Significance
Proteasomes are ATP-dependent proteases that occur in all three domains of life, and are the principal molecular machines for the regulated degradation of intracellular proteins. The eukaryotic 26S proteasome has been extensively characterized. However, its evolutionary precursor, the archaeal proteasome–ATPase complex, remains poorly understood. The inherent instability of these primordial protein complexes has so far hindered attempts for detailed structure determination. Using cryo-EM single-particle analysis, we were able to determine the structure of an archaeal PAN-proteasome, which is a complex of the proteolytic core and the ATPase PAN (proteasome-activating nucleotidase). The structures reported here not only provide insights into the functional cycle of PAN-proteasomes, they reveal a fundamental mechanism of ATPase operation.
Keywords: PAN, proteasome, ATPase cycle, cryo-EM, archaea
Abstract
Proteasomes occur in all three domains of life, and are the principal molecular machines for the regulated degradation of intracellular proteins. They play key roles in the maintenance of protein homeostasis, and control vital cellular processes. While the eukaryotic 26S proteasome is extensively characterized, its putative evolutionary precursor, the archaeal proteasome, remains poorly understood. The primordial archaeal proteasome consists of a 20S proteolytic core particle (CP), and an AAA-ATPase module. This minimal complex degrades protein unassisted by non-ATPase subunits that are present in a 26S proteasome regulatory particle (RP). Using cryo-EM single-particle analysis, we determined structures of the archaeal CP in complex with the AAA-ATPase PAN (proteasome-activating nucleotidase). Five conformational states were identified, elucidating the functional cycle of PAN, and its interaction with the CP. Coexisting nucleotide states, and correlated intersubunit signaling features, coordinate rotation of the PAN-ATPase staircase, and allosterically regulate N-domain motions and CP gate opening. These findings reveal the structural basis for a sequential around-the-ring ATPase cycle, which is likely conserved in AAA-ATPases.
Proteasomes are ATP-dependent self-compartmentalizing proteases that occur in all three domains of life, and are responsible for selective protein degradation in crowded cellular environments (1, 2). By degrading proteins that are misfolded, mistranslated, damaged, or no longer needed by the cell, proteasomes play a pivotal role in cellular protein homeostasis, stress response, protein quality control, cell cycle control, regulation of metabolism, and other essential processes (3, 4). Recent extensive studies have elucidated in detail the structure of yeast and mammalian 26S proteasomes, and provided conformational insights into their functional cycle (5–11). However, in case of archaeal proteasomes, neither the architecture nor the mechanistic principles are clearly understood.
Archaeal proteasomes are minimal complexes, composed of a 20S proteolytic core (CP), and an ATPase module (12). They lack an elaborate regulatory complex (RP), as is present in 26S proteasomes, and the ATPase alone drives protein degradation. These proteasomes possess the compositional simplicity of prokaryotic ATP-dependent proteases [such as ClpXP, ClpAP, HslUV, etc. (13)], along with key architectural features of the 26S proteasome, and can thus be regarded as evolutionary precursors of the 26S proteasome (14). However, detailed structural characterizations of archaeal proteasomes (CP-ATPase complexes) have not been published so far, in part because of difficulties in sample preparation that suggested a dynamic nature of these complexes.
The archaeal CP is an ∼700-kDa barrel-shaped protein complex composed of four stacked homoheptameric rings (15). The outer rings comprise α-subunits, whose N-terminal extensions form a gate that blocks protein diffusion into the proteolytic chamber (16). The inner rings comprise β-subunits that harbor the proteolytic sites (17). In the absence of a dedicated RP, the CP collaborates with a network of AAA-ATPases (18, 19), among which the proteasome activating-nucleotidase (PAN) was the first discovered with CP-activating properties (20). PAN is a chaperone present in many archaeal species (21), and exhibits ∼41–45% sequence homology to the Rpts (regulatory particle AAA-ATPases) of the 26S proteasome (20). Although the structure of full-length PAN is hitherto unsolved, crystal structures of domains of Methanocaldococcus janaschii and Archaeoglobus fulgidus PAN show that six PAN protomers associate to form three two-stranded coiled coils, a hexameric oligonucleotide/oligosaccharide binding (OB) ring, and a hexameric AAA ring (22, 23). At the C-terminal end of each PAN protomer is an HbYX motif (where Hb is a hydrophobic residue, Y is tyrosine, and X is any amino acid), which is supposedly involved in CP activation (24). However, the physical association and mode of interaction between PAN and CP lack detailed structural characterization. An in-depth analysis of the PAN-proteasome structure is therefore essential to understand archaeal protein degradation in further detail.
Here we report the cryo-EM structure of A. fulgidus PAN-proteasome (PAN-CP complex) in the presence of adenosine 5ʹ-O-(3-thiotriphosphate) (ATPγS). With the PAN-AAA ring resolved at ∼4.9 Å, and the CP resolved at ∼4.1 Å, it was possible to unambiguously model the PAN-proteasome structure to near-atomic resolution. We identified five distinct conformational states, which show sequential rotation of the hexameric PAN ATPase ring, alongside movements of the PAN N domain and CP gate. These structures not only provide invaluable insights into the functional cycle of the PAN-proteasome, they also elucidate a sequential around-the-ring ATPase cycle that is likely conserved in AAA-ATPases.
Results
Assembly of PAN-Proteasomes.
For structural analysis, recombinant PAN and CP from A. fulgidus were separately expressed in Escherichia coli, and PAN-proteasomes were reconstituted in vitro (Materials and Methods and SI Appendix, Fig. S1 A and B). Assembly reactions tested by negative stain EM showed complex formation in presence of ATP and its slowly hydrolysable analog, ATPγS (SI Appendix, Fig. S1C). However, in the presence of ATPγS, intact complexes were more frequently observed. In the presence of ADP or ADP-AlFx (ATP hydrolysis transition-state analog), PAN-CP complexes were not observed, indicating that ATP is essential for complex formation, and once hydrolyzed, the complex falls apart. This reflects a transient nature of PAN-CP interaction, and agrees with previous biochemical studies (25). However, it is unlike the 26S proteasome, which remains intact even in the presence of ADP or ADP-AlFx (7, 8). It is difficult to assess the stability of PAN-proteasomes since they occur in extremophilic organisms, and such conditions cannot be faithfully reproduced in vitro. Nevertheless, the absence of a dedicated RP, with clamping subunits, does predispose PAN-proteasomes to dissociation (26).
Cryo-EM Structure of the PAN-Proteasome.
PAN-proteasomes assembled in the presence of ATPγS, were vitrified and imaged using an FEI Titan Krios transmission electron microscope (TEM), equipped with a Gatan K2 direct electron detector. The micrographs showed predominantly double-capped particles, where two PAN hexamers assembled on either side of a CP (SI Appendix, Fig. S1D), resembling the negative stain MjPAN-TaCP complexes reported earlier (25). Two-dimensional analyses showed side views of mainly two types: (i) a “straight view,” with two PAN hexamers arranged collinear with a sevenfold symmetric CP; and (ii) a “bent view,” where both PAN hexamers appeared slightly inclined toward the CP (SI Appendix, Fig. S1E). Each PAN protomer interacts with the CP via its C-terminal domain and the N-terminal domain is free. Elements of secondary and tertiary structure were clearly resolved in the CP but less well resolved in PAN, and the PAN coiled coils appeared smeared. We initially reconstructed a 3D density map of the double-capped PAN-proteasomes applying C2 symmetry (Fig. 1A). Although the global resolution of our map was 4.47 Å, PAN was at a significantly lower resolution compared with the CP, and the resolution markedly decreased between the CP, PAN AAA ring, OB ring, and coiled coils (SI Appendix, Fig. S2A).
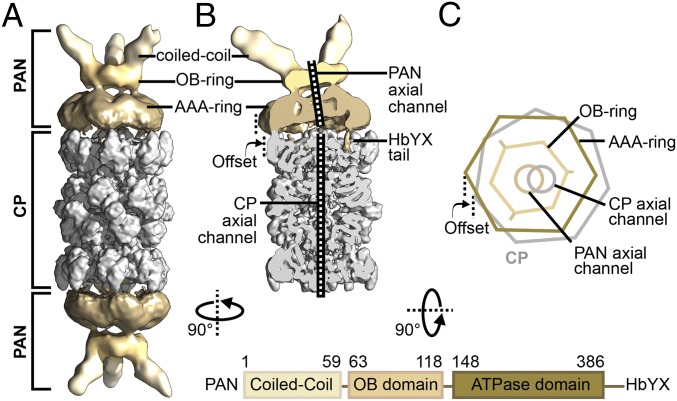
Fig. 1.
Architecture of the PAN-proteasome. (A) Cryo-EM reconstruction of double-capped PAN-proteasomes, with applied C2 symmetry. (B) Cutaway view of PSC particle reconstruction. The density maps in both A and B are filtered according to local resolution, with the CP shown in gray and PAN shown in sand. (C) Simplified illustration of a PAN-proteasome top view. The sevenfold symmetric CP is shown in gray, while the different domains of homohexameric PAN are shown in shades of sand. Mismatch of symmetry between PAN and CP leads to an off-axis location of PAN on top of the CP and the axial pores of PAN and CP are clearly misaligned. The domain architecture of PAN is depicted below.
To allow for deviations from C2 symmetry, the double-capped particles were density subtracted in silico into two pseudosingle-capped (PSC) particles and treated independently. An initial 3D reconstruction of the PSCs resolved at 4.43 Å, and connections between PAN and CP became visible (Fig. 1B and SI Appendix, Fig. S2A). Compared with the CP, PAN is positioned at an offset and the axial channels of PAN and CP are not aligned (Fig. 1 B and C). Such an arrangement arises from the symmetry mismatch between CP and PAN, and has been noted previously for other symmetry-mismatched complexes (27, 28).
Interestingly, our PSC reconstruction differs considerably from the MjPAN-MjCP complexes reported earlier, where PAN appears completely different in structure and is positioned centrally on the CP axis (29).
Conformational Variability of PAN-Proteasomes.
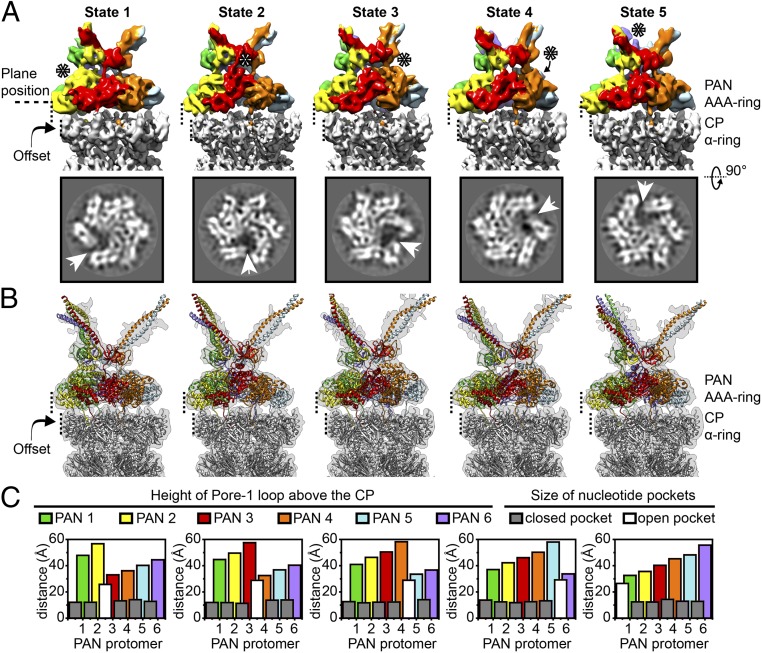
Although the use of PSCs significantly improved particle alignment, the resolution of PAN was significantly lower than the CP (SI Appendix, Fig. S2A). To resolve the structural heterogeneity of PAN, the PAN AAA ring along with the OB domain (together referred to as AAAob) was density subtracted from PSCs, aligned against a single reference, and classified further (SI Appendix, Fig. S3). Within the ensemble of AAAob, we observed the existence of five distinct conformational states (Fig. 2A and SI Appendix, Fig. S3). Each state was further cleaned in silico (SI Appendix, SI Materials and Methods and Fig. S4) to generate PSCs of the corresponding states (SI Appendix, Fig. S5A). The five states differed mainly in: (i) conformation of the AAA ring and (ii) disposition of the OB ring (SI Appendix, Table S1 and Movie S1). In each state, the pore helices of the PAN AAA ring adopted a spiral staircase conformation, with a “split site” formed between the subunit at the lowest position and the one in the highest (Fig. 2A). Linkers between the ATPase domain and OB domain were visible for most of the subunits and the OB ring appeared to perch on them. Keeping the offset position of PAN fixed with respect to the CP, the AAA staircase was rotated one subunit per state, giving rise to five distinct split-site positions. The states were unequally populated and AAAob densities refined to varying resolutions (SI Appendix, Fig. S5B). However, in all of the states, helices were clearly visible, thereby allowing us to build pseudoatomic models (Fig. 2B).
Fig. 2.
The PAN-proteasome exists in five rotated spiral-staircase conformations. (A) In the row above, density maps of AAAob from the five conformational states are displayed. For a fixed position of the PAN offset on the CP, the states differ in conformation of their AAA ring, and inclination of the N domain (OB ring and coiled coils). PAN is colored according to protomer, and in each state, the protomer occupying the highest position is indicated by asterisks. AAAob densities are placed in the context of the respective CP (in gray) to emphasize the fixed position of the offset. In the row below, slices through the indicated plane position are displayed. The existence of a split site is clearly visible in the slices and is further highlighted by white arrowheads. (B) Models of different states of the PAN-proteasome are superimposed on the respective PSC densities. In the models, PAN is colored according to protomer, and the CP is in gray. (C) Plot of pore loop height and nucleotide pocket depth for each conformational state (SI Appendix, SI Materials and Methods). Height of the tip of pore helices (α5 helix from each PAN protomer) above the plane of the CP is plotted for every PAN protomer, and is colored coherently. The size of a nucleotide pocket is plotted as an overlay. In all of the states, there is a distinct open pocket (colored white) between the highest and lowest pore helices, while the other pockets (closed) are of similar size, and colored gray.
In a PAN AAA staircase, the N-terminal tips of the α5 (pore-1) helices, and in turn the pore-1 loops, exhibit a uniform elevation above the plane of the CP (Fig. 2C and SI Appendix, SI Materials and Methods and Table S2). However, the distance between two adjacent tips remains constant at ∼10 Å, except at the split site. Similarly, there are five nucleotide pockets of similar size, while the one between the highest and lowest PAN protomers (at the split site) is significantly wider (Fig. 2C and SI Appendix, Table S2). Spiral-staircase conformations are typical of hexameric ring-shaped ATPases and have been reported earlier for p97 (30), Hsp104 (31), Vps4 (32), YME1 (33), Rpts (6), etc. It is hypothesized that the ATPase cycle progresses via rotation of the spiral staircase (33). Here, five rotated spiral-staircase conformations have been identified in a hexameric ATPase, revealing a nearly complete ATPase cycle.
Interestingly, the plane of the OB ring adjusts with the conformation of the AAA ring, and slants from the highest pore helix. Thus, with rotation of the AAA staircase, the OB plane shows a wobbling motion, with the coiled coils biased toward the same direction (SI Appendix, Table S1 and Movie S1). The relatively low resolution of the coiled coils reflects their high flexibility, which in conjunction with their unusual surface charge distribution probably facilitates substrate capture.
In all of the five states of the PAN-proteasome, an interesting feature is the discontinuity of PAN and CP axial channels (SI Appendix, Table S1), which arises due to the ∼10-Å offset between PAN and CP. Considering these structures to be snapshots in the proteasomal catalytic cycle, we presume there exist additional conformational states [as in the 26S proteasome (11)], where the axial channels align and ease substrate passage.
Structure of the PAN AAA Ring Reveals Intersubunit Signaling Features.
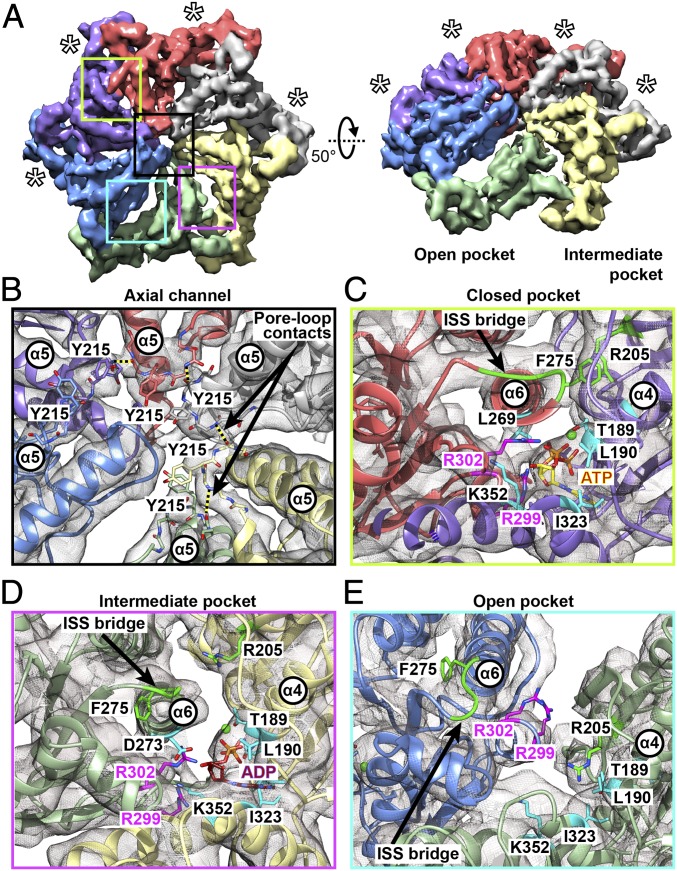
Although the AAAob densities from the five states were conformationally distinct, the geometry of their AAA rings was surprisingly similar, and could be superimposed after appropriate rotation. For each state, we therefore density subtracted PSCs to their respective AAA rings, and refined them (SI Appendix, Fig. S5C).The refined densities were subsequently locked in silico in the proper rotational register, and merged. Refinement of our ensemble of AAA rings resolved at 4.85 Å (Fig. 3A and SI Appendix, Fig. S2B), and bulky side chains could be seen in the best-resolved parts. The 4.85-Å density map was used to refine the AAA-ring model to near-atomic precision.
Fig. 3.
Structural basis for intersubunit communication in PAN. (A) A 4.85-Å cryo-EM map of the PAN AAA ring, colored according to constituent protomers. The closed pockets are indicated by asterisks. (Left) Top view of the AAA ring. (Right) The 50°-rotated view shows the spiral-staircase arrangement, highlighting the open and intermediate pockets. (B) Detailed view of the PAN axial channel, showing the arrangement of pore-1 loop tyrosines (Y215). Pore-loop contacts between the I216 backbone O and K214 backbone N are indicated by dashed lines. (C) Detailed view of a closed nucleotide pocket, where the ISS bridge (green) of the left subunit (coral) extends toward the right subunit (violet), establishing contact between F275 and R205. Interaction between the arginine finger (R302) and L269 in the pore-2 helix (α6) brings the α6 helix close to the bound nucleotide. (D) Detailed view of the intermediate pocket, where the ISS bridge (green) of the left subunit (light green) is partially withdrawn, disrupting the interaction between F275 (of the light-green subunit) and R205 (of the pale-yellow subunit). (E) Detailed view of the open pocket, where the ISS bridge of the left subunit (blue) is completely withdrawn, and forms a part of the α6 helix. There is no interaction between F275 and R205 of the adjacent (light-green subunit). In all cases, the arginine fingers (R299 and 302) are consistently colored magenta, and other crucial residues in the nucleotide pockets are colored cyan. The regions depicted in detail in C–E are indicated by boxes on the AAA ring in A, the color of the box matching the outline of the detailed view.
Close inspection of the AAA-ring density map enabled us to identify two major intersubunit contacts, which are also consistently present in all of the individual states: (i) the pore-1 loops of adjacent subunits are connected via hydrogen bonds between the backbone oxygen of I216 and backbone nitrogen of K214. These “pore-loop contacts” appear as an inner railing of the AAA staircase (Fig. 3B); (ii) a phenylalanine residue (F275) at the C-terminal end of the pore-2 helix (α6) of one subunit contacts an arginine residue (R205) in the β8 strand of the anticlockwise adjacent subunit, forming a bridge above the nucleotide binding pocket (Fig. 3C). Incidentally, F275 forms a part of a loop comprising D273-G274-F275, which is strictly conserved in many AAA-ATPases including the Rpts of the 26S proteasome (9, 33). It is believed that this loop is critically involved in intersubunit signaling (ISS) and communicates the nucleotide state between protomers (34).
Interestingly, the bridges formed by F275-D276-P277 (referred to here as “ISS bridges”) showed three distinct geometries in the PAN AAA ring: (i) there are four strong ISS bridges that occur above closed nucleotide pockets. In those pockets, the interaction of the arginine finger (R302) with the L269 (in the α6 helix) brings the α6 helix in close proximity to the bound nucleotide (Fig. 3C); (ii) there is one ISS bridge, partially withdrawn from the anticlockwise adjacent subunit. In that pocket, the arginine finger (R299) is disengaged from the bound nucleotide (Fig. 3D); (iii) the ISS motif is incorporated into the C-terminal end of the α6 helix, thereby elongating it (Fig. 3E). Such a situation is observed in the open pocket, where a nucleotide density is barely visible, and both arginine fingers (R299 and R302) are retracted. Thus, based on ISS bridge geometries, three types of nucleotide pockets can be identified––open, closed, and intermediate, although model-based measurements (SI Appendix, Table S2) indicate the occurrence of pockets of only two size ranges.
Three types of nucleotide pockets (open, closed, and intermediate) are possibly associated with three coexisting nucleotide states, as is found in the mitochondrial ATPase YME1 (33). Interestingly, PAN and YME1 share another common feature: the incorporation of ISS motif into the pore-2 helix above the open pocket. In analogy with YME1, we therefore modeled ATP in the closed pockets, ADP in the intermediate pocket, and left the open pocket nucleotide-free. However, we believe the intermediate pocket in our AAA-ring density contains a mixture of ATP(γS) and ADP.
Pore-loop contacts in AAA-rings have not been reported earlier. In case of PAN, an ∼10-Å separation between adjacent α5 helix tips possibly juxtaposes the pore loops to mutually interact. We suppose these contacts restrict flexibility of the pore loops and position the pore-loop tyrosine (Y215) in proper register to interact with substrate. Both the ISS contacts and pore-loop contacts are nonexistent at the split site, and with rotation of the spiral staircase, these contacts are created and destroyed.
HbYX-Mediated Interactions Between PAN and CP.
The C-terminal HbYX motif of PAN comprises residues M396-F397-V398 and is responsible for interaction with the CP. In our PSC reconstructions, HbYX insertions were visible and invariant for all PAN protomers in all five states, with one HbYX tail weaker than the others. To analyze HbYX contacts in detail, PSCs for the five states were density subtracted to 20Sαβ and refined with C1 symmetry (SI Appendix, Fig. S5D). In each density map, six of the seven inter-α subunit pockets were occupied by HbYX insertions. Since 20Sαβ densities from the five states showed similar HbYX occupancies, we have merged them to improve resolution. The merged 20Sαβ reconstruction resolved at 4.1 Å, and bulky side chains were visible in most parts of the density (Fig. 4A and SI Appendix, Fig. S2C). Side-chain information of the merged 20Sαβ was therefore utilized to refine the atomic models of the PAN-bound CP (SI Appendix, SI Materials and Methods and Fig. S6A).
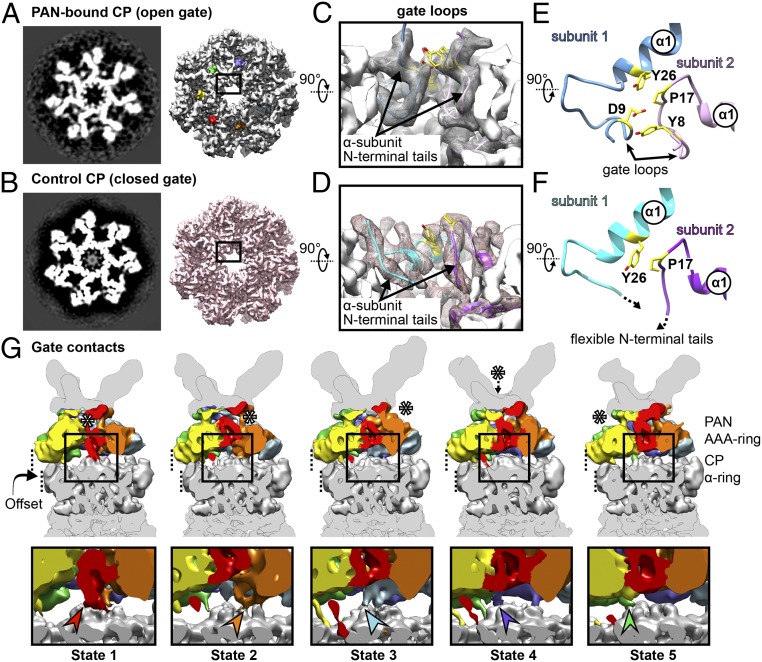
Fig. 4.
PAN binding leads to stabilization of the CP gate. Slice (Left) and isosurface (Right) of (A) PAN-bound CP and (B) control CP are shown from the top. In the PAN-bound CP, the axial channel is clear, with finger-like projections surrounding the axial pore. In the control CP, the axial channel appears clear in the isosurface, but is occupied by a smeared density in the slice. (C) Close-up view of the open gate shows gate loops formed by αN-terminal tails. These tails are flexible, and hence missing in the control CP (D). Model showing segments of two adjacent subunits of (E) PAN-bound CP and (F) control CP. The pore of the CP axial channel is lined by P17 reverse turns. In the PAN-bound CP, the residues Y8 and P17 from subunit 2 (pink) interact with D9 and Y26 from subunit 1 (blue), thereby forming an intersubunit quartet. In the control CP, intersubunit contacts are observed between Y26 and P17, although gate loops are missing. (G) In the row above, cutaway views of the PAN-CP interface (filtered to 9 Å) are displayed for the states 1 through 5. The interface densities are placed in the context of the respective PSCs, and colored as in Fig. 2. Asterisks indicate the lowest PAN protomer. The αN-terminal tail that forms a gate contact with PAN is colored according to the PAN protomer it contacts. The boxed regions are magnified in the row below, whereby the gate contacts are indicated by arrowheads of corresponding color.
Within the PAN-bound CP, backbone interactions are observed between the C-terminal HbYX tail of PAN and the α2 helix of CP α-subunit (SI Appendix, Fig. S6B). The backbone nitrogen of V398 (PAN) interacts with the backbone oxygen of G80 (CP α-subunit) while the backbone oxygen of M396 (PAN) interacts with the backbone nitrogen of V82 (CP α2 helix). To analyze PAN-induced structural changes in the CP, a dataset of recombinant A. fulgidus CP was acquired as control. The control 20Sαβ, refined with C7 symmetry, resolved to 3.7 Å, and side chains were clearly visible throughout the entire density map (Fig. 4B and SI Appendix, Fig. S2C). We generated a model of the control CP using the crystal structure of A. fulgidus 20Sαβ (Protein Data Bank ID code: 1J2Q; ref. 35). Interestingly, residuewise RMSD calculations show no significant structural variation in the α-ring or inter-α-subunit pockets of the control and PAN-bound CP (SI Appendix, Fig. S6C).
Open Gates in PAN-Proteasomes.
In all five states of the PAN-proteasome, 20Sαβ refinements exhibit CP gates in open conformation (SI Appendix, Fig. S5D). From the isosurface view, the gate of the control CP also appears open, although in the slice, a smeared density is observed obstructing the pore (Fig. 4B). This indicates that in the control CP, the flexible αN-terminal tails form a plug, blocking the axial channel. Such a CP gate conformation has been observed previously for the Thermoplasma acidophiulm CP, and is regarded as a closed gate (16).
In both PAN-bound and control CP, the mouth of the CP axial channel features a reverse turn of residues S16-P17-D18. However, in the PAN-bound CP, an interesting feature is the “gate loop” that is formed involving residues 8–17 at the αN terminus (Fig. 4C and Movie S2). These loops are unique to the open gate and are not observed in the control CP (Fig. 4D). Due to gate-loop formation, Y8 of one α-subunit is in close proximity to P17 of the same subunit, and D9 of the adjacent α-subunit. A cluster is thereby formed involving Y8 and P17 of one subunit and Y26 and D9 of the adjacent subunit (Fig. 4E). Such a cluster formation has been observed previously for activated T. acidophilum and Saccharomyces cerevisiae CP, and has been implicated in stabilization of the open gate (9, 36, 37). In the closed gate, we observe intersubunit contacts between P17 and Y26, which probably stabilize the S16-P17-D18 reverse turn in the axial pore (Fig. 4F and Movie S2).
All seven CP α-subunits in a PAN-proteasome show gate-loop formation. However, for the α-subunit closest to the lowest PAN subunit, the N-terminal tail extends toward PAN, establishing a contact. To characterize this unique feature, we density subtracted PSCs for each state to their respective interface models (comprising PAN AAA ring and CP α-ring) and refined them (SI Appendix, Fig. S5E). The connections between PAN and CP however remained poorly resolved, and for comparison of interstate conformational variation in the gate region, each interface reconstruction was down-filtered to 9 Å (Fig. 4G). Interestingly, a contact between the lowest PAN subunit and the N-terminal tail of the CP α-subunit was consistently visible in all five states of the PAN-proteasome (Fig. 4G). The contact rotates with rotation of the split site. Since this contact forms between the CP gate and PAN, we have named it as “gate contact.” However, the residues involved in contact formation could not be unambiguously identified.
Such gate contacts are not observed in case of the 26S proteasome. However, previous studies with ClpXP have shown that the N-terminal loops of ClpP form hydrophobic contacts with pore-2 loops of ClpX (38). Likewise, in case of the complex of PA26-HbYX chimera with CP, the N-terminal tails of the CP have been found to contact activation loops of PA26 (36).
Discussion
ATP Hydrolysis Cycle Around the PAN AAA Ring.
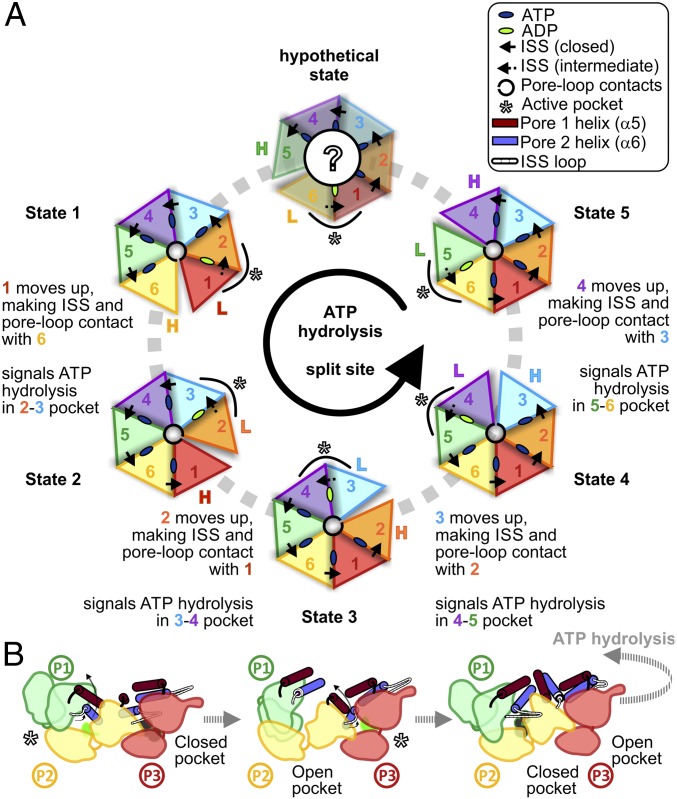
We have identified intersubunit contacts between PAN protomers, which reflect the occupancy state of the nucleotide binding pocket. Since the pattern of intersubunit interactions is repeatedly present in all of our five states, we can use our results to propose a model for ATP hydrolysis around the PAN AAA ring (Fig. 5). The intermediate pocket with ADP, and a partially withdrawn ISS bridge, is probably the active pocket, where ATP hydrolysis is taking place. Due to ATP hydrolysis in the P1-P2 pocket, arginine finger residues of P1 lose contact with the nucleotide, and the α6 (pore-2) and in turn α5 (pore-1) helices move upward. The ISS bridge of P1 is incorporated into its α6 helix, and a split site (open pocket) is formed between P1 and P2, with P1 occupying the highest position and P2, the lowest position in the AAA spiral staircase. The detachment of P1 ISS bridge from P2 signals the commencement of ATP hydrolysis in the P2-P3 pocket, and the cycle continues sequentially in a counterclockwise manner around the AAA ring. In parallel, ATP is reloaded in the open pocket (now P1-P2). This ensures that when ATP in the P2-P3 pocket is hydrolyzed, and the P2 pore helices move up to the highest position in the staircase, the P1-P2 pocket acquires a closed conformation, with the P1 ISS bridge reestablishing contact with P2. Rotation of the spiral staircase leads to rhythmic motions of the pore helix that, in turn, enable the pore-loop tyrosines to propel substrates into the CP.
Fig. 5.
Structure-based model for the ATPase cycle in PAN-proteasomes. (A) Cartoon representations of the five ATPase states identified, along with a putative state hypothesized (hypothetical state). In each state, the subunit in the highest and lowest positions is indicated with colored H and L, respectively, and the active (ADP) pocket is indicated by asterisks. (B) Putative scheme of the conformational changes that drive the ATPase cycle around the AAA ring. ATP hydrolysis in the active pocket causes withdrawal of the ISS loop, and upward movement of pore helices. Detachment of ISS contact signals the counterclockwise adjacent subunit to hydrolyze ATP.
Various sequential, concerted, and stochastic models currently exist that aim to link ATP hydrolysis by AAA-ATPases with substrate unfolding and translocation (31–33, 39–42). However, the bulk of information available is still insufficient to understand the mechanisms completely. The present study establishes that ATP hydrolysis occurs around the PAN AAA ring, and the model proposed depicts the most energetically favorable route.
Gate Transitions in a PAN-Proteasome.
Our results show that PAN binding stabilizes the CP gate in an open conformation. Since gate opening is considered an outcome of HbYX tail insertion within inter-α-subunit pockets, conformational changes at or propagating from the HbYX binding site are likely to signal CP activation. Interestingly, our rmsd estimates do not show a significant difference in the inter-α-subunit pockets or the α-ring of the control and PAN-bound CP (SI Appendix, Fig. S6C).
Previous structural and biochemical studies have reported varied molecular mechanisms of CP activation (36, 43–45). However, the consensus is the shifting of a P17 reverse turn that lines the entry pore of the CP axial channel. The reverse turn is relocated by either an ∼4° rigid-body rotation of the CP α-ring that results from induced-fit conformational changes in the inter-α-subunit pocket (45), or by subtle direct contacts between the penultimate HbYX residue (Y/F) and G19 on the CP α-subunit (44). In our case, with the control CP resolved at 3.7 Å, and the PAN-bound CP resolved at 4.1 Å, it would be problematic to interpret the 1.5-Å shifts we observed in the rmsd (SI Appendix, Fig. S6C). We prefer the latter model, whereby only subtle signals are involved, without major induced-fit conformational changes.
In PAN-proteasomes, a unique feature of CP gates is the gate contacts, which rotate in concert with the AAA staircase. This hints at an allosteric communication of ATP hydrolysis state between PAN and the CP, and likely plays a role in assisting substrate translocation through the axial channel. The function of gate contacts is yet unknown. However, contacts of a similar nature have been reported previously in ClpXP. There, the contacts vary dynamically with the nucleotide state of individual ClpX subunits, control ATP-hydrolysis rates, and facilitate efficient protein unfolding (38). Thus, gate contacts might be primitive means of bipartite communication in protease-activator complexes that have been lost with evolution.
Materials and Methods
Sample Preparation and Cryo-EM.
A. fulgidus PAN and CP were recombinantly produced in E. coli, and PAN-proteasomes were reconstituted in vitro in presence of ATPγS. Plunge-frozen samples were imaged in a Titan Krios TEM, equipped with a K2 direct electron detector (details in SI Appendix, SI Materials and Methods).
Image Processing and Model Building.
Acquired movie frames were processed as described in SI Appendix, SI Materials and Methods. Classification and reconstruction was performed in Relion 2.1 (46), and structural models were generated by molecular dynamics flexible fitting (47) (details in SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank M. Wehmer and M. Eisele for discussion. This work was supported by SFB-1035/Project A01.
Footnotes
The authors declare no conflict of interest.
Data deposition: Cryo-EM density maps and atomic coordinates have been deposited in the Electron Microscopy Data Bank (www.ebi.ac.uk/pdbe/emdb) and the Protein Data Bank (PDB) (www.rcsb.org), respectively [Accession codes: PAN-proteasome in state 1 (EMDB: EMD-0212; PDB: 6HE8), state 2 (EMDB: EMD-0213; PDB: 6HE9), state 3 (EMDB: EMD-0214; PDB: 6HEA), state 4 (EMDB: EMD-0215; PDB: 6HEC), state 5 (EMDB: EMD-0216; PDB: 6HED), AAA ring of PAN-proteasomes (EMDB: EMD-0209; PDB: 6HE4), PAN-bound CP (EMDB: EMD-0210; PDB: 6HE5) and control A. fulgidus CP (EMDB: EMD-0211; PDB: 6HE7)].
See Commentary on page 350.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817752116/-/DCSupplemental.
References
- 1.Voges D, Zwickl P, Baumeister W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 2.Zwickl P, Baumeister W, Steven A. Dis-assembly lines: The proteasome and related ATPase-assisted proteases. Curr Opin Struct Biol. 2000;10:242–250. doi: 10.1016/s0959-440x(00)00075-0. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: Snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. 2014;1843:13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweitzer A, et al. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc Natl Acad Sci USA. 2016;113:7816–7821. doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehmer M, et al. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci USA. 2017;114:1305–1310. doi: 10.1073/pnas.1621129114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding Z, et al. High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res. 2017;27:373–385. doi: 10.1038/cr.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisele MR, et al. Expanded coverage of the 26S proteasome conformational landscape reveals mechanisms of peptidase gating. Cell Rep. 2018;24:1301–1315.e5. doi: 10.1016/j.celrep.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, et al. Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome. Nat Commun. 2018;9:1360. doi: 10.1038/s41467-018-03785-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Peña AH, Goodall EA, Gates SN, Lander GC, Martin A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science. 2018 doi: 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat Rev Microbiol. 2011;10:100–111. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivares AO, Baker TA, Sauer RT. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol. 2016;14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volker C, Lupas AN. Molecular evolution of proteasomes. Curr Top Microbiol Immunol. 2002;268:1–22. doi: 10.1007/978-3-642-59414-4_1. [DOI] [PubMed] [Google Scholar]
- 15.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 16.Religa TL, Sprangers R, Kay LE. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328:98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- 17.Seemüller E, et al. Proteasome from Thermoplasma acidophilum: A threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 18.Forouzan D, et al. The archaeal proteasome is regulated by a network of AAA ATPases. J Biol Chem. 2012;287:39254–39262. doi: 10.1074/jbc.M112.386458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barthelme D, Sauer RT. Identification of the Cdc48•20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
- 21.Benaroudj N, Goldberg AL. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat Cell Biol. 2000;2:833–839. doi: 10.1038/35041081. [DOI] [PubMed] [Google Scholar]
- 22.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Pathare GR, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci USA. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickell S, et al. Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci USA. 2009;106:11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beuron F, et al. At sixes and sevens: Characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 29.Medalia N, et al. Architecture and molecular mechanism of PAN, the archaeal proteasome regulatory ATPase. J Biol Chem. 2009;284:22952–22960. doi: 10.1074/jbc.M809643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gates SN, et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science. 2017;357:273–279. doi: 10.1126/science.aan1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su M, et al. Mechanism of Vps4 hexamer function revealed by cryo-EM. Sci Adv. 2017;3:e1700325. doi: 10.1126/sciadv.1700325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puchades C, et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science. 2017;358:eaao0464. doi: 10.1126/science.aao0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hänzelmann P, Schindelin H. Structural basis of ATP hydrolysis and intersubunit signaling in the AAA+ ATPase p97. Structure. 2016;24:127–139. doi: 10.1016/j.str.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Investigations on the maturation and regulation of archaebacterial proteasomes. J Mol Biol. 2003;327:75–83. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- 36.Förster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 37.Förster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife. 2017;6:e25754. doi: 10.7554/eLife.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YC, Snoberger A, Schupp J, Smith DM. ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function. Nat Commun. 2015;6:8520. doi: 10.1038/ncomms9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordova JC, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadtmueller BM, et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem. 2010;285:13–17. doi: 10.1074/jbc.C109.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, et al. Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 2010;29:692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.