Abstract
Haematococcus pluvialis is a freshwater species of Chlorophyta, family Haematococcaceae. It is well known for its capacity to synthesize high amounts of astaxanthin, which is a strong antioxidant that has been utilized in aquaculture and cosmetics. To improve astaxanthin yield and to establish genetic resources for H. pluvialis, we performed whole-genome sequencing, assembly, and annotation of this green microalga. A total of 83.1 Gb of raw reads were sequenced. After filtering the raw reads, we subsequently generated a draft assembly with a genome size of 669.0 Mb, a scaffold N50 of 288.6 kb, and predicted 18,545 genes. We also established a robust phylogenetic tree from 14 representative algae species. With additional transcriptome data, we revealed some novel potential genes that are involved in the synthesis, accumulation, and regulation of astaxanthin production. In addition, we generated an isoform-level reference transcriptome set of 18,483 transcripts with high confidence. Alternative splicing analysis demonstrated that intron retention is the most frequent mode. In summary, we report the first draft genome of H. pluvialis. These genomic resources along with transcriptomic data provide a solid foundation for the discovery of the genetic basis for theoretical and commercial astaxanthin enrichment.
Keywords: genome sequencing, assembly, annotation, astaxanthin, Haematococcus pluvialis
Introduction
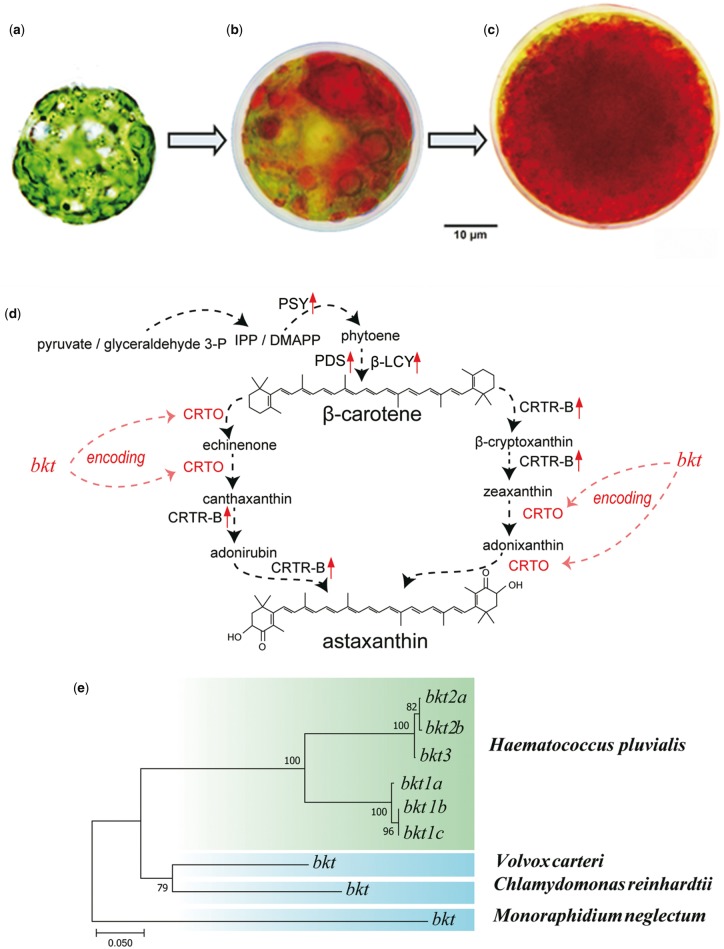
Haematococcus pluvialis is a unicellular green alga and is considered as the best natural resource for astaxanthin, which is a high-value carotenoid with strong biological activity for the food, feed, and pharmaceutical industries (Ambati et al. 2014). It has an interesting life cycle with a remarkable division between green motile and red immobile stages (fig. 1a–c). It enters the green motile stage under favorable environmental conditions. During their vegetative growth, H. pluvialis cells are spherical, ellipsoidal, or pear-shaped with flagella and chloroplasts (fig. 1a). When exposed to unfavorable environmental or stress conditions, H. pluvialis cells develop into red immobile cells (also called cysts; fig. 1c) by losing their flagella, increasing their cell size, forming thick cell walls, and accumulating astaxanthin (Shah et al. 2016).
Fig 1.
—The life cycle of Haematococcus pluvialis and the phylogeny of bkt genes for astaxanthin biosynthesis. (a) Green motile cell, (b) cell under stress, and (c) red immobile cell. (d) The pathway of astaxanthin biosynthesis (modified from Grunewald et al. 2001) revealed the important roles of CRTO (β-carotene ketolase), which was encoded by the bkt genes. (e) A total of six bkt genes were identified in the genome assembly of H. pluvialis, suggesting that multiple gene duplications occurred during genome evolution. In contrast, only a single bkt homologous sequence was identified in each closely related species such as Volvox carteri, Chlamydomonas reinhardtii, and Monoraphidium neglectum.
Transcriptomics-, metabolomics- and proteomics-based studies have revealed proteins involved in astaxanthin biosynthesis under stress conditions, such as high irradiation, nitrogen deprivation, or nutrient starvation (Kim et al. 2011; Su et al. 2014; Gao et al. 2015). However, because of limited genome information, how H. pluvialis regulates astaxanthin biosynthesis at the DNA level remains unclear. Meanwhile, these genomic resources will help to breed novel strains of H. pluvialis that could have higher astaxanthin yield. We were thus prompted to perform whole-genome sequencing, assembly, and annotation of this economically important microalga. In addition, carotene biosynthetic genes cooperate with β-carotene ketolase (CRTO) and hydroxylase (CRTR-B) to synthesize astaxanthin (fig. 1d) under high irradiation and salinity, which are the most common stresses that occur during H. pluvialis cultivation (Boussiba and Vonshak 1991). We therefore performed additional transcriptome sequencing on stressed cells to reveal additional genes that are potentially involved in the synthesis, accumulation, and regulation of astaxanthin production.
Materials and Methods
Sample Materials, Genomic DNA Extraction, and Genome Assembly
Haematococcus pluvialis 192.80 was purchased from the SAG Culture Collection of Algae (Göttingen, Germany). The alga cells were cultivated in ESP Ag medium as we reported previously (Zheng et al. 2017; see more details in the following section on Total RNA Isolation), and genomic DNA was isolated from cultured cells using Qiagen GenomicTip100 (Qiagen, Germantown, MD, USA). We applied the traditional whole-genome shotgun sequencing strategy (Lin et al. 2016) and built seven diverse paired-end libraries, including three short-insert libraries (250, 500, and 800 bp) and four long-insert libraries (2, 5, 10, and 20 kb). About 83.1 Gb of raw reads were generated from the seven libraries using the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA). After removing the low-quality (containing 10 or more Ns and low-quality bases with quality scores ≤7) and redundant reads, we obtained about 60.6 Gb of clean data for further de novo assembling. In addition, the clean reads from the 500- and 800-bp libraries were employed in the estimation of the genome size of H. pluvialis (see detailed methods in Li et al. 2010), which was about 935.3 Mb.
To assemble the whole-genome sequence, we employed the SOAP-denovo2 software (Luo et al. 2015) (with -k 65) to build contigs and primary scaffolds by utilizing reads from the short-insert libraries (250, 500, and 800 bp). Subsequently, reads from the long-insert libraries (2, 5, 10, and 20 kb) were mapped onto the contigs to shape corresponding scaffolds. The Gapcloser (in the package of SOAP-denovo2) was employed to fill the gaps in the scaffolds.
Genome Annotation
Before annotating gene structures of the H. pluvialis genome, we identified repeat sequences using multiple programs including Tandem Repeats Finder (Benson 1999), LTR_FINDER (Xu and Wang 2007), RepeatProteinMask, and RepeatMasker (Chen 2004). Tandem Repeats Finder was employed to search for tandem repeats in our genome assembly using the following parameters: Match = 2, Mismatch = 7, Delta = 7, PM = 80, PI = 10, Minscore = 50, and MaxPerid = 2,000. A de novo repeat library was built by the LTR_FINDER (version 1.0.6; parameter: -w 2). Subsequently, the RepeatMasker was utilized to align our genome sequences onto the Repbase TE (version 3.2.9; Jurka et al. 2005) to search the known repeat sequences as well as map onto the de novo repeat libraries to identify novel types of repeat sequences.
We then performed annotation of the H. pluvialis genome assembly with three approaches, including homology-based, transcriptome-based, and ab initio annotation. We selected several representative species, including Paramecium tetraurelia (Aury et al. 2006), Saccharomyces cerevisiae (Kellis et al. 2004), Symbiodinium kawagutii and Symbiodinium minutum (Lin et al. 2015), Chlamydomonas eustigma (Hirooka et al. 2017), Chromochloris zofingiensis (Roth et al. 2017), and Micromonas pusilla (Worden et al. 2009) to perform the homology annotation. The protein sequences from abovementioned species were aligned onto our genome sequences utilizing the TblastN (Mount 2007) with E-value ≤ 1e−5. Genewise 2.2.0 (Birney et al. 2004) was subsequently employed to predict possible gene structures based on all TblastN results. Total RNA was extracted from control cells (sample ID: LLMT4, 5, and 6; see more details in the following section on Total RNA Isolation) for subsequent transcriptome sequencing using an Illumina HiSeq 4000 platform. We utilized Cufflinks (version 2.2.1; Trapnell et al. 2010) to identify the preliminary genes. Moreover, Augustus (Stanke et al. 2006) and Genscan (Cai et al. 2014) were selected for ab initio annotation using the repeat-masked genome sequences. Finally, we employed GLEAN software (Elsik et al. 2007) to integrate all genes predicted from the three annotation procedures.
Total RNA Isolation and Transcriptome Assembly
Haematococcus pluvialis cells were cultured in 250 ml Erlenmeyer flasks with ESP Ag medium, statically incubated at 22 °C under a light intensity of 25 μM photons/m2/s with a 12-h light/12-h dark cycle (Zheng et al. 2017). When in the logarithmic phase, these cells were mixed and divided into six groups. Triplicated groups of HLST (HLSTA, HLSTB, and HLSTC) were treated with high irradiation (550 μM photons/m2/s) and high salinity (45 mM of sodium acetate, as referred by Su et al. 2014) with a pH value of 7.0 for 1.5 h, whereas triplicated groups of LLMT (LLMT4, LLMT5, and LLMT6) were used as controls (25 μM photons/m2/s, ESP Ag medium, pH 7.0). Total RNA from each sample was isolated and the corresponding cDNA library was separately constructed for subsequent sequencing (RNA-seq) on an Illumina HiSeq 4000 platform. Paired-end raw reads were then processed by removal of adapters and low-quality sequences using SOAPnuke software (v. 1.5.6; Chen et al. 2018) with default parameters. The remaining clean data were mapped onto the assembled genome with HISAT (Kim et al. 2015).
Transcript quantification in each sample [fragments per kilobase per million (FPKM); Mortazavi et al. 2008] was realized using RSEM (Li and Dewey 2011). Differentially expressed genes (DEGs) between treatment and control groups were identified using DESeq2 (Love et al. 2014) with log2 (ratio) ≥1 and the adjusted P value Padj ≤ 0.05 as threshold. Finally, pathway enrichment analysis was performed with these up- and down-regulated DEGs according to Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Ogata et al. 1999). We finally generated a total of 309,962,820 high-quality clean reads. The total mapping ratio of each library to the genome assembly ranged from 83.71% to 85.65%, and the number of transcribed genes in each sample was predicted to range from 22,243 to 22,609 (see more details in supplementary tables S1 and S2, Supplementary Material online). FPKM values of all transcripts in the six samples are provided in supplementary table S3, Supplementary Material online.
Evolutionary Status of Haematococcus pluvialis
To determine the evolutionary position of H. pluvialis, we performed a whole-genome phylogenetic analysis on H. pluvialis and other 13 related algae, of which complete gene sets or transcriptional data are available. These examined species included four Prasinococcales species (M.pusilla, GenBank Accession number: GCF_000151265.2; Micromonas commode, GCF_000090985.2; Ostreococcus tauri, GCF_000214015.2; and Ostreococcus lucimarinus, GCF_000092065.1), four Trebouxiophyceae species (Chlorella variabilis, GCF_000147415.1; Auxenochlorella protothecoides, GCF_000733215.1; Coccomyxa subellipsoidea, GCF_000258705.1 and Ettlia oleoabundans, GEEU00000000.1), and five Chlamydomonadales species (Volvox carteri, GCF_000143455.1; Chlamydomonas reinhardtii, GCF_000002595.1; Monoraphidium neglectum, GCF_000611645.1; Chlamydomonas eustigma, BEGY00000000.1; and Oophila amblystomatis, GFMX00000000.1). The whole-genome gene sets from H. pluvialis and other data were aligned by BLAST (version 2.2.6; Mount 2007) to check their homology and to generate a sequence similarity matrix. OrthoMCL was used to distinguish gene families from the sequence similarity matrix, and Markov Chain Clustering (MCL) with default parameters was applied.
We identified single-copy orthologues among all the target species, and these orthologues were aligned with MUSCLE version 3.7 (Edgar 2004). All alignments were combined to form a super alignment file. We first applied the maximum likelihood (ML) method to estimate the phylogenetic topology, which was implemented in PhyML version 3.0 (Guindon et al. 2010). To confirm the topology from ML, we also unutilized Bayesian inference (BI) to estimate the phylogenetic tree again, which was performed in MrBayes (version 3.2.2; Ronquist et al. 2012).
Identification of bkt genes in H. pluvialis Genome Assembly and Other Species
The previously identified three BKT protein sequences (BKT1: CAA60478.1, BKT2: BAA08300.1, and BKT3: AAT35555.1) were downloaded from the NCBI. We utilized BLAST version 2.2.6 (Mount 2007) to search the regions of bkt genes and their encoding sequences were further predicted by Exonerate version 2.2.0 (Slater and Birney 2005). All the possible bkt copies were translated to proteins and aligned together using MEGA version 7 (Kumar et al. 2016) with the Muscle module. We then classified bkt genes based on sequence similarity. Furthermore, the three BKT proteins were employed to predict the corresponding bkt orthologs in the three phylogenetically closed species including V. carteri, C. reinhardtii, and M. neglectum, which were identified by the phylogenetic analysis. Finally, we interpreted and aligned all extracted encoding sequences from H. pluvialis, V. carteri, C. reinhardtii, and M. neglectum and constructed a phylogenetic topology using the ML method, with 1,000 replicates implemented by PhyML to obtain corresponding node supports.
Long-Read Sequencing of a Mixed Transcriptome Sample
As earlier described, H. pluvialis cells in the logarithmic phase with or without the 1.5-h treatment and about 106 cells from both groups were collected through centrifugation. Total RNA was extracted using the RNA fast 200 kit (Fastagen, Shanghai, China). Only those RNAs with high RNA integrity numbers (RIN > 9.0) were used for subsequent cDNA preparation. Long-read sequencing by PacBio Isoform (Pacific Biosciences, Menlo Park, CA, USA) was performed on the mixture of all the cDNA samples to obtain full-length transcripts without the uncertainty of assembly from short reads.
Results and Discussion
Genome Assembly and Annotation
We generated a draft genome assembly with 669.0 Mb in total length and 7,855 scaffolds (>2,000 bp) with a high scaffold N50 of 288.6 kb (table 1). We further utilized the Benchmarking Universal Single-Copy Orthologs (BUSCO) software (Simao et al. 2015) to examine the completeness of our present assembly. The results demonstrated 59% complete and partial eukaryote BUSCO orthologues. It seems that the genome of H. pluvialis is very complicated, we therefore added the PacBio transcriptome data to fill up more gene regions. We finally used the de novo assembled transcripts (sequenced by HiSeq) to map the final gene set from both the genome assembly and the transcriptomes, and observed that this set covered 90% of transcripts (supplementary table S1, Supplementary Material online). We further identified and classified repeat sequences, which account for about 32.2% of the assembled genome (table 1).
Table 1.
Summary of the genome assembly and annotation
| Genome assembly | Parameter |
|---|---|
| Contig N50 size (kb) | 8.2 |
| Scaffold N50 size (kb) | 288.6 |
| Assembled genome size (Mb) | 669.0 |
| Genome coverage (×) | 83.1 |
| Longest scaffold (bp) | 1,782,609 |
| Genome annotation | |
| Number of protein-coding genes | 18,545 |
| Transposable elements content (%) | 32.2 |
The complete gene set is composed of 18,545 genes, with an average of 8.7 kb in length. All protein sequences of the GLEAN results were mapped onto the public TrEMBL, Swiss-Prot (Bairoch and Apweiler 2000; Boeckmann et al. 2003), InterProScan (Finn et al. 2014), and KEGG (Ogata et al. 1999) databases using BlastP software with E-value ≤ 1e-5. Finally, approximately 75.8% of the predicted genes have at least one related function assignments from these public databases.
Evolutionary Status of H. pluvialis
In total, 260 single-copy gene families including 3,640 genes were obtained from the 14 representative algal species. These genes from each species were concatenated together and constituted a super-length nucleotide data set to yield 1,147,546 aligned sites. Our phylogenetic analysis supports three main groups among the examined species, which is consistent with the putative classification of Prasinococcales, Trebouxiophyceae, and Chlamydomonadales. The H. pluvialis clade obviously located into the group of Chlamydomonadales and displayed a closer relationship with V.carteri, C.reinhardtii, and M.neglectum than other species. Thus, at the first time, we demonstrated the phylogenetic position of H. pluvialis at a whole-genome level. Interestingly, the H. pluvialis clade displays the longest branch length in Chlamydomonadales (supplementary fig. S1, Supplementary Material online). Since the branches are in units of substitutions per site with calculation by MrBayes (version 3.2.2; Ronquist et al. 2012), the result suggests that H. pluvialis has a faster nucleotide substitution rate during the evolution of flagellated green algae.
Genomic Basis for the High Production of Astaxanthin in H. pluvialis
Haematococcus pluvialis is well known for its capacity to produce large amounts of astaxanthin, a strong antioxidant for aquaculture and cosmetics. Commercially, more than 40 g of astaxanthin can be extracted from 1 kg of dry cells (Lorenz and Cysewski 2000), leading this species to be an ideal resource for astaxanthin production. A previous study (Grunewald et al. 2001) reported that the putative pathway of astaxanthin biosynthesis (fig. 1d) consists of two key enzymes (CRTO and CRTR-B). Interestingly, CRTO is encoded by β-carotene ketolase gene (bkt). Three bkt genes were previously confirmed to be upregulated when the H. pluvialis cells were induced in different stress conditions (Huang et al. 2006). Therefore, the bkt genes were considered as one of the major contributors to the rapid accumulation of large amounts of astaxanthin.
In our current study, we employed the three reported BKT protein sequences (Huang et al. 2006) as queries to search our genome assembly. Interestingly, we identified six bkt copies in H. pluvialis genome with a wide distribution in different scaffolds. However, only one corresponding ortholog was identified in each of the phylogenetically closed species (V. carteri, C. reinhardtii, and M. neglectum). Among the six bkt copies in H. pluvialis, we determined that three copies are more similar to bkt1, thus named them as bkt1a, bkt1b, and bkt1c respectively; since two copies are similar to bkt2, we defined them as bkt2a and bkt2b; the rest one is bkt3. Through the phylogenetic analysis (fig. 1e), we observed that the six copies of bkt genes clustered into a single clade, which is separated from the single bkt ortholog from V. carteri, C. reinhardtii, and M. neglectum. In addition, these bkt copies in H. pluvialis can be divided into two groups, in which one consists of bkt1s, while the other contains bkt2s and bkt3. In fact, among the nucleotide sequences, bkt2s and bkt3 are highly similar and display slight site changes in their protein sequence alignments; however, bkt1 presents relatively more site changes, especially within the initial 220 bp (supplementary fig. S3, Supplementary Material online).
Transcriptome Profiling and Validation of Novel Potential Genes for Astaxanthin Production
Isopentenyl pyrophosphate (IPP) is a key intermediate of carotenoid synthesis, and there are two independent pathways that produce IPP in algae cells, namely the mevalonate pathway (MVA) in the cytosol and nonmevalonate (MEP) in chloroplasts (Shah et al. 2016). Earlier investigations have shown that the MVA pathway has been lost in many green algae (Chlorophyta) and red algal taxa. Previously, only two MVA pathway enzymes (acetyl-CoA C-acetyltransferase [ACAT] and hydroxymethylglutaryl-CoA synthase [HMGS]) were identified in the H. pluvialis transcriptomes (Gwak et al. 2014). Here, by searching our genome and transcriptome data, we could only find these two genes for the MVA pathway too. The loss of hydroxymethylglutaryl-CoA reductase (HMGR), mevalonate kinase (MVK), phosphomevalonate kinase (PMK), and mevalonate diphosphate decarboxylase (MVD) in the genome of H. pluvialis confirms that as a member of the Chlorophyta, H. pluvialis exclusively uses the MEP pathway to synthesize IPP. This might be a common phenomenon in the green algae (Lohr et al. 2012; Qiu et al. 2016).
The molecular mechanisms underlying astaxanthin synthesis in H. pluvialis (Shah et al. 2016) and stress factors such as light intensity (He et al. 2018; Ma et al. 2018), salinity (Han et al. 2013), temperature (Hong et al. 2015), and chemical substances (Gao et al. 2015; Zhao et al. 2015) have been extensively studied. Here, we further examined the effects of high irradiation and salinity on the accumulation of astaxanthin in H. pluvialis and identified some novel genes that may participate in this process. By comparing with the control (LLMT) groups, we identified 1,121 DEGs (log2 [ratio] ≥1 and Padj ≤0.05) in the treated (HLST) groups, with 482 up-regulated and 639 down-regulated (supplementary table S4, Supplementary Material online). Subsequently, the KEGG enrichment analysis clustered all these DEGs into 103 KEGG pathways, including carotenoid biosynthesis (ko00906), terpenoid backbone biosynthesis (ko00900), and biosynthesis of unsaturated fatty acids (ko01040) (supplementary table S5 and fig. S2, Supplementary Material online).
There were six DEGs enriched into the carotenoid biosynthesis pathway, including phytoene synthase (PSY), phytoene desaturase (PDS), and beta-carotene hydroxylase (CrtR-b). All these DEGs were up-regulated in the treated HLST groups (supplementary table S6, Supplementary Material online). Except for CrtR-b, the rate-limiting enzyme CRTO (Fraser et al. 1997, Choi et al. 2006) was also eliminated from the DEG list, with a slight elevation of 30–70%. The upregulation of all six genes in the pathway of astaxanthin biosynthesis is consistent with the findings of previous reports (Han et al. 2013; He et al. 2018), in which high light and salinity enhance astaxanthin production in H. pluvialis.
Long-Read Reference Transcriptome of H. pluvialis
A total of 157,416 isoforms were identified after removing the chloroplast, mitochondrial, and ribosomal transcripts. The length of these sequences among this data set ranged from 50 bp to 14,995 bp, with a N50 of 5,106 bp. An isoform-level reference transcriptome set of 18,483 transcripts with high confidence was generated.
Alternative splicing and polyadenylation can contribute to the diversity of transcripts (Abdel-Ghany et al. 2016; Wang et al. 2016). We therefore employed AStalavista (Foissac and Sammeth 2007, 2015) to identify the five main modes of alternative splicing, including intron retention, exon skipping, alternative 3′ splice site, alternative 5′ splice site, and mutually exclusive exons. Interestingly, the final results revealed that the intron retention was the most abundant mode, while no mutually exclusive exon was identified in these H. pluvialis transcripts (supplementary table S7, Supplementary Material online).
Conclusions
We report the first whole-genome sequencing, assembly, and annotation of the astaxanthin-producing green microalga, H. pluvialis. This draft genome assembly is a valuable genetic resource for elucidating the deep genetic basis for astaxanthin production. In our present study, we observed a remarkable expansion of the bkt gene family in the H. pluvialis genome, which may contribute to high astaxanthin yield. Transcriptome sequencing highlighted that several important pathways were involved in astaxanthin synthesis. These genomic and transcriptomic data may be utilized in elucidating the molecular mechanisms underlying astaxanthin yield and accumulation, which in turn will facilitate breeding of novel strains with significantly higher astaxanthin content.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
Z.H. and Q.S. designed the project. C.B., Y.L., and Y.H. assembled and annotated the genome. Y.L. and C.B. performed the evolution analysis. Q.L., M.T., and Y.Z. collected the samples and prepared the quality control. Y.H., Q.L., and M.T performed the transcriptome analysis. M.T., Y.Z., C.W., J.L., B.J., J.c.L., Z.L., and J.X. participated in data analysis and figure preparation. C.B., M.T., Q.S., Q.L., Z.H., Y.L., and Y.H. prepared the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (Nos 31470431, 31400313, 31470389, and 31700309), Guangdong Natural Science Foundation for Major Cultivation Project (Nos 2014A030308017 and 2017A030310255), and Shenzhen Grant Plan for Science & Technology (Nos JSGG20130411160539208, NYSW20140327010012, CKCY2016042710211071, and JCYJ20170302144605664).
Literature Cited
- Abdel-Ghany SE, et al. 2016. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun. 7:11706.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati RR, Phang SM, Ravi S, Aswathanarayana RG.. 2014. Astaxanthin: sources, extraction, stability, biological activities and its commercial applications—a review. Mar Drugs 12(1):128–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury JM, et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444(7116):171–178. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R.. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28(1):45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. Genewise and genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31(1):365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussiba S, Vonshak A.. 1991. Astaxanthin accumulation in the green alga Haematococcus pluvialis. Plant Cell Physiol. 32(7):1077–1082. [Google Scholar]
- Cai Y, Gonzalez JV, Liu Z, Huang T.. 2014. Computational systems biology methods in molecular biology, chemistry biology, molecular biomedicine, and biopharmacy. Biomed Res Int. 2014:746814.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. 2004. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 5(1):4.10.1–4.10.14. [DOI] [PubMed]
- Chen Y, et al. 2018. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SK, Matsuda S, Hoshino T, Peng X, Misawa N.. 2006. Characterization of bacterial beta-carotene 3,3′-hydroxylases, CrtZ, and P450 in astaxanthin biosynthetic pathway and adonirubin production by gene combination in Escherichia coli. Appl Microbiol Biotechnol. 72(6):1238–1246. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, et al. 2007. Creating a honey bee consensus gene set. Genome Biol. 8(1):R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42(Database issue):D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac S, Sammeth M.. 2015. Analysis of alternative splicing events in custom gene datasets by AStalavista. Methods Mol Biol 1269:379–392. [DOI] [PubMed] [Google Scholar]
- Foissac S, Sammeth M.. 2007. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 35(Web Server issue):W297–W299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Miura Y, Misawa N.. 1997. In vitro characterization of astaxanthin biosynthetic enzymes. J Biol Chem. 272(10):6128–6135. [DOI] [PubMed] [Google Scholar]
- Gao Z, et al. 2015. Transcriptome analysis in Haematococcus pluvialis: astaxanthin induction by salicylic acid (SA) and jasmonic acid (JA). PLoS One 10(10):e0140609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald K, Hirschberg J, Hagen C.. 2001. Ketocarotenoid biosynthesis outside of plastids in the unicellular green alga Haematococcus pluvialis. J Biol Chem. 276(8):6023–6029. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Gwak Y, et al. 2014. Comparative analyses of lipidomes and transcriptomes reveal a concerted action of multiple defensive systems against photooxidative stress in Haematococcus pluvialis. J Exp Bot. 65(15):4317–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. et al. 2013. Astaxanthin in microalgae: pathways, functions and biotechnological implications. Algae 28(2):131–147. [Google Scholar]
- He B, et al. 2018. Transcriptome analysis in Haematococcus pluvialis: astaxanthin induction by high light with acetate and Fe(2). Int J Mol Sci.19(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka S, et al. 2017. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc Natl Acad Sci U S A. 114(39):E8304–E8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong ME, et al. 2015. Enhanced autotrophic astaxanthin production from Haematococcus pluvialis under high temperature via heat stress-driven Haber-Weiss reaction. Appl Microbiol Biotechnol. 99(12):5203–5215. [DOI] [PubMed] [Google Scholar]
- Huang JC, Chen F, Sandmann G.. 2006. Stress-related differential expression of multiple beta-carotene ketolase genes in the unicellular green alga Haematococcus pluvialis. J Biotechnol. 122(2):176–185. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1-4):462–467. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES.. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428(6983):617–624. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-K, et al. 2011. Transcriptomic analysis of Haematococcus lacustris during astaxanthin accumulation under high irradiance and nutrient starvation. Biotechnol. Bioprocess Eng. 16(4):698. [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. 2010. The sequence and de novo assembly of the giant panda genome. Nature 463(7279):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, et al. 2016. The seahorse genome and the evolution of its specialized morphology. Nature 540(7633):395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, et al. 2015. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350(6261):691–694. [DOI] [PubMed] [Google Scholar]
- Lohr M, Schwender J, Polle JE.. 2012. Isoprenoid biosynthesis in eukaryotic phototrophs: a spotlight on algae. Plant Sci. 185-186:9–22. [DOI] [PubMed] [Google Scholar]
- Lorenz RT, Cysewski GR.. 2000. Commercial potential for Haematococcus microalgae as a natural source of astaxanthin. Trends Biotechnol. 18(4):160–167. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2015. Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 4:30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R, et al. 2018. Gene expression profiling of astaxanthin and fatty acid pathways in Haematococcus pluvialis in response to different LED lighting conditions. Bioresour Technol. 250:591–602. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B.. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628. [DOI] [PubMed] [Google Scholar]
- Mount DW. 2007. Using the basic local alignment search tool (BLAST). CSH Protoc. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- Ogata H, et al. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27(1):29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Yoon HS, Bhattacharya D.. 2016. Red algal phylogenomics provides a robust framework for inferring evolution of key metabolic pathways. PLoS Curr. 8. doi: 10.1371/currents.tol.7b037376e6d84a1be34af756a4d90846. ecurrents.tol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS, et al. 2017. Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proc Natl Acad Sci U S A. 114(21):E4296–E4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MM, Liang Y, Cheng JJ, Daroch M.. 2016. Astaxanthin-producing green microalga Haematococcus pluvialis: from single cell to high value commercial products. Front Plant Sci. 7:531.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6(31): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server issue):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, et al. 2014. Metabolomic and network analysis of astaxanthin-producing Haematococcus pluvialis under various stress conditions. Bioresour Technol. 170:522–529. [DOI] [PubMed] [Google Scholar]
- Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, et al. 2016. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun. 7:11708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, et al. 2009. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324(5924):268–272. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang H.. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35(Web Server issue):W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. 2015. Enhanced astaxanthin production from a novel strain of Haematococcus pluvialis using fulvic acid. Process Biochem. 50(12):2072–2077. [Google Scholar]
- Zheng Y, Li Z, Tao M, Li J, Hu Z.. 2017. Effects of selenite on green microalga Haematococcus pluvialis: bioaccumulation of selenium and enhancement of astaxanthin production. Aquat Toxicol. 183:21–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.