Abstract
The molecular complexity of triple-negative breast cancers (TNBCs) provides a challenge for patient management. We set out to characterise this heterogeneous disease by combining transcriptomics and genomics data, with the aim of revealing convergent pathway dependencies with the potential for treatment intervention. A Bayesian algorithm was used to integrate molecular profiles in two TNBC cohorts, followed by validation using five independent cohorts (n = 1,168), including three clinical trials. A four-gene decision tree signature was identified which robustly classified TNBCs into six subtypes. All four genes in the signature (EXO1, TP53BP2, FOXM1 and RSU1) are associated with either genomic instability, malignant growth, or treatment response. One of the six subtypes, MC6, encompassed the largest proportion of tumours (~50%) in early diagnosed TNBCs. In TNBC patients with metastatic disease, the MC6 proportion was reduced to 25%, and was independently associated with a higher response rate to platinum-based chemotherapy. In TNBC cell line data, platinum-sensitivity was recapitulated, and a sensitivity to the inhibition of the phosphatase PPM1D was revealed. Molecularly, MC6-TNBCs displayed high levels of telomeric allelic imbalances, enrichment of CD4+ and CD8+ immune signatures, and reduced expression of genes negatively regulating the mitogen-activated protein kinase (MAPK) signalling pathway. These observations suggest that our integrative classification approach may identify TNBC patients with discernible and theoretically pharmacologically tractable features that merit further studies in prospective trials.
Keywords: Breast cancer, triple-negative breast cancer (TNBC), decision tree, integrative analysis
Introduction
Triple-negative breast cancers (TNBCs), defined by the lack of oestrogen receptor (ER), progesterone receptor (PgR) and human epidermal growth factor receptor 2 (HER2) expression, display remarkable molecular complexity and heterogeneous clinical behaviour (1). The overall prognosis of women with TNBC after metastatic relapse is significantly poorer when compared to that of women with other breast cancer subtypes (2). Chemotherapy remains the only systemic therapeutic approach for TNBC patients. Although subpopulations can be identified that are more responsive to chemotherapy, such as those with BRCA1 mutations, its effectiveness remains limited in an unselected TNBC population. Many agents are presently in clinical development, including PARP inhibitors and platinum salts (3–5), MEK inhibitors (6), and immunological agents (7). For some, predictive biomarkers have been recognised, while for others appropriate molecular features enabling optimal patient selection are still lacking (8,9).
Several studies have sought to take advantage of molecular profiling to classify TNBCs and subsequently provide clinically relevant information (10–13). One of the most recent classifications, TNBCtype-4, is based on gene expression and classifies TNBC into four subtypes. Among those, basal-like 1 (BL1) TNBCs were shown to achieve significantly higher pathological complete response (pCR) rates (49%) compared to all other subtypes (31%) when treated with neoadjuvant chemotherapy. However, classification approaches making use of multi-omics data are warranted to identify patients that may benefit from treatment beyond the standard-of-care (12).
TNBCs are characterised by extensive genomic instability (14,15). Measures capturing overall levels of genomic instability, like a chromosomal instability gene signature (CIN70), are informative for outcome prediction in ER-negative breast cancer patients (16). Similarly, genomic signatures, including Scars of Chromosomal Instability measures (SCINS) (17), Number of Telomeric Allelic Imbalance (NtAI) (18), the Homologous Recombination Deficiency (HRD) score (19), and HRDetect (20), are indicative of BRCAness and predictive of response to chemotherapy, particularly in the neoadjuvant setting (21). In metastatic TNBC trials (TNT (22) and TBCRC009 (23)), these methods were not able to identify patients specifically responding to platinum-based chemotherapy, suggesting some refinement of such approaches are required.
Here, we developed a four-gene decision tree signature based on integrated transcriptomics and genomics data that robustly classifies TNBC into 6 subtypes across 1,168 TNBCs. In TNBC patients with metastatic disease, our classification identified a subgroup of tumours sensitive to platinum-based chemotherapy. Molecular characteristics of this subgroup included increased number of allelic imbalanced aberrations in their telomeres, decreased inactivation of MAPK signalling, and enrichment of CD4+ and CD8+ immune signatures.
Materials and methods
Clinical sample data
TNBCs from the previously described Guy’s TNBC (E-MTAB-5270 and E-MTAB-2626) (24) were selected based on IHC status of HER2, ER and PgR. For 88 TNBCs, patient matched Affymetrix GeneChip Human Exon 1.0ST gene expression and Affymetrix SNP 6.0 copy number data were available.
METABRIC TNBC is a subset of the METABRIC study (EGAD00010000164) (10). Triple-negative status was based on IHC-assessed ER and HER2 status. Patient matched Illumina Human HT-12 v3 gene expression and Affymetrix SNP 6.0 copy number data were available for 112 TNBCs and processed, as reported previously (24).
A triple-negative subset of TCGA BRCA (25) was used as the TCGA TNBC cohort. For 95 TNBCs, defined by ER and HER2 status, gene expression was available on the Agilent 244K Custom Gene Expression array.
The TNBC616 cohort is a compilation of 24 different breast cancer cohorts (Supplementary Table S1). A total of 3,495 breast cancers were obtained, of which 616 were defined as triple-negative based on HER2, ER and PgR status. Details are provided in the online Supplementary Information.
Clinical trial data
The PrECOG 0105 cohort comprised 80 patients enrolled in a single-arm, phase II, early-stage study assessing the efficacy of neoadjuvant treatment using gemcitabine and carboplatin plus iniparib (26) (NCT00813956) (5). Response to treatment was stratified using the residual cancer burden (RCB) index (27). Patients with RCB > 1 were considered treatment-unresponsive; those with an RCB of 0 were deemed to have pCR, while those with an RCB of 1 had a partial response. Affymetrix HuGene 1.0st microarray gene expression was available and processed using the affy Bioconductor package (28) following RMA normalisation (29).
Sanofi Phase II (NCT00540358) (3) and Sanofi Phase III (NCT00938652) (4) are both two-arm clinical trials in TNBC patients with metastatic disease testing the efficacy of gemcitabine and carboplatin, with or without iniparib. A total of 123 and 519 patients in Phase II and Phase III, respectively, were randomly assigned to either arm. From the 74 (Phase II) and 319 (Phase III) patients for which gene expression data from the primary lesions were obtained, objective response was available for 61 and 224 patients. Gene expression was available on the Affymetrix Human Genome U133 Plus 2.0 microarray and was processed similarly to the PrECOG 0105 data.
Affymetrix SNP 6.0 copy number data
Raw Affymetrix SNP 6.0 data was normalised, allele-specific signal intensity measures were generated, and log R-ratio and B-allele frequencies were obtained using PennCNV-Affy (30,31). Allele specific copy number data was obtained using the ASCAT algorithm (32).
Cancer cell lines data
Transcriptomics and drug response data for TNBC cell lines (n = 16) was acquired from the Genomics of Drug Sensitivity in Cancer (GDSC) (33). The area under the dose response curve (AUC) values represent the relative sensitivity (low AUC) and resistance (high AUC) for each drug within each cell line. Associations between drug response and MC6 were assessed using Mann-Whitney U tests.
Statistical methods
Detailed information on the integrative analysis and statistical methods are described in the online Supplementary Information. In brief, genomics and transcriptomics data was integrated independently in Guy’s TNBC and METABRIC TNBC using the COpy Number and EXpression In Cancer (CONEXIC) algorithm (34). The identified four-gene signature required for TNBC subtyping was normalised before applied to additional cohorts.
Results
Integrative analysis of transcriptomics and genomics data identifies a four-gene decision tree signature
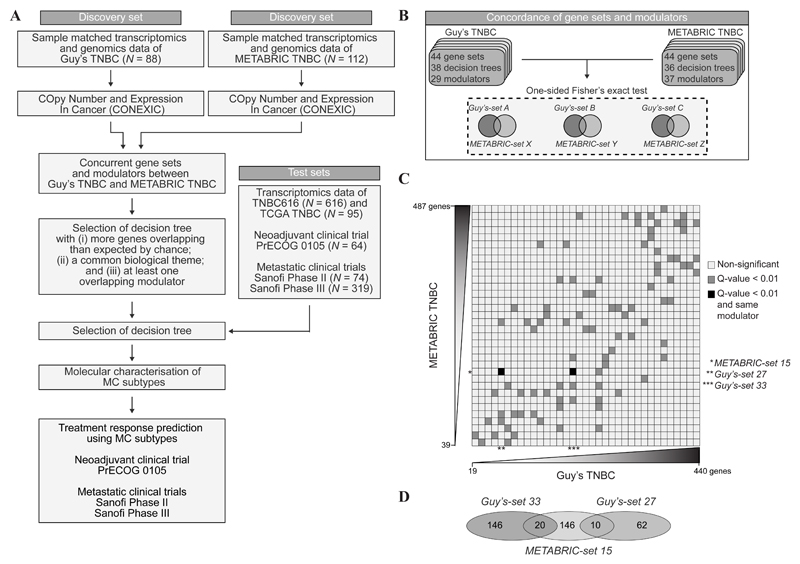
To determine if combined transcriptomics and genomics data could lead to a robust TNBC classification, we applied the previously published CONEXIC algorithm (34) to two independent TNBC cohorts; Guy’s TNBC (n = 88) (24) and TNBCs from the METABRIC study (METABRIC TNBC; n = 112) (10) (Figure 1A and Supplementary Table S2). CONEXIC constructs gene sets, defined as groups of genes which share similar expression patterns across all samples. As a next step, CONEXIC builds decision trees for each of these gene sets, using gene expression levels of copy number driven genes (modulators, copy number >=5). Within Guy’s TNBC and METABRIC TNBC, CONEXIC identified 38 and 36 gene sets with decision trees, respectively (Supplementary Tables S3-S6). We next sought to identify concurrent gene sets between the 38 Guy’s and 36 METABRIC gene sets by comparing all 1,368 possible gene set combinations for overlapping genes (Figure 1B) and identified 92 sets with statistically significant concurrent genes (Figure 1C, grey squares), including 3 gene sets that shared a common modulator in their respective decision trees, namely EXO1 (Figure 1C, black squares, and Figure 1D). In Guy’s-set 33 (Supplementary Table S7) and METABRIC-set 15 (Supplementary Table S8), genes involved in the inactivation of the mitogen-activated protein kinase (MAPK) pathway were significantly enriched. As no overlapping GO-terms were identified between Guy’s-set 27 (Supplementary Figure S9) and METABRIC-set 15, we focused on Guy’s-set 33 and METABRIC-set 15 (Supplementary Figure S1).
Figure 1.
Identification of concurrent cancer gene sets with CONEXIC. (A) Research design and workflow. After selecting a decision tree, a molecular characterisation of the subtypes was performed in two discovery and five validation sets. Treatment response to DNA damaging agents was assessed in three clinical trials. (B) The number of gene sets, decision trees and modulators identified by the CONEXIC algorithm in Guy’s TNBC and METABRIC TNBC. Overlap in gene set composition between the two cohorts was assessed using a one-sided Fisher’s exact test. (C) Level plot of concurrent gene sets between Guy’s TNBC and METABRIC TNBC. Dark grey boxes indicate gene sets with significantly overlapping genes. Black boxes, as dark grey boxes, in addition to overlapping modulators. Gene sets are ordered by size, as indicated by the scales. (D) Venn diagram depicting the number of common genes between Guy’s-set 27, Guy’s-set 33, and METABRIC-set 15.
A four-gene decision tree signature consistently classifies TNBCs into six subtypes in test cohorts
We applied both decision tree signatures to a total of 1,168 TNBCs, including (i) 95 TNBCs of TCGA BRCA (25); (ii) TNBC616 (see Supplementary Table S1 and online Supplementary Information); (iii) PrECOG 0105 (n = 64) (5), and both a phase II and a phase III trial in TNBC patients with metastatic disease; (iv) Sanofi Phase II (n = 74) (3) and (v) Sanofi Phase III (n = 319) (4) (Figure 1A). Transcriptomics data in each of the trials was derived from treatment naïve primary breast cancer tissue.
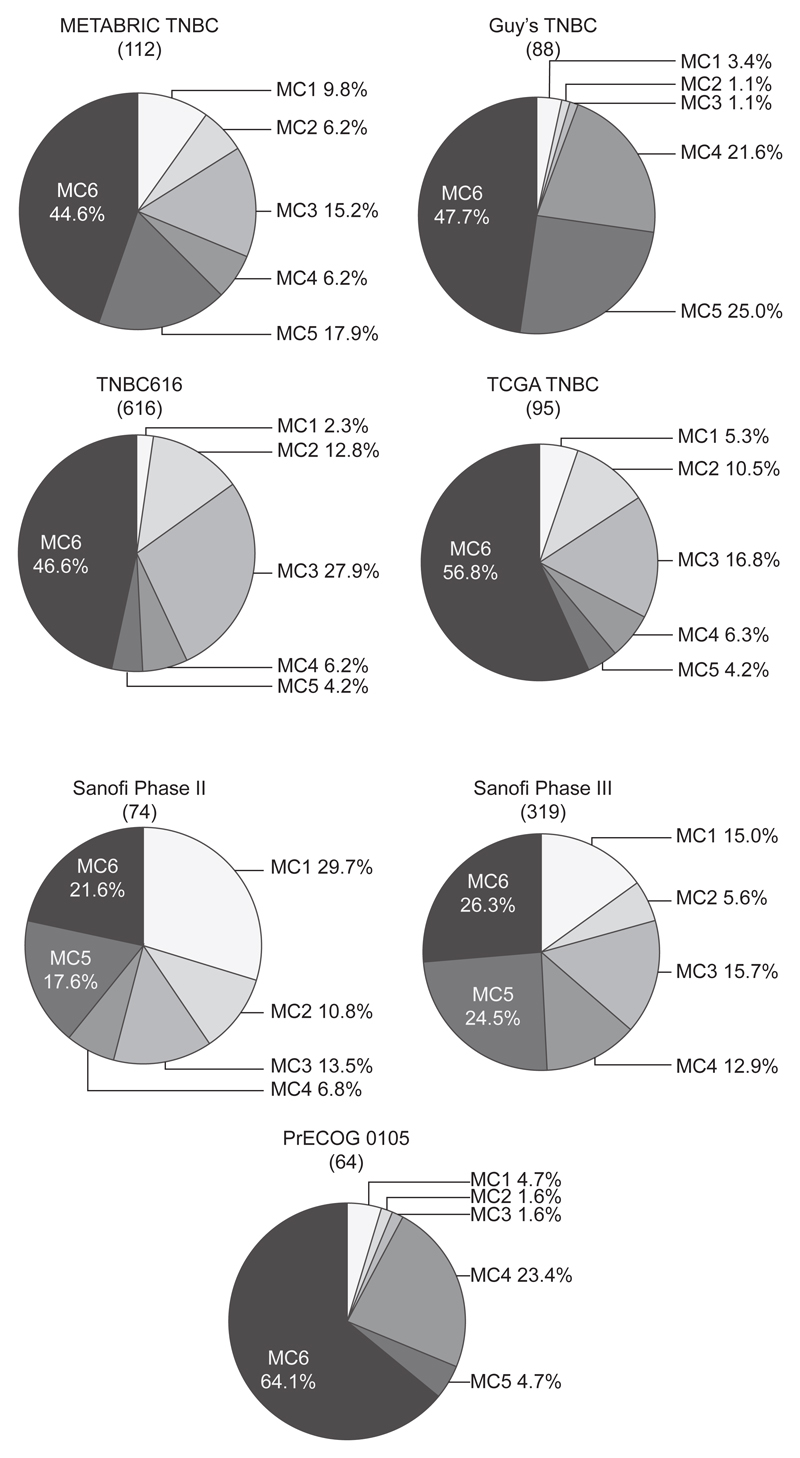
The classification defined by the decision tree of Guy’s-set 33, which is based on the expression levels of five genes (ST8SIA1, EXO1, NEK2, C8orf46 and MMS22L), nominated TNBCs in some cohorts, such as TCGA BRCA and Sanofi III, mostly as one class (Supplementary Figure S2) and could not be tested in TNBC616 as expression levels for MMS22L and C8orf46 were not available for assessment. Further investigations based on the Guy’s-set 33 decision tree were therefore not performed. In contrast, applying the METABRIC-set 15 decision tree signature based on the expression levels of four genes (TP53BP2, EXO1, FOXM1 and RSU1), established six subtypes, named MC1 to MC6, including one large subtype and five of varying sizes (Figure 2). The most frequent subtype (MC6) comprised 44.6%, 47.7%, 46.6% and 56.8% of all patients in METABRIC TNBC, Guy’s TNBC, TNBC616 and TCGA TNBC, respectively. In the PrECOG 0105 trial, the proportion of the MC6 subtype increased to 64%, whereas in TNBC patients with metastatic disease (Sanofi Phase II and III), its proportion reduced to ~24%. In contrast, the frequency of MC1-TNBCs increased from ~7.2% in the non-trial cohorts to ~22.4% in the metastatic setting. No difference in overall survival or risk of developing metastasis was observed when the four genes were tested separately in a univariate or multivariate Cox survival analysis, neither do the different MC subtypes demonstrate any association with increased or decreased overall and progression free survival or risk of developing metastasis (Supplementary Table S10-S13).
Figure 2.
TNBC cohorts classified using the METABRIC-set 15 four-gene decision tree signature. Pie charts illustrating the proportion of the MC subtypes in four primary invasive TNBC cohorts, namely METABRIC TNBC, Guy’s TNBC, TNBC616 and TCGA TNBC, as well as three clinical TNBC studies, including Sanofi Phase II, Sanofi Phase III and PrECOG 0105. The total number of tumours in each cohort is listed in brackets. The percentage for each subgroup is shown next to the respective pies.
Molecular characterisation of MC subtypes
MC subtypes are non-synonymous with other breast cancer classifications
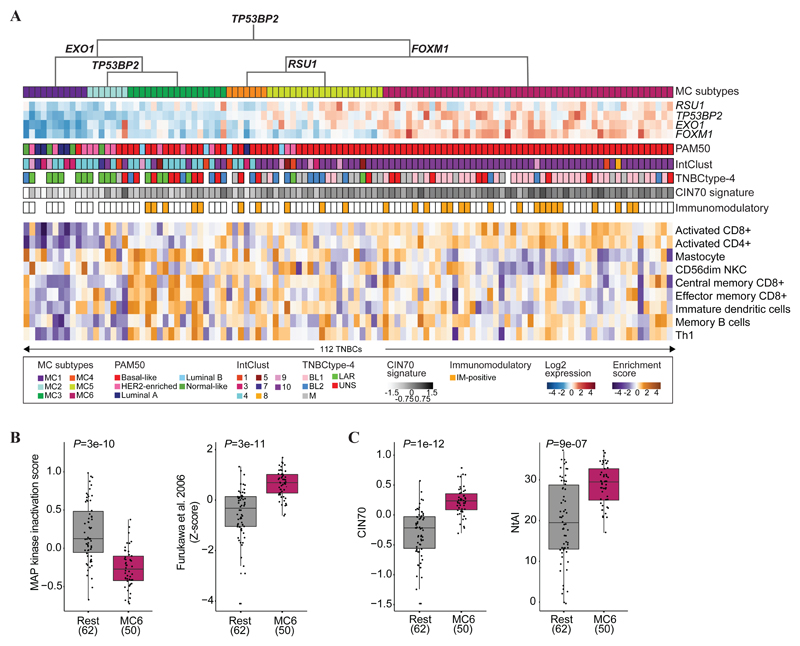
Currently, TNBCs are classified by TNBCtype-4 (12), the PAM50 subtypes (35–37) and the IntClust breast cancer classification (10). We sought to evaluate if the four-gene decision tree classification converges to these ones (Figure 3A and Supplementary Table S14 and S15). MC6-TNBCs consisted exclusively of PAM50 basal-like cases (100%, P = 2e-08, Fisher’s exact test), although only 57% of the basal-like TNBCs were classified as MC6. MC1-TNBCs were enriched for Luminal A (36%, P = 3e-04, Fisher’s exact test) and Normal-Like (36%, P = 3e-03, Fisher’s exact test), and MC2-TNBCs for HER2-enriched (57%, P = 2e-03, Fisher’s exact test). TNBCs of the remaining three MC subtypes fell across all PAM50 subtypes. Except for three TNBCs, MC6-TNBCs belonged to the IntClust 10 group (78%, P = 1e-02, Fisher’s exact test), whereas TNBCs of other MC subtypes were classified mostly as IntClust 4, 5, 3 and 1, although no significant enrichments were observed. MC6-TNBCs were found to be enriched for the BL1 subtype (61%, P = 1e-06, Fisher’s exact test), whereas MC1-TNBCs and MC2-TNBCs were primarily of the LAR subtype (86% and 67%, P = 1e-04 and 1e-02, respectively, Fisher’s exact test). Thus, the MC subtypes diverged from established breast cancer classifications
Figure 3.
Molecular characterisation of the MC subtypes in METABRIC TNBC. (A) The four-gene decision tree underlying the MC subtypes is shown, followed by a heat map representing the expression levels of the four modulatory genes. The coloured bars represent sample-specific characteristics, including PAM50, IntClust, TNBCtype-4, CIN70 signature and the immunomodulatory status. The heatmap at the bottom illustrates the enrichment of immune gene signatures. (B) (left) MAPK inactivation scores, calculated by summarising the expression levels of DUSP4, DUSP5, DUSP6, DUSP10 and SPRED2, in MC6-TNBCs (magenta) and the remaining TNBCs (grey). Significance was assessed using Wilcoxon rank-sum tests. (right) Z-scores for the DUSP6 gene set were obtained using ssGSEA. Significance was assessed using a Wilcoxon rank-sum test. (C) Boxplots displaying the CIN70 signature (left) and NtAI (right) in MC6-TNBCs (magenta) and the remaining TNBCs (grey). Significance was assessed using Wilcoxon rank-sum tests.
MC subtypes differ in their immune gene enrichment
Based on Lehmann’s immunomodulatory (IM) classification (12), 37% of the MC6-TNBCs were IM-positive, MC1- and MC2-TNBCs were exclusively IM-negative, and MC3-, MC4- and MC5-TNBCs were mostly IM-negative (74%) (Figure 3A). Next, we performed a gene set enrichment analysis (38) on gene expression data from METABRIC TNBC to further deconvolute immune cell enrichment across the MC subtype. Using 28 different immune gene sets (39), activated CD4+ and CD8+ immune signatures were found enriched in MC6-TNBCs (Q = 3e-08, and 1e-04, respectively, Mann-Whitney U test), and low in MC1-TNBCs (Q = 2e-05 and 1e-05, Mann-Whitney U test) (Figure 3A and Supplementary Table S16). MC6-TNBC had lower mastocytes and CD56dim natural killer cell activation in comparison to other MC subtypes (Q = 5e-03, and 3e-02, respectively, Mann-Whitney U test). In MC3-TNBCs, increased central memory CD8+, effector memory CD8+ and immature dendritic cells expression levels were observed (Q = 4e-02, 4e-02 and 4e-02, respectively, Mann-Whitney U test). MC2-TNBCs had reduced expression of the memory B cell signature (Q = 4e-02, Mann-Whitney U test), whereas in MC1-TNBCs, Th1 expression was reduced (Q = 4e-02, Mann-Whitney U test). This clearly demonstrates a variety of immune cell features across the MC subtypes.
Pathway deregulation in MC subtypes
DUSP4, DUSP5, DUSP6, DUSP10 and SPRED2 were amongst the METABRIC-set 15 gene set (Figure 1D and Supplementary Table S8) and are involved in the negative regulation of the MAPK signalling pathway. We investigated their expression levels across the MC subtypes and found that MC6-TNBCs had the lowest expression of these genes in comparison to the other subtypes (P < 0.05, Mann-Whitney U test) (Figure 3B and Supplementary Figure S3A). Genomic regions encompassing DUSP4, DUSP5 and DUSP6 were deleted (Supplementary Table S17), potentially resulting in decreased expression levels. To further corroborate the lack of negative regulation of MAPK inactivation, a set of genes previously reported under control of this inhibitory mechanism (40) was interrogated in the MC subtypes and found increased in MC6-TNBCs compared to TNBCs of other MC subtypes (P < 0.05, Mann-Whitney U test) (Figure 3B and Supplementary Figure S3B). These results demonstrate the highly selective features of MC6-TNBCs.
In MC5-TNBCs, genes highly expressed were enriched for ErbB signalling (Q = 6e-03, Hypergeometric test), particularly through the epidermal growth factor receptor pathway (P = 1e-02, Fisher’s exact test). MC4-TNBCs were enriched for PI3K/Akt signalling (Q = 5e-03, Hypergeometric test) and DNA replication (P = 2e-02, Fisher’s exact test). Although not enriched for Lehmann’s IM classification, many genes upregulated in MC3-TNBCs are involved in innate and adaptive immune response (P = 7e-04 and 1e-03, respectively, Fisher’s exact test), and chemokine signalling (Q = 3e-03, Hypergeometric test). MC2-TNBCs were enriched for metabolic processes (Q = 2e-28, Hypergeometric test), and MC1-TNBCs for steroid hormone mediated signalling (P = 3e-02, Fisher’s exact test), particularly oestrogen and androgen receptor signalling (P = 7e-04 and 2e-02, respectively, Fisher’s exact test). The unique pathway activities underlying the MC subtypes further demonstrate the molecular complexity of TNBC and further strengthen our TNBC classification.
Increased levels of telomeric allelic imbalanced aberrations in MC6-TNBCs
Next, we investigated the levels of genomic instability. MC6-TNBCs showed significantly higher levels of chromosomal instability (P < 0.05, Mann-Whitney U test) (Figure 3C and Supplementary Figure S3C) as defined by the CIN70 gene expression signature (16,41). MC1-TNBCs had the lowest levels of chromosomal instability. To further decipher this genomic instability, we investigated diverse genomic instability measurements based on copy number data (17,18). MC6-TNBCs displayed a high burden of allelic imbalanced aberrations in their telomeres (P = 9e-07, Mann-Whitney U test) (Figure 3C). MC5-TNBCs exhibited a medium burden. In contrast, the remaining MC subtypes did not harbour specific types of copy number aberrations.
MC6 identifies TNBCs responsive to platinum-based chemotherapy in Sanofi Phase III clinical trial
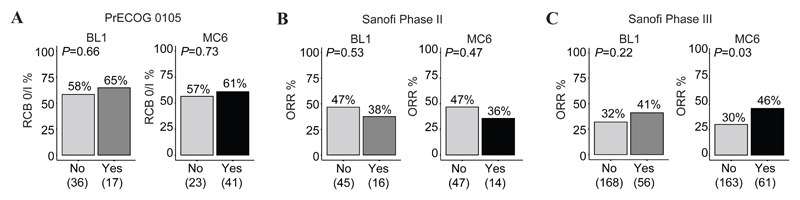
Given the selective features of MC6-TNBCs, we next assessed whether this subtype carried any predictive value in treatment response to DNA damaging agents. Using three clinical trials (Figure 1A), TNBCs from each cohort were dichotomised as either being MC6-TNBCs or non-MC6 (referred to as remaining TNBCs). We included the TNBCtype-4 classification as a comparator, as BL1-TNBCs were previously shown to be responsive to neoadjuvant chemotherapy (12). In the neoadjuvant PrECOG 0105 trial, the RCB 0/I pathological response rate in BL1-TNBCs (11/17, 65%) was similar to that of MC6-TNBCs (25/41, 61%) (Figure 4A). However, out of the 64 patients enrolled in the PrECOG 0105 trial, the MC6 subtype identified 39% of the responders with an accuracy of 55%, in contrast to the BL1 subtype, identifying 17%, with an accuracy of 41%. In the metastatic TNBC Sanofi Phase II trial, neither the BL1 nor the MC6 subtype was predictive of the overall response rate (ORR) (Figure 4B and Supplementary Table S18). However, in the metastatic TNBC Sanofi Phase III trial, the MC6 subtype was a significant predictor of treatment response in a multivariate model (OR = 2.41, CI = 1.01 to 5.81, P < 0.05), with an ORR in MC6-TNBCs of 46% (28/61), compared to an ORR of 30% (49/163) in the remaining TNBCs (Figure 4C). The ORR in BL1-TNBCs was 41% (23/56), as compared with 32% (54/168) in the remaining TNBCs. In contrast to BL1, the MC6 subtype did reach significance in the univariate and in the multivariate model (OR = 1.78, CI = 0.76 to 4.19, P = 0.18) (Supplementary Table S19).
Figure 4.
Response rates for the MC6 and BL1 subtypes in clinical trials. (A) Response to neoadjuvant platinum-based chemotherapy, as assessed by the RCB index, in BL1-TNBCs and MC6-TNBCs obtained from the PrECOG 0105 clinical trial. (B) ORR in Sanofi Phase II and (C) Sanofi Phase III. Response rates are dichotomised by being BL1 or not (left), and MC6 or not (right). Subtypes were assessed as predictors of treatment response using a multivariate logistic regression, by including age and race for Sanofi II, and age, race, and grade for Sanofi III into the models.
MC6 cell lines are sensitive to cisplatin
To test the hypothesis that MC6-TNBCs are responsive to platinum-based chemotherapeutics, we exploited drug sensitivity profiles of 16 TNBC tumour cell lines generated as part of the GDSC project (33). We found that the median AUC of cisplatin in the MC6 TNBC cell lines (n = 4) was 0.89, compared to 0.92 in cell lines from other MC subtypes (n = 11) (P = 5.6e-02, Mann-Whitney U test), suggesting an enhanced sensitivity to platinum salts in the MC6 subtype. With the aim of identifying novel alternative therapeutic sensitivities, we also assessed the associations between >200 GDSC drug response profiles and the MC6 subtype (Supplementary Table S20). We found that amongst the GDSC drug sensitivity profiles, MC6 TNBC cell lines displayed the greatest sensitivity to CCT007093 (42), a small molecule inhibitor of the DNA-damage activated phosphatase, PPM1D (43) (P = 7.7e-03, Mann-Whitney U test, Supplementary Figure S4).
Discussion
With the purpose of classifying molecular heterogeneous TNBCs into clinically relevant subtypes, our multi-omics integrative approach differs from previously published methods (10–13). Our classification was constructed using data exclusively from TNBCs, strengthening the exploration of their molecular complexity in more detail. Robustness of our approach was increased by applying the CONEXIC algorithm (34) to two independent TNBC cohorts to then identify concurrent gene sets and modulatory genes. Guy’s-set 33 and METABRIC-set 15 shared similarities in both their gene set and modulatory genes. The latter represented well known cancer-associated genes potentially contributing to tumour progression, and thereby substantiating our analytical approach to identify candidate drivers with effects on gene expression patterns. However, upon validation of both decision trees, only METABRIC-set 15 was taken forward due to the consistently reproducible MC subtypes. Of note, the molecular features of primary tumours of patients that will be developing metastasis seem to be distinct, as was reflected by the change in proportion of the MC1 and MC6 subtype between non-trial genomic and metastatic TNBC cohorts. The MC6 subtype identified metastatic TNBC patients who showed improved response to platinum-based chemotherapy in the Sanofi Phase III, but not in the neoadjuvant PrECOG 0105 trial (5). In early stage TNBCs, which are more likely to respond to platinum-based chemotherapy, the MC6 subtype is not able to differentiate between those that do, and do not respond to this treatment. However, in the metastatic setting, where patients are less responsive to platinum-based chemotherapy, MC6 appears to be more discriminative as a predictor. In contrast, genomic signatures lack predictive value in the metastatic setting (22,23).
The four-gene decision tree signature consists of TP53BP2, RSU1, FOXM1 and EXO1. The expression of TP53BP2, a known regulator of apoptosis and cell growth, has been reported to be copy number dependent, and associated with poor response to chemotherapy in TNBC (14). RSU1, Ras suppressor-1, is involved in the RAS signal transduction pathway and was proposed as a biomarker for metastasis in breast cancers (44). The proto-oncogenic transcription factor FOXM1 has frequently been shown to mediate cell proliferation, survival, migration, progression and tumourigenesis in TNBC (45); can modulate cisplatin sensitivity by regulating the expression of EXO1 in ovarian cancer (46); and is part of a recently identified KRAS-associated signature in colorectal cancer (47). All four genes provide biological rationales for being good candidates for classification approaches.
To further substantiate the finding that MC6-TNBCs were more sensitive to platinum-based chemotherapeutics, cisplatin drug response profiles were assessed across 16 TNBC cell lines categorised by MC subtypes. In line with findings from Sanofi Phase III, MC6 cell lines appeared overall more sensitive to cisplatin than those of other subtypes. In addition, MC6 TNBC cell lines were found to exhibit enhanced sensitivity to CCT007093, a chemical inhibitor of PPM1D (42). In parallel, MC6 TNBC cell lines displayed lower expression of DUSP10 and SPRED2 as a part of the MAPK inactivation signature; both are negative regulators of p38. Loss of DUSP4, DUSP5 and DUSP6 in TNBC has been previously reported by others (48–50). Thus, we hypothesise that the sensitivity to CCT007093 may be a result of p38-dependent cell death, thereby pointing to a potential PPM1D dependency for this group of TNBCs. Given that PPM1D modulates the activity of a series of substrates including p38, ATM, CHK1 and CHK2 (43), this association further demonstrates the connectivity between genomic instability and TNBC.
The proposed decision tree is an ideal classification approach to be performed with relative ease using other platforms, including qPCR and NanoString, due to the small number of genes required. However, there are some limitations, as in its current form, the four-gene decision tree signature relies on the distribution of each of the four genes and careful standardisations experiments would need be implemented. Further validation of our classification across independent clinical trials are warranted to investigate if association with treatment effect to platinum-based chemotherapy is specific or rather reflects a combinatorial effect with gemcitabine in Sanofi Phase III.
The management of TNBC, especially in the metastatic setting, can be complex. With single-agent chemotherapy still considered the standard-of-care, targeted therapeutic strategies are required, which will rely on appropriate biomarkers for optimal patient selection. The unique molecular features of the MC subtypes reflect the intrinsic heterogeneity of TNBC and revealed targetable pathways, such as the p38 MAPK signalling pathway in MC6-TNBC. The same subset also showed elevated levels of genomic instability in telomeric regions, a type of genomic instability which may be the result of escaping from telomere crisis. EXO1, a modulatory gene, may further contribute to this process (51,52). Further characterisations of the MC subtypes are warranted to establish their association with genomic signatures from whole-genome sequence data (53,54).
In conclusion, we sought to decipher the complex nature of TNBC, with the goal of informing patient selection for current and future treatment strategies. We showed that a four-gene decision tree signature based on copy number dependent genes classifies TNBCs into six subtypes. Given the current lack of selection criteria for TNBC patients with metastatic disease, this classification warrants further testing in randomised metastatic TNBC trials, such as TNT (22).
Supplementary Material
Acknowledgements
Patient samples and data were provided by King’s Health Partners Cancer Biobank, London, UK, which is supported by the Experimental Cancer Medicine Centre at King’s College London and the Department of Health via the National Institute for Health Research Comprehensive Biomedical Research Centre award. This paper represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London, and Cancer Research UK King's Health Partners Centre at King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. We thank Assistant Professor Uri David Akavia for his help with CONEXIC and Dr Syed Haider for critical reading and suggestions.
Financial information: This work was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas (Jelmar Quist); Breast Cancer Now Programme Grants at King’s College London (Hasan Mirza, Andrew N. J. Tutt and Anita Grigoriadis) and The Institute of Cancer Research (Christopher J. Lord); and Cancer Research UK King's Health Partners Centre at King’s College London.
Footnotes
Conflict of interest disclosure statement: The authors declare no conflict of interest.
References
- 1.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–90. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68(9):3108–14. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 3.O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364(3):205–14. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 4.O'Shaughnessy J, Schwartzberg L, Danso MA, Miller KD, Rugo HS, Neubauer M, et al. Phase III study of iniparib plus gemcitabine and carboplatin versus gemcitabine and carboplatin in patients with metastatic triple-negative breast cancer. J Clin Oncol. 2014;32(34):3840–7. doi: 10.1200/JCO.2014.55.2984. [DOI] [PubMed] [Google Scholar]
- 5.Telli ML, Jensen KC, Vinayak S, Kurian AW, Lipson JA, Flaherty PJ, et al. Phase II Study of Gemcitabine, Carboplatin, and Iniparib As Neoadjuvant Therapy for Triple-Negative and BRCA1/2 Mutation-Associated Breast Cancer With Assessment of a Tumor-Based Measure of Genomic Instability: PrECOG 0105. J Clin Oncol. 2015;33(17):1895–901. doi: 10.1200/JCO.2014.57.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infante JR, Papadopoulos KP, Bendell JC, Patnaik A, Burris HA, 3rd, Rasco D, et al. A phase 1b study of trametinib, an oral Mitogen-activated protein kinase kinase (MEK) inhibitor, in combination with gemcitabine in advanced solid tumours. Eur J Cancer. 2013;49(9):2077–85. doi: 10.1016/j.ejca.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016;34(21):2460–7. doi: 10.1200/jco.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28(7):1145–53. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 10.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One. 2016;11(6):e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18(2):123–33. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lips EH, Michaut M, Hoogstraat M, Mulder L, Besselink NJ, Koudijs MJ, et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. 2015;17(1):134. doi: 10.1186/s13058-015-0642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71(10):3447–52. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watkins J, Weekes D, Shah V, Gazinska P, Joshi S, Sidhu B, et al. Genomic Complexity Profiling Reveals That HORMAD1 Overexpression Contributes to Homologous Recombination Deficiency in Triple-Negative Breast Cancers. Cancer Discov. 2015;5(5):488–505. doi: 10.1158/2159-8290.CD-14-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birkbak NJ, Wang ZC, Kim JY, Eklund AC, Li Q, Tian R, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2(4):366–75. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107(10):1776–82. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23(4):517–25. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol. 2014;15(7):747–56. doi: 10.1016/S1470-2045(14)70160-3. [DOI] [PubMed] [Google Scholar]
- 22.Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24(5):628–37. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isakoff SJ, Mayer EL, He L, Traina TA, Carey LA, Krag KJ, et al. TBCRC009: A Multicenter Phase II Clinical Trial of Platinum Monotherapy With Biomarker Assessment in Metastatic Triple-Negative Breast Cancer. J Clin Oncol. 2015;33(17):1902–9. doi: 10.1200/JCO.2014.57.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braso-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22(11):1303–13. doi: 10.1038/nm.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendeleyev J, Kirsten E, Hakam A, Buki KG, Kun E. Potential chemotherapeutic activity of 4-iodo-3-nitrobenzamide. Metabolic reduction to the 3-nitroso derivative and induction of cell death in tumor cells in culture. Biochem Pharmacol. 1995;50(5):705–14. doi: 10.1016/0006-2952(95)00189-7. [DOI] [PubMed] [Google Scholar]
- 27.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25(28):4414–22. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 28.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36(19):e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107(39):16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–61. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143(6):1005–17. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 37.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furukawa T, Kanai N, Shiwaku HO, Soga N, Uehara A, Horii A. AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene. 2006;25(35):4831–9. doi: 10.1038/sj.onc.1209494. [DOI] [PubMed] [Google Scholar]
- 41.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 42.Rayter S, Elliott R, Travers J, Rowlands MG, Richardson TB, Boxall K, et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27(8):1036–44. doi: 10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 43.Oghabi Bakhshaiesh T, Majidzadeh AK, Esmaeili R. Wip1: A candidate phosphatase for cancer diagnosis and treatment. DNA Repair (Amst) 2017;54:63–6. doi: 10.1016/j.dnarep.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Giotopoulou N, Valiakou V, Papanikolaou V, Dubos S, Athanassiou E, Tsezou A, et al. Ras suppressor-1 promotes apoptosis in breast cancer cells by inhibiting PINCH-1 and activating p53-upregulated-modulator of apoptosis (PUMA); verification from metastatic breast cancer human samples. Clin Exp Metastasis. 2015;32(3):255–65. doi: 10.1007/s10585-015-9701-x. [DOI] [PubMed] [Google Scholar]
- 45.Hamurcu Z, Ashour A, Kahraman N, Ozpolat B. FOXM1 regulates expression of eukaryotic elongation factor 2 kinase and promotes proliferation, invasion and tumorgenesis of human triple negative breast cancer cells. Oncotarget. 2016;7(13):16619–35. doi: 10.18632/oncotarget.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Wang Y, Wang Y, Yin X, He Y, Chen L, et al. FOXM1 modulates cisplatin sensitivity by regulating EXO1 in ovarian cancer. PLoS One. 2014;9(5):e96989. doi: 10.1371/journal.pone.0096989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pek M, Yatim S, Chen Y, Li J, Gong M, Jiang X, et al. Oncogenic KRAS-associated gene signature defines co-targeting of CDK4/6 and MEK as a viable therapeutic strategy in colorectal cancer. Oncogene. 2017;36(35):4975–86. doi: 10.1038/onc.2017.120. [DOI] [PubMed] [Google Scholar]
- 48.Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med. 2012;18(7):1052–9. doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4(2):232–45. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craig DW, O'Shaughnessy JA, Kiefer JA, Aldrich J, Sinari S, Moses TM, et al. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol Cancer Ther. 2013;12(1):104–16. doi: 10.1158/1535-7163.MCT-12-0781. [DOI] [PubMed] [Google Scholar]
- 51.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18(3):175–86. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue Y, Marvin ME, Ivanova IG, Lydall D, Louis EJ, Maringele L. Rif1 and Exo1 regulate the genomic instability following telomere losses. Aging Cell. 2016;15(3):553–62. doi: 10.1111/acel.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.