Abstract
The role of the Pseudomonas aeruginosa OxyR-controlled antioxidants alkyl hydroperoxide reductase CF (AhpCF) and catalase B (KatB) was evaluated in biofilm versus planktonic culture upon exposure to H2O2. AhpCF was found to be critical for survival of biofilm bacteria while KatB was more important for survival of planktonic free-swimming organisms.
Keywords: Pseudomonas aeruginosa, Hydrogen Peroxide, OxyR, Biofilms, Alkylhydroperoxide Reductase, Catalase
1. Introduction
Pseudomonas aeruginosa is an important opportunistic pathogen of humans, predominantly immunocompromised individuals suffering from burn [1], cancer chemotherapy [2], organ transplantation [3] various pneumonias [4]) or patients afflicted with the autosomal genetic disorder, cystic fibrosis (CF) [5, 6]. During CF airway infection, neutrophil titers rise nearly 1500-fold and remain so during the course of the disease [7]. The infection process triggers activation of a respiratory burst, creating an oxidative stress for resident bacteria that predominantly include P. aeruginosa. One potentially toxic product of the respiratory burst is hydrogen peroxide (H2O2, [8, 9]. Within the phagolysosomal vacuole, H2O2 levels can reach concentrations as high as 100 mM [10], a concentration that easily kills P. aeruginosa in both planktonic culture and in biofilms, the latter of which are highly organized, typically adherent, antibiotic-resistant communities [11–15]. In fact, stimulated neutrophils release even significant antimicrobial concentrations of H2O2 (~12 μM) in the extracellular milieu [16].
The major response of P. aeruginosa to H2O2 is governed by the global transactivator, OxyR [17, 18]. This response is generally assumed to be predominantly a defensive strategy because the three major OxyR-regulated genes encode the cytoplasmic antioxidant enzymes catalase B (KatB), alkylhydroperoxide reductase CF (AhpCF), and the periplasmic AhpB [17, 18]. In addition, an ankyrin-like protein AnkB (encoded by the ankB gene), part of a two-gene operon with katB, also contributes to H2O2 resistance by an unappreciated mechanism. An oxyR mutant possesses some very distinct phenotypes, the most unusual is an inability to form single colonies on aerobic but not anaerobic rich media (e.g., Luria-Bertani agar) [17, 19]. When oxyR mutant bacteria are streaked on aerobic Luria agar plates, oxyR mutant bacteria are incapable of growing as isolated colonies because the medium actually generates H2O2. In fact, we have shown that <107 oxyR mutant bacteria cannot grow in aerobic L-broth unless catalase is provided or cultures are grown in the absence of oxygen. This mutant is also impaired virulence properties in mouse and Drosophila melanogaster infection models [20], and is more susceptible to killing by human neutrophils [20]. Still, the most remarkable phenotype of a P. aeruginosa oxyR mutant is an exquisite sensitivity to H2O2, despite the fact that the organism possesses near wild-type catalase activity [17, 18]. In this study, we evaluated the potentially different roles of OxyR-controlled antioxidant enzymes in planktonic versus biofilm resistance to the important biocide, H2O2.
2. Materials and Methods
2.1. Bacterial growth conditions
Planktonic free-living P. aeruginosa were routinely grown aerobically in either trypticase soy broth (TSB) or Luria-Bertani broth (LB). Media were supplemented with 200 μg ml−1 carbenicillin when plasmid maintenance was required. Planktonic cultures were grown at 37°C with shaking at 300 rpm for 18 h. For culture of biofilm cells, aerobic, stationary phase bacteria were diluted 1:50 into 1% TSB and 0.2 ml was used to inoculate Stovall flow chambers (Stovall Life Sciences, Greensboro, NC). After a 1 h incubation at room temperature, media flow was initiated at a rate of 0.17 ml min−1 and biofilms were allowed to develop for 3 days. For enumeration of colony forming units (CFU) on agar surfaces, media were solidified with 1.5% Bacto agar.
2.2. H2O2 susceptibility measurements
All bacteria were grown aerobically in TSB for 18 h at 37°C. Cells were diluted to a final O.D.600 nm of 0.01 in 5 ml of TSB broth containing 0.8% low-melting-point agarose (SeaPlaque). Suspensions were distributed evenly on 15 × 100 mm TSB agar plates and allowed to solidify. Filter paper disks (7 mm) impregnated with 10 μl of 8.8 M H2O2 were placed on the top-agar surface and the plates were incubated at 37°C for 24 h. Mature, 3 day-old biofilms were treated with different concentrations of H2O2 for 30 min before staining with a viability stain composed of SYTO 9 (green fluorescence) and propidium iodine (red fluorescence) (BacLight, InVitrogen). Biofilm images were obtained using an LSM 510 confocal microscope. The live/dead ratios of the bacteria embedded within biofilms were calculated using the 3-dimensional reconstruction component for LSM (V.1.4.2) software.
2.3. Catalase assays
Planktonic and biofilm bacteria were harvested by centrifugation at 4°C. The cell pellets were then resuspended in ice cold 50 mM potassium phosphate, pH 7.0 (KPi) and disrupted by intermittent sonication on ice. Catalase activity in cell extracts was determined by following the decomposition of 19.5 mM H2O2 in KPi at 240 nm using a Spectronics Genesys 5 spectrophotometer [11, 12, 19]. The Bradford protein assay was used to estimate protein concentration with bovine serum albumin (fraction V) serving as the standard [21]. The definition of one unit (U) of activity is defined as that which decomposes 1 μmol of H2O2 min−1 mg protein−1.
2.4. H2O2 minimum inhibitory concentration (MIC) measurements
Aerobic stationary phase cultures were diluted 5000-fold into 5 ml of fresh TSB containing increasing concentrations of H2O2. The bacteria were then grown under aerobic conditions for an additional 18 h on a roller wheel at 70 rpm after which the MICs were recorded.
2.5. ß-galactosidase assays
For planktonic cultures, stationary phase aerobic bacteria grown in TSB were used while mature, 3 day-old biofilm bacteria were collected from flow cells by scraping the biofilms into ice cold Z-buffer [22]. Samples were then assayed for ß-galactosidase activity [22]. All assays were performed at least in triplicate, and the values recorded as the mean +/− standard error.
3. Results and Discussion
3.1. H2O2 susceptibility of P. aeruginosa oxyR mutant bacteria in planktonic culture: role of KatB, AhpB and AhpCF.
Planktonic oxyR mutant bacteria harboring plasmids expressing either katB, ahpB or ahpCF were first grown in TSB to stationary phase and H2O2 sensitivity profiles were measured. Fig. 1A demonstrates that provision of the catalase, KatB (lane 5), helped protect the oxyR mutant against H2O2 more effectively than the alkyl hydroperoxide reductase, AhpCF (lane 6). The periplasmic enzyme, AhpB, provided no protection compared to the aforementioned enzymes (lane 4). Moreover, MIC measurements clearly showed that KatB is more critical for protection of planktonic bacteria against this biocide (Fig. 1B, lanes 4 vs. 5,6) while oxyR mutant bacteria are >400-fold more sensitive to H2O2 in planktonic culture than their wild-type counterparts (Fig.1B, lanes 2 vs. 1).
Fig. 1. Sensitivity of P. aeruginosa compared with mutants strains to H2O2.
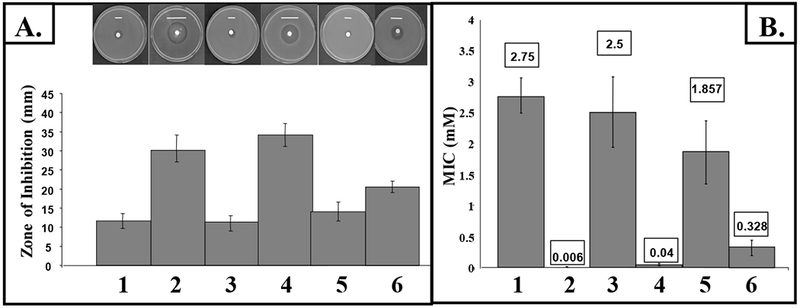
(A) Equivalent titers of aerobic stationary phase bacteria grown in TSB broth were added to molten top-agarose and poured onto TSB agar plates. Filter paper disks (7 mm) impregnated with 8.8 M H2O2 were then placed on the solidified top agarose. Small white bars, the precise width of the killing zone, are placed just above each zone for ease of viewing. (B) Approximately 106 stationary phase bacteria were sub-cultured into fresh TSB containing increasing concentration of H2O2 after which the MICs were determined 24 hr post-exposure. Lane 1, wild-type; lane 2, oxyR; lane 3, oxyR-poxyR; lane 4, oxyR + pahpB; lane 5, oxyR + pkatB; lane 6, oxyR + pahpCF.
We next examined the role of OxyR-regulated antioxidant enzymes KatB, AhpCF and AhpB in biofilm susceptibility to H2O2. Aerobically grown, stationary phase bacteria were diluted 50-fold in 1% TSB and used as an inoculant for cultivation of mature biofilms for 3 days in a flow-through cultivation system [19]. We have previously shown that oxyR mutant organisms, that are devoid of KatB, AhpB and AhpCF activity, are very sensitive to H2O2 in the low millimolar range during biofilm culture [19]. We performed additional biofilm experiments, comparing the H2O2 sensitivity profiles of oxyR mutant bacteria harboring stable plasmids containing katB, ahpB or ahpCF, respectively. In contrast to wild-type bacteria, the vast majority (~88%) of oxyR mutant bacteria were easily killed by a brief 30 min treatment of mature biofilms with 70 mM H2O2 (Fig. 2), a level needed to cause significant killing of highly refractory P. aeruginosa in biofilm [19]. We have previously shown that such high concentrations of H2O2 can trigger mechanical dispersion of mature biofilms due to vigorous oxygen bubbling mediated by the very high catalase activity of P. aeruginosa [19, 23], thus explaining the somewhat contorted structure of H2O2-treated biofilms (lower panels, Fig. 2). The oxyR mutant expressing AhpB was still very sensitive to H2O2, and comparable in sensitivity with oxyR mutant bacteria (Fig. 2). In contrast, provision of AhpCF restored biofilm resistance to H2O2, in fact, at levels even greater than wild-type bacteria (4.14 ± 1.35 live/dead ratio of treated cells). Surprisingly, and in contrast to planktonic bacteria, the oxyR mutant expressing KatB was nearly as sensitive to H2O2 as the oxyR mutant.
Fig. 2. Confocal laser scanning microscopic images of H2O2-treated biofilms.
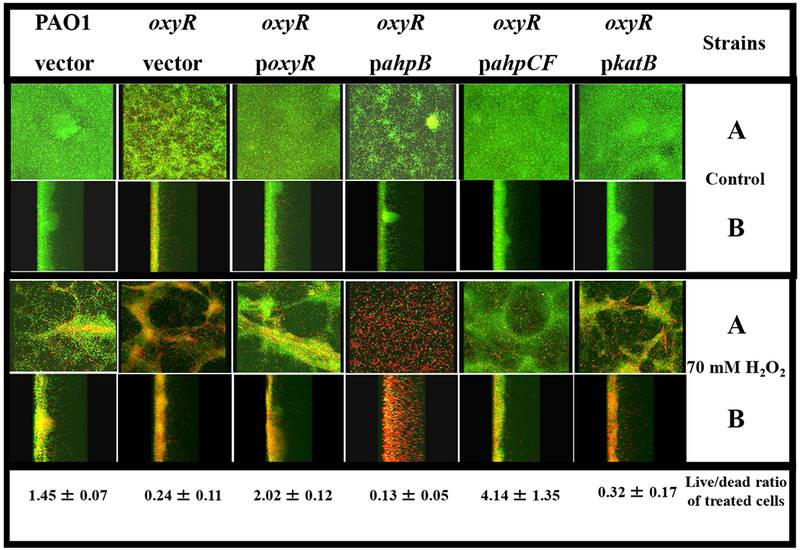
Biofilms were grown in confocal “friendly” flow chambers as previously described by Lequette and Greenberg [30]. Briefly, bacteria were grown aerobically in L-broth at 37°C until the stationary growth phase, diluted 1:50 into 1% TSB and a 0.2 ml suspension used to inoculate flow cells (Stovall Life Sciences, Inc., Greensboro, NC). Flow was initiated at a rate of 0.17 ml min−1 after the bacteria were allowed to attach for 1 hr. After a 3 day incubation at room temperature (~23°C), biofilms were treated with 70 mM H2O2 for 30 min. The biofilms were then stained with a live/dead viability stain composed of SYTO 9 and propidium iodine (Molecular Probes, Inc., Eugene, OR). Biofilm images were obtained using an LSM 510 confocal microscope (Carl Zeiss, Inc., Germany). The excitation and emission wavelengths for green fluorescence were 488 nm and 500 nm, while those for red fluorescence were 490 nm and 635 nm, respectively. All biofilm experiments were repeated at least 3 times. The live/dead ratios of the biofilms were calculated using the 3D for LSM (V.1.4.2) software (Carl Zeiss, Inc., Germany). The panels labeled A are top/bottom images while those in B are saggital images. The top third panel depicts the strains used in these studies. The live to dead ratio of treated bacteria is given on the bottom panel for each strain.
3.2. Protection of wild-type P. aeruginosa biofilms from H2O2: role of KatB vs. AhpCF.
We next elected to determine whether the protective effect of AhpCF in oxyR mutant bacteria could also be observed in wild-type bacteria. To test this hypothesis, we constructed isogenic single ahpCF and katB mutants in wild-type strain PAO1 and monitored biofilm susceptibility to H2O2. Fig. 3B (in contrast to untreated bacteria in Fig. 3A) demonstrates that the ahpCF mutant in biofilms is more sensitive to H2O2 relative to an isogenic katB mutant. Moreover, the ratio of live versus dead bacteria was nearly 6 times greater in the katB mutant relative to ahpCF mutant bacteria after exposure to 70 mM H2O2 (Fig. 3C).
Fig. 3. Sensitivity of P. aeruginosa ahpCF versus katB mutants to H2O2 in biofilms.
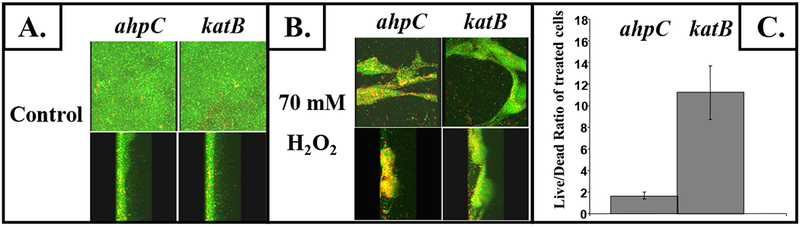
The same conditions used in Fig. 2 were used to grown ahpC and katB mutant biofilms. Confocal laser scanning microscopic images of top and saggital views of mature 3-day old biofilms were obtained in (A) untreated bacteria and (B) after treatment with 70 mM of H2O2 for 30 min followed by viability staining. (C) The live/dead ratios of the treated biofilms were calculated using the 3D for LSM (V.1.4.2) software (Carl Zeiss, Inc., Germany).
3.3. Differential expression of the ahpCF and katB genes in planktonic vs. biofilm bacteria treated with H2O2
To help explain why AhpCF provided better protection of biofilm bacteria against H2O2 while KatB provided better protection of planktonic organisms, we tested the hypothesis that the expression of katB and ahpCF differ during these two distinct growth modes by constructing lacZ reporter fusions to both genes. For planktonic culture, we used stationary phase organisms. Although unattached to surfaces like biofilm bacteria, such organisms represent a metabolically less active form than exponentially growing, mid-log phase cells and, as such, could arguably represent biofilm-like cells. This is due to their significantly reduced metabolic activity and antibiotic resistance properties [24]. We discovered that katB expression is upregulated at least 4.1-fold when organisms are exposed to 5 to 25 mM H2O2 in planktonic culture but only moderately by 5 mM H2O2 in biofilms (Fig. 4A vs. B). In contrast, ahpCF expression was inducible by a factor of ~1.6-fold in biofilms while there was no significant induction in planktonic culture (Fig. 4D vs. C).
Fig. 4. Expression of katB and ahpCF in plantonic vs. biofilm culture in response to various concentrations of H2O2.
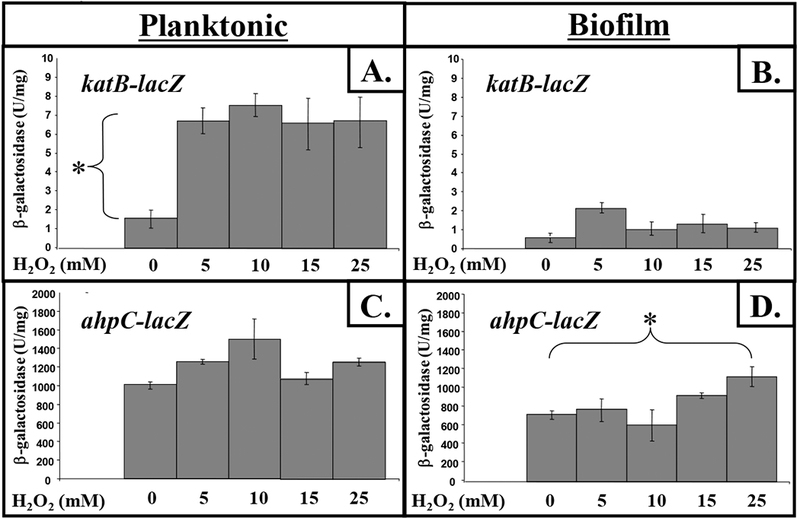
Planktonic, stationary phase bacteria or mature, 3-day old biofilm organisms were exposed to various concentrations of H2O2 for 30 min after which β–galactosidase activity was determined in cell extracts (n=3).
3.4. Catalase-like activity of AhpCF
Finally, because provision of ahpCF in trans afforded protection of biofilm bacteria from H2O2 exposure, we formulated a hypothesis that AhpCF in P. aeruginosa possesses the capacity to degrade H2O2 in addition to organic peroxides. This hypothesis is not unprecedented, for AhpCF in the related organism Xanthomonas campestris (AhpC 86%, AhpF 76% identical to P. aeruginosa AhpCF) was not only inducible in a katA mutant, but possessed catalase activity that rescued the H2O2-sensitivity phenotype of the mutant [25]. Similarly, in Escherichia coli, AhpCF is important for scavenging low concentrations of endogenously generated H2O2 while catalases KatE and KatG provide protection at higher concentrations [26]. This is likely due to the recently discovered fact that AhpCF in the related organism Salmonella typhimurium is able to utilize H2O2 as a substrate with a KM ~100-fold less than that of organic hydroperoxides [27]. To test the hypothesis that AhpCF could function catalatically, we transformed lac-based plasmids harboring constitutively expressed ahpCF into an oxyR katA mutant, that possesses no detectable catalase activity [17, 18], and measured catalase activity in planktonic culture. We discovered that the oxyR katA double mutant harboring ahpCF possessed catalase activity albeit at low levels (0.9 ± 0.1 U mg−1). The low catalase activity is likely due to the fact that all AhpCF proteins examined thus far requires NADH as a cofactor [27, 28]. As mentioned above, the X. campestris AhpCF is inducible in a katA mutant. This is similar to our results demonstrating that expression of ahpC was inducible in biofilm bacteria but not in plantonic organisms (Fig. 4C and D). Therefore, we postulated whether catalase levels under both conditions might be inversely correlative with ahpCF expression. To test this, we measured catalase activity in both planktonic vs. biofilm culture. We found that all biofilm bacteria harbored lower catalase activity compared to their planktonic counterparts, even though it was not significantly different in oxyR mutant bacteria relative to wild-type bacteria (Fig. 5). This is consistent with the fact that mature biofilm bacteria are essentially starved for iron, as evidenced by very low catalase and increased Mn-containing superoxide dismutase activities [29]. Thus, lower expression of catalase in biofilms mediated by alternations in overall iron metabolism relative to planktonic bacteria may be one factor that allows for increased expression of AhpCF in response to H2O2.
Fig. 5. Catalase activity in plantonic vs. biofilm culture.
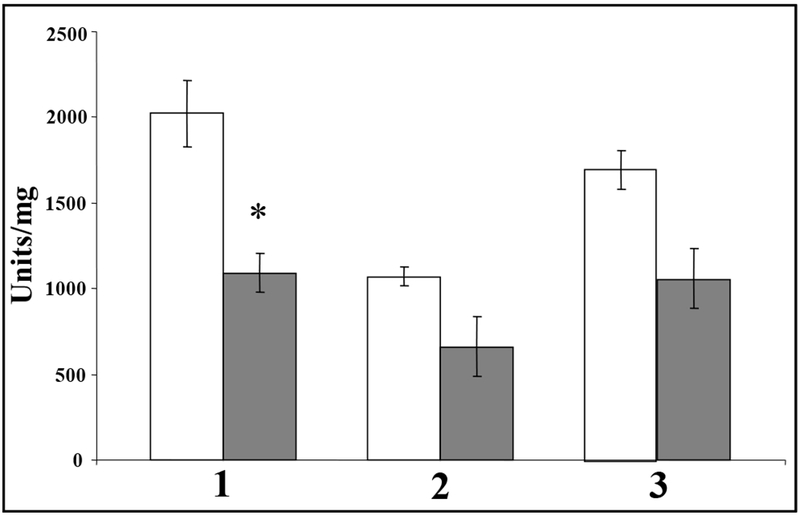
All bacteria were harvested from either stationary phase planktonic (white bars) or 3-day old biofilm (grey bars) culture in either TSB or 1% TSB, respectively. Cell extracts were assayed for catalase activity as described in the materials and methods. Lane 1, wild-type; lane 2, oxyR; lane 3, oxyR-poxyR.
4. Conclusions
In this study, we have discovered that OxyR-regulated gene products in the important opportunistic pathogen P. aeruginosa differ in levels of protection conferred when grown in sessile, organized communities known as biofilms versus planktonic, free-swimming culture. The alkyl hydroperoxidase, AhpCF, was found to be critical for optimal resistance to H2O2 in biofilms, while the 240 kDa tetrameric catalase, KatB, is more critical for protection of planktonic bacteria against this biocide. This was, in part, due to differences in the transcriptional response to H2O2 in biofilms; transcription of ahpCF is increased in biofilm culture while that of katB is more upregulated in planktonic organisms. Moreover, AhpCF is capable of detoxifying H2O2 more effectively in biofilms, when organisms have less catalase than their planktonic counterparts. Future studies are designed to better understand the molecular basis for differential regulation of OxyR-controlled genes in planktonic vs. biofilm bacteria.
Acknowledgements
This work was supported by National Institutes of Health Grant GM-69845 to D.J.H.
REFERENCES
- [1].Holder IA (1993) P. aeruginosa burn infections: pathogenesis and treatment. Pseudomonas aeruginosa as an opportunistic pathogen In, Infectious Agents and Pathogenesis. Plenum Press, N.Y. 275–295. [Google Scholar]
- [2].Simon A, Bode U and Beutel K (2006) Diagnosis and treatment of catheter-related infections in paediatric oncology: an update. Clin Microbiol Infect 12, 606–620. [DOI] [PubMed] [Google Scholar]
- [3].Dua S, Chalermskulrat W, Miller MB, Landers M and Aris RM (2006) Bilateral hematogenous Pseudomonas aeruginosa endophthalmitis after lung transplantation. Am J Transplant 6, 219–224. [DOI] [PubMed] [Google Scholar]
- [4].Lau GW, Hassett DJ and Britigan BE (2005) Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol 13, 389–397. [DOI] [PubMed] [Google Scholar]
- [5].Govan JRW and Harris GS (1986) Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol. Sci 3, 302–308. [PubMed] [Google Scholar]
- [6].Boucher RC (2007) Cystic fibrosis: a disease of vulnerability to airway surface dehydration. Trends Mol Med 13, 231–240. [DOI] [PubMed] [Google Scholar]
- [7].Britigan BE, Hayek MB, Doebbeling BN and Fick RB Jr. (1993) Transferrin and lactoferrin undergo proteolytic cleavage in the Pseudomonas aeruginosa-infected lungs of patients with cystic fibrosis. Infect Immun 61, 5049–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hassett DJ and Imlay JA (2006) Oxidative stress systems in bacteria: four model systems. Eds., Nickerson C and Schurr MJ. Klewer Academic-Plenum Publishers, N.Y.-Boston-Dordrecht-London–Moscow: Chapter 14 pp. 544–573. [Google Scholar]
- [9].Storz G and Imlay JA (1999) Oxidative stress. Curr Opin Microbiol 2, 188–194. [DOI] [PubMed] [Google Scholar]
- [10].Iyer GYN, Islam MF and Quastel JH (1961) Biochemical aspects of phagocytosis. Nature 192, 535–541. [Google Scholar]
- [11].Brown SM, Howell ML, Vasil ML, Anderson AJ and Hassett DJ (1995) Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol 177, 6536–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ma J-F, Ochsner UA, Klotz MG, Nanayakkara VK, Howell ML, Johnson Z, Posey J, Vasil ML, Monaco JJ and Hassett DJ (1999) Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol 181, 3730–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hassett DJ, Ma J-F, Elkins JG, McDermott TR, Ochsner UA, West SEH, Huang C-T, Fredericks J, Burnett S, Stewart PS, McPheters G, Passador L and Iglewski BH (1999) Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol 34, 1082–1093. [DOI] [PubMed] [Google Scholar]
- [14].Hassett DJ, Elkins JG, Ma J-F and McDermott TR (1999) Pseudomonas aeruginosa biofilm sensitivity to biocides: use of hydrogen peroxide as model antimicrobial agent for examining resistance mechanisms. Meth. Enzymol 310, 599–608. [DOI] [PubMed] [Google Scholar]
- [15].Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Hoiby N and Givskov M (2005) Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151, 373–383. [DOI] [PubMed] [Google Scholar]
- [16].Test ST and Weiss SJ (1984) Quantitative and temporal characterization of the extracellular H2O2 pool generated by human neutrophils. J. Biol. Chem 259, 399–405. [PubMed] [Google Scholar]
- [17].Hassett DJ, Alsabbagh E, Parvatiyar K, Howell ML, Wilmott RW and Ochsner UA (2000) A protease-resistant catalase, KatA, that is released upon cell lysis during stationary phase, is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J. Bacteriol 182, 4557–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K and Hassett DJ (2000) Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB, ahpB, and ahpCF. J. Bacteriol 182, 4533–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Panmanee W, Gomez F, Witte D, Vijay P, Britigan BE and Hassett DJ (2008) The Peptidoglycan-Associated Lipoprotein, OprL, Helps Protect a Pseudomonas aeruginosa Mutant Devoid of the Transactivator, OxyR, From Hydrogen Peroxide-Mediated Killing During Planktonic and Biofilm Culture. J Bacteriol 190, 3658–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lau GW, Britigan BE and Hassett DJ (2005) Pseudomonas aeruginosa OxyR is required for full virulence in rodent and insect models of infection and for resistance to human neutrophils. Infect Immun 73, 2550–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- [22].Miller JH (1992) Procedures for working with lac In, A Short Course in Bacterial Genetics. Plainview, New York: Cold Spring Harbor Press; 72–74. [Google Scholar]
- [23].Hassett DJ, Charniga L, Bean KA, Ohman DE and Cohen MS (1992) Antioxidant defense mechanisms in Pseudomonas aeruginosa: resistance to the redox-active antibiotic pyocyanin and demonstration of a manganese-cofactored superoxide dismutase. Infect. Immun 60, 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stewart PS (2002) Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 292, 107–113. [DOI] [PubMed] [Google Scholar]
- [25].Charoenlap N, Eiamphungporn W, Chauvatcharin N, Utamapongchai S, Vattanaviboon P and Mongkolsuk S (2005) OxyR mediated compensatory expression between ahpC and katA and the significance of ahpC in protection from hydrogen peroxide in Xanthomonas campestris. FEMS Microbiol Lett 249, 73–78. [DOI] [PubMed] [Google Scholar]
- [26].Seaver LC and Imlay JA (2001) Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183, 7173–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Parsonage D, Karplus PA and Poole LB (2008) Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci U S A 105, 8209–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Reynolds CM, Meyer J and Poole LB (2002) An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry 41, 1990–2001. [DOI] [PubMed] [Google Scholar]
- [29].Bollinger N, Hassett DJ, Iglewski BH, Costerton JW and McDermott TR (2001) Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J Bacteriol 183, 1990–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lequette Y and Greenberg EP (2005) Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol 187, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


