Significance
Ovarian cancer has the propensity for early dissemination of microscopic metastases, making early detection challenging. Development of treatments for patients with advanced/recurrent chemotherapy-resistant disease remains an unmet need. We sequenced primary, synchronous bilateral ovarian cancer (SBOC), metastatic, and recurrent ovarian tumors. We found primary and metastatic tumors to be remarkably similar and SBOC to be clonally related in all cases, suggesting early dissemination to represent an intrinsic feature of ovarian cancer. c-MYC and PIK3CA amplifications were prevalent and maintained in progression to metastasis and enriched in tumor recurrence. c-MYC amplified chemotherapy-resistant cell lines and xenografts were sensitive to JQ1 and GS-626510, suggesting Bromodomain and Extra-Terminal motif (BET) inhibitors as effective targeted agents in patients with c-MYC–amplified recurrent/chemotherapy-resistant ovarian tumors.
Keywords: ovarian carcinoma, whole-exome sequencing, BET inhibitors, bilateral ovarian tumors
Abstract
Ovarian cancer remains the most lethal gynecologic malignancy. We analyzed the mutational landscape of 64 primary, 41 metastatic, and 17 recurrent fresh-frozen tumors from 77 patients along with matched normal DNA, by whole-exome sequencing (WES). We also sequenced 13 pairs of synchronous bilateral ovarian cancer (SBOC) to evaluate the evolutionary history. Lastly, to search for therapeutic targets, we evaluated the activity of the Bromodomain and Extra-Terminal motif (BET) inhibitor GS-626510 on primary tumors and xenografts harboring c-MYC amplifications. In line with previous studies, the large majority of germline and somatic mutations were found in BRCA1/2 (21%) and TP53 (86%) genes, respectively. Among mutations in known cancer driver genes, 77% were transmitted from primary tumors to metastatic tumors, and 80% from primary to recurrent tumors, indicating that driver mutations are commonly retained during ovarian cancer evolution. Importantly, the number, mutation spectra, and signatures in matched primary–metastatic tumors were extremely similar, suggesting transcoelomic metastases as an early dissemination process using preexisting metastatic ability rather than an evolution model. Similarly, comparison of SBOC showed extensive sharing of somatic mutations, unequivocally indicating a common ancestry in all cases. Among the 17 patients with matched tumors, four patients gained PIK3CA amplifications and two patients gained c-MYC amplifications in the recurrent tumors, with no loss of amplification or gain of deletions. Primary cell lines and xenografts derived from chemotherapy-resistant tumors demonstrated sensitivity to JQ1 and GS-626510 (P = 0.01), suggesting that oral BET inhibitors represent a class of personalized therapeutics in patients harboring recurrent/chemotherapy-resistant disease.
Due to the lack of effective screening programs, epithelial ovarian cancer (EOC) remains the most lethal gynecologic malignancy, with more than two-thirds of EOC patients diagnosed with advanced-stage disease (i.e., abdominal carcinomatosis) (1). While the majority of patients initially respond to either primary surgical cytoreduction followed by platinum-based chemotherapy or neoadjuvant chemotherapy followed by cytoreduction, the development of chemotherapy-resistant disease results in only a 20 to 30% 5-y survival rate (2). This poor prognosis underscores the need for a better understanding of the molecular drivers contributing to early metastases and chemotherapy resistance.
Recent whole-exome sequencing (WES) and whole-genome sequencing (WGS) studies focusing on primary chemonaive high-grade serous carcinoma (HG-SC) (3) and chemotherapy-resistant tumor cells collected from patients developing ascites, demonstrated that HG-SC, the most common histologic type of ovarian cancer (4), is characterized by TP53 mutations in up to 96% of the tumors, by high genomic instability, and by germline or somatic defects in homologous recombination repair (HRR) genes in about 50% of patients. Reversion of BRCA1 or BRCA2 mutations in individual patients and recurrent promoter fusion associated with overexpression of the drug efflux pump MDR1 were also observed in a handful of patients with recurrent chemotherapy-resistant disease (4). WGS results, however, were not able to demonstrate any recurrent event in the over 800 gene fusions potentially capable of producing a fused transcript (4).
While extensive genomic data for primary chemonaive ovarian cancer are present in the literature (3), very limited data are currently available for metastatic ovarian cancer or for ovarian tumors exposed to the selective pressure of chemotherapy (4). Accordingly, we used WES of tumor and germline DNA from ovarian cancer patients to evaluate genomic differences among primary, metastatic, and recurrent chemotherapy-resistant tumors obtained from fresh biopsy samples. In addition, to evaluate their evolutionary history, we also performed WES of 13 left–right synchronous bilateral ovarian cancer (SBOC) pairs from patients with bilateral tumors. Lastly, because recurrent amplifications of chromosome 8q23-24 encompassing c-MYC were frequent in primary and metastatic tumors and enriched in recurrent cancers, we assessed the activity of GS-626510, a novel Bromodomain and Extra-Terminal motif (BET) inhibitor, against primary ovarian cancer cell lines and xenografts derived from chemotherapy-resistant disease.
Results
The Genetic Landscape of Primary, Metastatic, and Recurrent Ovarian Cancer.
We analyzed tumors and matched normal samples from 77 patients. These included 64 unilateral primary tumors and 13 matched pairs of tumors from patients with SBOC. We also sequenced 41 metastatic and 17 recurrent tumors. The majority of patients (55/77) had high-grade serous papillary histology. There were also 5 patients with endometrioid tumors, 5 patients with clear-cell tumors, 2 patients with dedifferentiated tumors, and 10 patients with mixed-histology tumors. The clinical features of these patients are presented in SI Appendix, Table S1. Tumor samples were sequenced using the NimbleGen/Roche capture reagent, followed by 74 base paired-end DNA sequencing on the Illumina HiSeq platform (5) to an average of 195 independent reads per base. Ninety-five percent of targeted bases had 20 or more independent reads. Normal samples were sequenced to a mean of 101 reads per base, with 96% having 10 or more independent reads, sufficient for high-quality germline calls (SI Appendix, Table S2). Tumor purity was estimated from B-allele frequency of somatic single-nucleotide variants (SNVs) and regions of loss of heterozygosity (LOH) and copy number variation (CNV). Tumors with low (<40%) or indeterminate purity were excluded from analysis. At the high levels of coverage achieved, there was no significant relationship between tumor purity and the number of somatic variants detected.
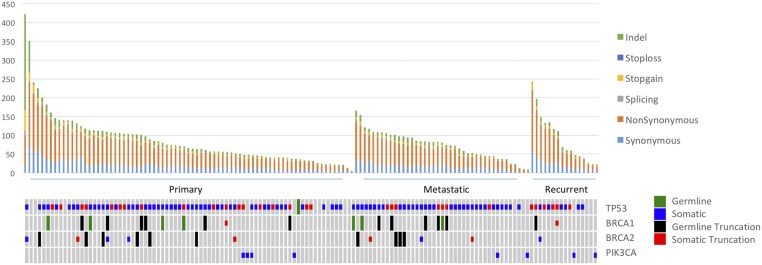
Somatic mutations were identified with MuTect2 and likely damaging mutations were identified (premature termination, splice site, and indel mutations and missense mutations at phylogenetically conserved sites; see Materials and Methods). The burden of mutation in individual genes was assessed by Mutsig and Oncodrive FM. TP53 was the most commonly mutated gene across all settings (Fig. 1 and SI Appendix, Fig. S1), with 86% of all primary tumors containing a somatic TP53 mutation. In agreement with previously reported WES and WGS analyses (3, 4), 91% (50/55) of the serous tumors contained a somatic TP53 mutation. Most other subtypes also demonstrated high TP53 somatic mutation rates: 4 of 5 endometrioid, 2 of 2 dedifferentiated, and 8 of 10 mixed-histology tumors. In contrast, clear cell tumors, had a 40% (2/5) TP53 mutation rate (SI Appendix, Table S3).
Fig. 1.
Distribution of somatic mutations. Each tumor was analyzed for somatic mutation burden and mutation type. Recurrent tumors also contained the highest non–synonymous-to-synonymous ratio of somatic mutations (3.59:1), followed by metastatic tumors (3.45:1) and primary tumors (3.23:1). For patients with two primary tumors, only the right tumor was included in this distribution.
In gene burden analysis of somatic mutations in primary tumors, two additional genes had q values of <0.1: MACF1, which was mutated in 8 of 77 of patients (with a q value of <10−4), and NF1, mutated in 4 of 77 tumors (with a q value of 8 × 10−2). These results confirm NF1’s status as a significantly mutated gene in ovarian cancer, as found in previous WES studies (4). MACF1, which has not been previously identified as a significantly mutated cancer gene, is a cytoskeletal protein known to interact with the Wnt pathway (6). We also found that 7 of 77 primary and 4 of 41 metastatic tumors contained at least one somatic mutation in a mismatch repair gene associated with Lynch syndrome, including three patients with mutations in MSH3, three patients with MLH3 mutations, two patients with PMS1 mutations, and additional mutations in MSH2 and MLH1. Most of these tumors had a mixed or endometrioid histology and a significantly higher somatic point mutation burden than tumors without MMR somatic mutations (mean somatic SNV count of 112.5 vs. overall mean of 66.9, P = 0.0033 by Wilcoxon rank test, excluding a single hypermutated tumor) (SI Appendix, Table S4). Additionally, two synchronous tumors with MMR mutations, both from the same patient, had two and four MMR mutations, respectively. The right ovarian tumor had two MMR mutations and 263 somatic mutations, while the left tumor had four somatic MMR mutations and 1267 somatic mutations.
PIK3CA Mutation Enrichment.
PIK3CA demonstrated significant copy number amplification and a high burden of known pathogenic somatic SNVs [H1047R (found in two recurrent tumors), E542K, E545K, and C420R]. Of the five PIK3CA mutations present in primary–recurrent tumor pairs, four PIK3CA mutations were absent in at least one corresponding primary tumor, while all five were present in recurrent tumors (P = 0.048). Two of the five mutations in primary–metastatic pairs were present in both tumors. Amplifications were found in 69 to 88% of various tumor classes, and pathogenic somatic SNVs were found in 2 to 18%. Recurrent tumors had the highest burden of both somatic SNVs and copy number gains (SI Appendix, Tables S5 and S6 a–c), suggesting a role in progression. In addition, we found several other mutations that may represent clinically pathogenic mutations, including N345K and R93Q that, which have been shown to be oncogenic-activating mutations in functional studies (7), and an additional two (G106V and V344M), which were predicted to be damaging SNVs by Combined Annotation–Dependent Depletion (CADD) and Radial Support Vector Machine (RadialSVM) analysis.
Comparison of Mutation Characteristics Among Matched Tumors.
To characterize similarities between primary tumors and their matched metastatic and recurrent tumors, we analyzed matched pairs of primary and metastatic or recurrent tumors. Recurrent tumors had a nonsignificantly higher burden of mutations not found in the corresponding primary tumor (mean = 49.8 unique mutations) compared with metastatic tumors (mean = 19.4 mutations, P = 0.13) (SI Appendix, Table S7). We analyzed number and mutation signature in matched pairs of primary and metastatic or recurrent tumors to investigate somatic mutation processes in different settings while controlling for differences between individual patients (SI Appendix, Fig. S2). Overall, substitutions were distributed across all six possible base substitutions, consistent with previously described mutation signatures in breast and ovarian cancer (8). Despite containing a mixture of shared and unique somatic mutations, the overall distribution of substitution types remains essentially the same, indicating little genetic alteration accumulation from primary tumors to metastatic and recurrent tumors. As depicted in SI Appendix, Fig. S3, primary and metastatic tumors contained overlapping repertoires of somatic exomic mutations, with many cases demonstrating a number of identical variants larger or comparable with that of private somatic events (see below and SI Appendix, Fig. S3).
Mutation Transmission from Primary Tumors to Metastatic and Recurrent Tumors.
Among 41 pairs of primary and metastatic tumors, an average of 60% (54.3/91.3) of the somatic mutations found in the primary tumor were found in the corresponding metastatic tumor. Similarly, among 16 pairs of primary and recurrent tumors (excluding one hypermutated recurrent tumor), primary tumors transmitted 54% of mutations (42.1/77.3) to the corresponding recurrent tumor. Somatic mutations in known cancer-related genes were more frequently transmitted than nondriver mutations, with 77% and 81% of mutations in known cancer genes found in primary tumors also found in matched metastases and recurrent tumors, respectively (P = 2.2e-3 for metastatic tumors, P = 0.016 for recurrent tumors). There were few somatic mutations in known ovarian cancer genes in metastases (three mutations in 41 patients) and recurrent tumors (five mutations in 16 patients) that were absent in primary tumors (SI Appendix, Table S7). Among these, two metastatic tumors had mutations in NF1 [a gene previously implicated in ovarian cancer (4)] that were not present in primary tumors. Based on the low rate of new mutations in ovarian cancer-implicated genes, this event was unlikely to occur by chance alone (P = 0.016). Among two matched metastatic, recurrent, and primary trios, each metastatic and recurrent pair of tumors shared a high proportion of somatic SNVs (83%), again suggesting that key mutations transmitted to metastatic tumors were likely to be retained during tumor recurrence.
Germline Analysis.
We analyzed normal samples (n = 77) from tumor–normal pairs for germline mutations in known ovarian cancer-predisposition genes. We also ran a parallel analysis on a control panel of 6,226 healthy patient exomes to estimate the baseline prevalence of these mutations in a healthy population. Twenty-five percent (19/77) of patients had pathogenic mutations (found in ClinVar) in genes of the HRR pathway; these included 10 patients with mutations in BRCA1, 6 patients with mutations in BRCA2, and 3 patients with a mutation in either CHEK2, PALB2, or BRIP1. The number of patients with mutation in BRCA1 or BRCA2 was significantly enriched compared with controls (21% vs. 0.7%, P < 2.2e-16, odds ratio = 37.6) (SI Appendix, Fig. S4 and Table S8 a–c). There were additional somatic point mutations in HRR genes in primary tumors. All of the 16 patients with BRCA1 or BRCA2 mutations developed somatic loss of the mutant allele (13 patients with copy number deletions and three patients with LOH). Five patients with germline BRCA1 or BRCA2 mutations developed at least one somatic mutation in an HRR gene. Eight other patients had one or two exclusively somatic mutations in HRR genes (SI Appendix, Table S8b). An additional eight patients had pathogenic or damaging germline mutations in other genes in the BROCA gene set (SI Appendix, Table S9).
Analysis of CNVs.
We analyzed CNVs across 64 primary, 41 metastatic, and 17 recurrent tumors from 77 patients. CNVs were identified by comparing tumor sample depth of coverage to the corresponding normal sample depth of coverage. We applied Genomic Identification of Significant Targets in Cancer (GISTIC) to calculate the significance of recurring amplifications and deletions in each group to detect recurring variations unlikely to be caused by chance alone (Fig. 2). Chromosomes 3q26 (69 to 88%), 8q23-24 (74 to 82%), and 14q11 (57 to 71%), were found to be the most significant amplifications among all groups. Specifically, the 3q26 amplification, for which PIK3CA has been previously validated as a driver gene (9), was found to be closely associated with PIK3CA point mutations: seven of eight tumors containing point mutations in PIK3CA also contained the corresponding 3q26 amplification.
Fig. 2.
Recurrent amplifications and deletions in primary, metastatic, and recurrent settings. Multiple significantly recurrent amplifications (A) and deletions (B) were detected across all groups. Significant amplifications corresponding to PIK3CA and c-MYC were found to have the highest prevalence in recurrent tumors, followed by metastatic tumors and primary tumors.
Among 17 matched pairs of primary and recurrent tumors, four patients had gained 3q26 (PIK3CA) amplifications in recurrence, with no patients losing amplifications or gaining deletions (SI Appendix, Fig. S5 A–C). Similarly, two patients gained 8q23-24 amplifications [which are likely to be driven by c-MYC (10, 11)] in the recurrent setting with no losses, demonstrating the higher likelihood of enrichment of amplifications rather than deletions or amplification loss in progression to recurrence (SI Appendix, Fig. S5 A, B, and D and Table S6b). Overall, the 3q26 and 8q23-24 chromosomes demonstrated a nonsignificantly greater prevalence of amplification in recurrent tumors (88% and 82%, respectively), compared with metastatic tumors (71% and 74%, respectively) and primary tumors (69% and 78%, respectively). The 14q11 amplification, previously identified as a recurring CNV in ovarian cancer (3, 4), also demonstrated a similar enrichment in the recurrent setting (71%) compared with the metastatic (61%) and primary (57%) settings. The most significant deletions, all of which were present in all three settings, were 1p36 (ARID1A and RPL22); 4p16 (FGFR3, a known tumor suppressor in ovarian cancer); 7p22 (CARD11, activator of NFκB); 8p21-23; 9q34 (NOTCH1); 11p15 (HRAS); and 19p13 (GNA11 and STK11). Many of these regions have been published in the past as known breast or ovarian cancer amplifications, which supports the validity of this CNV screen (12) (SI Appendix, Table S6 a and c).
Evolution and Clonal Relationship of Bilateral Ovarian Cancer.
About 25% of patients with ovarian carcinoma at the time of diagnosis (i.e., surgical staging) have tumors in both ovaries (13). However, the origin and the relationship of bilateral tumors have not been conclusively established. To determine whether right and left tumors arise independently as two distinct synchronous tumors or represent primary and metastatic tumors, we compared somatic SNVs in left and right tumors of 13 patients. We found that, on average, SBOC shared 68.5% of somatic mutations, with a mean of 43 (range, 6 to 115) mutations shared. Pooled together, there were a total of 735 somatic mutations in lower-mutation count tumors, of which 516 were shared between left and right (70.2%). Through Monte Carlo simulation of each left–right pair, we estimated that the shared mutations in each of the 13 pairs were highly unlikely to occur by chance alone (P < 1e-08), which was sufficient to establish that each pair of bilateral tumors arose from a common somatic origin (Fig. 3). Moreover, 90% of cancer driver mutations in these bilateral tumors were shared by both tumors (excluding a single hypermutated tumor). Among shared mutations, corresponding CNVs at the point mutation loci were also highly shared, with 80% of CNVs shared between left and right tumors. Lastly, to characterize the clonal architecture of SBOC, cancer cell fractions adjusted for tumor purity were clustered using PyClone, and the possible phylogenetic trees were reconstructed using ClonEvol. We found all 13 SBOCs to contain subclonal populations that were private to one of the tumors. Of interest, in all bilateral samples in which a consensus evolution model was successfully generated (7/13), a branching structure was revealed, suggesting that they all share a common ancestry before evolving independently (SI Appendix, Fig. S6).
Fig. 3.
Comparison of left and right tumors. Thirteen patients had bilateral primary tumors from the left and right ovaries analyzed. The number of mutations unique to left and right tumors is represented in blue and orange, respectively. All bilateral tumor pairs shared at least six somatic mutations (gray), establishing a common origin (P < 1e-8).
GS-626510 Activity in Preclinical Models of Chemotherapy-Resistant HG-SC.
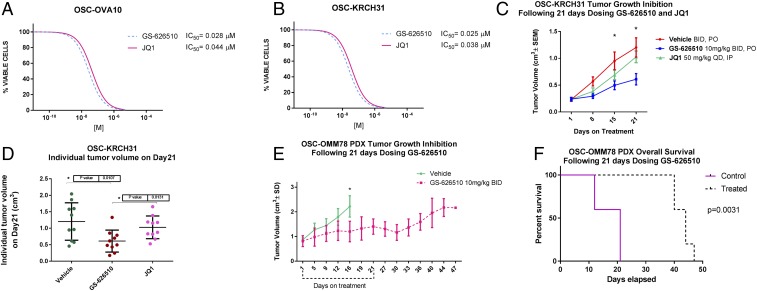
c-MYC gain of function in ovarian cancer has previously been reported in comprehensive genetic analyses (3, 4). Given the high prevalence of MYC amplifications in primary tumors (74%), metastasis (78%), and chemotherapy-resistant recurrence (82%)—suggesting a key role of MYC amplification in ovarian cancer—we evaluated the effect of a novel orally bioavailable BET inhibitor (i.e., GS-626510; Gilead Sciences Inc.) and JQ1 (GS-589903, a previously characterized BET-inhibitor) (14) on cell growth of seven primary ovarian cancer cell lines, including two fully sequenced primary tumors obtained from chemotherapy-resistant patients (i.e., KRCH31 and OVA10), demonstrating c-MYC amplification by WES and increased c-MYC expression by qRT-PCR but no amplification in any of the BRD2, BRD3, BRD4, and BRDT genes. We found chemotherapy-resistant ovarian tumors to be highly sensitive to the exposure to GS-626510 and JQ1, with IC50 values in the range of 0.025 to 0.04 μM (14) (Fig. 4 A and B). Next, we evaluated the activity of GS-626510 in xenografts and patient-derived xenografts (PDXs) in two models of chemotherapy-resistant ovarian cancer in vivo. We found GS-626510 to be active against the ovarian cancer xenograft (i.e., KRCH31) (Fig. 4 C and D) as well as the PDX model (i.e., OMM78, Fig. 4 E and F). Indeed, twice-daily oral administration of GS-626510 showed a significant tumor growth inhibition after 21 d of treatment (P = 0.003 and P = 0.0005 for xenografts and PDXs, respectively) and significantly improved overall survival in the PDX model of chemotherapy-resistant disease (Fig. 4, P = 0.003). Pharmacodynamics studies in KRCH31 xenografts demonstrated a significant on-target effect of both JQ1 and GS-626510 on c-MYC (SI Appendix, Fig. S7 and Table S10). Of interest, in our experimental conditions, the novel BET inhibitor GS-626510 was more potent than JQ1 in head-to-head in vivo experiments (P = 0.01).
Fig. 4.
GS-626510 and JQ1 inhibited cell proliferation in primary ovarian carcinoma cell lines in vitro and in ovarian cancer xenografts and PDXs in vivo. (A and B) IC50 for GS-626510 and JQ1 of primary carcinoma cell lines incubated for 72 h with varying concentrations of GS-626510 and JQ1. (C and D) Tumor growth inhibition of OSC-KRCH31, a chemotherapy-resistant tumor cell line established from a patient experiencing progressive disease in vivo. (E) Tumor growth inhibition of OSC-OMM78, a chemotherapy-resistant PDX established from a patient experiencing progressive disease in vivo. (F) Overall survival of animals harboring OSC-OMM78 PDX and treated with GS-626510 vs. placebo control. *P < 0.05.
Discussion
We analyzed by WES the genetic landscape of primary, metastatic, and recurrent ovarian cancer, most with HG-SC histology, obtained from fresh surgical biopsy samples. We also subjected 13 SBOC tumors to WES to evaluate their clonal relationship and evolutionary history. Our results demonstrate that despite displaying a mixture of shared and unique somatic mutations, the overall distribution of substitution types in matched tumor signatures was very similar in primary, metastatic, and recurrent tumors. Importantly, among mutations in known cancer genes, 77% were transmitted from primary tumors to metastatic tumors, and 80% were transmitted from primary to recurrent tumors. These data are consistent with the results of reports demonstrating that while primary ovarian tumors may exhibit individual evolutionary trajectories and diverse genomic tapestries (15, 16), the large majority of driver mutations originally present are retained throughout tumor evolution, with little accumulation of new somatic mutations and copy number alterations during transcoelomic metastasis (15, 16). Taken together, these findings support recent evidence suggesting that cancer spreading to the peritoneal cavity or the other ovary may take place very early during ovarian cancer natural history using the primary tumor preexisting metastatic ability to rapidly disseminate to the adipocyte-rich omentum (17) rather than relying on the acquisition of additional driver mutations, as described in tumor evolution models of other human solid tumors (18, 19). Importantly, these findings, combined with the capability of ovarian tumor-associated macrophages to drive spheroid formation and independent tumor growth at early stages of transcoelomic metastasis, as recently demonstrated by our group (20), may pose formidable clinical challenges for the early detection of ovarian cancer through active screening/surveillance.
Previous studies using a variety of molecular analyses have attempted to determine the relationship between synchronous ovarian tumors (21, 22). While some of these reports using cytogenetic analysis indicated that bilateral ovarian tumors develop through metastatic spreading (21), others provided inconsistent conclusions, with up to 24% of synchronous ovarian tumors found to be polyclonal when evaluated using somatic mitochondrial DNA variants (22). Of interest, a recent study of 12 SBOC tumors analyzed using next-generation sequencing found that all 12 cases were clonally related (23). Our comprehensive WES study of 13 bilateral ovarian tumors is consistent with this recent data. Together with the 12 patients in the Yin et al. (23) study, our results unequivocally demonstrate the uniform monoclonality of ovarian tumors and further support the conclusion that cancer cell dissemination to the other ovary may occur early, when primary carcinoma is still relatively small (<100 million cells), and form through pelvic spread rather than independent multifocal oncogenesis.
Mutations in HRR genes are of particular interest in ovarian cancer patients, because poly (ADP-ribose) polymerase (PARP) inhibitors, a promising new class of targeted therapies, are hypothesized to induce synthetic lethality in HRR-deficient cells and, accordingly, may be therapeutically effective specifically in patients harboring BRCA gene mutations or with platinum-sensitive disease (20). In our study, 50.6% (39/77) of patients were found to harbor a germinal or somatic damaging mutation in an HRR gene implicated in ovarian cancer predisposition. These data are consistent with previous observations (3, 4) and suggest that evaluation of HRR deficiency may represent a cost-effective approach for the identification of ovarian cancer patients potentially benefitting the most from PARP-inhibitor treatment.
Identification of targeted agents effective against platinum-resistant ovarian cancer remains an unmet medical. HG-SC is a disease driven not by recurrent somatic point mutations but by genomic instability as documented in previous studies demonstrating high copy number gains and losses (3, 4). Accordingly, TP53 was the only gene found somatically mutated in a large number of ovarian cancer samples, with few other genes, including NF1, MACF1, and BRCA1/2, showing a lower prevalence of mutations. Of interest, in addition to abrogating the tumor suppressor functions of wild-type p53, ovarian cancer-associated p53 mutations like the one identified in our study have been recently suggested to confer the mutant protein with new activities (i.e., mutp53 gain of function) that can contribute actively to both tumor progression and increased resistance to treatment (24). Importantly, in addition to recurrent TP53 mutations in primary, metastatic, and recurrent tumors, recurrent PIK3CA mutations in the kinase domain (H1047R) were found in two patients with recurrent disease. These data suggest a potential progressive PIK3CA pathway activation/dependence in ovarian cancer during evolution and/or chemotherapy exposure.
We also detected multiple recurrent focal deletions across primary, metastatic, and recurrent tumors. CNV analysis allowed the identification of 3q26, 8q23-24, and 14q11 as the most significantly amplified regions across all ovarian cancer groups tested. The 3q26 and 8q23-24 amplifications encompass PIK3CA and c-MYC, respectively, two genes previously identified in WES ovarian cancer studies and implicated in survival and chemoresistance (3, 4). These results raise the possibility that treatments targeting genes and pathways such as PIK3CA and c-MYC may prove efficacious in treating chemotherapy-resistant disease. Importantly, the establishment and characterization of multiple primary ovarian cancer cell lines, including two fully sequenced (WES) ovarian tumor models originated from patients with progressive disease, provided us with the opportunity to test the activity of BET inhibitors in both in vitro and in vivo experiments of chemotherapy-resistant ovarian cancer.
Despite its abundant amplification in cancer and its known driver function, c-MYC is difficult to inhibit with small molecules due to the lack of targetable ligand sites. Additionally, c-MYC is typically up-regulated through amplification rather than targetable gain-of-function mutations (25). Importantly, BET inhibition has been recently developed as a novel strategy for treating c-MYC–driven cell proliferation because BET protein inactivation may consistently down-regulate c-MYC transcription (26).
Accordingly, GS-626510 and JQ1 target c-MYC by reversibly binding the BET proteins BRD2, BRD3, BRD4, and BRDT, preventing protein–protein interaction between BET proteins and acetylated histones or transcription factors (14). We showed that GS-626510, a novel oral BET inhibitor, has remarkable activity against chemotherapy-resistant cell lines not only in vitro but also in vivo against xenografts and PDXs established from patients harboring chemotherapy-resistant tumors. These preclinical data with JQ1 and GS-626510 in primary ovarian cancer cell lines and xenografts confirm and expand the results of recent in-tumor shRNA genetic screens revealing c-MYC overexpression as a therapeutic target in chemotherapy-resistant tumors through the use of BRD4 inhibitors (27, 28). While previous studies have found that some ovarian tumors may contain BRD4 amplifications and that such tumors may respond to BET inhibition (29), the prevalence of these amplifications in ovarian cancer tends to be low (30). Our data did not show any significant point mutations or CNVs in any BET genes, suggesting that the activity of JQ1 and GS-626510 in our tumor models was most likely related to c-MYC amplification that may be targetable by BET inhibition along with BRD4 amplifications. Consistent with this view, pharmacodynamics studies in KRCH31 xenografts demonstrated a significant on-target effect of both JQ1 and GS-626510 on c-MYC. Transcription down-regulation of c-MYC using a variety of novel BET inhibitors is currently being tested in clinical trials (25).
In conclusion, our results define the genetic landscape of primary, metastatic, and recurrent HG-SC and provide insight into the origins of bilateral ovarian tumors. Some of the genes and pathways frequently mutated in recurrent chemotherapy-resistant disease, such as PIK3CA and c-MYC, may represent immediate targets using existing PIK3CA and BET inhibitor targeted agents currently in phase I/II clinical trials. Preclinical in vitro and in vivo validation studies with the potent BET inhibitor GS-626510 in chemotherapy-resistant ovarian cancer cell lines and xenografts further support this view. These findings provide a useful starting point for further work to define the best therapeutic approaches to the treatment of chemotherapy-resistant disease.
Materials and Methods
Patients and Specimens.
The collection of the specimens and the study protocol were approved by the Azienda Socio Sanitaria Territoriale Spedali Civili di Brescia and the Yale University Human Investigation Committees. Prior to surgical staging, patients were consented for tumor banking in accordance with the Declaration of Helsinki. DNA was extracted from 64 primary tumors, including 13 matched pairs from patients with bilateral ovarian tumors, metastatic tumors (n = 41), and recurrent tumors (n = 17) from 77 patients. Most of the tumors (84%) had HG-SC histology. The 1988 International Federation of Gynecology and Obstetrics staging system was used, and histology was further evaluated by board-certified pathologists to confirm the diagnosis. See SI Appendix, Materials and Methods for details.
WES.
DNA was extracted from frozen samples by standard methods. Genomic DNA was captured on the NimbleGen 2.1M human exome array and subjected to 74 base paired-end reads on the Illumina HiSEq 2000 instrument as previously described (5). Sequence reads were mapped to the reference genome (hg19) using the ELAND program. Reads outside the targeted sequences were discarded, and statistics on coverage were collected from the remaining reads using in-house Perl scripts.
Somatic Single-Nucleotide Mutation Calling.
For matched tumor–normal pairs, somatic point mutations were called by MuTect2. The output from MuTect2 was further filtered to remove false-positive calls. All C>A mutations were excluded from analysis in two tumors that contained possible oxidative damage, as evidenced by high C>A mutation count. The same transformation was applied to their respective paired tumors for mutation count comparisons. Somatic indels were called by an in-house pipeline, and all indels were manually curated. For unmatched tumors, SAMtools was used to call variant bases appended with quality scores. Among these, variants with a frequency >2 × 10−5 in the Exome Aggregation Consortium database (exac.broadinstitute.org) were excluded. See SI Appendix, Materials and Methods for details.
Somatic Copy Number Mutation Calling.
The ratio of normalized coverage depth between normal and primary, metastatic, or recurrent tumor samples was calculated for each exome capture probe. The distribution of coverage depth ratio was evaluated for each individual tumor–normal pairs, and only samples showing strong clustering at discrete values were included in CNV analysis. This quality-control procedure was previously detailed in ref. 31. LOH calling and purity estimation were performed as previously described (12).
Evaluation of Subclonality in SBOC Samples.
Variant allele fraction of somatic mutations called by MuTect2 and allele-specific CNVs called by Sequenza (32) were provided to PyClone (33) to cluster cancer cell fractions using a hierarchical bayesian clustering model. Clusters containing at least two mutations were used to infer a consensus clonal evolution model for bilateral ovarian tumors using ClonEvol (34).
qRT-PCR.
RNA isolation and qRT-PCR was performed using standard protocols on the AB 7500 RealTime PCR instrument. Primer sequences are described in SI Appendix, Materials and Methods.
Cell Lines.
Primary ovarian carcinoma cell line establishment is described in detail in SI Appendix, Materials and Methods. Source-patient characteristics are described in SI Appendix, Table S1.
Drugs.
GS-626510 and JQ1 were obtained from Gilead Science Inc. They were dissolved in DMSO (Sigma-Aldrich) to create a 10 mM stock solution for the in vitro studies.
Cell Viability Assay.
To determine dose response, cells were treated with scalar amounts of each drug ranging from 0.001 μM to 5 μM and then counted by flow cytometry. Further information is provided in SI Appendix, Materials and Methods.
Xenograft Implantation and in Vivo Drug Study.
Briefly, the cell line KRCH31 and the OMM78 PDX were xenografted in female CB17/lcrHsd-Prkd/scid mice s.c. into the lower abdomen area. Mice were triaged into treatment groups when tumor was established. Dosing began upon reaching target size and was delivered orally twice daily for GS-626510 and i.p. once daily for JQ1 for a total of 21 d. On day 21, after last dose administration, animals were either killed or followed up for survival. All mice were housed and treated in accordance with the policies set forth by the Institutional Animal Care and Use Committee at Yale University.
Pharmacodynamics Experiments.
Immunohistochemistry was performed on the tumor tissues excised from KRCH31 xenografted animals after 21 d of twice-daily oral treatment with GS-626510 (10 mg/kg) or daily i.p. treatment with JQ1 (50 mg/kg) as previously described (35).
Statistical Analysis.
The IC50 values of the cell lines were compared using one-way analysis of variance. Grouped mean IC50 values were compared using two-tailed Student’s t test. All statistical analysis was performed using Prism 6 software (GraphPad Prism Software Inc.). P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the NIH (U01 CA176067-01A1), the Deborah Bunn Alley Ovarian Cancer Research Foundation, The Honorable Tina Brozman Foundation, the Discovery to Cure Foundation, the Stand Up to Cancer Foundation, and the Guido Berlucchi Foundation (to A.D.S.); and by Gilead Sciences Inc.. This investigation was also supported by the Research Grant CA-16359 from the NIH National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814027116/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Matulonis UA, et al. Ovarian cancer. Nat Rev Dis Primers. 2016;2:16061. doi: 10.1038/nrdp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patch AM, et al. Australian Ovarian Cancer Study Group Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 5.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen HJ, et al. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 2006;20:1933–1945. doi: 10.1101/gad.1411206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dogruluk T, et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res. 2015;75:5341–5354. doi: 10.1158/0008-5472.CAN-15-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421, and erratum (2013) 502:258. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto H, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkiewicz AK, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown JR, et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res. 2012;18:3791–3802. doi: 10.1158/1078-0432.CCR-11-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micci F, et al. Tumor spreading to the contralateral ovary in bilateral ovarian carcinoma is a late event in clonal evolution. J Oncol. 2010;2010:646340. doi: 10.1155/2010/646340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashashati A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231:21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JY, et al. Tumor evolution and intratumor heterogeneity of an epithelial ovarian cancer investigated using next-generation sequencing. BMC Cancer. 2015;15:85. doi: 10.1186/s12885-015-1077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nieman KM, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 19.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 20.Yin M, et al. Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. J Clin Invest. 2016;126:4157–4173. doi: 10.1172/JCI87252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pejovic T, et al. Bilateral ovarian carcinoma: Cytogenetic evidence of unicentric origin. Int J Cancer. 1991;47:358–361. doi: 10.1002/ijc.2910470308. [DOI] [PubMed] [Google Scholar]
- 22.Van Trappen PO, et al. Somatic mitochondrial DNA mutations in primary and metastatic ovarian cancer. Gynecol Oncol. 2007;104:129–133. doi: 10.1016/j.ygyno.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Yin X, et al. Clonality, heterogeneity, and evolution of synchronous bilateral ovarian cancer. Cancer Res. 2017;77:6551–6561. doi: 10.1158/0008-5472.CAN-17-1461. [DOI] [PubMed] [Google Scholar]
- 24.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Gao P. Small molecules remain on target for c-Myc. eLife. 2017;6:e22915. doi: 10.7554/eLife.22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baratta MG, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proc Natl Acad Sci USA. 2015;112:232–237. doi: 10.1073/pnas.1422165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi X, et al. Loss of TRIM33 causes resistance to BET bromodomain inhibitors through MYC- and TGF-β-dependent mechanisms. Proc Natl Acad Sci USA. 2016;113:E4558–E4566. doi: 10.1073/pnas.1608319113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhyasen GW, et al. BRD4 amplification facilitates an oncogenic gene expression program in high-grade serous ovarian cancer and confers sensitivity to BET inhibitors. PLoS One. 2018;13:e0200826. doi: 10.1371/journal.pone.0200826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones DH, Lin DI. Amplification of the NSD3-BRD4-CHD8 pathway in pelvic high-grade serous carcinomas of tubo-ovarian and endometrial origin. Mol Clin Oncol. 2017;7:301–307. doi: 10.3892/mco.2017.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao S, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci USA. 2013;110:2916–2921. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Favero F, et al. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth A, et al. Clonal genotype and population structure inference from single-cell tumor sequencing. Nat Methods. 2016;13:573–576. doi: 10.1038/nmeth.3867. [DOI] [PubMed] [Google Scholar]
- 34.Dang HX, et al. ClonEvol: Clonal ordering and visualization in cancer sequencing. Ann Oncol. 2017;28:3076–3082. doi: 10.1093/annonc/mdx517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonazzoli E, et al. Inhibition of BET bromodomain proteins with GS-5829 and GS-626510 in uterine serous carcinoma, a biologically aggressive variant of endometrial cancer. Clin Cancer Res. 2018;24:4845–4853. doi: 10.1158/1078-0432.CCR-18-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.