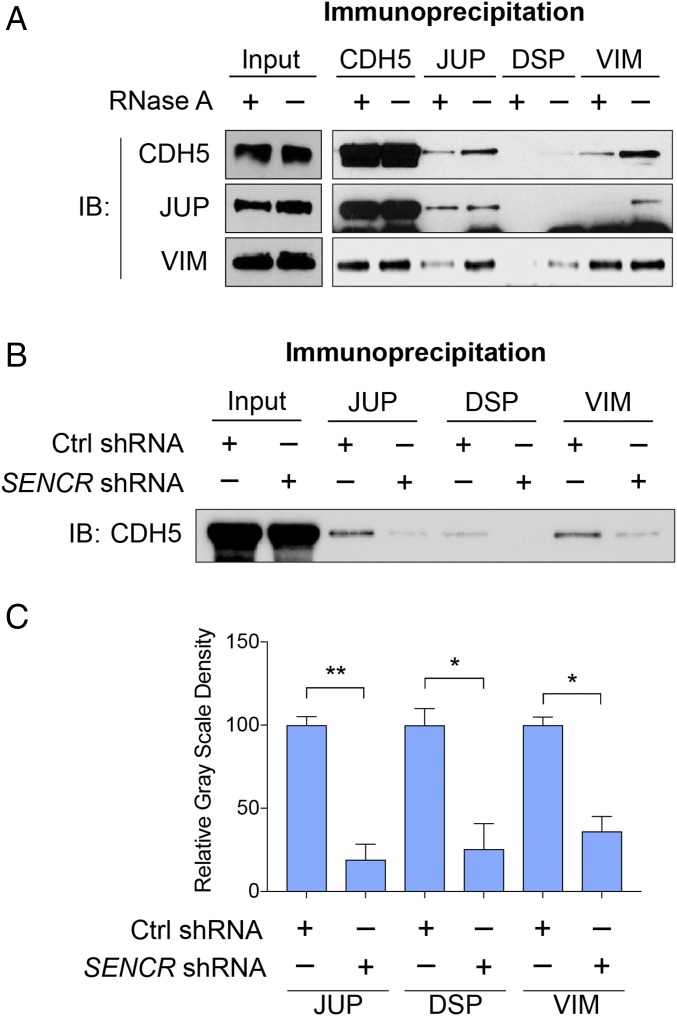
CKAP4 localizes to the plasma membrane of epithelial cells (24). To determine whether a role exists for CKAP4-SENCR in EC membrane integrity, we first performed coimmunoprecipitation (co-IP) in HUVEC ± RNase A treatment. Immunoblot results showed that interactions between CDH5 and plakoglobin (JUP), desmoplakin (DSP), and vimentin (VIM) are partially RNA-dependent (Fig. 4A). To investigate the role of SENCR in the adherens junction protein complex, we performed co-IP with HUVEC ± SENCR shRNA using antibodies targeting CDH5, JUP, DSP, and VIM. Immunoblot results demonstrated that SENCR knockdown impaired interactions between CDH5 with various adherens junction proteins (Fig. 4 B and C). These findings suggest that SENCR is involved in the maintenance of EC adherens junction protein complex integrity.
Fig. 4.
Knockdown of SENCR impairs EC adherens junction. (A) Co-IP of whole-cell lysate (HUVEC) ± RNase A treatment using indicated antibodies. (B) Co-IP of whole-cell lysate (HUVEC) ± SENCR shRNA using indicated antibodies and immunoblotting with anti-CDH5 antibody (n = 3). A representative image is shown. (C) Quantitative grayscale density of B. *P < 0.05, **P < 0.01.
SENCR Maintains Adherens Junction Integrity by Binding CKAP4.
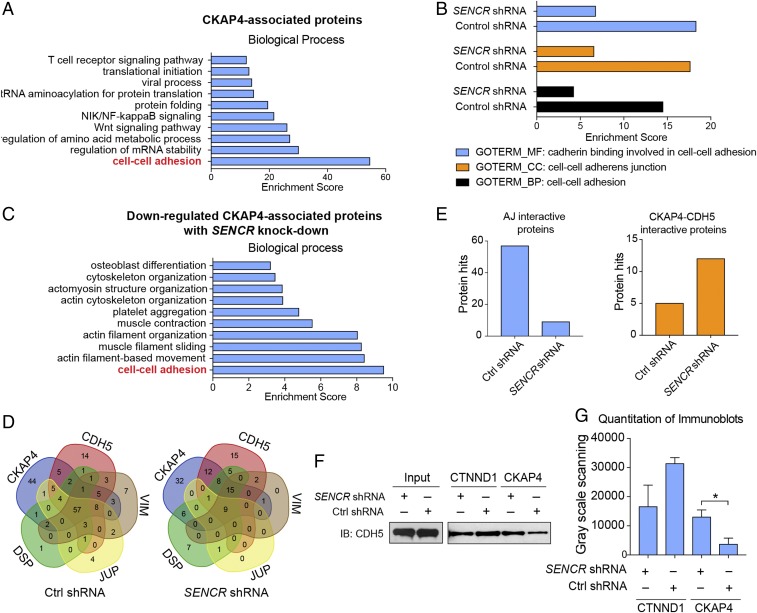
Although CKAP4 has been shown to interact with several proteins, no studies have defined CKAP4–protein interactions in EC (25). We therefore performed an unbiased IP/mass spectrometry experiment in HUVEC to define CKAP4 interacting proteins. The mass spectrometry output was analyzed by DAVID (26) and the results showed that CKAP4-associated proteins strongly correlated with cell–cell adhesion (Fig. 5A and Dataset S2). We repeated the same experiment ± SENCR shRNA knockdown and found that the correlation with adhesion/junctional proteins was reduced (Fig. 5B). When only CKAP4-associated proteins down-regulated with SENCR silencing were considered, a strong correlation with cell–cell adhesion as a biological process term was observed (Fig. 5C).
Fig. 5.
CKAP4 mediates EC adherens junction protein–protein association. (A) Hierarchy of Gene Ontology (GO) term (biological process) following mass spectrometry output data of CKAP4-associated proteins enriched by IP with anti-CKAP4 antibody. (B) GO term comparison within each category of molecular function (MF), cellular component (CC), and biological process (BP) following mass spectrometry output data using anti-CKAP4 antibody in HUVEC ± SENCR shRNA. (C) GO term hierarchy within BP category following mass spectrometry output data of down-regulated CKAP4-associated proteins in HUVEC with SENCR knockdown. (D) Multi-Venn diagram comparing interacting proteins with CDH5, JUP, DSP, VIM, and CKAP4 following mass spectrometry of each respective IP from whole-cell lysate (HUVEC). (E) Summary of data in D emphasizing the unique up-regulation of CKAP4–CDH5 association versus other adherens junction interactive proteins. (F) IP with indicated antibodies using HUVEC whole-cell lysate ± SENCR shRNA and immunoblotting with CDH5 (n = 3). (G) Quantitative grayscale density for F. *P < 0.05.
To further explore the interaction between CKAP4 and adherens junction proteins in HUVEC ± SENCR, co-IP was performed using antibodies targeting CKAP4, CDH5, JUP, DSP, and VIM. Enriched proteins from each co-IP were subjected to mass spectrometry analysis and the results demonstrated that CKAP4, CDH5, JUP, DSP, and VIM share 57 common-associated proteins in HUVEC with control shRNA. In contrast, the amount of shared common-associated proteins was sharply reduced to nine in HUVEC with SENCR knockdown (Fig. 5D and Dataset S3). Notably, the amount of common proteins only associated with CKAP4 and CDH5 was increased (from 5 to 12) after SENCR knockdown, further supporting an increased affinity between CKAP4 and CDH5 upon knockdown of SENCR RNA (Fig. 5E).
CTNND1 is a membrane-associated protein that functions to anchor CDH5 at the adherens junction of cells (17, 27). Because SENCR knockdown results in increased CDH5 internalization, we considered whether an effect on CTNND1–CDH5 association occurs with SENCR knockdown. Indeed, the CTNND1–CDH5 association was reduced upon SENCR knockdown concomitant with an increase in CDH5–CKAP4 association (Fig. 5 F and G). We repeated this experiment under LSS conditions and similar results were observed (SI Appendix, Fig. S4A). To validate the increased CDH5–CKAP4 interaction, co-IP/mass spectrometry was performed. Remarkably, while most other CDH5-associated proteins were reduced with SENCR knockdown, a notable increase in association was observed with CKAP4 (SI Appendix, Fig. S4B). These results suggest that loss of SENCR in EC impairs the adherens junction and, by extension, membrane integrity through augmented CDH5–CKAP4 association. The CTNND1 in surface versus cytosolic fraction with SENCR knockdown was also investigated and results revealed a similar trend as CDH5 (SI Appendix, Fig. S4C vs. Fig. 6A).
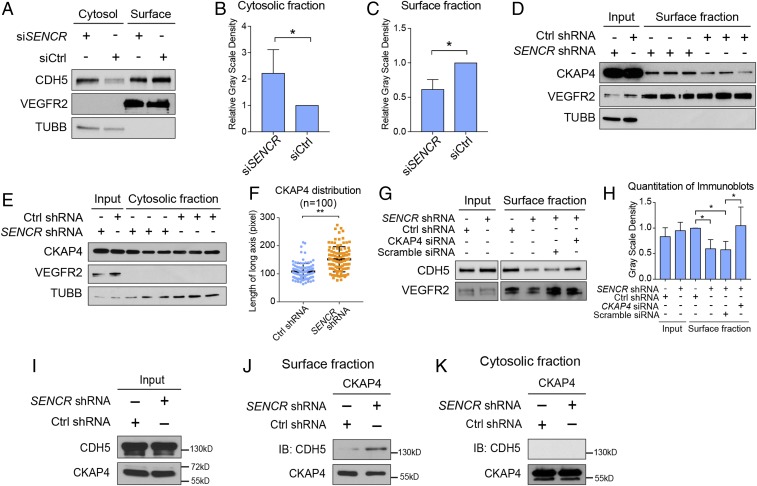
Fig. 6.
CKAP4–CDH5 association is enhanced in HUVEC with SENCR knockdown. (A) Immunoblot of CDH5 in cytosolic versus surface fraction of HUVEC ± SENCR shRNA. Anti-VEGFR2 antibody was used as surface fraction marker; anti-TUBB antibody was used as cytosolic fraction marker. (B and C) Quantitation of CDH5 level in cytosolic vs. surface fraction in HUVEC ± SENCR shRNA (n = 3). (D) Immunoblot of surface fraction of HUVEC ± SENCR shRNA using anti-CKAP4 antibody (n = 3). Anti-VEGFR2 and anti-TUBB antibodies were used as surface and cytosolic fraction markers, respectively. (E) Immunoblot of cytosolic fraction of HUVEC ± SENCR shRNA using anti-CKAP4 antibody (n = 3). (F) Quantitative measure of length of CKAP4 distribution along the long axis of HUVEC ± SENCR shRNA (n = 100). (G) Immunoblot of surface fraction of CDH5 in HUVEC ± SENCR/CKAP4 double knockdown (n = 4). (H) Quantitative grayscale density for G was summarized and shown. (I) Immunoblot of CDH5 and CKAP4 for surface versus cytosolic fraction input in HUVEC ± SENCR shRNA. (J) Co-IP of CKAP4-enriched proteins using surface fraction of HUVEC ± SENCR shRNA and immunoblot with anti-CDH5 and anti-CKAP4 antibody. (K) Co-IP of CKAP4 enriched proteins using cytosolic fraction of HUVEC ± SENCR shRNA and immunoblot with anti-CDH5 and anti-CKAP4 antibody. *P < 0.05, **P < 0.01.
SENCR Knockdown Enhances Cell Surface CKAP4–CDH5 Association.
To investigate the underlying mechanism of enhanced CKAP4–CDH5 association with SENCR knockdown, cell surface protein fractionation was performed. The results demonstrated a reduced level of cell surface CDH5 protein and an increased internalized CDH5 following SENCR knockdown (Fig. 6 A–C). Findings also revealed elevated CKAP4 at the cell surface following SENCR knockdown with little change in the cytosolic fraction (Fig. 6 D and E). Consistent with these results, immunofluorescence microscopy showed more dispersed CKAP4 cytoplasmic localization in HUVEC with SENCR knockdown (SI Appendix, Fig. S4 D–F). Quantitatively, the CKAP4 distribution along the long axis of SENCR knockdown EC was significantly greater than control cells (Fig. 6F). Importantly, the level of cell surface CDH5 was restored upon simultaneous knockdown of both CKAP4 and SENCR (Fig. 6 G and H). To further corroborate the cell surface localization of CDH5–CKAP4, co-IP/immunoblot was performed using cell surface and cytosolic fractions. Results showed enhanced CDH5–CKAP4 interaction in the cell surface fraction of SENCR knockdown EC (Fig. 6 I and J). However, there was no CDH5–CKAP4 association detected in the cytosolic fraction (Fig. 6K). These results suggest that following SENCR knockdown, a pool of CKAP4 redistributes to the cell surface and displaces CTTND1 binding to CDH5; this then allows for a CKAP4–CDH5 interaction that favors internalization of CDH5, thereby destabilizing adherens junction and membrane integrity.
The RBD1 Domain of CKAP4 Interacts with the JMD of CDH5.
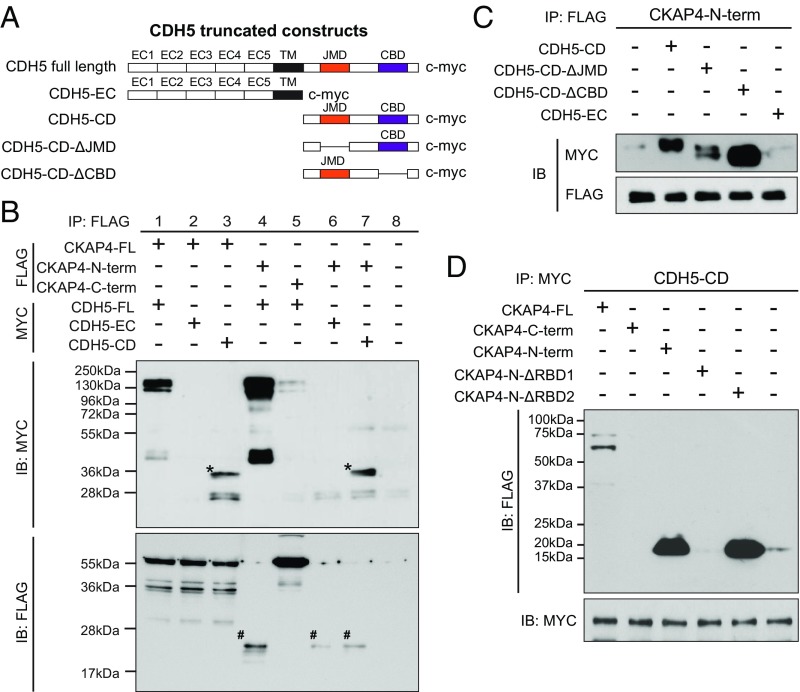
Structure–function studies were carried out to gain insight into the underlying mechanism of CDH5–CKAP4 association. A series of truncated CDH5 expression constructs was generated, including the extracellular domain (CDH5-EC), cytosolic domain (CDH5-CD), cytosolic domain with deleted JMD (CDH5-CD-ΔJMD), and cytosolic domain with deleted catenin binding domain (CDH5-CD-ΔCBD) (Fig. 7A). HEK-293 cells were cotransfected with each CDH5 truncated mutant and various CKAP4 constructs (CKAP4-N, CKAP4-N-ΔRBD1, CKAP4-N-ΔRBD2) for co-IP/immunoblot. Results demonstrated an association between CDH5-CD and CKAP4 N-terminal (cytoplasmic domain) (Fig. 7B). No interaction was seen between the extracellular domain of CDH5-EC and the C-terminal domain of CKAP4 (Fig. 7B). To further map the interaction domain mediating CDH5–CKAP4 association, CDH5-CD-ΔJMD or CDH5-CD-ΔCBD was cotransfected with CKAP4-N. The results showed significant impairment of the interaction between CDH5-CD-ΔJMD and CKAP4-N, suggesting the importance of the JMD in CDH5–CKAP4 association (Fig. 7C). Co-IP/immunoblot results of CDH5-CD and CKAP4-N-ΔRBD1 or CKAP4-N-ΔRBD2 demonstrated that the interaction between CKAP4 and CDH5 was reduced in CKAP4-N-ΔRBD1 (Fig. 7D). Collectively, these results suggest that that the JMD of CDH5 and the RBD1 domain of CKAP4 are essential for protein–protein interaction.
Fig. 7.
CKAP4–CDH5 interaction occurs through cytosolic domains. (A) Schematic for generating MYC-tagged CDH5 expression constructs and truncated CDH5 plasmids. (B) Co-IP of CKAP4 and CDH5 association. FLAG-tagged CKAP4 full length (CKAP4-FL), CKAP4 N-terminal truncated (CKAP4-N-term), and CKAP4 C-terminal truncated (CKAP4-C-term) and MYC-tagged CDH5 full length (CDH5-FL), CDH5 extracellular domain (CDH5-EC), and CDH5 cytosolic domain (CDH5-CD) were cotransfected in HEK-293T and protein–protein interactions were detected by immunoblot. An asterisk (*) indicates correct size band for CDH5-CD product. A pound sign (#) indicates correct size band for CKAP4-N-term. (C) Co-IP of CKAP4 N-terminal (FLAG-tagged) and CDH5 truncated proteins (MYC-tagged). IP was performed with anti-FLAG antibody and immunoblot was performed using anti-MYC antibody. (D) Co-IP of CDH5-CD (MYC-tagged) and CKAP4 truncated proteins (FLAG-tagged). IP was performed with anti-MYC antibody and immunoblot was performed using anti-FLAG antibody.
Discussion
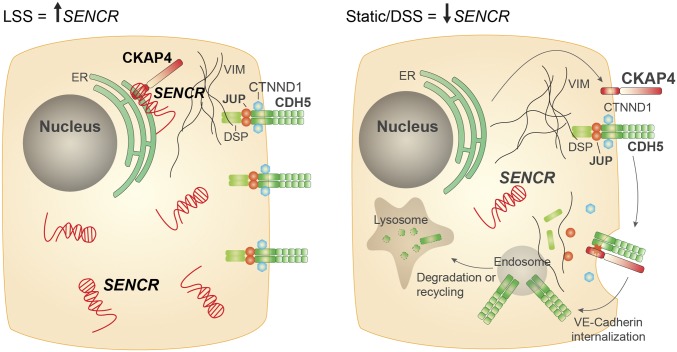
Evidence is provided for SENCR functioning as an LSS-responsive lncRNA that facilitates EC membrane integrity at the adherens junction. Loss-of-function studies suggest that SENCR controls the localization of CDH5 at the adherens junction while restricting CKAP4 to the cytosol, presumably within the endoplasmic reticulum (ER) (28). CKAP4 directly binds SENCR through a noncanonical RBD. Reduced SENCR results in radial distribution of CKAP4, which appears to foster displacement of a key CDH5-binding protein, CTNND1, thereby promoting a CKAP4–CDH5 association. The elevated CKAP4–CDH5 interaction prevents normal localization of CDH5 at the EC membrane, thus resulting in a defective adherens junction and leading to heightened EC permeability. We surmise that under LSS conditions, the CKAP4–SENCR association functions to indirectly stabilize CDH5 at the adherens junction, thus maintaining normal EC membrane homeostasis (Fig. 8).
Fig. 8.
Hypothetical model of SENCR regulating CDH5 internalization. EC subjected to LSS have higher expression of SENCR (Left) vs. EC under static or DSS flow conditions (Right). SENCR interacts with CKAP4 near the rough ER. In this scenario, the adherens junction is stable and membrane integrity is intact (Left). Reduced levels of SENCR, shown in this report to occur under static flow conditions or within the DSS region of mouse aorta liberates a pool of CKAP4 to localize at the cell membrane, where it displaces CTNND1, promoting CDH5 internalization and impairing adherens junction (Right).
Normal EC function is maintained by a gene program under the regulation of LSS, which antagonizes vascular disease (14). Previous studies have demonstrated LSS-responsive lncRNAs, including LINC00341 (29), MANTIS (9), LEENE (10), and LISPR1 (30). SENCR is shown here to be induced by LSS in multiple EC types; however, DSS, which promotes vascular disease (15), showed no observable impact on SENCR RNA level in cultured HUVEC. The onset of LSS-induced SENCR was delayed compared with KLF2, suggesting KLF2, a central regulator of LSS-responsive gene expression in EC (31), may mediate augmented SENCR expression. Because of the lack of evidence for a SENCR ortholog in rodents, we generated a humanized mouse model of SENCR using the piggyBac transposon system. Importantly, and consistent with in vitro studies, SENCR RNA was more abundantly expressed in LSS regions of the aorta than in DSS regions. These findings suggest that SENCR may function in concert with other LSS-induced genes to antagonize vascular disease formation. The pervasive transcription of lncRNAs in cells suggests there are likely to be numerous lncRNA–protein associations at or near the EC membrane. In support of this concept, we noted that the interacting proteome at the adherens junction is sensitive to RNase treatment of cells as well as SENCR knockdown. Future studies should fully disclose lncRNA associations and mechanisms of action at the adherens junction.
The role of lncRNAs in membrane integrity has been limited to tight junctions, primarily those in EC of the blood–brain barrier. For example, NEAT1 and TUG1 were proposed to function as competing endogenous RNAs to reduce the expression level of ZO1 and CLDN5, resulting in increased barrier permeability (32, 33). Here, we provide an example of an lncRNA effecting changes in the adherens junction of EC. However, in contrast to NEAT1 and TUG1, SENCR does not appear to function as a competing endogenous RNA because the profile of down-regulated genes with reduced SENCR expression failed to show a common microRNA binding sequence in SENCR. Instead, we provide strong evidence for SENCR physically associating with the RNA-binding protein, CKAP4. We propose that this interaction stabilizes membrane-bound CDH5, a key component of the adherens junction because, upon SENCR knockdown, membrane-bound CDH5 redistributes, thus eliciting elevated EC membrane permeability. CKAP4 localization expands toward the EC membrane where it binds CDH5 via a highly conserved (SI Appendix, Fig. S5) noncanonical RBD1, presumably by displacing the interaction between CTNND1 and CDH5. Preliminary studies indicate that SENCR interacts with mouse CKAP4 in a humanized mouse model, highlighting the utility of BAC engineering mice for the functional appraisal of protein-binding lncRNAs. Whether SENCR has any effect on tight junctions remains an open question.
CDH5 is a principal component of the adherens junction and plays an important role in regulating vascular permeability (34). The localization of CDH5 at the cell membrane is stabilized by CTNND1, which associates with the JMD of CDH5 (17). The CDH5 JMD functions as a CTNND1-binding site as well as an endocytic sensor (35). Association of CTNND1 and CDH5 masks the endocytic sensor and stabilizes CDH5 membrane retention (27). Phosphorylation of CTNND1 increases the binding affinity of CDH5 and prevents CDH5 from being internalized or recycled (27). However, dephosphorylation or knockdown of CTNND1 destabilizes the association with CDH5 and triggers CDH5 internalization, resulting in adherens junction impairment (36). Data herein suggest SENCR knockdown disrupts the interaction between CDH5 and CTNND1 by promoting a CDH5–CKAP4 association. We suggest other lncRNAs may be involved in maintaining or perturbing the adherens junction given the discovery of a noncanonical RBD within CKAP4.
CKAP4 (also known as P63, CLIMP-63, and ERGIC-63) was originally identified as an ER/Golgi intermediate compartment (ERGIC) localizing protein that functions as a microtubule-binding protein and anchors ER membranes to the actin-cytoskeleton (28, 37). CKAP4 is a type II transmembrane protein comprising an N-terminal intracellular domain, a single transmembrane domain, and a C-terminal extracellular domain (38). The N terminus of CKAP4 comprises two critical functional domains: an ER anchoring domain (2–21 aa) and a microtubule-binding domain (36–59 aa) (39). While the role of CKAP4 in tumor cells and vascular smooth muscle cells has been reported (24, 40, 41), its function in vascular EC was, until now, unexplored. Here, we show a previously unrecognized, noncanonical RBD (6–15 aa) within the ER anchoring domain of CKAP4 binds SENCR. This suggests that SENCR may function to anchor CKAP4 at the ER membrane because loss in SENCR results in some CKAP4 mislocalization toward the cell membrane. The precise function of the SENCR–CKAP4 complex at the ER membrane will be an important area of future investigation.
In summary, SENCR is an LSS-induced lncRNA that helps maintain a nonpermeable EC membrane. Reduced levels of SENCR RNA displaces some CKAP4 to the EC membrane where CKAP4 encounters and binds CDH5 resulting in CDH5 internalization and a perturbation at the adherens junction. Whether changes in EC or vascular smooth muscle cell SENCR RNA contribute to disease progression awaits further study. The SENCR-BAC mice reported here offer a unique opportunity to address this and other related questions.
Materials and Methods
Cell Culture.
HUVEC were cultured in Medium 200 (#M200500; Thermo-Fisher) supplied with low-serum growth supplement (#S00310; Thermo-Fisher). HPAEC and HCAEC were purchased from a commercial source (#CC-2530 and #CC-2585; Lonza) and cultured with VascuLife EnGS endothelial medium kit (#LL-0002; Lifeline). The HEK-293FT cell line was purchased (#R70007; Thermo-Fisher) and cultured with DMEM (#11965–092; Thermo-Fisher) plus 10% FBS (#A3160902; Thermo-Fisher). All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
RNA Isolation, Reverse Transcription, and Real-Time qPCR.
Total RNA was isolated using RNeasy Mini Kit (#74104; Qiagen) according to the manufacturer’s guidance. Reverse transcription was performed using Bio-Rad iScript cDNA synthesis kit (#1708891; Bio-Rad) after RNA quantitation and DNase I treatment. Real-time qPCR was performed using iTaq Universal SYBR Green Supermix (#1725121; Bio-Rad) with primers listed in Dataset S4.
RNA-FISH.
RNA-FISH for cells on coverslips was performed following the manufacturer’s protocol (https://www.biosearchtech.com/support/resources/stellaris-protocols).
Generation of Humanized SENCR Mice.
A BAC clone (RP11-744N12) was acquired from the Children’s Hospital Oakland Research Institute to produce humanized SENCR mouse lines, as described previously (42). Briefly, a BAC vector (pBACe3.6) was modified into RP11-744N12 (227,863 bp) containing the human SENCR and FLI1 gene loci by recombineering. A cassette with the piggyBac terminal sequences was electroporated into BAC containing Escherichia coli cells and uptake was selected for using Spectinomycin selection. C57BL/6J mouse zygotes were injected with the piggyBac retrofitted BAC and PB transposase mRNA. Mouse experiments reported were approved by the University of Rochester Institutional Animal Care and Use Committee.
En Face Preparation and Immuno-RNA-FISH.
En face preparations of mouse aorta were prepared following an established method (43). Immuno-RNA-FISH was carried out according to a previously defined method (44). Briefly, following fixation with 4% paraformaldehyde, mouse aortae were cut longitudinally and permeabilized with 1× PBS and 0.1% Triton X-100. Samples were then washed twice with 2× SSC and 10% formamide and then hybridized in Stellaris RNA-FISH hybridization buffer (#SMF-HB1-10; Biosearch) with Stellaris RNA-FISH probe for SENCR at 37 °C overnight in hybridization oven. Samples were washed twice in 2× SSC and 10% formamide and refixed with 4% paraformaldehyde. Anti-CDH5 antibody (#555289 for mouse; Cell Signaling) was then incubated with samples at room temperature for 1 h. After two washes in PBS-T, tissues were incubated with Alexa Fluor 488-conjugated secondary antibody for 1 h at room temperature. Aortae were counterstained with DAPI and mounted for confocal microscopy.
Transfection.
Transfection of siRNAs was performed with Lipofectamine RNAiMAX reagent (#13778-075; Thermo-Fisher) according to the manufacturer’s protocol. All plasmid transfections were performed with Lipofectamine 3000 reagent (#L3000-008; Thermo-Fisher) according to the manufacturer’s manual.
Plasmid Constructs and Lentivirus Packaging.
Full-length SENCR cDNA was synthesized by IDT and cloned into pcDNA3.1(+) vector (#V79020; Thermo-Fisher). Human CKAP4 expression plasmid was purchased from Addgene (#80977; Addgene) and subcloned into pcDNA3.1(+) with 3xFLAG tag at the C terminus. Truncated CKAP4 constructs were generated by DNA synthesis from IDT. Human CDH5 expression plasmid was purchased from Addgene (#85144; Addgene) and subcloned into pcDNA3.1/myc-His expression vector (#V80020; Thermo-Fisher). Truncated CDH5 protein constructs were generated using primers in Dataset S4. SENCR siRNAs were designed based on human SENCR (NR_038908) using siRNA designing program, SiExplorer. SiRNA sequences are listed in Dataset S4. We validated siRNA efficiency and selected siRNA-1 to produce shRNA using pmiRZip-based (#MZIP1-PA-1; SBI) lentiviral expression vector. ShRNA expression vectors were cotransfected with pMD2.g and psPAX2 into HEK-293FT cell line for producing lentivirus.
Cell Permeability Assay.
Cell permeability assay for EC monolayer was performed with In Vitro Vascular Permeability Assay (FITC-Dextran) kit (#ECM644; Millipore) following the manufacturer’s protocol.
TEER Assay.
TEER was used to define EC monolayer integrity using an ECIS (electric-substrate impedance sensing) system (Applied BioPhysics), as previously described (22, 45). Cells were seeded in eight-chambered electrode arrays (8W10E+) pretreated with cysteine and gelatin. The arrays were mounted on the ECIS device in a 37 °C incubator for normal cell culture. TEER was measured over time.
Biotinylated Cell Surface Protein Labeling Assay.
HUVECs were cultured in 10-cm culture dishes 24 h before biotinylation. Cells were washed with cold 1× PBS and labeled with EZ-link Sulfo-NHS-SS-Biotin (#21331; Thermo-Fisher) on ice for 30 min. Uncoupled biotin was washed twice with cold 1× PBS. Cells were incubated at 37 °C for 30 min to allow internalization of biotinylated proteins. Cells were washed three times with cold 1× PBS and lysed with Nonidet P-40 Cell Lysis Buffer. After brief sonication and centrifugation, supernatants were transferred to fresh tubes and incubated with NeutraAvidin Agarose (#29200; Thermo-Fisher) in a cold room overnight. Beads were washed 3× with cold 1× PBS and samples were heated at 70 °C for 10 min with 2× sample loading buffer to release proteins for downstream Western blotting and/or mass spectrometry.
CDH5 Internalization Assay.
HUVECs were cultured on coverslips for 24 h before initiating the experiment. Anti-CDH5 clone BV6 antibody (#ALX-803–305-C100; Enzo) was incubated with cells on ice bath for 30 min. Cells were then rinsed with ice cold Medium 200 to remove uncoupled antibodies. Next, cells were incubated at 37 °C for 30 min to allow internalization from cell surface. Cells were washed with 1× PBS (pH 2.7) containing 25 mM glycine and 3% bovine serum album (BSA), rinsed, and fixed for fluorescent-conjugated secondary antibody incubation. Images were taken using Olympus IX81 confocal microscope.
Western Blotting.
Cells were washed twice with cold 1× PBS and lysed with Cell Lysis Buffer (#9803; Cell Signaling) supplied with complete and EDTA-free protease inhibitor mixture (#4693132001; Sigma-Aldrich). Protein concentration was quantified by DC Protein Assay kit II (#5000112; Bio-Rad) and denatured by boiling in NuPAGE LDS sample buffer (#NP0008; Thermo-Fisher). Protein samples were loaded in 4–12% SDS/PAGE gels with 1× Tris-Glycine running buffer. Membrane transfer was done using PVDF membranes. Membranes were then blocked in 5% nonfat milk and incubated with appropriate antibodies. After the final wash step, membranes were visualized by adding SuperSignal West Pico PLUS chemiluminescent substrate (#34580; Thermo-Fisher) and processed with film developer.
Immunofluorescence Microscopy.
Cells were cultured on 22 × 22-mm coverslips in six-well plates. After treatment, cells were washed twice with prewarmed 1× PBS and fixed in 4% paraformaldehyde for 10 min at room temperature. Coverslips were rinsed 2× in 1× PBS and permeabilized with 1× PBS containing 0.1% Triton X-100. Cells were then blocked with 3% BSA for 20 min at room temperature. Primary antibodies were then added to coverslips at dilutions of 1:50. Coverslips were incubated at room temperature for 40 min and rinsed twice with 1× PBS before addition of fluorescence-conjugated secondary antibodies. After the final wash step, cells were mounted with ProLong Gold anti-fade reagent with DAPI (#P36935; Thermo-Fisher) for confocal microscopy. Images were acquired with a confocal microscope using uniform parameters across the entire image. For immuno-RNA-FISH images, we increased the gain and contrast uniformly across each image using the same parameters across each individual image (Fig. 1H).
Immunoprecipitations.
Cells were cultured in 10-cm dishes to 90% confluency. Cell lysates were collected by scraping cells in Nonidet P-40 Cell Lysis Buffer [50 mM Hepes, pH, 7.5, 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% Nonidet P-40 (vol/vol), 0.5 mM DTT and protease inhibitor mixture]. After brief sonication, cell lysates were centrifuged and supernatants were transferred to a fresh 1.5-mL microfuge tube. Antibodies (Anti-FLAG #F1804; Sigma; MYC tag #60003-2-Ig; Proteintech; Anti-CKAP4 #16686-1-AP; Proteintech; Anti-CTNND1 #66208-1-Ig; Proteintech; Anti-CDH5 #2500; Cell Signaling; Anti-JUP #2309; Cell Signaling; Anti-DSP #E2715; Santa Cruz; Anti-VIM #5741; Cell Signaling) were then added to cell lysates and incubated on a rotator in cold room for 2 h. Prewashed Dynabeads protein G were added to the lysate-antibody mix, incubated for 2 h in a cold room, and then treated with a magnetic rack to pull down interacting proteins. The beads were then washed 4–5× with Nonidet P-40 Cell Lysis Buffer. Beads were then boiled in NuPAGE LDS sample buffer (#NP0008; Thermo-Fisher) and Western Blotting done with target antibodies.
In Vitro Transcription and RNA 3′ Biotinylation.
In vitro transcription was performed using MEGAscript T7 transcription kit (#AM1334; Thermo-Fisher) following the manufacturer’s protocol with PCR products containing T7 promoter. RNA was extracted by phenol/chloroform/isoamyl alcohol (25:24:1) and precipitated with 3 M sodium acetate (pH, 5.2) and 100% ethanol. To label RNA for downstream RNA pull-down assay, RNAs were 3′ end-labeled with a biotinylation kit (#20160; Thermo-Fisher) following the manufacturer’s instructions.
RNA Pull-Down and Mass Spectrometry.
Biotinylated RNA was heated to 95 °C in RNA structure buffer (10 mM Tris⋅HCl pH, 7.0, 0.1 M KCl, 2 mM MgCl2) and immobilized on MyOne Streptavidin C1 Dynabeads (#65001; Thermo-Fisher). Beads were washed 3× using 1× PBS-T to remove unbound RNA and then incubated in Nonidet P-40 Cell Lysis Buffer at 4 °C for 2 h. Beads were then washed 4× with 1× PBS-T and boiled in SDS/PAGE sample buffer for 10 min. Samples were run in 4–12% precast gels (#NP0321BOX; Thermo Fisher) and delivered to the University of Rochester Medical Center Mass Spectrometry Resource laboratory of the University of Rochester for mass spectrometry analysis.
RNA IP.
HUVECs were cultured in 10-cm dishes to 90% confluency. Cells were rinsed twice with 1× PBS and lysed with Polysome Lysis Buffer. Cell lysate was briefly sonicated and centrifuged and the supernatants were transferred to fresh tubes. Antibodies to CKAP4 (#16686-1-AP; Proteintech) and FLAG (#F1804; Sigma) were added to cell lysates and incubated at 4 °C overnight. Prewashed Dynabeads protein G (#10003D; Thermo-Fisher) were then added to lysate-antibody mix and incubated 2 h at 4 °C. Beads were subjected to a magnetic rack and washed 4× with Polysome Lysis Buffer and another 4× with 1 M urea-containing Polysome Lysis Buffer. Beads were treated with proteinase K at 55 °C for 30 min on a thermomixer. RNA was extracted with phenol-chloroform-IAA (25:24:1) and precipitated with ethanol. RNA was pelleted, air-dried, and dissolved in RNase-free water for reverse transcription and downstream qPCR.
RNA EMSA.
RNA EMSA was performed based on an established protocol (46). Briefly, radioactive-labeled RNA probes were produced by in vitro transcription incorporated with α-32P-UTP using MEGAscript T7 transcription kit (#AM1334; Thermo-Fisher). Proteins were acquired by transfecting HEK-293T with FLAG-tagged expression plasmids and purified by anti-FLAG antibody. Labeled RNA and purified protein were incubated in binding buffer [40 mM Tris⋅HCl pH, 8.0, 30 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.01% (wt/vol) Nonidet P-40] at 37 °C for 30 min. Next, 50 mg/mL heparin was added and incubated at room temperature for 10 min. Samples were treated with RNase T1 (1 U/μL) for 10 min at room temperature and run in a 6% bis-acrylamide gel containing 0.5× TBE buffer. Gels were dried (Gel Drying System, #1651790; Bio-Rad) and then exposed to X-ray film at −80 °C overnight.
RNA-Sequencing.
HUVECs (passage 2) were cultured in Medium 200 with low-serum growth supplement in 6-cm culture dishes. Lentivirus expressing SENCR shRNA (shSENCR) or scrambled shRNA (shCtrl) were generated based on pLV-CMV-EF1-GFP. HUVECs were transduced with lentivirus and cultured in medium containing puromycin until all cells expressed GFP. Cells were then seeded in triplicate in 6-cm culture dishes and grown to 80% confluency. Total RNA was isolated using TRIzol following the manufacturer’s protocol and RNA was dissolved in RNase-free water. TruSeq stranded mRNA library kit (Illumina) was used to generate each cDNA library. RNA-seq was performed at a depth of ∼20 million reads per sample using HiSeq2500 (Illumina) Single with 100 base pair read length. All pre- and postrun analyses were performed at the University of Rochester Genomics Research Center (https://www.urmc.rochester.edu/research/rochester-genomics-center.aspx). RNA-seq data have been deposited in the Gene Expression Omnibus (accession no. GSE122490).
Statistical Analysis.
Paired t test was used for comparisons between experimental and control conditions or one- and two-way ANOVA for multiple group comparisons. All data analysis was performed in GraphPad Prism 7. Results are expressed as mean ± SD. A value of P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Chen Yan (University of Rochester) for helpful suggestions on experimental design; Dr. Pieter De Jong (Children’s Hospital Oakland Research Institute) for engineering the piggyBac clone carrying SENCR and FLI1; and Jonathan Neumann (University of California, Davis) for generating several founder mice carrying the BAC. The work was supported by Grants NIH R01 HL112793, HL117907, and HL138987 (to J.M.M.); NIH R01 HL122686 (to X.L.); NIH R01 HL130870 (to A.R.); and AHA 17POST3360938 (to Q.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE122490).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810729116/-/DCSupplemental.
References
- 1.Miano JM, Long X. The short and long of noncoding sequences in the control of vascular cell phenotypes. Cell Mol Life Sci. 2015;72:3457–3488. doi: 10.1007/s00018-015-1936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 3.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melé M, Rinn JL. “Cat’s cradling” the 3D genome by the act of lncRNA transcription. Mol Cell. 2016;62:657–664. doi: 10.1016/j.molcel.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9:e1471. doi: 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leisegang MS, et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation. 2017;136:65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao Y, et al. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat Commun. 2018;9:292. doi: 10.1038/s41467-017-02113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neumann P, et al. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun. 2018;9:237. doi: 10.1038/s41467-017-02431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dejana E, Tournier-Lasserve E, Weinstein BM. The control of vascular integrity by endothelial cell junctions: Molecular basis and pathological implications. Dev Cell. 2009;16:209–221. doi: 10.1016/j.devcel.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Komarova YA, Kruse K, Mehta D, Malik AB. Protein interactions at endothelial junctions and signaling mechanisms regulating endothelial permeability. Circ Res. 2017;120:179–206. doi: 10.1161/CIRCRESAHA.116.306534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traub O, Berk BC. Laminar shear stress: Mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler Thromb Vasc Biol. 1998;18:677–685. doi: 10.1161/01.atv.18.5.677. [DOI] [PubMed] [Google Scholar]
- 15.Davies PF. Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nat Clin Pract Cardiovasc Med. 2009;6:16–26. doi: 10.1038/ncpcardio1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dejana E, Orsenigo F, Lampugnani MG. The role of adherens junctions and VE-cadherin in the control of vascular permeability. J Cell Sci. 2008;121:2115–2122. doi: 10.1242/jcs.017897. [DOI] [PubMed] [Google Scholar]
- 17.Chiasson CM, Wittich KB, Vincent PA, Faundez V, Kowalczyk AP. p120-catenin inhibits VE-cadherin internalization through a Rho-independent mechanism. Mol Biol Cell. 2009;20:1970–1980. doi: 10.1091/mbc.E08-07-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell RD, et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34:1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boulberdaa M, et al. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol Ther. 2016;24:978–990. doi: 10.1038/mt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekker RJ, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Krüppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasan B, et al. TEER measurement techniques for in vitro barrier model systems. J Lab Autom. 2015;20:107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slavin SA, Leonard A, Grose V, Fazal F, Rahman A. Autophagy inhibitor 3-methyladenine protects against endothelial cell barrier dysfunction in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2018;314:L388–L396. doi: 10.1152/ajplung.00555.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar M, Gromiha MM, Raghava GP. SVM based prediction of RNA-binding proteins using binding residues and evolutionary information. J Mol Recognit. 2011;24:303–313. doi: 10.1002/jmr.1061. [DOI] [PubMed] [Google Scholar]
- 24.Kimura H, et al. CKAP4 is a Dickkopf1 receptor and is involved in tumor progression. J Clin Invest. 2016;126:2689–2705. doi: 10.1172/JCI84658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuchi A, Fumoto K, Kimura H. The Dickkopf1-cytoskeleton-associated protein 4 axis creates a novel signalling pathway and may represent a molecular target for cancer therapy. Br J Pharmacol. 2017;174:4651–4665. doi: 10.1111/bph.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Nanes BA, et al. p120-catenin binding masks an endocytic signal conserved in classical cadherins. J Cell Biol. 2012;199:365–380. doi: 10.1083/jcb.201205029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang TS, et al. LINC00341 exerts an anti-inflammatory effect on endothelial cells by repressing VCAM1. Physiol Genomics. 2017;49:339–345. doi: 10.1152/physiolgenomics.00132.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Josipovic I, et al. Long noncoding RNA LISPR1 is required for S1P signaling and endothelial cell function. J Mol Cell Cardiol. 2018;116:57–68. doi: 10.1016/j.yjmcc.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Gimbrone MA, Garcia-Cardena G. Vascular endothelium, hemodynamics, and the pathobiology of atherosclerosis. Cardiovasc Pathol. 2013;22:9–15. doi: 10.1016/j.carpath.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J, et al. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2240–2254. doi: 10.1016/j.bbadis.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Cai H, et al. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget. 2015;6:19759–19779. doi: 10.18632/oncotarget.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bazzoni G, Dejana E. Endothelial cell-to-cell junctions: Molecular organization and role in vascular homeostasis. Physiol Rev. 2004;84:869–901. doi: 10.1152/physrev.00035.2003. [DOI] [PubMed] [Google Scholar]
- 35.Xiao K, et al. p120-catenin regulates clathrin-dependent endocytosis of VE-cadherin. Mol Biol Cell. 2005;16:5141–5151. doi: 10.1091/mbc.E05-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariner DJ, et al. Identification of Src phosphorylation sites in the catenin p120ctn. J Biol Chem. 2001;276:28006–28013. doi: 10.1074/jbc.M102443200. [DOI] [PubMed] [Google Scholar]
- 37.Vedrenne C, Klopfenstein DR, Hauri HP. Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell. 2005;16:1928–1937. doi: 10.1091/mbc.E04-07-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweizer A, Ericsson M, Bächi T, Griffiths G, Hauri HP. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J Cell Sci. 1993;104:671–683. doi: 10.1242/jcs.104.3.671. [DOI] [PubMed] [Google Scholar]
- 39.Schweizer A, Rohrer J, Hauri HP, Kornfeld S. Retention of p63 in an ER-Golgi intermediate compartment depends on the presence of all three of its domains and on its ability to form oligomers. J Cell Biol. 1994;126:25–39. doi: 10.1083/jcb.126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrads TP, et al. CKAP4/p63 is a receptor for the frizzled-8 protein-related antiproliferative factor from interstitial cystitis patients. J Biol Chem. 2006;281:37836–37843. doi: 10.1074/jbc.M604581200. [DOI] [PubMed] [Google Scholar]
- 41.Razzaq TM, et al. Functional regulation of tissue plasminogen activator on the surface of vascular smooth muscle cells by the type-II transmembrane protein p63 (CKAP4) J Biol Chem. 2003;278:42679–42685. doi: 10.1074/jbc.M305695200. [DOI] [PubMed] [Google Scholar]
- 42.Jung CJ, et al. Comparative analysis of piggyBac, CRISPR/Cas9 and TALEN mediated BAC transgenesis in the zygote for the generation of humanized SIRPA rats. Sci Rep. 2016;6:31455. doi: 10.1038/srep31455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ko KA, Fujiwara K, Krishnan S, Abe JI. En face preparation of mouse blood vessels. J Vis Exp. 2017:55460. doi: 10.3791/55460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih JD, Waks Z, Kedersha N, Silver PA. Visualization of single mRNAs reveals temporal association of proteins with microRNA-regulated mRNA. Nucleic Acids Res. 2011;39:7740–7749. doi: 10.1093/nar/gkr456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami M, et al. The FGF system has a key role in regulating vascular integrity. J Clin Invest. 2008;118:3355–3366, and correction (2009) 119:2113. doi: 10.1172/JCI35298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rio DC. Electrophoretic mobility shift assays for RNA-protein complexes. Cold Spring Harb Protoc. 2014;2014:435–440. doi: 10.1101/pdb.prot080721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.