Abstract
Cytochrome c oxidase (complex IV, CIV) is known in mammals to exist independently or in association with other respiratory proteins to form supercomplexes (SCs). In Saccharomyces cerevisiae, CIV is found solely in a SC with cytochrome bc1 (complex III, CIII). Here, we present the cryo-EM structure of S. cerevisiae CIV in a III2IV2 SC at 3.3 Å resolution. While overall similarity to mammalian homologues is high, we found notable differences in the supernumerary subunits Cox26 and Cox13; the latter exhibits a unique arrangement that precludes CIV dimerization as seen in bovine. A conformational shift in the matrix domain of Cox5A – involved in allosteric inhibition by ATP – may arise from its association with CIII. The CIII–CIV arrangement highlights a conserved interaction interface of CIII, albeit one occupied by complex I in mammalian respirasomes. We discuss our findings in the context of the potential impact of SC formation on CIV regulation.
Introduction
Cytochrome c oxidase (or complex IV, CIV) is the last enzyme in the mitochondrial electron transport chain whose activity drives oxidative phosphorylation, the process by which cells produce most of their ATP1. CIV is a complex multi-subunit protein embedded in the inner membrane of mitochondria. Its catalytic core, conserved across kingdoms, is composed of three subunits encoded in the mitochondrial DNA. CIV catalyses the reduction of molecular oxygen to water, conserving the released energy as coupled proton transfers across the membrane, through a mechanism that is still highly debated2. In addition, mitochondrial forms of CIV have supernumerary subunits (11 in humans), which are encoded in the nuclear genome. The supernumerary subunits might have a role in CIV assembly or stability, or supercomplex (SC) formation, and there is an increasing interest in understanding the role of specific isoforms in optimising CIV core catalytic function in different tissues or physiological states3,4.
CIV can exist as a monomer, a dimer, or in a SC with complexes I and/or III in varying stoichiometries (I–III2–IV1-4 and III2–IV1-2) with relative abundances depending on species5,6 and in mammals, tissue-type, cellular energy requirement and disease state7–10. Several structures of complex I-containing SCs have been published recently11–13 that provide details of their interaction.
Despite its lack of complex I (CI), Saccharomyces cerevisiae has been extensively used as a model system for the study of mitochondrial respiratory chains, given its unique genetic amenability14. In the case of CIV, S. cerevisiae is the only system that contains the supernumerary subunits of so much interest (including isoforms), whilst allowing site-directed mutagenesis of the mitochondrial DNA. This uniquely permits the investigation of fundamental aspects of its catalytic mechanism as well as the identification of putative regulatory elements within supernumerary subunits. In addition, S. cerevisiae CIV is known to form SCs with cytochrome bc15 (or complex III, CIII), so it represents an ideal opportunity to investigate the role of SC formation on CIV activity and regulation.
High resolution structural information for the mitochondrial form of the enzyme is limited to crystal structures of dimeric bovine CIV215 and more recently a cryo-EM structure of human CIV16 within a respirasome. Here, we present the cryo-EM structure of S. cerevisiae CIV in a III2IV2 SC at 3.3 Å resolution.
Results
Architecture of the III2IV2 SC
The III2IV2 SC was purified from S. cerevisiae mitochondrial membranes after solubilisation with glyco-diosgenin (GDN) and successive metal affinity and gel filtration chromatography (Supplementary Fig. 1). The purified SC is active and reduces molecular oxygen in the presence of exogenous cytochrome c at a rate of 30.4 ± 1.3 e.s-1 using decylubiquinol as a reductant. When no cytochrome c was added, no oxygen reduction was observed (Supplementary Fig. 1).
The cryo-EM map (Table 1) reveals a CIII dimer at the core of the SC flanked by a CIV monomer on either side (Supplementary Fig. 2), as seen in previous low resolution models17,18. The core of the map was resolved to <3 Å, but the two CIV monomers could only be resolved to 5–8 Å, consistent with the dynamic nature of CIV within other SC structures11–13,19. To address this problem, we used subtracted experimental particle images, focusing only on masked refinements for each CIV monomer as previously described20. This procedure increased the resolution of the CIVa and CIVb monomers to 3.31 and 3.38 Å, respectively, revealing new atomic details (Supplementary Fig. 2 and Table 1). The improvement in resolution confirms that the two CIV monomers in the III2IV2 SC are identical.
Table 1. Cryo-EM data collection, refinement and validation statistics.
| III2IV2 SC (EMD-0262, PDB 6HU9) |
CIVa (EMD-0269) |
CIVb (EMD-0268) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 130,000 | ||
| Voltage (kV) | 300 | ||
| Electron exposure (e–/Å2) | 1.645 | ||
| Defocus range (μm) | -1.6 to -3.6 | ||
| Pixel size (Å) | 1.048 | ||
| Symmetry imposed | C1 | ||
| Initial particle images (no.) | 98,968 | ||
| Final particle images (no.) | 44,915 | ||
| Map resolution (Å) | 3.31 | 3.31 | 3.38 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | - | - | - |
| Map sharpening B factor (Å2) | -60.26 | -69.04 | -79.03 |
| Refinement1 | |||
| Initial model used (PDB code) | 1KYO, 1V54 | ||
| Model resolution (Å)2 | 3.35 | ||
| FSC threshold | 0.50 | ||
| Model resolution range (Å) | 3.35 | ||
| Model composition | |||
| Nonhydrogen atoms | 63,033 | ||
| Protein residues | 7,636 | ||
| Ligands | 69 | ||
| B factors (Å2)3 | |||
| Protein | 59.9 (141.4 – 20.7) | ||
| Ligand | 54.9 (149.7 – 35.5) | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.01 | ||
| Bond angles (°) | 1.32 | ||
| Validation3 | |||
| MolProbity score | 2.32 | ||
| Clashscore | 5.62 | ||
| Poor rotamers (%) | 8.49 | ||
| Ramachandran plot | |||
| Favored (%) | 95.61 | ||
| Allowed (%) | 4.35 | ||
| Disallowed (%) | 0.04 |
The model was refined against a map generated by merging the map from the 3D refinement (III2IV2 SC) with the two maps generated after particle subtraction and 3D refinement focused on CIVa or CIVb.
Determined by phenix.mtriage
Determined by phenix.molprobity (values in parenthesis denote highest and lowest values)
The merged map (Fig. 1) allowed us to build 44 protein subunits for the III2IV2 SC, corresponding to all 20 protein subunits of the obligatory CIII dimer, including Qcr10, which is absent from all yeast structures published to date, and 12 protein subunits for each CIV monomer; the 11 classically described subunits21,22 and the recently assigned Cox2623,24. The presence of all polypeptides forming the III2IV2 SC was confirmed by mass spectrometry analyses (Supplementary Table 1). Mass spectrometry also revealed the presence of the respiratory supercomplex factors Rcf1 and Rcf2 in the final protein preparation, but those proteins are absent from the cryo-EM structure. This observation supports the role of Rcf1 and Rcf2 in yeast SC formation25 but suggests that they are not essential for SC stability, which is similar to findings in mammalian systems. The high resolution of the map allowed the confident modelling of 96% of all amino acids in the structure (Table 1). All prosthetic groups and metal cofactors are well defined, and 44 lipids including 8 cardiolipin molecules can be confidently modelled.
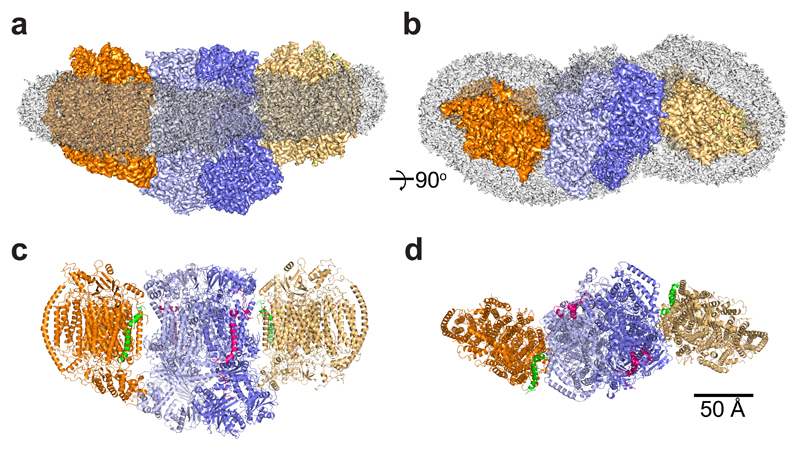
Fig. 1. Overall structure of the S. cerevisiae III2IV2 supercomplex.
a, Side and top view of the III2IV2 supercomplex (SC) merged cryo-EM map with overall SC dimensions of 289x157x114 Å. CIII and CIV are represented in blue and orange, each monomer being represented by a different shade. The transparent grey density displays the extent of the GDN detergent belt. b, Side and top view of the III2IV2 SC model. CIII and CIV are represented in the same colours as in a, with Cox26 (green) and Qcr10 (red) highlighted.
Atomic resolution structure of the S. cerevisae CIV
The yeast CIV structure comprises 12 subunits (Fig. 2a, Supplementary Table 1). Cox1-3 form the catalytic core and are surrounded by 9 supernumerary subunits (Cox4-9, Cox12, Cox13, Cox26). The structure of yeast CIV is remarkably similar to mammalian systems, with a calculated RMSD against bovine26 and human16 enzymes of 0.95 and 1.10 Å, respectively, but there are notable differences as discussed below. Subunit composition is also very similar: all but one S. cerevisiae subunit (Cox26) have homologues in mammals and conversely, all mammalian subunits but three (COX7B, COX8 and NDUFA4) have yeast homologues. Yeast Cox5 (homologous to mammalian COX4) exists as one of two isoforms depending on oxygen level27. In normoxic conditions, almost all CIV is assembled with the Cox5A isoform, although low levels of Cox5B are also expressed. To enhance the homogeneity of our protein preparations, we used a COX5B knockout mutant strain (ΔCOX5B) that only expresses the Cox5A isoform.
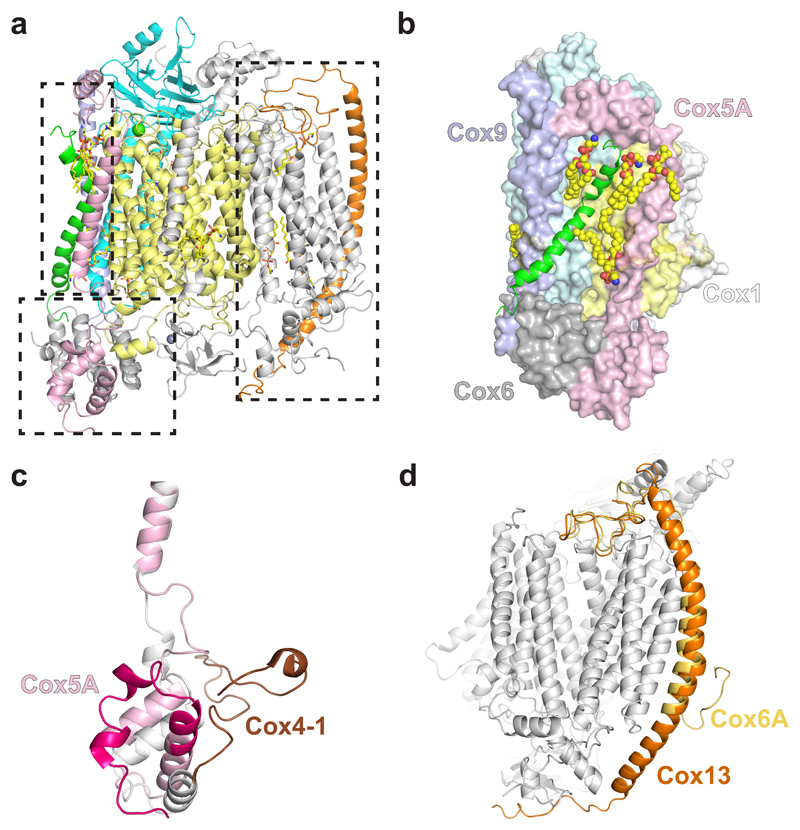
Fig. 2. Structure of S. cerevisiae CIV.
a, Atomic model of CIV showing all 12 subunits. Cox1 is shown in yellow, Cox2 in cyan, Cox5A in pink, Cox26 in green, Cox9 in purple and Cox13 in orange. b, Interactions of Cox26 with Cox1, Cox2, Cox6, Cox9 (coloured as in a) and lipids (spacefill representation). c, Alignment of Cox5A (pink) with its bovine homologue (COX4-1 – grey and brown, PDB 1V54). The first ~50 amino acids are highlighted by a darker shade. d, Differences in length and shape of the transmembrane helix of Cox13 with its bovine homologue (COX6A, PDB 1V54).
Cox1 and Cox2 house the prosthetic groups responsible for electron transfer and O2 reduction, namely the dinuclear CuA centre, haem a, haem a3 and CuB. All prosthetic groups are well resolved in the EM map, as well as the characteristic HPEVY ring of amino acids formed by covalent linkage between Nε2-His241 and Cε-Tyr245 in Cox1 (Supplementary Fig. 3). Clear densities were observed at the expected magnesium and calcium/sodium binding sites in Cox1 (modelled as a Ca2+ ion28) and the zinc binding site in Cox4. The density map also confirms the predicted positioning of the residues forming the D-, K- and H-channels described as putative proton pathways in Cox1 of A1-type oxidases29.
Cox26, recently assigned as a subunit of S. cerevisiae CIV23,24, is composed of a single transmembrane helix with a kink towards its C-terminus. This arrangement enables extensive hydrophobic interactions with Cox1 and Cox2 (Fig. 2b), which are further supported by two lipids held against the core of the enzyme. In the matrix, Cox26 interacts at its N-terminus with Cox6 and Cox9 via a network of electrostatic and hydrophobic interactions (Fig. 2b). Deletion of Cox26 does not prevent CIV assembly, nor does it have any significant effect on its activity23,24. However, the position of Cox26 in the SC, and the interactions it makes with the subunits above, suggest Cox26 could stabilise the region of CIV at the interface with CIII, supporting a role in formation or stability of the III2IV2 SC, even though no direct connection with CIII is evident from the structure.
A notable difference between yeast and mammalian CIV is the conformation of the N-terminal domain of Cox5A (Fig. 2c) which is shifted towards CIII in the S. cerevisiae SC to form protein-protein interactions further described below. This region is implicated in allosteric inhibition of CIV by ATP30–32 and Ser43, a residue which was shown by mutagenesis to contribute to this allosteric effect32, is at the interface with CIII. More detailed biochemical analyses will be required to determine whether SC formation has a role in the ATP feedback inhibition mechanism.
Another difference is seen in Cox13, the most peripheral subunit of the SC (Fig. 1c, Fig. 2a,d). Cox13 is characterised by a single extended and bow-shaped transmembrane α-helix, protruding from the membrane into the matrix and the intermembrane space (IMS), with a π bulge between residues Ser55 and Leu60. The N- and C- termini of Cox13 are oriented towards the core of the complex, interacting with Cox1 and Cox3 on the IMS and Cox4 on the matrix side. Its mammalian homologue, COX6A, has a shorter N-terminus that folds back into the membrane, providing the dimerization interface observed in the bovine crystal structure (Fig. 2d). The relevance of the dimeric state of CIV is still disputed, and refinement of CIV within the human respirasome16 showed that NDUFA4 would prevent dimerization via COX6A. Yeast does not have a NDUFA4 homologue but the peculiar arrangement of Cox13 observed here in the yeast SC would hinder CIV dimerization via this subunit.
Complete structure of S. cerevisiae CIII
In S. cerevisiae, each CIII monomer consists of 10 subunits. Cytochrome b, cytochrome c1 and Rip1 form the catalytic core containing two haems B, one haem C and a [2Fe-2S] cluster, respectively. All prosthetic groups are visible in the density map along with a molecule of ubiquinone in two apparent configurations at the Qi site residing between the two cytochrome b subunits (Supplementary Fig. 4, Fig. 3). This is a common feature in structures of CIII of various origins, including yeast, whenever the hydrophobic substrate was not completely extracted from the preparation, and it is due to the higher affinity of ubiquinone to the Qi site as compared to Qo. Apart from the presence of Qcr10 and changes resulting from interactions with CIV (see below), our model is consistent with the CIII structures described previously33.
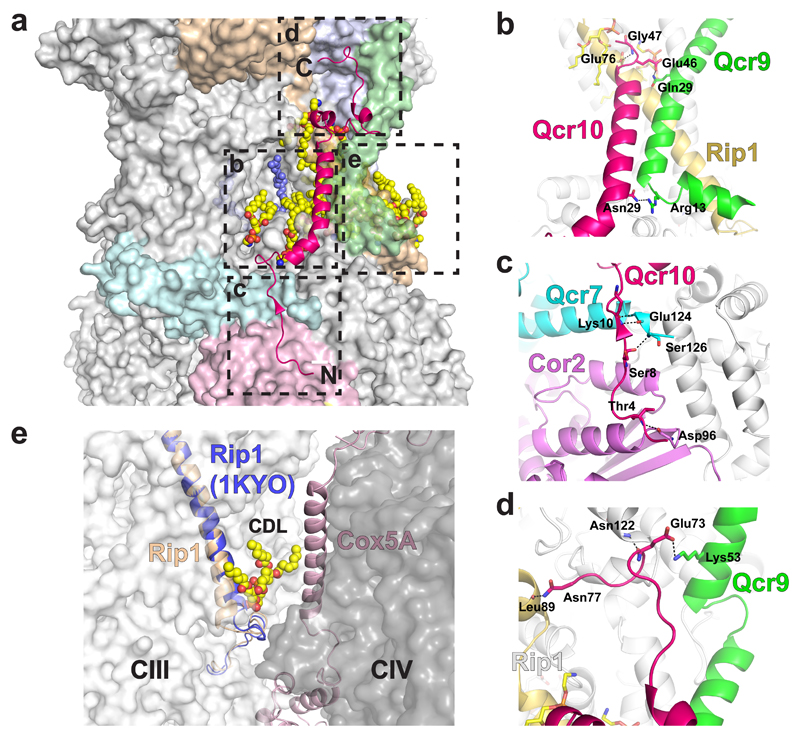
Fig. 3. Interactions of Qcr10 with other subunits of CIII, and that of Rip1 with a lipid at the interface with CIV.
a, Position of Qcr10 (pink ribbon) in the CIII structure with its N- and C-termini highlighted. Other subunits of CIII that interact with Qcr10 are highlighted in colours. A molecule of ubiquinone at the Qi site is shown as blue spheres. The boxes indicate specific regions of interaction that are highlighted in the other panels. b, Qcr10 (pink) forms interactions with the transmembrane helix of Rip1 (yellow) and with Qcr9 (green) at both the matrix and IMS faces of the membrane. c, The N-terminal tail of Qcr10 interacts with Cor2 (purple) and Qcr7 (cyan) in the matrix. d, Interactions in the IMS between Qcr10 and Cytc1 (grey), Rip1 (yellow) and Qcr9 (green). e, A slight shift in the N-terminus of Rip1 (yellow) compared to the yeast X-ray structure (blue) accommodates interactions with a cardiolipin that goes on to interact with Cox5A of CIV (light pink), forming part of the interface between the two complexes.
Despite low sequence identity, the transmembrane helix of Qcr10 occupies a position similar to that of subunit 11 in mammalian structures34, making extensive interactions with Qcr9, and with Rip1 on the IMS side of the inner mitochondrial membrane (IMM) supported by a lipid (Fig. 3). In the matrix, the extended N-terminus of Qcr10 interacts with Cor2, and Qcr7 of the other monomer, while in the IMS, Qcr10 extends to interact with cytochrome c1. These observations are in line with the proposed role of Qcr10 in CIII assembly and Rip1 stability35.
A conformational change is observed at the N-terminus of the transmembrane helix of Rip1 due to interactions with a cardiolipin molecule that also interacts with Cox5A (Fig. 3 and below) suggesting SC formation could have a role in stabilising this part of CIII. However, this change at the N-terminus does not seem to impact the head domain of Rip1, which maintains its known flexibility.
The CIII-CIV interface
Whilst the CIII dimer is symmetrical and no differences can be seen in the two CIII–CIV interfaces, the alignment of the two halves of the merged SC structure on CIII reveals a deviation of up to 3 Å at the extreme periphery of CIV (Supplementary Fig. 5). This most likely arises from the intrinsic flexibility of the SC and explains why a 3D refinement with C2 symmetry failed to improve the SC resolution (see Methods).
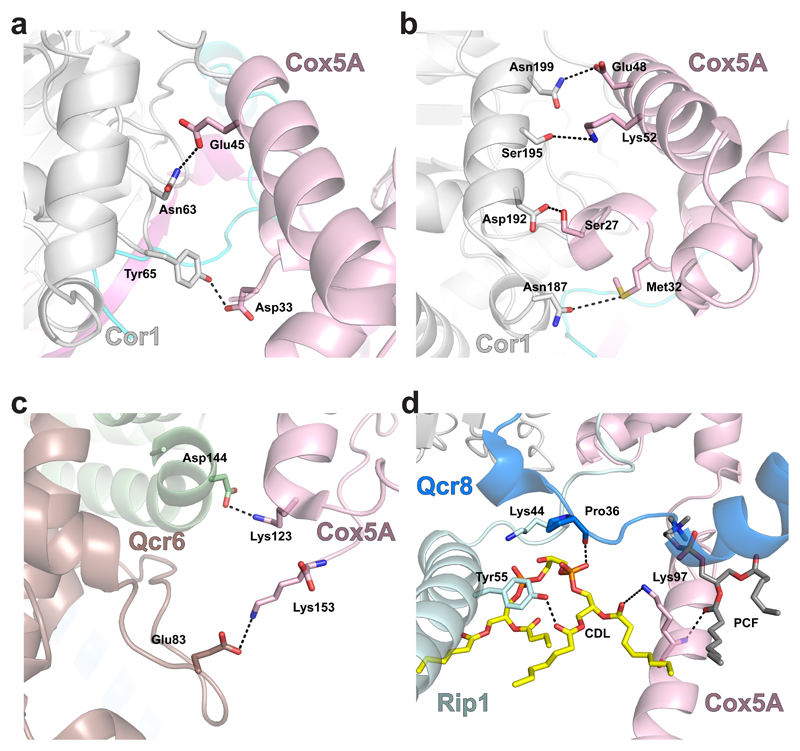
The CIII–CIV interface reveals protein-protein interactions on either side of the IMM, and interactions via bridging lipids in the membrane region itself. The majority of interactions occur on the matrix side between Cor1 and the N-terminus of Cox5A, facilitated by the conformational shift in the latter as described above (Fig. 4a). In the IMS, the C-terminal domain of Cox5A is in position to interact with both the C-terminus of Qcr6 and a loop region between helices 6 and 7 of cytochrome c1 (Fig. 4b-c). Within the membrane, Cox5A contacts the N-terminal helix of Rip1 and Qcr8 via a cardiolipin molecule and another lipid modelled as phosphocholine (Fig. 4d). Two other cardiolipins indirectly support the CIII–CIV interface highlighting their crucial role in SC formation36.
Fig. 4. Protein-protein interactions between CIV and CIII.
a and b, View from the matrix side of protein-protein interactions involving Cor1 (white) and Cox5A (pink). Residues that make inter-subunit interactions are indicated and their interactions are shown as dashed lines. c, Interactions between cytochrome c1 (Cytc1, brown) and Qcr6 (pale green) with Cox5A (pink) in the IMS. d, A cardiolipin molecule (CDL, yellow) and a phosphocholine (PCF, grey) within the membrane interacts with residues of Cox5A (pink), Rip1 (pale blue) and Qcr8 (blue).
Previous work indicates that mammalian COX7A2L is required for the formation and stability of the CIII–CIV SC in mammals37 which is consistent with published respirasome structures11,12. S. cerevisiae has no homologue of COX7A2L so it must follow a different mechanism of CIII–CIV SC formation than the one proposed in mammals. In addition, our work suggests that the III2IV2 SC in yeast doesn’t require any other proteins to maintain its stability, the interaction between Cox5A and Cor1, two highly conserved subunits, and phospholipids being sufficient to stabilise the SC. However, no mammalian structure of the III2IV1/2 SC is currently available, and in the absence of CI, it cannot be excluded that the CIII–CIV interface is different in a III2IV1/2 supercomplex compared to the respirasome. Mammalian CIV has an additional subunit, COX7B, in front of COX4-1 (the mammalian homolog of Cox5A). However, superimposition of the bovine CIV structure onto the yeast III2IV2 SC suggests a similar interaction between CIII and CIV as the one observed in yeast remains possible (Supplementary Fig. 6).
Discussion
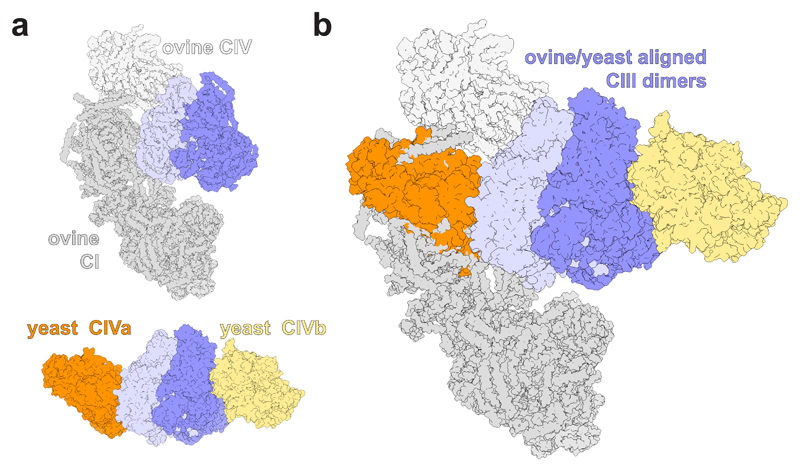
The functional role of SC formation is still unclear38,39. Reactive oxygen species (ROS) prevention by steric inhibition of one half of CIII by CIV in the respirasome has been proposed40. In the yeast III2IV2 SC, the symmetry of CIII is maintained and no obvious interactions are apparent that could stabilise the hinge region of Rip1, whose flexibility allows the movement of the head group domain, which is implicated in ROS production. However, it is noteworthy that the same subunits of CIII are involved in SC formation in all SC structures resolved so far (Fig. 5). The homologues of yeast CIII subunits Cor1, Rip1, Qcr6 and Qcr8 interact with CI in the mammalian respirasome structures. These are the same subunits that interact with CIV in our III2IV2 structure, highlighting a conservation of the CIII interaction interface, albeit one occupied by CI in mammals and by CIV in S. cerevisiae, which lacks CI. Additionally, in the CI-containing Y. lipolytica respirasome, where the CIII symmetry is apparently maintained, the suggested CIII–CIV arrangement is similar to the one observed in the yeast III2IV2 SC6. This suggests that SC formation may serve to stabilise the active CIII monomer(s) by other proteins.
Fig. 5. Alignment of the mammalian I1III2IV1 respirasome with the III2IV2 SC from S. cerevisiae.
a, The structures of the tight ovine I1III2IV1 respirasome11 (PDB 5J4Z, displayed in shades of grey) and the yeast III2IV2 SC (coloured as in Figure 1). b, These structures were aligned on their CIII dimers. The view in both a and b is from the IMS.
Finally, modulation of CIV activity in response to energy requirements by differential expression of isoforms such as Cox5 in yeast41 (COX4 in mammals4) and COX7A in mammals42 has been recognised. From the yeast structure presented here, and that of mammalian respirasomes, it seems that these subunits form a substantial part of the interface between proteins in SCs. Therefore, biochemical and biophysical studies investigating these isoforms, and the effect of allosteric sites identified within supernumerary subunits on CIV core catalytic subunits, must take into account SC formation. With homologues of many mammalian supernumerary subunits, extensive genetic amenability and in light of the SC structures now available, S. cerevisiae offers a powerful system to study how these factors modulate CIV activity and respiratory SC formation.
Methods
Yeast strain and cell growth
A ΔCOX5B S. cerevisiae strain only expressing the Cox5A isoform of CIV (α ade2 leu2 trp1 ura3 cox5B::KanMx4) with a six-histidine-tag at the C-terminus of COX13 was constructed from W303-1B43,44. Yeast cells were grown in YPGal (1% yeast extract, 2% peptone and 2% galactose) medium at 28°C in 2 L baffled flasks with shaking at 200 rpm as described in Meunier et al.43. Cells were harvested in late log phase by centrifugation at 6500 rpm for 5 minutes at 4°C. Cells were washed by resuspension in 50 mM KPi, pH 7.0, and centrifuged again. Cell pellets were stored at -80°C until use.
Preparation of mitochondrial membranes
Mitochondrial membranes were prepared essentially as described previously43. Briefly, thawed yeast cells were resuspended in 30 mL 650 mM D-mannitol, 50 mM KPi, 5 mM EDTA, pH 7.4 containing 0.1 mM PMSF. Glass beads (425-600 μm diameter) were added and cells were broken by mechanical lysis using a bead-beater cell disruptor. Cell debris was removed by centrifugation at 5600 × g for 20 minutes at 4°C, and the supernatant was centrifuged at 120,000 × g for 50 minutes at 4°C to harvest the mitochondrial membranes. The membranes were then resuspended and homogenised in 50 mM KPi, 100 mM KCl, 10 mM MgCl2, 150 μM CaCl2, 0.1 mM PMSF, pH 7.4, and centrifuged as in the previous step. The pellet was then washed by cycles of resuspension/centrifugation in 50 mM KPi, 2 mM EDTA, 0.1 mM PMSF, pH 7.4, until the measure of the absorbance of the supernatant at 260 nm was below 1. Finally, the membranes were resuspended in a minimal volume of 50 mM HEPES, pH 8.0, and stored at -80°C until use.
Membrane solubilisation and SC purification
Membranes were diluted in 50 mM HEPES, 150 mM NaCl, 1 mM PMSF, pH 8.0 to a CIII concentration of 45 nM and protein complexes were solubilised for 1 hour on ice by the addition of 1% GDN (glyco-diosgenin, Anatrace). After solubilisation, 350 mM NaCl (to make 500 mM final) and 5 mM imidazole are added. Insoluble material was removed by centrifugation at 120,000 × g for 30 minutes at 4°C. Solubilised proteins were then loaded overnight in a cold room at a flow rate of approx. 0.6 mL/min using a peristaltic pump onto a 5 mL HisTrap HP column (GE Healthcare) previously equilibrated with 2 column volumes (CV) of 50 mM HEPES, 500 mM NaCl, 5 mM imidazole, 0.05% GDN, pH 8.0. After loading, the column was washed with 3 CV of 50 mM HEPES, 500 mM NaCl, 5 mM imidazole, 0.05% GDN, pH 8.0, and then with 5 CV of 50 mM HEPES, 150 mM NaCl, 0.05% GDN, 5 mM imidazole, pH 7.2. Bound proteins were eluted with 50 mM HEPES, 150 mM NaCl, 0.05% GDN, 100 mM imidazole, pH 7.2. The eluted proteins were concentrated by centrifugation using 100 kDa MWCO centrifugal concentrators (GE Healthcare). The resulting sample was then further purified by gel filtration, using an Äkta Pure 25 (GE Healthcare) operated at 4°C with UV detection at 280 nm and automated fraction collection, by loading on a Superose 6 Increase column (GE Healthcare) equilibrated with 50 mM HEPES, 150 mM KCl, 0.05% GDN, pH 7.2. Fractions containing SCs were pooled and concentrated as above, and reapplied once to the same column.
Analytical methods
Resulting fractions were then concentrated and analysed for haem content and purity using UV-visible difference spectroscopy and BN-PAGE. Final protein concentration was determined by the Pierce BCA protein assay (Thermo Scientific) as per the manufacturer’s protocol.
UV-visible difference spectroscopy
UV-visible difference spectra were recorded between 400 and 700 nm using a home built spectrophotometer. Protein samples were diluted as necessary in 50 mM HEPES, 0.1% UDM, pH 8.0. Spectra were measured from sodium dithionite reduced minus oxidised spectra, using absorption coefficients (ΔƐ) of 26 mM-1 cm-1 (604-621 nm) and 28 mM-1 cm-1 (562-578 nm) for CIV and CIII respectively.
Gel electrophoresis
Protein samples for BN-PAGE analysis were mixed with BN-PAGE sample buffer (final concentration, 50 mM BisTris, 50 mM NaCl, 10% (w/v) glycerol, 0.001% Ponceau S, pH 7.2) as per the manufacturer’s instruction (Invitrogen). Pre-cast 3-12% BisTris gels (Novex) were used throughout, and run at 150 V for 90-120 minutes. The running buffer contained 50 mM BisTris, 50 mM Tricine, pH 6.8, and the cathode buffer was made by addition of 0.002% Coomassie G-250 to the running buffer. Gels were destained overnight using 10% (v/v) acetic acid, followed by multiple exchanges of water.
Mass spectroscopy
50 µg protein of purified III2IV2 SC sample were diluted 1:4 in reducing sample buffer consisting of 12% (w/v) SDS, 6% (v/v) β-mercaptoethanol, 30% (w/v) glycerol, 0.05% Coomassie blue G-250 (Serva), 150 mM Tris-HCl pH 7.0 and incubated for 15 min at 37°C. Samples were loaded onto a 10% Tricine-SDS polyacrylamide gel and the electrophoresis was performed according to Schägger45 until the entire amount of protein entered into the gel (~30 min; 50 V). A molecular mass marker (Precision plus protein standards, dual colour, BioRad) was used to monitor the correct loading of proteins into the gel. Gel-spots containing SC III2IV2 were cut and prepared for MS identification, following the in-gel trypsin digestion protocol described previously46. Tryptic peptides were separated by liquid chromatography and analysed by tandem mass spectrometry (LC-MS/MS) in a Q-Exactive 2.0 Orbitrap Mass Spectrometer equipped with an Easy nLC1000 nano-flow ultra-high pressure liquid chromatography system (Thermo Fisher Scientific). MS was run twice for each sample. MS raw data files were analysed using the MaxQuant software (v1.5.0.25) using the settings detailed in Guerrero-Castillo et al.47, except for the search against a compiled version of the S. cerevisiae protein database including the pig trypsin and other protein contaminants, such as human keratins.
Activity measurements
Steady-state oxygen consumption rates were measured using a Clark-type oxygen electrode (Oxygraph, Hansatech), operated at 25°C. Assays were conducted with purified protein at 15 nM, in 10 mM KPi, 50 mM KCl, pH 6.6, supplemented with 0.05% GDN, 500 units/mL SOD and 250 units/mL catalase. SC activity was measured in the presence and absence of 50 μM equine heart cytochrome c. The reaction was initiated by addition of 40 μM decylubiquinol. The reaction was stopped by addition of 1 mM KCN and the resulting rate was used as a baseline. Turnover numbers were calculated from linear fitting of the oxygen consumption rate using Origin (OriginLab, Northampton, MA) following the formula T.N. (e s-1) = O2 consumption gradient (M.s-1) x 4 / [CcO] (M). The result presented is the average of four independent experiments (± standard deviation) from two independent protein preparations for cryo-EM.
Cryo-electron microscopy
Purified III2IV2 SC (9 mg/mL total protein) was diluted 1:3 in 50 mM HEPES, 150 mM NaCl, 0.05 % GDN, pH 7.2 and applied to glow discharged UltrAuFoil R1.2/1.3 grids (Quantifoil). Grids were blotted for 8.5 seconds at 4°C and 100% humidity, and then rapidly frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher) and stored in liquid nitrogen. Preliminary imaging was done using a Polara microscope operated at 300 kV and equipped with a Quantum energy filter (Gatan) with a post-GIF K2 Summit direct electron detector (Gatan) operating in counting mode. The primary data were collected using a Titan Krios microscope (Thermo Fisher) operated at 300 kV and equipped with a Quantum energy filter (Gatan) (electron Bio-Imaging Centre, Diamond Light Source, Oxfordshire, UK). The images were collected with a post-GIF K2 Summit direct electron detector (Gatan) operating in counting mode at a nominal magnification of 130,000x, corresponding to the pixel size of 1.048 Å. An energy slit with a width of 20 eV was used during data collection. The dose rate on the specimen was set to 6.58 electrons per Å2 per s and a total dose of 52.64 e/Å2 was fractionated over 32 frames. Data were collected using EPU software (Thermo Fisher) with a nominal defocus range set from -1.6 μm to -3.6 μm. A total of 2740 micrographs were collected.
Image processing
Frame alignment and exposure weighting were performed with MOTIONCOR248. Contrast transfer function parameters of the motion-corrected micrographs were estimated with CTFFIND4.149. Micrographs were screened manually to remove those with excessive specimen drift, overfocus or ice defects. 98,968 particles were selected from the 2634 remaining micrographs using reference-free particle picking with Gautomatch v0.53 (written by Dr Kai Zhang, https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/), using a 360×360 Å2 box size. Particles were sorted using several iterations of reference-free 2D classification in cryoSPARC50, resulting in a final particle number of 52,257. An initial model was build using ab-initio reconstruction in cryoSPARC, which was refined using heterogeneous refinement in RELION v3.051. The III2IV2 SC accounted for 44,915 (86%) of the particles present (14% represented the III2IV1 supercomplex), and these particles were then used for homogeneous refinement in RELION v3.0 resulting in a 3.31 Å resolution map, based on the FSC-gold standard. Local resolution was calculated using ResMap52, revealing a range of resolutions in the map, with the core of CIII resolved to 2.9-3.2 Å, whereas the peripheral edges of the two CIV proteins were resolved to 5-8 Å.
To increase the resolution of the two CIV monomers, we used a particle subtraction approach20. In short, a soft mask was generated around III2IVb and used to subtract density from the particles, resulting in a new set of particles which was used for focussed 3D refinement of CIVa. This process was repeated with a soft mask around III2IVa to refine CIVb. This resulted in an increased resolution of the two CIV monomers to 3.31 Å and 3.38 Å, with a homogeneous distribution of resolution throughout the protein (Supplementary Fig. 2). The two CIV maps were then aligned to the original map of the SC and a merged map was generated using UCSF Chimera53.
Model building
The three individual maps described above were used for all model building using real space refinement (Table 1) in Coot54. A high resolution crystal structure of dimeric CIII (PDB 1KYO)55, and a yeast homology model22 were used as starting references for model building. All maps displayed clearly interpretable features such as bulky side chains, metal clusters, haem ligands, cardiolipin and ubiquinone. These features enabled unambiguous assignment of amino-acids in all chains, except some flexible N- and C-termini. Notably, the C-terminus of Rip1 of CIII has weak density (residues 95-215), and this is attributed to its characteristic flexibility. Its Fe-S cluster is clearly visible in the map. Finally, the N-terminal residues of Qcr6 (1-73) in CIII, are not resolved, a feature common to all yeast CIII crystal structures, possibly due to the high composition of charged residues in this region. Additional densities in the map indicated the presence of long carbon chains which were modelled as di-palmitoyl-phosphatidylethanolamine (PEF), diacyl-glycero-phosphocholine (PCF) and cardiolipin (CDL) molecules based on map interpretation and similarities with previous structures where these ligands were found. Lipid tails were truncated according to the density maps. The three models (dimeric CIII, and two CIV monomers) where then individually refined using the real space refine tool in Phenix56, using secondary structure restraints. Geometry definitions for the ligands were defined from values in the CCP4 ligand library57. Additional bond and distance restraints were implemented on specific molecules based on previously published high resolution structures. For initial refinement in Phenix, Ramachandran and rotamer constraints were also used. The models were then visually inspected in Coot for additional corrections. A final real space refinement was performed in Phenix by disabling rotamer constrains resulting in an increase of the model-to-map fit. To confirm the validity of the map, a final real space refinement was performed to the complete III2IV2 SC using the merged map described above. The final model contains 7,636 protein residues and 69 ligands. For the CIII dimer we modelled four B-haems and one ubiquinone molecule in the two cytochrome b subunits, one C-haem for each cytochrome c1 subunit, one [2Fe-2S] iron-sulphur cluster for each Rip1 subunit, eight cardiolipin molecules, twelve PEF and six PCF molecules. For each CIV we modelled a haem a, haem a3 and CuB in Cox1 as well as one calcium and one magnesium ions. In Cox2 we modelled a dinuclear CuA centre and in Cox4 one zinc ion. Each CIV contains additionally eight PEF molecules and one PCF molecule. Refinement and model statistics are summarized in Table 1. Map and molecule representations in the figures were prepared by PyMOL (https://pymol.org/) and UCSF Chimera.
Supplementary Material
Acknowledgements
We would like to thank Diamond Light Source for access and support to the cryo-EM facilities at the UK National electron bio-imaging centre (eBIC, proposal EM14704-36) funded by the Wellcome Trust, the Medical Research Council UK and the Biotechnology and Biological Sciences Research Council. We would also like to thank D. Houldershaw and the computer support group at Birkbeck. This work was supported by the Medical Research Council UK (Career Development Award MR/M00936X/1 to AM), the Birkbeck Wellcome Trust Institutional Strategic Support Fund (105628/Z/14/Z to AM) and a Wellcome Trust grant to the Birkbeck EM facility (079605/2/06/2).
Footnotes
Data availability
All relevant data are included in the manuscript and/or are available from the corresponding authors upon reasonable request. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-0262 (III2IV2 SC), EMD-0269 (CIVa), EMD-0268 (CIVb). The coordinates of the atomic model of the III2IV2 SC built from a combination of the three maps have been deposited in the Protein Data Bank (PDB) under accession code 6HU9.
Author contributions
A.M. designed and supervised the research. B.M. produced the yeast mutant strain. A.M.H. did all the protein work with contribution from Y.Z. A.C.-O. and S.A. performed mass spectrometry analysis. N.L. and A.M.H. performed all microscopy work. A.M.H. and N.P. processed the cryo-EM images. N.P. built the model with inputs from A.M. and A.M.H. A.M., A.M.H. and N.P. wrote the manuscript with contributions from all authors.
Competing interests
The authors declare no competing interests.
References
- 1.Rich PR, Maréchal A. The mitochondrial respiratory chain. Essays Biochem. 2010;47:1–23. doi: 10.1042/bse0470001. [DOI] [PubMed] [Google Scholar]
- 2.Rich PR. Mitochondrial cytochrome c oxidase: catalysis, coupling and controversies. Essays Biochem. 2010;47:1–23. doi: 10.1042/BST20160139. [DOI] [PubMed] [Google Scholar]
- 3.Pierron D, et al. Cytochrome c oxidase: evolution of control via nuclear subunit addition. Biochim Biophys Acta. 2012;1817:590–597. doi: 10.1016/j.bbabio.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horvat S, Beyer C, Arnold S. Effect of hypoxia on the transcription pattern of subunit isoforms and the kinetics of cytochrome c oxidase in cortical astrocytes and cerebellar neurons. J Neurochem. 2006;99:937–951. doi: 10.1111/j.1471-4159.2006.04134.x. [DOI] [PubMed] [Google Scholar]
- 5.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1773–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies KM, Blum TB, Kühlbrandt W. Conserved in situ arrangement of complex I and III2 in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proc Natl Acad Sci USA. 2018;115:3024–3029. doi: 10.1073/pnas.1720702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schägger H, Pfeiffer K. The ratio of oxidative phosphorylation complexes I-V in Bovine Heart Mitochondria and the Composition of Respiratory Chain Supercomplexes. J Biol Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 8.Acín-Pérez R, et al. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Greggio C, et al. Enhanced respiratory chain supercomplex formation in response to exercise in human skeletal muscle. Cell Metab. 2017;25:301–311. doi: 10.1016/j.cmet.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 10.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial Respiratory Chain Supercomplexes Are Destabilized in Barth Syndrome Patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 11.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 12.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 13.Guo R, et al. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell. 2017;170:1247–1257. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 14.Lasserre JP, et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis Model Mech. 2015;8:509–526. doi: 10.1242/dmm.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 16.Zong S, et al. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 2018;28:1026–1034. doi: 10.1038/s41422-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinemeyer J, Braun H-P, Boekema E, Kouril R. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 18.Mileykovskaya E, et al. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun C, et al. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature. 2018;557:123–126. doi: 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai XC, et al. Sampling the conformational space of the catalytic subunit of human ɣ-Secretase. Elife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geier BM, et al. Kinetic properties and ligand binding of the eleven subunit cytochrome c oxidase from Saccharomyces cerevisiae isolated with a novel large scale purification method. Eur J Biochem. 1995;227:296–302. doi: 10.1111/j.1432-1033.1995.tb20388.x. [DOI] [PubMed] [Google Scholar]
- 22.Maréchal A, et al. Yeast cytochrome c oxidase: a model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochim Biophys Acta. 2012;1817:620–628. doi: 10.1016/j.bbabio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levchenco M, et al. Cox26 is a novel stoichiometric subunit of the yeast cytochrome c oxidase. Biochim Biophys Acta. 2016;1863:1624–1632. doi: 10.1016/j.bbamcr.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Strecker V, et al. Supercomplex-associated Cox26 protein binds to cytochrome c oxidase. Biochim Biophys Acta. 2016;1863:1643–1652. doi: 10.1016/j.bbamcr.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strogolova V, et al. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukihara T, et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci USA. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burke PV, et al. Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J Biol Chem. 1997;272:14705–14712. doi: 10.1074/jbc.272.23.14705. [DOI] [PubMed] [Google Scholar]
- 28.Maréchal A, Iwaki M, Rich PR. Structural changes in cytochrome c oxidase induced by binding of sodium and calcium ions: an ATR-FTIR study. J Am Chem Soc. 2013;135:5802–5807. doi: 10.1021/ja4005706. [DOI] [PubMed] [Google Scholar]
- 29.Rich PR, Maréchal A. Functions of the hydrophilic channels in protonmotive cytochrome c oxidase. J R Soc Interface. 2013;10:1–14. doi: 10.1098/rsif.2013.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold S, Kadenbach B. Cell respiration is controlled by ATP, an allosteric inhibitor of cytochrome c oxidase. Eur J Biochem. 1997;249:350–354. doi: 10.1111/j.1432-1033.1997.t01-1-00350.x. [DOI] [PubMed] [Google Scholar]
- 31.Acín-Pérez R, Gatti DL, Bai Y, Manfredi G. Protein phosphorylation and preventation of cytochrome c oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab. 2011;13:712–719. doi: 10.1016/j.cmet.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess KC, et al. A mitochondrial CO2-adenylyl cyclase-cAMP signalosome controls yeast normoxic cytochrome c oxidase activity. FASEB J. 2014;28:4369–4380. doi: 10.1096/fj.14-252890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunte C, et al. Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 35.Brandt U, Uribe S, Schägger H, Trumpower BL. Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex. J Biol Chem. 1994;269:12947–12953. [PubMed] [Google Scholar]
- 36.Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem Phys Lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cogliati S, et al. Mechanism of super-assembly of respiratory complexes III and IV. Nature. 2016;539:579–582. doi: 10.1038/nature20157. [DOI] [PubMed] [Google Scholar]
- 38.Milenkovic D, Blaza JN, Larsson NG, Hirst J. The enigma of the respiratory chain supercomplex. Cell Metab. 2017;25:765–776. doi: 10.1016/j.cmet.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Fedor JG, Hirst J. Mitochondrial supercomplexes do not enhance catalysis by quinone channeling. Cell Metab. 2018;28:525–531. doi: 10.1016/j.cmet.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letts JA, Sazanov LA. Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport cChain. Nat Struct Mol Biol. 2017;24:800–808. doi: 10.1038/nsmb.3460. [DOI] [PubMed] [Google Scholar]
- 41.Allen LA, Zhao X-J, Caughey W, Poyton RO. Isoforms of yeast cytochrome c oxidase subunit V affect the binuclear reaction center and alter the kinetics of interaction with the isoforms of yeast cytochrome c. J Biol Chem. 1995;270:110–118. doi: 10.1074/jbc.270.1.110. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 43.Meunier B, Maréchal A, Rich PR. Construction of histidine-tagged yeast mitochondrial cytochrome c oxidase for facile purification of mutant forms. Biochem J. 2012;444:199–204. doi: 10.1042/BJ20120116. [DOI] [PubMed] [Google Scholar]
- 44.Dodia R, Meunier B, Kay CWM, Rich PR. Comparisons of subunit 5A and 5B isozymes of yeast cytochrome c oxidase. Biochem J. 2014;464:335–342. doi: 10.1042/BJ20140732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 46.Heide H, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16:538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Guerrero-Castillo S, Cabrera-Orefice A, Huynen MA, Arnold S. Identification and evolutionary analysis of tissue-specific isoforms of mitochondrial complex I subunit NDUFV3. Biochim Biophys Acta. 2017;1858:208–217. doi: 10.1016/j.bbabio.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 51.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lange C, Hunte C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci USA. 2002;99:2800–2805. doi: 10.1073/pnas.052704699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.