Abstract
Native mass spectra of large, polydisperse biomolecules with repeated subunits, such as lipoprotein Nanodiscs, can often be challenging to analyze by conventional methods. The presence of tens of closely spaced, overlapping peaks in these mass spectra can make charge state, total mass, or subunit mass determinations difficult to measure by traditional methods. Recently, we introduced a Fourier Transform-based algorithm that can be used to deconvolve highly congested mass spectra for polydisperse ion populations with repeated subunits and facilitate identification of the charge states, subunit mass, charge-state-specific, and total mass distributions present in the ion population. Here, we extend this method by investigating the advantages of using overtone peaks in the Fourier spectrum, particularly for mass spectra with low signal-to-noise and poor resolution. This method is illustrated for lipoprotein Nanodisc mass spectra acquired on three common platforms, including the first reported native mass spectrum of empty “large” Nanodiscs assembled with MSP1E3D1 and over 300 noncovalently associated lipids. It is shown that overtone peaks contain nearly identical stoichiometry and charge state information to fundamental peaks but can be significantly better resolved, resulting in more reliable reconstruction of charge-state-specific mass spectra and peak width characterization. We further demonstrate how these parameters can be used to improve results from Bayesian spectral fitting algorithms, such as UniDec.
Keywords: Native Mass Spectrometry, Nanodisc, Fourier Transform, Deconvolution
Table of Contents Graphic
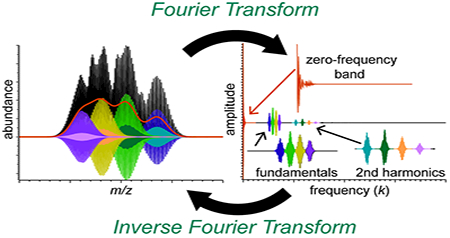
Introduction
Native electrospray ionization mass spectrometry (ESI-MS) can be a powerful tool for investigating the stoichiometry of large, multi-subunit biomolecular assembly ions [1–20]. However, as polydispersity, size, and complexity of these biomolecular ions increase, accurate mass and charge determination can become very challenging, because salt or other co-solute adduction [21, 22] and a high density of mass spectral peaks [5, 23–30] reduce practical resolution. Although separation methods such as chromatography and ion mobility separation can improve resolution of ion subpopulations differing sufficiently in chemical properties or shape [23, 31–39], many types of analytes can still be difficult to separate with these methods, necessitating development of new methods for determining charge states and assembly stoichiometries. In addition to charge-stripping or -reducing approaches, which can increase peak spacing [33, 40], fitting-based mass deconvolution algorithms have been shown to greatly facilitate charge state determination and mass analysis [24, 41–44]. However, to obtain accurate results from these fitting algorithms, the user is typically required to input estimates for some initial parameters, such as mass and charge state ranges and widths of the mass spectral peaks, that are close to the true values. It can thus be highly advantageous to have accurate estimates of these parameters before implementing the algorithms in order to obtain reliable results. Alternatively, Fourier Transform-based deconvolution approaches require minimal data processing and parameter guessing [25, 45, 46]. For example, the recently introduced program iFAMS (interactive Fourier-Transform Analysis for Mass Spectrometry) requires only linear data interpolation and specification of the minimum allowed number of data points separating frequency-domain peaks [25].
Here, we extend our previously-introduced Fourier-Transform-based method for characterizing polydisperse assembly ion mass spectra [25], focusing on the use of information contained in higher harmonic peaks in the Fourier domain, including mass spectral peak shape information. We illustrate the use of higher harmonics in studying lipid-protein Nanodiscs, which are self-assembled discoidal non-covalent assemblies consisting of a phospholipid bilayer surrounded by two amphipathic helical membrane scaffold proteins (MSPs) [47]. Nanodiscs have been used in numerous biochemical applications to study isolated, embedded membrane proteins and protein complexes [47, 48] as well as in native IM-MS to study lipid binding to peripheral and transmembrane protein complexes [29, 49, 50].
A common occurrence in native ESI-MS analysis of Nanodiscs and other large, polydisperse ions, is a relatively large baseline in the raw mass spectrum [1, 25, 26, 37, 51, 52]. This results from the overlap from the superposition of many closely-spaced peaks and can occur even for mass spectra acquired on high-resolution quadrupole–time-of-flight (QTOF) instruments [25, 26, 52]. In such cases, it can be difficult to intuit what part of the mass spectral signal constitutes the baseline and what part contains information that can be used to determine ion masses and charge states. Data pre-processing software for many commercial mass spectrometers, including some Orbitrap and Fourier Transform-Ion Cyclotron Resonance (FT-ICR) mass spectrometers, often performs baseline subtraction, apodization, or other signal processing that can alter peak shapes before data is displayed to the user [53]. After first discussing how Fourier Transform (FT) can be used to extract the information-rich portion of mass spectra for assembly ions, we show how analysis of harmonic peaks in the Fourier spectrum can be used to mitigate artifacts resulting from overlap or low signal-to-noise of peaks in the Fourier spectrum. Previously analyzed native Nanodisc mass spectra acquired on Orbitrap and FT-ICR mass spectrometers [1] are used as benchmarks. Advantages of this FT method are then illustrated for a mass spectrum of native-like “large” Nanodiscs containing over 300 lipids acquired on a QTOF mass spectrometer, which represents an extreme example of mass spectral congestion and is also the first reported mass spectral analysis of this type of “empty” Nanodisc. Finally, we show how information learned from the higher harmonic Fourier analysis can be used to characterize mass spectral peak shapes as well as to determine values for input parameters to improve the quality of results obtained from a Bayesian deconvolution method, UniDec [24].
Methods
Nanodisc preparation.
Nanodiscs containing dimyristoylphosphatidylcholine (DMPC) or dipalmitoylphosphatidylcholine (DPPC) were prepared according to a method adapted from that of Sligar and coworkers [54]. Briefly, all lipids (Avanti Polar Lipids Inc., Alabaster, AL, USA) were prepared as 5 mg/mL solutions in chloroform, dried until opaque with dry nitrogen gas, and resuspended to a final concentration of 50 mM in a pH 7.4 aqueous buffer containing 100 mM sodium cholate (Sigma-Aldrich, St. Louis, MO, USA), 20 mM Tris (Bio-Rad, Hercules, CA, USA), 100 mM sodium chloride, and 0.5 mM ethylenediaminetetraacetic acid (EDTA). Membrane scaffold protein MSP1D1 or MSP1E3D1 (Sigma-Aldrich, St. Louis, MO, USA), for “small” or “large” Nanodiscs, respectively, was reconstituted in pH 7.4 aqueous buffer (20 mM Tris, 100 mM sodium chloride, 0.5 mM EDTA, 0.01% sodium azide) to a concentration of ∼200 μM. Lipid suspensions were mixed with MSP1D1 solutions and additional buffer to a final concentration 50 μM in MSP1D1, 4.0 mM in DMPC, and 25 mM in cholate. MSP1E3D1 Nanodiscs were prepared in a similar fashion, differing only in the final lipid concentration, which was 9.0 mM for DPPC. The samples were incubated for 1 h at room temperature for DMPC (37 °C for DPPC). Nanodisc self-assembly was initiated by 1000:1 (vol:vol) dialysis into the Tris buffer, and BioBeads SM-2 (Bio-Rad, Hercules, CA, USA) were added to the dialysis buffer after being previously rinsed and sonicated three times in methanol followed by three times in the reconstitution buffer. Nanodisc samples were removed from dialysis after 24 hours and were purified by size exclusion chromatography. Fractions containing Nanodiscs were pooled together and concentrated to a final concentration ~10 µM in Nanodiscs. Concentration was determined by UV absorbance of the MSP1D1 or MSP1E3D1 protein, and divided by two (due to the presence of the two scaffold proteins) to obtain Nanodisc concentration.
Native electrospray ionization mass spectrometry.
Native nanoelectrospray ionization (nESI) mass spectra were acquired on an Orbitrap-EMR (Thermo Scientific, Bremen, Germany), Synapt G2-Si Quadrupole Time-of-Flight (QTOF; Waters-MS Technologies, Manchester, UK), or SolariX 15-Tesla FT-ICR (Bruker Daltonics, Bremen, Germany) mass spectrometer. All instrumental parameters are described in greater detail in the Supplementary Material.
Computational work.
All FT-based analysis was performed using the Prell group’s home-built program, iFAMS (interactive Fourier Analysis for Mass Spectra) v. 4.2. All mass spectra are symmetrized before FT is performed to yield real-valued Fourier spectra for ease of graphical presentation. Signal-to-noise is calculated as the maximum amplitude of a peak divided by the root-mean-square white noise at baseline in a neighborhood of the peak. Unless otherwise specified, all other data analysis was performed using Igor Pro v. 6.37 (WaveMetrics, Inc., Lake Oswego, OR, USA).
Theory
The principle of the Fourier Transform (FT)-based mass spectrum analysis method used here for deconvolving heterogeneous mass populations was previously described in detail [25, 45, 46] and therefore will only be briefly explained here. The key concept to the FT algorithm is that a distribution of mass spectral peaks for an individual charge state z of an analyte containing various numbers of a repeated subunit with mass ms can be described as a comb of equally-spaced peaks multiplied by a mass spectral envelope function. Mathematically the mass spectrum (s(m/z)) can be decomposed into three separate functions (Figure 1A): a comb function (c(m/z)) with spacing ms/z between adjacent delta functions, a peak shape function (p(m/z)) that is convolved with the comb function and describes the typical shape of the peaks in the comb, and an envelope function (e(m/z)) that describes the relative abundances of the peaks in the comb, i.e., the stoichiometry distribution. This relationship can be described symbolically in the following way:
| 1. |
where the symbols * and × represent convolution and multiplication, respectively.
Figure 1.
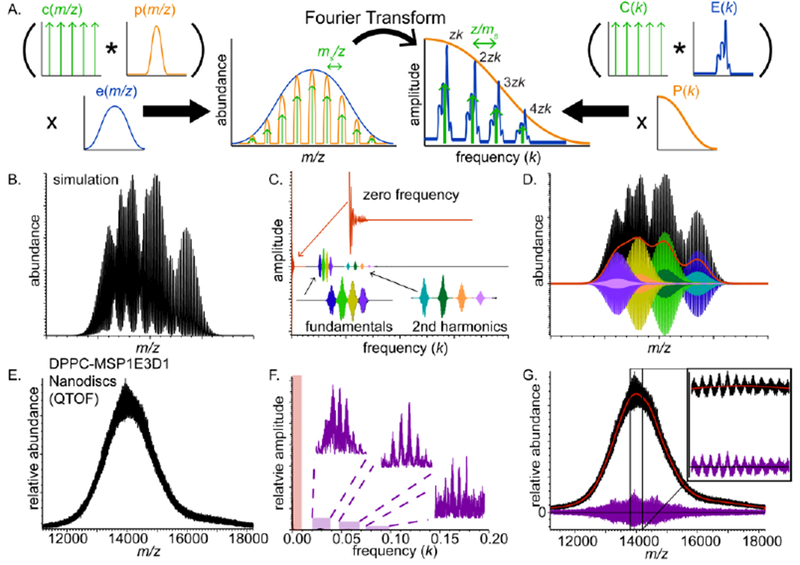
(A) Graphical depiction of mathematical decomposition of a nESI mass spectrum (left) and its Fourier Transform (right) for a polydisperse ion population with a repeated subunit. (B) Simulated mass spectrum, (C) corresponding Fourier spectrum, and (D) reconstructed FT-baseline (red) and charge-state-specific mass spectra with colors identical to their corresponding Fourier-domain peaks from C. (E) DPPC-MSP1E3D1 Nanodisc mass spectrum acquired on an QTOF mass spectrometer, (F) corresponding Fourier spectrum with highlighted zero-frequency band (red) and harmonic peak series (violet), and (G) reconstructed low-information signal (red) and FT-baseline-subtracted mass spectral signal (violet)
The FT of Eq. 1 is (hereafter referred to as the “Fourier spectrum”):
| 2. |
where k = z/Δm is the frequency corresponding to peak spacing Δm/z the mass spectrum. For a given charge state, the peaks in S(k) each have the shape E(k), which is the FT of e(m/z), and the comb of peaks in S(k) decays as P(k), which is the FT of p(m/z). The FT of the entire mass spectrum, which may include several charge states, is the sum of the Fourier spectra corresponding to each charge state present. The fundamental peaks in the Fourier spectrum for each charge state are spaced by 1/ms, thus ms is found by computing the reciprocal of the fundamental peak spacing. The charge states present in the ion population are determined by multiplying the frequencies of the fundamental peaks in the Fourier spectrum by ms, and the stoichiometry distribution e(m/z) can be found for a particular charge state by inverse Fourier Transforming its corresponding fundamental peak in the Fourier spectrum. The relationship between these characteristics of the mass spectrum and its Fourier Transform is illustrated for one charge state series in Fig. 1A.
P(k), which describes the decay of the harmonic frequency peaks for a given charge state, is the FT of p(m/z), the average peak shape for the mass spectral peaks for that charge state. The inverse Fourier Transform of P(k) is therefore p(m/z), the width of which reflects the width of each peak in the comb associated with the chosen charge state.
Results and Discussion
In extremely congested mass spectra with poor resolution, a significant baseline is often observed that may have a complex shape. Beyond a constant or linear baseline subtraction, other forms of baseline fitting and subtraction are often used in mass spectral analysis software that can model curved baseline shapes. These algorithms include fitting a baseline to a polynomial [1] or smoothing a spliced sequence of step functions that measure local minima in the raw mass spectrum [42], and data points with negative abundance values after baseline fitting and subtraction are often removed or set to zero abundance. These methods can be highly effective when adjacent peaks of interest are well-separated and the baseline originates primarily from chemical interferents, such as salt or small molecule clusters [41, 55]. In cases where many peaks of interest overlap strongly, a significant baseline, perhaps even larger in magnitude than the modulation depth of the spectrum, can arise due to the superposition of the tails of these peaks [26, 56, 57]. In such instances, it is not always easy to assess to what extent the above-described baseline subtraction methods distort the true peak shapes, centroids, or relative abundances of ions in the mass spectrum.
For example, Figure 2 shows a QTOF native mass spectrum of “large”-diameter (12.9 nm wide [58]) Nanodiscs assembled using membrane scaffold protein MSP1E3D1 and DPPC lipids. This and other sizes of Nanodiscs are widely used in biochemical studies of isolated membrane protein complexes [47, 50, 59, 60]. A very large, curved baseline is observed that results from the overlap of many poorly-resolved peaks attributed to individual charge states and lipid stoichiometries. Poorly-resolved mass spectra of polydisperse native-like ion populations [27, 28], such as this, motivate a careful analysis of the baseline and signal modulation to determine what portion of the mass spectral signal can be used to reconstruct ion mass, charge, and stoichiometry information, and how this can be done reliably. After establishing the self-consistency of an FT-based method for analyzing similar Nanodisc mass spectra acquired with significantly higher resolution and other instruments, results from this method for this lower-resolution mass spectrum in Fig. 2 are presented below.
Figure 2.
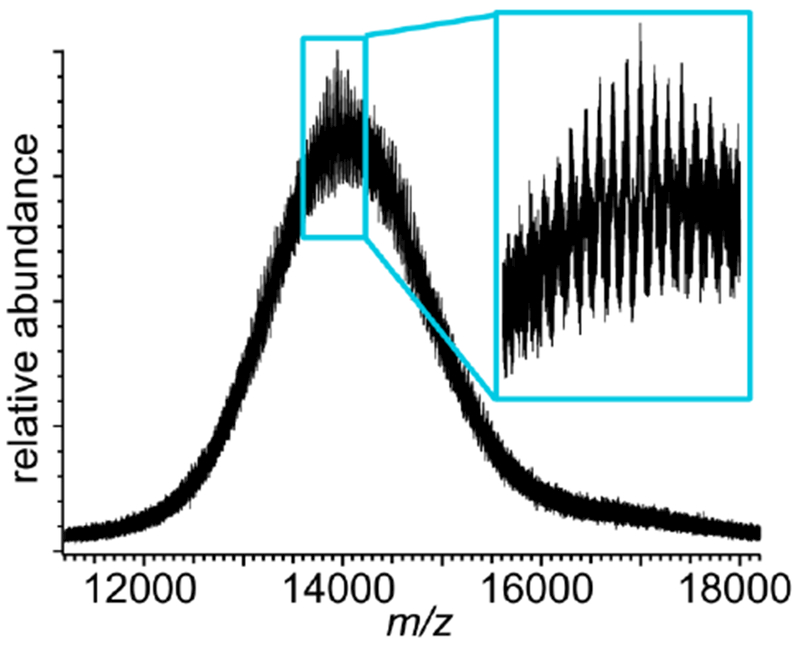
Mass spectrum of lipoprotein Nanodisc ions assembled with DPPC lipid and membrane scaffold protein MSP1E3D1. Inset shows signal modulation in m/z range 13,600–14,200
FT-based approaches for baseline characterization and noise filtering in mass spectra.
Fourier filtering is a well-known procedure in signal processing and many spectroscopic techniques that can facilitate detection and characterization of periodic signals even in a noisy environment as well as removal of low-frequency baselines [61–63]. A raw signal is first Fourier transformed, signals at frequencies of interest are identified, and these signals are extracted using frequency windows before inverse Fourier transforming to yield the filtered signal in the original domain. Figure 1 illustrates a simulated mass spectrum for a population of assembly ions containing a range of subunit stoichiometries and charge states as well as the corresponding FT spectrum. Signal in the Fourier spectrum is observed near zero frequency and at several series of equally-spaced peaks corresponding to the fundamental and higher harmonics belonging to each charge state (Fig. 1C). Because FT is a linear operation, the Fourier spectrum is a sum of each charge-state-specific signal in the Fourier domain. White noise and other non-periodic signals in the mass spectrum are spread out across all frequencies in the Fourier spectrum. While the spacing and shapes of the fundamental and harmonic peaks contain information about charge state, subunit mass, and stoichiometry distribution [25], it is extremely challenging to extract this information from the frequency band around 0 in the Fourier spectrum because this band contains a superposition of signals from all of the charge states. This band is thus much less information-rich than the other peaks in the Fourier spectrum, and it is reasonable to define its inverse Fourier Transform (IFT) as the “low-information signal” of the mass spectrum (see Fig. 1D, red trace).
The remaining signal after subtracting the low-information signal in the mass spectrum thus contains the most useful information, and it typically takes on both positive and negative values that oscillate about zero (see Fig. 1D). It should be noted that this signal is generally not the same as the signal obtained by subtracting a low-order polynomial baseline or by using other common mass spectral domain baseline fitting approaches [24, 42]. Its FT corresponds to the entire Fourier spectrum outside the zero-frequency band. As shown for the simulated, noise-free mass spectrum in Fig. 2, for each charge state, the envelope of the IFT of each harmonic has identical shape but different scaling, and the low-information signal resembles the sum of these envelopes (Fig. 1D). A similar decomposition of the experimental mass spectrum from Fig. 2, which contains white noise, is shown in Fig. 1E-G.
Fourier filtering, which is available in iFAMS, has the potential advantage over simpler low- or high-pass filtering methods in that noise outside and between frequency bands of interest can be eliminated, and signal at several frequencies can be characterized simultaneously. As implemented in iFAMS, Fourier filtering is equivalent to removal of all signal and noise outside the zero-frequency band and Fourier domain peaks. Figure 3 shows the low-information signal and Fourier-filtered mass spectrum reconstructions for previously reported mass spectra of DMPC-MSP1D1 NDs ionized using nESI on an Orbitrap–EMR and 15 Tesla FT-ICR [1], and for DPPC-MSP1E3D1 NDs acquired using a QTOF (same data as in Fig. 2). The FT-ICR and Orbitrap mass spectra have nearly completely resolved peaks and very small low-information signals, whereas the QTOF mass spectrum exhibits a very large low-information signal, in agreement with visual intuition. The root-mean-square (RMS) random noise in the Fourier spectrum for the QTOF data is essentially constant as a function of frequency, whereas it decreases with increasing frequency (up to the Nyquist frequency) for the FT-ICR and Orbitrap data. These differences in mass spectral resolution and frequency-dependence of the RMS random noise can be explained in part by noting that Orbitrap pre-processing software combines magnitude- and absorption-mode data to improve apparent mass spectral peak resolution [53], and both the Orbitrap and FT-ICR data (which is analyzed here in magnitude mode) are apodized during pre-processing to remove ringing artifacts caused by a finite sampling period. The small depth of modulation for the Fourier-filtered QTOF mass spectrum (Fig. 3C, blue trace) as compared to the FT-baseline subtracted mass spectrum in Fig. 1G (violet trace) is consistent with the relatively large random white noise and illustrates the utility of Fourier-filtering in removing this type of noise. A comparison of Fourier filtering with other common noise filtering techniques (Savitzky-Golay, moving-average, and median filters) is shown in the Supplementary material (Supplementary Figure S1) and illustrates that Fourier filtering typically removes much more low- and high-frequency noise from the mass spectrum than do these other filters and has the additional advantage of leaving Fourier-domain peak amplitudes unaltered.
Figure 3.
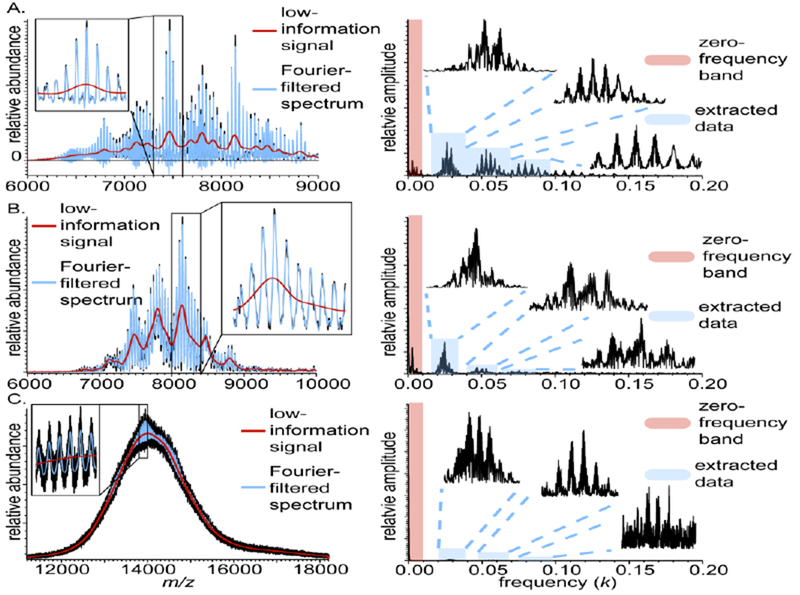
Mass spectra (left) and corresponding Fourier spectra (right) for native-like DMPC-MSP1D1 Nanodiscs measured on (A) Orbitrap and (B) FT-ICR mass spectrometers, and (C) a DPPC-MSP1E3D1 Nanodisc mass spectrum measured on a QTOF mass spectrometer. Fourier-filtered mass spectra (blue overlays) are calculated using the first three harmonics, as well as the Fourier baseline (red) the corresponding to the zero-frequency band. Detailed peak structure illustrated in insets
When multiple harmonics are resolved in the Fourier spectrum, there are in principle multiple pathways by which to determine the subunit mass, charge state distribution, and charge-state-specific mass spectra for this type of ion population, though different pathways may have unique advantages for real data exhibiting Fourier peak overlap or low signal-to-noise. In addition to the spacing and shape of individual peaks in the Fourier spectrum, the relative scaling of the harmonics contains information about the shape and width of peaks in the mass spectrum. Extraction of information from harmonic peaks is described below for Nanodisc mass spectra.
Consistency of subunit mass and charge state determinations using different Fourier-domain harmonic peak series.
In our previous report, FT-based analysis of mass spectra with well-resolved fundamental peaks in the Fourier domain was illustrated [25]. In all three Fourier spectra shown in Fig. 3, the fundamental peaks overlap significantly, thus stoichiometry information obtained by directly inverse Fourier transforming the fundamental peaks may be unreliable, because overlap of signal from adjacent Fourier-domain peaks and use of frequency windows that are too narrow are potential sources of error in Fourier filtering. Supplementary Figure S2 illustrates the relationship between Fourier-domain peak separation, IFT window width, and reconstructed mass spectral ringing artifacts (which are less than 5% of the maximum signal when Fourier-domain peak separation is at least ~1.5 times the sum of adjacent peak widths). A potential solution to these problems is to use higher harmonic peaks rather than fundamental peaks to determine subunit stoichiometry distributions, because they are more widely spaced and less to prone to overlap, though it is important to note that higher harmonic peaks can have lower signal-to-noise than their corresponding fundamentals. Thus, a trade-off between overlap-induced artifacts and artifacts introduced by noise must be considered when performing this analysis.
Figure 4 illustrates this approach, where charge-state-specific mass spectrum reconstructions for the fundamental, second, and third harmonics are shown for the baseline-resolved Orbitrap mass data from Figure 3A. The fundamental peaks have extensive overlap, but charge states 15–23+ are still found using iFAMS, in agreement with previously published results using UniDec [1]. A subunit mass of 678.5 ± 3.6 Da is calculated using iFAMS, which is close to the known mass of DMPC (677.993 Da). These results are also consistent with the second harmonic series, which also indicate charge states 15–23+ and a more accurate and precise subunit mass of 678.2 ± 1.0 Da. Using the third harmonics, iFAMS calculates similar subunit mass (677.3 ± 0.8 Da) but only identifies charge states 16–21+, due to overlap of the higher charge states with the 4th harmonics. Thus, overlap of two different charge states from two different harmonic series can in certain cases represents a limitation in the higher harmonic analysis. These results are summarized in Table 1.
Figure 4.
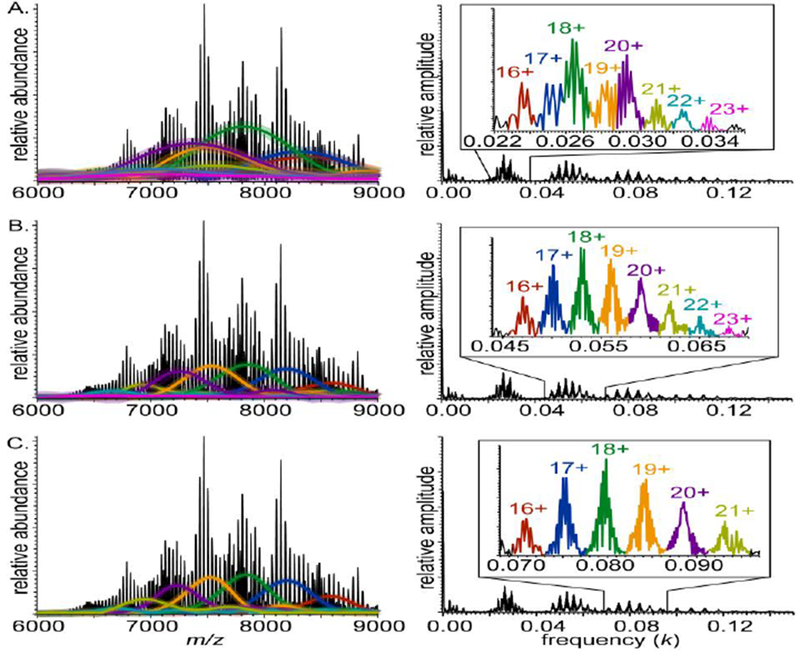
Mass spectrum of DMPC-MSP1D1 Nanodiscs acquired on an Orbitrap mass spectrometer (left) and corresponding Fourier spectrum (right) for (A) fundamentals, (B) second harmonics, and (C) third harmonics. IFT of the charge-state specific peaks in Fourier spectra (insets) are shown as overlaid envelope functions of the same color in mass spectra. Faint color surrounding envelope functions represents uncertainty of envelope functions calculated from average noise amplitude in the Fourier domain
Table 1.
Lipid Mass, Charge States, and Lipid Stoichiometry Statistics Determined for Native-Like Nanodisc Ions Using FT-Based Approach.
| Analyte (Instrument) | Harmonic | Measured Subunit Mass (Da) | z | Lipid Stoichiometry |
|---|---|---|---|---|
| DPPC-MSP1E3D1 (QTOF) | Fundamental | 733. ± 2. | 18 | 273 ± 36† |
| 19 | 290 ± 32† | |||
| 20 | 304 ± 28† | |||
| 21 | 316 ± 29† | |||
| 22 | 325 ± 30† | |||
| 23 | 340 ± 41† | |||
| 24 | 353 ± 47 | |||
| 2nd | 733.0 ± 0.8 | 18 | 275 ± 41* | |
| 19 | 294 ± 29 | |||
| 20 | 305 ± 26 | |||
| 21 | 315 ± 26 | |||
| 22 | 327 ± 33 | |||
| 23 | 352 ± 46* | |||
| 24 | 383 ± 56* | |||
| Average of fundamental and 2nd harmonic | N/A | 18 | 274 ± 39 | |
| 19 | 292 ± 31 | |||
| 20 | 304 ± 27 | |||
| 21 | 316 ± 28 | |||
| 22 | 326 ± 32 | |||
| 23 | 346 ± 45 | |||
| 24 | 372 ± 55 | |||
| DMPC-MSP1D1 (Orbitrap) | Fundamental | 678.5 ± 3.6 | 16 | 125 ± 15† |
| 17 | 129 ± 24† | |||
| 18 | 137 ± 19† | |||
| 19 | 143 ± 22† | |||
| 20 | 149 ± 20† | |||
| 21 | 158 ± 22† | |||
| 22 | 166 ± 23 | |||
| 23 | 179 ± 23 | |||
| 2nd | 678.2 ± 1.0 | 16 | 128 ± 17 | |
| 17 | 136 ± 14 | |||
| 18 | 143 ± 12 | |||
| 19 | 146 ± 14 | |||
| 20 | 150 ± 16 | |||
| 21 | 154 ± 20 | |||
| 22 | 162 ± 25 | |||
| 23 | 178 ± 28 | |||
| 3rd | 677.3 ± 1.2 | 16 | 127 ± 17 | |
| 17 | 137 ± 12 | |||
| 18 | 141 ± 12 | |||
| 19 | 145 ± 12 | |||
| 20 | 150 ± 15 |
indicates Fourier-domain peaks with signal-to-noise less than 10:1
indicates Fourier-domain peaks spaced by less than 1.5 times the sum of their apparent standard deviations
While the charge states and subunit mass measurements are similar for the fundamental, second, and third harmonics, a notable difference can be seen in their reconstructed mass spectral envelope functions, particularly when using the fundamentals. The reconstructed charge-state-specific envelope functions using the fundamental peaks are visibly wider than those for the 2nd and 3rd harmonics, a result attributed to strong overlap of the fundamental peaks in the Fourier spectrum (Fig. 4). The higher harmonic peaks in the Fourier spectrum are more widely spaced and are better resolved, leading to narrower corresponding charge-state-specific envelope reconstructions (Fig. 4). The corresponding standard deviations in the number of lipids present (Table 1) are consistently lower than those found using the fundamental peaks, and lipid statistics are nearly identical when using the 2nd and 3rd harmonics for charge states 16–21+. Notably, all of these Fourier-domain peaks have signal-to-noise greater than 10:1 and inter-peak separation exceeding 1.5 times the sum of adjacent peak widths. Subunit statistics for Fourier-domain peaks that meet these two criteria are therefore likely to be accurate in general even for mass spectra for which only one set of harmonic peaks meets them, as is often the case in the other Nanodisc spectra presented here. Similar results for mass spectra exhibiting even greater overlap of the fundamental peaks are illustrated in Supplementary Figure S3 and Supplementary Table 1.
Figure 5 shows results for the much more congested QTOF mass spectrum from Fig. 2, in which the mass spectral peaks are far from baseline-resolved. iFAMS analysis of the fundamental frequencies indicates charge states 18–24+ and average sub-unit mass of 732. ± 2. Da, which is within one standard deviation of the average mass of DPPC (733.562 Da). However, the 19 and 20+ fundamental peaks overlap strongly, so their corresponding charge-state-specific mass spectra cannot be reliably reconstructed from these peaks alone. By contrast, the second harmonic peak sequence is baseline-separated. The same range of charge states, 18–24+, is identified, and a more accurate and precise average sub-unit mass of 733.0 ± 0.8 Da is determined. The average number and standard deviation in the number of lipid subunits for these charge states determined using the fundamental and second harmonic peaks are very similar. Together with the results for DMPC-MSP1D1 Nanodisc ions described above, these results show that higher harmonic frequencies can often beneficial in determining charge states, subunit mass, and charge-state-specific mass spectra for polydisperse assembly ion populations. Similar results were obtained for the spectrum in Fig. 3B and are shown in Supplementary Figure S4.
Figure 5.
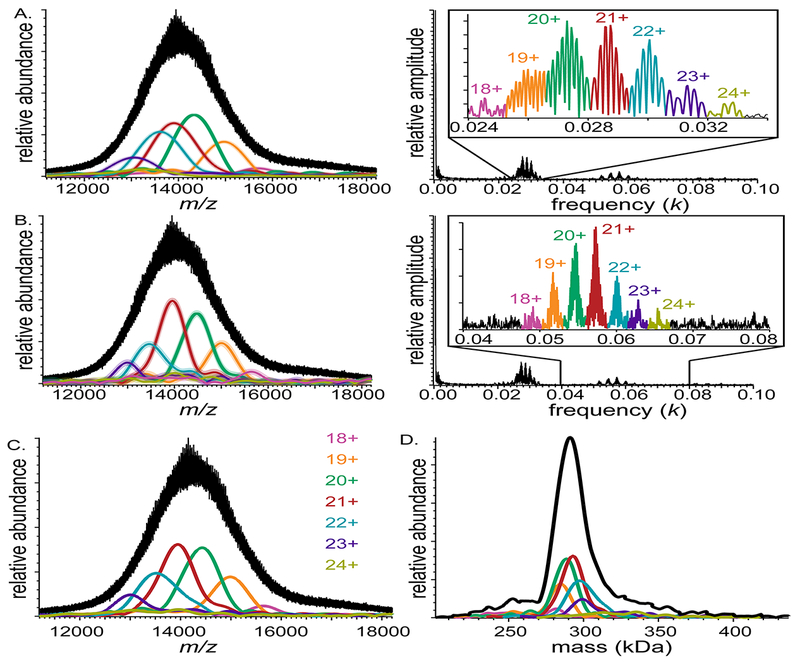
Mass spectrum of DPPC-MSP1E3D1 Nanodiscs acquired on a QTOF mass spectrometer (left) and corresponding Fourier spectrum (right) for (A) fundamentals and (B) second harmonics. IFT of the charge-state specific peaks in Fourier spectra (insets) are shown as overlaid envelope functions of the same color in mass spectra. Faint color surrounding envelope functions represents uncertainty of envelope functions calculated from average noise amplitude in the Fourier domain. (C) Harmonic-averaged reconstruction of envelope functions. (D) Zero-charge spectrum (black), calculated from harmonic-averaged spectra for all charge states
In addition to signal overlap and artifacts introduced by using overly narrow Fourier frequency windows (see Fig. S2), artifacts in reconstructions of charge-state-specific envelope functions can occur if the white noise present in the Fourier spectrum obscures the true shape of higher harmonic peaks that have relatively low signal-to-noise. Averaging the shape of two or more reconstructed spectra from different harmonics belonging to the same charge state is a potential strategy for reducing artifacts of this type. “Harmonic-average” charge-state-specific mass spectra for the data from Fig. 2 were reconstructed by directly averaging the IFT of the fundamental and higher harmonic peaks for each charge state (Fig. 5C). The harmonic-averaged spectral envelopes widths are slightly wider than those for the second harmonics, but contain fewer periodic “ripples” in the tails of the spectra due to averaging with the data from the fundamentals. A “zero-charge” mass spectrum for the entire ion population was also calculated from these harmonic-averaged mass spectra (Fig. 5D) to illustrate the mass distribution for the entire ion population. (Zero-charge spectra for the Orbitrap and FT-ICR spectrum in Figs. 3A and 3B can be found in Supplementary Figures S5 and S6.) The average ion mass found by this method is ~290 kDa, which, assuming two scaffold proteins are present per ion, means that the DPPC-MSP1E3D1 Nanodiscs in the ion population represented in Fig. 2 have an average of ~306 lipids. Using this number, an average condensed-phased area per lipid head group in the Nanodisc of ~60 A2 can also be estimated, assuming that each leaflet has a diameter of 2 nm smaller than the diameter of the assembly [58]. This result agrees well with previous computational simulations of model DPPC bilayers [64].
Characterizing peak width and unresolved adductions in the mass spectrum and Fourier domain.
When baseline-resolved mass spectral peaks can be attributed to only one charge state, peak broadening caused by unresolved adductions (such as solvent molecules and small cosolute ions) for each peak can in principle be determined from the width of the peaks in the mass spectrum and (separately-measured) instrumental resolving power. If peaks from more than one charge state overlap or there is a large baseline, peak width determination directly from the mass spectrum can be more challenging. Statistics for the average peak shape for each charge state instead can be determined from analysis of the higher harmonic peaks in the Fourier spectrum, which have amplitudes that decay as the FT of the average mass spectral peak shape for the corresponding charge state (P(k); see Fig. 1A). Then, p(m/z) for each charge state is simply the IFT of P(k). However, for a typical ESI mass spectrum with multiple overlap charge states, it is typically infeasible to determine this zero-frequency contribution for individual charge states, but it may be reasonable to assume that the zero-frequency amplitude has a fixed relationship to the amplitudes of all the other harmonic peaks. For example, if p(m/z) is Gaussian in shape, as in the simulated spectrum in Supplementary Figures S7A and S7B, its Fourier Transform, P(k), is also a Gaussian, and the FWHM of P(k) is inversely proportional to that of p(m/z). The FWHM of p(m/z) can then be determined from P(k) without knowing its amplitude at zero frequency, provided enough harmonics have sufficient signal-to-noise to confidently fit P(k) to a Gaussian.
The baseline-resolved Orbitrap mass spectrum in Fig. 4A has roughly Gaussian peaks and is in this respect similar to the modeled spectrum in Supplementary Figure S7B. Average peak-width statistics for each charge state can thus be readily estimated from reconstruction of P(k). This is shown in Fig. 6A for charge states 17, 18, and 19+ using the second, third, and fourth harmonics (the fundamentals, as well as fifth and higher order harmonics, could not be used due to overlap). Using these parameters, fitting a Gaussian to the higher harmonics results in a reconstructed average FWHM, in m/z, of 8.1 ± 0.1, 7.9 ± 0.1, and 7.5 ± 0.1 for the 17, 18, and 19+ charge states respectively. Note that these and all peak width uncertainties reported below reflect fitting to a forced Gaussian shape and do not include uncertainty reflecting exact, i.e., non-Gaussian, peak shapes. These peak width values are close to forced-Gaussian FWHM measurements determined directly from the mass spectrum (7.8 ± 0.6, 6.4 ± 0.6, and 6.3 ± 0.6 for charge states 17, 18, and 19+), with the small discrepancies likely arising from the slightly asymmetric shape of the mass spectral peaks. This result is consistent with a small degree of salt adduction and the slight deviation of the data from the Gaussian fits shown in Fig. 6A.
Figure 6.
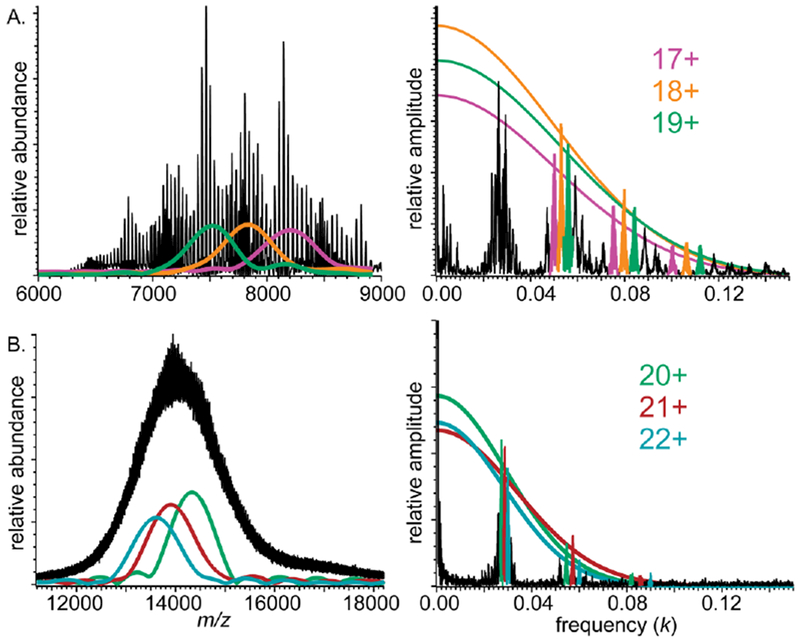
Mass spectra (left) and corresponding Fourier spectra (right) of (A) DMPC-MSP1D1 Nanodiscs acquired using an Orbitrap mass spectrometer and (B) DPPC-MSP1E3D1 Nanodiscs acquired using a QTOF mass spectrometer. Charge-state-specific mass spectral envelope functions (left), and Gaussian frequency decay functions (P(k), right) are shown with same color
A similar analysis was performed for the poorly resolved QTOF spectrum from Fig. 2 assuming Gaussian shape peaks (Fig. 6B). The FWHM, in m/z, for charge states 20, 21, and 22+ are found to be 13.7 ± 0.2, 12.2 ± 0.1, and 13.6 ± 1.0, respectively. This is within error of values found by measuring the most abundant peak in each charge state after an initial smoothing and baseline subtraction (13. ± 1., 12. ± 1., and 13. ±1., respectively). Notably, both the directly measured and FT-reconstructed peak FWHM are less than half the inter-peak spacing for each charge state (~17–18, in m/z, for these ions).
Potential errors in peak-width determination can occur when using this method if the mass spectral peak shapes are very poorly resolved or highly non-Gaussian, especially if a large baseline is subtracted before peak shape analysis. The extent of these potential errors was investigated using model spectra with peak FWHM ranging from 115% to 180% of the inter-peak spacing for each charge state. For example, Supplementary Figure 7A shows a mass spectrum in which the peak FWHM is 115% of the inter-peak spacing. The FT approach described above yields a FWHM, in m/z, of 6.5, whereas direct measurement of the FWHM in the mass spectrum after baseline subtraction is ~5.1, slightly smaller than the true value (5.9). When the true FWHM are increased, the resulting mass spectra have even larger baselines. If these curved (i.e., polynomial-fitted, not FT) baselines are subtracted, the resulting spectrum is ostensibly baseline-resolved, and directly measuring the FWHM of the peaks leads to nearly identical measurements for the FWHM (5.3, 5.4, and 5.6, respectively), increasingly far from the correct values (7.1, 8.2, and 9.4), but all very similar to half the inter-peak spacing (5.1). This error trend arises from the fact that the signal modulation with respect to the low-information signal in the mass spectrum asymptotically approaches a simple sinusoid at the fundamental frequency as the width of the individual peaks increases. In sharp contrast, Fourier transforming the model mass spectra, with or without subtracting the curved baseline, results in much more accurate FWHM measurements because the shape of P(k) is easily measured in the Fourier spectra. These results illustrate the robustness of this method and indicate one should exercise caution in interpreting mass spectra after curved baseline subtraction, especially as relates to peak width and resolution, if peak widths are similar to or larger than inter-peak spacing. Peak width analysis can be even more challenging in the case that the mass spectral peak shape is far from Gaussian (see Supplementary Material, especially Figure S7C and S7D).
Use of parameters determined from FT algorithm to improve results of Bayesian mass spectral fitting.
Polydisperse mass populations with repeated subunits from native mass spectra, such as the Nanodisc mass spectra investigated here, can in many cases be deconvolved using other strategies besides Fourier Transform, such as Bayesian fitting, the approach implemented in UniDec/MetaUniDec [1, 24, 65], or MaxEnt [55]. These fitting algorithms typically require the user to input some initial parameters, such as charge state, peak width, and mass estimates, and a modeled data set based on those parameters is iteratively fit to the experimental data set until convergence is achieved. (The MaxEnt algorithm further assumes that the stoichiometry distribution for each charge state is identical, which may not be the case for some polydisperse ion populations, such as the Nanodiscs investigated here; see Table 1.) When using these fitting algorithms, choosing the right initial parameters can therefore be highly advantageous for extracting accurate information, especially when the mass spectra have low signal-to-noise or the goodness-of-fit function used in the algorithm contains multiple local extrema. These effects are illustrated for simulated DPPC-MSP1E3D1 Nanodisc mass spectra in Supplementary Figure S8 with signal-to-white-noise ratio 20:1 and a realistic increase in average lipid content of 10 lipids per charge state. Despite the low signal-to-noise ratio, nearly perfect reconstruction of the exact zero-charge mass spectrum is achieved using the FT approach alone, and dramatic improvement of reconstructions using UniDec is observed when the charge state range is limited using output from iFAMS and a Fourier-filtered mass spectrum is used as input.
Shown in Figure 7 are two deconvolutions for the experimental DPPC-MSP1E3D1 Nanodisc mass spectrum from Fig. 2 using UniDec’s Bayesian deconvolution algorithm. The initial parameters for both analyses were chosen as follows: a charge state range of 15–27+ and a mass range for the entire complex of 250–350 kDa. These values were chosen to simulate a typical scenario in native MS in which the approximate charge state and mass range of a population of assembly ions can be estimated, but the repeated sub-unit mass is unknown. Figs. 7A, 7B, and 7C show results from using the Bayesian algorithm with no initial guess for the lipid mass and an initial peak FWHM, in m/z, of 8.9, which is obtained from the mass spectrum using the peak width tool in UniDec after performing an initial baseline subtraction. Overall, the results are similar, but not identical, to those from the FT-based approach (see Fig. 4 and Table 1), though a number of high-intensity orphan peaks, presumably artifacts, are found in the zero-charge mass spectrum with these input parameters at masses below 270 kDa and above 300 kDa.
Figure 7.
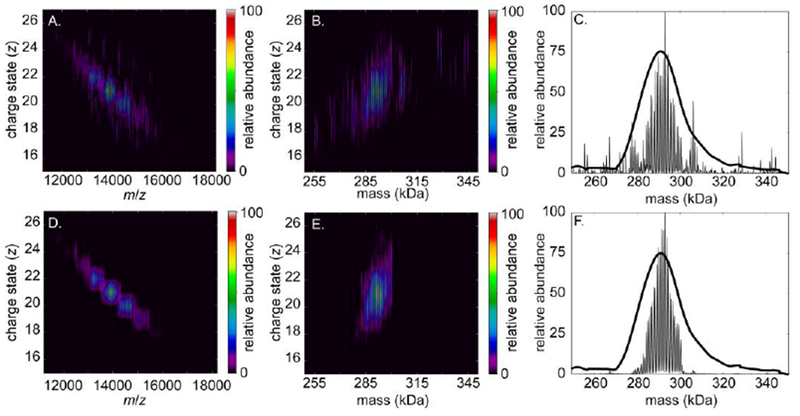
Deconvolved QTOF mass spectra vs. charge (left), mass vs. charge (center) and zero-charge mass spectra (right) of DPPC-MSP1E3D1 Nanodiscs determined using UniDec. Mass spectral data are the same as in Fig. 2. A, B, and C result from using “naïve” input parameters for subunit mass, charge state range, peak width, and total mass range, whereas D, E, and F result from using values obtained from the FT-based method (see text). Smooth black trace in C and F represents zero-charge mass spectrum reconstructed using FT approach (see Fig. 5D)
Figs. 7D, 7E, and 7F show results from UniDec for the same mass spectrum, but using input mass spectra and parameters determined from the different FT-based approaches described above: the Fourier-filtered spectrum as the input spectrum, a subunit mass of 733 Da determined using iFAMS and a peak FWHM of 13 determined from P(k). With these input parameters, the spurious peaks in the zero-charge mass spectrum are nearly eliminated and charge states and mass estimates for the ion population agree much more closely with results obtained using the FT analysis above. The zero-charge mass spectrum reconstructed with UniDec is somewhat narrower than the one reconstructed using the FT approach, consistent with the non-overlapping charge-state-specific mass spectra reconstructed in UniDec (Fig. 7D). This demonstrates that information obtained from the Fourier analysis can be used as input parameters in Bayesian fitting algorithms to improve the quality of the results. Similar results were found for UniDec processing of the Orbitrap and FT-ICR spectra, as shown in Supplementary Figures S9 and S10.
Conclusions
As the composition and structure of larger and more heterogeneous assembly ions probed with mass spectrometry continue to increase in complexity, analysis of their mass spectra demands more powerful approaches for assigning charge state, mass, and stoichiometry. Developing instruments with increased resolving power is one potential strategy, but access to these higher resolution instruments may not always be available for some laboratories, and it may be desirable to perform tandem experiments, such as ion mobility spectrometry and surface-induced dissociation, that are not widely available in high-resolution instruments. Furthermore, resolution limitations associated with current instruments will inevitably arise again for future instruments as the size and complexity of ions continues to increase, even with the advantages of charge-stripping or –reduction techniques [33, 40]. The results presented here illustrate how the Fourier Transform-based analysis strategy can allow one to work within current instrumental limitations when studying assembly ions with repeated subunits. Both experimental and computational results indicate that reliable and self-consistent charge state, subunit mass, and subunit stoichiometry determinations can be made using this strategy, especially if Fourier-domain peaks are well resolved (with inter-peak spacing at least ~1.5 times the sum of adjacent peak widths) and have signal-to-noise of at least 10:1. This deconvolution method can be used alone or in combination with Bayesian deconvolution techniques, for which it can provide input parameter values, such as charge state range, subunit mass, mass range and mass spectral peak width, to improve results. Furthermore, information learned through FT-based analysis can help avoid potential errors in analyses associated with mass spectral domain deconvolution algorithms, including error attributed to curved baseline subtraction or parameter estimation. The general principles discussed here are valid for many other types of assembly ions with repeated subunits, including biomolecular assemblies, polymers, and inorganic cluster ions, due to the analogous form of their nESI mass spectra. An updated version of iFAMS, used here to analyze mass spectra with the FT approach, is freely available to the public as an open-source Python program available for download at https://github.com/seanpatcleary/iFAMS.
Supplementary Material
Table 2.
Mass Spectral Peak Widths for Native-Like Nanodisc Ions Determined Using P(k) and Directly from Mass Spectrum
| Analyte (Instrument) | z | FWHM found using P(k) | FWHM from Mass Spectrum |
|---|---|---|---|
| DPPC-MSP1E3D1 (QTOF) | 20+ | 13.7 ± 0.2 | 13. ± 1. |
| 21+ | 12.2 ± 0.1 | 12. ± 1. | |
| 22+ | 13.6 ± 1.0 | 13. ± 1. | |
| DMPC-MSP1D1 (Orbitrap) | 17+ | 8.1 ± 0.1 | 7.8 ± 0.6 |
| 18+ | 7.9 ± 0.1 | 6.4 ± 0.6 | |
| 19+ | 7.5 ± 0.1 | 6.3 ± 0.6 |
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health under Award Number R21AI125804 (to J.S.P.) and R01GM103479 and S10RR028893 (to J.A.L.) and the American Society for Mass Spectrometry Postdoctoral Research Award (to H.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank the Amgen Pharmacokinetics and Drug Metabolism group (PKDM) in South San Francisco for instrument time on the Orbitrap EMR.
References
- 1.Campuzano IDG, Li H, Bagal D, Lippens JL, Svitel J, Kurzeja RJM, Xu H, Schnier PD, Loo JA: Native ms analysis of bacteriorhodopsin and an empty nanodisc by orthogonal acceleration time-of-flight, orbitrap and ion cyclotron resonance. Anal. Chem. 88, 12427–12436 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrera NP, Isaacson SC, Zhou M, Bavro VN, Welch A, Schaedler TA, Seeger MA, Miguel RN, Korkhov VM, van Veen HW, Venter H, Walmsley AR, Tate CG, Robinson CV: Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat. Methods. 6, 585–587 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benesch JLP, Ruotolo BT: Mass spectrometry: Come of age for structural and dynamical biology. Curr. Opin. Struct. Biol. 21, 641–649 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Cui WD, Gross ML, Blankenship RE: Native mass spectrometry of photosynthetic pigment-protein complexes. FEBS Lett. 587, 1012–1020 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marty MT, Zhang H, Cui WD, Blankenship RE, Gross ML, Sligar SG: Native mass spectrometry characterization of intact nanodisc lipoprotein complexes. Anal. Chem. 84, 8957–8960 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewing SA, Donor MT, Wilson JW, Prell JS: Collidoscope: An improved tool for computing collisional cross-sections with the trajectory method. J. Am. Soc. Mass Spectrom. 28, 587–596 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donor MT, Ewing SA, Zenaidee MA, Donald WA, Prell JS: Extended protein ions are formed by the chain ejection model in chemical supercharging electrospray ionization. Anal. Chem. 89, 5107–5114 (2017) [DOI] [PubMed] [Google Scholar]
- 8.Li HL, Wolff JJ, Van Orden SL, Loo JA: Native top-down electrospray ionization-mass spectrometry of 158 kda protein complex by high-resolution fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 86, 317–320 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laganowsky A, Reading E, Hopper JTS, Robinson CV: Mass spectrometry of intact membrane protein complexes. Nat. Protoc. 8, 639–651 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, Dagan S, Wysocki VH: Protein subunits released by surface collisions of noncovalent complexes: Nativelike compact structures revealed by ion mobility mass spectrometry. Angew. Chem., Int. Ed. 51, 4336–4339 (2012) [DOI] [PubMed] [Google Scholar]
- 11.Sterling HJ, Kintzer AF, Feld GK, Cassou CA, Krantz BA, Williams ER: Supercharging protein complexes from aqueous solution disrupts their native conformations. J. Am. Soc. Mass Spectrom. 23, 191–200 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heck AJR, van den Heuvel RHH: Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 23, 368–389 (2004) [DOI] [PubMed] [Google Scholar]
- 13.Loo JA: Electrospray ionization mass spectrometry: A technology for studying noncovalent macromolecular complexes. Int. J. Mass Spectrom. 200, 175–186 (2000) [Google Scholar]
- 14.Pukala TL, Ruotolo BT, Zhou M, Politis A, Stefanescu R, Leary JA, Robinson CV: Subunit architecture of multiprotein assemblies determined using restraints from gas-phase measurements. Structure. 17, 1235–1243 (2009) [DOI] [PubMed] [Google Scholar]
- 15.Robinson CV, Chung EW, Kragelund BB, Knudsen J, Aplin RT, Poulsen FM, Dobson CM: Probing the nature of noncovalent interactions by mass spectrometry. A study of protein−coa ligand binding and assembly. J. Am. Chem. Soc. 118, 8646–8653 (1996) [Google Scholar]
- 16.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT: Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 (2010) [DOI] [PubMed] [Google Scholar]
- 17.Konijnenberg A, Butterer A, Sobott F: Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta, Proteins Proteomics 1834, 1239–1256 (2013) [DOI] [PubMed] [Google Scholar]
- 18.Abzalimov RR, Kaltashov IA: Electrospray ionization mass spectrometry of highly heterogeneous protein systems: Protein ion charge state assignment via incomplete charge reduction. Anal. Chem. 82, 7523–7526 (2010) [DOI] [PubMed] [Google Scholar]
- 19.Zhou M, Wysocki VH: Surface induced dissociation: Dissecting noncovalent protein complexes in the gas phase. Acc. Chem. Res. 47, 1010–1018 (2014) [DOI] [PubMed] [Google Scholar]
- 20.Salbo R, Bush MF, Naver H, Campuzano I, Robinson CV, Pettersson I, Jørgensen TJD, Haselmann KF: Traveling-wave ion mobility mass spectrometry of protein complexes: Accurate calibrated collision cross-sections of human insulin oligomers. Rapid Commun. Mass Spectrom. 26, 1181–1193 (2012) [DOI] [PubMed] [Google Scholar]
- 21.Pan J, Xu K, Yang X, Choy W-Y, Konermann L: Solution-phase chelators for suppressing nonspecific protein−metal interactions in electrospray mass spectrometry. Anal. Chem. 81, 5008–5015 (2009) [DOI] [PubMed] [Google Scholar]
- 22.McKay AR, Ruotolo BT, Ilag LL, Robinson CV: Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 128, 11433–11442 (2006) [DOI] [PubMed] [Google Scholar]
- 23.Trimpin S, Plasencia M, Isailovic D, Clemmer DE: Resolving oligomers from fully grown polymers with ims−ms. Anal. Chem. 79, 7965–7974 (2007) [DOI] [PubMed] [Google Scholar]
- 24.Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP, Robinson CV: Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleary SP, Thompson AM, Prell JS: Fourier analysis method for analyzing highly congested mass spectra of ion populations with repeated subunits. Anal. Chem. 88, 6205–6213 (2016) [DOI] [PubMed] [Google Scholar]
- 26.Hopper JTS, Yu YTC, Li DF, Raymond A, Bostock M, Liko I, Mikhailov V, Laganowsky A, Benesch JLP, Caffrey M, Nietlispach D, Robinson CV: Detergent-free mass spectrometry of membrane protein complexes. Nat. Methods 10, 1206–1208 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trimpin S, Clemmer DE: Ion mobility spectrometry/mass spectrometry snapshots for assessing the molecular compositions of complex polymeric systems. Anal. Chem. 80, 9073–9083 (2008) [DOI] [PubMed] [Google Scholar]
- 28.Larriba C, de la Mora JF, Clemmer DE: Electrospray ionization mechanisms for large polyethylene glycol chains studied through tandem ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 25, 1332–1345 (2014) [DOI] [PubMed] [Google Scholar]
- 29.Zhang YX, Liu L, Daneshfar R, Kitova EN, Li CS, Jia F, Cairo CW, Klassen JS: Protein-glycosphingolipid interactions revealed using catch-and-release mass spectrometry. Anal. Chem. 84, 7618–7621 (2012) [DOI] [PubMed] [Google Scholar]
- 30.Fouquet T, Sato H: Extension of the kendrick mass defect analysis of homopolymers to low resolution and high mass range mass spectra using fractional base units. Anal. Chem. 89, 2682–2686 (2017) [DOI] [PubMed] [Google Scholar]
- 31.Causon TJ, Hann S: Theoretical evaluation of peak capacity improvements by use of liquid chromatography combined with drift tube ion mobility-mass spectrometry. J. Chromatogr. A. 1416, 47–56 (2015) [DOI] [PubMed] [Google Scholar]
- 32.Arthur KL, Turner MA, Reynolds JC, Creaser CS: Increasing peak capacity in nontargeted omics applications by combining full scan field asymmetric waveform ion mobility spectrometry with liquid chromatography - mass spectrometry. Anal. Chem. 89, 3452–3459 (2017) [DOI] [PubMed] [Google Scholar]
- 33.Bagal D, Zhang H, Schnier PD: Gas-phase proton-transfer chemistry coupled with tof mass spectrometry and ion mobility-ms for the facile analysis of poly(ethylene glycols) and pegylated polypeptide conjugates. Anal. Chem. 80, 2408–2418 (2008) [DOI] [PubMed] [Google Scholar]
- 34.Kintzer AF, Thoren KL, Sterling HJ, Dong KC, Feld GK, Tang II, Zhang TT, Williams ER, Berger JM, Krantz BA: The protective antigen component of anthrax toxin forms functional octameric complexes. J. Mol. Biol. 392, 614–629 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV: Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 (2008) [DOI] [PubMed] [Google Scholar]
- 36.Clemmer DE, Jarrold MF: Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 32, 577–592 (1997) [Google Scholar]
- 37.Hoi KK, Robinson CV, Marty MT: Unraveling the composition and behavior of heterogeneous lipid nanodiscs by mass spectrometry. Anal. Chem. 88, 6199–6204 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ewing MA, Glover MS, Clemmer DE: Hybrid ion mobility and mass spectrometry as a separation tool. J. Chromatogr. A. 1439, 3–25 (2016) [DOI] [PubMed] [Google Scholar]
- 39.Dwivedi P, Wu C, Matz LM, Clowers BH, Siems WF, Hill HH: Gas-phase chiral separations by ion mobility spectrometry. Anal. Chem. 78, 8200–8206 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laszlo KJ, Munger EB, Bush MF: Folding of protein ions in the gas phase after cation-to-anion proton-transfer reactions. J. Am. Chem. Soc. 138, 9581–9588 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H, Ojha PC, McClean S, Black ND, Hughes JG, Shaw C: Heuristic charge assignment for deconvolution of electrospray ionization mass spectra. Rapid Commun. Mass Spectrom. 17, 429–436 (2003) [DOI] [PubMed] [Google Scholar]
- 42.Morgner N, Robinson CV: Massign: An assignment strategy for maximizing information from the mass spectra of heterogeneous protein assemblies. Anal. Chem. 84, 2939–2948 (2012) [DOI] [PubMed] [Google Scholar]
- 43.van Breukelen B, Barendregt A, Heck AJR, van den Heuvel RHH: Resolving stoichiometries and oligomeric states of glutamate synthase protein complexes with curve fitting and simulation of electrospray mass spectra. Rapid Commun. Mass Spectrom. 20, 2490–2496 (2006) [DOI] [PubMed] [Google Scholar]
- 44.Stengel F, Baldwin, Andrew J., Bush, Matthew F., Hilton, Gillian R., Lioe, H., Basha, E., Jaya, N., Vierling, E., Benesch, Justin L. P.: Dissecting heterogeneous molecular chaperone complexes using a mass spectrum deconvolution approach. Chem. Biol. 19, 599–607 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prebyl BS, Cook KD: Use of fourier transform for deconvolution of the unresolved envelope observed in electrospray ionization mass spectrometry of strongly ionic synthetic polymers. Anal. Chem. 76, 127–136 (2004) [Google Scholar]
- 46.Danis PO, Huby FJ: The computer-assisted interpretation of copolymer mass spectra. J. Am. Soc. Mass Spectrom. 6, 1112–1118 (1995) [DOI] [PubMed] [Google Scholar]
- 47.Denisov IG, Sligar SG: Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 117, 4669–4713 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG: Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cong X, Liu Y, Liu W, Liang X, Russell DH, Laganowsky A: Determining membrane protein–lipid binding thermodynamics using native mass spectrometry. J. Am. Chem. Soc. 138, 4346–4349 (2016) [DOI] [PubMed] [Google Scholar]
- 50.Marty MT, Hoi KK, Gault J, Robinson CV: Probing the lipid annular belt by gas-phase dissociation of membrane proteins in nanodiscs. Angew. Chem.-Int. Edit 55, 550–554 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bechara C, Noell A, Morgner N, Degiacomi MT, Tampe R, Robinson CV: A subset of annular lipids is linked to the flippase activity of an abc transporter. Nat. Chem. 7, 255–262 (2015) [DOI] [PubMed] [Google Scholar]
- 52.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJR: Interrogating viral capsid assembly with ion mobility–mass spectrometry. Nat. Chem. 3, 126–132 (2011) [DOI] [PubMed] [Google Scholar]
- 53.Lange O, Damoc E, Wieghaus A, Makarov A: Enhanced fourier transform for orbitrap mass spectrometry. Int. J. Mass Spectrom. 369, 16–22 (2014) [Google Scholar]
- 54.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG: Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 (2004) [DOI] [PubMed] [Google Scholar]
- 55.Mann M, Meng CK, Fenn JB: Interpreting mass-spectra of multiply charged ions. Anal. Chem. 61, 1702–1708 (1989) [Google Scholar]
- 56.Marty MT, Zhang H, Cui WD, Gross ML, Sligar SG: Interpretation and deconvolution of nanodisc native mass spectra. J. Am. Soc. Mass Spectrom. 25, 269–277 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu J, Trnka MJ, Roh S-H, Robinson PJJ, Shiau C, Fujimori DG, Chiu W, Burlingame AL, Guan S: Improved peak detection and deconvolution of native electrospray mass spectra from large protein complexes. J. Am. Soc. Mass Spectrom. 26, 2141–2151 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bayburt TH, Sligar SG: Membrane protein assembly into nanodiscs. FEBS Lett. 584, 1721–1727 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EAW, Dafforn TR, Baldus M, Killian JA: Detergent-free isolation, characterization, and functional reconstitution of a tetrameric k+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. U. S. A. 111, 18607–18612 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao Y, Cao E, Julius D, Cheng Y: Trpv1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 534, 347–351 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schultze M, Ramasesha K, Pemmaraju CD, Sato SA, Whitmore D, Gandman A, Prell JS, Borja LJ, Prendergast D, Yabana K, Neumark DM, Leone SR: Attosecond band-gap dynamics in silicon. Science. 346, 1348–1352 (2014) [DOI] [PubMed] [Google Scholar]
- 62.Forbes AMG: Fourier transform filtering - a cautionary note. J. Geophys. Res.-Oceans 93, 6958–6962 (1988) [Google Scholar]
- 63.Mosierboss PA, Lieberman SH, Newbery R: Fluorescence rejection in raman-spectroscopy by shifted-spectra, edge-detection, and fft filtering techniques. Appl. Spectrosc. 49, 630–638 (1995) [Google Scholar]
- 64.Pandey PR, Roy S: Headgroup mediated water insertion into the dppc bilayer: A molecular dynamics study. J. Phys. Chem. B. 115, 3155–3163 (2011) [DOI] [PubMed] [Google Scholar]
- 65.Li J, Richards MR, Bagal D, Campuzano IDG, Kitova EN, Xiong ZJ, Privé GG, Klassen JS: Characterizing the size and composition of saposin a lipoprotein picodiscs. Anal. Chem. 88, 9524–9531 (2016) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


