Abstract
Background
Computed Tomography (CT) and spirometry are the current standard methods for assessing lung anatomy and pulmonary ventilation, respectively. However, CT provides limited ventilation information and spirometry only provides global measures of lung ventilation. Thus, a method that can enable simultaneous examination of lung anatomy and ventilation is of clinical interest.
Purpose
To develop and test a 4D respiratory-resolved sparse lung MRI (XD-UTE: eXtra-Dimensional Ultra-short TE imaging) approach for simultaneous evaluation of lung anatomy and pulmonary ventilation.
Study Type
Prospective
Population
23 subjects (11 volunteers and 12 patients, mean age=63.6±8.4).
Field Strength/Sequence
3T MR; a prototype 3D golden-angle radial UTE sequence, a Cartesian breath-hold volumetric-interpolated examination (BH-VIBE) sequence.
Assessment
All subjects were scanned using the 3D golden-angle radial UTE sequence during normal breathing. 10 subjects underwent an additional scan during alternating normal and deep breathing. Respiratory-motion-resolved sparse reconstruction was performed for all the acquired data to generate dynamic normal-breathing or deep-breathing image series. For comparison, BH-VIBE was performed in 12 subjects. Lung images were visually scored by three experienced chest radiologists and were analyzed by two observers who segmented the left and right lung to derive ventilation parameters in comparison with spirometry.
Statistical Tests
Nonparametric paired two-tailed Wilcoxon signed-rank test; Intra-class correlation coefficient, Pearson correlation coefficient.
Results
XD-UTE achieved significantly improved image quality compared both to Cartesian BH-VIBE and to radial reconstruction without motion compensation (P<0.05). The global ventilation parameters (a sum of the left and right lung measures) were in good correlation with spirometry in the same subjects (correlation coefficient=0.724). There were excellent correlations between the results obtained by two observers (intra-class correlation coefficient ranged from 0.8855 to 0.9995).
Conclusion
Simultaneous evaluation of lung anatomy and ventilation using XD-UTE is demonstrated, which have shown good potential for improved diagnosis and management of patients with heterogeneous lung diseases.
Keywords: XD-UTE, XD-GRASP, lung MRI, lung ventilation, free breathing, golden-angle radial
Introduction
Computed Tomography (CT) and spirometry are currently the standard methods for assessing lung anatomy and pulmonary ventilation, respectively. On the one hand, CT offers excellent spatial morphologic information, but it requires radiation exposure and provides limited ventilation information. On the other hand, pulmonary function tests, measured by a spirometer, are often performed to make a diagnosis of airway obstruction. However, these tests only provide global measures of lung ventilation. Without spatially-localized information concerning regional ventilation, a standard pulmonary function test is inadequate for assessing disease heterogeneity (1).
MRI is a promising imaging modality for simultaneous evaluation of lung anatomy and pulmonary function. Compared to CT, MRI has several attractive features, such as the lack of radiation exposure, superior soft-tissue contrast, and the possibility for a multi-contrast functional evaluation. Compared to spirometry, MRI can offer spatially-resolved ventilation information for evaluation of disease heterogeneity. However, despite encouraging results demonstrated in many studies, a vast majority of lung MR exams have been performed in a research environment only. Major challenges associated with lung MRI include low proton density in the inflated lung that leads to low signal-to-noise ratio (SNR) (2), the short T2* of the lung parenchyma resulting from numerous air-tissue interfaces (2,3), long acquisition times, and artifacts from respiration and heart motion.
Multiple approaches can be used to improve the imaging performance of lung MRI. For example, MRI using hyperpolarized gases is one of the most useful approaches to visualize lung airspaces (4,5) and to assess lung function (6,7). With gaseous contrast agents, such as helium-3 or xenon-129, the low signal due to low proton density of the lung can be overcome by using an external polarization device, thus offering a unique opportunity for a comprehensive evaluation of different lung diseases. In addition to hyperpolarized gases, several recent studies have also shown that MRI with additional oxygen enhancement can further improve the imaging of ventilation for assessment of lung function (8,9). MR sequences with an ultra-short echo time (UTE sequences) are another effective way of imaging lung structure (10–13). Although UTE-MRI cannot directly image the lung airspace as in hyperpolarized MRI, it enables visualization of short T2* components in the lung with increased signal intensity. For this reason, UTE-MRI may be a more attractive technique for the screening of lung abnormalities, such as lung nodules or cystic fibrosis (14,15), particularly given the increased cost of gaseous contrast agents and the complex imaging procedure associated with hyperpolarized MRI.
Despite recent advances in imaging techniques, respiratory motion remains a major challenge for lung MRI. Although external respiratory monitoring devices (e.g., a respiratory bellow) can be used to track respiratory motion for gated data acquisitions (16), respiratory gating reduces scan efficiency and requires a cumbersome setup procedure prior to the start of imaging. This issue has been addressed by self-gating, and many works have shown that the imaging performance of lung MRI can be increased with retrospective respiratory-gating, where a respiratory motion signal is obtained from acquired data (17–20). Sparse imaging (e.g., rapid MRI exploiting image sparsity) is an alternative approach to handle respiratory motion. Although it was originally introduced to MRI for accelerated data acquisitions (21), it has been shown that sparsity can also be leveraged to reconstruct extra motion dimensions, using sparsity-enforcing reconstruction algorithms to fill in missing data in continuously acquired datasets sorted into distinct motion states (22). Thus, motion compensation can be performed intrinsically during image reconstruction, and more importantly, the new motion dimensions can also provide additional physiological information of potential clinical value. This relatively new imaging technique, called XD-GRASP (eXtra-Dimensional Golden-angle RAdial Sparse Parallel MRI), combines the self-navigation and motion-sorting properties of radial sampling with the acceleration capability of sparsity-based reconstruction (22). Since its original development, XD-GRASP has been tested in several clinical applications including liver imaging (23) and cardiovascular imaging (24–26).
The feasibility of extending motion-resolved sparse reconstruction to 4D time-resolved lung imaging, by combining the XD-GRASP approach with a self-navigated 3D golden-angle radial UTE sequence, was demonstrated in a preliminary study with good imaging performance (27). In a recent study, Jiang and colleagues have also applied the idea of motion-resolved sparse imaging to UTE-MRI of the lung, and have tested its performance in assessing various lung diseases (28). However, both of these two studies aimed to reconstruct high-resolution, high-quality anatomical lung images only. Tibiletti et al have also demonstrated the performance of XD-GRASP for lung imaging in small animal MRI (29), but it is not trivial to study ventilation of the lung in animal. In this study, we aimed to move one step further to extend this framework, so that it can be used for assessing not only lung anatomy, but also pulmonary ventilation in clinical studies. Our hypothesis was that this new framework, termed as XD-UTE in this work, can not only serve as a potential alternative to both CT and spirometry for lung examination, but provide further insight into the heterogeneity of lung ventilation.
Methods
Subjects
This prospective study was HIPAA-compliant, and was approved by our institutional review board. A total of 23 subjects, including 12 patients with known lung diseases (7 male, 5 female, mean age=63.6±8.4) and 11 healthy adult volunteers (6 male, 5 female, mean age=33.5±11.7), were recruited to participate. The patients were recruited only if they were able to cooperate with the breathing instructions designed for this study. Written, informed consent was obtained from all subjects prior to the MR scans.
Imaging Sequence
A prototype golden-angle radial imaging sequence that employs center-out half-spoke 3D radial acquisitions with ramp sampling was used for data acquisitions (13). The sequence was adapted from the self-navigating dual-echo UTE sequence described in (13) to a single-echo acquisition scheme, simply by disabling the acquisition of the second echo. Two-fold (2x) oversampling was employed in each readout without increasing scan time. As shown in Figure 1a, the golden-angle sampling trajectory is segmented into multiple interleaves that are rotated by a golden angle based on a spiral phyllotaxis pattern (30). Each Interleaf consistently starts with a measurement oriented along the superior-inferior (SI) direction (red lines in Figure 1a) for self-navigation, and is preceded by CHESS fat saturation. Imaging was performed on a clinical 3.0 T MR scanner (MAGNETOM Prisma, Siemens Healthineers, Germany) without external gating or triggering.
Figure 1.
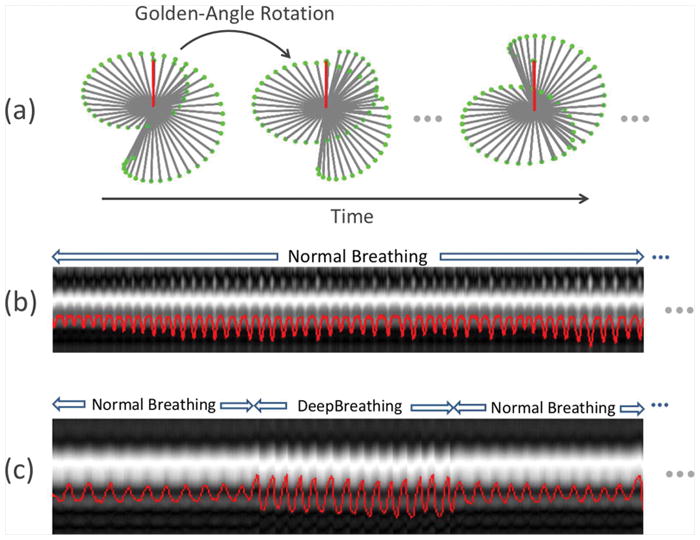
(a) Sampling trajectories for the self-navigated 3D golden-angle UTE radial sequence, in which data acquisition is segmented into multiple interleaves that rotate by a golden angle. Half-spokes are acquired in a center-out fashion to achieve an ultra-short echo time. Each interleaf starts with a projection oriented along the superior-inferior direction (red lines) for self-navigation. (b-c) Two exemplary respiratory motion signals superimposed on their corresponding projection profiles, one for the NB-UTE acquisition (b), and the other for the NDB-UTE acquisition alternating between normal and deep breathing (c).
Data Acquisitions
Free-Breathing UTE MR Data Acquisition
Two types of UTE MR acquisitions were performed. The first occurred during normal breathing (referred to as NB-UTE hereafter) and this was performed in all the 23 subjects. The second occurred during alternating normal and deep breathing (referred to as NDB-UTE hereafter) and was performed in 10 subjects (7 volunteers and 3 patients). During the NB-UTE scan, the subjects were asked to breathe normally and consistently. Relevant imaging parameters set in the scanner console included: TR/TE=3.3/0.05ms, FOV=250×250×250mm3 (500×500×500mm3 with 2x oversampling in 3D radial imaging), readout points=256 (512 with 2x oversampling), matrix size=256×256×256 (512×512×512 with 2x oversampling in 3D radial imaging), acquired voxel size=~0.98×0.98×0.98mm3, RF excitation angle=6°, and bandwidth=600Hz/Pixel. A total of 122,520 half-spokes were acquired in each subject in 8 minutes, including 2042 golden-angle interleaves with 60 spokes each.
For the NDB-UTE scan, 10 subjects were trained, before scanning, to follow a special breathing pattern alternating between normal and deep breathing. Specifically, through a monitor set up for functional MRI studies, subjects were asked to breathe normally for one minute, followed by a consistent deep breathing maneuver for another one minute, during which they were instructed to inhale as deeply as possible then exhale as rapidly as possible for multiple times. These one-minute-normal and one-minute-deep breathing cycles were repeated 4 times, leading to a total of 8-minute scan time. Relevant imaging parameters of the NDB-UTE acquisitions were the same as in the NB-UTE case, except for a smaller matrix size of 128×128×128 (256×256×256 with 2x oversampling in 3D radial imaging) and thus a lower spatial resolution (~1.95×1.95×1.95mm3). NDB-UTE acquisitions were acquired with a reduced spatial resolution because of the need to reconstruct a high-temporal-resolution image series (see below), in which spatial morphological visualization is less important.
Breath-Hold MR Data Acquisition
Conventional fat-saturated Volumetric Interpolated Breath-Hold Examination (BH-VIBE) was performed for comparison in 7 volunteers and 5 patients, using a routine clinical imaging protocol in an axial orientation. Relevant imaging parameters included: in-plane FOV=350×350mm2, in-plane matrix=166×256, voxel size=2.1×1.4mm2, reconstructed slice thickness=3mm, and TR/TE=2.8–3.1ms/1.25–1.41ms. BH-VIBE images were acquired with a lower spatial resolution because of the need to acquire each data set covering the whole lung during one breath hold (~15 seconds), which necessitated a trade-off between resolution and volumetric coverage.
Spirometry Measurement
Spirometry measurements were performed in 6 subjects (4 volunteers and 2 patients) out of the 10 subjects who underwent the NDB-UTE scan. The test was performed using a conventional spirometer (FlowMIR, MIR USA, Inc.) in a sitting position. The subjects were asked to follow a breathing pattern similar to what they had during the NDB-UTE MRI scans.
XD-UTE Reconstruction
Respiratory Motion Detection
A respiratory motion signal was first extracted from the self-navigation spokes in each acquired 3D radial data set with the following steps. First, the missing half of the SI half-spoke was zero-filled, and a fast Fourier transform (FFT) was performed to generate z-direction projection profiles oriented along the SI direction. Second, a respiratory motion signal was extracted using a motion detection algorithm previously described in (31). Specifically, a principal component analysis was performed in the projection profiles from each coil element. The first principal component from each coil element was selected and pooled together for a clustering procedure that finds the dominant signal variation pattern. Given that respiratory motion is the most common signal pattern across all coils, this dominant signal was selected as the respiratory motion signal. This motion detection algorithm was designed to exclude “poor” coil elements that do not have a good representation of respiratory motion, and to select a signal without relying on frequency information, as described in (31). Since the alternating breathing windows for the NDB-UTE acquisitions were known, motion detection was performed using only the deep breathing projection profiles in the NDB-UTE data.
Image Reconstruction
For visualization of lung anatomy, the NB-UTE data were reconstructed with 4 respiratory phases spanning from expiration to inspiration (matrix size=256×256×256×4, voxel size≈0.98×0.98×0.98 mm3), using the extracted respiratory motion signals. The number of respiratory phases was selected based on prior experiences in implementing the XD-GRASP technique (22,25,26). To retrospectively evaluate imaging performance with different acceleration rates, and to determine the most suitable acquisition time, all NB-UTE datasets were reconstructed with different durations of acquisition, corresponding to different acceleration rates. Specifically, each data set was reconstructed with data acquired during the first one minute (XD-UTE-NB-1min: 15,300 spokes), the first three minutes (XD-UTE-NB-3min: 45,900 spokes), the first five minutes (XD-UTE-NB-5min: 76,560 spokes) and all eight minutes (XD-UTE-NB-8min: all 122,520 spokes). For comparison, all datasets were also reconstructed using all acquired spokes with only gridding reconstruction without any motion compensation, thus simply averaging the effect of motion (referred to as Ave-UTE).
For assessment of pulmonary ventilation, two dynamic image series, including one normal breathing image series (XD-UTE-PF-NB) and one deep breathing image series (XD-UTE-PF-DB), were reconstructed with 30 respiratory phases spanning from expiration to inspiration and then back to expiration covering a full respiratory cycle. XD-UTE-PF-DB images were reconstructed using the deep breathing data (net scan times≈4 minutes) from the NDB-UTE acquisitions, with a voxel size of ~1.95×1.95×1.95mm3. XD-UTE-PF-NB images were reconstructed using the first half of the data from the NB-UTE acquisitions (net scan times=4 minutes). In order to match the spatial resolution of XD-UTE-PF-DB, the XD-UTE-PF-NB reconstruction was performed with a voxel size of ~1.95×1.95×1.95mm3 by discarding part of the high-frequency component in each radial spoke.
All reconstructions were performed using the XD-GRASP framework that solves the following optimization problem:
Here, d is the 4D dynamic image series to be reconstructed, S is the corresponding multidimensional radial k-space data, D is a density compensation filter used for 3D radial image reconstruction (13,32), and T is the sparsifying transform applied along the respiratory dimension with a regularization parameter λ. E represents an encoding operator that incorporates coil sensitivities (C), a non-uniform FFT (NUFFT) operator (F) and the density compensation filter (D). The density compensation filter was separated into two square root terms, one applied in a pre-compensation step before iteration and the other one applied during each iteration, to ensure that E and its Hermitian transpose EH are adjoint.
Image reconstruction was implemented in MATLAB (Mathworks, Natick, MA) with a nonlinear conjugate gradient algorithm. All reconstructions were performed on a Linux server equipped with 256 GB memory and two 6-GB NVIDIA graphics processing unit cards. First-order finite differences were employed along the dynamic dimension as the sparsifying transform. Coil sensitivity maps were estimated from the static 3D image volume reconstructed from all spokes using the adaptive combination approach (33). The NUFFT operation was implemented using a gpuNUFFT toolbox presented in (34) for acceleration of reconstruction speed. For each type of reconstruction, a regularization parameter was empirically selected by comparing different values in terms of visual image quality and temporal blurring using gridding images as a surrogate reference, as previously described in (35). Once the value was selected, it was then fixed for all datasets that were acquired and reconstructed with the same protocol.
Visual Image Quality Assessment
Image quality was scored visually to compare XD-UTE-NB images with different acquisition times (1-min, 3-min, 5-min and 8-min), AVE-UTE and BH-VIBE. For XD-UTE-NB, the end-expiratory phases were used for the assessment. After pooling and randomizing all images (XD-UTE-NB: 92 cases including different acceleration rates, AVE-UTE: 23 cases, BH-VIBE: 12 cases), three board-certified chest radiologists (W.H.M., F.G., and R.R. with 13, 12 and 3 years of post-fellowship experience in chest MRI, respectively) blinded to the reconstruction and acquisition schemes scored the visualization of great vessels (i.e., aorta and pulmonary artery), large airways, segmental arteries, segmental bronchovascular structures, subsegmnetal vessels and pleura region using a 6-point Likert-type scale as follows: 5 = excellent visualization; 4 = good visualization; 3 = acceptable visualization. 2 = slightly limited to acceptable; 1 = poor visualization; 0 = structure not visible. In addition, the readers also scored the overall artifact level, including the blurring effect and streaks, for each case using a 4-point Likert-type scale as follows: 4 = no artifact; 3 = minor artifact not affecting image assessment; 2 = moderate artifact degrading visualization of structures, but still diagnostic; 1 = severe artifact leading to non-diagnostic images. The reported scores were averaged over the three readers to yield mean ± standard deviation for each assessment category and each reconstruction/acquisition scheme.
Pulmonary Ventilation Analyses
For spirometry measurements, the forced expiratory volume during the first second (FEV1) and the forced vital capacity (FVC) were obtained, from which the FEV1/FVC ratio was then calculated. For both XD-UTE-PF-NB and XD-UTE-PF-DB images, the left and right lungs were semi-automatically segmented by two readers using FireVoxel, an image processing software platform available at http://cai2r.net/resources/software. The volume of each lung was computed in each respiratory phase, and the left and right lung ventilation parameters, including the tidal volume (TV), residual volume (RV), lung capacity (LC), and FEV1/FVC ratio, were then obtained. The global FEV1 was calculated as the sum of right and left lung FEV1, and the global FVC was calculated as the sum of right and left lung FVC. The global FEV1/FVC ratio, which was calculated as global FEV1/global FVC, was then compared with the FEV1/FVC ratio measured with spirometry.
Statistical Analyses
For visual image quality assessment, a non-parametric paired two-tailed Wilcoxon signed-rank test was performed for statistical comparison using an online tool (http://vassarstats.net/wilcoxon.html), with a P-value less than 0.05 indicating statistical significance. Bland-Altman analysis was used to assess inter-reader variability in image quality scoring. For lung ventilation measurements, the intra-class correlation was used to compare inter-reader variability. The coefficient of variance was calculated to assess the scan-rescan variability, and the Pearson correlation coefficient was calculated to compare the FEV1/FVC measured using XD-UTE MRI and spirometry. Statistical analyses for lung ventilation measurements were performed in SPSS (SPSS Inc, Chicago, IL, USA).
Results
Figure 1b–c show two exemplary respiratory motion signals superimposed on their corresponding projection profiles, one for the NB-UTE acquisition (b) and the other for the NDB-UTE acquisition alternating normal and deep breathing (c).
Table 1 summarizes readers’ scores for each reconstruction/acquisition scheme, averaged over the two readers. Both XD-UTE-NB-8min and XD-UTE-NB-5min were significantly better than AVE-UTE in all assessment categories. XD-UTE-NB-8min was significantly better than BH-VIBE in all assessment categories; and XD-UTE-NB-5min was significantly better than BH-VIBE except for the visualization of the great vessels and the artifact level. However, XD-UTE-NB-5min was significantly worse than XD-UTE-NB-8min in all assessment categories. The image quality of both XD-UTE-3min and XD-UTE-NB-1min was significantly lower compared to that of XD-UTE-NB-8min. Bland-Altman analyses assessing the inter-reader variability of image scoring are shown in Figure S1 to Figure S3 in the Supplementary Materials.
Table 1.
Summary of readers’ scores for each reconstruction/acquisition scheme. Both XD-UTE-NB-8min and XD-UTE-NB-5min were significantly better than AVE-UTE in all assessment categories. XD-UTE-NB-8min was significantly better than BH-VIBE in all assessment categories; and XD-UTE-NB-5min was significantly better than BH-VIBE except for the visualization of the great vessels and the artifact level. However, XD-UTE-NB-5min was significantly worse than XD-UTE-NB-8min in all assessment categories. The scores of both XD-UTE-3min and XD-UTE-NB-1min were significantly lower compared to XD-UTE-NB-8min.
| Large Arteries | Large Airways | Segmental Arteries | Segmental Bronchovascular Structures | Subsegmnetal Vessels | Pleura Region | Artifact Level | |
|---|---|---|---|---|---|---|---|
| BH-VIBE (n=12) | 2.73±0.66 | 2.28±0.61 | 2.21±0.53 | 1.19±0.53 | 1.22±0.52 | 2.11±0.69 | 2.64±0.70 |
| AVE-UTE (n=23) | 2.40±0.65 | 2.60±0.69 | 2.19±0.63 | 1.41±0.70 | 1.30±0.61 | 2.04±0.57 | 2.07±0.54 |
| XD-UTE-NB-8min (n=23) | 3.28±0.79 ab | 3.60±0.75 ab | 3.40±0.82 ab | 2.83±1.13 ab | 2.33±0.85 ab | 3.03±0.65 ab | 3.26±0.60 ab |
| XD-UTE-NB-5min (n=23) | 2.82±0.70 bc | 3.32±0.88 abc | 3.01±0.84 abc | 2.32±1.03 abc | 1.86±0.78 abc | 2.70±0.74 abc | 2.51±0.62 bc |
| XD-UTE-NB-3min (n=23) | 2.35±0.63 c | 2.66±0.75 c | 2.24±0.75 bc | 1.56±0.75 c | 1.26±0.61 c | 1.96±0.75 c | 1.78±0.40 c |
| XD-UTE-NB-1min (n=23) | 1.72±0.58 c | 2.06±0.77 c | 1.65±0.63 c | 0.98±0.52 c | 0.83±0.48 c | 1.67±0.57 c | 1.46±0.49 c |
Scoring criterial:
For arteries, airways, and pleura:
5 = Excellent visualization;
4 = Good visualization;
3 = Acceptable visualization;
2 = Slightly limited to acceptable;
1 = Poor visualization;
0 = Structure not visible.
For overall artifact level:
4 = No artifact;
3 = Minor artifact not restricting image assessment;
2 = Moderate artifact degrading visualization of structures, but still diagnostic;
1 = Severe artifact leading to non-diagnostic images.
XD-UTE-NB is significantly better than the BH-VIBE;
XD-UTE-NB is significantly better than the UTE-AVE;
The scores of XD-UTE-NB-5min, XD-UTE-NB-3min or XD-UTE-NB-1min were significantly lower than XD-UTE-NB-8min.
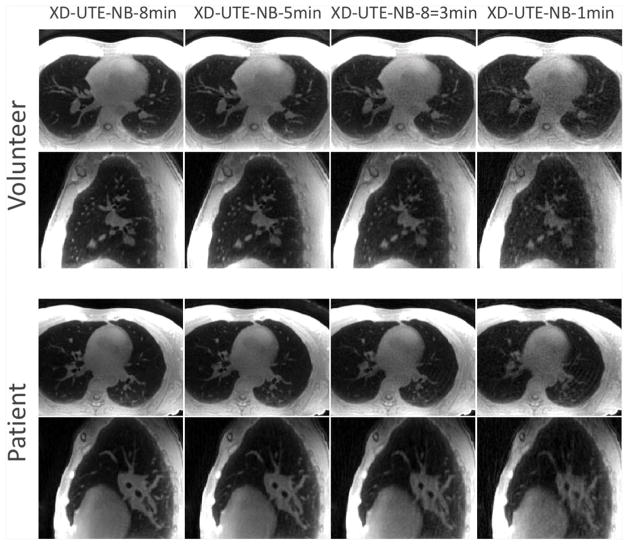
Figure 2 shows a comparison of XD-UTE-NB images reconstructed with different durations of acquisitions in a volunteer (upper rows) and a patient with emphysema (bottom rows). Among four different types of reconstructions, XD-UTE-NB-8min achieved the best visual image quality and the delineation of lung anatomy. With the decrease of acquisition duration, image quality degraded with compromised visualization of the lung structure. XD-UTE-NB-1min achieved significantly degraded image quality with increased residual artifacts, blurring and noise.
Figure 2.
A comparison of XD-UTE-NB images reconstructed with different quantities of data, and, hence, different acceleration rates in a volunteer (upper rows) and a patient with emphysema (bottom rows). Among the four different types of reconstructions, XD-UTE-NB-8min achieved the best visual image quality and delineation of lung anatomy. With increasing acceleration rate, image quality degraded, with compromised visualization of lung structure. XD-UTE-NB-1min showed significantly degraded image quality, with increased residual artifacts, blurring and noise.
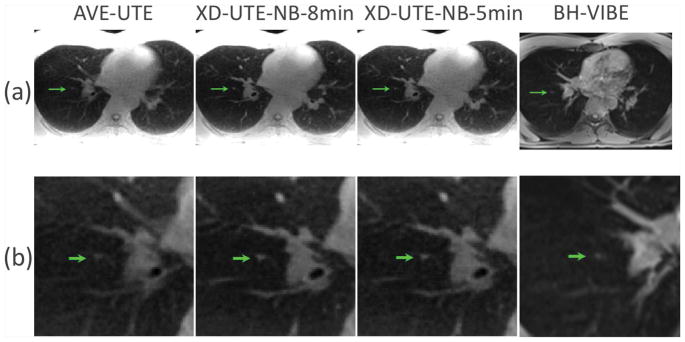
Figure 3 shows a comparison of AVE-UTE, XD-UTE-NB-8min, XD-UTE-NB-5min and BH-VIBE images in a 30-year-old man with a suspected lung nodule. Both XD-UTE-NB-8min and XD-UTE-NB-5min achieved improved image quality with better delineation of the lung anatomy compared to AVE-UTE and BH-VIBE. The suspected lung nodule, indicated by the green arrows, was better captured in the XD-UTE-NB results compared to the AVE-UTE and BH-VIBE results, as confirmed in the zoomed images in Figure 3b.
Figure 3.
A comparison of AVE-UTE, XD-UTE-NB-8min, XD-UTE-NB-5min and BH-VIBE images in a 30-year old man with a suspected lung nodule. Both XD-UTE-NB-8min and XD-UTE-NB-5min achieved improved image quality as compared with the other two cases, with greater delineation of the lung anatomy compared to AVE-UTE and BH-VIBE. The lung nodule, indicated by the green arrows, was better displayed in the XD-UTE-NB results as compared to the AVE-UTE and BH-VIBE images.
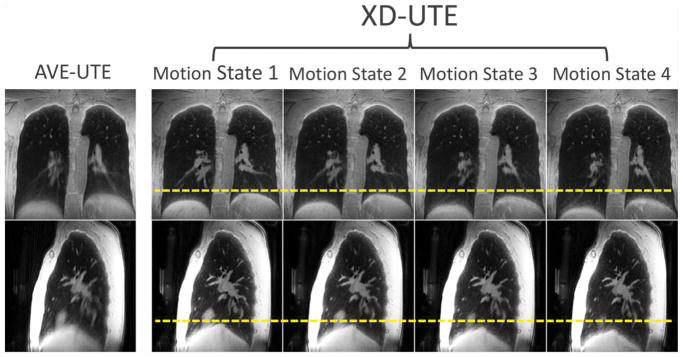
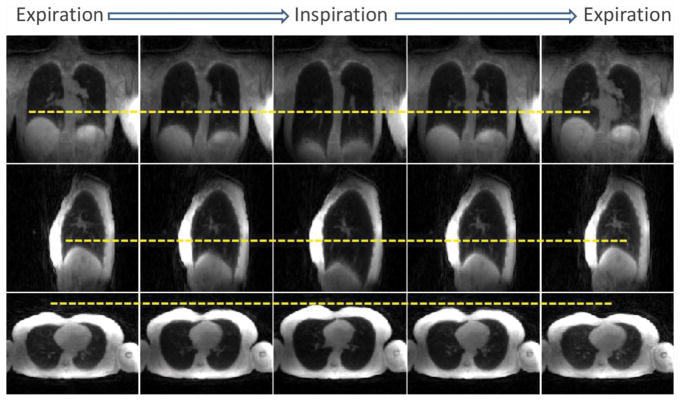
Figure 4 shows XD-UTE-NB lung images for four respiratory phases, in comparison with AVE-UTE. This demonstrates that respiratory motion can be resolved (yellow dashed lines) by reconstructing an extra motion dimension. Moreover, the intrinsic motion compensation in the XD-UTE-NB reconstruction improves image quality
Figure 4.
An example of XD-UTE-NB lung images in different respiratory phases, in comparison with AVE-UTE. This demonstrates that respiratory motion can be resolved (note varying diaphragm position with respect to yellow dashed lines) by reconstructing an extra motion dimension to improve image quality.
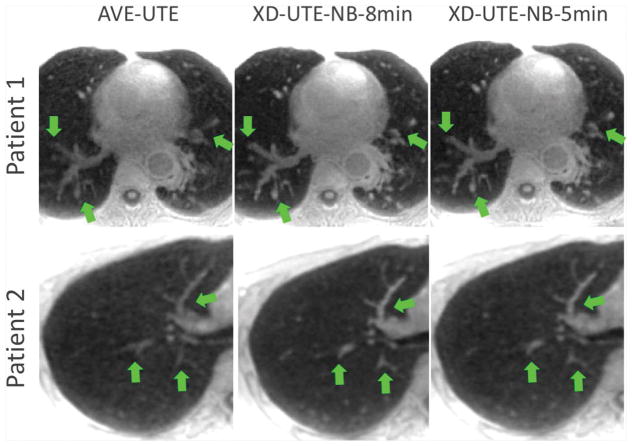
A similar comparison of AVE-UTE and XD-UTE-NB images in two additional patients are shown in Figure 5. As described above, XD-UTE-NB images show improved overall image quality and increased sharpness compared to the averaged reconstruction AVE-UTE (see structure shown by the green arrows).
Figure 5.
A comparison of AVE-UTE and XD-UTE-NB images in two additional patients. XD-UTE-NB images show improved overall image quality and sharpness compared to the averaged reconstruction without motion compensation (green arrows).
Figure 6 shows motion-resolved lung images during deep breathing (XD-UTE-PF-DB) in a representative volunteer. Despite reduced image quality compared to XD-UTE-NB, due to deep breathing and a lower spatial resolution, the images provide dynamic ventilation information that can be used to obtain pulmonary ventilation parameters. The differential positions of the diaphragm with respect to the yellow dashed lines indicate that respiratory motion can be resolved with XD-UTE reconstruction. Figure S4 (Supplementary Materials) shows the same images as in Figure 6 zoomed to the lung area in both coronal and sagittal planes. It should be noted that the deep-breathing data were acquired with a lower spatial resolution and were reconstructed with a higher temporal resolution than the 4 respiratory-phase XD-UTE-NB images.
Figure 6.
Motion-resolved lung images during deep breathing (XD-UTE-PF-DB) in a representative subject. Despite reduced image quality and lower spatial resolution compared to normal breathing, images obtained during deep breathing provide dynamic functional information that can be used to obtain pulmonary functional parameters. The varying position of the diaphragm with respect to the yellow dashed lines indicates that respiratory motion can be resolved using XD-UTE reconstruction.
Table 2 summarizes the XD-UTE lung ventilation parameters that were averaged over all subjects and two independently observers. There was an excellent inter-reader agreement in right, left, and global measurements of the tidal volume, residual volume, lung capacity, and FEV1/FVC, with an intra-class correlation coefficient ranging from 0.8855 to 0.9995. The scan-rescan variability in the right, left, and global measures of PF-MR, available from 4 subjects who underwent a second MR scan, resulted in coefficients of variance ranging from 6.5% to 15.3% for different PF-MR parameters. The average FEV1, FVC and FEV1/FVC results from spirometry, averaged over the six subjects who underwent the measurement, were 2.73±0.96, 3.39±1.50 and 75.72±11.25, respectively. There was a moderate to good agreement between the FEV1/FVC ratio measured using the XD-UTE MRI and the spirometer (76.04±17.79 verse 75.72±11.25, correlation coefficient=0.724). A scatter plot of the global FEV1/FVC from both MRI and spirometry in the six subjects are shown in Figure S5 in the Supplementary Materials.
Table 2.
MR measures of pulmonary function for right lung, left lung, and the sum of both lungs (global measurements) in all subjects. Inter-reader agreement was computed with intra-class correlation (ICC), ranging from 0.8855 to 0.9995. Scan-rescan variability was assessed with coefficient of variance (CV) as noted in the table.
| Mean | ICC | Scan-Rescan CV | |
|---|---|---|---|
| Right Tidal Vol (L) | 0.25 (0.15–0.56) | 0.9977 (0.9867–0.9996) | 6.5% |
| Right Residual Vol (L) | 1.23 (0.77–1.7) | 0.9966 (0.9805–0.9994) | 9.8% |
| Right Lung Capacity (L) | 2.51 (1.49–3.71) | 0.9995 (0.9971–0.9999) | 13.6% |
| Right FEV1/FVC (%) | 74.5 (47–100) | 0.9737 (0.8559–0.9954) | 12.3% |
| Left Tidal Vol (L) | 0.22 (0.14–0.53) | 0.9985 (0.9910–0.9997) | 7.2% |
| Left Residual Vol (L) | 1.18 (0.9–1.61) | 0.9971 (0.9834–0.9995) | 7.5% |
| Left Lung Capacity (L) | 2.43 (1.61–3.61) | 0.9996 (0.9979–0.9999) | 15.3% |
| Left FEV1/FVC (%) | 76 (65.6–99) | 0.8855 (0.4778–0.9794) | 15% |
| Global Tidal Vol (L) | 0.46 (0.26–1.1) | 0.9991 (0.9950–0.9999) | 7% |
| Global Residual Vol (L) | 2.5 (2.0–3.3) | 0.9977 (0.9869–0.9996) | 8% |
| Global Lung Capacity (L) | 5.05 (3.56–7.32) | 0.9981 (0.9892–0.9997) | 13.7% |
| Global FEV1/FVC (%) | 75 (57.5–99) | 0.9562 (0.7693–0.9923) | 13.1% |
Discussion
This work developed and tested a respiratory motion-resolved 4D lung MRI framework called XD-UTE that combines a self-navigated golden-angle radial UTE sequence with XD-GRASP reconstruction. XD-UTE allows efficient respiratory motion detection and generation of motion-resolved lung images with good image quality. More importantly, our initial experience suggests that XD-UTE can be used for simultaneous evaluation of lung structure and pulmonary ventilation. The high-spatial-resolution anatomical lung MR images can serve as an alternative radiation-free approach to CT for detection of lung abnormalities, particularly for patients who need frequent follow-up screening. Meanwhile, the high-temporal-resolution dynamic lung images can provide spatial information that complements conventional pulmonary function tests with spirometry, for studying disease heterogeneities.
The golden-angle reordering scheme enables retrospective evaluation of reconstruction performance with different quantities of data corresponding to different scan durations. For normal breathing acquisitions, we compared the overall quality of lung images reconstructed with data acquired in 8 minutes, 5 minutes, 3 minutes and 1 minute. The results suggest that 8-minute acquisitions are sufficient to reconstruct high-quality lung images. While a 5-minute scan yielded reduced image quality, it may still be adequate for clinical use based on the image quality scores provided by the readers. On the other hand, 3-minute and 1-minute acquisition times were insufficient to achieve diagnostic image quality.
XD-UTE was performed with two continuous acquisitions in this work: one normal-breathing acquisition reconstructed with a high spatial resolution but a low temporal resolution for morphological evaluation, and the other normal-deep breathing acquisition reconstructed with a low spatial resolution but a high temporal resolution for assessment of lung ventilation. For morphological assessment in which dynamic information is not needed, on the one hand, XD-UTE images were reconstructed with 4 respiratory phases to provide sufficient image quality for visualization of lung structure. For assessment of lung ventilation, on the other hand, an increased number of respiratory phases were reconstructed to cover a full respiratory cycle. For the alternated 1-min normal breathing and 1-min deep breathing scan (NDB-UTE), we noted that the motion pattern in the normal breathing data were not predictable because the subjects needed to take a break after a deep-breathing scan segment and prepare for the next segment. Thus, we elected to perform a separate normal breathing scan (NB-UTE) for morphological evaluation.
Despite reduced image quality due to high temporal resolution/high acceleration rate and the required deep breathing pattern, the reconstructed images sufficed to compute lung volume over time in our study. The initial results suggest that the image quality is adequate to segment the left lung and right lung for calculation of the lung ventilation. Although MRI and spirometry were performed in different positions (MRI was performed in a supine position while spirometry was performed in a sitting position, which is the default standard setting in our pulmonary function test lab), there was still a moderate to good correlation of the lung ventilation measurements between them. The correlation is expected to be further improved if a MRI-compatible spirometer (36) is available for comparison in future work.
Unlike previous motion-resolved lung MRI studies (28,29), we have demonstrated the feasibility of applying XD-UTE for evaluation of both lung anatomy and ventilation in this work. In addition, our XD-UTE technique uses a self-navigated golden-angle UTE sequence, which enables more efficient respiratory motion detection and leads to faster image reconstruction speed. Furthermore, XD-UTE was tested in a cohort of both patients and volunteers, and all images were evaluated on a clinical basis by experienced chest radiologists. Finally, we retrospectively evaluated reconstructions with different quantities of data, and suggested appropriate acquisition durations suitable for clinical use.
We acknowledge the following limitations in this study, which will be addressed in future work. First, we did not control for particular clinical conditions in our patient recruitment. Future studies to evaluate XD-UTE in patients with specific clinical indications (i.e., lung nodules, cystic fibrosis, or chronic obstructive pulmonary disease) would be necessary to assess the performance of this framework for well-defined clinical tasks. Second, self-navigation spokes were acquired with half-projections. Thus, motion detection was performed by first zero-filling the half-spokes to generate projection profiles. An approach for better motion detection is to acquire the self-navigation spokes with full-projection while acquiring other spokes with half-projection, or using a dual-echo acquisition scheme as previously shown in (13), but at the cost of increased scan times. Third, although we selected BH-VIBE imaging as our reference approach in this work, additional comparisons with CT images, or other respiratory gated UTE acquisitions, such as the gated PETRA sequence (16), would be very interesting and useful to further evaluate the performance of XD-UTE, and this will be explored in future work. Preliminary results comparing XD-UTE with CT in patients with cystic fibrosis were recently reported in an abstract presented at the ISMRM meeting (37). Fourth, the half-spoke acquisition is required to achieve ultra-short echo time in a UTE sequence. However, this acquisition scheme significantly reduces scan efficiencies compared to standard radial sequences, since only half of the data can be acquired within a given scan time. It would be interesting to explore a stack-of-spirals acquisition scheme for lung imaging as shown in (38). By acquiring different image partitions with varying TEs, this sequence is expected to achieve ultra-short echo time and improved imaging efficiency simultaneously. Lastly, although one would like to reconstruct an image series with only one deep-breathing cycle (e.g., 5–8 seconds), this is very challenging due to the relatively slow imaging speed in MRI, particularly given our desired spatiotemporal resolution. Thus, we designed our current breathing pattern and have elected to acquire data during multiple breathing cycles, followed by post-processing to synchronize them together as a composited respiratory cycle. The current 1-min normal-breathing and 1-min deep-breathing protocol used in this study may not be the optimal pattern, and we have not compared different breathing protocols. Thus, additional work to optimize the breathing protocol for ventilation evaluation would be necessary to ensure that patients can easily cooperate with breathing instructions and that robust ventilation measurements can be derived.
In conclusion, the proposed self-navigated XD-UTE framework represents a promising approach for free-breathing non-contrast lung MRI with self-navigation for efficient motion detection and reconstruction of motion-resolved lung images. It has the potential for imaging lung anatomy and ventilation simultaneously.
Supplementary Material
Acknowledgments
Grant Support
This work was supported in part by the NIH (P41 EB017183 and R01 EB018308) and was performed under the rubric of the Center for Advanced Imaging Innovation and Research (CAI2R), an NIBIB Biomedical Technology Resource Center.
The authors would like to thank Dr. Henry Rusinek from the New York University School of Medicine for the support in using the Firevoxel software.
References
- 1.Cazzola M, Hanania NA, MacNee W, Rudell K, Hackford C, Tamimi N. A review of the most common patient-reported outcomes in COPD--revisiting current knowledge and estimating future challenges. Int J Chron Obstruct Pulmon Dis. 2015;10:725–738. doi: 10.2147/COPD.S77368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatabu H, Alsop DC, Listerud J, Bonnet M, Gefter WB. T2* and proton density measurement of normal human lung parenchyma using submillisecond echo time gradient echo magnetic resonance imaging. European journal of radiology. 1999;29(3):245–252. doi: 10.1016/s0720-048x(98)00169-7. [DOI] [PubMed] [Google Scholar]
- 3.Stock KW, Chen Q, Hatabu H, Edelman RR. Magnetic resonance T2* measurements of the normal human lung in vivo with ultra-short echo times. Magnetic resonance imaging. 1999;17(7):997–1000. doi: 10.1016/s0730-725x(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 4.Salerno M, Altes TA, Mugler JP, 3rd, Nakatsu M, Hatabu H, de Lange EE. Hyperpolarized noble gas MR imaging of the lung: potential clinical applications. European journal of radiology. 2001;40(1):33–44. doi: 10.1016/s0720-048x(01)00347-3. [DOI] [PubMed] [Google Scholar]
- 5.van Beek EJ, Wild JM, Kauczor HU, Schreiber W, Mugler JP, 3rd, de Lange EE. Functional MRI of the lung using hyperpolarized 3-helium gas. Journal of magnetic resonance imaging : JMRI. 2004;20(4):540–554. doi: 10.1002/jmri.20154. [DOI] [PubMed] [Google Scholar]
- 6.Qing K, Mugler JP, 3rd, Altes TA, et al. Assessment of lung function in asthma and COPD using hyperpolarized 129Xe chemical shift saturation recovery spectroscopy and dissolved-phase MRI. NMR Biomed. 2014;27(12):1490–1501. doi: 10.1002/nbm.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song EJ, Kelsey CR, Driehuys B, Rankine L. Functional airway obstruction observed with hyperpolarized (129) Xenon-MRI. J Med Imaging Radiat Oncol. 2018;62(1):91–93. doi: 10.1111/1754-9485.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruger SJ, Fain SB, Johnson KM, Cadman RV, Nagle SK. Oxygen-enhanced 3D radial ultrashort echo time magnetic resonance imaging in the healthy human lung. NMR Biomed. 2014;27(12):1535–1541. doi: 10.1002/nbm.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zha W, Kruger SJ, Johnson KM, et al. Pulmonary ventilation imaging in asthma and cystic fibrosis using oxygen-enhanced 3D radial ultrashort echo time MRI. Journal of magnetic resonance imaging : JMRI. 2018;47(5):1287–1297. doi: 10.1002/jmri.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Togao O, Tsuji R, Ohno Y, Dimitrov I, Takahashi M. Ultrashort echo time (UTE) MRI of the lung: assessment of tissue density in the lung parenchyma. Magnetic resonance in medicine. 2010;64(5):1491–1498. doi: 10.1002/mrm.22521. [DOI] [PubMed] [Google Scholar]
- 11.Grodzki DM, Jakob PM, Heismann B. Ultrashort echo time imaging using pointwise encoding time reduction with radial acquisition (PETRA) Magnetic resonance in medicine. 2012;67(2):510–518. doi: 10.1002/mrm.23017. [DOI] [PubMed] [Google Scholar]
- 12.Johnson KM, Fain SB, Schiebler ML, Nagle S. Optimized 3D ultrashort echo time pulmonary MRI. Magnetic resonance in medicine. 2013;70(5):1241–1250. doi: 10.1002/mrm.24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delacoste J, Chaptinel J, Beigelman-Aubry C, Piccini D, Sauty A, Stuber M. A double echo ultra short echo time (UTE) acquisition for respiratory motion-suppressed high resolution imaging of the lung. Magn Reson Med. 2018;79(4):2297–2305. doi: 10.1002/mrm.26891. [DOI] [PubMed] [Google Scholar]
- 14.Ohno Y, Koyama H, Yoshikawa T, et al. Standard-, Reduced-, and No-Dose Thin-Section Radiologic Examinations: Comparison of Capability for Nodule Detection and Nodule Type Assessment in Patients Suspected of Having Pulmonary Nodules. Radiology. 2017;284(2):562–573. doi: 10.1148/radiol.2017161037. [DOI] [PubMed] [Google Scholar]
- 15.Dournes G, Menut F, Macey J, et al. Lung morphology assessment of cystic fibrosis using MRI with ultra-short echo time at submillimeter spatial resolution. Eur Radiol. 2016;26(11):3811–3820. doi: 10.1007/s00330-016-4218-5. [DOI] [PubMed] [Google Scholar]
- 16.Dournes G, Grodzki D, Macey J, et al. Quiet Submillimeter MR Imaging of the Lung Is Feasible with a PETRA Sequence at 1. 5 T. Radiology. 2015;276(1):258–265. doi: 10.1148/radiol.15141655. [DOI] [PubMed] [Google Scholar]
- 17.Fischer A, Weick S, Ritter CO, et al. SElf-gated Non-Contrast-Enhanced FUnctional Lung imaging (SENCEFUL) using a quasi-random fast low-angle shot (FLASH) sequence and proton MRI. NMR Biomed. 2014;27(8):907–917. doi: 10.1002/nbm.3134. [DOI] [PubMed] [Google Scholar]
- 18.Tibiletti M, Paul J, Bianchi A, et al. Multistage three-dimensional UTE lung imaging by image-based self-gating. Magn Reson Med. 2016;75(3):1324–1332. doi: 10.1002/mrm.25673. [DOI] [PubMed] [Google Scholar]
- 19.Higano NS, Hahn AD, Tkach JA, et al. Retrospective respiratory self-gating and removal of bulk motion in pulmonary UTE MRI of neonates and adults. Magn Reson Med. 2017;77(3):1284–1295. doi: 10.1002/mrm.26212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veldhoen S, Weng AM, Knapp J, et al. Self-gated Non-Contrast-enhanced Functional Lung MR Imaging for Quantitative Ventilation Assessment in Patients with Cystic Fibrosis. Radiology. 2017;283(1):242–251. doi: 10.1148/radiol.2016160355. [DOI] [PubMed] [Google Scholar]
- 21.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 22.Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med. 2016;75(2):775–788. doi: 10.1002/mrm.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandarana H, Feng L, Ream J, et al. Respiratory Motion-Resolved Compressed Sensing Reconstruction of Free-Breathing Radial Acquisition for Dynamic Liver Magnetic Resonance Imaging. Investigative radiology. 2015;50(11):749–756. doi: 10.1097/RLI.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng L, Axel L, Latson AL, Xu J, Sodickson DK, Otazo R. Compressed sensing with synchronized cardiorespiratory sparsity for free-breathing cine MRI: initial comparative study on patients with arrhythmias. Proceedings of the 17th Annual SCMR Scientific Sessions; New Orleans, LA, USA. 2014; O. 17. [Google Scholar]
- 25.Piccini D, Feng L, Bonanno G, et al. Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. Magn Reson Med. 2017;77(4):1473–1484. doi: 10.1002/mrm.26221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng L, Coppo S, Piccini D, et al. 5D whole-heart sparse MRI. Magn Reson Med. 2018;79(2):826–838. doi: 10.1002/mrm.26745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng L, Delacoste J, Chandarana H, et al. Four-Dimensional Respiratory Motion-Resolved Sparse Lung MRI. Proceedings of the 24nd Annual Meeting of ISMRM; Singapore. 2016. p. 2918. [Google Scholar]
- 28.Jiang W, Ong F, Johnson KM, et al. Motion robust high resolution 3D free-breathing pulmonary MRI using dynamic 3D image self-navigator. Magnetic resonance in medicine. 2018;79(6):2954–2967. doi: 10.1002/mrm.26958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tibiletti M, Bianchi A, Kjorstad A, Wundrak S, Stiller D, Rasche V. Respiratory self-gated 3D UTE for lung imaging in small animal MRI. Magn Reson Med. 2017;78(2):739–745. doi: 10.1002/mrm.26463. [DOI] [PubMed] [Google Scholar]
- 30.Piccini D, Littmann A, Nielles-Vallespin S, Zenge MO. Spiral phyllotaxis: the natural way to construct a 3D radial trajectory in MRI. Magn Reson Med. 2011;66(4):1049–1056. doi: 10.1002/mrm.22898. [DOI] [PubMed] [Google Scholar]
- 31.Feng L, Huang C, Shanbhogue K, Sodickson DK, Chandarana H, Otazo R. RACER-GRASP: Respiratory-weighted, aortic contrast enhancement-guided and coil-unstreaking golden-angle radial sparse MRI. Magnetic resonance in medicine. 2018;80(1):77–89. doi: 10.1002/mrm.27002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielles-Vallespin S, Weber MA, Bock M, et al. 3D radial projection technique with ultrashort echo times for sodium MRI: clinical applications in human brain and skeletal muscle. Magn Reson Med. 2007;57(1):74–81. doi: 10.1002/mrm.21104. [DOI] [PubMed] [Google Scholar]
- 33.Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magnetic resonance in medicine. 2000;43(5):682–690. doi: 10.1002/(sici)1522-2594(200005)43:5<682::aid-mrm10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 34.Knoll F, Schwarzl A, Diwoky C, Sodickson DK. gpuNUFFT - An Open Source GPU Library for 3D Regridding with Direct MATLAB Interface. Proceedings of the 22nd Annual Meeting of ISMRM; Milan, Italy. 2014. p. 4297. [Google Scholar]
- 35.Feng L, Grimm R, Block KT, et al. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med. 2014;72(3):707–717. doi: 10.1002/mrm.24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciet P, Tiddens HA, Wielopolski PA, et al. Magnetic resonance imaging in children: common problems and possible solutions for lung and airways imaging. Pediatr Radiol. 2015;45(13):1901–1915. doi: 10.1007/s00247-015-3420-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delacoste J, Beigelman C, Feng L, et al. Assessment of Cystic Fibrosis Disease Using UTE Imaging with XD-GRASP Reconstruction: A Comparison with CT. Proceedings of the 25th Annual Meeting of ISMRM; Honolulu, HI, USA. 2014. p. 829. [Google Scholar]
- 38.Kumar S, Rai R, Stemmer A, et al. Feasibility of free breathing Lung MRI for Radiotherapy using non-Cartesian k-space acquisition schemes. The British journal of radiology. 2017;90(1080):20170037. doi: 10.1259/bjr.20170037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.