Abstract
Microtubules are central elements of the eukaryotic cytoskeleton that often function as part of branched networks. Current models for branching include nucleation of new microtubules from severed microtubule seeds or from gamma-tubulin recruited to the side of a pre-existing microtubule. Here, we found that microtubules can be directly remodeled into branched structures by the microtubule-remodeling factor SSNA1 (or also NA14/DIP13). The branching activity of SSNA1 relies on its ability to self-assemble into fibrils in a head-to-tail fashion. SSNA1 fibrils guide protofilaments of a microtubule to split apart to form daughter microtubules. We further found that SSNA1 localizes at axon branching sites and has a key role in neuronal development. SSNA1 mutants that abolish microtubule branching in vitro also fail to promote axon development and branching when overexpressed in neurons. We have therefore, discovered a mechanism for microtubule-branching and implicated its role in neuronal development.
Introduction
Cell-shape control is critical in a number of physiological processes. Microtubules, the major cytoskeletal component determining cell shape, are mostly nucleated at the centrosome in proliferating cells. During specialized cell-shaping events, such as mitosis or cell polarization, cytoskeletal remodeling is thought to be driven by local nucleation of microtubules using a centrosome-independent mechanism1–3.
Neuronal cells are a distinctive example of cells with highly complex morphologies. Neurons are shaped in an extremely polarized fashion with a unique-shaped axon protruding from the cell body and stretching over long distances. Individual cells develop branch points from their axons to connect to neighboring cells, creating an intricate communication network in the nervous system. As the shape of axons is determined by microtubules, these branch points require remodeling of microtubules to split the cytoskeletal path into separate branches1–5. As the centrosome is not necessary for the morphological development of the axon6, it is possible that axonal transformation occurs in a locally regulated manner within the axon. At axon branching points, the local destabilization and fragmentation of microtubules is mediated by the microtubule-severing enzyme spastin, which is leading to the emergence of short microtubules7. However, the process of splitting the microtubule networks and, specifically, how the local rearrangement of spastin-processed tubulin oligomers or short microtubule fragments occurs has been enigmatic.
Due to its importance in various cell activities, the microtubule cytoskeleton has been well characterized in vitro. In the classical view, microtubules are considered as cylindrical polymers made of ~13 protofilaments. However, in living cells, it has been suggested that microtubules form higher-order branched networks to regulate their distribution within complex cytoskeletal networks1,8,9. The branched networks could be envisioned either through the attachment of new microtubule modules onto the side of an existing microtubule, or through direct branching of microtubules. So far, only one example of branching microtubule network has been shown, involving the microtubule nucleators augmin and γ-tubulin, which allow microtubules to grow out from nucleation points that attach to the side of existing microtubules8,10–13. There was, however, no evidence that protofilaments in a single microtubule can split apart to form a branched structure. Particularly at axon branch sites, augmin is less likely to be involved in generating branched microtubule networks, but has rather been implicated in determining microtubule orientation by crosslinking adjacent microtubules within an axon14.
Here we focused on the protein SSNA1, a microtubule-binding protein implicated in the dynamic assembly of microtubules. SSNA1 is found at centrosomes or basal bodies in sperm cells15,16, and at the midbody in dividing cells17. A recent study reported that SSNA1 accelerates neuronal development by promoting axon elongation and branch formation17. These observations collectively suggest a versatile role of SSNA1 for microtubule remodeling. However, the nature of its activity in controlling microtubule dynamics is unclear.
We now demonstrate that SSNA1 is a powerful microtubule-nucleating and -branching factor. In vitro reconstitution of SSNA1-mediated microtubule nucleation showed an induction of branched microtubules, where new daughter microtubules directly branch out from existing microtubules. SSNA1 attaches along single protofilaments, guiding them to grow away from a microtubule and template a branched microtubule. Mutation of residues essential for oligomerization and the microtubule-branching activity of SSNA1, which we designed by structure-guided in vitro experiments, lead to defective axonal branching in primary neurons, showing that the simple scaffolding mechanism of SSNA1 can lead to vast morphological changes in neurons.
Results
SSNA1 localizes at axon branching sites in primary neurons
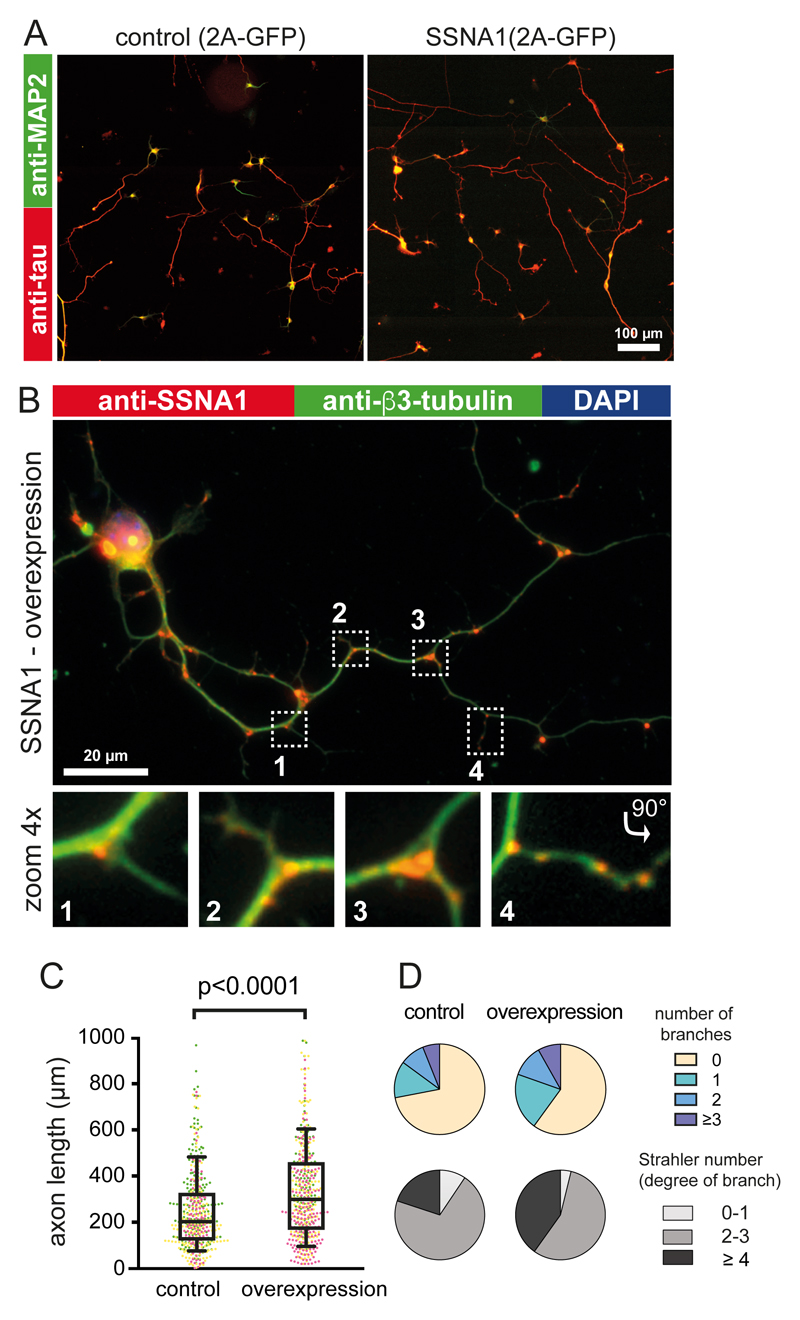
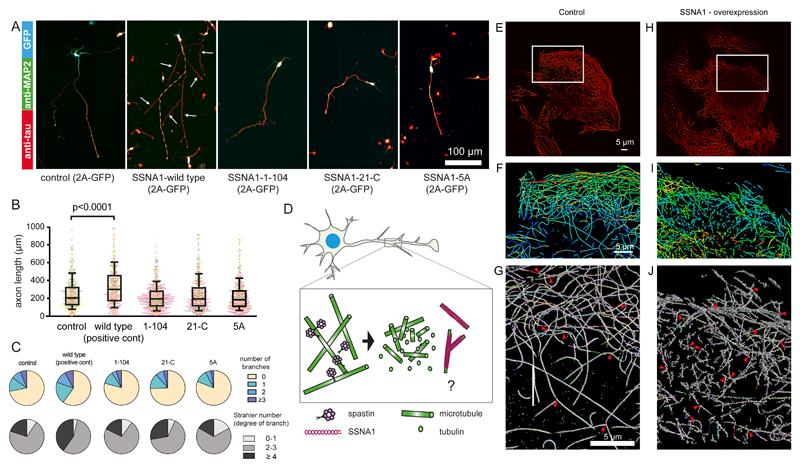
A previous study implicated SSNA1 in the promotion of axon branching17, but the underlying mechanism remained elusive. To investigate how SSNA1 plays a role in neuron development, we transduced wild-type murine primary hippocampal neurons with lentiviral particles encoding GFP-2A-mouse_SSNA1. Overexpression of SSNA1 led to the promotion of axon outgrowth (Fig. 1A, C) in agreement with a previous report17. In addition, we found a striking accumulation of SSNA1 at axon branches (Fig. 1B, 1D, S1), which was also observed at secondary branch sites that emanate from an already existing axon branch (Fig. 1B, panel “4”, Fig. S1). In agreement with its localization, SSNA1 overexpression led to increased and more complex branching as characterized by the Strahler number (Fig. 1D). Since SSNA1 localizes at the cytosolic compartments where microtubule are dynamic15–17, we hypothesized that SSNA1-clusters at branching sites in neurons might facilitate local microtubule nucleation.
Fig. 1. The effect of SSNA1 overexpression on primary hippocampal neurons.
(A) Immunostaining of MAP2 (green) and Tau (red) in control (GFP over-expression) and mSSNA1 WT over-expression. (B) Immunostaining of SSNA1 (red) and β3-tubulin (green) in mSSNA1 WT overexpressing neurons shows the localization of SSNA1 at axon branch sites. (C) Scatter dot plots of axon length under over-expression of SSNA1. The longest protrusion from soma was defined as axon, and cells with very short protrusions were also included in the counting, so that under-developed neurons could be assessed as well. The promotion of axon development occurs only in over-expression of WT SSNA1. Experiments were performed in triplicates, shown in magenta, green and yellow. Every cell is represented by a single point: Control (n = 505 cells), wild type (n = 499 cells), pooled from 3 independent experiments, and the overlaid box-and-whisker plots cover 50% (boxes) and 90% (whiskers) of the entire population, with median values indicated as lines within the boxes. The results show statistical significance (p < 0.0001) as tested using the Kruskal-Wallis test, followed by Dunn’s multiple comparison post-hoc test. (D) Pie graphs showing the distribution of the number of branches under over-expression conditions (control (n=496 cells), wild type (n=490 cells) pooled from 3 independent experiments) and Strahler number (degree of sub-branch formations on the existing branches), control (n=266 cells), wild type (n=289 cells) pooled from 3 independent experiments. Distributions of the branches and the Strahler number in SSNA1 expressed neurons differ significantly from control (GFP over-expression) according to χ2 two-sample test (χ2= 20.7, p < 0.01 and 18.6, p < 0.005, respectively). See supplementary table 3 for source data.
SSNA1 induces direct microtubule branching
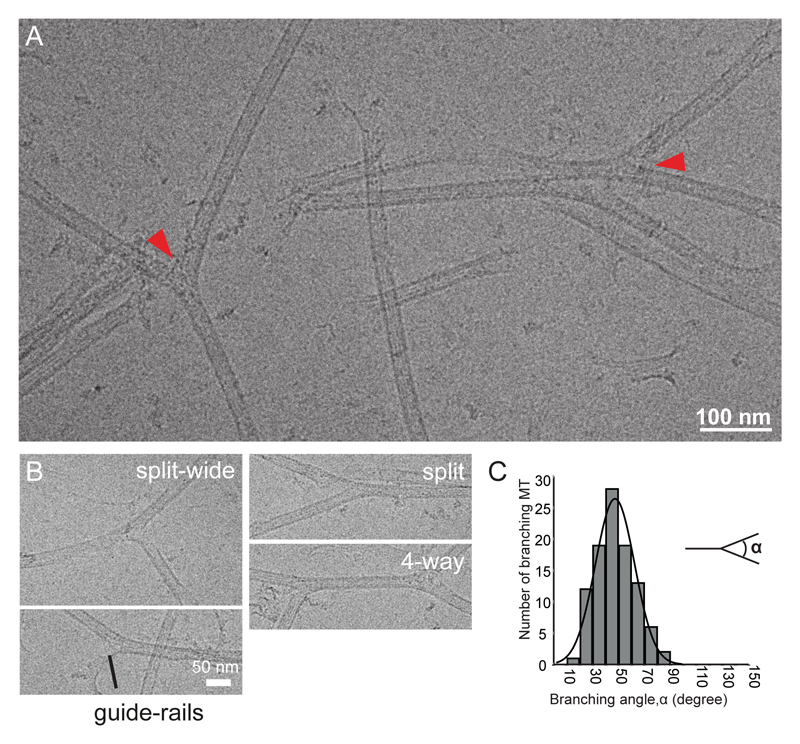
To assess the influence of SSNA1 on microtubules, we prepared recombinant SSNA1 (from Chlamydomonas reinhardtii, Fig. S2A-C) and tested its interaction using cryo-EM (Fig. 2A, Fig. S2D). CrSSNA1induced a formation of direct microtubule branches that split from a single microtubule (Fig. 2A, arrowheads), in contrast to an attachment of a second microtubule on the microtubule surface. Branching occurs by splitting the lattice of the microtubule, and protofilaments of mother microtubules directly continue into the outer surface of the branched microtubule. The bending angle was variable (Fig. 2B-C, 47°±15°, Fig. S2D-E), which suggests a rather flexible junction, in contrast to the more rigid, 70° Arp2/3-mediated actin-branching18,19. Moreover, microtubules occasionally formed fork-like structures with several branches or junctions (Fig. 2B, S2D). This has so far not been observed in any other system, and further underpins the uniqueness of SSNA1-mediated microtubule branching.
Fig. 2. Characterization of in vitro reconstituted microtubule branching.
(A) Cryo-EM image of branched microtubules. Arrowheads show examples of branching points. Microtubules were stabilized with 1mM GMPCPP. (B) Snapshots of branching microtubules. “Guide-rail” depicts thin lines of density often seen at the split of the branch point. (C) Distribution of branching angles (47±15°, n = 99 branch points). These experiments were performed three independent times.
Cryo-ET shows diverging microtubules with a break in the microtubule lattice
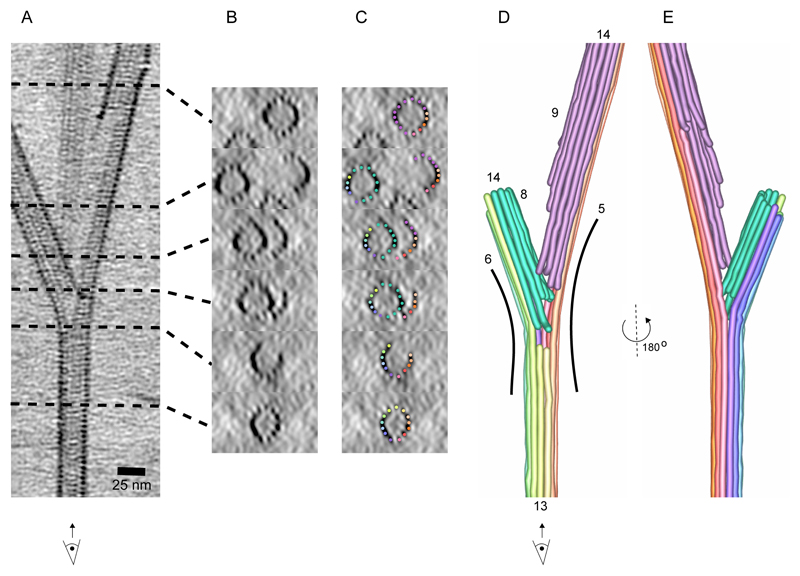
To further understand the organization of microtubule branches, we performed cryo-electron tomography on branched microtubules (Fig. 3, S2F). Even though SSNA1 itself was not detectable due to the resolution limit of tomographic reconstructions, the microtubule lattice was clearly visible (Fig. 3A-B) and facilitated a tracing of individual protofilaments at the branch (Fig. 3C-E). The tracing showed that two branching microtubules shared a subset of protofilaments with their mother microtubule. In addition, we traced newly assembled protofilaments that were not connected to the mother microtubule (Fig. 3D), as the number of protofilaments doubles compared to the mother microtubule. This shows a discontinuity in the microtubule lattice at the splitting point of the branch.
Fig. 3. The cryo-ET reconstruction of SSNA1-mediated microtubule branch.
(A) A 25-nm slice of a tomographic reconstruction highlighting branching point of a microtubule. With this view, individual protofilaments and tubulin units are visible, but the SSNA1 density is too thin to be visualized. (B) Cross-sections of the branched microtubules in A. (C) Individual protofilaments are overlaid with color represented in the segmentation in D. (D) Tracing of protofilaments of the 3D density map in A. Individual protofilaments are colored in rainbow-color coding. The newly formed protofilaments from the branched microtubules are colored in green (left) and in pink (right). Number of protofilaments in this particular branched microtubule are counted to be 13 (mother microtubule), 14 (left branched microtubule) and 14 (right, branched microtubule). 13 mother protofilaments are split to 6 to the left and 5 to the right side of branched microtubules. (E) 180° rotated segmentation of the branched microtubule.
SSNA1 self-clusters and nucleates microtubules
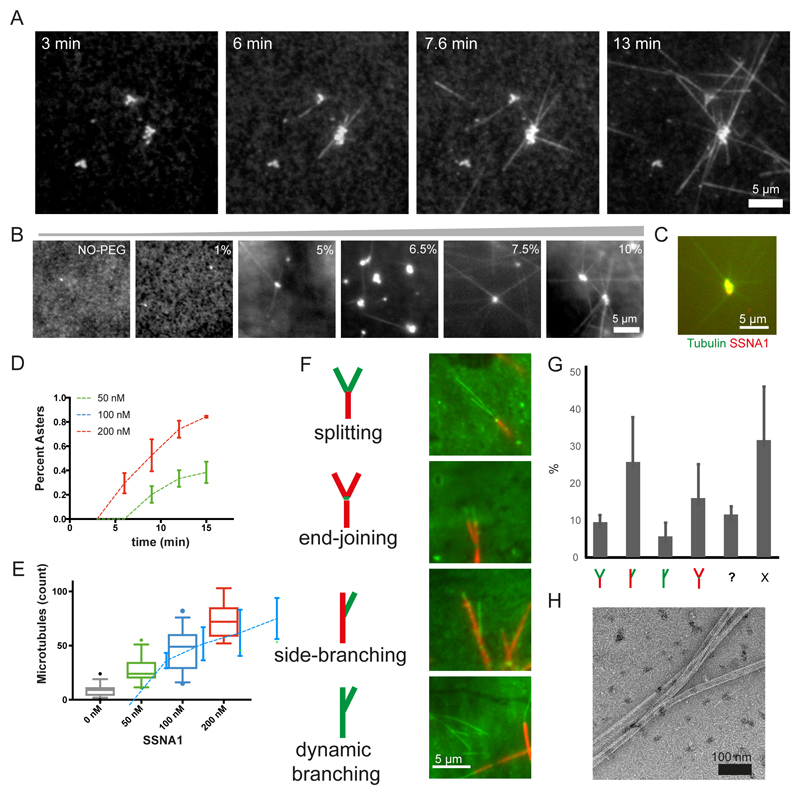
To explore the dynamic behavior of SSNA1 causing this unique action to microtubules, we tested the interaction of SSNA1 with unpolymerized tubulin using fluorescence microscopy (Fig. 4A). Considering the average cellular concentration of SSNA1 187 nM20, we mixed 200 nM CrSSNA1 and 8 μM tubulin in the presence of polyethylene glycol (PEG)21. Above a concentration of 5% PEG (Fig. 4B), we observed condensates of CrSSNA1 clustering with tubulin (Fig. 4A-C). Interestingly, several microtubules emerged from these CrSSNA1-tubulin clusters (Fig. 4A-B), reminiscent of aster formation seen during microtubule nucleation1,22, and thus indicating that CrSSNA1 nucleates microtubules. This process was effective at a CrSSNA1 concentration of as little as 50 nM (Fig. 4D-E). Nucleation events were confined to the local condensates of CrSSNA1 and tubulin did not polymerize without SSNA1, highlighting the effect of the CrSSNA1 condensates and the requirement of a high local concentration. The number of growing asters and microtubules (Fig. 4D-E) correlated well with the concentration of CrSSNA1. We also observed new microtubules emerging from already formed microtubules (movie S1 and S2).
Fig. 4. Nucleation and branching of microtubules mediated by CrSSNA1 under various conditions.
(A) Aster-like formation of microtubules (20% HiLyte488 tubulin) occurs within 3 min after mixing tubulin with lower concentration (200 nM) of SSNA1 (upper) under conditions mimicking molecular crowding (7.5% PEG, typically used as a crowding agent), where tubulin alone does not form any polymers. Microtubules propagate out from tubulin concentrate, serving as a nucleation center. These experiments were performed three independent times with similar results. (B) 200 nM SSNA1 and 8 μM tubulin self-associate forming clusters in the presence of PEG with the concentration >~5%. (C) SSNA1 antibody recognizes the microtubule nucleation center. (D) Plot of the percentage of the concentrates growing into asters with microtubules as a function of time (min) in the presence of 50, 100 and 200 nM CrSSNA1. Error bars are mean +/- SD from n=3 independent experiments. As little as 50 nM of CrSSNA1 is sufficient to observe aster formation in the presence of 7.5% PEG. (E) Counts of microtubules observed per field of view, in the presence of different concentrations of CrSSNA1. Box plots cover 50% of the entire population (boxes) and 90% (whiskers) with median values indicated as lines within the boxes. Sample Size: 0 nM: n=42 fields of view, 50nM: n=29, 100nM: n=30, 200nM: n=25. Data were pooled from 3 independent experiments except for the first point (0 nM) for which data were pooled from 4 independent experiments. (F) Green colored dynamic microtubules on red-microtubule GMPCPP seeds in the presence of higher concentration of SSNA1 (30 μM) without molecular crowding agents to achieve globally concentrated conditions. ‘branch-like’ nucleation is observed. Branches were categorized as ‘splitting’, ‘end-joining’, ‘side-branching’ or ‘dynamic branching’ and their ratio (n = 895 branches, mean +/- S.D. pooled from 3 independent experiments) are shown in (G). “?” shows the bundled microtubules, making it difficult to categorize. “X” shows microtubules without branching. Branch-like nucleation can be seen from the locally concentrated SSNA1, condition described in (A-E), however observations of individual microtubules are challenging due to the high local protein concentrations. (H) A negative-stain EM image of SSNA1-mediated branched microtubules in the presence of 200 nM SSNA1 and 7.5% PEG, representative of three independent experiments. See supplementary table 3 for source data.
SSNA1 induces microtubule nucleation from mother microtubules
To understand and assess SSNA1-mediated nucleation from template microtubules, we mixed (3 or 30 μM) CrSSNA1 with tubulin in the presence of GTP with GMPCPP-stabilized microtubule seeds23. Several microtubules were able to grow out from the ends or the wall of pre-existing microtubules (Fig. 4F-G, S3A-B, movies S3-S4), agreeing with the cryo-EM observation (Fig. 2A). By differentially labelling pre-existing microtubules (red) and newly polymerized dynamic microtubules (green), we could categorize the branching events to (1) “splitting”, in which newly formed microtubules split from the end of a pre-formed microtubule, (2) “end-joining”, showing three pre-existing microtubules connected through newly formed tubulin oligomers, (3) “side-branching”, seen as new microtubules coming out from the side of pre-existing microtubules and (4) “dynamic branching”, in which newly generated dynamic microtubules form a branch. The “side-branching” is reminiscent of local microtubule nucleation mediated by augmin and γ-tubulin in cell extracts8,13. However, in contrast to the augmin-mediated mechanism, the formation of CrSSNA1-mediated branching did not require γ-tubulin, indicating that SSNA1 works by a novel mode of action.
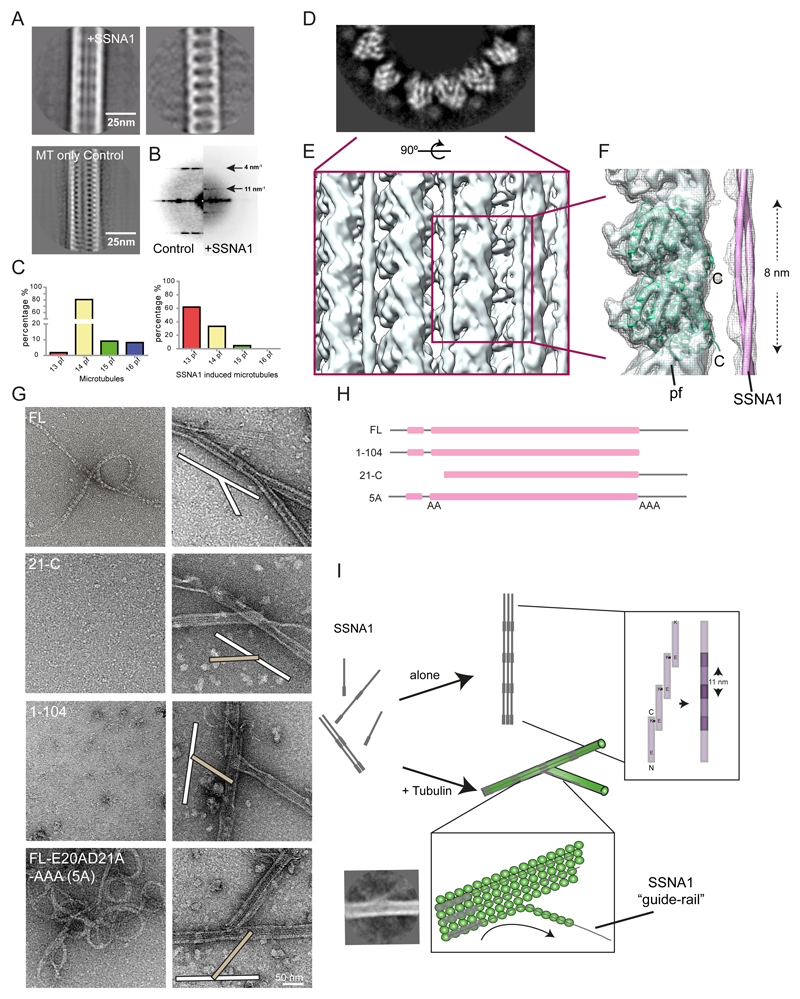
SSNA1 forms a fibril-like assembly on the surface of the microtubule with 11-nm periodicity
Although cryo-ET did not visualize the decoration of SSNA1 on microtubules, we observed that the free ends of microtubules are often extended with thin fibrils (Fig. S2D, red arrowheads). These fibrils extend from splitting microtubules, seemingly to work as a ‘guide-rail’ for the growth of branched microtubules (Fig. 2B; S2D, “guide-rail”). Computational averages of the cryo-EM images of microtubules allowed the visualization of CrSSNA1 directly attached to the surface of microtubules, revealing a ladder-like pattern (Fig. 5A, compare to “microtubule only control”) with a periodicity of ~11 nm (Fig. 5B). We observed that CrSSNA1 facilitated preferential assembly of 13-protofilament microtubules similar to doublecortin24 and EBs25. In contrast, 14-protofilament microtubules are predominantly polymerized in the absence of SSNA1 (Fig. 5C).
Fig. 5. Molecular characterization of the branching action of SSNA1.
(A) Left - Representative class average of the SSNA1-induced microtubules, Right – SSNA1 decoration emphasized by computationally subtracting microtubule densities35 and Left-bottom – Average of microtubules without decoration for comparison. (B) The power spectrum of microtubule class averages shows an additional 11 nm periodicity in the presence of SSNA1. (C) Distribution of protofilament numbers of microtubules reconstituted from brain tubulin in the absence (left) and in the presence of CrSSNA1 (right) shifting the majority from 14- to 13-protofilament microtubules. (D) Grey-scale density map of the plus-end-on view of the SSNA1-microtubule 3D reconstruction. SSNA1 decoration and the secondary structures of tubulin density are well resolved. (E) Rendering of the microtubule surface decorated with SSNA1. The resolution of the microtubule surface (~10 Å) is not as high as the core (< 8 Å) due to the SSNA1 decoration. (F) Tubulin atomic model (PDB ID: 3jal) fitted to the map. The SSNA1 coiled-coil fibril is indicated as a tube representation. Note that the periodical feature of SSNA1 is averaged out because of the symmetrical mismatch between tubulin dimers (8 nm) and SSNA1 fibrils (11 nm). (G) Morphological observation of SSNA1 and its branching activity. Left, observation of the purified protein at 0 h incubation (i.e. immediately after purification), and Right: a magnified view of the copolymerized microtubules. Microtubule branching was observed with FL, while other protein fragments do not facilitate branching. For the proteins that do not cause the branching, examples of typical crossing of microtubules (white and beige bars at the scheme within the image), instead of branching are shown. Detailed observations are available in Fig. S5A. (H) Graphical scheme of SSNA1 constructs used in (G). (I) Scheme of the SSNA1 self-assembly and microtubule nucleation mediated by SSNA1. While SSNA1 oligomers alone can also undergo a slow self-assembly process, the SSNA1 oligomers interact with tubulin dimers to promote their co-polymerization. The polymerized SSNA1 may further act as a guide-rail (bottom inlet) for protofilament splitting, resulting in microtubule branch formation. A class average indicating the guide-rail mechanism is shown. Other class averages are available in Fig. S2E.
SSNA1 forms a head-to-tail fibril with 11-nm periodicity and covers on the C-terminus tail of microtubules
SSNA1 is a 14-kDa protein predicted to adapt a tropomyosin-like single parallel coiled-coil configuration26,27 (Fig. S3E). Fibril formation has previously been observed as a result of head-to-tail self-assembly27,28. Accordingly, we observed that CrSSNA1 readily forms short fibrillar appearances with occasional long fibril formations (Fig. 5G, “FL”). Furthermore, the shorter fibrils of CrSSNA1 were converted into longer, organized bundles of fibrils after ~24 h incubation (Fig. S3F). A closer look at these bundles revealed a striped, knob-like pattern, which leads to the formation of a sheet (Fig. S3F, 24 h) with a 11-nm periodicity (Fig. S3F, inlet), and the inter-fibril distance of 3.5 nm. This pattern is comparable to that observed on the microtubule surface (Fig. 5A), indicating that the fibrils are covering microtubules along their long axis giving a 11-nm spaced ladder-like pattern.
To further characterize the interaction between SSNA1 and microtubules, we obtained a cryo-EM 3D structure of CrSSNA1 in complex with microtubules (Fig. 5D-F, S4A) with an overall resolution of 6.1Å (Fig. S4B). Due to the symmetry mismatch between microtubules (4- or 8-nm periodicity) and SSNA1 (11-nm periodicity), SSNA1 was averaged out, and the fibril appeared as a ‘cloud’ of protein density running parallel to the microtubule surface, with an apparent local resolution of ~11Å (Fig. S4C). However, it was possible to visualize thin lines of additional densities running parallel to the microtubule surface (Fig. 5E-F), which we interpreted as SSNA1 filaments. The SSNA1 filaments run between two protofilaments, proximal to the unstructured, highly acidic C-terminal tails (E-hooks) of tubulin (Fig. 5F labelled “C”). Removal of E-hooks resulted in weakening of SSNA1 crosslinking with microtubules as determined by EDC (~49% less crosslinked; Fig. S4E). E-hooks create a negative electrostatic cloud by their periodical arrangement on the microtubule surface29, which could attract the SSNA1 fibrils. This could explain why SSNA1 interacts with microtubules despite the symmetry mismatch. It also suggests that the head-to-tail assembly of SSNA1 fibrils could guide protofilament assembly and microtubule polymerization by covering and neutralizing the E-hooks as shown previously30.
The head-to-tail fibril formation of SSNA1 is essential for microtubule branching
Based on the observation that SSNA1 fibrils appear to guide the protofilaments of microtubules, we hypothesized that the microtubule branching activity is mediated by the formation of long SSNA1 fibrils that curve away and guide the protofilament out of the lattice (guide-rail, Fig. 2B, S2D). To test this, we created a series of truncated SSNA1 fragments that abolish fibril formation. Based on a PHYRE2 analysis31 and previous reports27, we found that SSNA1 contains a well conserved alpha-helical region (residues 6-104) followed by an unstructured C-terminal tail (Fig. S3E). A series of N-terminal truncations showed that the first 19 residues were not necessary for fibril formation, as CrSSNA1(20-111, 20-C) formed cable-like bundled fibrils, which were less ordered compared to full-length protein (CrSSNA1 FL), but displayed a ~11 nm pattern (Fig. 5G-H, S5A, Table S1). In contrast, CrSSNA1(21-C), a truncation missing one more residue E20, was unable to form fibrils (Fig. 5G-H, S5A, Table S1). This observation correlates with the ability of CrSSNA1(20-C), but not CrSSNA1(21-C) to mediate microtubule branching (Fig. S5A, Table S1). The key role of residue E20 for fibril-formation and microtubule-branching activity of SSNA1 was further underpinned by point mutations E20A and E20A/D21A, which drastically reduced microtubule branch formation (Fig. S5A, Table S1). These mutants may form fibrils, but with much lower frequency and without a distinct higher-order organization.
In the C-terminal region of SSNA1, three distinctive lysine residues (K105, K106, and K107) mark the beginning of the unstructured C-terminus. CrSSNA1 truncations 1-104 and 1-105 (Fig. 5G-H, S5A, Table S1) showed that CrSSNA1(1-104) can no longer form fibrillar oligomers, or induce microtubule branching (Fig. 5G-H), while CrSSNA1(1-105) was purified as fibrils and showed microtubule branching activity (Fig. S5A) at similar efficiency as CrSSNA1-FL. These results indicate that the positive charge of the lysine residues is essential for the ability of CrSSNA1 to form fibrils. We confirmed this by generating a triple point mutant, K105A/K106A/K107A, which indeed abolished fibril formation and microtubule branching (Fig. S5A, Table S1) for both, the full length and the 1-107 fragment. Altogether, our mutational analyses indicate that the key interaction for longitudinal fibril formation is mediated by E20 of one unit and the C-terminal tail (K105-107) of the adjacent interacting unit (Fig. 5I). To confirm this, we created mutants in which the charges of residues E20/D21 and K105/K106/K107 were swapped. When two of the opposite charges were swapped (E20K/D21K/K105E/K106E, or E20K/D21K/K106E/K107E), both, cable-like formation and microtubule branching activity of SSNA1 was retained. In contrast, swapping of the two negative residues at the N-terminus and the three positive residues at the C-terminus (E20K/D21K/K105E/K106E/K107E), resulting in change of net charge from +1 to -1, abolished microtubule branching (Fig. S5B). However, this construct was still able to form SSNA1 fibrils and cable-like structures, indicating that the microtubule branching activity depended not only on fibril formation of SSNA1, but also on the presence of an extra negative charge at the unstructured SSNA1 C-terminus. This was confirmed by the mutant E20A/D21A/K105A/K106A/K107A, termed 5A, showing a complete loss of microtubule branching activity (Fig. 5G).
Microtubule-branching deficient SSNA1 mutants abolished the promotion of axon branches
Having gained insights into the molecular organization of SSNA1 and its effect on microtubule nucleation and branching, we hypothesized that the promotion of axon growth and branching observed in neurons overexpressing wildtype SSNA1 might be altered when microtubule-branching deficient SSNA1 versions are expressed. Our results indeed showed that, in contrast to the SSNA1 wildtype (Fig. 1, 6A-C), SSNA1 with mutations abolishing microtubule branching in vitro also failed to promote the growth of axons or axon branches (Fig. 6A-C, S6A-E) in primary neurons. Notably, a dominant negative effect was also observed when the 5A mutant was overexpressed for the number of total neurite processes (Fig. 6C), showing a decreased number of major and minor branches. This dominant-negative effect was also found when the two negative residues at the N-terminus and the three positive residues at the C-terminus were swapped (swap-KK/EEE). Notably, swapping only two opposite charges and leaving the third C-terminal lysine intact (swap-KK/EE) could still promote axon growth (Fig. S6D-E). Together these findings show that the ability of SSNA1 to induce fibril formation and microtubule branching at the molecular level correlates with its function of mediating axon branching and development, suggesting the intriguing possibility that it locally generates branched microtubules at axon branch sites.
Fig. 6. Effect of various SSNA1 constructs on primary hippocampal neurons and fibroblast cells.
(A) Immunostaining of MAP2 (green), Tau (red) and GFP (blue, expression control) in mSSNA1 over-expressing cells. For the SSNA1 WT, the axon is guided with a dotted line. (B) Scatter dot plots of axon length under over-expression of various SSNA1. Control and WT profiles shown in Fig. 1 are placed as a negative and positive control, respectively. The promotion of axon development occurs only in over-expression of WT SSNA1, while no apparent effect was observed for the constructs that fail to mediate microtubule branching. Every cell is represented by a single point, control (n=496 cells), wild type (n=490), 1-104 (n=788), 21-C (n=610), 5A (n=642) from 3 independent experiments, shown in magenta, green and yellow, and the overlaid box-and-whisker plots cover 50% (boxes) and 90% (whiskers) of the entire population, with median values indicated as lines within the boxes. (C) Pie graphs showing the distribution of the number of branches and Strahler number under different over-expression conditions. GFP expression control and SSNA1 WT overexpression profiles in Fig. 1 are placed as controls. (D) Schematic model describing how SSNA1-mediated microtubule nucleation could contribute to axon branch formation. Spastin has been shown to localize at axon branches7 and to interact with SSNA136. Taken together with our finding of SSNA1 localization at axon branches, it is possible that the two proteins work sequentially by spastin severing microtubules to provide tubulin oligomers, and SSNA1 nucleating microtubules at the branching site. (E) DNA-PAINT image of a 500-nm slice of the microtubule network in SSNA1 over-expressing cells. (F) Zoomed-in view from E - the object is colored in a rainbow code according to the depth. (G) Individually recognized microtubules are highlighted in various colors. 3-way intersections are indicated with red arrowheads. (H-J) Corresponding view of a 500-nm slice of the microtubule network in untreated cells (control). For analysis, 3 independent SSNA1 over-expressing and control cells were assessed each, containing the total microtubule lengths of 5700 μm, 7900 μm and 1900 μm and 7700 μm, 8500 μm, 7700 μm, respectively. See supplementary table 3 for source data.
Morphological change of microtubule networks in non-neuronal cells
To test if the function of SSNA1 is conserved in different cell types, we used fibroblasts, which are structurally less specialized than neurons, and tested if overexpression of SSNA1 has the capacity of changing the microtubule organization (Fig. 6E-J). Super-resolution light microscopy with DNA-PAINT showed that individual microtubules are well-resolved in the control cells with a wide-ranging network (Fig. 6H-J). In contrast, microtubules were rather short in SSNA1-overexpressing cells (Fig. 6E-G), suggesting that SSNA1 can promote nucleation, generating more but shorter microtubules. We also occasionally found microtubules forming 3-way intersections, as if one microtubule emerged out of another in both control and SSNA1-overexpressing cells (Fig. 6G, 6H, arrowheads). These events occurred more often in SSNA1-overexpressing cells (2.8 ± 1.2 occurrences/100 μm microtubule) than control cells (1.0 ± 0.35 occurrences/100 μm microtubule). Although the limited resolution in light microscopy prevented us from discerning if microtubules branched with a shared lattice or if two microtubules only attached to each other, the observations were consistent with our in vitro studies by electron microcopy.
Discussion
During cell polarization, the dynamics and distribution of the microtubule cytoskeleton is tightly regulated. Though the centrosome has a major role as an MTOC in less differentiated cells, the inactivation of centrosomes in neurons does not affect axon growth, a process strongly dependent on microtubule assembly6. Thus far, the molecular mechanisms regulating axonal microtubule nucleation, especially in the form of branching, have remained a mystery. Here, we show that SSNA1 accumulates at axon branches and promotes axon branching in primary neurons, and can nucleate microtubules in vitro. Mutations interfering with SSNA1 in vitro nucleation activity also affect the occurrence of axon branches in neurons. Together, these results suggest that SSNA1 could act as a microtubule nucleator at axon branch sites.
Strikingly, our work revealed that SSNA1 independently mediates microtubule branching by causing protofilaments to splay apart from the lattice. To our knowledge, no other microtubule-binding protein shows this activity. When tested EB3 and ch-TOG, known regulators of microtubule dynamics, under the same conditions, did not induce branching (Fig. S6F-G). In addition, we showed that microtubule branching requires co-polymerization of SSNA1 with microtubules, as well as specific amino-acid interactions. Taken together, these results indicate that the observed branching activity is highly specific to SSNA1.
Our work has uncovered a surprising example of how co-polymerization of a simple coiled-coil protein with tubulin can induce global remodeling of the microtubule network. Association of SSNA1 may reinforce longitudinal connections of tubulin oligomers, facilitate protofilament formation, and act as polymerization seed for microtubule formation. Concomitantly, the preference of SSNA1 for lateral connections may facilitate the lateral connections between microtubule protofilaments. Polymerized SSNA1 may precede microtubule protofilaments, guiding protofilaments out of the microtubule axis, thus providing a template for a new microtubule branch (‘guide-rail’ mechanism). In a cellular context, however, the situation is more complex as SSNA1 activity is likely modulated by other factors. Further experiments are necessary to test this mode of action of SSNA1 for microtubule branching in cells.
Our in vitro reconstitutions showed that SSNA1 self-assembles into clusters together with tubulin at high local concentration. As axons are densely packed with cytoskeletal components, this organization is a plausible prerequisite for the physiological function of SSNA1, allowing SSNA1 to concentrate locally, self-assemble and become a microtubule nucleation center at designated locations. Alternatively, the requirement of high local concentration of SSNA1 may be a means to limit the microtubule-remodeling activity of SSNA1 to specific subcellular areas such as axon branches, the midbody in dividing cells, and the base of cilia.
As microtubules are much less dynamic in axons compared to less polarized cells32–34, SSNA1-mediated branching may be restricted to locally destabilized sites of the microtubule cytoskeleton. Interestingly, SSNA1 interacts with spastin, a protein important for the initiation of axon branching and thought to increase the pool of soluble tubulin through microtubule fragmentation7. It is tempting to speculate that short, spastin-severed microtubules (i.e. tubulin oligomers) provide the building blocks for SSNA1-mediated microtubule nucleation and branching. Thus, the synergistic action of spastin and SSNA1 could facilitate the formation of axonal branch points (Fig. 6D). Further investigation of SSNA1 activity in situ will bring valuable insights into the initiation and organization of axon branches. In particular, it will be interesting to explore whether SSNA1-mediated microtubule branching is a direct driving force for axon branching, or an intermediate state during early stages of neuronal morphogenesis. Considering the diverse sites at which SSNA1 is localized in various cell types17,27, the microtubule-branching mechanism discovered here could have broad implications for understanding the regulation of various microtubule functions, providing new clues to previously unanswered questions about the cytoskeleton and intracellular transport.
Methods
Protein preparation and purification
The DNAs of CrSSNA1 and mouse SSNA1 were obtained by gene synthesis (GeneArt, ThermoFisher) and cloned into self-generated LIC (ligation-independent cloning) vectors. The SSNA1 fragments were prepared as hexahistidine (His) fusion proteins with a TEV-protease recognition site. The proteins were expressed in E. coli BL21(DE3) (Merck, Darmstadt, Germany) by inducing with 0.4 mM IPTG (Carl Roth, Karlsruhe, Germany) for overnight at 18 °C. Cells were sonicated in lysis buffer (50 mM Na-phosphate buffer pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 5mM β-mercaptoethanol) supplemented with protease inhibitors (1 mM Pepstatin A, 1mM AEBSF and 1mM Leupeptin) and clarified. Soluble fraction was purified by Ni-NTA affinity chromatography. The His tag was either removed by TEV cleavage or left on the protein. Biochemical analysis did not show any differences with or without the tag. For CrSSNA1 (1-104), an additional step of size-exclusion chromatography (Superdex 200, GE Healthcare) was applied. CD spectra were obtained on a JASCO 715 CD Spectrometer equipped with peltier-thermostat, at 4 °C, 25 °C and 37 °C. Tubulin was purified from porcine brains (The Bayerische Landesanstalt für Landwirtschaft (Poing, Germany)) according to the protocol37 or purchased from Cytoskeleton, Inc. The oligomerizations of SSNA1 proteins were followed by SSNA1 variants for 0 h (immediately after purification), 24 h and 48 h.
The DNA for mouse ch-TOG (a.a 1-505) and human EB3 (a.a. 1-281) were obtained from Mammalian Gene Collection (MGC, Source BioScience LifeSciences). mTOG protein was expressed in E.coli BL21(DE3)pLysS by inducing 0.5mM IPTG and cells were grown overnight at 16 °C. Cells were sonicated in lysis buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidazole,1mM DTT) supplemented with protease inhibitors (1mM Pepstatin A, 1 mM Leupeptin,1mM PMSF) and clarified. The protein was purified using Ni-affinity chromatography followed by ion exchange chromatography (Hi Trap S, GE Healthcare) and size exclusion chromatography (Superdex 200, GE Healthcare). His tag was removed by 3C protease. EB3 was expressed in E.coli BL21(DE3). Cells were sonicated in lysis buffer (20 mM Pipes, 1 mM MgCl2, 1 mM EGTA 500 mM NaCl, pH 6.8, 1mM DTT) supplemented with protease inhibitors (1mM Pepstatin A, 1 mM Leupeptin, 1mM PMSF) and clarified. The protein was purified using Ni-affinity chromatography followed by size exclusion chromatography (Superdex 200; GE Healthcare). His tag was removed by 3C protease.
Electron microscopy of SSNA1
SSNA1 (0.05 mg/ml) of different time points of incubation (0, 24 and 48 h) were applied on manually prepared carbon-coated grids and stained with 1% (w/v) uranyl-acetate. The specimens were observed using a CM200 (Thermo Fisher Scientific) at 160 kV with a nominal magnification of 50,000x, corresponding to 2.16 Å/pixel.
Electron microscopy of SSNA1-microtubules complex and image processing
For screening conditions that were also used for light microscopic observations, 8 or 15 μM of tubulin was mixed with 0.1 to 30μM of SSNA1 in BRB80 buffer (80mM Pipes-KOH pH 6.8, 1 mM MgCl2, 1 mM EDTA) supplemented with 1 mM GTP or GMPCPP for 5 min, and directly applied on an EM grid for negative staining. The cluster of the microtubules observed in light microscopic environments were only partially preserved under the negative stain condition, due to the fixation process of the sample for negative stain EM. The center of the cluster is not visible due to high electron densities.
For cryo-EM, we used 15 μM of tubulin and two to five times excess of SSNA1 to maximize the decoration with the protein. Microtubules were stabilized using non-hydrolyzable GTP analogue mimicking GTP-bound conditions GMPCPP. After 5 min, 5 μl of sample without dilution was applied to glow-discharged grids with holey carbon (Quantifoil, Cu, R1.2/1.3) and vitrification was carried out in liquid ethane using a home-made manual plunger. The cryo-EM specimens were observed by a Tecnai F20 (Thermo Fisher Scientific) at 200kV with a magnification of 29,000x using a Falcon2 direct detector (Thermo Fisher Scientific), corresponding to 3.46 Å/pixel with a defocus ~-2.5 μm. The total 98 images with the dose of 50 electrons per Å2 were used during image collection.
For counting the numbers of the protofilament of the microtubules, we referred a well-known specific interference pattern (moiré pattern) of the microtubules38 observed under cryo-EM.
For the measurement of the branching angles, we used 99 branched microtubules. Using FIJI software, 2-connector segments were drawn with each segment approximately 50 nm, placing the junction of the segments at the center of the branching points. Then the 2 segments were aligned along the direction of the 2 branched microtubules. Examples of branches with various angles are shown in Fig. S2D.
For the initial analysis visualizing the 11-nm periodicity of SSNA1 on the microtubule surface, a dataset using F20 (described above) was used. EMAN239 e2helixboxer scheme was used to extract the segments of microtubules. The box size was set to 256 pixels corresponding to 886 Å with 90% overlap and 6160 segments were extracted in total. For classification and averaging of the images, RELION240 software was used.
The dataset for 3D reconstruction was collected using a Titan Arctica (Thermo Fisher Scientific) at 200 kV with a magnification of 92 kx with a Falcon3 direct detector (Thermo Fisher Scientific), corresponding to 1.6 Å/pixel, and Titan Krios (Thermo Fisher Scientific) with SerialEM software at 300kV with a magnification of 105 kx, corresponding to 1.34 Å/pix with a K2 Summit direct electron detector (Gatan). Final reconstruction included in this report was carried out using the dataset taken with Titan Krios. For the dataset that were included in the final reconstruction, 762 images were collected with defocus varying from -1.5 μm to -3.5 μm. The detector was operated in counting mode with dose rate of 10.1 electrons/pixels/s. A total exposure time of 6 sec, corresponding to an accumulative dose of 34.08 electron/Å2 was fractionated into 24 movie frames with 0.25 s exposure time and a dose of 1.42 electron/Å2 for each frame. The movie frames were aligned, and averaged using UCSF Motioncorr2 program41.
For the 2D classification of branched microtubules, RELION2 was used. 226 branched microtubules without overlapping with other microtubules were selected from the dataset recorded with Titan Krios and boxed out with a box size of 1000 pixels, corresponding to 1340 Å. The branch angles were variable causing structural heterogeneity, limiting the resolution of averages.
For image analysis leading to the 3D reconstruction of the microtubule-SSNA1 complex, quality, defocus and astigmatism of each micrograph were assessed using CTFFIND442. Out of 762 images, 478 images containing microtubules were selected for further processing. 1774 selected microtubules were segmented with a box size of 480 Å with 90% of overlap. As microtubules with 13-protofilaments were the majority, we chose to process 13-protofilament microtubules further. 13-protofilament microtubules contain a seam that breaks the helical continuity of tubulin dimers, which is a building block. To circumvent this problem, a specially designed package described in43 was used in combination with the method described in44–46. Briefly, multi-reference alignment was performed using 20 Å low-pass-filtered, 2D projections of a microtubule with 13- protofilament as reference. The package uses a reference which was computationally synthesized using the atomic structure of tubulin decorated by kinesin. The alignment revealed polarity and the position of the seam by following the segmented boxes that position along a single microtubule. After the determination of the seam, re-segmentation of the microtubules from the micrograph was performed using the alignment information and with the box size of 600 Å and every 80 Å as an interval. FREALIGN47 package was implemented in the package for refinement with options of helical analysis. For this, the known helical parameter of a microtubule with 13-protofilament was used (helical_rise: -9.37308 Å, helical_twist: 27.692 degree, helical_subunit: 13). Afterwards the methods in references44–46 was implemented for refinement as this method follows the consistency of patterns within individual microtubules, independent of kinesin decorated pattern as a reference of alignment. We however observed that the SSNA1 decoration on the microtubules affected the accuracy of the particle alignment as well as the seam detection as indicated in the local resolution estimation shown in Fig. S4. While the reconstruction of the microtubules could be further improved by a more laborious strategy, SSNA1 on the contrary cannot be better resolved because of the existing symmetry mismatch with the microtubules. The resulting reconstruction is nevertheless informative as it allows us to visualize a long SSNA1 fibril attached along a protofilament of microtubules. The global resolution was determined to be 6.1 Å by calculating the Fourier Shell Correlation (FSC) of two independent reconstructions. However, we note that the alpha and beta tubulins are not sufficiently separated and the higher resolutions are only effective in the core of tubulin. The reconstruction was filtered based on local resolution estimation by “blocres” scheme48 with a scan box size of 50 pixel.
Cryo-Electron tomography of SSNA1-microtubules complex and image processing
10 nm BSA-coated gold (Aurion, The Netherlands) was used as fiducial marker. 4 μl of sample was mixed with 1 μL fiducial marker and then was applied to glow-discharged grids (Quantifoil, Cu, R2/2). Plunge freezing was immediately followed using Vitrobot (Thermo-Scientific). Tomographic tilt-series were collected on a Titan Krios (Thermo-Scientific) operated at an acceleration voltage of 300 kV, equipped with a Gatan K2 Summit direct electron detector, with magnification of 64,000x corresponding to 2.23 Å /pixel. Images were collected in a sequential manner, starting at 0° and increasing to +59° with 1° increment. After acquiring +59°, stage was returned to 0° and tilt series was collected till -59° with 1° increment as well. Each tilt series was collected with defocus value set between 3-7 μm. Images were acquired as movies in counting mode using dose rate of 4.7 electron/pixels/sec. The total accumulative dose of the tilt-series was 112.46 electron/Å2. The movie frames were aligned using UCSF Motioncorr2 program.
Tomogram reconstruction was performed using the IMOD package49. Tilt series were aligned using fiducial gold markers and further binned by a factor of 4 (final pixel size of 8.92 Å/pixel). Tomograms were reconstructed by back projection and simultaneous iterative reconstruction technique (SIRT) with seven iterations in IMOD.
Subtilisin Treatment of Microtubules and crosslinking
20 μM taxol-stabilized microtubules were mixed with 7.4 μM subtilisin (Sigma Aldrich) and incubated for 0-60 min at 37 °C. The reaction was stopped by adding 2.5 mM PMSF. In 10 min, subtilisin completes the cleavage of beta tubulin E-hooks and the cleavage of alpha tubulin E-hooks follows. For the cross-linking assay, 5 μM microtubules were mixed with 25 μM of SSNA1, and 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Fisher) was added to a final concentration of 5 mM. Samples were incubated at room temperature for 1 h. The densities of SDS-PAGE were measured using Fiji.
Light microscopy of in vitro SSNA1-mediated microtubule nucleation
Flow cells were assembled with cover glass and passivated cover slips as described before50. The use of the GODCAT oxygen scavenging system, common for microtubule growth observation, blocked the effects of CrSSNA1 on microtubule polymerization in our assays. Therefore, instead we used the PCA/PCD/Trolox oxygen scavenging system51, which contains 10 nM protocatechuate 3,4-dioxygenase from the Pseudomonas species, 2.5 mM 3,4-dihydroxybenzoic acid ‘PCA’ and 1mM Trolox, (Sigma). Total internal reflection fluorescence microscopy was performed on a DeltaVision Elite imaging system (GE Healthcare). The formation of ‘asters’, the conditions used were: 8 μM tubulin (20% HiLyte488 Tubulin, Cytoskeleton), 50-200 nM CrSSNA1, 0-10 % PEG and 2 mM GTP. Further experiments containing PEG were performed in the presence of 7.5 % PEG. For the detection of the localization of CrSSNA1, after 5 min of the incubation of the mixture of the samples, an anti-SSNA1 antibody was added, then an anti-rabbit antibody labeled with Alexa Fluor 568 (Life Technology) was added for the visualization of the antibody. We observed that PEG causes formations of concentrate of SSNA1, which is detectable with > 4 % of PEG, and can mediate microtubule formation with > 5 % PEG, well agreed with other proteins previously reported to nucleate microtubules21. With any of the abovementioned conditions, it is confirmed that spontaneous formations of microtubules do not occur without SSNA1. As little as 50 nM CrSSNA1 was effective to mediate a microtubule formation in the presence of 7.5% PEG.
To mimic the nucleation event, seeds were used as template. The seeds were formed by incubating 30 μM of tubulin with 15 % of atto565-labeled tubulin in the presence of 0.5 mM GMPCPP at 37 °C for 30 min then centrifuged at 15800xg for 8 min to remove excess GMPCPP. Pellets were dissolved in BRB80 buffer. 1 μM seeds were mixed with 15 μM tubulin containing 20% HiLyte488 tubulin, 2 mM GTP and 3-30 μM CrSSNA1, then the microtubule growth was immediately observed. Snapshots were taken after 30 min of incubation. Movies were made by acquiring one frame every 15-20 s for 15-20 min. All experiments were performed at least 3 times independently. We observed branch formations both from templated GMPCPP stabilized microtubules as well as dynamic microtubules. At 3μM, branch-like microtubules started appearing (20%, 115 out of 559 microtubules) and at 30 μM, 50% (448 out of 895 microtubules) had branch-like protrusion of microtubules. For categorizing the types of branches, the snapshots of microtubules in the presence of 30μM SSNA1 was used, all the microtubules (n=895) were selected out of 89 snapshots from 3 independent experiments (n=47, 21, 21) and categorized into ‘splitting’- dynamic microtubules with GTP are growing out from the end of the preformed microtubules, ‘end joining’ – two pre-existing microtubules are annealed through dynamic tubulin oligomers, ‘side branching’- dynamic microtubules are growing out of the wall of the pre-existing microtubules, ‘dynamic branching’ – newly formed dynamic microtubules branch out, ‘indistinguishable’ and ‘no-branch’ – microtubules without branching.
Mouse hippocampal primary neuron cultures
Animal care and use for this study were performed in accordance with the recommendations of the European Community (2010/63/UE). Experimental procedures were specifically approved by the ethics committee of the Institut Curie CEEA-IC #118 (authorization n°04395.03 given by National Authority) in compliance with the international guidelines. The study is compliant with all relevant ethical regulations regarding animal research.
Mouse hippocampal neurons were cultured as described previously 52. Briefly, wild type dams at 17.5 days of pregnancy were sacrificed using cervical dislocation, the embryos were decapitated and their hippocampi dissected. Hippocampi were digested with 0.25% trypsin-EDTA (ThermoFisher, 15090046) for 20 min at 37°C, followed by mechanical dissociation with glass pipettes. Dissociated neurons were then plated in plating medium (MEM supplemented with 10% FBS and 0.6% w/v glucose (sigma G-8769)) on coverslips coated with poly-D-lysine (#354210, Corning). After four hours after plating, media were replaced with neurobasal media containing 1% glutamate, 2% B27 and with/without lentivirus.
Overexpression of SSNA1 in cultured mouse hippocampal neurons
Mouse SSNA1(FL) SSNA1(1-104), SSNA1(21-119, corresponding to 21-C), SSNA1(E20A/E21A/K105A/K112A/K117A, corresponding to 5A) SSNA1 swap-KK/EE (E20K/E21K/K105E/K112E) and swap-KK/EEE (E20K/E21K/K105E/K112E/K117E) were cloned into modified lentiviral vector pTRIP using one step sequence and ligation independent cloning method. The pTRIP vector contains 2A peptide sequence between EGFP and SSNA1 sequence so that the expression of protein constructs can be assured with the expression of GFP signals without tagging. Lentiviral particles for the mouse SSNA1 constructs were produced as described53. Briefly, lentiviral vectors along with viral packaging vectors (psPAX2 and pCMV-VSV-G) were co-transfected in Lenti-X-293T cells using TransIT®-293 transfection reagent (Mirus Bio LLC). The virus-containing medium was filtered and stored at -80 °C. Amount of virus to be used for experiments was determined by adding different volumes of virus to the neurons. Mouse hippocampal neurons were cultured as described in52. On DIV0 (days in vitro 0), four hours after plating, neurons were transduced with lentiviruses encoding for different SSNA1 constructs. On DIV3, neurons were fixed as described in54.
Immunofluorescence analyses of primary neuron
Primary neurons fixed on DIV3 were stained with anti-MAP2 and Tau1 antibodies. Cells were then incubated with anti-mouse Alexa Fluor 647 and anti-rabbit Alexa Fluor 568. Nuclei were stained with DAPI (0.02 μg/ml, ThermoFisher Scientific). For immunostaining of mSSNA1, anti-SSNA1 and anti-beta III tubulin were used. Antibody information is provided in table S2. Cells were mounted using ProLong Gold anti-fade (ThermoFisher Scientific). Cells were imaged on a Zeiss Axio Imager.M2 with 20x or 40x objectives. Acquired images were analyzed using FIJI55. Cells with very short axons were included in the analysis so that under-developed neurons could be assessed as well. Note for dataset overexpressing swap-KK/EE and swap-KK/EEE, primary neurons were prepared at a different time causing the change in general growth profiles of axons. Control (GFP transfected) was used as a standard for comparison of promotion or reduction of axon development of different mutants. Axons, defined as the longest protrusion from the soma, were selected using the Simple Neurite Tracer plugin56. The collateral branches longer than 15 μm were defined as major branches. The total number of collateral branches and the total length of all the branches for each axon was determined by tracing of the neuron morphology. In each image the position of the cell bodies was determined by segmentation of the nuclei: after applying a Gaussian filter and subtracting the background, the image was thresholded using Otsu’s method. In the over expression experiments, the neurons were screened for transduction efficiency, as measured by EGFP expression. The branch network was obtained by segmenting and combining the intensity in the Tau1 and MAP2 fluorescence images: for each channel, the neurites were highlighted by mapping the curvature of the image (Compute Curvature plugin) and thresholding this quantity using Otsu’s method. The resulting binary mask was then skeletonized to outline all the neurites detected. By overlapping the traced axons with this image only the neurites branching from the axons were kept and measured. The branching complexity of each neuron was summarized with the Strahler number57,58 (Strahler Analysis plugin), using the location of its corresponding cell body to mark the root branch, which is the start point of the axon.
Immunostaining for DNA-PAINT
DNA-labeled antibodies were prepared as previously reported59. In brief, 300 μl of 1 mg/ml secondary donkey anti-rat antibody (Jackson ImmunoResearch, 711-005-152) was reacted with 10x mole excess Maleimide-PEG2-succinimidyl ester crosslinker (Sigma-Aldrich, 746223), then 10x mole excess of DNA was added to the antibody-crosslinker. Final usage concentration was 10 μg/ml.
Mouse embryonic fibroblast cells were transfected with pTRIP_2A_EGFP vector using Lipofectamine® 2000 (Thermo Fisher Scientific). Cells were fixed and stained as described previously59. then were incubated at 4 °C overnight with primary rat α-Tubulin (YL1/2) antibody. Antibody information is provided in table S2. 10 μg/ml DNA-labeled secondary antibody was added and incubated for 1h. Samples were then incubated for 5 min with 90-nm gold particles (Cytodiagnostics, G-90-100) at 1:10 ratio in PBS, then residual gold was washed away. Cells were kept at 4 °C until they were used for imaging within 48 h.
DNA-PAINT
Fluorescence imaging was carried out on an inverted Nikon Eclipse Ti microscope (Nikon Instruments) with the Perfect Focus System, applying an objective-type TIRF configuration with an oil-immersion objective (Apo SR TIRF 100x, NA 1.49, Oil). Two lasers were used for excitation: 561 nm (200 mW, Coherent Sapphire) or 488 nm (200 mW, Toptica iBeam smart). The laser beam was passed through a cleanup filter (ZET488/10x or ZET561/10x, Chroma Technology) and coupled into the microscope objective using a beam splitter (ZT488rdc or ZT561rdc, Chroma Technology). Fluorescence light was spectrally filtered with two emission filters (ET525/50m and ET500lp for 488 nm excitation and ET600/50 and ET575lp for 561 nm excitation, Chroma Technology) and imaged on a sCMOS camera (Andor Zyla 4.2) without further magnification, resulting in an effective pixel size of 130 nm after 2x2 binning. Camera Readout Sensitivity was set to 16-bit, Readout Bandwith to 200 MHz.
Transfected cells were screened using 488 nm laser excitation at 0.01 kW/cm2. The excitation was switched to 561 nm, focal plane and TIRF angle were readjusted and imaging was subsequently performed using ~1.5 kW/cm2 561 nm laser excitation. Imager strand concentration varied dependent on the measurement from 2 nM – 5 nM Cy3b-P1 and was adjusted to minimize double-binding events. Imaging was performed in 1xPCA (Sigma-Aldrich, 37580-25G-F) / 1xPCD (Sigma-Aldrich, P8279-25UN) / 1xTrolox (Sigma-Aldrich, 238813-1G) in Buffer C (PBS + 500 mM NaCl) and imaged for 20,000-40,000 frames at 200 ms exposure time. 3D imaging was performed using a cylindrical lens in the detection path as previously reported60.
Super-resolution data analysis
Raw data movies were reconstructed with the Picasso software 59. Drift correction was performed with a redundant cross-correlation and/or gold particles as fiducials. Using Picasso, the localization information was converted to an image volume with isotropic pixel sampling of 10 nm. The volumes were denoised by applying a Gaussian filter with a standard deviation of 30 nm. The topology of the microtubules was derived using stretching open active contour modeling, as implemented in the SOAX software61. 3 independent SSNA1-overexpressing fibroblasts and control cells were assessed each, containing the total tube lengths (i.e. microtubule lengths) of 7700 μm, 8500 μm and 7700 μm for control cells and 5700 μm, 7900 μm and 1900 μm for SSNA1-overexpressing cells. In each cells, the occurrences of the 3-way intersections were counted to be 0.96, 0.78 and 1.2 per 100 μm for control cells and 1.6, 3.2 and 4.2 per 100 μm for SSNA1-overexpressing cells. As the expression level of SSNA1 varies between individual cells, the transfected cells were selected based on the signal of GFP, which was co-expressed with SSNA1. Three independent cells containing the strongest signals out of >500 cells have been selected.
Data availability
The cryo-EM structure of the SSNA1-microtubule is available through EMDB with the accession code EMD-4188. The additional tomography images are available in Figshare (https://figshare.com/articles/Microtubule_branch_png/6809795). Source data for Fig. 1, 4, 6 and Supplementary Fig. 6 have been provided as Supplementary Table 3. Other data supporting the findings of this study such as the cryo-tomography data are available from the corresponding author on reasonable request.
Code availability
Morphology analysis of the neurons (total number of collateral branches and total length of all the branches for each axon) was performed using Fiji, with the help of scripts written ad-hoc for the task. All scripts are available from the authors upon request.
Statistics and Reproducibility
All microtubule nucleation assays and TIRF based assays were performed independently at least three times unless otherwise stated. Similar results were observed in all the replicates performed. Primary neuron preparation was performed from three independent mice.
χ2 two-sample test was performed to determine the significance of comparison made between two data set. Kruskal-Wallis test, followed by Dunn’s multiple comparison post-hoc test was performed to test the significance across multiple independent samples. Reproducibility was conformed.
Supplementary Material
Acknowledgements
We thank Carsten Grashoff, Esben Lorentzen, and Julia von Blume for assistance at various stages of the project, Elena Conti, Wolfgang Baumeister and the imaging-, biochemistry- and cryo-EM core facilities for resources and infrastructure (Max Planck Institute of Biochemistry). We thank Andrew Carter (University of Cambridge) and Charlotte Kelley (Max Planck Institute of Biochemistry) for careful proofreading of the manuscript and helpful discussions. We thank PICT-IBiSA@Orsay and Sophie Leboucher (Institut Curie, Orsay) for technical assistance. We are grateful to Günther Woehlke (Technical University Munich, Germany) for combined efforts of purification of tubulin, and to Christian Gonzalez-Billault and Cristopher Villablanca (University of Chile, Santiago) for help with the analysis of neurons. We also acknowledge Charles Sindelar and Rui Zhang for providing software package for microtubule analysis and giving guidance for the usage of them. The EM map of the SSNA1-microtubule complex is deposited with EMDB ID: EMD-4188. NM, RJ and CB acknowledge the Max Planck Society for the financial support. This study was supported by the DFG through grants within GRK1721, MI 1745/1, and the European Research Council (ERC-CoG-724209 to NM, ERC-StG-680241 to RJ). NM is a recipient of EMBO Young Investigator award and Boehringer Ingelheim Foundation Plus 3 Program grant. RJ is supported by DFG Emmy Noether Program (DFG JU 2957/1-1). CJ is supported by the Institut Curie, the CNRS, the ANR-10-LBX-0038, ANR-10-IDEX-0001-02 PSL⋆, ANR-12-BSV2-0007, INCA 2013-1-PLBIO-02-ICR-1 and 2014-PLBIO-11-ICR-1. AHC acknowledges LISBOA-01-0145-FEDER-007660 and FCT.
Footnotes
Author contribution
N.B. and N.M. performed electron and light microscopy, designed mutant constructs and performed biochemistry experiments, analyzed the data and prepared the figures. S.B. and M.M.M. performed experiments with neuron primary culture. N.B. and A.H.C. performed light microscopy experiments. N.B. C.B. and M.T. cloned and purified proteins. N.M., N.B., H.N., S.B., M.M.M. and C.J. analyzed neuron data and G.C. facilitated the automation of neuron analysis. N.M., N.B. and H.N. performed electron microscopic data collection and analyzed the data. H.N. and T.S. performed super-resolution light microscopy experiments and N.M. H.N. G.C. C.B and R.J. analyzed the data. Experiments were designed by N.M. and the manuscript was written by N.M. with contributions from the other authors.
References
- 1.Ishihara K, Nguyen PA, Groen AC, Field CM, Mitchison TJ. Microtubule nucleation remote from centrosomes may explain how asters span large cells. Proc Natl Acad Sci USA. 2014;111:17715–17722. doi: 10.1073/pnas.1418796111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meunier S, Vernos I. Acentrosomal Microtubule Assembly in Mitosis: The Where, When, and How. Trends Cell Biol. 2016;26:80–87. doi: 10.1016/j.tcb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Petry S, Vale RD. Microtubule nucleation at the centrosome and beyond. Nat Cell Biol. 2015;17:1089–1093. doi: 10.1038/ncb3220. [DOI] [PubMed] [Google Scholar]
- 4.Kalil K, Dent EW. Branch management: mechanisms of axon branching in the developing vertebrate CNS. Nat Rev Neurosci. 2014;15:7–18. doi: 10.1038/nrn3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis TL, Courchet J, Polleux F. Cell biology in neuroscience: Cellular and molecular mechanisms underlying axon formation, growth, and branching. J Cell Biol. 2013;202:837–848. doi: 10.1083/jcb.201305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stiess M, et al. Axon extension occurs independently of centrosomal microtubule nucleation. Science (New York, NY) 2010;327:704–707. doi: 10.1126/science.1182179. [DOI] [PubMed] [Google Scholar]
- 7.Yu W, et al. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker F, Oriola D, Dalton B, Brugués J. Autocatalytic microtubule nucleation determines the size and mass ofXenopus laevisegg extract spindles. Elife. 2018;7 doi: 10.7554/eLife.31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murata T, et al. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 11.Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamasaki T, et al. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J Cell Biol. 2013;202:25–33. doi: 10.1083/jcb.201304031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sánchez-Huertas C, et al. Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat Commun. 2016;7 doi: 10.1038/ncomms12187. 12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfannenschmid F, et al. Chlamydomonas DIP13 and human NA14: a new class of proteins associated with microtubule structures is involved in cell division. J Cell Sci. 2003;116:1449–1462. doi: 10.1242/jcs.00337. [DOI] [PubMed] [Google Scholar]
- 16.Lai CK, et al. Functional characterization of putative cilia genes by high-content analysis. Mol Biol Cell. 2011;22:1104–1119. doi: 10.1091/mbc.E10-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyal U, Renvoisé B, Chang J, Blackstone C. Spastin-interacting protein NA14/SSNA1 functions in cytokinesis and axon development. PLoS ONE. 2014;9:e112428. doi: 10.1371/journal.pone.0112428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchoin L, et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 19.Volkmann N, et al. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 20.Itzhak DN, Tyanova S, Cox J, Borner GH. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5 doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodruff JB, et al. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 2017;169:1066–1077.e10. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 22.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 23.Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bechstedt S, Brouhard GJ. Doublecortin recognizes the 13-protofilament microtubule cooperatively and tracks microtubule ends. Dev Cell. 2012;23:181–192. doi: 10.1016/j.devcel.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georges des A, et al. Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat Struct Mol Biol. 2008;15:1102–1108. doi: 10.1038/nsmb.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos-Morales F, Infante C, Fedriani C, Bornens M, Rios RM. NA14 is a novel nuclear autoantigen with a coiled-coil domain. J Biol Chem. 1998;273:1634–1639. doi: 10.1074/jbc.273.3.1634. [DOI] [PubMed] [Google Scholar]
- 27.Price HP, et al. The orthologue of Sjögren's syndrome nuclear autoantigen 1 (SSNA1) in Trypanosoma brucei is an immunogenic self-assembling molecule. PLoS ONE. 2012;7:e31842. doi: 10.1371/journal.pone.0031842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez-Rodríguez M, et al. Characterization of the structure and self-recognition of the human centrosomal protein NA14: implications for stability and function. Protein Eng Des Sel. 2011;24:883–892. doi: 10.1093/protein/gzr050. [DOI] [PubMed] [Google Scholar]
- 29.Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Crevenna AH, Kunze I, Mizuno N. Structural basis for the extended CAP-Gly domains of p150(glued) binding to microtubules and the implication for tubulin dynamics. Proc Natl Acad Sci USA. 2014;111:11347–11352. doi: 10.1073/pnas.1403135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witte H, Bradke F. The role of the cytoskeleton during neuronal polarization. Curr Opin Neurobiol. 2008;18:479–487. doi: 10.1016/j.conb.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 33.Conde C, Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 34.Kapitein LC, Hoogenraad CC. Building the Neuronal Microtubule Cytoskeleton. Neuron. 2015;87:492–506. doi: 10.1016/j.neuron.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 35.Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc Natl Acad Sci USA. 2007;104:20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Errico A, Claudiani P, D'Addio M, Rugarli EI. Spastin interacts with the centrosomal protein NA14, and is enriched in the spindle pole, the midbody and the distal axon. Hum Mol Genet. 2004;13:2121–2132. doi: 10.1093/hmg/ddh223. [DOI] [PubMed] [Google Scholar]
- 37.Shelanski ML, Gaskin F, Cantor CR. Microtubule assembly in the absence of added nucleotides. Proc Natl Acad Sci USA. 1973;70:765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chrétien D, Kenney JM, Fuller SD, Wade RH. Determination of microtubule polarity by cryo-electron microscopy. Structure (London, England : 1993) 1996;4:1031–1040. doi: 10.1016/s0969-2126(96)00110-4. [DOI] [PubMed] [Google Scholar]
- 39.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang Z, et al. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. Elife. 2014;3:e04686. doi: 10.7554/eLife.04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R, Nogales E. A new protocol to accurately determine microtubule lattice seam location. J Struct Biol. 2015;192:245–254. doi: 10.1016/j.jsb.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alushin GM, et al. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grigorieff N. Frealign: An Exploratory Tool for Single-Particle Cryo-EM. Meth Enzymol. 2016;579:191–226. doi: 10.1016/bs.mie.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 50.Crevenna AH, et al. Side-binding proteins modulate actin filament dynamics. Elife. 2015;4 doi: 10.7554/eLife.04599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat Cell Biol. 2015;17:907–916. doi: 10.1038/ncb3188. [DOI] [PubMed] [Google Scholar]
- 52.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 53.Lahaye X, et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Magiera MM, Janke C. Investigating tubulin posttranslational modifications with specific antibodies. Methods Cell Biol. 2013;115:247–267. doi: 10.1016/B978-0-12-407757-7.00016-5. [DOI] [PubMed] [Google Scholar]
- 55.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Longair MH, Baker DA, Armstrong JD. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- 57.Hollingworth T, Berry M. Network analysis of dendritic fields of pyramidal cells in neocortex and Purkinje cells in the cerebellum of the rat. Philos Trans R Soc Lond, B Biol Sci. 1975;270:227–264. doi: 10.1098/rstb.1975.0008. [DOI] [PubMed] [Google Scholar]
- 58.Vormberg A, Effenberger F, Muellerleile J, Cuntz H. Universal features of dendrites through centripetal branch ordering. PLoS Comput Biol. 2017;13:e1005615. doi: 10.1371/journal.pcbi.1005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 60.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science (New York, NY) 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu T, et al. SOAX: a software for quantification of 3D biopolymer networks. Sci Rep. 2015;5 doi: 10.1038/srep09081. 9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM structure of the SSNA1-microtubule is available through EMDB with the accession code EMD-4188. The additional tomography images are available in Figshare (https://figshare.com/articles/Microtubule_branch_png/6809795). Source data for Fig. 1, 4, 6 and Supplementary Fig. 6 have been provided as Supplementary Table 3. Other data supporting the findings of this study such as the cryo-tomography data are available from the corresponding author on reasonable request.