Significance
Mutations in the LAMP-2 gene are associated with Danon disease. Although dysregulation of autophagy has been described in Danon disease, the mechanisms by which LAMP-2 deficiency leads to autophagy dysregulation remain elusive. Autophagy ends with fusion between autophagosomes and late endosomes/lysosomes for degradation. It has been shown that STX17 is essential for autophagic fusion in noncardiomyocytes. Here, we demonstrate that LAMP-2B promotes autophagosome–lysosome fusion in cardiomyocytes (CMs), independently of STX17. LAMP-2B deficiency in CMs causes defects in autophagic fusion and functional abnormalities, recapitulating the phenotype of Danon CMs. Gene correction of the LAMP-2 mutation restores CM function. Our results thus reveal a significant autophagosome–lysosome fusion mechanism in CMs and provide a foundation for gene correction therapy in patients with Danon disease.
Keywords: LAMP-2B, autophagy, cardiomyopathy, Danon disease, autophagosome–lysosome fusion
Abstract
Mutations in lysosomal-associated membrane protein 2 (LAMP-2) gene are associated with Danon disease, which often leads to cardiomyopathy/heart failure through poorly defined mechanisms. Here, we identify the LAMP-2 isoform B (LAMP-2B) as required for autophagosome–lysosome fusion in human cardiomyocytes (CMs). Remarkably, LAMP-2B functions independently of syntaxin 17 (STX17), a protein that is essential for autophagosome–lysosome fusion in non-CMs. Instead, LAMP-2B interacts with autophagy related 14 (ATG14) and vesicle-associated membrane protein 8 (VAMP8) through its C-terminal coiled coil domain (CCD) to promote autophagic fusion. CMs derived from induced pluripotent stem cells (hiPSC-CMs) from Danon patients exhibit decreased colocalization between ATG14 and VAMP8, profound defects in autophagic fusion, as well as mitochondrial and contractile abnormalities. This phenotype was recapitulated by LAMP-2B knockout in non-Danon hiPSC-CMs. Finally, gene correction of LAMP-2 mutation rescues the Danon phenotype. These findings reveal a STX17-independent autophagic fusion mechanism in human CMs, providing an explanation for cardiomyopathy in Danon patients and a foundation for targeting defective LAMP-2B–mediated autophagy to treat this patient population.
Autophagy plays a crucial role in cell homeostasis and function (1–4). Two types of autophagy have been well studied. Chaperone-mediated autophagy (CMA) is a process of chaperon-dependent selection of cytosolic proteins that are translocated into the lysosome for degradation. Macroautophagy (referred to as autophagy hereafter) is mediated by double-membrane autophagosomes that enclose cytosolic cargoes, followed by fusion with late endosomes/lysosomes for degradation. STX17 localized to autophagosomes is essential for autophagosome–lysosome fusion as it interacts with SNAP29 and VAMP8 localized to late endosomes/lysosomes (5, 6). ATG14 localized to autophagosomes enhances autophagic fusion by interacting with the STX17–SNAP29 complex (7). Whether autophagosome–lysosome fusion in cardiomyocytes (CMs) is primarily mediated by STX17 remains elusive.
Mutations in the X-linked LAMP-2 gene are associated with Danon disease. Symptoms of Danon disease include hypertrophic/dilated cardiomyopathy, heart failure, muscle weakness, retinopathy, and mental retardation (8–11). The mean ages in years of diagnosis of cardiomyopathy and death are 13 and 19 in men and 30 and 35 in women (11). No specific or effective therapeutics have yet been identified for Danon disease (12), which might be due to a lack of defined molecular mechanisms of disease pathogenesis. Autophagic dysregulation has been described in muscle tissues and hiPSC-CMs derived from patients with Danon disease (9, 12, 13). However, the molecular mechanisms by which LAMP-2 deficiency causes autophagy dysregulation and disease pathogenesis are poorly understood.
Alternative splicing of pre-LAMP-2 mRNA produces three isoforms: LAMP-2A, LAMP-2B, and LAMP-2C. The LAMP-2 isoforms share an identical N-terminal domain but have distinct transmembrane and cytosolic domains at the C terminus (14). LAMP-2A functions as an essential receptor in CMA (15). Deletion of all three LAMP-2 isoforms in mice causes defects in autophagy (16). Hubert et al. (17) reported that LAMP-2A is required for autophagy by playing a role in localization of STX17 to autophagosomes in mouse embryonic fibroblasts (MEFs). Unlike the biological function of LAMP-2A, those of LAMP-2B and LAMP-2C remain elusive.
Using hiPSC-CMs and genome editing-based approaches, we identified LAMP-2B, the major LAMP-2 isoform expressed in CMs, as required for autophagic fusion in human CMs. The cytosolic CCD of LAMP-2B is required for it to promote formation of the ATG14–VAMP8 complex, as well as to mediate autophagic fusion. LAMP-2B is able to suppress accumulation of autophagosomes caused by knockdown of STX17 in non-CMs. Knockout of STX17 in hiPSC-CMs caused neither accumulation of autophagosomes nor affected the ability of LAMP-2B to promote autophagy. Knockout of LAMP-2 or LAMP-2B in hiPSC-CMs dramatically decreased colocalization of ATG14 with VAMP8, autophagosomal fusion with late endosomes/lysosomes, mitochondrial and contractile function, and reduced adenosine triphosphate (ATP) content. hiPSC-CMs derived from patients with Danon disease recapitulated this phenotype. Correction of the LAMP-2 mutation in Danon hiPSC-CMs restores normal autophagy and mitochondrial function, as well as improves contractility. Given the similarity of phenotype between LAMP-2B knockout and Danon hiPSC-CMs, we conclude that LAMP-2B deficiency is sufficient and necessary to cause the phenotypes observed in Danon CMs. Our findings thus provide insights into the nature of autophagy in human CMs and Danon pathogenesis.
Results
LAMP-2B Is the Predominant LAMP-2 Isoform Expressed in Human and Mouse CMs.
While half of LAMP-2 knockout mice display an almost normal life span (16), patients with Danon disease are susceptible to cardiac death or require heart transplantation at a young age (11), suggesting species-specific effects on pathogenesis of Danon disease. To investigate these mechanisms in a human model, we generated hiPSCs from skin fibroblasts derived from two unrelated control males (hereafter referred to as CUSO-1 and CUSO-2) and three unrelated males with Danon disease (hereafter referred to as MD-111, MD-186, and MD-506) (18). The three patients, MD-111, MD-186, and MD-506, carried LAMP-2 frameshift (c. 1082 delA, exon 8), nonsense (c.247C > T, exon 3), and splicing (c.64+1 G > A IVS-1) mutations, respectively (SI Appendix, Fig. S1A and Table S1). These hiPSC lines had normal karyotypes and demonstrated pluripotency by expressing pluripotency markers and capacity to differentiate into derivatives of three germ layers (SI Appendix, Fig. S1 B–D). We generated hiPSC-CMs with greater than 90% purity from each line for subsequent studies using established protocols (19, 20) (SI Appendix, Fig. S1 E and F). LAMP-2 protein was not detectable in Danon hiPSC-CMs (SI Appendix, Fig. S1G), consistent with previous findings that mutations in the LAMP-2 gene led to complete LAMP-2 deficiency in patients with Danon disease (9), could be due to a nonsense-mediated RNA decay (NMD) mechanism (21). Differentiation of Danon hiPSCs into hiPSC-CMs was not significantly different from control lines (SI Appendix, Fig. S1H). Monolayer hiPSC-CMs expressed CM markers (SI Appendix, Fig. S1 I–K) and beat spontaneously (Movie S1). Importantly, these hiPSC-CMs expressed a high ratio of MYH7 to MYH6, comparable to human heart tissues (SI Appendix, Fig. S1L). α-Actinin–positive sarcomeric structures in Danon hiPSC-CMs were comparable to those in controls (SI Appendix, Fig. S1M). hiPSC-CMs on day ∼50 after induction were used in this study if the time point was otherwise not specified.
The pathological hallmark of Danon disease is accumulation of glycogen and vacuoles in the patient’s CMs (8). Compared with control hiPSC-CMs, Danon hiPSC-CMs displayed increased Periodic Acid-Schiff stain (PAS)-positive glycogen storage and accumulation of vacuoles (SI Appendix, Fig. S2 A–C), suggesting that this hiPSC-CM platform is appropriate to characterize the mechanisms. Consistent with a previous study (13), no significant increase in apoptosis, as assayed by TUNEL staining, was observed in any examined Danon hiPSC-CM line cultured for either 50 or 100 days, compared with controls (SI Appendix, Fig. S2D). In addition, levels of reactive oxygen species (ROS) assayed by MitoSOX in Danon hiPSC-CMs were comparable to those in control hiPSC-CMs (SI Appendix, Fig. S2E). These data suggest that LAMP-2 deficiency did not induce apoptosis in Danon hiPSC-CMs.
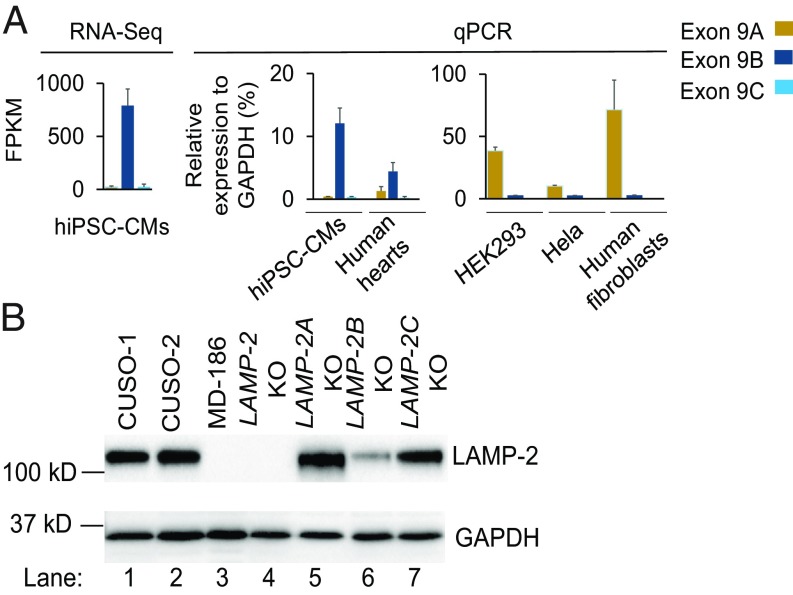
The three LAMP-2 isoforms, LAMP-2A, LAMP-2B, and LAMP-2C, share an identical lysosomal domain at their N terminus, but have distinct transmembrane and C-terminal cytosolic domains composed of 11 amino acids (SI Appendix, Fig. S3A). RNA-seq and real-time PCR (qPCR) analysis demonstrated that LAMP-2B was predominantly expressed in both human and mouse CMs (Fig. 1A and SI Appendix, Fig. S3B). In contrast, LAMP-2A was highly enriched in non-CMs such as HEK293, HeLa, and human skin fibroblasts (Fig. 1A). To examine biological functions of LAMP-2 in human CMs, we used CRISPR/Cas9 genome editing technology (22) to generate isogenic LAMP-2 knockout (LAMP-2 KO) hiPSC lines by deleting exon 2 in the control CUSO-1 hiPSC line, as well as LAMP-2 isoform-specific knockout lines, LAMP-2A KO, LAMP-2B KO, and LAMP-2C KO, in the control CUSO-2 hiPSC line by targeting exon 9A, exon 9B, and exon 9C, respectively (SI Appendix, Fig. S4). The LAMP-2 KO hiPSC line contains a frameshift mutation and does not express LAMP-2 protein (Fig. 1B). Use of these isogenic hiPSC-CM lines can avoid the interference of genetic background in elucidating the functions of LAMP-2 isoforms. In hiPSC-CMs, deletion of the LAMP-2B isoform decreased total LAMP-2 protein expression by more than 70%. The total LAMP-2 protein levels in LAMP-2A KO and LAMP-2C KO hiPSC-CMs were comparable to that in wild-type hiPSC-CMs (Fig. 1B and SI Appendix, Fig. S3C). These data strongly indicate that LAMP-2B is the predominant isoform expressed in CMs.
Fig. 1.
LAMP-2B is the predominant LAMP-2 isoform expressed in human CMs. (A) Expression of the three LAMP-2 isoforms in different types of cells was examined by RNA-seq or qPCR. RNAs from three human hearts were used for qPCR. FPKM, fragments per kilobase of exon per million reads. (B) Immunoblotting analysis of LAMP-2 expression in indicated hiPSC-CMs. Quantification of LAMP-2 from three independent experiments is shown in SI Appendix, Fig. S3C.
Deletion of LAMP-2B in Human CMs Causes Defects in Autophagosome–Lysosome Fusion.
Deletion of all three LAMP-2 isoforms in mice causes accumulation of autophagosomes (16). We examined which isoform caused this phenotype. First, we determined levels of microtubule-associated protein light chain 3 (LC3), and LC3-II, the membrane form of LC3 and an autophagosomal marker, by immunoblotting. The amount of LC3-II is correlated with autophagosome number (1). Levels of LC3-II were similar in HEK293, human fibroblasts, and hiPSC-CMs under regular conditions, whereas addition of bafilomycin A1, a blocker of autophagosome–lysosome fusion and/or lysosomal degradation, increased LC3-II levels more in hiPSC-CMs than in HEK293 and human fibroblasts (SI Appendix, Fig. S5A), suggesting that autophagic turnover occurs at a faster rate in human CMs. Thus, the hiPSC-CM platform is a suitable model to study autophagy. Next, we examined whether autophagy was altered in Danon hiPSC-CMs wherein the LAMP-2 protein was not detectable. Compared with control hiPSC-CMs, Danon and LAMP-2 KO hiPSC-CMs exhibited increased levels of LC3-II, especially under starvation conditions (SI Appendix, Fig. S5 B–E). Interestingly, LC3-II levels in skin fibroblasts derived from Danon patients were comparable to those in controls (SI Appendix, Fig. S5B), indicating that autophagosomes in CMs might be removed by mechanisms distinct from those in fibroblasts. Consistent with immunoblotting, greater numbers of LC3 puncta, which represent autophagosomes or related membrane structures, accumulated in Danon hiPSC-CMs under starvation conditions (SI Appendix, Fig. S5F). These data suggest that LAMP-2 deficiency causes accumulation of autophagosomes in human CMs, as observed in LAMP-2 KO mice (16).
Next, we sought to determine which LAMP-2 isoform participates in autophagy. LAMP-2B deficiency led to accumulation of LC3-II in hiPSC-CMs, whereas LAMP-2A or LAMP-2C deficiency did not (Fig. 2A and SI Appendix, Fig. S5G). Levels of LC3-II in LAMP-2B KO hiPSC-CMs were comparable to those in Danon and LAMP-2 KO hiPSC-CMs (Fig. 2B and SI Appendix, Fig. S5H, lanes 3, 4, 5, 8, 9, and 10). We performed further rescue experiments using LAMP-2B KO hiPSC-CMs with adenovirus carrying either LAMP-2A or LAMP-2B. Adenoviral infection did not change hiPSC-CM morphology (SI Appendix, Fig. S6A). Forced expression of LAMP-2B suppressed the LC3-II accumulation caused by LAMP-2B deficiency. In contrast, overexpression of LAMP-2A did not decrease LC3-II accumulation (Fig. 2C and SI Appendix, Fig. S6B, lanes 2–4). These data suggest that LAMP-2B, but not LAMP-2A or LAMP-2C, is required for autophagy in human CMs.
Fig. 2.
LAMP-2B deficiency is sufficient to cause defects in autophagosome–lysosome fusion. (A) Autophagic flux in control and LAMP-2 isoform-specific knockout hiPSC-CMs under regular and starvation conditions was assayed. Cells were cultured in either regular or starvation medium with or without 400 nM bafilomycin A1 for 4 h followed by immunoblotting analysis of indicated proteins. Quantification of LC3-II/GAPDH from three independent experiments is shown in SI Appendix, Fig. S5G. (B) Autophagic flux in indicated hiPSC-CMs was assayed. Quantification of LC3-II/GAPDH from three independent experiments is shown in SI Appendix, Fig. S5H. (C) LAMP-2B KO hiPSC-CMs were infected with adenovirus carrying either LAMP-2A (Ad-LAMP-2A) or LAMP-2B (Ad-LAMP-2B). Three days later, autophagic flux was assayed. Quantification of LC3-II/GAPDH from three independent experiments is shown in SI Appendix, Fig. S6B. (D and E) Monitoring autophagic flux in indicated hiPSC-CMs by using mRFP-EGFP-LC3. Indicated hiPSC-CMs were infected with adenovirus carrying mRFP-EGFP-LC3. hiPSC-CMs were cultured in regular or starvation medium for 4 h followed by imaging. Representative confocal images and statistical analysis of GFP+, RFP+, and GFP−, RFP+ puncta are shown in D and E, respectively. Fifteen to 20 cells for each cell line per condition were analyzed. Data are presented as mean + SD. *P < 0.05 as assessed by Student’s t test. (Scale bars, 10 μm.) (F and G) Representative confocal images of indicated hiPSC-CMs stained for LC3 (green), LysoTracker (red), and nuclei (blue). hiPSC-CMs were starved for 4 h, followed by immunostaining and confocal imaging sequentially. Statistical analysis of colocalization of LC3 puncta and LysoTracker in hiPSC-CMs (20 cells for each group) in G. Data are presented as mean + SD for LC3 puncta/lysosomes, or mean − SD for LC3 puncta alone. (Scale bars, 5 μm.)
This accumulation of autophagosomes could be due to increased induction of autophagy and/or blocked autophagosome–lysosome fusion. To distinguish between these possibilities, we performed autophagic flux assays by using an mRFP-EGFP-LC3 tandem construct wherein the GFP signal can be quenched by the acidic lysosomal pH (23–25). Thus, autophagosomes are marked by both RFP and GFP signals (yellow). After fusion with lysosomes to become autolysosomes, only RFP signals (red) can be observed. If induction of autophagy is increased, both yellow puncta and red puncta are increased. However, if fusion between autophagosomes and lysosomes is blocked, only yellow punctae are increased. Danon and LAMP-2B KO hiPSC-CMs showed more yellow puncta but fewer red puncta compared with control hiPSC-CMs under both regular and starved conditions (Fig. 2 D and E), indicating that autophagosome fusion/maturation into autolysosome is blocked in Danon or LAMP-2B KO hiPSC-CMs. However, a similar fraction of autolysosomes was observed in Danon and LAMP-2B KO hiPSC-CMs (Fig. 2 D and E), suggesting LAMP-2B–independent autophagic fusion mechanisms in CMs. Second, we measured autophagic flux by immunoblotting analysis of LC3-II. When autophagic fusion was blocked by bafilomycin A1 in hiPSC-CMs under regular or starved conditions, levels of LC3-II in Danon or LAMP-2B KO hiPSC-CMs were similar to those in control hiPSC-CMs (Fig. 2 A–C), indicating normal induction of autophagic flux. Finally, knockout of LAMP-2B in hiPSC-CMs caused a reduction in colocalization of LC3 puncta with LysoTracker-labled late endosomes/lysosomes assayed by confocal imaging, which was also observed in Danon hiPSC-CMs (Fig. 2 F and G). Taken together, these results suggest that LAMP-2B deficiency in CMs cause accumulation of autophagosomes by blocking autophagosome–lysosome fusion.
The Cytosolic CCD of LAMP-2B Is Required for It To Promote the Formation of the ATG14–VAMP8 Complex.
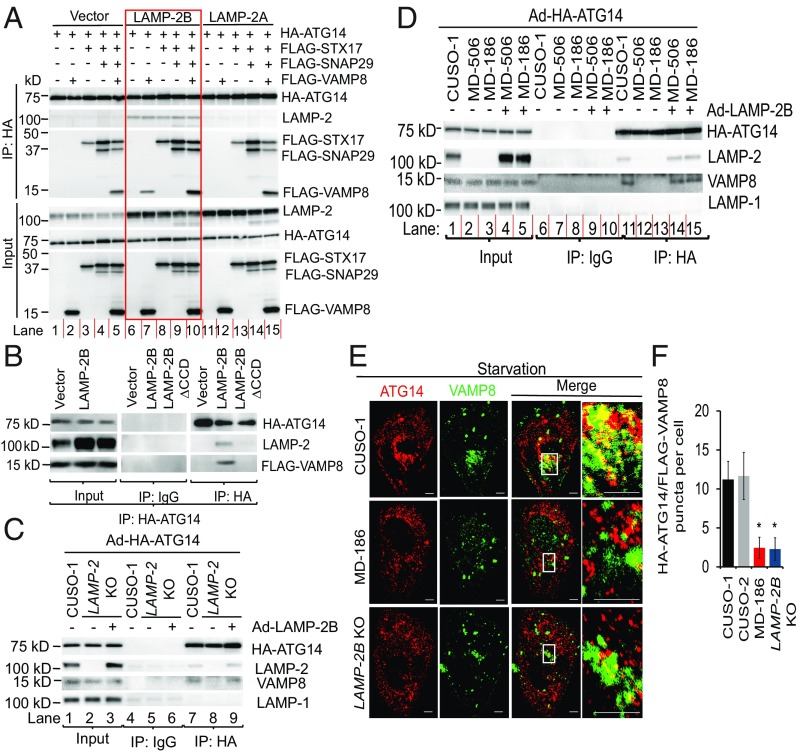
In non-CMs, VAMP8 is required for autophagic fusion (5). STX17 acts as a key player in fusion by interacting with SNAP-29 and VAMP8 (6). ATG14 on autophagosomes promotes fusion by directly binding to the STX17–SNAP29 binary complex (7). Because LAMP-2B deficiency leads to defects in autophagic fusion, we hypothesized that LAMP-2B would promote fusion by interacting with these proteins. To test this hypothesis, we performed coimmunoprecipitation assays in HEK293 cells wherein basal and starvation-induced autophagy has been well-characterized (26). HA-ATG14 formed a complex with FLAG-STX17 and FLAG-SNAP29 (Fig. 3A, lanes 3–5), but not with FLAG-VAMP8 alone in HEK293 cells in which LAMP-2B is barely expressed (Fig. 3A, lane 2). Strikingly, HA-ATG14 was precipitated with VAMP8 when LAMP-2B was overexpressed, but overexpression of LAMP-2A did not lead to formation of the ATG14–VAMP8 complex (Fig. 3A, lanes 2, 7 and 12). Deletion of the C-terminal cytosolic CCD of LAMP-2B, composed of 11 amino acids (LAMP-2B∆CCD), completely abolished the interaction between ATG14 and VAMP8 (Fig. 3B). ATG14 and VAMP8 formed a complex in hiPSC-CMs, which natively express LAMP-2B (Fig. 3C, lane 7 and Fig. 3D, lane 11). The ATG14–VAMP8 complex was also disrupted in LAMP-2 KO hiPSC-CMs and hiPSC-CMs derived from patients with Danon disease (Fig. 3C, lane 8 and Fig. 3D, lanes 12 and 13). Conversely, overexpression of LAMP-2B restored the ATG14–VAMP8 complex in LAMP-2 KO or Danon hiPSC-CMs (Fig. 3C, lane 9 and Fig. 3D, lanes 14 and 15). Finally, colocalization of HA-ATG14 with FLAG-VAMP8 was significantly decreased in both LAMP-2B KO and Danon hiPSC-CMs (Fig. 3 E and F). These data indicate that LAMP-2B promotes formation of the ATG14–VAMP8 complex.
Fig. 3.
The CCD of LAMP-2B is required for it to promote formation of the ATG14–VAMP8 complex. (A) HEK293 cells were transfected with HA-ATG14 plus empty vector, LAMP-2B shown in red box, or LAMP-2A with or without FLAG-STX17, FLAG-SNAP29, and FLAG-VAMP8. Two days later, cells were lysed and immunoprecipitated with anti-HA antibody. Immunoblotting was performed with anti-HA, anti-LAMP-2, and anti-FLAG antibodies. (B) HEK293 cells were transfected with HA-ATG14 and FLAG-VAMP8 with empty vector, LAMP-2B, or LAMP-2B∆CCD. Two days later, cells were lysed and immunoprecipitated with anti-HA antibody, followed by immunoblotting with anti-LAMP-2, anti-FLAG, or anti-HA antibodies. (C and D) Control, LAMP-2 KO (C), or Danon (D) hiPSC-CMs were infected with adenovirus carrying HA-ATG14 (Ad-HA-ATG14) with or without coinfection with adenovirus carrying LAMP-2B (Ad-LAMP-2B). Three days later, CMs were lysed and immunoprecipitated with anti-HA antibody, followed by immunoblotting analysis for indicated proteins. (E and F) Representative confocal images of indicated hiPSC-CMs stained for HA-ATG14 (red) and FLAG-VAMP8 (green) in E. hiPSC-CMs were infected with adenovirus carrying HA-ATG14 or FLAG-VAMP8. Three days later, hiPSC-CMs were starved for 2 h followed by immunostaining for HA-ATG14 and FLAG-VAMP8. White boxes are enlarged in Insets. Quantification of colocalization of ATG14 and VAMP8 is shown in F. n = 17–24 CMs, Student’s t test, *P < 0.0001. Data are presented as mean ± SD. (Scale bars, 5 μm.)
The CCD of LAMP-2B Is Required To Promote Autophagosome–Lysosome Fusion in a STX17-Independent Manner.
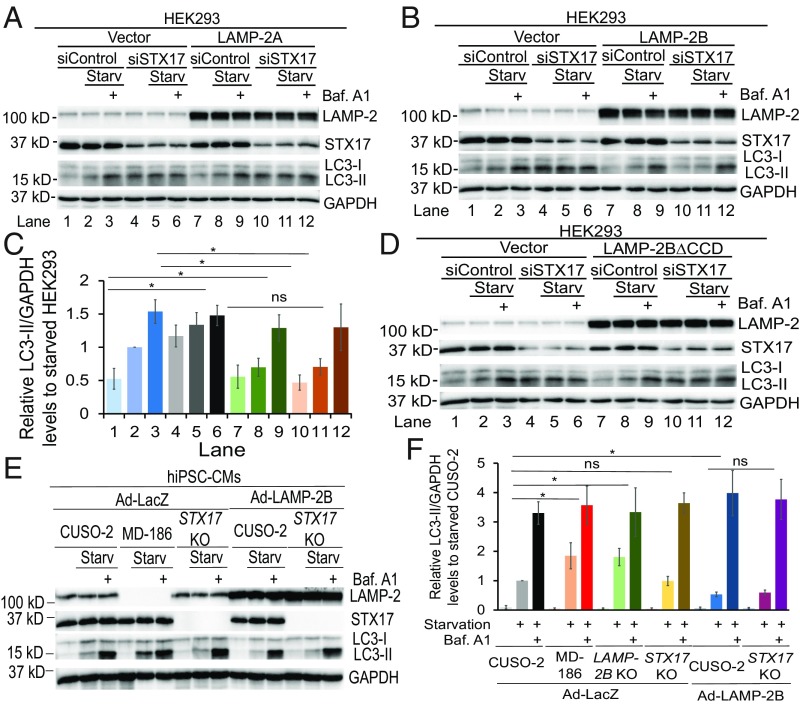
LAMP-2B promotes formation of the ATG14–VAMP8 complex. We hypothesized that LAMP-2B could therefore promote autophagic fusion. In HEK293 cells, small interfering RNA (siRNA)-mediated knockdown of STX17 caused significant accumulation of LC3-II (Fig. 4 A–C, lanes 4 and 5 versus 1 and 2), confirming the essential role of STX17 in autophagic fusion (6, 7). Expression of LAMP-2B, but not LAMP-2A, suppressed the accumulation of LC3-II caused by knockdown of STX17 (Fig. 4 A–C and SI Appendix, Fig. S7A, lanes 4 and 5 versus 10 and 11). Importantly, expression of LAMP-2B led to similar LC3-II levels in control siRNA- and STX17 siRNA-treated HEK293 cells (Fig. 4 B and C, lanes 7 and 8 versus 10 and 11). Addition of bafilomycin A1 to block fusion led to similar LC3-II levels in both control siRNA- and STX17 siRNA-treated cells expressing either vector or LAMP-2B (Fig. 4 B and C, lanes 3, 6, 9, and 12). These data suggest that LAMP-2B promotes autophagic fusion, rather than inhibiting the induction of autophagy. In contrast, expression of LAMP-2B∆CCD could not suppress the accumulation of LC3-II caused by STX17 knockdown (Fig. 4D and SI Appendix, Fig. S7B, lanes 10 and 11 versus 4 and 5). These data suggest that the CCD of LAMP-2B is essential for it to decrease the accumulation of LC3-II caused by knockdown of STX17 in HEK293 cells.
Fig. 4.
The CCD is required for LAMP-2B to promote autophagosome–lysosome fusion, independently of STX17. Starv, starvation. (A–D) Immunoblotting analysis of indicated proteins in HEK293 cells. HEK293 cells transfected with empty vector, LAMP-2A, LAMP-2B, or LAMP-2B∆CCD were treated with siRNAs against luciferase (siControl) or STX17. Three days later, cells were cultured in regular or starvation medium for 4 h with or without 400 nM of bafilomycin A1 (Baf. A1). Western blotting was performed with indicated antibodies. Densitometry quantification of LC3-II/GAPDH from three independent experiments in A is shown in SI Appendix, Fig. S7A; B is shown in C; and D is shown in SI Appendix, Fig. S7B, with data being normalized to siControl-treated cells under starved conditions in each experiment. Data are presented as mean ± SD. *P < 0.05 as assessed by Student’s t test. ns, not significant. (E and F) Immunoblotting analysis of indicated proteins in indicated hiPSC-CMs. hiPSC-CMs infected with adenovirus carrying LacZ (Ad-LacZ) or LAMP-2B (Ad-LAMP-2B). Autophagic flux was assayed as described in A–D. Densitometry quantification of LC3-II/GAPDH is shown in F, with data being normalized to control hiPSC-CMs under starved conditions in each experiment. Data are presented as mean ± SD. *P < 0.05 as assessed by Student’s t test. n = 4. ns, not significant.
To determine whether STX17 is essential for LAMP-2B to promote autophagic fusion in human CMs, we deleted exon 4 of STX17 gene in hiPSCs (STX17 KO) using CRISPR/Cas9 technology. The excision of exon 4 results in a frame-shift mutation. Only the first 66 amino acids of STX17 are assumed to be produced, with no detectable expression (SI Appendix, Fig. S7 C–E). Surprisingly, neither knockdown or knockout of STX17 caused accumulation of LC3-II in hiPSC-CMs under either regular or starvation conditions (Fig. 4 E and F and SI Appendix, Fig. S7 F–H). Of note, in hiPSC-CMs, LAMP-2B deficiency caused a significant increase in LC3-II under starvation conditions (Fig. 4F), implying that LAMP-2B plays a more important role than STX17 in autophagic fusion in human CMs. Overexpression of LAMP-2B reduced LC3-II levels similarly in both wild-type and STX17 KO hiPSC-CMs (Fig. 4 E and F). These data indicate that STX17 is not essential for autophagy in CMs. LAMP-2B functions in autophagic fusion independently of STX17.
ATG14 and VAMP8 Are Essential for LAMP-2B to Promote Autophagosome–Lysosome Fusion in Human CMs.
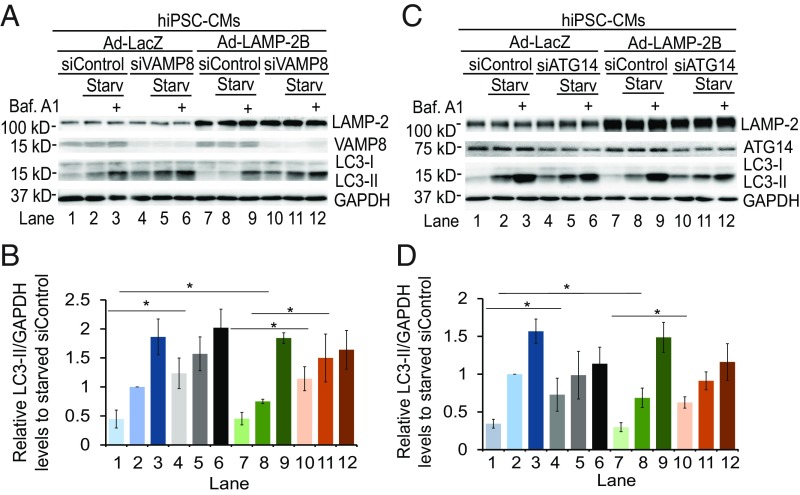
The CCD of LAMP-2B is required not only for it to promote formation of the ATG14–VAMP8 complex, but also to complete the fusion step of autophagy. Therefore, we examined whether ATG14 and/or VAMP8 are required for LAMP-2B to promote autophagic fusion. Knockdown of VAMP8 caused accumulation of LC3-II in HEK293 cells and hiPSC-CMs under regular and starvation conditions (Fig. 5 A and B and SI Appendix, Fig. S8 A and B, lanes 4 and 5), confirming an essential role of VAMP8 in fusion (5). Overexpression of LAMP-2B significantly decreased accumulation of LC3-II, especially under starvation conditions (Fig. 5 A and B and SI Appendix, Fig. S8 A and B, lanes 2 and 8). However, knockdown of VAMP8 totally abolished the ability of LAMP-2B to suppress accumulation of LC3-II (Fig. 5 A and B and SI Appendix, Fig. S8 A and B, lanes 7 and 8 versus 10 and 11). Down-regulation of VAMP8 did not affect lysosomal biogenesis in hiPSC-CMs as assayed by flow cytometry analysis of LysoTracker intensity (SI Appendix, Fig. S8 C and D), consistent with previous findings that knockdown of VAMP8 did not affect lysosomal biogenesis and function (5). These data suggest that VAMP8 is required for LAMP-2B to promote autophagy-specific fusion in both non-CMs and CMs.
Fig. 5.
Both VAMP8 and ATG14 are required for LAMP-2B to promote fusion between autophagosomes and endosomes/lysosomes for degradation. Immunoblotting analysis of indicated proteins in hiPSC-CMs. hiPSC-CMs infected with adenovirus carrying LacZ (Ad-LacZ) or LAMP-2B (Ad-LAMP-2B) were treated with siRNAs against luciferase, VAMP8 (in A and B) or ATG14 (in C and D). Three days later, cells were cultured in regular or starvation medium for 4 h with or without 400 nM bafilomycin A1 (Baf. A1). Western blotting was performed with anti-LAMP-2, anti-LC3, anti-VAMP8 or anti-ATG14, and anti-GAPDH antibodies. Quantification of LC3-II/GAPDH from three independent experiments is shown in B and D, with data being normalized to siControl-treated cells under starved conditions in each experiment. Data are presented as mean ± SD. *P < 0.05 as assessed by Student’s t test. Starv, starvation.
ATG14 is a key player not only in autophagic fusion by interacting with STX17-SNAP29 (7) but also in induction of autophagy (27, 28). Knockdown of ATG14 in HEK293 or hiPSC-CMs increased LC3-II levels under regular conditions compared with cells treated with control siRNA (Fig. 5 C and D and SI Appendix, Fig. S8 E and F, lane 1 versus 4), suggesting that ATG14 is required for clearance of autophagosomes. However, knockdown of ATG14 also decreased LC3-II levels caused by bafilomycin A1 treatment under starvation conditions, suggesting that ATG14 also participated in induction of autophagy, especially under starvation conditions. Importantly, knockdown of ATG14 completely abolished the effect of LAMP-2B overexpression on decreasing LC3-II levels (Fig. 5 C and D and SI Appendix, Fig. S8 E and F, lanes 7 and 8 versus 10 and 11). These data suggest that ATG14 is required for LAMP-2B to promote autophagic fusion.
LAMP-2 Mutation Is Sufficient To Cause Danon Phenotype.
To better understand the mechanisms of action of LAMP-2, we examined global gene expression in six hiPSC-CM lines. This RNA-seq analysis identified 420 differentially expressed genes, which demonstrated a greater than 1.5-fold change in expression between control and Danon hiPSC-CMs (SI Appendix, Fig. S9A). Global gene expression in LAMP-2 KO hiPSC-CMs is very similar to that in Danon hiPSC-CMs (SI Appendix, Fig. S9B). Gene ontology analysis demonstrated that 150 differentially expressed genes with known functions were involved in metabolic processes (SI Appendix, Fig. S9C), suggesting potential metabolic abnormalities in LAMP-2–deficient hiPSC-CMs. Mitochondria have a central role in the regulation of cellular metabolism. In CMs, autophagy is crucial for mitochondrial homeostasis by removal of unhealthy mitochondria with decreased membrane potential (29). Removal of depolarized mitochondria by autophagy is initiated by accumulation of PTEN-induced putative kinase 1 (PINK1) on the outer membrane (29, 30). As LAMP-2 KO and Danon hiPSC-CMs exhibit defects in autophagosome–lysosome fusion, we hypothesized that LAMP-2 deficiency could lead to mitochondrial and metabolic defects. To test this hypothesis, we analyzed mitochondrial membrane potential using flow cytometry with tetramethylrhodamine methyl ester (TMRM). The mitochondrial membrane potential of Danon and LAMP-2 KO hiPSC-CMs was significantly lower than that of control hiPSC-CMs (SI Appendix, Fig. S9D), indicating mitochondrial depolarization. Immunoblotting analysis revealed that PINK1 was accumulated in the mitochondria of Danon and LAMP-2 KO hiPSC-CMs (SI Appendix, Fig. S9E). Finally, electron microscopy analysis demonstrates that some autophagosomes contained mitochondria (SI Appendix, Fig. S2C), suggesting dysregulation of both general and mitochondrial autophagy in Danon hiPSC-CMs. These data indicate that removal of depolarized mitochondria by autophagy is impaired in LAMP-2–deficient hiPSC-CMs.
Next, we assessed mitochondrial morphology and function in control, Danon, and LAMP-2 KO hiPSC-CMs. Mitochondrial fragmentation was more evident in LAMP-2–deficient hiPSC-CMs than control (SI Appendix, Fig. S9F). Accumulation of the short isoform of optic atrophy 1 (S-OPA1) is associated with mitochondrial fission and fragmentation (31). Levels of S-OPA1 were increased in LAMP-2–deficient hiPSC-CMs (SI Appendix, Fig. S9 G and H). LAMP-2–deficient hiPSC-CMs also produced significantly lower levels of ATP than control hiPSC-CMs (SI Appendix, Fig. S10A). Next, we analyzed mitochondrial function by measuring the oxygen consumption rate (OCR) of hiPSC-CMs cultured in one of three different energy sources: glucose, galactose, and lactate. OCR of basal respiration, ATP production, and maximal respiratory capacity (MRC) were significantly decreased in Danon hiPSC-CMs cultured with each of these three energy sources (SI Appendix, Fig. S10 B and C). Taken together, these results demonstrate that LAMP-2 deficiency profoundly impaired mitochondrial function in CMs.
While LAMP-2–deficient hiPSC-CMs uniformly exhibit mitochondrial abnormalities, certain phenotypes observed in hiPSC-CMs vary from patient to patient (SI Appendix, Fig. S10C). These data are consistent with the clinical observation that Danon patients display variations in disease onset and severity (11). An individual patient’s genetic background may contribute to the variations in clinical features. Patient-derived hiPSCs could potentially have numerous other genetic differences besides LAMP-2 mutations, which might also contribute to progression of Danon cardiomyopathy. To determine whether the LAMP-2 mutation is sufficient and necessary to cause Danon cardiomyopathy, we corrected the LAMP-2 mutation in MD-186 hiPSCs using a CRISPR-based approach. A single guide RNA was designed to target exon 3 near the mutation site (c.247C > T). The T base responsible for the point mutation was replaced with C through homology-directed repair between the mutated gene segment and a 100-mer single-strand DNA oligonucleotides containing normal LAMP-2 sequence (SI Appendix, Fig. S11A). Corrected hiPSC (hereinafter referred to as MD-186C) clones were confirmed by sequencing and immunoblotting for LAMP-2 protein (SI Appendix, Fig. S11 B and C). First, immunoblotting revealed similar LC3-II levels in mutation-corrected MD-186C hiPSC-CMs, compared with control (Fig. 6 A and B). Autophagic flux assays by the mRFP-EGFP-LC3 construct demonstrated that correction of the LAMP-2 mutation restored autophagosome–lysosome fusion (Fig. 6 C and D). Additionally, while Danon hiPSC-CMs produced significantly lower levels of ATP than control hiPSC-CMs, correction of the LAMP-2 mutation significantly increased ATP content, reaching a similar level to control (Fig. 6E). Finally, OCR of basal respiration, ATP production, and MRC were significantly increased in MD-186C hiPSC-CMs to levels comparable to control (Fig. 6F). These data indicate that correction of the LAMP-2 mutation (c.247C > T) is sufficient to completely rescue Danon phenotype.
Fig. 6.
Correction of the LAMP-2 mutation rescues functional abnormalities in human CMs. (A and B) Autophagic flux measured by LC3-II in control, MD-186, and corrected MD-186C hiPSC-CMs was assayed. Quantification of LC3-II/GAPDH from three independent experiments is shown in B. Data are presented as mean ± SD. *P < 0.05 as assessed by Student’s t test. ns, not significant. (C and D) Monitoring autophagic flux in indicated hiPSC-CMs by using mRFP-EGFP-LC3. hiPSC-CMs were infected with adenovirus carrying mRFP-EGFP-LC3. hiPSC-CMs were cultured in regular or starvation medium for 4 h followed by imaging. Representative fluorescent images and statistical analysis of GFP+/RFP+ and GFP−/RFP+ puncta are shown in C and D, respectively. Twenty cells for each condition were analyzed. Data are presented as mean ± SD. *P < 0.05 as assessed by Student’s t test. ns, not significant. (E) Cellular ATP content in control, MD-186, and MD-186C hiPSC-CMs was measured. Student’s t test, *P < 0.005, n = 3. Data are presented as mean ± SD. ns, not significant. (F) OCR of control, MD-186, and MD-186C hiPSC-CMs to indicate mitochondrial function was measured using the Seahorse XF Cell Mito Stress Test Kit. Representative time course data for indicated hiPSC-CMs are shown at Left. Data are presented as mean ± SD. Statistical analysis of OCR are shown at Right. Student’s t test, *P < 0.05, n = 3. Data are presented as mean ± SD. ns, not significant.
Taken together, these data indicate that LAMP-2 deficiency is sufficient and necessary to cause Danon cardiomyopathy.
LAMP-2B Deficiency in Human CMs Causes Mitochondrial and Contractile Abnormalities.
To examine whether LAMP-2B is responsible for the mitochondrial defects of Danon disease, we next assessed mitochondrial morphology and function in LAMP-2B KO hiPSC-CMs. LAMP-2B KO, LAMP-2 KO, or Danon hiPSC-CMs produced significantly lower levels of ATP than control hiPSC-CMs. However, knockout of LAMP-2A or LAMP-2C did not decrease ATP production (Fig. 7A). Next, OCR of basal respiration, ATP production, and MRC were significantly decreased in LAMP-2B KO hiPSC-CMs, reaching a similar level to Danon hiPSC-CMs. However, deletion of LAMP-2A or LAMP-2C in hiPSC-CMs did not cause mitochondrial dysfunction (Fig. 7 B and C). Taken together, these results demonstrate that LAMP-2B is specifically responsible for metabolic defects in Danon hiPSC-CMs.
Fig. 7.
LAMP-2B deficiency causes mitochondrial and contractile abnormalities in human CMs. (A) ATP levels in indicated hiPSC-CMs is shown. n = 3, Student’s t test, *P < 0.01. Data are presented as mean + SD. (B and C) Mitochondrial function indicated by OCR of hiPSC-CMs was measured using the Seahorse XF Cell Mito Stress Test Kit. Representative time course data for indicated hiPSC-CMs are shown. Data are expressed as mean ± SD. Quantification of OCR is shown in C. n = 3. Data are presented as mean + SD, Student’s t test, *P < 0.05 versus control. ns, not significant. (D) Maximal tension generated by myofibrils isolated from hearts of control and patients with Danon disease. Nine to 16 myofibrils were isolated from cardiac tissue from each patient. Three donors and three Danon disease hearts were analyzed, respectively. Each point represents the maximal tension of a single myofibril of an individual patient. Data are presented as mean ± SD. P values were obtained from a one-way ANOVA with a Tukey’s multiple comparison test, *P < 0.05. (E–G) Twitch force produced by single hiPSC-CMs on day 60. Force of control and LAMP-2 KO hiPSC-CMs measured from the six to seven independent experiments (with ∼25 cells each) are plotted together in E. Force of control and Danon hiPSC-CMs measured from the three independent experiments (with ∼25 cells each) are plotted together in F. Twitch force of Danon (MD-186) and genetics-corrected (MD-186C) hiPSC-CMs measured from the four to five independent experiments (with ∼25 cells each) are plotted together in G. Each data point represents the average force per post of an individual cell. Lines represent average force per post for each group. The reported P value was obtained from a one-way ANOVA with a Bonferroni post hoc test, *P < 0.05.
Having established the importance of the LAMP-2B isoform for Danon disease, we next investigated whether we could recapitulate whole-heart phenotypes of Danon disease using hiPSC-CMs. Danon patients develop cardiomyopathy, which is often accompanied by impaired contractile function (10). Myofibrils isolated from hearts of patients with Danon disease generated reduced maximal tension compared with myofibrils from control hearts (Fig. 7D). We therefore examined whether LAMP-2 KO or Danon hiPSC-CMs could recapitulate the contractile abnormalities of Danon hearts. To characterize contractile force produced by single hiPSC-CMs, we used a platform of micropost arrays (32). LAMP-2 KO or Danon hiPSC-CMs generated less contractile force per post than control hiPSC-CMs (Fig. 7 E and F and SI Appendix, Fig. S12 A and B). Importantly, genetic correction of the LAMP-2 mutation (c.247C > T), which restored the LAMP-2 expression (SI Appendix, Fig. S11 B and C), significantly increased contractile force generation of hiPSC-CMs (Fig. 7G). These data suggest that LAMP-2B, accounting for >70% of LAMP-2 protein in CMs, is required for CM as well as whole heart function.
Discussion
We have identified previously undefined biological and molecular roles for LAMP-2B in the control of fusion between autophagosomes and endosomes/lysosomes via an interaction among ATG14, VAMP8, and LAMP-2B. LAMP-2B deficiency caused defects in autophagy in hiPSC-CMs by disrupting this STX17-independent autophagosome–lysosome fusion in human CMs, leading to Danon cardiomyopathy (SI Appendix, Fig. S13). Gene correction of the LAMP-2 mutation rescues the Danon phenotype (Figs. 6 and 7G), implying a potential therapy for this devastating disease.
Autophagic turnover occurs at a faster rate in CMs than in non-CMs (SI Appendix, Fig. S5A), suggesting an important role for autophagy in cardiac homeostasis. Here, we identified a distinct autophagic pathway in CMs mediated by LAMP-2B (SI Appendix, Fig. S13). In non-CMs where LAMP-2A is predominant, STX17 acts as an essential molecule to mediate fusion by interacting with SNAP29 and VAMP8. In these cells, knockdown of STX17 caused accumulation of LC3-II. In CMs, knockout of STX17 failed to do so (Fig. 4 E and F), suggesting a unique mechanism in CMs that is distinct from that mediated by STX17. Our studies demonstrate that LAMP-2B mediates STX17-independent autophagic fusion by interacting with ATG14 and VAMP8 through its C-terminal CCD. However, we cannot completely exclude the possibility that LAMP-2B promotes autophagic fusion by interacting with other molecules. Knockout of LAMP-2B did not completely block fusion between autophagosomes and lysosomes (Fig. 2 D and E), indicating that LAMP-2B–independent pathways are also involved in this process in CMs. We also found that deficiency of LAMP-2B rather than LAMP-2A or LAMP-2C causes metabolic and autophagic abnormalities in human CMs. This finding is strongly supported by clinical data. Although most Danon patients carry mutations that result in deficiency of all three LAMP-2 isoforms, some Danon patients carry mutations that only lead to LAMP-2B deficiency (9, 11), supporting the notion that LAMP-2B deficiency is both necessary and sufficient for Danon pathologenesis.
LAMP-2B interacts with ATG14 and VAMP8 through its CCD tail. Interestingly, the CCD of ATG14 interacts with the helix motif of STX17 (7). This interaction may stabilize the STX17–SNAP29 complex, which is important for its fusion activity by further interacting with the helix domain of VAMP8 (7). It would be interesting to determine whether the interaction between the CCD of LAMP-2B and the VAMP8 helical domain allows the complex to be more susceptible to interacting with autophagosomal proteins such as ATG14 for fusion. Both ATG14 and VAMP8 are required for LAMP-2B to promote autophagy-specific fusion in CMs. However, it is also likely that LAMP-2B regulates cardiac autophagy by interacting with other molecules besides ATG14 and VAMP8. In addition, the mechanism by which LAMP-2B is recruited to interact with VAMP8 and ATG4 is not known.
Intensive studies have conclusively demonstrated that LAMP-2A is essential for CMA by serving as a receptor for heat shock proteins (15). It was previously reported that LAMP-2A is required for autophagy in MEFs (17). However, our studies, as well as those by others, do not support this conclusion. First, overexpression of LAMP-2A in different types of cells did not change LC3-II levels (ref. 33; Figs. 2C and 4A). Second, LAMP-2A deficiency in hiPSC-CMs did not cause accumulation of LC3-II under either regular or starvation conditions (Fig. 2A). Third, LAMP-2 deficiency in skin fibroblasts derived from Danon patients did not cause accumulation of LC3-II under either regular or starvation conditions, compared with control fibroblasts (SI Appendix, Fig. S5B), despite LAMP-2A representing the dominant isoform in these cells. Therefore, LAMP-2A is dispensable for autophagosome-mediated autophagy.
Using Danon hiPSC-CMs, it was previously reported that 20–25% of Danon hiPSC-CMs underwent apoptosis (34). Subsequently, the same group used Danon hiPSC-CM lines to characterize mitochondrial function that was depressed (35). However, whether the observed decreased mitochondrial function is due to CM apoptosis in their Danon hiPSC-CM samples or due to some other mechanisms remains unclear. Controversially, significant apoptosis in Danon hiPSC-CMs was not detected in a more recent study (13). Consistently with these findings by Yoshida et al. (13), we also did not detect significant CM apoptosis in Danon or LAMP-2 KO hiPSC-CM lines cultured for 50 or 100 days (SI Appendix, Fig. S2D). In addition, if 20–25% of CMs as described by Hashem et al. (34) were apoptotic in hearts of patients with Danon disease, it is hard to imagine that these patients could survive beyond their teens. Therefore, CM apoptosis is unlikely to act as the primary contributor to Danon cardiomyopathy.
Mitochondrial dysfunction is often observed in heart failure and ischemic heart disease. Therefore, improving mitochondrial function in CMs has been regarded as a potential therapy to improve the function of the failing heart (36). Mitochondrial function is impaired in LAMP-2B KO or Danon hiPSC-CMs, suggesting that further studies using the LAMP-2B KO or Danon hiPSC-CM platform could lead to novel mechanistic insights into the effects of mitochondrial dysfunction on the pathogenesis of cardiomyopathy, and could serve as a foundation to develop novel metabolism-focused therapies for heart failure.
Aside from cardiomyopathy, muscle weakness, retinopathy, and intellectual disabilities have been observed in patients with Danon disease (11). It is not known whether all of these clinical symptoms are due to defects in autophagic fusion mediated by LAMP-2B. Importantly, LAMP-2B is able to promote autophagic fusion not only in CMs, but also in HEK293 cells, suggesting a universal function for LAMP-2B in autophagy. Studies using neuronal progenitor cells or neurons derived from Danon, LAMP-2A KO, LAMP-2B KO, and LAMP-2C KO hiPSCs could provide novel insights into mental disabilities. These studies as well as our findings presented here could lead to the development of therapeutics for cardiomyopathy, retinopathy, and mental retardation.
Materials and Methods
Experimental Model and Subject.
The human subjects portion of this study was reviewed and approved by the Colorado Multiple Institutions Review Board (COMIRB no. 06–0452). Human hearts from healthy donors and patients with Danon Disease were obtained from a tissue bank maintained by the Division of Cardiology at the University of Colorado (COMIRB no. 01–568). All patients were followed by the University of Colorado Heart Failure Program and offered participation in the research protocol. All research involving animals complied with protocols approved by the Institutional Animal Care and Use Committee of University of Colorado.
Generation, Cardiac Differentiation, and Genome Editing of hiPSCs.
Human skin fibroblasts were reprogrammed into hiPSCs using either retroviral or Sendai viral vectors carrying Oct3/4, SOX2, KLF4, and C-MYC. hiPSCs were induced into CMs using a monolayer method (19), followed by lactate selection (20). CRISPR-mediated editing reagents were delivered into hiPSCs by electroporation. Details are included in SI Appendix, Materials and Methods.
Analysis of Glycogen, Vacuole Accumulation, and Cell Death.
hiPSC-CMs were analyzed by PAS staining, electron microscopy imaging, and TUNEL assays. Details are included in SI Appendix, Materials and Methods.
Starvation Assays.
For starvation, cells were washed with DPBS (Dulbecco’s phosphate-buffered saline) three times and incubated in starvation medium at 37 °C for 1, 2, or 4 h. For CMs, glucose-free DMEM (Gibco) was used as starvation medium; for non-CMs, the composition of the starvation medium is as described (26). To block autophagy flux, 400 nM bafilomycin A1 (Cayman) was added to the starvation medium.
siRNA Treatments.
Negative control and gene-specific siRNAs were purchased from Dharmacon. siRNAs were delivered into the target cells with Lipofectamine RNAiMAX (Invitrogen, for non-CMs) or Lipofectamine 3000 (Invitrogen, for CMs) reagents according to the manufacturer’s protocols. Cells were incubated with siRNA for 48 h before switching to regular culture medium for recovery. Cells were lysed at 72 h after infection for downstream protein analysis.
Examination of Gene Expression.
For immunostaining, Cells were fixed in 2% paraformaldehyde for 30 min at room temperature and then washed three times with DPBS. For immunoblotting, cells were washed with ice-cold DPBS twice before lysing in ice-cold lysis buffer. For transcriptome analysis, total RNA was extracted with TRIzol reagent (Invitrogen) and purified with the RNeasy Plus Universal Mini Kit (Qiagen), followed by qPCR analysis and deep sequencing. All RNA-seq data have been deposited in the Gene Expression Omnibus. Accession numbers for those experiments reported in this paper are GSE71405 for NMCMs (37), GSE102792 for adult mouse cardiomyocytes, and GSE108429 for hiPSC-CMs. Details are provided in SI Appendix, Materials and Methods.
Adenoviral Infection.
Adenovirus was generated using the ViraPower Adenoviral Gateway Expression Kit (Invitrogen). hiPSC-CMs were infected with a multiplicity of infection (MOI) of 5. Details are provided in SI Appendix, Materials and Methods.
Coimmunoprecipitation Assays.
Whole-cell lysates were collected after removal of cell debris by centrifugation. Five hundred micrograms of lysate was then incubated with the primary antibody and Dynabeads Protein G (Invitrogen) overnight at 4 °C to pull down protein complexes. Details are provided in SI Appendix, Materials and Methods.
Metabolic Assays.
Mitochondrial morphology and membrane potential were assayed with MitoTracker Orange (Molecular Probes) or TMRM (Molecular Probes). For mitochondrial function assays, Seahorse XF Cell Mito Stress tests were performed according to manufacturer’s instructions. Cellular ATP levels were measured by the ATP Bioluminescence Assay Kit HS II (Roche). ROS amount in hiPSC-CMs was analyzed with MitoSox (Molecular Probes). Details are provided in SI Appendix, Materials and Methods.
Imaging mRFP-EGFP-LC3 in hiPSC-CMs.
hiPSC-CMs were infected with Ad-mRFP-EGFP-LC3 at a MOI of 5. Four days later, half of cells were washed with DPBS three times and treated with starved medium for 4 h. Cells were incubated in DPBS for confocal imaging. Details are provided in SI Appendix, Materials and Methods.
Measurement of Human Heart Myofibril Mechanics.
Human hearts from patients with Danon disease were obtained from a tissue bank maintained by the Division of Cardiology at the University of Colorado. Hearts were collected at the time of orthotropic cardiac transplantation. Control hearts were obtained from unused donor hearts that could not be used for transplantation. Myofibril mechanics were quantified using the fast solution switching technique (38). Details are provided in SI Appendix, Materials and Methods.
Measurement of CM Contractility by Micropost Arrays.
hiPSC-CMs on day 50 were shipped to University of Washington. One week later, CMs were seeded on microposts for experiments in a blinded manner. Micropost experiments were blindedly conducted in the N.J.S. laboratory. Arrays of microposts were used to calculate the twitch force of individual cells following established protocols (32). Details are provided in SI Appendix, Materials and Methods.
Quantification and Statistical Analysis.
For quantification of Western blotting, gel images were quantified using densitometry analysis in ImageJ (NIH). Statistical analysis of quantified gels was conducted in GraphPad Prism (GraphPad Software Inc.), using Student’s t test. P values <0.05 were considered significant. When comparing the mean values of the twitch force between cell lines, a one-way ANOVA with a Bonferroni post hoc test was used. When comparing the mean values of the maximal tension of myofibrils between control and patients with Danon disease, a one-way ANOVA function in GraphPad with Tukey’s multiple comparison test was used. The resulting P value <0.05 was considered a significant difference between group means. Details are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by funds from the Boettcher Foundation, American Heart Association Grant 13SDG17400031, the University of Colorado Department of Medicine Outstanding Early Career Scholar Program, and NIH Grant R01HL133230 (to K.S.). N.J.S. was supported by a National Science Foundation Grant CBET-1509106. A.L. was supported by an NIH postdoctoral fellowship (F32HL126332). W.E.K. was supported by a postdoctoral fellowship from the University of Colorado Consortium for Fibrosis Research & Translation and Cardiology training Grant T32HL007822. This research was also supported by Grants P30CA046934 and P30AR057212 and the Flow Cytometry, Genomics Shared Resources at the University of Colorado Anschutz Medical Campus.
Footnotes
Conflict of interest statement: E.M.S. is the recipient of a research grant from Novartis Pharmaceuticals that is not related to the present study.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE102792 and GSE108429).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808618116/-/DCSupplemental.
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kitsis RN, Peng CF, Cuervo AM. Eat your heart out. Nat Med. 2007;13:539–541. doi: 10.1038/nm0507-539. [DOI] [PubMed] [Google Scholar]
- 3.Lavandero S, Chiong M, Rothermel BA, Hill JA. Autophagy in cardiovascular biology. J Clin Invest. 2015;125:55–64. doi: 10.1172/JCI73943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shirakabe A, Ikeda Y, Sciarretta S, Zablocki DK, Sadoshima J. Aging and autophagy in the heart. Circ Res. 2016;118:1563–1576. doi: 10.1161/CIRCRESAHA.116.307474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Diao J, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danon MJ, et al. Lysosomal glycogen storage disease with normal acid maltase. Neurology. 1981;31:51–57. doi: 10.1212/wnl.31.1.51. [DOI] [PubMed] [Google Scholar]
- 9.Nishino I, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 10.Maron BJ, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301:1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’souza RS, et al. Danon disease: Clinical features, evaluation, and management. Circ Heart Fail. 2014;7:843–849. doi: 10.1161/CIRCHEARTFAILURE.114.001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nascimbeni AC, Fanin M, Angelini C, Sandri M. Autophagy dysregulation in Danon disease. Cell Death Dis. 2017;8:e2565. doi: 10.1038/cddis.2016.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida S, et al. Characteristics of induced pluripotent stem cells from clinically divergent female monozygotic twins with Danon disease. J Mol Cell Cardiol. 2018;114:234–242. doi: 10.1016/j.yjmcc.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Eskelinen EL, et al. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic. 2005;6:1058–1061. doi: 10.1111/j.1600-0854.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- 15.Cuervo AM. Chaperone-mediated autophagy: Selectivity pays off. Trends Endocrinol Metab. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 17.Hubert V, et al. LAMP-2 is required for incorporating syntaxin-17 into autophagosomes and for their fusion with lysosomes. Biol Open. 2016;5:1516–1529. doi: 10.1242/bio.018648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tohyama S, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Fanin M, et al. Generalized lysosome-associated membrane protein-2 defect explains multisystem clinical involvement and allows leukocyte diagnostic screening in Danon disease. Am J Pathol. 2006;168:1309–1320. doi: 10.2353/ajpath.2006.050646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda Y, et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res. 2015;116:264–278. doi: 10.1161/CIRCRESAHA.116.303356. [DOI] [PubMed] [Google Scholar]
- 25.Li DL, et al. Doxorubicin blocks cardiomyocyte autophagic flux by inhibiting lysosome acidification. Circulation. 2016;133:1668–1687. doi: 10.1161/CIRCULATIONAHA.115.017443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musiwaro P, Smith M, Manifava M, Walker SA, Ktistakis NT. Characteristics and requirements of basal autophagy in HEK 293 cells. Autophagy. 2013;9:1407–1417. doi: 10.4161/auto.25455. [DOI] [PubMed] [Google Scholar]
- 27.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 29.Dorn GW., 2nd Mitochondrial dynamism and heart disease: Changing shape and shaping change. EMBO Mol Med. 2015;7:865–877. doi: 10.15252/emmm.201404575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wai T, et al. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science. 2015;350:aad0116. doi: 10.1126/science.aad0116. [DOI] [PubMed] [Google Scholar]
- 32.Beussman KM, et al. Micropost arrays for measuring stem cell-derived cardiomyocyte contractility. Methods. 2016;94:43–50. doi: 10.1016/j.ymeth.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashem SI, et al. Brief report: Oxidative stress mediates cardiomyocyte apoptosis in a human model of Danon disease and heart failure. Stem Cells. 2015;33:2343–2350. doi: 10.1002/stem.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashem SI, et al. Impaired mitophagy facilitates mitochondrial damage in Danon disease. J Mol Cell Cardiol. 2017;108:86–94. doi: 10.1016/j.yjmcc.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Brown DA, et al. Expert consensus document: Mitochondrial function as a therapeutic target in heart failure. Nat Rev Cardiol. 2017;14:238–250. doi: 10.1038/nrcardio.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong MY, et al. Histone deacetylase activity governs diastolic dysfunction through a nongenomic mechanism. Sci Transl Med. 2018;10:eaao0144. doi: 10.1126/scitranslmed.aao0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.