Abstract
Plastid genome (ptDNA) data of Glaucophyta have been limited for many years to the genus Cyanophora. Here, we sequenced the ptDNAs of Gloeochaete wittrockiana, Cyanoptyche gloeocystis, Glaucocystis incrassata, and Glaucocystis sp. BBH. The reported sequences are the first genome-scale plastid data available for these three poorly studied glaucophyte genera. Although the Glaucophyta plastids appear morphologically “ancestral,” they actually bear derived genomes not radically different from those of red algae or viridiplants. The glaucophyte plastid coding capacity is highly conserved (112 genes shared) and the architecture of the plastid chromosomes is relatively simple. Phylogenomic analyses recovered Glaucophyta as the earliest diverging Archaeplastida lineage, but the position of viridiplants as the first branching group was not rejected by the approximately unbiased test. Pairwise distances estimated from 19 different plastid genes revealed that the highest sequence divergence between glaucophyte genera is frequently higher than distances between species of different classes within red algae or viridiplants. Gene synteny and sequence similarity in the ptDNAs of the two Glaucocystis species analyzed is conserved. However, the ptDNA of Gla. incrassata contains a 7.9-kb insertion not detected in Glaucocystis sp. BBH. The insertion contains ten open reading frames that include four coding regions similar to bacterial serine recombinases (two open reading frames), DNA primases, and peptidoglycan aminohydrolases. These three enzymes, often encoded in bacterial plasmids and bacteriophage genomes, are known to participate in the mobilization and replication of DNA mobile elements. It is therefore plausible that the insertion in Gla. incrassata ptDNA is derived from a DNA mobile element.
Keywords: Glaucophyta, Archaeplastida, plastid genome, horizontal gene transfer, phylogenomics
Introduction
Viridiplants (land plants and green algae), red algae, and glaucophytes belong to the Archaeplastida supergroup (Adl et al. 2012), which includes plastid-bearing lineages that putatively share a unique common ancestor that established an endosymbiotic association with a cyanobacterium between the late Paleoproterozoic and Mesoproterozoic eras (1.5–2.0 Ga) (Yoon et al. 2004; Parfrey et al. 2011; Shih and Matzke 2013; Sánchez-Baracaldo et al. 2017). However, independent phylogenomic surveys of nuclear sequences have consistently failed to recover the Archaeplastida clade with strong support, or even to resolve the three archaeplastidian groups in an exclusive clade (e.g., Yabuki et al. 2014; Burki et al. 2016; Janouškovec et al. 2017; Brown et al. 2018; Heiss et al. 2018). These results challenge the hypothesis that viridiplants, red algae, and glaucophytes constitute an exclusive monophyletic group and leave open other alternatives to explain the origin of their host (i.e., the nucleo-cytoplasm component) ancestor. Some alternative scenarios propose the single origin of a primary plastid in one lineage followed by independent establishment of plastids in the different Archaeplastida lineages via secondary endosymbiosis (e.g., Kim and Maruyama 2014; Stiller 2014). Another possibility is that the three Archaeplastida groups descend from a common host along with other lineages that lost the primary plastid (i.e., Archaeplastida is paraphyletic). Whereas the single origin of the Archaeplastida host is still contentious, the results derived from plastid data strongly suggest that the photosynthetic organelles of the Archaeplastida have a common origin. Evidence comes not only from plastid phylogenomics (e.g., Deschamps and Moreira 2009; Criscuolo and Gribaldo 2011; Ponce-Toledo et al. 2017; Reyes-Prieto et al. 2018) but also from the presence of a relatively well conserved plastid gene cluster (5′-rpoB-rpoC1-rpoC2-rps2-atpI-atpH-atpG-atpF-atpD-atpA-3′) found across the Archaeplastida and in lineages with secondary plastids (Stoebe and Kowallik 1999), common enzyme replacements in plastid-localized pathways (Reyes-Prieto and Bhattacharya 2007; Reyes-Prieto and Moustafa 2012), and the unique origin of components of the plastid TIC/TOC protein import machinery (McFadden and van Dooren 2004; Steiner et al. 2005).
Regardless of the Archaeplastida host origin conundrum, investigations into the tempo and mode of evolution of the primary plastids using their genome sequences are relevant to understand the subsequent spread and diversification of photosynthetic organelles across the eukaryote lineage. For instance, by identifying the earliest primary plastid branch, we can assess the directionality of evolutionary patterns (e.g., genome expansions or compactions, gene transfer events or architectural changes) followed by the organelle genomes independently of the nuclear genome of the host lineages. Diverse phylogenetic analyses considering different plastid loci and disparate taxonomic samples have recovered, alternatively, each of the three different Archaeplastida groups as the earliest plastid branch (Rodríguez-Ezpeleta et al. 2005; Deschamps and Moreira 2009; Janouskovec et al. 2010; Criscuolo and Gribaldo 2011; Price et al. 2012; Ponce-Toledo et al. 2017). However, most of these studies have included glaucophyte data only from Cyanophora paradoxa and, to a lesser extent, a single Glaucocystis species (e.g., Price et al. 2012). The inclusion of a broader glaucophyte plastid data set, such as additional Cyanophora and Glaucocystis species and sequences from other genera, is important to estimate more robust (i.e., less sensitive to stochastic errors) plastid phylogenies.
Genomes of primary plastids have a collection of universally conserved genes (Sánchez Puerta et al. 2005; Lee, Cho, et al. 2016; Reyes-Prieto et al. 2018) that have been extensively used in different subset combinations to investigate the evolution of diverse Archaeplastida groups, such as land plants (e.g., Parks et al. 2012; Xi et al. 2012; Ruhfel et al. 2014), green (e.g., Hamaji et al. 2013; Turmel et al. 2015; Leliaert et al. 2016; Lemieux et al. 2016; Fang et al. 2017), and red algae (Janouškovec et al. 2013; Lee, Cho, et al. 2016; Muñoz-Gómez et al. 2017). In contrast, plastid phylogenomics has not been used for studies of glaucophyte diversity. This is largely due to the rareness of glaucophytes in nature, their restricted habitat (apparently limited to freshwater environments), and the apparent species-depauperate status of the group: There are only 14 described species, distributed in 4 different genera, with vouchers available in scientific collections; table 1) (Kies and Kremer 1986; Jackson et al. 2015; Price et al. 2017). Regardless of the considerable historical attention that glaucophyte plastids have received due to putative ancestral traits shared with free-living cyanobacteria, such as the vestigial peptidoglycan wall between the organelle membranes (Hall and Claus 1963; Pfanzagl et al. 1996; Löffelhardt and Bohnert 2001; Steiner and Löffelhardt 2011) and similar composition of their photosynthetic apparatuses (Klein et al. 1981; Steiner and Löffelhardt 2011; Misumi and Sonoike 2017), their genomes have long been ignored. Despite the establishment of inexpensive high-throughput DNA sequencing technologies as standard procedures, since the publication of the plastid genome of C.paradoxa (strain UTEX LB 555) more than 20 years ago (Stirewalt et al. 1995), only the partial ptDNA sequence of C.kugrensii NIES-763 (Smith et al. 2014) and four mitochondrial genomes (mtDNA) of different genera (Price et al. 2012; Jackson and Reyes-Prieto 2014) have been made available in public data repositories. Plastid sequence data from Glaucocystis nostochinearum (strain UTEX 64; denominated Gla.geitleri in Takahashi et al. 2016) were reported but are not yet publicly available (Price et al. 2012). Additionally, only few individual plastid loci (e.g., psbA, psaB and rrs) of Gloeochaete and Cyanoptyche have been sequenced and used in phylogenetic analyses (Chong et al. 2014; Takahashi et al. 2014, 2016).
Table 1.
Glaucophyte Species Described
| Taxa | Authority | Plastid Genome GenBank Accessiona |
|---|---|---|
| Order Cyanophorales | Kies and Kremer 2006 | |
| Cyanophora biloba | Kugrens et al. 1999 | MG601103 |
| Cyanophora cuspidata | Takahashi and Nozaki 2014 | |
| Cyanophora paradoxa | Korshikov 1924 | U30821 |
| Cyanophora kugrensii | Takahashi and Nozaki 2014 | KM198929b |
| Cyanophora sudae | Takahashi and Nozaki 2014 | MG601102 |
| Order Glaucocystales | Bessey 1907 | |
| Glaucocystis geitleri | Koch 1964 | |
| Glaucocystis cingulata | Bohlin 1897 | |
| Glaucocystis nostochinearum | Itzigsohn in Rabenhorst 1868 | |
| Glaucocystis miyajii | Takahashi and Nozaki 2016 | |
| Glaucocystis oocystiformis | Prescott 1944 | |
| Glaucocystis bhattacharyaec | Takahashi and Nozaki 2016 | |
| Glaucocystis incrassata | Takahashi and Nozaki 2016 | MF167425 |
| Order Gloeochaetales | Kies and Kremer 2006 | |
| Gloeochaete wittrockiana | Lagerheim 1883 | MF167426 |
| Cyanoptyche gloeocystis | Pascher 1929 | MF167427b |
| Putative speciesd | ||
| Peliaina cyanea | Pascher 1929 | |
| Strobilomonas cyaneus | Schiller 1954 | |
| Cyanophora tetracyaneaa | Korshikov 1941 | |
| Archaeopsis monococca | Skuja 1954 | |
| Glaucocystopsis africana | Bourrelly 1960 | |
| Glaucocystis simplex | Tarnogradskij 1959 | |
| Chalarodora azurea | Pascher 1929 |
Taxa with plastid genomes sequenced.
Partial sequence.
Glaucocystis sp. strain BBH, plastid genome sequenced in this work (GenBank accession MF167424), was identified as Glaucocystis bhattacharyae in Price et al. (2017).
Taxa with no isolates in culture collections neither microscopy or sequence data available.
In contrast to the limited availability of glaucophyte ptDNAs, the GenBank collection contains 102 ptDNAs sequenced from red algae and 2,733 from viridiplants, including 127 from diverse green algal species (as of November 2018). Producing additional glaucophyte plastid data is relevant not only to investigate the evolution of Archaeplastida and their photosynthetic organelles, but also to explore in detail the intrinsic diversity within this rare algal group. Here, we present the plastid genomes of four glaucophyte species from the genera Glaucocystis (two species), Gloeochaete, and Cyanoptyche. For the first time, these new data allow comparative studies of ptDNAs from each of the four glaucophyte genera available in public culture collections (table 1).
Materials and Methods
DNA Extraction, Sequencing, Assembling, and Annotation
Total DNA extracted from Cyanoptyche gloeocystis (strain SAG 4.97), Gloeochaete wittrockiana (strain SAG 48.84), Gla.incrassata (strain SAG 229-2), and Glaucocystis sp. BBH (this strain is referred to as Gla.bhattacharyae in Price et al. 2017) was sequenced using Illumina technology producing 60 × 106, 82 ×106, 94 ×106, and 37 × 106 100-base reads from ∼450-nt-insert paired-end libraries, respectively. Illumina reads were assembled using Ray v2.2.0 (Boisvert et al. 2010), SPAdes v 3.10.1 (Bankevich et al. 2012), or NOVOplasty v1.2.5 (Dierckxsens et al. 2017). Additionally, for Cyanopt.gloeocystis, four PacBio SMRT cells were produced. The PacBio reads were corrected and assembled using the RS HGAP Assembly.2 protocol from the SMRT Analysis Software v2.2.0 (Pacific Biosciences of California). All DNA sequencing was performed at Genome Québec (McGill University). Contigs containing plastid genes were identified by TBlastN searches using available sequences from Cyanophora species as queries (Smith et al. 2014) and were assembled into scaffolds comprising complete genomes using read-mapping approaches with Geneious v8 (Biomatters). Transfer RNA genes were predicted with the tRNAscan-SE Search Server (Lowe and Chan 2016). The presence of transfer-messenger RNAs was evaluated with the program ARAGORN (Laslett and Canback 2004). Plastid genomes were aligned with the progressive algorithm implemented in MAUVE v 2.4.0 using the default options (Darling et al. 2004). Genome maps were initially generated using GenomeVx (Conant and Wolfe 2008) and then manually edited.
Phylogenetic Inference
To prepare multiple sequence alignments (MSAs) of plastid-encoded proteins, we used as a reference a MSA of 97 proteins previously assembled to investigate the evolutionary connections between cyanobacteria and primary plastids (Ponce-Toledo et al. 2017). First, we revised the plastid repertoire in Archaeplastida to select a subset of 42 plastid-encoded proteins that minimized the amount of missing data (i.e., gaps and regions with ambiguous alignments) in our MSA (see supplementary table S1, Supplementary Material online). Then, we identified by BlastP searches orthologous sequences from the 4 glaucophytes reported here, 19 red algae and 15 viridiplants (supplementary table S1, Supplementary Material online). Finally, the new sequences and the selected 42 original sequence sets (Ponce-Toledo et al. 2017) were aligned individually with MAFFT v7 (Katoh and Standley 2013) and manually refined. Resulting multiple protein alignments were concatenated to produce a MSA 9,039 residues long.
Maximum likelihood (ML) phylogenetic trees were estimated with IQ-TREE v1.5.6 (Nguyen et al. 2015) using the protein substitution model LG+R9 (the best-fitting model according to ModelFinder [Kalyaanamoorthy et al. 2017] as well as the mixture models LGFX and LG+C40+F (these latter two models are not evaluated by ModelFinder). Branch support was assessed in all tree searches with 5,000 ultrafast bootstrap replicates (Hoang et al. 2018). Bayesian node posterior probabilities were assessed with PhyloBayes-MPI v1.6 (Rodrigue and Lartillot 2014) using the CAT-GTR empirical mixture model and four discrete Gamma rate categories. Convergence of two independent Monte Carlo Markov chains was monitored using the program tracecomp (Rodrigue and Lartillot 2014) until the likelihood <0.1 and the effective sample size (SES) >200. Following chain convergence, which occurred at 5,000 cycles (910,000 generations), node posterior probabilities were calculated sampling every 10 trees after discarding the first 1,000 cycles as burnin.
Additional ML phylogenetic trees from individual protein alignments prepared with MAFFT v7 were estimated with IQ-TREE using the best-fitting model selected by ModelFinder in each case (see supplementary fig. S3, Supplementary Material online).
Tree Topology Testing
To evaluate alternative hypotheses of the Archaeplastida plastid branching history, we generated 13 arbitrary competing topologies with TreeGraph 2 v2.13.0-748 (Stöver and Müller 2010) using as reference the best ML tree estimated under the LG+C40+F model. Competing topologies included trees placing alternatively red algae or viridiplants as the earliest diverging branch, and diverse trees where the Archaeplastida clade was arbitrarily disrupted. All evaluated trees in Newick format are available in supplementary table S8, Supplementary Material online. We used the site-wise log likelihoods of the 13 alternative trees calculated with IQ-TREE (LG+C40+F substitution model) to estimate the approximately unbiased (AU) test P values of the competing topologies with Consel v0.20 (Shimodaira and Hasegawa 2001; Shimodaira and Goldman 2002).
Pairwise Sequence Distances and Synonymous and Nonsynonymous Substitution Rates
A set of 19 plastid loci was selected to estimate genetic distances within each of the three Archaeplastida groups (supplementary tables S2–S4, Supplementary Material online). In order to estimate genetic distances, plastid nucleotide sequence sets from glaucophytes (4 species), red algae (17 species), and viridiplants (21 species) were aligned independently with MAFFT v7. Alignments of protein-coding sequences were revised to confirm the correct alignment of the codon positions. All multiple alignments were manually edited to discard gaps and ambiguous regions. Sequence distances using the Kimura 2-parameter (K2P) substitution model were estimated with MEGA v7 (Kumar et al. 2016).
Results
Genome Assembly
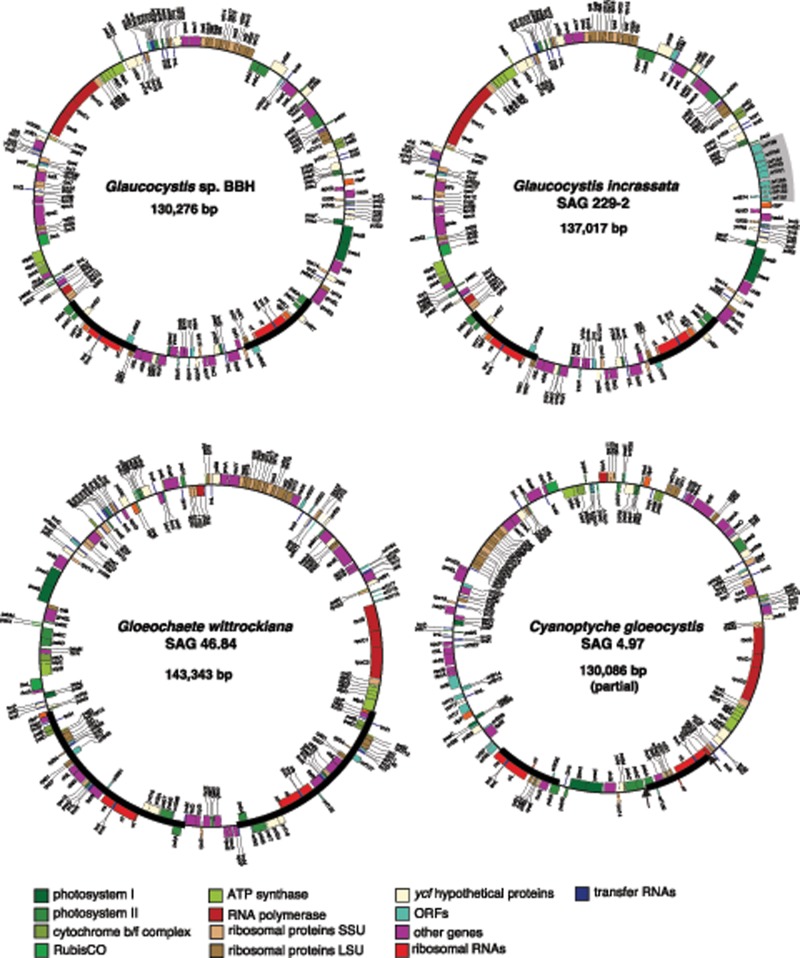
The sequence data obtained using Illumina technology allowed us to completely assemble the plastid genomes of Glaucocystis sp. BBH, Gla.incrassata, G.wittrockiana (fig. 1A–C and supplementary table S5, Supplementary Material online), and partially assemble the ptDNA of Cyanopt.gloeocystis (fig. 1D). For Cyanopt.gloeocystis, the presence of short repeats in intergenic regions precluded the full assembly of the ptDNA, but the use of low coverage (15×) SMRT reads (PacBio) allowed us to bridge the Illumina contigs into two scaffolds (9.3 and 120.7 kb, respectively) (fig. 1D). The three complete ptDNAs are circular mapping and present an inverted repeat (IR) and two single-copy regions (i.e., a quadripartite structure), with three ribosomal RNAs (rRNAs: 5S, 16S, and 23S) encoded in the IR region (fig. 1). The 24.6-kb IR of the G.wittrockina ptDNA contains 21 protein-coding genes, 4 unidentified open reading frames (ORFs), and 9 tRNAs, double the size and at least triple the coding capacity of the IRs in other glaucophyte plastid genomes (table 2).
Fig. 1.
—Maps of the sequenced glaucophyte plastid genomes. Circular representations of the complete ptDNAs of Glaucocystis sp. (strain BBH), Glaucocystis incrassata (SAG 229-2; the gray box indicates an insertion of putative HGT origin), and Gloeochaete wittrockiana (SAG 46.84). The partial sequence of the Cyanoptyche gloeocystis (SAG 4.97) ptDNA is depicted as a circle for illustration purposes; two sequence gaps of unknown length are indicated with black arrowheads. IRs in each map are indicated with thick black lines.
Table 2.
General Characteristics of Glaucophyte Plastid Genomes
| Glaucocystissp. BBH | Glaucocystis incrassata (SAG 229-2) | Gloeochaete wittrockiana (SAG 46.84) | Cyanoptyche gloeocystis(SAG 4.97) | Cyanophora paradoxa (UTEX LB555) | |
|---|---|---|---|---|---|
| GenBank accession | MF167424 | MF167425 | MF167426 | MF167427 | U30821 |
| Length (bp) | 130,276 | 137,017 | 143,343 | 130,086a | 135,599 |
| IR length (bp) | 10,582 | 10,538 | 24,788 | 9,348 | 11,285 |
| Protein-coding genes in the IRb | 7 | 7 | 21 | 5 | 4 |
| Noncoding DNA (bp; %) | 20,257 (15.5) | 21,243 (15.5) | 26,439 (18.44) | 24,431 (18.7) | 26,951 (19.9) |
| Introns | ND | ND | ND | 1 | 1 |
| GC content (%) | 33.4 | 33.6 | 29.6 | 30.6 | 30.5 |
| Mean intergenic space (bp)c | 148 | 152 | 199 | 179 | 199 |
| Unique protein-coding genesb | 137 | 137 | 129 | 121 | 136 |
| Unknown ORFs (>100 bp) | 11 | 20d | 18 | 20 | 8 |
| Unique RNA-coding genes | |||||
| rRNA | 3 | 3 | 3 | 3 | 3 |
| tRNA | 32 | 31 | 31 | 29 | 31 |
| tmRNA | ND | ND | ND | 1 | 1 |
| rnpB gene | 1 | 1 | 1 | 1 | 1 |
Partial sequence.
Excluding unknown ORFs.
RNA genes were not considered in the estimation.
Excluding ten ORFs in the 7.9-kb insertion.
RNA-Coding Genes
The majority of tRNAs detected in glaucophyte ptDNAs are shared between all species investigated (supplementary table S6, Supplementary Material online). In the case of completely sequenced ptDNAs, the tRNA collections (36–40 genes, respectively) are sufficient to decode all amino acids used in plastid-encoded proteins (supplementary table S9 from S10, Supplementary Material online). In the plastid scaffolds of Cyanopt.gloeocystis, no tRNAs were detected for glutamic acid, asparagine and tyrosine, but the corresponding codons are regularly used in the Cyanopt.gloeocystis plastid coding regions (supplementary table S9 from S10, Supplementary Material online). It is possible that these three tRNA genes are encoded on the missing section of the Cyanopt.gloeocystis ptDNA, but the import of tRNA molecules from the cytosol into the plastid, proposed to occur in some nonphotosynthetic angiosperms (Alkatib et al. 2012), cannot be discarded as an alternative. The only intron identified in the new ptDNA sequences is of the group IB type and is localized in the trnL gene (UAA anticodon) of Cyanopt.gloeocystis. An intron of the same type is also present in the trnL gene of Cyanophora species (Stirewalt et al. 1995; Reyes-Prieto et al. 2018).
The ptDNA of Cyanopt.gloeocystis contains the gene ssrA, which encodes a putative transfer-messenger RNA (tmRNA). This gene is also present in Cyanophora species, but no homologs were detected in the Glaucocystis or Gloeochaete plastid genomes. tmRNAs are mediators of the trans-translation process that rescues stalled ribosomes during protein translation (Gueneau de Novoa and Williams 2004; Janssen and Hayes 2012). The sequence of a tmRNA comprises a tRNA-like domain that can be exclusively aminoacylated with alanine but lacks the corresponding anticodon triplet. Additionally, tmRNAs have a relatively long loop that includes an ORF encoding a polypeptide of 13–16 residues (supplementary fig. S1, Supplementary Material online). The addition of this polypeptide to the arrested protein chain directs the release of the ribosome and tags the failed protein for degradation. An alignment of Cyanoptyche and Cyanophora ssrA sequences with cyanobacterial homologs shows that the glaucophyte genes contain the region encoding the polypeptide tag, as well as segments (i.e., stems and pseudoknots) predicted to be important in stabilizing the secondary structure of tmRNAs in cyanobacteria (supplementary fig. S1, Supplementary Material online) (Williams 2002; Gueneau de Novoa and Williams 2004). It is unknown if the Cyanoptyche and Cyanophora tmRNAs are involved in a plastid trans-translation system.
All-glaucophyte ptDNAs have the gene rnpB, which encodes the RNA component of Ribonuclease P, a ribonucleoprotein responsible for the maturation of the 5′ end of tRNA molecules. Conversely, the gene smpB, encoding the protein section of Ribonuclease P, is absent in all sequenced glaucophyte ptDNAs. Nonetheless, it has been experimentally demonstrated that the C.paradoxa rnpB transcript has endonuclease activity (i.e., it is a ribozyme) and is able to process the 5′ ends of tRNA molecules in the absence of the protein component (Li et al. 2007).
Protein-Coding Genes
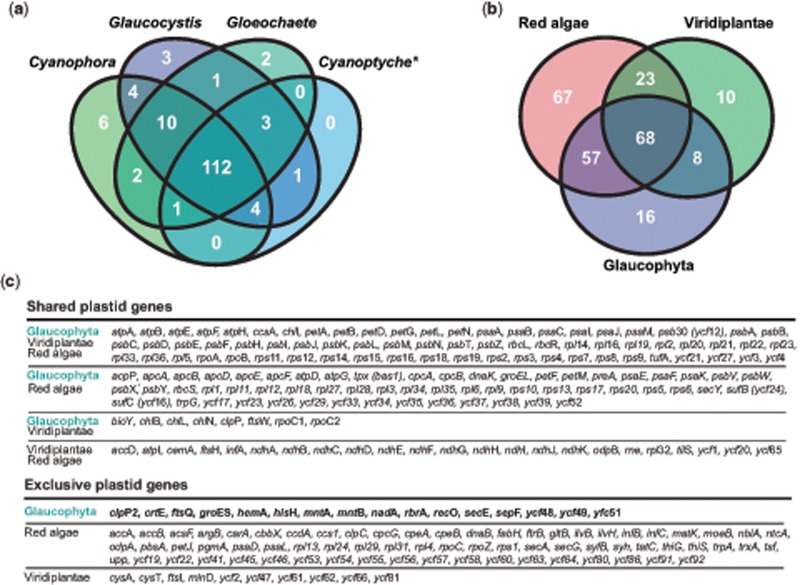
The number of unique plastid protein-coding genes (excluding unknown ORFs) varies from 137 in the two Glaucocystis species to 121 in Cyanopt.gloeocystis (supplementary table S10 no S11, Supplementary Material online), although it is possible that some plastid genes were not detected in the latter due to the incomplete assembly of this ptDNA. The gene collections of the individual glaucophyte ptDNAs are larger than the repertoires of most green algal and land plant counterparts (the vast majority of which have between 50 and 100 genes), but smaller than the typical red algal plastid genomes (between 160 and 210 genes). The gene complement of the glaucophyte ptDNAs is highly conserved, comprising 112 coding regions shared between representatives of the 4 genera and 18 genes that are present in at least 3 of the taxa examined (fig. 2A and supplementary table S10 no S11, Supplementary Material online). Only 11 genes, 6 of them in Cyanophora, are exclusively present in a single genus (fig. 2A). If we consider the combined gene repertoire of the 4 glaucophyte genera, there are 149 protein-coding genes that constitute the all-glaucophyte plastid collection. Almost 90% of the genes (133) in the all-glaucophyte set have putative orthologs in plastid genomes of red algae and viridiplants, but only 68 of them are universally shared by the 3 Archaeplastida lineages (fig. 2B and C). Glaucophytes share more plastid genes exclusively with red algae (57) than with viridiplants (8), whereas the latter two groups share 23 genes that are apparently absent from glaucophyte ptDNAs (fig. 2B and C).
Fig. 2.
—Protein-coding genes shared by different plastid genomes. The Venn diagram in (a) summarizes the number of protein-coding genes shared between the ptDNAs of Cyanophora, Glaucocystis, Gloeochaete, and Cyanoptyche (*the ptDNA of Cyanoptyche gloeocystis SAG 4.97 is partially sequenced). ORFs of unknown function exclusive to a single genus were not considered in the comparison. (b) The shared plastid protein-coding genes between the three Archaeplastida lineages. Note that the rbcL and rbcS genes from red algae are of different origin (proteobacterial) to the glaucophyte and viridiplants (both of cyanobacterial provenance). Table (c) lists the names of the plastid genes shared between the Archaeplastida lineages and those exclusive to each group.
The all-glaucophyte set also contains 16 genes thus far not identified in any other plastid genomes. This group includes genes present in all-glaucophyte ptDNAs, such as nadA (subunit A of the quinolinate synthetase), hemA (glutamyl-tRNA reductase), clpP2 (similar to clpP1, also plastid encoded, that encodes the proteolytic subunit of the CLP protease), ycf48 (putative assembly factor of photosystem II), and ycf51 (hypothetical protein of the DUF2518 family of unknown function) (fig. 2C). Genes exclusive to glaucophytes but not present in all genera investigated are groES (cochaperone GroES), rbrA (symerythrin; a rubrerythrin-like protein of the ferritin-like superfamily), ycf49 (a hypothetical protein of the DUF2499 family), hisH (glutamine amidotransferase subunit of the imidazole glycerol phosphate synthase; histidine biosynthesis), ftsQ (“cell division” protein FtsQ), secE (the subunit SecE of the Sec-translocase), recO (DNA repair protein RecO), crtE (geranyl-geranyl diphosphate synthase involved in carotenoid production), sepF (protein putatively involved in the formation of the Z-ring during cell division), and the pair mntA and mntB (encoding subunits of a putative manganese/zinc ABC-transporter) only present in Cyanophora species.
Conserved Gene Clusters and Rearrangements
Genome alignments of C.paradoxa ptDNA with the novel Glaucocystis sp. BBH, Cyanopt.gloeocystis, and Gloeochaete wittrockiana sequences indicate that a number of inversions and translocations have occurred during the diversification of the Glaucophyta, but there is no evidence of major architectural changes (supplementary fig. S7, Supplementary Material online). Some collinear gene blocks are conserved among glaucophyte ptDNAs, including two ribosomal protein clusters (5′-rps12-rps7-tufA-rps10-3′ and 5′-rpl3-rpl23-rpl2-rps19-rpl22-rps3-rpl16-rps17-rpl14-rpl5-rps8-rpl6-rpl18-rps5-3′) and the ensemble 5′-rpoB-rpoC1-rpoC2-rps2-atpH-atpG-atpF-atpD-atpA-petG-ycf24-ycf16-3′ (except in G.wittrockiana, where petG-ycf24-ycf16 are not part the cluster) (supplementary fig. S7, Supplementary Material online). Conservation of gene order in this latter cluster is found only in ptDNAs (i.e., it is not observed in extant cyanobacterial genomes) and this has been suggested as evidence for the single endosymbiotic origin of the primary plastids (Stoebe and Kowallik 1999; Löffelhardt 2014).
A 7.9-kb Insertion in the Glaucocystis incrassata ptDNA Might Be Derived from a DNA Mobile Element
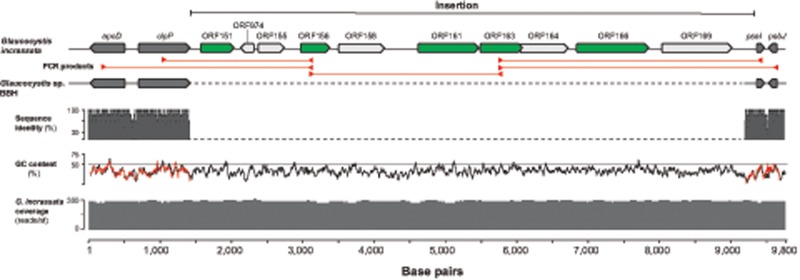
The alignment of the complete ptDNAs from the two Glaucocystis species not only showed that the gene order is practically identical (supplementary fig. S2, Supplementary Material online) but also revealed a 7.9-kb region between the clpP and psaI genes in the Gla.incrassata ptDNA that is absent in Glaucocystis sp. BBH (supplementary fig. S2, Supplementary Material online). To determine whether the 7.9-kb insertion in Gla.incrassata was an assembly artifact, we compared the read coverage of the insertion with flanking known-plastid sequences. The average read coverage of the insertion (295 ± 50 reads per nucleotide) is similar to the Gla.incrassata ptDNA as a whole (286 ± 68 reads per nucleotide) (fig. 3). Moreover, the read coverage of the complete Gla.incrassata mitochondrial genome (190 ± 55 reads per nucleotide) and a sample of 20 nuclear genes (6 ± 2 reads per nucleotide) suggests that the insertion is not a misassembled fragment originating from these latter genomic compartments. To confirm that the insertion originated from the Gla.incrassata ptDNA, we carried out PCR experiments using primer pairs matching sequences of the ptDNA genes flanking the insertion and internal regions of the insertion itself (the position of the different primer pairs used are indicated in fig. 3). Although we were unable to amplify the complete 7.9-kb region in a single PCR, Sanger sequencing and assembly of five partial amplicons allowed us to reconstruct the entire region (fig. 3). The resulting sequence perfectly matched the Illumina assembly, indicating that the insertion does indeed reside in the Gla.incrassata ptDNA. The 7.9-kb stretch accounts for most of length difference observed between the Gla.incrassata and Glaucocystis sp. BBH ptDNAs (supplementary fig. S2, Supplementary Material online).
Fig. 3.
—A putative insertion in the plastid genome of Glaucocystis incrassata (SAG 229-2). The 7.9-kb insertion between the typical plastid clpP and psaI genes includes ten ORFs, of which five (highlighted in green) encode putative proteins similar to known bacterial sequences (see text, supplementary figs. S2–S4, Supplementary Material online, for more details). Red arrowheads mark the position of primers designed to amplify flanking regions and fragments of the insertion. The red lines represent the five fragments amplified and sequenced using Sanger sequencing to corroborate contigs generated from Illumina reads. The sequence identity bar graph illustrates the similarity between the homologous regions of Gla. incrassata and Glaucocystis sp. BBH. The GC content percentage of the Gla. incrassata and Glaucocystis sp. BBH sequences is indicated with black and red lines, respectively.
The Gla.incrassata insertion contains ten ORFs of which only five encode proteins similar (BlastX E value < 1.0 e−10) to sequences available in public repositories (fig. 3). Four of these latter putative proteins present similarity to bacterial homologs, such as DNA primase/helicases (ORF 166), peptidoglycan aminohydrolases (ORF 163), and serine recombinases (ORFs 151 and 161). A homolog of the putative Gla.incrassata serine recombinases was also detected in the Cyanopt.gloeocystis ptDNA sequenced in this work (supplementary figs. S3B and S4, Supplementary Material online). In the case of the fifth ORF (ORF 156), the only two BlastX hits (∼30% sequence similarity) from GenBank were hypothetical proteins predicted in the ptDNAs of the green algae Prasiola crispa (Trebouxiophyceae) and Ettlia pseudoalveolar (Chlorophyceae). To further investigate the origin of ORFs 163, 166, 151, and 161, we used conceptual protein translations as BlastP queries to recover homologs from diverse biological groups and prepared multiple protein alignments for ML analyses. The putative serine recombinases of Gla.incrassata (ORFs 151 and 161) and Cyanopt.gloeocystis were resolved in a single clade (72% bootstrap support [BS]) that branches among bacterial and diatom sequences, but with no well-supported association to any of these groups (supplementary fig. S3B, Supplementary Material online). Similarly, for peptidoglycan aminohydrolase (ORF 163; supplementary fig. S3A, Supplementary Material online) and the DNA primase/helicase (ORF 166; supplementary fig. S3C, Supplementary Material online), the branching position of the Gla.incrassata sequences is not unambiguously resolved.
Although we cannot infer the origin of the Gla.incrassata ptDNA insertion based solely on the limited phylogenetic signal of the individual ORFs, the functions of the proteins that could be identified provide some insights into the possible source. For instance, peptidoglycan aminohydrolases (ORF 163), DNA primases (ORF 166), and serine recombinase (ORF 151 and ORF 161) are often encoded in bacterial plasmids and bacteriophage genomes (Ilyina et al. 1992; Laverde Gomez et al. 2014; Johnson 2015). It is known that these three types of enzymes play roles in the mobilization (peptidoglycan aminohydrolases) and replication (DNA primase and serine recombinase) of such DNA mobile elements (Ilyina et al. 1992; Regamey and Karamata 1998; DeWitt and Grossman 2014; Laverde Gomez et al. 2014; Rutherford and Van Duyne 2014). As such, it is possible that the insertion in the Gla.incrassata ptDNA is derived from a bacterial plasmid or phage sequence that became integrated into the plastid chromosome.
Phylogenomics: History of the Primary Plastids and Relationships between Glaucophyte Lineages
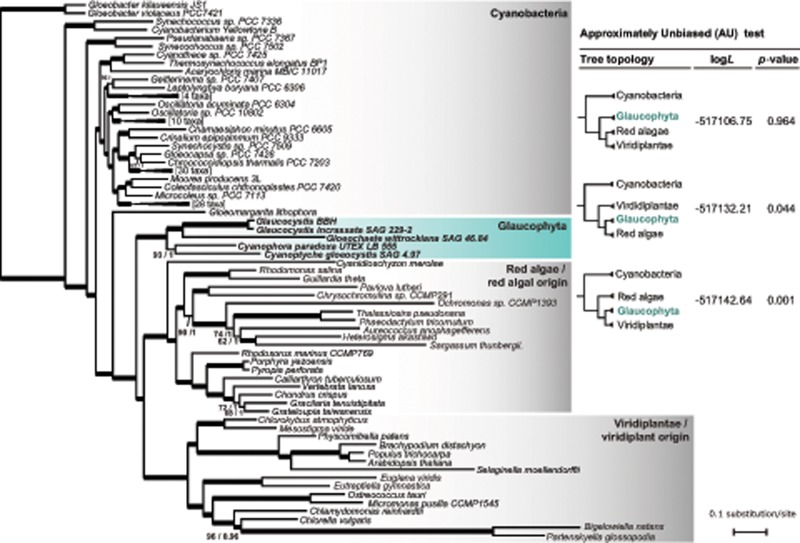
A ML estimation with the substitution model LG+C40+R (ln L = −517,106.76; fig. 5) produced a tree with a higher likelihood than the LG4X (ln L = −533,824.14) and LG+9R (ln L = −534,986.92) models, but overall the resulting topologies are very similar with only minor differences (supplementary fig. S5, Supplementary Material online). The Bayesian estimation with the mixture model CAT-GTR G4 produced a tree topology largely consistent with the ML analyses, (supplementary fig. S5, Supplementary Material online). Importantly, in all cases, a monophyletic Glaucophyta (100% BS, 1.0 posterior probabilities [PP]) was resolved as the earliest branching plastid lineage, with red algae and viridiplants (including corresponding lineages with plastids of secondary origin) united in a single clade (100% BS and 1.0 PP) (fig. 4 and supplementary fig. S5, Supplementary Material online). To further scrutinize the branching position of Glaucophyta, we also used the AU test (Shimodaira and Goldman 2002) to evaluate alternative phylogenetic hypotheses. A chosen set of competing topologies comprised trees with red algae (tree 1) or viridiplants (tree 2) as the earliest diverging archaeplastidian branch, and topologies where either the Glaucophyta (trees 3–7) or the Archaeplastida (trees 8–13) monophyly was disrupted (the 13 evaluated trees are available in supplementary table S8, Supplementary Material online). Consistent with the ML and Bayesian estimations, the AU test identified the tree with Glaucophyta in the earliest plastid branching position (tree 3) as the best hypothesis (P value = 0.964) but did not reject (P < 0.01) the tree with viridiplants as the earliest diverging group (P value = 0.044) (fig. 4). In contrast, a tree with red algae branching first was rejected (P value = 0.001), as were the other ten alternative hypotheses tested (supplementary table S8, Supplementary Material online). Within Glaucophyta, the genera duos Cyanophora–Cyanoptyche and Glaucocystis–Gloeochaete, respectively, branch as sister taxa with moderate to strong support (93–99% BS) (supplementary fig. S5, Supplementary Material online). The same phylogenetic relationships between the four glaucophyte genera were obtained previously in phylogenies estimated from complete mitochondrial genomes (Jackson and Reyes-Prieto 2014).
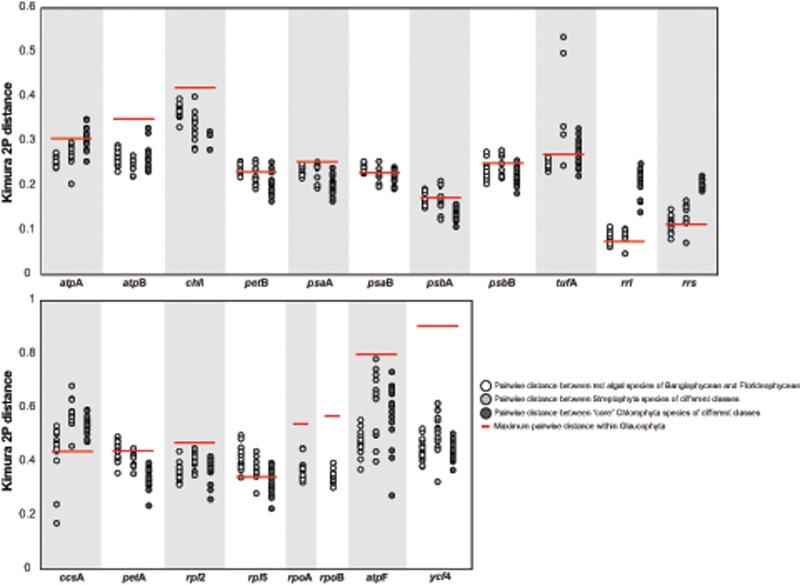
Fig. 5.
—K2P distances estimated with diverse plastid genes. Pairwise corrected distances were calculated independently for each major Archaeplastida group (list of the selected taxa, corresponding classes, subclasses, orders, and details of estimated values are provided in supplementary tables S2–S4, Supplementary Material online). Each dot in the plot represents a pairwise K2P distance between two species. Red bars highlight the maximum pairwise distance within Glaucophyta.
Fig. 4.
—Multiloci phylogenetic analysis. A maximum likelihood phylogenetic tree was estimated from a set of 42 plastid-encoded proteins with IQ-Tree v1.56 using the substitution model LG+C40+R (see supplementary table S3, Supplementary Material online, for details). Branch support was evaluated with 5,000 ultrafast BS as implemented in IQ-Tree. Bayesian posterior PP were estimated with PhyloBayes-MP v 1.6 under the CAT-GTR+G4 model running two independent Monte Carlo Markov chains until convergence (see Materials and Methods section for details). Thick lines represent branches with BS proportion values ≥99 and PP 1.0. Numbers near branches indicate BS proportion values <99 and corresponding PP, respectively. Branch lengths are proportional to the number of substitutions per site as indicted by the scale bar. The results of the AU test are detailed in the inset table.
Plastid Sequence Divergence within Glaucophyta
Previous comparative studies (i.e., phylogenies, genetic distance, and DNA barcoding analyses) of organelle genes have provided some insights into species diversity within the genera Cyanophora and Glaucocystis (Chong et al. 2014; Smith et al. 2014; Takahashi et al. 2014, 2016), but almost nothing is known about the amount of genetic distance between the different glaucophyte lineages above the species level. To investigate the magnitude of sequence divergence within Glaucophyta, we estimated genetic distances between species from different genera using a sample of 19 plastid genes that included widely used molecular markers (supplementary table S2, Supplementary Material online). For comparison, we also used the same plastid gene set to quantify the level of sequence divergence within the red (supplementary table S3, Supplementary Material online) and green (supplementary table S4, Supplementary Material online; only 17 genes producing reliable multiple alignments were considered in this case) algal lineages. The taxon sampling was designed to include a set of representative species from the major groups recognized within red algae (Yang et al. 2016) and green algae (Leliaert et al. 2012), respectively (supplementary table S7, Supplementary Material online). The K2P distances were estimated separately for glaucophytes (4 species from 4 genera), red algae (17 species from 7 different classes), and green algae (21species from 15 different classes) (details in supplementary table S7, Supplementary Material online). The K2P distance average values showed substantial variability in the amount of sequence divergence among the different loci (supplementary fig. S6, Supplementary Material online). For instance, in the three Archaeplastida groups, the genes encoding subunits of the photosystems tend to accumulate fewer substitutions than atpF, ycf4, ccsA, petA, and the two genes encoding ribosomal proteins (supplementary fig. S6, Supplementary Material online). Nonetheless, in most cases, the mean K2P distance values estimated with the same gene (for example atpA) are similar in the three Archaeplastida lineages (supplementary fig. S6, Supplementary Material online).
The maximum K2P distances estimated between any glaucophyte pair frequently (14/19 genes) involved G.wittrockiana (supplementary table S2, Supplementary Material online). To contrast the sequence divergence of the four Glauocophyta genera, we first compared the highest distance within this group (supplementary table S2, Supplementary Material online) against the sequence divergence between representatives of the red algal classes Bangiophyceae and Florideophyceae. We found that in 12/19 evaluated genes, the maximum distance estimated between two glaucophyte species is higher than at least 91% (in eight cases this value is 100%) of the K2P values calculated for any Bangiophyceae–Florideophyceae pair (supplementary table S3, Supplementary Material online and fig. 5). Only in two gene comparisons (rpl5 and rrs) is the maximum distance between two glaucophyte species lower than at least 50% of the Bangiophyceae versus Florideophyceae estimates (supplementary table S3, Supplementary Material online). Further, comparisons against pairwise distances estimated within the Streptophyta revealed that in 12/17 evaluated genes (viridiplant genes rpoA and rpoB did not produce reliable multiple alignments and were not included in this comparison), the maximum sequence divergence within glaucophyte species is higher than at least 64% (in 7 cases this value is 100%) of the distances estimated between representatives of the different Streptophyta classes considered (supplementary table S4, Supplementary Material online and fig. 5). Only in five genes was the maximum interglaucophyte distance lower than the majority (at least 80% of the pairwise estimations) of comparisons between Streptophyta classes. Finally, in 12 of the 17 genes tested, the maximum inter-glaucophyte distance exceeds at least 62% (in 8 cases this value is 100%) of the pairwise estimates between species of different “core” Chlorophyta classes (supplementary table S4, Supplementary Material online and fig. 5).
Discussion
Several inversions and translocations of gene clusters have occurred among the plastid genomes of the different glaucophyte genera. However, the basic circular-mapping architecture seems relatively simple when compared with the embellishments observed in plastid genomes of certain Archaeplastida subgroups. For example, in all glaucophyte ptDNAs sequenced thus far there is no evidence of chromosome linearization, fragmentation, expansion of intergenic regions (e.g., proliferation of repetitive sequences or transposable elements) or presence of intron-rich sequences as observed in some green (Smith and Keeling 2015; Turmel et al. 2015; Brouard et al. 2016; Lemieux et al. 2016; Del Cortona et al. 2017; Gaouda et al. 2018) and red algal taxa (Perrineau et al. 2015; Muñoz-Gómez et al. 2017). In addition to this relatively simple architecture, the gene repertoire of the glaucophyte ptDNAs is highly conserved, with only a few genes occurring exclusively in any individual genome. Based on the exclusive plastid gene content of each archaeplastidian group and the sets of shared genes, we can predict that the Last Plastid Common Ancestor of the Archaeplastida (LPCA) had a collection of at least ∼251 protein-coding genes. Then, if we consider this collection as the ancestral plastid gene complement of the archaeplastidians, then the red algal set of ∼215 genes most closely resembles the repertoire of the LPCA, with only ∼35 genes “lost.”
Genetic distance comparisons performed in this study indicate that several plastid loci have higher sequence divergence values between species of different glaucophyte genera than between red algal species from the Bangiophyceae and Florideophyceae (groups that diverged circa 1 Ga [Yang et al. 2016]) and between species of different classes within the “core” Chlorophyta group (the age of this clade is between 700 and 900 Myr [De Clerck et al. 2012]). These data suggest that the four glaucophyte genera may represent plastid lineages of ancient divergence, but it is also possible that glaucophyte plastid genes accumulate substitutions at higher rates than red algal orthologous sequences (i.e., glaucophyte plastids have fast evolving genomes). Further analyses including data of more glaucophyte taxa, ideally from putative species recently reported (e.g., Chalarodora azurea), and thorough investigations of divergence times are needed to discern between the two scenarios.
The cyanobacterial origin of primary plastids predicts that the genes contained in plastid genomes of Archaeplastida originated from a cyanobacterial genome. This working hypothesis seems to be true for most plastid genes, but there are known cases of plastid sequences of noncyanobacterial origin that were likely acquired from other genomic sources. The apparent noncyanobacterial origin of the ten ORFs identified in a 7.9-kb insertion of the Gla.incrassata ptDNA suggests that those coding regions were horizontally transferred into the plastid chromosome from foreign sources. Overall, single-locus phylogenetic analyses of the four ORFs with recognizable homologs in public databases were insufficient to resolve the origin of those coding regions. However, the putative enzymatic capabilities of some of the encoded proteins suggest that the 7.9-kb insertion originated from a mobile DNA element. It is known that serine recombinases (ORFs 151 and 161), DNA primases (ORF 166), and peptidoglycan aminohydrolases (ORF 163) participate in the mobilization of diverse transposable elements (e.g., transposons), phage genomes, and bacterial plasmids into bacterial chromosomes (Ilyina et al. 1992; DeWitt and Grossman 2014; Laverde Gomez et al. 2014; Stark 2014). Hence, we hypothesize that these three enzyme types are encoded within the same sequence stretch because they were all inserted into the Gla.incrassata ptDNA as part of a DNA mobile element of unclear origin. The presence of sequences acquired from DNA mobile elements have been reported also in ptDNAs of diatoms (Ruck et al. 2014), green (Leliaert and Lopez-Bautista 2015; Brouard et al. 2016), and red algae (Janouškovec et al. 2013; Lee, Kim, et al. 2016; Muñoz-Gómez et al. 2017). Additionally, glaucophytes are not the only group with plastid-encoded DNA recombinases. For instance, the plastid genomes of some pennate diatoms and the dinoflagellate Kryptoperidinium foliaceum, which possesses plastids of diatom origin, encode serine recombinases likely recruited by horizontal gene transfer (HGT) from plasmids of putative bacterial origin localized within the host cells (Hildebrand et al. 1992; Imanian et al. 2010; Brembu et al. 2014; Ruck et al. 2014). Moreover, the green algae Oedogonium cardiacum (Chlorophyceae), Roya anglica (Zygnematophyceae) (Brouard et al. 2008; Civáň et al. 2014), and diverse stramenopile algae (Cattolico et al. 2008; Imanian et al. 2010; Brembu et al. 2014) encode tyrosine recombinases, a different type of site-specific DNA recombinase, in their plastid genomes. Further investigation is required to determine if the DNA recombinases encoded in ptDNAs of disparate algal lineages are actually involved in site-specific recombination mechanisms promoting the integration of foreign sequences into plastid chromosomes.
Plastid genomes are less prone than mitochondrial and nuclear counterparts to capture foreign sequences via HGT (Keeling and Palmer 2008; Keeling 2010), but there are several known HGT cases in plastids genomes. Prominent examples include the leuC/D operon of proteobacterial origin in the florideophyceaen red alga Gracilaria tenustipitata (Janouškovec et al. 2013), two genes of proteobacterial origin encoding the RuBisCO subunits in red algae (Delwiche and Palmer 1996), the gene rpl36 in cryptophytes and haptophytes (Rice and Palmer 2006), genes involved in the biosynthesis of vitamin K in cyanidiales (Gross et al. 2008), ORFs of possible mitochondrial origin in the green alga O.cardiacum (Brouard et al. 2008), diverse genes in diatom plastid genomes acquired from plasmids resident in both the nucleus and plastids of the same diatoms (Ruck et al. 2014), bacterial-derived genes encoding enzymes involved in DNA replication and mobilization (DNA polymerases, transposases, integrases, and primases) in the green algae Bryopsis plumosa and Tydemania expeditiones (Leliaert and Lopez-Bautista 2015), the DNA polymerase of the cryptophytes Rhodomonas salina and Teleaulax amphioxeia (Khan et al. 2007; Kim et al. 2015), genes involved in isoprenoid synthesis in the eustigmatophyte Monodopsis (Yurchenko et al. 2016) and intron sequences in the cryptophyte R.salina (Khan et al. 2007) and the diatom Seminvis robusta (Brembu et al. 2014). The 7.9-kb insertion in the plastid genome of G. incrassata appears to be another example suggesting that horizontal transfer of genetic material has contributed, if only rarely, to the evolution of plastid genomes.
If the three Archaeplastida lineages share a single nucleo-cytoplasm ancestor (i.e., the host component) that established a unique endosymbiosis with cyanobacteria (the plastid ancestor), then we would predict that nuclear and plastid phylogenomics should produce largely congruent results that reflect a common evolutionary history. Contrary to that prediction, the recurrent outcome of such analyses is incongruence, that is, nonmonophyly versus monophyly of the Archaeplastida in phylogenies from nuclear versus plastid data, respectively (see Mackiewicz and Gagat 2014 for review). Plastid phylogenomics cannot help us to directly solve the Archaeplastida nucleo-cytoplasm monophyly conundrum, but it can provide insights into the evolutionary rates and branching patterns during diversification of the plastid lineages and to finally identify the earliest diverging plastid lineage. Most previous investigations of the diversification of primary plastids included genomic data from only a single glaucophyte species, C.paradoxa, and in a few cases (e.g., Price et al. 2012) from two taxa. Our phylogenomic analyses are the first to include multigene plastid data from the four glaucophyte genera available in public collections. In all phylogenetic analyses, Glaucophyta were recovered as a monophyletic group, but the sister relationships resolved between the four genera (Cyanophora–Cyanoptyche and Gloeochaete–Glaucocystis) do not have full node support. These inter-genera sister relationships are consistent with our previous phylogenomic survey using mitochondrial genomes (Jackson and Reyes-Prieto 2014). Moreover, the presence of a tmRNA gene in Cyanophora and Cyanoptyche and the apparent loss of the type I intron in the trnL gene (this intron is widely conserved in cyanobacteria and plastid genomes [Kuhsel et al. 1990; Besendahl et al. 2000]) of Gloeochaete and Glaucocystis, respectively, are consistent with the affiliation of each of these two genera pairs in the plastid and mitochondrial phylogenies. However, the evidence is still based on a small taxonomic sample and inclusion of further complete organelle data from additional glaucophyte species (e.g., Ch.azurea) will be important to untangle the phylogenetic affiliations between the different glaucophyte lineages.
Our ML and Bayesian phylogenomic results suggest that the Glaucophyta is the first primary plastid lineage to diverge from the Archaeplastida stem, but given that the AU test did not reject a competing tree depicting viridiplants as the earliest diverging branch, alternative hypotheses are still open. Some previous investigations using different sets of plastid protein-coding genes have also resolved Glaucophyta as the earliest diverging archaeplastidian branch (Rodríguez-Ezpeleta et al. 2005 [fig. 1]; Qiu et al. 2012 [fig. 3A]; Li et al. 2014 [fig. S4]; Ponce-Toledo et al. 2017 [fig. 1 and fig. S1]), whereas other studies have recovered the two alternative evolutionary scenarios, with red algae (Janouskovec et al. 2010 [fig. 5]; Criscuolo and Gribaldo 2011 [fig. 2]; Price et al. 2012 [fig. S5]), or viridiplants as the first diverging lineage (Deschamps and Moreira 2009 [figs. 1, 3B, and 3C]; see Mackiewicz and Gagat [2014] for a thorough discussion of these conflicting results). Solving the branching history of primary plastids might rely not only on further analyses with expanded taxon sampling, including plastid data of new glaucophyte taxa and early-branching red algae, but also on the further development of substitution models that cope better with heterogeneous substitution rates in plastid sequences and differences in amino acid composition between lineages.
Genomic studies of additional glaucophyte representatives will be important not only to investigate the early diversification of primary plastids in more detail, as well as the phylogenetic history of Glaucophyta and the apparent high genetic divergence between glaucophyte genera, but also to explore whether the integration of foreign DNA sequences into plastid genomes, such as the insertion in Gla.incrassata, have played significant roles during the evolution of this rare algal group.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The present research work was supported by the Natural Sciences and Engineering Research Council of Canada (Grant No. 402421-2011), the Canada Foundation for Innovation (Grant No. 28276), and the New Brunswick Innovation Foundation (Grant No. RIF2012-006) awarded to A.R.-P. We sincerely thank Dr Aurora Nedelcu and Dr Gary Saunders for the thoughtful comments during the elaboration of this article. We also want to acknowledge the valuable suggestions of two anonymous reviewers.
Literature Cited
- Adl SM, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59(5):429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkatib S, Fleischmann TT, Scharff LB, Bock R.. 2012. Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res. 40(14):6713–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besendahl A, Qiu YL, Lee J, Palmer JD, Bhattacharya D.. 2000. The cyanobacterial origin and vertical transmission of the plastid tRNA(Leu) group-I intron. Curr Genet. 37(1):12–23. [DOI] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J.. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 17(11):1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembu T, et al. 2014. The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar Genomics. 16:17–27. [DOI] [PubMed] [Google Scholar]
- Brouard J-S, Otis C, Lemieux C, Turmel M.. 2008. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics 9(1):290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard J-S, Turmel M, Otis C, Lemieux C.. 2016. Proliferation of group II introns in the chloroplast genome of the green alga Oedocladium carolinianum (Chlorophyceae). PeerJ 4:e2627.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MW, et al. 2018. Phylogenomics places orphan protistan lineages in a novel eukaryotic super-group. Genome Biol Evol. 10(2):427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, et al. 2016. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc R Soc Lond B Biol Sci. 283 (1823):20152802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattolico R, et al. 2008. Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics 9(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Jackson C, Kim JI, Yoon HS, Reyes-Prieto A.. 2014. Molecular markers from different genomic compartments reveal cryptic diversity within glaucophyte species. Mol Phylogenet Evol. 76C:181–188. [DOI] [PubMed] [Google Scholar]
- Civáň P, Foster PG, Embley MT, Séneca A, Cox CJ.. 2014. Analyses of charophyte chloroplast genomes help characterize the ancestral chloroplast genome of land plants. Genome Biol Evol. 6:897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH.. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24(6):861–862. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2011. Large-scale phylogenomic analyses indicate a deep origin of primary plastids within Cyanobacteria. Mol Biol Evol. 28(11):3019–3032. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clerck O, Bogaert KA, Leliaert F.. 2012. Diversity and evolution of algae: primary endosymbiosis. Adv Bot Res. 64:55–86. [Google Scholar]
- Del Cortona A, et al. 2017. The plastid genome in cladophorales green algae is encoded by hairpin chromosomes. Curr Biol. 27(24):3771–3782. [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Palmer JD.. 1996. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol Evol. 13(6):873–882. [DOI] [PubMed] [Google Scholar]
- Deschamps P, Moreira D.. 2009. Signal conflicts in the phylogeny of the primary photosynthetic eukaryotes. Mol Biol Evol. 26(12):2745–2753. [DOI] [PubMed] [Google Scholar]
- DeWitt T, Grossman AD.. 2014. The bifunctional cell wall hydrolase CwlT is needed for conjugation of the integrative and conjugative element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol. 196(8):1588–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Leliaert F, Zhang Z-H, Penny D, Zhong B-J.. 2017. Evolution of the chlorophyta: insights from chloroplast phylogenomic analyses. J Syst Evol. 55:322–332. [Google Scholar]
- Gaouda H, et al. 2018. Exploring the limits and causes of plastid genome expansion in volvocine green algae. Genome Biol Evol. 10(9):2248–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Meurer J, Bhattacharya D.. 2008. Evidence of a chimeric genome in the cyanobacterial ancestor of plastids. BMC Evol Biol. 8:117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueneau de Novoa P, Williams KP.. 2004. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 32(Database issue):D104–D108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall WT, Claus G.. 1963. Ultrastructural studies on the blue-green algal symbiont in Cyanophora paradoxa Korschikoff. J Cell Biol. 19(3):551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji T, et al. 2013. Mitochondrial and plastid genomes of the colonial green alga Gonium pectorale give insights into the origins of organelle DNA architecture within the volvocales Verbruggen. PLoS One 8(2):e57177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss AA, et al. 2018. Combined morphological and phylogenomic re-examination of malawimonads, a critical taxon for inferring the evolutionary history of eukaryotes. R Soc Open Sci. 5(4):171707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M, et al. 1992. Nucleotide sequence of diatom plasmids: identification of open reading frames with similarity to site-specific recombinases. Plant Mol Biol. 19(5):759–770. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina TV, Gorbalenya AE, Koonin EV.. 1992. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J Mol Evol. 34(4):351–357. [DOI] [PubMed] [Google Scholar]
- Imanian B, Pombert J-F, Keeling PJ, Schleiermacher C, Stoye J.. 2010. The complete plastid genomes of the two ‘Dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS One 5(5):e10711.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C, Clayden S, Reyes-Prieto A.. 2015. The Glaucophyta: the blue-green plants in a nutshell. Acta Soc Bot Pol. 84(2):149–165. [Google Scholar]
- Jackson CJ, Reyes-Prieto A.. 2014. The mitochondrial genomes of the glaucophytes Gloeochaete wittrockiana and Cyanoptyche gloeocystis: multilocus phylogenetics suggests a monophyletic archaeplastida. Genome Biol Evol. 6(10):2774–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Horak A, Obornik M, Lukes J, Keeling PJ.. 2010. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 107(24):10949–10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2013. Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS One 8(3):e59001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2017. A new lineage of eukaryotes illuminates early mitochondrial genome reduction. Curr Biol. 27(23):3717–3724.e5. [DOI] [PubMed] [Google Scholar]
- Janssen BD, Hayes CS.. 2012. The tmRNA ribosome-rescue system. Adv Protein Chem Struct Biol. 86:151–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC. 2015. Site-specific DNA inversion by serine recombinases. Microbiol Spectr. 3:1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. 2010. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 365(1541):729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD.. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 9:605–618. [DOI] [PubMed] [Google Scholar]
- Khan H, et al. 2007. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 24(8):1832–1842. [DOI] [PubMed] [Google Scholar]
- Kies L, Kremer BP.. 1986. Typification of the Glaucocystophyta. Taxon 35(1):128–133. [Google Scholar]
- Kim E, Maruyama S.. 2014. A contemplation on the secondary origin of green algal and plant plastids. Acta Soc Bot Pol. 83(4):331–336. [Google Scholar]
- Kim JI, et al. 2015. The plastid genome of the cryptomonad Teleaulax amphioxeia. PLoS One 10(6):e0129284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Jaynes JM, Kent SS, Vernon LP.. 1981. Properties of the photosynthetic system and DNA of Cyanophora paradoxa cyanelles. Plant Physiol. 68(2):407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhsel MG, Strickland R, Palmer JD.. 1990. An ancient group I intron shared by eubacteria and chloroplasts. Science 250(4987):1570–1573. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B.. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverde Gomez JA, Bhatty M, Christie PJ.. 2014. PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone-responsive plasmid pCF10. J Bacteriol. 196(3):527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cho CH, et al. 2016. Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 14:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim KM, et al. 2016. Reconstructing the complex evolutionary history of mobile plasmids in red algal genomes. Sci Rep. 6:23744.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, Lopez-Bautista JM.. 2015. The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genomics 16:204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, et al. 2012. Phylogeny and molecular evolution of the green algae. CRC Crit Rev Plant Sci. 31(1):1–46. [Google Scholar]
- Leliaert F, et al. 2016. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. Sci Rep. 6:25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M.. 2016. Comparative chloroplast genome analyses of streptophyte green algae uncover major structural alterations in the Klebsormidiophyceae, Coleochaetophyceae and Zygnematophyceae. Front Plant Sci. 7:697.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lopes JS, Foster PG, Embley TM, Cox CJ.. 2014. Compositional biases among synonymous substitutions cause conflict between gene and protein trees for plastid origins. Mol Biol Evol. 31(7):1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Willkomm DK, Schön A, Hartmann RK.. 2007. RNase P of the Cyanophora paradoxa cyanelle: a plastid ribozyme. Biochimie 89(12):1528–1538. [DOI] [PubMed] [Google Scholar]
- Löffelhardt W, editor. 2014. The single primary endosymbiotic event In: Endosymbiosis. Vienna (Austria: ): Springer; p. 39–52. [Google Scholar]
- Löffelhardt W, Bohnert HJ.. 2001. The cyanelle (muroplast) of Cyanophora paradoxa: a paradigm for endosymbiotic organelle evolution In: Seckbach J, editor. Symbiosis. Mechanisms and model systems. Cellular origin, life in extreme habitats and astrobiology. Vol. 4 Dordrecht (The Netherlands: ): Springer; p. 111–130. [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P, Gagat P.. 2014. Monophyly of Archaeplastida supergroup and relationships among its lineages in the light of phylogenetic and phylogenomic studies. Are we close to a consensus? Acta Soc Bot Pol. 83(4):263–280. [Google Scholar]
- McFadden GI, van Dooren GG.. 2004. Evolution: red algal genome affirms a common origin of all plastids. Curr Biol. 14(13):R514–R516. [DOI] [PubMed] [Google Scholar]
- Misumi M, Sonoike K.. 2017. Characterization of the influence of chlororespiration on the regulation of photosynthesis in the glaucophyte Cyanophora paradoxa. Sci Rep. 7:46100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gómez SA, et al. 2017. The new red algal subphylum proteorhodophytina comprises the largest and most divergent plastid genomes known. Curr Biol. 27(11):1677–1684.e4. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Knoll AH, Katz LA.. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 108(33):13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A.. 2012. Separating the wheat from the chaff: mitigating the effects of noise in a plastome phylogenomic data set from Pinus L. (Pinaceae). BMC Evol Biol. 12(1):100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrineau M-M, Price DC, Mohr G, Bhattacharya D.. 2015. Recent mobility of plastid encoded group II introns and twintrons in five strains of the unicellular red alga Porphyridium. PeerJ 3:e1017.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanzagl B, et al. 1996. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 178(2):332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce-Toledo RI, et al. 2017. An early-branching freshwater cyanobacterium at the origin of plastids. Curr Biol. 27(3):386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DC, Steiner JM, Yoon HS, Bhattacharya D, Löffelhardt W.. 2017. Glaucophyta In: Archibald JM, Simpson AGB, Slamovits CH, editors. Handbook of the protists. Cham, Switzerland:Springer International Publishing; p. 23–87. [Google Scholar]
- Price DC, et al. 2012. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335(6070):843–847. [DOI] [PubMed] [Google Scholar]
- Qiu H, Yang EC, Bhattacharya D, Yoon HS.. 2012. Ancient gene paralogy may mislead inference of plastid phylogeny. Mol Biol Evol. 29(11):3333–3343. [DOI] [PubMed] [Google Scholar]
- Regamey A, Karamata D.. 1998. The N-acetylmuramoyl-l-alanine amidase encoded by the Bacillus subtilis 168 prophage SP beta. Microbiology 144(4):885–893. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Bhattacharya D.. 2007. Phylogeny of Calvin cycle enzymes supports Plantae monophyly. Mol Phylogenet Evol. 45(1):384–391. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Moustafa A.. 2012. Plastid-localized amino acid biosynthetic pathways of Plantae are predominantly composed of non-cyanobacterial enzymes. Sci Rep. 2:955.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Russell S, Figueroa-Martinez F, Jackson C.. 2018. Comparative plastid genomics of glaucophytes In: Jansen R, Chaw S-M, editors. Advances in botanical research. Plastid genome evolution. Vol. 85 San Diego (CA: ): Academic Press; p. 95–127. [Google Scholar]
- Rice DW, Palmer JD.. 2006. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 4(1):31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue N, Lartillot N.. 2014. Site-heterogeneous mutation-selection models within the PhyloBayes-MPI package. Bioinformatics 30(7):1020–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, et al. 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 15(14):1325–1330. [DOI] [PubMed] [Google Scholar]
- Ruck EC, Nakov T, Jansen RK, Theriot EC, Alverson AJ.. 2014. Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol Evol. 6(3):644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh J.. 2014. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 14(1):23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Van Duyne GD.. 2014. The ins and outs of serine integrase site-specific recombination. Curr Opin Struct Biol. 24:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Baracaldo P, Raven JA, Pisani D, Knoll AH.. 2017. Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc Natl Acad Sci U S A. 114:E7737–E7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Puerta MV, Bachvaroff TR, Delwiche CF.. 2005. The complete plastid genome sequence of the haptophyte Emiliania huxleyi: a comparison to other plastid genomes. DNA Res. 12(2):151–156. [DOI] [PubMed] [Google Scholar]
- Shih PM, Matzke NJ.. 2013. Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc Natl Acad Sci U S A. 110(30):12355–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Goldman N.. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51(3):492–508. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247. [DOI] [PubMed] [Google Scholar]
- Smith DR, Jackson CJ, Reyes-Prieto A.. 2014. Nucleotide substitution analyses of the glaucophyte Cyanophora suggest an ancestrally lower mutation rate in plastid vs mitochondrial DNA for the Archaeplastida. Mol Phylogenet Evol. 79:380–384. [DOI] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ.. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 112(33):10177–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark WM. 2014. The serine recombinases. Microbiol Spectr. 2(6):MDNA3-0046-2014. [DOI] [PubMed] [Google Scholar]
- Steiner JM, Löffelhardt W.. 2011. The photosynthetic apparatus of the living fossil, Cyanophora paradoxa In: Peschek GA, Obinger C, Renger G, editors. Bioenergetic processes of cyanobacteria. Dordrecht (The Netherlands: ): Springer; p. 71–87. [Google Scholar]
- Steiner JM, Yusa F, Pompe JA, Löffelhardt W.. 2005. Homologous protein import machineries in chloroplasts and cyanelles. Plant J. 44(4):646–652. [DOI] [PubMed] [Google Scholar]
- Stiller JW. 2014. Toward an empirical framework for interpreting plastid evolution. J Phycol. 50(3):462–471. [DOI] [PubMed] [Google Scholar]
- Stirewalt VL, Michalowski CB, Löffelhardt W, Bohnert HJ, Bryant DA.. 1995. Nucleotide sequence of the cyanelle genome from Cyanophora paradoxa. Plant Mol Biol Rep. 13(4):327–332. [Google Scholar]
- Stoebe B, Kowallik KV.. 1999. Gene-cluster analysis in chloroplast genomics. Trends Genet. 15(9):344–347. [DOI] [PubMed] [Google Scholar]
- Stöver BC, Müller KF.. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11(1):7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, et al. 2014. Five Cyanophora (Cyanophorales, Glaucophyta) species delineated based on morphological and molecular data. J Phycol. 50(6):1058–1069. [DOI] [PubMed] [Google Scholar]
- Takahashi T, et al. 2016. Delineation of six species of the primitive algal genus Glaucocystis based on in situ ultrastructural characteristics. Sci Rep. 6:29209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C.. 2015. Dynamic evolution of the chloroplast genome in the green algal classes pedinophyceae and trebouxiophyceae. Genome Biol Evol. 7(7):2062–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP. 2002. Descent of a split RNA. Nucleic Acids Res. 30(9):2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, et al. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci U S A. 109(43):17519–17524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki A, et al. 2014. Palpitomonas bilix represents a basal cryptist lineage: insight into the character evolution in Cryptista. Sci Rep. 4:4641.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EC, et al. 2016. Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci Rep. 6:21361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D.. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 21(5):809–818. [DOI] [PubMed] [Google Scholar]
- Yurchenko T, Ševčíková T, Strnad H, Butenko A, Eliáš M.. 2016. The plastid genome of some eustigmatophyte algae harbours a bacteria-derived six-gene cluster for biosynthesis of a novel secondary metabolite. Open Biol. 6(11):160249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.