Cryoelectron microscopy (cryo-EM) has revolutionized knowledge of protein remodeling and unfolding by ATPases of the AAA (ATPases associated with various cellular activities) family, including those associated with energy-dependent proteolysis. A study in PNAS by Majumder et al. (1) provides insight into evolutionary conserved functions of AAA-ATPases through cryo-EM single-particle analysis of the archaeal AAA-ATPase proteasome-activating nucleotidase (PAN) bound to 20S proteasomes [core particles (CPs)]. Five conformational states of PAN were identified in the presence of adenosine 5′-O-(3-thiotriphosphate) (ATPγS) that now guide understanding of its functional cycle in the conversion of nucleotide binding, hydrolysis, and release into the gripping, unfolding, and translocation of substrate proteins into the proteasomal CP for destruction. Particularly striking is the sequential cycle of the PAN ATPase that relies on nucleotide-dependent intersubunit signaling to trigger residues to grip and move substrate protein through its central pore. The mechanism can be described as protomers traversing down a spiral staircase of substrate engagement; the step from the bottom to the top provides the protomer its next turn in the sequential ATPase cycle that is interwoven with substrate engagement and unfolding. This around-the-ring ATP cycle is observed for related ATPases of the AAA+ superfamily of eukaryotes and bacteria that unfold proteins, revealing that the mechanism is ancient and conserved in all domains of life.
Proteasomes are energy-dependent proteases needed for proteostasis and the regulated turnover of proteins associated with metabolism, signaling, cell cycle control, stress responses, and other important cellular processes (2). Proteasomes are widespread and essential for survival of eukaryotes (3) and archaea (4) and are needed for the persistence of mycobacteria (5). This central function provides impetus for the design and use of proteasome inhibitors to treat cancers and infectious disease (6). Proteins targeted to the proteasome are often covalently linked to ubiquitin or ubiquitin-like protein modifiers (7). Unstructured regions of proteins can also trigger proteasome-mediated proteolysis.
Although peptide-bond hydrolysis is exergonic and the proteolytic active sites are relatively nonspecific, the proteasome system has evolved into a selective, energy-dependent proteolytic machine. This energy dependence, while costly, ensures that the machine is selective and processive in degrading proteins to peptides. This feature avoids mistakes and prevents the buildup of partial proteins that could otherwise perturb cell function (e.g., protein aggregation).
Proteasomes are energy dependent due to their self-compartmentalized architecture. The proteolytic active sites are housed within a 20S CP formed by four stacked heptameric rings, with the outermost rings of α-type subunits and the innermost rings of β-type subunits (8). The α-rings form the gated, narrow openings on each end of the cylinder that prevent the access of folded proteins into the proteolytic chamber formed by the β-type subunits. The gates are opened and the substrate protein is unfolded and translocated into the proteolytic chamber by AAA-ATPases that bind and hydrolyze ATP to generate the mechanical movement needed for these processes to occur. In eukaryotes, a 19S regulatory particle (RP) binds and regulates both ends of the CP to form the 26S proteasome. The RP is an elaborate structure of at least 19 subunits that recruits polyubiquitinated proteins and catalyzes their deubiquitination and unfolding during the proteolytic process. Most RP subunits are unique to eukaryotes, with the exception of the AAA-ATPase Rpt1 to Rpt6 and JAMM/MPN Rpn11 deubiquitinase subunits, which are related to the PAN and JAMM2 required for the proteasome-mediated proteolysis of ubiquitin-like modified proteins in archaea (9).
While archaea have led the way in understanding proteasomal CP structure and function (8), the AAA-ATPases associated with this energy-dependent proteolytic system have remained relatively elusive from a structural perspective due to their dynamic nature. A network of AAA-ATPases (e.g., PAN and Cdc48/VAT) are found to stimulate the energy-dependent hydrolysis of proteins by CPs in the archaeal system, but are not tightly bound to the CP (9–13). Furthermore, the archaeal AAA-ATPases are simultaneously in multiple conformational states that are altered by ATP cycling (14–18). This dynamic nature, coupled with low affinity, has complicated the capture of intact CPs in complex with AAA-ATPases for structural analysis in archaea. Negative-stain EM imaging of AAA-ATPases bound to CPs (19–21) and X-ray crystallography of PAN subdomains (22, 23) have provided glimpses into the mechanism but are limited when considering dynamic structures. Cryo-EM analysis of an archaeal Cdc48 engaged with internalizing itself as a substrate (18) provides promise that the dynamic nature of AAA-ATPases can be captured in the act of proteasome-mediated proteolysis in archaea.
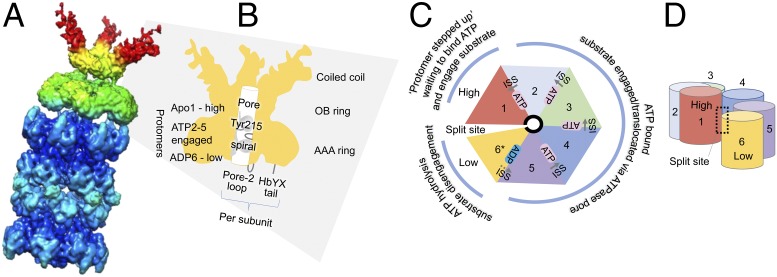
To advance the field, Majumder et al. (1) used cryo-EM to identify the structural states of archaeal PAN bound to CPs (Fig. 1). The archaeon of choice was Archaeoglobus fulgidus, a sulfate reducer from marine hydrothermal environments (24). The researchers purified the archaeal proteins from recombinant Escherichia coli and reconstituted the complex using ATPγS. CPs capped on each end by PAN were found to predominate the cryo-EM field. To enhance resolution, the team treated these doubly capped complexes as two pseudo–single-capped PAN-CPs, and thus were freed from the constraint of C2 symmetry when constructing the 3D density map. For more detailed analysis, the region encompassing the AAA and the oligonucleotide/oligosaccharide-binding fold rings of PAN (designated AAAob) was extracted from the map. From this heroic approach, five distinct conformational states of the AAAob region were identified. Contacts between PAN and the CP were also discerned.
Fig. 1.
Archaeal PAN-proteasome complex. (A) The 3D density map of the pseudo–single-capped PAN bound to the 20S CP as determined by Majumder et al. (1). Modified with permission from ref. 1. (B–D) Highlights of the PAN hexamer, including the N-terminal coiled-coil domain, oligonucleotide/oligosaccharide-binding fold (OB) ring, and AAA-ATPase ring. The spiral staircase of pore loop Tyr215 (needed for protein unfolding and translocation) and the bipartite determinants (pore-2 loop and HbYX tail) that interact with the CP are indicated. Protomer 1 is in a nucleotide-free (Apo) state, protomers 2–5 are ATP bound, and protomer 6 is the active pocket where ATP hydrolysis occurs (ADP; 6* in C). The split site formed in the spiral staircase between protomer 1 (highest) and protomer 6 (lowest) is indicated. In the next round of the ATPase cycle, protomer 6 steps up to the 1 position and the nucleotide pocket of protomer 5 (now the lowest position) becomes active in ATP hydrolysis. Intersubunit signaling (ISS) of strong (solid gray arrows in C), intermediate (dotted gray arrow in C), and split contacts is correlated with substrate engagement (e.g., Tyr215 availability) and protomer elevation in the ring.
Together, these exciting findings by Majumder et al. reveal that the fundamental mechanism of AAA+-ATPases is conserved in all domains of life and suggest an ancient origin for these protein unfolding machines.
Considering this and previous work, a model emerges of the PAN ATPase cycle that is used to grip and move substrate proteins through its central pore in a manner that resembles a ride down a spiral staircase (Fig. 1). The PAN protomer at the bottom actively hydrolyzes ATP and releases substrate into the CP, while the protomer at the top has just stepped up to the plate to obtain its ATP ticket for the next ride in gripping and dragging the substrate protein through the AAA tunnel. Thanks to these new structures, the intersubunit signaling contacts between the PAN protomers are now correlated with nucleotide occupancy and substrate engagement. This around-the-ring ATP cycle that drives protein unfolding and remodeling can be observed in related AAA+-ATPases of eukaryotes and bacteria, including those that are substrate-engaged (e.g., 26S proteasome and ClpB) (25, 26). Together, these exciting findings by Majumder et al. reveal that the fundamental mechanism of AAA+-ATPases is conserved in all domains of life and suggest an ancient origin for these protein unfolding machines.
Acknowledgments
Funds for this project were awarded to J.M.F. through the Bilateral Biotechnology and Biological Sciences Research Council/NSF Directorate for Biological Sciences program (NSF Grant 1642283); the US Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences, Physical Biosciences Program (Grant DOE DE-FG02-05ER15650); and the NIH (Grant R01 GM57498).
Footnotes
The author declares no conflict of interest.
See companion article on page 534.
References
- 1.Majumder P, et al. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc Natl Acad Sci USA. 2019;116:534–539. doi: 10.1073/pnas.1817752116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara T, et al. Proteasomes are essential for yeast proliferation. cDNA cloning and gene disruption of two major subunits. J Biol Chem. 1990;265:16604–16613. [PubMed] [Google Scholar]
- 4.Zhou G, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 6.Bibo-Verdugo B, Jiang Z, Caffrey CR, O’Donoghue AJ. Targeting proteasomes in infectious organisms to combat disease. FEBS J. 2017;284:1503–1517. doi: 10.1111/febs.14029. [DOI] [PubMed] [Google Scholar]
- 7.Maupin-Furlow JA. Prokaryotic ubiquitin-like protein modification. Annu Rev Microbiol. 2014;68:155–175. doi: 10.1146/annurev-micro-091313-103447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 9.Fu X, et al. Ubiquitin-like proteasome system represents a eukaryotic-like pathway for targeted proteolysis in archaea. MBio. 2016;7:e00379-16. doi: 10.1128/mBio.00379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson HL, Ou MS, Aldrich HC, Maupin-Furlow J. Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J Bacteriol. 2000;182:1680–1692. doi: 10.1128/jb.182.6.1680-1692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
- 12.Barthelme D, Sauer RT. Identification of the Cdc48•20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forouzan D, et al. The archaeal proteasome is regulated by a network of AAA ATPases. J Biol Chem. 2012;287:39254–39262. doi: 10.1074/jbc.M112.386458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snoberger A, Brettrager EJ, Smith DM. Conformational switching in the coiled-coil domains of a proteasomal ATPase regulates substrate processing. Nat Commun. 2018;9:2374. doi: 10.1038/s41467-018-04731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YC, Snoberger A, Schupp J, Smith DM. ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function. Nat Commun. 2015;6:8520. doi: 10.1038/ncomms9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwitz AA, et al. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem. 2007;282:22921–22929. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- 18.Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife. 2017;6:6. doi: 10.7554/eLife.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Medalia N, et al. Architecture and molecular mechanism of PAN, the archaeal proteasome regulatory ATPase. J Biol Chem. 2009;284:22952–22960. doi: 10.1074/jbc.M809643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barthelme D, Chen JZ, Grabenstatter J, Baker TA, Sauer RT. Architecture and assembly of the archaeal Cdc48*20S proteasome. Proc Natl Acad Sci USA. 2014;111:E1687–E1694. doi: 10.1073/pnas.1404823111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 24.Stetter KO. Archaeoglobus fulgidus gen. nov, sp. nov.: New taxon of extremely thermophilic Archaebacteria. Syst Appl Microbiol. 1988;10:172–173. [Google Scholar]
- 25.Yu H, et al. ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc Natl Acad Sci USA. 2018;115:E9560–E9569. doi: 10.1073/pnas.1810648115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de la Peña AH, Goodall EA, Gates SN, Lander GC, Martin A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science. 2018;362:eaav0725. doi: 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]