Abstract
Polygenic risk scores (PRSs) are typically constructed as weighted sums of risk allele counts of single nucleotide polymorphisms (SNPs) associated with a disease or trait. PRSs are typically constructed based on published results from Genome-Wide Association Studies (GWASs), the majority of which have been performed in large populations of European Ancestry (EA) individuals. While many genotype-trait associations have generalized across populations, the optimal choice of SNPs and weights for PRSs may differ between populations due to different linkage disequilibrium (LD) and allele frequency patterns. We compare various approaches for PRS construction, using GWAS results from both large EA studies and a smaller study in Hispanics/Latinos: the Hispanic Community Health Study/Study of Latinos (HCHS/SOL, n = 12,803). We consider multiple approaches for selecting SNPs and for computing SNP weights. We study the performance of the resulting PRSs in an independent study of Hispanics/Latinos from the Women’s Health Initiative (WHI, n = 3,582). We support our investigation with simulation studies of potential genetic architectures in a single locus. We observed that selecting variants based on EA GWASs generally performs well, except for blood pressure trait. However, use of EA GWASs for weight estimation was suboptimal. Using non-EA GWAS results to estimate weights improved results.
Keywords: genetic diversity, admixed populations, linkage disequilibrium
Introduction
Polygenic Risk Scores (PRSs, see Table 1 for acronyms and shorthands) summarize the genetic component of a disease or quantitative (continuous) trait and are routinely used in public health genetics research for a wide range of applications, such as improving disease and trait prediction (Morrison et al., 2007); studying the shared genetic basis between traits (Raffield et al., 2017); increasing power by integrating over multiple variants rather than one variant at a time; and Mendelian Randomization studies (Voight et al., 2012; Vimaleswaran et al., 2013) in which a PRS associated with one trait is used as an instrumental variable in a causal analysis of the association of the trait with another outcome. PRSs are typically constructed as the weighted sum of risk alleles of single nucleotide polymorphisms (SNPs) associated with the trait of interest, where SNPs and weights are usually selected based on findings from published studies, such as Genome-Wide Association Studies (GWASs) (Qi et al., 2012). However, most GWASs to date have been performed in studies of individuals of exclusively or predominantly European genetic ancestry (EA). This poses a difficulty for PRS construction in non-EA populations.
Table 1:
Acronyms and shorthands used in the manuscript.
| AA | African Ancestry |
| ADM | an admixed population |
| ADMn | an admixed population with n thousands individuals (n ∈ {5,000, 12,000g) |
| ADMp | an admixed population proportion of YRI ancestry equal to p (p ∈ {0.2, 0.4}) |
| ADMn,p | an admixed population with n thousands individuals and proportion of YRI ancestry equal to p. (n ∈ {5, 000, 12, 000}, p ∈ {0.2, 0.4}). |
| BMI | Body Mass Index |
| CEU | Utah Residents (CEPH) with Northern and Western European Ancestry; from 1000 genomes. |
| DBP | diastolic blood pressure |
| EA | European Ancestry |
| GWAS | Genome-Wide Association Study |
| HA | Hispanic American |
| HCHS/SOL | Hispanic Community Health Study/Study of Latinos |
| HGB | Hemoglobin concentration |
| HIP | Hip circumference |
| LD | Linkage Disequilibrium |
| MAP | Mean Arterial Pressure |
| Mbp | Mega (1,000,000) base-pairs |
| META | meta-analysis of GWASs, one in an EA population and the second in an admixed population |
| PCs | Principal Components |
| PLT | Platelet count |
| PP | Pulse Pressure |
| PRS | Polygenic Risk Score |
| RMSPE | Root Mean Squared Prediction Error |
| SBP | Systolic Blood Pressure |
| SNP | Single Nucleotide Polymorphism |
| WBC | White Blood Cell count |
| WC | Waist Circumference |
| WHI | Women’s Health Initiative |
| WHI-SHARe | Women’s Health Initiative SNP Health Association Resource |
| WHR | Waist-to-Hip Ratio |
| YRI | Yoruba in Ibadan, Nigeria; from 1000 genomes. |
Using EA GWAS to select SNPs and choose weights for PRSs in non-EA populations may seem like a reasonable approach: Hispanics/Latinos are admixed with European ancestry, and previous studies have shown that many genetic EA genetic associations generalize to Hispanics/Latinos (Qi et al., 2017; Graff et al., 2017). Furthermore, EA GWASs are typically very large, with tens or hundreds of thousands of individuals, therefore having large statistical power to detect the most strongly associated variants from genomic association regions and obtain precise estimates of effects sizes. In fact, Dudbridge (2013) studied power and prediction accuracy of PRSs and suggested that hundreds of thousands of individuals may be needed to estimate SNP effects. While such numbers are available in EA GWASs, they are not currently available in GWASs of diverse populations such as Hispanics/Latinos. For example, in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), there are fewer than 13,000 individuals who consented for genetic studies. Of these, about 7,000 individuals participated in a GWAS of diabetes (Qi et al., 2017) which is the largest published GWAS to date in Hispanics/Latinos. In contrast, the largest published GWAS of diabetes (DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al., 2014) meta-analyzed multiple EA studies (N =≈70,000 cases and controls), and a smaller number of population studies of other ancestries (including about 2,500 Mexicans).
There are a number of drawbacks to using EA GWAS for PRS construction in non-EA populations. Specifically, linkage disequilibrium (LD) patterns vary across populations, rendering different best available tag SNPs between populations; allele frequencies often differ across populations; and, at least for some traits, effect sizes differ between populations and allelic heterogeneity exists (The International HapMap Consortium, 2005; Musunuru et al., 2012). Admixed populations, such as Hispanics/Latinos, may have different genetic architecture and effect sizes at a genetic association region compared to an ancestral population due to gene-gene (epistasis) or gene-environment interactions, or because a causal variant is monomorphic in one ancestral population (Jain et al., 2017; Qi et al., 2017). Belsky et al. (2013) constructed a PRS for obesity based on EA GWAS results and found that its utility for an African American (AA) population was low, and much lower than that for an EA population. Martin et al. (2017) studied transferability of PRSs constructed based on single-ancestry GWASs to other ancestries, and demonstrated that scores inferred from EA GWASs may perform poorly in other ancestries. Others have recently shown that PRSs are highly associated with genetic ancestry and cautioned against using EA-based PRSs in diverse populations (Reisberg et al., 2017; Curtis, 2018). Collectively, these studies highlight the need to adapt PRS construction methods to diverse ancestries. Even across Hispanic/Latino populations, LD patterns and allele frequencies can differ. For example, at rs4133185, a SNP on chromosome 17, we see both different allele frequencies (Burkart et al., 2018) and distinct LD patterns (see Figures S1-S6 in the Supplementary Methods) across multiple background groups with distinct admixture histories in HCHS/SOL.
How to best construct a PRS for a study in a Hispanic/Latino population is still an open question. Should one use only the information published in a large, primarily EA study? Or, can we use results from a smaller, non-EA study? In particular, will incorporating information from these lower-powered (smaller sample size) non-EA GWAS in fact improve PRS construction, or will it instead introduce harmful variability? Here, we take a systematic, empirical approach to constructing PRSs for Hispanics/Latinos, based on published GWASs results from large EA population studies and medium-sized studies of Hispanics/Latinos. Specifically, we use GWAS findings from the HCHS/SOL to construct and evaluate PRSs in an independent study of Hispanic/Latina women from the Women’s Health Initiative (WHI). We support our results with simulations mimicking potential genetic architecture within a single, trait-associated genomic region.
Materials and Methods
Let yi be a quantitative trait measured on the ith participant, i = 1,…, n, and xi a k × 1 vector of covariates such as confounders. Let g1i,…,gPi be allelic counts or dosages of P independent variants associated with yi, and α1,…, αP their effect sizes, so that the additive linear model holds:
| (1) |
where ϵi are residual errors. An optimal PRS for yi is , the weighted sum with the causal genotypes and their true effects.
Issues in selection of SNPs for PRSs
In practice, we do not know which are the true causal genotypes for a trait, so we have to select a set of SNPs to use in our PRS. Often, the data we have at our disposal for selecting SNPs are derived from a genotyping platform that did not interrogate all sequence genotypes, but rather a reduced set of a few million (or fewer) variants. For PRS construction, we often have only a set of associated genotypes that likely tag a subset of the causal genotypes.
Let gp be a causal genotype in EA populations, and let be a tag SNP to gp that was detected in an association study, perhaps because gp was not genotyped at all. It is well known (Pritchard and Przeworski, 2001) that the size of trait association at is related to the LD of with gp, denoted by ρp, so that . Given a large enough dataset, we expect that the lead variant (the variant with strongest association in the region) will be the one with ∣ρp∣ closest to the maximal value 1, among all available (genotyped or imputed) variants.
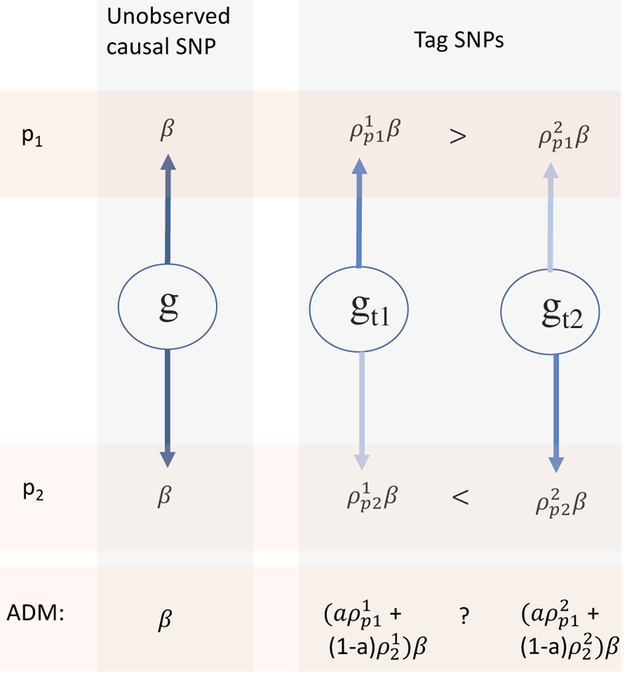
Further complicating this situation is the fact that tag SNPs may differ across populations. Assume the simple scenario of a single causal SNP in an association region, with the same effect size β in two ancestral populations P1 and P2. Suppose that in the admixed population (ADM) the proportion of genomic intervals containing the causal variant inherited from populations P1 and P2 is α and (1 − α), respectively. This is demonstrated in Figure 1, which shows that even if the same tag SNPs were available in the two ancestral populations P1 and P2, and we knew which SNPs were the best tags in each population, it is not clear which is the best tag SNP in ADM. This becomes even more complicated when there are multiple ancestral populations, when SNP availability differs due to different genotyping platforms, and when effect sizes differ between ancestral populations.
Figure 1:
Observed SNPs gt1, gt2 likely have different LDs ρ with the unobserved causal SNP g in ancestral populations P1 and P2, leading to distinct tag SNPs in the two populations: gt1 in P1 and gt2 in P2. In the admixed population (ADM), the associations between the observed tag SNPs and the unobserved causal SNP depend on a, the proportion of admixed haplotypes that inherited this region from ancestral population P1.
PRS construction
SNP selection.
Consider a GWAS performed in an EA study. Association results are available for d variants, with p-values and effect size estimates . Assume that almost all variants are also available in a GWAS of individuals with admixed ancestry, such as the HCHS/SOL, with corresponding p-values and effect size estimates , and . Based on the information from both EA and HCHS/SOL GWASs, we can perform fixed-effects meta-analysis (META) to produce meta-analytic p-values and effect sizes , and . We can also perform generalization analysis (Sofer et ah, 2017a), to test the composite null hypothesis that is rejected if an association exists in both the EA population and in the HCHS/SOL study, and get r-values for these variants .
We created lists of candidate SNPs for PRSs by filtering variants based on these measures (pe, pa, pm, rm) and with varying thresholds. We considered the p-value thresholds 5 × 10−8, 1 × 10−7, 1×10−6, 1×10−5, 1×10−4, 1×10−3, 0.01, 0.05, 0.5 and r-value thresholds 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 0.99. For generalization analysis, we initially took all variants with p-value < 10−6 in the EA GWAS, then performed generalization analysis using these SNPs to compute r-values. Therefore, by construction, smaller lists of SNPs are considered using this approach. Finally, we clumped the SNPs using PLINK (Purcell et al., 2007) to generate a list of SNPs in no or low LD (ρ2 < 0.2), where LD was evaluated using the 1000 Genomes EA population panel (1000 Genomes Project Consortium, 2012).
In simulation studies, which focused on the effects of LD and sample sizes, we selected only a single lead SNP from the region based on the same criteria described above (lead EA, lead in META, etc.), without setting a p-value threshold or clumping.
SNP weights.
The optimal PRS weights reflect the true size of association between each SNP in the PRS and the trait. In practice, the size of this association must be estimated. We considered the effect size estimates computed in the EA GWAS (be), which may be very accurate in the EA population but potentially less appropriate in the admixed population. Therefore, we also considered the effect size estimated in a trainig admixed population (ba) and the effect size estimated in fixed effects meta-analysis (bm). We also compared these to PRSs without weights (or αp = 1 ∀p) because unweighted PRSs are often used in practice (Qi et al., 2017; Raffield et al., 2017). We always oriented the alleles to represent trait-increasing alleles.
PRS evaluation.
To evaluate PRS approaches, we constructed PRSs in an independent validation dataset based on SNPs and weights according to the training dataset. Let the PRS for participant i in the validation dataset be
where is the selected set of SNPs (which is likely different than the true causal set S), and is the estimated effect of the jth SNP is the set. We considered two measures for evaluation. For simulation studies we used the Root Mean Squared Prediction Error (RMSPE), computed by
| (2) |
across the nv individuals in the validation dataset. In data analysis, we computed the variance explained by each PRS in a regression model adjusted for sex, age, and the first five principal components (PCs) of genetic ancestry. This was calculated by first fitting a model with these covariates, but without the PRS, and obtaining the residual variance denoted by , then fitting a model that also included the PRS and obtaining the residual variance . The estimated percent variance explained is 100 × .
Simulation study
Our simulation studies focused on the impact of LD and variability in the estimation of effect sizes, caused by small sample size and admixture, on PRSs, in admixed populations with two ancestral populations (compared to the three ancestral populations of Hispanics/Latinos), CEU (EA) and YRI (African ancestry, AA), for simplicity. Henceforth, we always refer to admixed populations as ADM. When distinction is needed, we add subscripts to denote sample sizes (an integer representing thousands of individuals) and proportions of AA admixture (a number between 0 and 1). We simulated genotypes in a 1Mbp genomic region for a large EA sample (nEA = 50,000), a moderately sized admixed sample (ADM12, with nADM12 = 12,000), and a small admixed sample (ADM5, with nADM5 = 5, 000). Admixture proportions where either 20% or 40% of YRI. These proportions were selected based on observed proportions of African ancestries in HCHS/SOL’s Dominican and Puerto-Rican background groups respectively (see Figure 2 in Conomos et al. (2016)). We simulated quantitative traits under a few potential genetic architectures, assuming one or two causal SNPs, which are either shared or different between the two ancestral populations, in the region. Each simulation setting was repeated 500 times. Details about simulating admixed populations genomic association regions and architectures, are provided in the Supplementary Material.
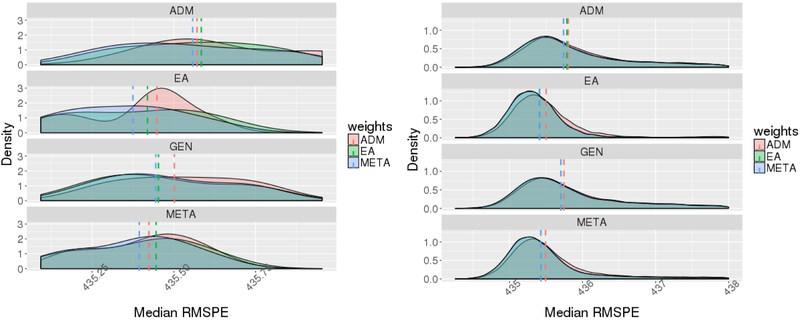
Figure 2:
The smoothed distribution of median root mean squared prediction errors (RMSPEs), where each median was computed over 500 repetitions of the same simulation setting, and the distribution is across all possible choices of causal SNP(s) in the locus. The left panel corresponds to the scenario in which there is a a single causal SNP in the locus, which is monomorphic in the African population, and the right panel corresponds to the scenario in which there are two causal SNPs, one of which is monomorphic in EA. In these figures, the training datasets were EA and ADM12,0.2, while the test dataset was ADM5, 0.4. Dashed vertical lines correspond to median of the plotted distribution. In the right panel, the lines corresponding to EA and meta-analysis (META) weights overlap.
Evaluating similarity between LD patterns
We evaluated the similarity between LD patterns of the simulated populations CEU, ADM0.2, and ADM0.4. For each SNP j = 1,…,617 in the simulated genomic region, we identified its best tag SNP in CEU, ADM0.2, and ADM0.4 by finding the SNP j′ ≠ j which had the highest LD (r2) with SNP j in that sample. Given one of the simulated admixed populations, we calculated the proportion of SNPs in the region that had a tag SNP with higher LD in CEU compared to second admixed populations (which had different admixture proportions), and the other way around.
Using the HCHS/SOL to develop Hispanic/Latino-specific PRSs Previously published EA GWASs and traits
We considered three groups of traits with previously published GWAS results: anthropometric, comprising of height, body mass index (BMI), hip circumference (HIP), waste circumference (WC), and waste-to-hip ratio (WHR) from the GIANT consortium GWAS; (Wood et al., 2014; Locke et al., 2015; Shungin et al., 2015) blood pressure traits, comprising of systolic and diastolic blood pressure (SBP, DBP), pulse pressure (PP = SBP-DBP), and mean arterial pressure (MAP=DBP+1/3PP), with GWASs performed by the International Consortium of Blood Pressure (ICBP) (Wain et al., 2011; International Consortium for Blood Pressure Genome-Wide Association Studies, 2011); and finally, blood count traits, including white blood cell count (WBC), platelet count (PLT), and hemoglobin concentration (HGB) (Tajuddin et al., 2016; Chami et al., 2016; Eicher et al., 2016). Note that for these blood count GWASs, we used just the EA GWAS results and not the transancestry results that were also available.
The Hispanic Community Health Study/Study of Latinos (HCHS/SOL)
The HCHS/SOL is a community-based cohort study following 16,415 self-identified Hispanic/Latino participants with initial visits between 2008 and 2011 (Sorlie et al., 2010). Participants were recruited into the study in four field centers (Chicago, IL, San Diego, CA, Bronx, NY, and Miami, FL) via a two-stage sampling scheme, by which community block units were first sampled, followed by households within the block units. Some or all household members were recruited. The sampling probabilities were set preferentially towards sampling Hispanics/Latinos (LaVange et al., 2010). In total, 12,803 study participants consented to genetic studies. Henceforth, we focus on this subset when describing the HCHS/SOL population. The HCHS/SOL participants are very diverse, and usually self-identify as belonging to one of six background groups: Central American, Cuban, Dominican, Mexican, Puerto Rican and South American. Genotyping, imputation, and quality control in the HCHS/SOL have been described in Conomos et al. (2016).
Genome-Wide Association Studies in the HCHS/SOL
HCHS/SOL analyses followed the standards developed by the HCHS/SOL Genetic Analysis Center and reported in Conomos et al. (2016); Sofer et al. (2016). Table S1 in the Supplementary Material summarizes the information about each of these GWASs, including sample sizes, specific covariates, trait transformations, and imputation reference panels. In brief, analyses used mixed models with variance components due to genetic relatedness, household, and block unit sharing, and were adjusted for sex, age, log-transformed sampling weights (to prevent potential selection bias due to the study design), and the first five principal components reflecting ancestry (to control for population stratification). Samples sizes ranged from 11,809 to 12,705. GWASs for blood count traits in the HCHS/SOL were reported in Schick et al. (2016); Hodonsky et al. (2017); Jain et al. (2017), and GWASs for blood pressure (BP) traits were reported in Sofer et al. (2017b).
The Women’s Health Initiative SNP Health Association Resource (WHI SHARe)
The WHI is a long-term health study following 161,808 postmenopausal women aged 50-79 years old who were recruited from 1993 through 1998 from 40 clinical centers throughout the United States (Hays et al., 2003). Ten of the 40 WHI clinical centers with expertise and access to specific minority groups (American Indian, black, Asian American or Pacific Islander, and Hispanic) were selected to serve as minority recruitment sites. Clinical information was collected by self-report and physical examination. A total of 5,469 self-identified Hispanic Americans (HA) were consented to genetic research and were eligible for WHI-SHARe. Due to budget constraints, we randomly selected a subsample of 3,642 (66.6%) HA women. DNA was extracted by the Specimen Processing Laboratory at the Fred Hutchinson Cancer Research Center from specimens that were collected at the time of enrollment. All participants provided written informed consent as approved by local human subjects committees. Genotype data are from the Affymetrix Genome-Wide Human SNP Array 6.0 that contains 906,000 single nucleotide polymorphisms (SNPs) and more than 946,000 probes for the detection of copy number variants. The genotype data were processed for quality control, including call rate, concordance rates for blinded and unblinded duplicates, and sex discrepancy, leaving 871,309 unflagged SNPs with a genotyping rate of 99.8% and 3586 HA women used in the current analysis. Genotype Imputation was carried out with MaCH. For imputation in HA samples, we used reference panel of HapMap III CEU + MEX (Mexican ancestry in Los Angeles, California) + YRI samples for a total of 1,387,466 SNPs (MAF > 1%), of which 1,368,178 SNPs met the quality threshold of r2 > 0.3 (Reiner et al., 2012).
Results
Simulations
Our simulations focused on a single, 1Mbp genomic region (locus), which contains 617 SNPs. We first evaluated the similarity of LD patterns between potential training samples (EA, ADM training dataset) and the test ADM dataset, and then evaluated the combined impact of LD, genetic architecture, and variability in effect estimates on PRS performance.
LD patterns in the admixed samples are more similar to each other than to EA samples
Table 2 reports how often each training sample (EA or training ADM) provided a tag SNP having higher LD with the true causal SNP in the test ADM dataset. The tag SNPs in CEU and the training ADM population were often the same (46-50% of the time). When they differed, the tag SNP identified in the training ADM population was usually a better tag of the causal SNP in the test ADM population.
Table 2:
The mean percentage of SNPs (empirical standard deviation (SD)) in the investigated locus for which the tag SNP in each of the training populations had higher LD with the causal SNP in the test population. Means and SDs were computed over 500 realizations of the simulated admixed populations. In each simulation repetition, the EA training data had 50,000 individuals. The subscripts in ADMn,p provide sample size n ∈ {5,12} (in thousands), and p ∈ {0.2, 0.4}, denoting proportion of YRI ancestry.
| Training ADM population |
Test population | EA better tag | Training ADM better tag | Same tag |
|---|---|---|---|---|
| ADM12,0.2 | ADM5,0.4 | 3% (0.3%) | 47% (0.4%) | 50% (0.4%) |
| ADM12,0.4 | ADM5,0.2 | 9% (0.6%) | 45% (0.6%) | 45% (0.3%) |
Performance of PRSs in the simulated test datasets
Our simulation study considered four different scenarios of genetic architecture at the locus, focusing only on the effect of LD, sample sizes, and admixture proportions, and assuming that effect sizes were the same across populations. Details are provided in the Supplementary Material. For each choice of causal SNP(s) we repeated the simulation 500 times, constructing 12 PRSs each time (all combinations of the four selection and three weight estimation approaches), and we recorded the median RMSPE for each PRS across the 500 repetitions.
Results are visualized in figures depicting the distributions of the median RMSPEs across the various choices of causal SNPs in each of the simulation scenarios. Figure 2 provides these distributions for two simulation scenarios, where the test dataset was a small admixed population with 40% African ancestry (ADM5,0.4) and the training datasets were a large EA sample and a moderately sized sample of admixed individuals with 20% African ancestry (ADM12,0.2,). Figures corresponding to all other scenarios and settings are provided in the Supplementary Material, Figures S7-S10.
In almost all simulation scenarios, SNP selection by ADM training dataset and weights calculated by ADM (regardless of SNP selection approaches) performed the worst. Only in the simulation scenario in which there were two causal SNPs with one being polymorphic only in YRI, computation of weights in the ADM training data is sometimes advantageous over EA weights. However, this was true only when ADM12, 0.2, was the training dataset, but not when ADM12,0.4, was the training dataset. Other than that, both EA and META SNP selections and weights constructions usually performed similarly, with a few more settings in which META weights outperformed EA weights. We do note that all types of PRS suffered from outlying scenarios: specific combinations of causal SNP(s) in which one PRS produced extremely large RMSPEs. However, on average we see better performance of PRSs based on the META and EA GWAS relative to the other two selection approaches when the causal SNPs’ effect sizes are the same across populations.
Performance of PRSs in the Women’s Health Initiative dataset
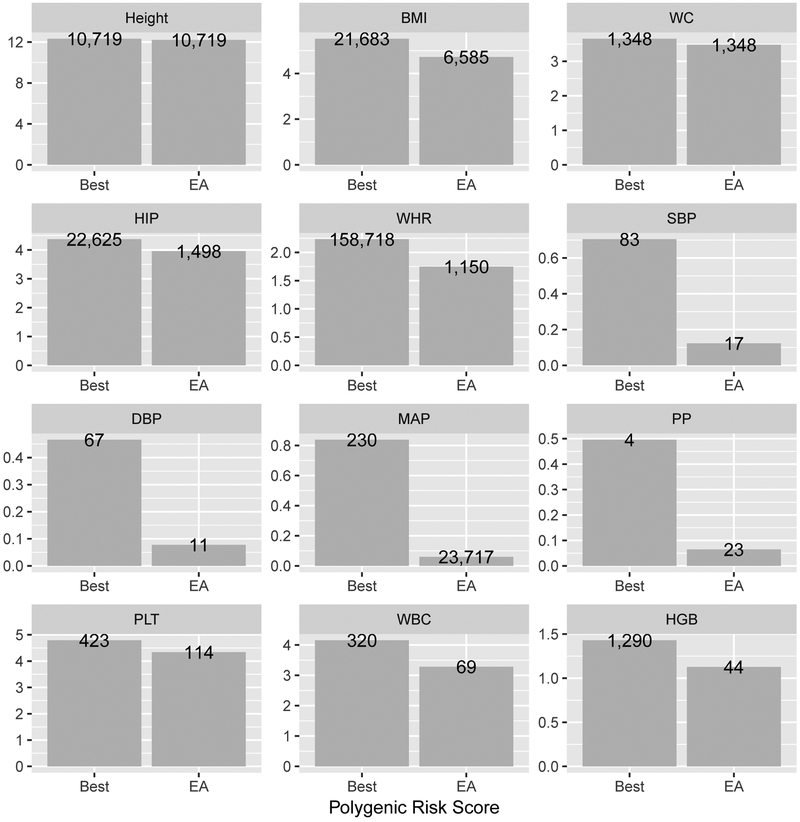
We constructed PRSs for 3,642 Hispanic/Latina women from the WHI based on combinations of results from EA and HCHS/SOL GWASs. Figure 3 demonstrates the highest variance explained obtained by the highest performing EA-based PRS (SNP selection and weight estimation by EA GWAS), and the highest variance explained by any of the approaches. Table 3 provides information about the best performing EA-based PRS, and the overall best performing PRS.
Figure 3:
Variance explained by the highest performing EA-based PRS and highest performing PRS across all approaches, for all investigated traits, in WHI Hispanic Americans. The numbers on the bars represent the number of SNPs used in the PRS. Table 3 provides more details about the PRSs, including p-value or r-value threshold, weights used, etc.
Table 3:
Characteristics and performance, in terms of variance explained, of the highest performing EA-based PRS and highest performing PRS across all approaches, for all investigated traits, in WHI Hispanic Americans. ‘Max N EA GWAS’ is the maximum number of participants used for estimating genetic associations in the EA GWAS that was used as a training dataset. ‘Fold change’ is the variance explained by the best performing PRS, divided by variance explained by the best performing EA-based PRS. The EA-based PRS selected SNPs based on EA GWAS results, with pruning based on EA populations from 1000 Genomes. The weights used in the PRSs were effect sizes from the EA GWASs. In the ‘best performing PRS’, SNPs were selected based on either EA GWASs, meta-analysis of EA and HCHS/SOL GWASs (META), or Generalization analysis (GEN) performed based on discovery in EA GWAS and generalization in the HCHS/SOL GWAS. SNP clumping was based on EA populations from 1000 Genomes. Weights were based on EA GWAS, Meta-analysis of EA and HCHS/SOL GWAS (Meta), HCHS/SOL GWAS (SOL), or ‘None’ - a simple sum of trait-increasing alleles.
| Best performing PRS | Best EA-based performing PRS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Max N EA GWAS |
Selection | Weights | Thresh- old |
# SNPs |
Variance ex- plained |
Thresh- old |
# SNPs |
Variance ex- plained |
Fold change |
| Anthropometric traits | ||||||||||
| Height | 253,280 | EA | Meta | 0.001 | 10,719 | 12.32 | 0.001 | 10,719 | 12.21 | 1.01 |
| BMI | 322,154 | EA | Meta | 0.05 | 21,683 | 5.52 | 0.01 | 6,585 | 4.72 | 1.17 |
| WC | 232,101 | EA | Meta | 0.001 | 1,348 | 3.65 | 0.001 | 1,348 | 3.48 | 1.05 |
| HIP | 213,038 | EA | Meta | 0.05 | 22,625 | 4.37 | 0.001 | 1,498 | 3.96 | 1.10 |
| WHR | 212,248 | EA | Meta | 0.5 | 158,718 | 2.23 | 0.001 | 1,150 | 1.75 | 1.27 |
| Blood count traits | ||||||||||
| PLT | 108,598 | EA | SOL | 0.001 | 423 | 4.79 | 1e-05 | 114 | 4.34 | 1.10 |
| WBC | 108,596 | EA | Meta | 0.001 | 320 | 4.16 | 1e-05 | 69 | 3.28 | 1.29 |
| HGB | 106,377 | EA | None | 0.01 | 1,290 | 1.43 | 1e-06 | 44 | 1.13 | 1.26 |
| Blood pressure traits | ||||||||||
| SBP | 69,909 | META | None | 1e-05 | 83 | 0.70 | 1e-07 | 17 | 0.12 | 5.83 |
| DBP | 69,899 | EA | SOL | 1e-05 | 67 | 0.47 | 5e-08 | 11 | 0.08 | 5.87 |
| MAP | 74,064 | META | None | 1e-04 | 230 | 0.84 | 0.05 | 23,717 | 0.06 | 14.0 |
| PP | 74,064 | GEN | SOL | 0.7 (r-value) | 4 | 0.50 | 1e-06 | 23 | 0.06 | 8.33 |
For anthropometric traits, EA GWASs sample sizes ranged from 212-253K individuals. The pattern of results was similar for all anthropometric traits, in that optimal PRSs had SNPs selected according to EA GWAS, with weights according to the meta-analysis of the EA and the HCHS/SOL GWASs. Optimal SNP selection threshold varied, but were quite high (0.001-0.5).
For blood count traits, EA GWASs sample sizes were 106-108K individuals. Optimal PRSs had SNPs selected according to EA GWAS, but optimal weights differed between traits (HCHS/SOL, meta-analysis, or no weights). Interestingly, the number of SNPs used in the optimal PRSs for blood count traits is much lower than the number of SNPs used in the optimal PRSs for anthropometric traits, while achieving similar variance explained percentages. This may reflect different genetic architecture: less SNPs with stronger effects for blood count compared to anthropometric traits, and potentially different effect sizes between population groups.
For blood pressure traits, EA GWASs sample sizes were 70-74K individuals. There was no consistent pattern for the highest performing PRSs, but clearly the EA-based PRSs performed poorly. All PRSs explained less than 1% of the variance of blood pressure traits.
Discussion
We studied several approaches for constructing PRSs in Hispanic/Latino populations, using GWAS results from independent studies in large populations of European ancestry (EA) and medium-sized GWASs in Hispanics/Latinos. We studied the performance of PRSs constructed using these approaches on an independent dataset. We investigated 12 traits in data analysis. Results differed by trait, and possibly by sample size of the discovery EA GWAS (the discovery GWASs in Hispanics/Latinos had about the same sample size for all traits). For example, for all anthropometric and blood count traits, which had sample sizes of more than 100K individuals in the EA discovery GWAS, optimal SNP selection was based on EA GWAS. However, results were not consistent in the blood pressure traits, which had smaller EA discovery sample sizes: around 70K individuals. However, for all traits, using estimated effect sizes from the EA GWAS as weights was never optimal. Our simulation studies were performed in simplified scenarios in which admixture arises from only two ancestral populations (unlike the HCHS/SOL, in which there are three ancestral populations), and the effect sizes were the same in both ancestries, and in the admixed populations themselves. The LD patterns between our simulated training and testing admixed populations were more similar to each other than to the LD pattern in the simulated EA population, despite the fact that the two admixed populations had different admixture proportions. Therefore, given large enough sample sizes, we expect that the training admixed population would have been a better reference for PRS construction in the testing admixed population compared to the EA population. However, in these simulations usually the PRSs with SNP selection using EA GWAS and weights using META performed best, and weights using the admixed training datasets usually performed poorly. These results are supported by Dudbridge (2013), who suggested that many thousands of individuals are required to effectively calculate effect sizes to be used as SNP weights.
Although the setup of our simulation study focused on a simplified scenario, we can draw a few conclusions by comparing the results to that of the data analysis. Based on simulation results, if the causal SNPs are the same and have the same effect sizes in all EA and Hispanic/Latino populations, we would expect that EA-based SNP selection combined with either EA or META weights would have optimal performance. This was the pattern of results for the anthropometric traits, however, not for blood count or blood pressure traits. Therefore, we hypothesize that there may be differences in effect sizes between population groups for those traits. This is in agreement with the work of Coram et al. (2017) who assumed different effect sizes between populations, and found that estimating effect for risk prediction purposes is useful in ethnically-matched population, while SNP selection using EA GWAS is generally appropriate. Moreover, for platelet count, diastolic blood pressure, and pulse pressure, EA selection worked better with HCHS/SOL weights. In simulations, this happened when either the LD of the selected SNP with the causal one was higher in ADM, or when the selected SNP was more frequent in ADM. Both phenomena can be related to natural selection (Slatkin (2008), and recent work from Guo et al. (2018), which investigated natural selection evidence for complex traits). It is an important avenue for future research to study the evidence of natural selection in admixed populations by phenotypes, and its implication for construction of PRSs. Finally, in simulation studies, SNP selection by META performed well, but not in data analysis. In Tables S6-S7 in the Supplementary Material, we demonstrate that the poor performance of META selection in the data analysis is due to the use of an EA reference panel for clumping. When we pruned SNPs based on base-pair distance instead of LD-clumped, META-based selection performed well, and better than EA-based PRSs (that clumped SNPs) for blood pressure and blood count traits, and worse for anthropometric traits.
Our study has a few limitations. First, we looked only at the performance of PRSs in independent validation datasets, so our results do not inform the construction of PRSs to be used in the same study (e.g., a Mendelian Randomization study). Furthermore, the independent validation study in our data analysis, WHI, only includes female participants, while our training studies, EA GWAS results and the HCHS/SOL, included both males and females. As gene-sex interactions likely exist, the PRSs constructed using the general population may not be optimal for women. However, this is unlikely to introduce any systematic biases to the SNP selection and SNP weights calculation procedures, so the relative performance of the PRS construction approaches should not be impacted. We did not investigate the entire literature for each trait and then investigate each of those loci separately, as is sometimes done in practice, but instead applied the same algorithm to each trait, based on two reference GWASs. While the first approach is useful for investigators who work with a single PRS and want to optimize it, it is also more case-dependent, and less generalizable. Our systematic approach is easier to apply on a number of traits and is appropriate for drawing general conclusions. Finally, we did not use multi-ethnic GWASs for PRS construction, but rather focused on EA and and Hispanic/Latino GWASs. Our goal was to more clearly delineate properties of the genetic architecture similarities or differences between populations. Another limitation of the current study is the lack of systematic investigation into generalizability of our results to other types of populations and varying sample sizes. It is a topic of future research as results from larger diverse studies become available. In the Supplementary Material, we report the results of a secondary analysis repeating the same data analysis reported in the manuscript, while evaluating PRSs on WHI African American women. Interestingly, the pattern of results is generally similar. This suggest that leveraging trans-ethnic information into PRS construction is beneficial.
Other recent methodological work on PRSs has been performed primarily in the context of EA populations. Shi et al. (2016) suggested to penalize the estimated effect sizes used in a PRS, by fitting an l1-penalized regression. It is a topic of future work to suggest an approach that reduces the computational burden of applying shrinkage estimation procedure in mixed models and test its utility for improving the effect size estimates in an admixed population training dataset. Vilhjálmsson et al. (2015) proposed LDpred for incorporating information from GWAS summary statistics and a reference panel to use information from multiple SNPs, rather than only the lead SNP, from an association region. While they demonstrated this method to be useful under specific priors for genetic architecture, their approach hinges on having a good reference panel. Different admixed populations differ in their admixture patterns, so the same reference panel may not be appropriate across the board. It will be interesting to study and potentially extend Vilhjálmsson et al. (2015)’s approach to admixed populations, despite the lack of training and testing samples with the same LD structure. Recently, Baker et al. (2018) proposed POLARIS, a method to construct PRSs while accounting for LD structure in the test dataset (i.e. not in a reference panel). It is a topic for future research to study the POLARIS approach for admixed and diverse populations. Other investigators worked on incorporating information from large studies in EA populations and a smaller admixed population, specifically focusing on trait prediction. Márquez-Luna et al. (2017) considered an approach that constructs a prediction model based on two PRSs, each constructed based on GWAS in a different population (EA and admixed population), and principal components of ancestry. They used either a validation dataset or cross validation to select LD parameters for clumping, p-value threshold for SNP selection, and also to select parameters for combining the two PRSs. We performed analyses mimicking their approach, by dividing the HCHS/SOL dataset to obtain training and validation datasets. However, this approach did not perform well, as determined by variance explained in WHI HA population. Details are provided in Section 2.2. of the Supplementary Material.
In summary, we reported a study about the construction of PRSs for use in general association studies performed in studies of admixed populations, specifically focusing on Hispanics/Latinos, based on results from independent GWASs in a large EA population, and in a small/medium Hispanic/Latino population. Our results indicate that current sample sizes of GWAS in Hispanics/Latinos are insufficient for good SNP selection, but utilizing EA GWAS for SNP selection and weights construction is useful. Importantly, we found that using only EA GWAS for constructing PRSs for blood pressure traits performs poorly in Hispanics/Latinos, with EA GWAS sample sizes of 70K individuals. While PRS construction depends on both sample size of discovery GWAS and genetic architecture, we hypothesize that as sample sizes in GWAS keep increasing, PRSs will become better, even when using only EA GWAS. We caution against using a reference panel that does not match a GWAS discovery population for SNP clumping. However, more flexible approaches for computing PRS weights are useful. We provide files with SNP selections and weights for our optimal PRSs validated in WHI HA and AA for the 12 traits investigated here, but also recommend that future investigators who construct PRSs for traits with relatively low sample sizes, attempt to follow our approach of training and testing on an independent dataset to select an optimal PRS. When computational resources are limited, we recommend to clump SNPs based on EA GWAS results and compare multiple weighting schemes.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of HCHS/SOL for their important contributions. The Hispanic Community Health Study/Study of Latinos is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I / N01-HC-65233), University of Miami (HHSN268201300004I / N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I / N01-HC-65235), University of Illinois at Chicago - HHSN268201300003I / N01-HC-65236 Northwestern Univ), and San Diego State University (HHSN268201300005I / N01-HC-65237). The following Institutes/Centers/Offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Funding support for the “Epidemiology of putative genetic variants: The Women’s Health Initiative” study is provided through the NHGRI grants HG006292 and HL129132. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSC271201100004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: https://www.whi.org/about/SitePages/Study%20Organization.aspx. T.S. was supported by the NHLBI (R01 HL120393-03S1 and 1R35HL135818) and NHGRI (R01HG005827). K.G. was supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1256082. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Supplementary Material
The Supplementary Material includes detailed information about the simulation studies and figures with results from simulations, results from the secondary analysis of PRS construction in WHI African American, and from comparisons to a method proposed by Márquez-Luna et al. (2017). SNP selection, alleles, and weights, for constructing the optimal PRSs for Hispanics/Latinos and African Americans, are provided in https://github.com/tamartsi/PRSs_for_admixed_populations.
References
- 1000 Genomes Project Consortium (2012). An integrated map of genetic variation from 1,092 human genomes. Nature, 491 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E, Schmidt KM, Sims R, O’Donovan MC, Williams J, Holmans P, Escott-Price V and Consortium w. t. G. (2018). POLARIS: Polygenic LD-adjusted risk score approach for set-based analysis of GWAS data. Genetic epidemiology, 42 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Sugden K, Williams B, Houts R, McCarthy J and Caspi A (2013). Development and evaluation of a genetic risk score for obesity. Biodemography and social biology, 59 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart KM, Sofer T, London SJ, Manichaikul A, Hartwig FP, Yan Q, Soler Artigas M, Avila L, Chen W, Thomas SD et al. (2018). A Genome-wide Association Study in Hispanics/Latinos Identifies Novel Signals for Lung Function. The Hispanic Community Health Study/Study of Latinos. American journal of respiratory and critical care medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chami N, Chen M-H, Slater AJ, Eicher JD, Evangelou E, Tajuddin SM, Love-Gregory L, Kacprowski T, Schick UM, Nomura A et al. (2016). Exome genotyping identifies pleiotropic variants associated with red blood cell traits. The American Journal of Human Genetics, 99 8–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos M, Laurie C, Stilp A, Gogarten S, McHugh C and Nelson e. a., SC (2016). Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. The American Journal of Human Genetics, 98 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram MA, Fang H, Candille SI, Assimes TL and Tang H (2017). Leveraging Multi-ethnic Evidence for Risk Assessment of Quantitative Traits in Minority Populations. The American Journal of Human Genetics, 101 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D (2018). Polygenic risk score for schizophrenia is more strongly associated with ancestry than with schizophrenia. bioRxiv 287136. [DOI] [PubMed] [Google Scholar]
- DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium Mahajan, A., Go MJ, Zhang W. et al. (2014). Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature genetics, 46 234244 URL http://europepmc.org/articles/PMC3969612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F (2013). Power and predictive accuracy of polygenic risk scores. PLoS Genet, 9 e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher JD, Chami N, Kacprowski T, Nomura A, Chen M-H, Yanek LR, Tajuddin SM, Schick UM, Slater AJ, Pankratz N et al. (2016). Platelet-related variants identified by exomechip meta-analysis in 157,293 individuals. The American Journal of Human Genetics, 99 40–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff M, Emery LS, Justice AE, Parra E, Below JE, Palmer ND, Gao C, Duan Q, Valladares-Salgado A, Cruz M et al. (2017). Genetic architecture of lipid traits in the Hispanic community health study/study of Latinos. Lipids in Health and Disease, 16 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wu Y, Zhu Z, Zheng Z, Trzaskowski M, Zeng J, Robinson MR, Visscher PM and Yang J (2018). Global genetic differentiation of complex traits shaped by natural selection in humans. Nature communications, 9 1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays J, Hunt JR, Hubbell FA, Anderson GL, Limacher M, Allen C and Rossouw JE (2003). The Women’s Health Initiative recruitment methods and results. Annals of epidemiology, 13 S18–S77. [DOI] [PubMed] [Google Scholar]
- Hodonsky CJ, Jain D, Schick UM, Morrison JV, Brown L, McHugh CP, Schurmann C, Chen DD, Liu YM, Auer PL et al. (2017). Genome-wide association study of red blood cell traits in Hispanics/Latinos: The Hispanic Community Health Study/Study of Latinos. PLoS Genetics, 13 e1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association Studies (2011). Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature, 478 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Hodonsky CJ, Schick UM, Morrison JV, Minnerath S, Brown L, Schurmann C, Liu Y, Auer PL, Laurie CA et al. (2017). Genome-wide association of white blood cell counts in Hispanic/Latino Americans: the Hispanic Community Health Study/Study of Latinos. Human Molecular Genetics, 26 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVange L, Kalsbeek W, Sorlie P, Avilés-Santa L, Kaplan R and Barnhart e. a., J (2010). Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Annals of epidemiology, 20 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J et al. (2015). Genetic studies of body mass index yield new insights for obesity biology. Nature, 518 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Luna C, Loh P-R, Consortium SAT. D S., Consortium STD. and Price AL. (2017). Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genetic epidemiology, 41 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD and Kenny EE (2017). Human demographic history impacts genetic risk prediction across diverse populations. The American Journal of Human Genetics, 100 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy M, Kane JP, Pankow JS, Devlin JJ, Willerson JT and Boerwinkle E (2007). Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities Study. American journal of epidemiology, 166 28–35. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Romaine SP, Lettre G, Wilson JG, Volcik KA, Tsai MY, Taylor HA Jr, Schreiner PJ, Rotter JI, Rich SS et al. (2012). Multi-ethnic analysis of lipid-associated loci: the NHLBI CARe project. PLoS One, 7 e36473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK and Przeworski M (2001). Linkage disequilibrium in humans: models and data. The American Journal of Human Genetics, 69 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR, Ridker PM, Hunter DJ, Willett WC, Rimm EB et al. (2012). Sugar-sweetened beverages and genetic risk of obesity. New England Journal of Medicine, 367 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Stilp AM, Sofer T, Moon J-Y, Hidalgo B, Szpiro AA, Wang T, Ng MC, Guo X, Chen Y-DI et al. (2017). Genetics of Type 2 Diabetes in US Hispanic/Latino Individuals: Results from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Diabetes db161150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffield LM, Louie T, Sofer T, Jain D, Ipp E, Taylor KD, Papanicolaou GJ, Avilés-Santa L, Lange LA, Laurie CC et al. (2017). Genome-wide association study of iron traits and relation to diabetes in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL): potential genomic intersection of iron and glucose regulation? Human Molecular Genetics, 26 1966–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Beleza S, Franceschini N, Auer PL, Robinson JG, Kooperberg C, Peters U and Tang H (2012). Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. The American Journal of Human Genetics, 91 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisberg S, Iljasenko T, Läll K, Fischer K and Vilo J (2017). Comparing distributions of polygenic risk scores of type 2 diabetes and coronary heart disease within different populations. PloS one, 12 e0179238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick UM, Jain D, Hodonsky CJ, Morrison JV, Davis JP, Brown L, Sofer T, Conomos MP, Schurmann C, McHugh CP et al. (2016). Genome-wide association study of platelet count identifies ancestry-specific loci in Hispanic/Latino Americans. The American Journal of Human Genetics, 98 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Park J-H, Duan J, Berndt ST, Moy W, Yu K, Song L, Wheeler W, Hua X, Silverman D et al. (2016). Winner’s Curse Correction and Variable Thresholding Improve Performance of Polygenic Risk Modeling Based on Genome-Wide Association Study Summary-Level Data. PLoS genetics, 12 e1006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE et al. (2015). New genetic loci link adipose and insulin biology to body fat distribution. Nature, 518 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M (2008). Linkage disequilibrium?understanding the evolutionary past and mapping the medical future. Nature Reviews Genetics, 9 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer T, Heller R, Bogomolov M, Avery C, Graff M and North e. a, KE (2017a). A Powerful Statistical Framework for Generalization Testing in GWAS, with Application to the HCHS/SOL. Genetic Epidemiology, 41 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer T, Shaffer J, Graff M, Qi Q, Stilp A and Gogarten e. a., SM (2016). Meta-Analysis of Genome-Wide Association Studies with Correlated Individuals: Application to the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Genetic epidemiology, 40 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer T, Wong Q, Hartwig FP, Taylor K, Warren HR, Evangelou E, Cabrera CP, Levy D, Kramer H, Lange LA, Horta BL, Consortium CB, Kerr KF, Reiner AP and Franceschini N (2017b). Genome-Wide Association Study of Blood Pressure Traits by Hispanic/Latino Background: the Hispanic Community Health Study/Study of Latinos Scientific Reports, In print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie P, AvilÉs-Santa L, Wassertheil-Smoller S, Kaplan R, Daviglus M and Giachello e. a, AL (2010). Design and implementation of the Hispanic Community Health Study/Study of Latinos. Annals of epidemiology, 20 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajuddin SM, Schick UM, Eicher JD, Chami N, Giri A, Brody JA, Hill WD, Kacprowski T, Li J, LyytikÄinen L-P et al. (2016). Large-scale exome-wide association analysis identifies loci for white blood cell traits and pleiotropy with immune-mediated diseases. The American Journal of Human Genetics, 99 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium (2005). A haplotype map of the human genome. Nature, 437 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, Genovese G, Loh P-R, Bhatia G, Do R et al. (2015). Modeling linkage disequilibrium increases accuracy of polygenic risk scores. The American Journal of Human Genetics, 97 576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimaleswaran KS, Berry DJ, Lu C, Tikkanen E, Pilz S, Hiraki LT, Cooper JD, Dastani Z, Li R, Houston DK et al. (2013). Causal relationship between obesity and vitamin d status: bi-directional mendelian randomization analysis of multiple cohorts. PLoS Med, 10 e1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Peloso GM, Orho-Melander M, Frikke-Sohmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T et al. (2012). Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. The Lancet, 380 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain L, Verwoert G, O’Reilly P, Shi G, Johnson T and Johnson e. a., AD (2011). Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nature genetics, 43 1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z et al. (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nature genetics, 46 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.