Abstract
Pancreatic cancer, the fourth leading cause of cancer death in the United States, has a negative prognosis because metastasis occurs before symptoms manifest. Although combination therapies are showing improvements in treatment, the survival rate for pancreatic cancer five years post diagnosis is only 8%, stressing the need for new treatments. The receptor for advanced glycation end products (RAGE) has recently emerged as a chemotherapeutic target in KRAS driven pancreatic cancers both for treatment and in chemoprevention. RAGE appears to be an important regulator of inflammatory, stress and survival pathways that lead to carcinogenesis, resistance to chemotherapy, enhanced proliferation and the high metastatic potential of pancreatic cancer. RAGE expression has been demonstrated in pancreatic cancer tumors but not in adjacent epithelial tissues. Its presence is associated with increased proliferation and metastasis. In an effort to identify novel inhibitors of RAGE among our collection of marine-derived secondary metabolites, a cell-based screening assay utilizing flow cytometry was developed. This effort led to the identification of scalarin as the active compound in a marine sponge identified as Euryspongia cf. rosea. Scalarin is a sesterterpene natural product isolated previously from a different marine sponge. Scalarin reduces the levels of RAGE and inhibits autophagy in the PANC-1 and MIA PaCa-2 pancreatic cancer cell lines. Its IC50 for cytotoxicity ranges between 20–30 μM in the AsPC-1, PANC-1, MIA PaCa-2 and BxPC-3 pancreatic cancer cell lines. Inhibition of autophagy limits tumor growth and tumorigenesis in pancreatic cancer, making scalarin an interesting compound that may merit further study.
Keywords: Marine Natural Products, Pancreatic Cancer, RAGE, autophagy
Introduction
The American Cancer Society estimates that about 55,440 new cases of pancreatic cancer will be diagnosed in the US in 2018. Of those patients, only 8% will survive 5 years post diagnosis. Pancreatic cancer is the fourth most lethal cancer due to its rapid proliferation, high rate of metastasis and its delayed detection, as most patients remain asymptomatic until the cancer has metastasized [1]. Current treatments remain only moderately effective as evidenced by the low long-term survival rate.
The receptor for advanced glycation end products (RAGE) is a member of the immunoglobulin family of receptors. RAGE is a type I transmembrane protein that has three extracellular immunoglobulin-like domains [2]. While mRNA suggests the possibility that many different RAGE isoforms exist, only two functional alternatively spliced isoforms are currently known: the full-length transmembrane form which can initiate signal transduction; and a soluble form that can act as a decoy receptor [2]. The full length form can also be proteolytically cleaved to generate a soluble form with similar decoy function to the alternatively spliced soluble RAGE [2]. RAGE expression can be increased by an accumulation of its ligands and inflammatory mediators. RAGE has the ability to bind a myriad of ligands, including glycation end products (AGE), S100/calgranulin proteins and the high mobility group box 1 (HMGB1), leading to a variety of functions including stress responses, metabolic processes, and neuronal growth. Because many of the ligands for RAGE accumulate in tissues as the body ages or in instances of disease and inflammation, RAGE is considered a pattern recognition receptor.
Soluble RAGE is also thought to be a biomarker for chronic inflammation. RAGE activation can lead to activation of the nuclear factor kappa B (NFκB), a transcription factor involved in the regulation of apoptosis, proliferation and inflammation; MAP kinases that regulate stress response; STAT3 another regulator of inflammation and cellular proliferation; and adhesion molecules (Reviewed in [2,3,4]).
RAGE has emerged as an extremely important molecule in pancreatic cancer etiology. Elevated levels of its ligands HMGB1 [2] and S100P [5] are found in pancreatic cancer, and their expression is associated with survival and strong metastatic potential [6,2]. Promotion of metastasis by S100P occurs through its interaction with RAGE [7,8]. Expression of RAGE itself is elevated in pancreatic cancers [9] and pancreatic cancer cell lines [10,11] but not in adjacent epithelial tissues. The expression of RAGE has been associated with enhanced tumor formation and resistance to chemotherapy. Silencing of RAGE through siRNA in a pancreatic cancer cell line led to reduced growth of tumors when these cells were injected into mice; in vitro, silencing RAGE led to apoptosis while over-expression promoted survival [12]. High levels of soluble RAGE (which antagonizes membrane-bound RAGE) in the sera of smokers were associated with a decreased risk of developing pancreatic cancer [13]. Lower levels of soluble RAGE are found among pancreatic cancer patients than the general population [14]. Silencing of RAGE through siRNA in a pancreatic cancer cell line led to decreased autophagy [12], suggesting a link between RAGE signaling and autophagy in pancreatic cancer. RAGE has been considered to be a switch between apoptosis and autophagy following oxidative injury [15,16]. Autophagy is a conserved pathway to eliminate proteins through lysosomal degradation and is thought to be a caspase-independent manner of programmed cell death [17]. While its role in cancer depends on the tissue of origin, elevated levels of basal autophagy are found in many pancreatic cancer cell lines and primary tumors, and inhibition of autophagy limits tumor growth and tumorigenesis [18]. Similarly, expression of the autophagy marker LC3-II was shown in pancreatic cancers and correlated with low survival rates [19], and silencing of RAGE through siRNA decreases LC3-II levels in pancreatic cancer cells [12]. Taken together, all these studies suggest that RAGE is an important regulator of inflammatory, stress and survival pathways that contribute to resistance to chemotherapy, enhanced proliferation and high metastatic potential of pancreatic cancer.
The oceans are a rich source of bioactive natural products [20]. The uniqueness, chemical diversity and structural complexity of marine natural products represent an unexploited source of lead structures for use as biological probes or in drug discovery and development. In an effort to discover small molecule inhibitors of RAGE expression in pancreatic cancer cells, the Harbor Branch Oceanographic Institute library of enriched fractions obtained from marine organisms (the HBOI Peak Library) was screened using a fluorescent cell based assay to detect compounds that reduced levels of expression of RAGE in PANC-1 pancreatic carcinoma cells.
Materials and Methods
Reagents
Enriched fractions (peak) or pure compounds were obtained from the Harbor Branch Oceanographic Institute Peak or Pure Compound Libraries. The scalarin stock solution was at a concentration of 1 mg/mL in methanol. Methanol and isopropanol used in the experiments were purchased from Fisher Scientific, Fair Lawn, NJ. The 3-[4,5-Dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide (MTT) used for cell viability assays was purchased from Sigma Chemical Co., St. Louis, MO.
Cell Culture
The human pancreatic cancer cell lines PANC-1 (CRL-1469), AsPC-1 (CRL-1682), BxPC-3 (CRL-1687) and MIA PaCa-2 (CRL-1420) cell lines were obtained from ATCC, grown, aliquoted and maintained in liquid nitrogen. Aliquots of AsPC-1, PANC-1, and BxPC-3 were thawed and grown in RPMI-1640 supplemented with 10% Fetal Bovine Serum, 0.11 mg/mL sodium pyruvate, 4.5 g/L D-glucose, 18 mM HEPES Buffer, 100 U/mL penicillin G sodium, 100 μg/mL streptomycin sulfate, 0.25 μg/mL amphotericin B, 2 mM L-glutamine and 50 μg/mL gentamicin (Complete RPMI). The MIA PaCa-2 cell line was grown in RPMI-1640 supplemented with 10% Fetal Bovine Serum, 2.5% horse sera, 100 U/mL penicillin G sodium, 100 μg/mL streptomycin sulfate, 0.25 μg/mL amphotericin B, and 50 μg/mL gentamicin. Cells were maintained in a humidified incubator at 37°C and 5% CO2. Cells were kept in culture for 10 weeks (20 passages) when a new aliquot was thawed.
Isolation and Purification of scalarin
A specimen of a marine sponge identified as Euryspongia cf. rosea, Family Dysideidae, Order Dictyoceratida (See supplementary for details) was collected off Chub Cay Bahamas using the Johnson Sea Link submersible at a depth of 597 m (HBOI Sample ID 8-XII-84-2-010). The sample was frozen immediately after collection and stored at −20°C until extraction. 164 g of the frozen sponge was freeze-dried and then ground and extracted sequentially using a Dionex Accelerated Solvent Extractor in 3 steps (Step 1: heptane; Step 2: EtOH: EtOAc (5:1 v/v) and Step 3: CH3OH:H2O (5:1 v/v)) at 100 °C with 3 static cycles per step. 1.02 g of the EtOH:EtOAc (5:1 v/v) extract was separated using an Isco Combiflash™ Rf4. The material was adsorbed onto Celite and solvent removed for solid sample loading. A 26 g C-18 RP Rf Gold Column (Isco Teledyne) was used for the separation. The total run time was 32.2 column volumes (CV) over 23.8 minutes with a flow rate of 35 ml/minute. The column was eluted with gradient elution as follows: Solvent A: H2O, Solvent B: CH3CN, Solvent C: CH3OH; t=0 minutes A:B (8:2 v/v) hold for 1.2 CV then elute with a linear gradient ending at 100% B at 15 CVs; hold at 100% B for 5 CV then wash with 100% C for 3 CVs. The pressure was maintained at 200 psi over the course of the elution. Eluates between 15 CV and 18 CV (Tubes 58–68) were combined and after solvent removal yielded 29.2 mg of a fraction containing scalarin 1 as a major component. Further purification by semi-preparative HPLC on a Phenomenex Luna C18 column (10 mm × 250 mm, 10 μ particle size) using isocratic elution with H2O:CH3CN (85.5:14.5 v/v) at 3 ml/minutes resulted in the purification of 5.5 mg of scalarin that was used in the biological studies. Interpretation of the full 1D and 2D NMR data set for 1 and comparison to published values identified the active compound as scalarin 1 [21].
RAGE Inhibition Screening Assay
PANC-1 cells were plated in a U-bottom plate and allowed to adhere overnight. Media was removed and replaced with media containing treatment which consisted of either media alone, 30μM curcumin, 5μg/mL HBOI test compounds/fractions or their respective vehicle controls for 24 hours. Gates were set up against an IgG isotype antibody using the methanol solvent control. Curcumin and any compound that may autofluoresce had their own IgG isotype control. Cells were harvested by adding cell dissociation buffer (0.05% collagenase I in trypsin/EDTA solution) containing 7-aminoactinomycin D (7AAD) to each well. 7AAD is cell impermeable and only enters cells that are dead or in the final steps of cell death. Cells were pelleted by centrifugation, fixed with 4% paraformaldehyde in PBS, and permeabilized with ice cold methanol. Cells were labeled with primary antibody against RAGE (Millipore AB5601; EMD Millipore, Billerica, MA) or isotype IgG (005-000-003; Jackson ImmunoResearch, West Grove, PA) followed by phycoerythrin conjugated secondary against goat IgG (705-116-147; Jackson ImmunoResearch, West Grove, PA). Cells were analyzed with a FACSCanto flow cytometer with a plate attachment (BD Biosciences, San Jose, CA) acquiring 30,000 events. Data obtained was normalized to solvent controls for ease of comparison between runs using Microsoft Excel (Redmond, WA).
Western blotting
Cells were seeded at normal density, and allowed to adhere overnight, then treated with 10, 5, 2.5, 1.25 or 0.625 μg/mL scalarin, 30 μM curcurmin or methanol for 24h for RAGE westerns. For LC3-II westerns, cells were treated with 10 μg/mL scalarin or incubated in media without serum for 0.5, 2, 6, 24, or 48h. Control samples were treated with either 10 μM manzamine A (a marine natural product known to inhibit autophagy [22]) or methanol for 48h. At the end of the incubation, media and trypsinized cells were pooled and pelleted by centrifugation. Cells were lysed in lysis buffer (10 mM Tris-Cl pH 7.5, 100 mM NaCl, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, Halt Protease Inhibitor Cocktail, 1 mM Na3VO4, 1 mM NaF) for 30 minutes at 4°C, followed by centrifugation to pellet the cell debris. The supernatant containing the protein was transferred to a new tube, quantitated using BCA Protein Assay Kit (Pierce, Rockford, IL) and stored at −80°C. Protein (20 μg) was run in a pre-cast denaturing 15% SDS-PAGE gel (Bio-Rad, Hercules CA), which was then transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules CA), and blocked with 5% non-fat milk in Tris-buffered saline containing Tween-20 (TBST) buffer for one hour at room temperature. After repeated washing, the membrane was incubated with primary antibody overnight at 4 °C, repeatedly washed and incubated for one hour at room temperature with horseradish peroxidase conjugated secondary antibody. Detection of proteins was done with chemiluminescence (Amersham Biosciences, Piscataway, NJ), followed by imaging with the ChemiDoc MP System and densitometry analysis using Image Lab 4.1 software (Bio-Rad, Hercules, CA). Primary antibodies used were RAGE (SAB1401326; Sigma, St. Louis MO) and LC3-II (3868; Cell Signaling Technologies, Beverly, MA). Peroxidase-conjugated anti-rabbit IgG secondary antibody was obtained from Jackson ImmunoResearch (West Grove, PA). All antibodies were used at manufacturer’s recommended dilutions. RAGE westerns were repeated four times; LC3-II westerns were repeated three times.
Intracellular Staining
To determine the expression levels of selected signal transduction molecules affected by scalarin, PANC-1 and MIA PaCa-2 cells were treated for 24 hours with 10 μg/mL scalarin or methanol (vehicle control). At the end of treatment, cells were trypsinized, fixed with 4% paraformaldehyde and permeabilized with ice cold methanol. Cells were labeled with primary antibodies specific for the active (phosphorylated) form of NFκB (PE Mouse anti-NFκB p65 (pS529); 558423; BD Pharmingen, San Diego, CA, USA), phosphorylated Erk 1/2, phosphorylated STAT3ser727, S100P, Bcl-XL (Cell Signaling Technologies, Danvers, MA, USA), mouse IgG or Rabbit IgG isotypes (Jackson ImmunoResearch, West Grove, PA, USA) for 1 hour at room temperature. Cells were labeled with a secondary phycoerythrin conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA) for all antibodies except NFκB for 30 min at room temperature. Antibodies were used at the manufacturer’s recommended dilutions for this application. Cells were washed and resuspended in 2% FBS in PBS and analyzed via flow cytometry in a BD FACSCanto equipped with a plate adapter (BD Biosciences, San Jose, CA), acquiring 20,000 events.
Cytotoxicity Assay (MTT)
12,000 cells were plated on a 96-well tissue culture plate. Cells were allowed to adhere for 24 hours. At the end of this incubation, 100 μl of medium was removed from each test well and 100 μl of medium containing treatment was added. Treatment consisted of a range of final concentrations from 0.078 to 20 μg/mL scalarin or media with methanol. The cells were then incubated for 72 hours at 37°C and 5% CO2. After this incubation, 75 μL of 5 mg/mL MTT were added to each well, and the cells were further incubated for 3 hours at 37°C. The plates were centrifuged for 10 minutes at 800 rpm. The supernatant was removed and 200 μL acidified isopropanol (1:500 solution of hydrochloric acid to isopropanol) were added to each well. The plates were shaken for 15 minutes and then the absorbance of these solutions were measured at 570 nm with a plate reader (NOVOstar, BMG Labtech Inc., Durham, NC). The resulting absorbances were normalized against methanol treated cells using Microsoft Excel. Determination of the dose at which 50% inhibition is found (IC50) was done using a non-linear regression curve fit with GraphPad Prism 5 software (La Jolla, CA).
Caspase Cleavage
To determine induction of apoptosis, a commercially available kit for measuring the cleavage of caspase 3/7 (G0890; Promega, Madison, WI) was used following the manufacturer’s protocol. Briefly, 10,000 cells were plated in a 96-well flat bottom white plate. Cells were allowed to adhere overnight then treated with media, 10 μg/mL scalarin, 10 μM manzamine A (positive control) or vehicle control for 24 hours. Some wells received the same treatments along with 100 ng/mL killer TRAIL (Alexis Biochemicals, San Diego, CA). At the end of treatment an equal volume of pro-luminescent caspase 3/7 substrate was added to the wells and incubated at room temperature with mild shaking for 30–60 minutes. In cells with active caspase 3/7, the tetrapeptide sequence DEVD is cleaved to release aminoluciferin, and the resulting luminescence was read with a plate reader (NOVOstar, BMG Labtech Inc., Durham, NC). Luminescence results are presented as fold induction compared to the vehicle control treated sample.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
To determine induction of apoptosis, a commercially available kit (G3250; Promega, Madison, WI) was used to measure fragmented DNA by incorporating fluorescein 12-dUTP at 3′-OH DNA ends with the Terminal Deoxynucleotidyl Transferase enzyme following the manufacturer’s protocol. Briefly, 8,000 cells were plated on a 96-well black plate with clear bottom. Cells were allowed to adhere overnight then treated with media, 10 μg/mL scalarin, 10 μM manzamine A (positive control) or vehicle control for 24 hours. Some wells received the same treatments along with 100 ng/mL killer TRAIL (Alexis Biochemicals, San Diego, CA). At the end of treatment, cells were fixed with 4% paraformaldehyde and permeabilized with ice cold methanol. Cells were equilibrated for five minutes and then treated with 25 μl of a reaction mix containing the nucleotide mix and the enzyme to label the fragmented DNA for 60 minutes at 37°C. Cells were also stained with NucBlue Live Cell Stain Hoechst 33342 (Molecular Probes R37605, Eugene, OR). The reaction was stopped by the addition of 20mM EDTA. The resulting fluorescence was visualized using the FITC and DAPI filters under a 10X objective in the ImageXpress Micro XLS Widefield High-Content Analysis System (Molecular Devices, San Jose, CA).
Statistics
Statistical analysis of the data sets to determine mean, standard deviation, and standard error of the mean was performed using Microsoft Excel. Experiments were repeated a minimum of three times. Data sets were compared using the Student’s T Test. A p value ≤ 0.05 was considered significant. Outliers were detected through the Grubbs’ Test.
Results
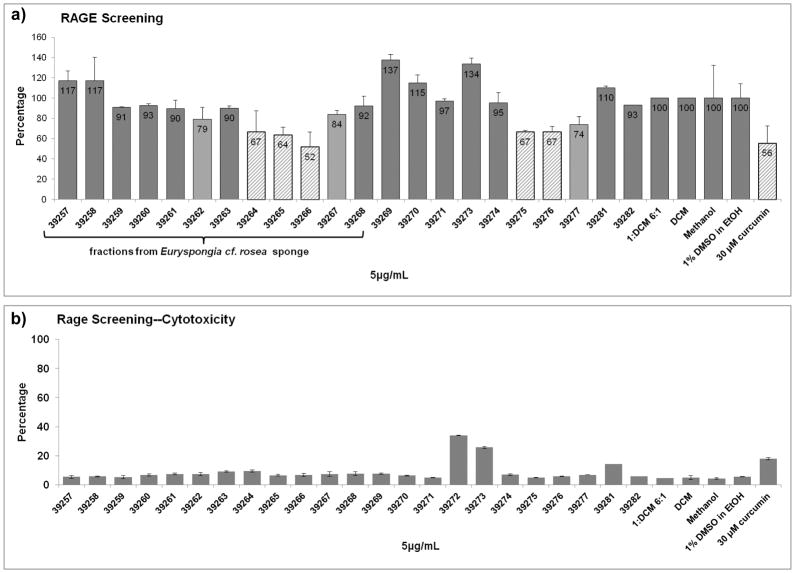
To identify novel inhibitors of RAGE among our collection of marine-derived secondary metabolites, we established a cell-based screening assay utilizing flow cytometry. Of the pancreatic cancer cell lines maintained by our labs (AsPC-1, MIA PaCa-2, PANC-1 and BxPC-3) PANC-1 and MIA PaCa-2 are known to express high levels of RAGE [11,10]. This led us to select the PANC-1 cell line for the screening assay. Curcumin, a natural product that inhibits RAGE [23], was used as a positive control. For the screen, cells were treated with marine-derived samples or controls for 24h then labeled with fluorescent RAGE antibody and subjected to flow cytometric analysis. Cells were also labeled with a cell impermeable nucleic acid stain to measure cytotoxicity which allowed us to eliminate compounds that caused a decrease in RAGE by killing the cells. Samples that showed ≥ 30% inhibition of RAGE expression (Figure 1a) and exhibited ≤ 20% cytotoxicity (Figure 1b) at a concentration of 5 μg/mL were considered hits.
Figure 1. Screening Data.
a) A screening assay was set up to measure effects on the levels of RAGE in the PANC-1 pancreatic carcinoma cell line after treatment with marine samples. Levels were normalized to their respective solvent controls for ease of comparison. Samples showing ≥ 30% inhibition of RAGE expression and ≤ 20% cytotoxicity were considered hits. Fractions from Euryspongia cf. rosea sponge showed reduction of RAGE expression. b) Cytotoxicity was measured concomitantly by using the membrane impermeable DNA stain 7-amino actinomycin D (7AAD). Cells were exposed to this after treatment and prior to permeabilization. All of the fractions presented less than 20 % cytotoxicity. Screening results were confirmed by repetition.
Three fractions from the sponge Euryspongia cf. rosea showed activity in the screening assay. The hit fractions (in diagonal line fill) and related fractions (fractions from the same organism that showed some activity but did not meet our criteria for a hit; in light grey fill) are shown in Figure 1a. The active compound was purified using bioassay-guided fractionation and was identified as the marine compound scalarin [24] whose structure is shown in Figure 2. While this is a known compound, this is the first report of its inhibition of RAGE in pancreatic cancer cells.
Figure 2. Structure of Scalarin.
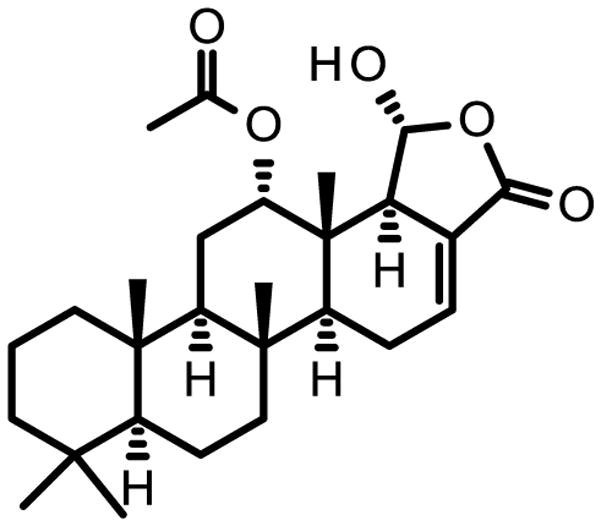
The active compound in the fractions was identified to be scalarin, a known compound for which inhibition of RAGE is a novel activity.
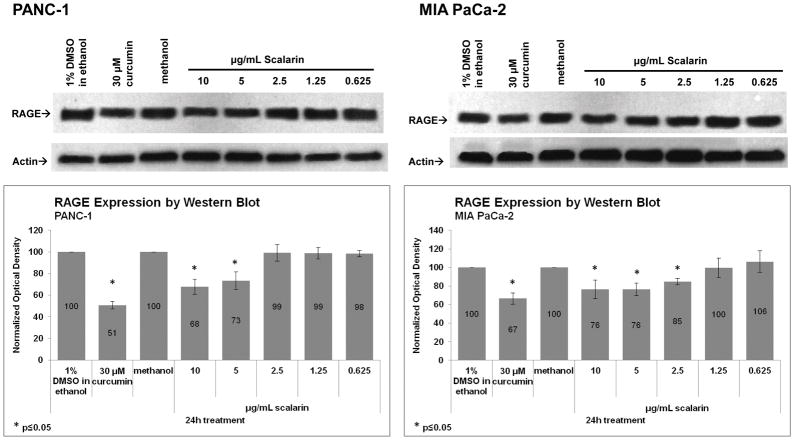
To confirm the decrease in RAGE expression, the expression of RAGE in PANC-1 and MIA PaCa-2 cells treated for 24h with scalarin (10, 5, 2.5, 1.25, and 0.625 μg/mL) or controls was determined using western blotting. As shown in Figure 3, the ability of scalarin to reduce levels of RAGE expression was confirmed in both the PANC-1 and MIA PaCa-2 cell lines. Densitometry analysis showed that this decrease was significant in PANC-1 cells treated with 10 and 5 μg/mL scalarin and in MIA PaCa-2 cells treated with 10, 5 and 2.5 μg/mL scalarin.
Figure 3. Confirmation of RAGE Inhibition by Western Blotting.
The expression of RAGE in PANC-1 and MIA PaCa-2 cells treated with 10, 5, 2.5, 1.25, and 0.625 μg/mL (22.5, 11.3, 5.6, 2.8 and 1.4 μM) scalarin or controls for 24h was ascertained using western blotting. The ability of scalarin to reduce levels of RAGE expression was confirmed in the PANC-1 and MIA PaCa-2 cell lines. Densitometry analysis showed that this decrease was significant in PANC-1 cells treated with 10 and 5 μg/mL scalarin and in MIA PaCa-2 cells treated with 10, 5 and 2.5 μg/mL scalarin. Western blot for 1 representative experiment is shown. Graph shows the average densitometry ± standard deviation for 4 experiments.
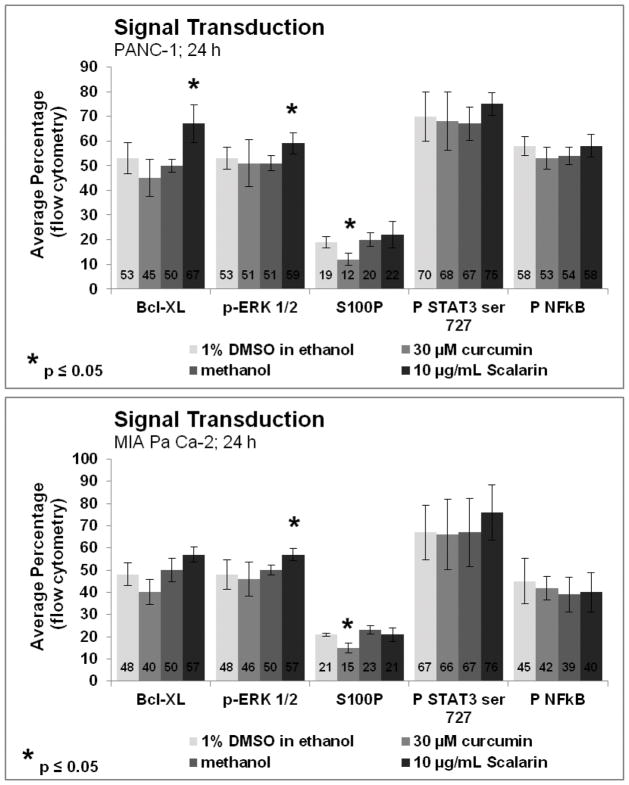
Because RAGE is known to be an important modulator of different signaling pathways, scalarin was assayed to determine if it caused a reduction in the activation of NFκB, STAT3, and Erk-1/2 or a change in the expression of S100P or Bcl-XL. Treatment of PANC-1 and MIA PaCa-2 cells with 10 μg/mL scalarin for 24 hours led toa significant increase in Bcl-XL levels in PANC-1 cells, but the increase in MIA PaCa-2 cells failed to be significant (p≤ 0.08). Cells treated with this compound had modest but significant increases in phosphorylated Erk 1/2 levels in both cell lines. No significant differences in S100P, phosphorylated STAT3ser727 or phosphorylated NFκB were seen for either cell line (Figure 4).
Figure 4. Effects of Scalarin on Signal Transduction Molecules.
RAGE activation can lead to activation of NFκB, MAP kinases and STAT3, thus we expected to see reduction in levels of these molecules after its inhibition. Similarly, the levels of S100P and the anti-apoptotic molecule Bcl-XL can be affected by RAGE expression. No significant differences in S100P, phosphorylated STAT3ser727 or phosphorylated NFκB were seen for either cell line. A very modest, but statistically significant, increase in levels of phosphorylated Erk1/2 was seen in both cell lines. A slight increase in levels of Bcl-XL was seen in both cell lines, but was only statistically significant in the PANC-1 cells. Data shown is the average ± standard deviation of 4 independent experiments.
The fractions containing scalarin were chosen as hits because they showed little cytotoxicity in the screening assay. Nevertheless, an effort was made to determine the concentration of scalarin needed to see 50% cytotoxicity after 72h of treatment (IC50) in the four pancreatic cancer cell lines maintained in the lab. As shown in Table 1, scalarin has only moderate cytotoxicity with its IC50 between 20–30 μM in all cell lines and showed the greatest cytotoxicity against PANC-1 cells. In comparison, curcumin exhibits more cytotoxicity in these cell lines, with its IC50 between 5–13 μM.
Table 1. IC50 for cytotoxicity of Scalarin in four pancreatic cancer cell lines.
The concentration of scalarin needed to induce 50% cytotoxicity (IC50) was determined using a nonlinear regression curve fit in the pancreatic cancer cells lines AsPC-1, BxPC-3, MIA PaCa-2 and PANC-1. Data shown is the average ± standard deviation of 3 independent experiments. Data for curcumin was previously published [30] and is provided here for ease of comparison.
| IC50 μM ± S.D. | ||||
|---|---|---|---|---|
| Compound | AsPC-1 | BxPC-3 | MIA PaCa-2 | PANC-1 |
| scalarin | 27 ± 0.5 | 29 ± 0.5 | 24 ± 0.7 | 19 ± 0.5 |
| curcumin | 13 ± 3.7 | 5.1 ± 1.2 | 5.1 ± 1.1 | 12 ± 1.4 |
Kang et al. showed that RAGE promoted cell survival through increased autophagy and reduced apoptosis [11]. Therefore, it was possible that a RAGE inhibitor would lead to an increase in sensitivity to apoptosis. To test this possibility, PANC-1 and MIA PaCa-2 pancreatic cancer cells were treated with 10 μg/mL scalarin in the presence or absence of 100ng/mL killer TRAIL. As shown in Figure 5, treatment of MIA PaCa-2 cells or PANC-1 cells with scalarin neither induced apoptosis nor sensitized cells to undergo any further TRAIL-induced apoptosis as measured through caspase 3/7 cleavage or DNA fragmentation (TUNEL) than cells that just received killer TRAIL.
Figure 5. Effects of Scalarin on Apoptosis.
PANC-1 and MIA PaCa-2 pancreatic cancer cells were plated, allowed to adhere overnight, then treated for 24 hours with 10 μg/mL scalarin, methanol (solvent control) or 10μM manzamine A (positive control), in the presence or absence of 100 ng/mL killer TRAIL. At the end of the incubation apoptosis was measured by following caspase 3/7 cleavage or TUNEL. Data shown is the average ± standard error of the mean of at least 3 independent experiments.
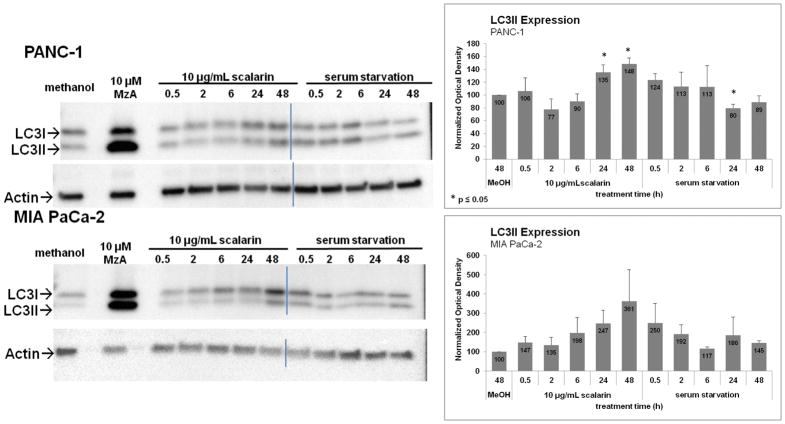
Finally, inhibition of RAGE in pancreatic cancer cells is expected to reduce autophagy. To determine whether scalarin had any effect on autophagy, cells were cultured in media containing no glutamate or FBS (serum starvation media) for 0.5,2,6,24 and 48 hours, treated with 10 μg/mL scalarin for these same time points, or treated for 48h with methanol (solvent control) or 10μM manzamine A (positive control). Protein from these cells was isolated and used in a western blot using LC3-II antibody (Figure 6). Serum starvation induces autophagy that is resolved within the period tested, causing an accumulation followed by a reduction of LC3-II (lower band). Cells treated with scalarin showed an accumulation in LC3-II that failed to resolve by 48h, which suggests that scalarin inhibits autophagy.
Figure 6. Inhibition of Autophagy by Scalarin.
PANC-1 and MIA PaCa-2 pancreatic cancer cells were plated, allowed to adhere overnight, then cultured in media containing no glutamate or FBS (serum starvation media) or treated with 10 μg/mL scalarin for 0.5, 2, 6, 24 and 48 hours or for 48h with methanol (solvent control) or 10 μM manzamine A (positive control). Cells were lysed and subjected to Western Blotting for LC3-II, a known marker for autophagy. Serum starvation induces autophagy, which resolves within 48h. Inhibitors of RAGE inhibit autophagy resulting in an accumulation of autophagic vesicles. Scalarin resulted in an unresolved accumulation of LC3-II in both cell lines, suggesting it inhibits autophagy. Western blot for 1 representative experiment is shown. Graph shows the average densitometry ± standard deviation for three independent experiments.
Discussion
By screening our collection of marine-derived secondary metabolites we have identified novel inhibitors of RAGE with the potential to be chemotherapeutic for pancreatic cancer. Fractions from the sponge Euryspongia cf. rosea showed activity in the cell based screening assay, leading to the identification of scalarin [24]. Structurally related compounds have shown anti-microbial activity against Staphylococcus aureus [25] and cytotoxic activity against leukemia [25], breast, colon, prostate [26] and cervical [27] cancer cell lines. Inhibition of RAGE in pancreatic cancer cells is a new activity for scalarin.
Scalarin, at concentrations of 11.3 and 22.5 μM, significantly reduced the level of RAGE in the PANC-1 pancreatic cancer cell line; while the level in the MIA PaCa-2 cell line was significantly reduced at 5.6, 11.3 and 22.5 μM. While this compound has only moderate activity it is comparable to curcumin, as treatment with 30 μM curcumin was required to reduce RAGE expression 30–50% in these cell lines.
Even though scalarin and curcumin show similar inhibitory activity against RAGE, curcumin was more cytotoxic in these cell lines. Scalarin showed little cytotoxicity in the screening assay and its IC50 for cytotoxicity ranges between 20–30 μM in the AsPC-1, PANC-1, MIA PaCa-2 and BxPC-3 pancreatic cancer cell lines, a desirable property if scalarin is used as an adjuvant to other chemotherapies. Our tests showed that scalarin does not induce apoptosis. Manzamine A, a marine natural compound known to inhibit autophagy [22] has low cytotoxicity, but is able to sensitize AsPC-1 cells to undergo TRAIL-induced apoptosis [28]. Scalarin, however, does not share this characteristic.
RAGE is an important regulator of inflammatory, stress and survival pathways that lead to carcinogenesis, resistance to chemotherapy, enhanced proliferation and the high metastatic potential of pancreatic cancer. Our focused effort to determine the effects of scalarin in different signaling pathways associated with inflammation (phosphorylated NFκB and phosphorylated STAT3), stress (phosphorylated Erk 1/2), inhibition of apoptosis (Bcl-XL) and its own ligand (S100P), which are known to be regulated by RAGE, were not impacted by scalarin. It is possible that scalarin could show effects at other time points or at other steps in these signaling cascades.
RAGE is an important regulator of autophagy in pancreatic cancer. Indeed, inhibition of RAGE through scalarin appears to lead to inhibition of autophagy in PANC-1 and MIA PaCa-2 as observed by following LC3-II by western blotting. Inhibition of RAGE [12] and autophagy [29] has resulted in the decrease of tumor size in mice models, suggesting that scalarin may merit further studies.
Supplementary Material
Acknowledgments
Funding for this project was provided by the National Institutes of Health R21CA176222 awarded to Dr. Guzmán. Research Excellence funds from the Harbor Branch Oceanographic Institute Foundation (HBOIF) awarded to Dr. Guzmán and Dr. Wright contributed to finalizing this publication. We thank Dr. Peter McCarthy for proofreading the article. This is HBOI contribution number 2167.
Footnotes
Conflicts of interest: None.
Compliance with Ethical Standards
This study was funded by the National Institutes of Health R21CA176222. All authors—Guzmán, Pitts, Diaz and Wright—declare that they have no conflicts of interest. Only invertebrate not-sentient marine sponges were used in this study. Projects using invertebrates are not subject to review by the Institutional Animal Care and Use Committee of Florida Atlantic University. This article does not contain studies with human participants or vertebrate animals performed by any of the authors.
References
- 1.American Cancer Society. [Accessed (05/11/18)];Cancer Facts and Figures. 2018 https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2018.html.
- 2.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 3.Han SH, Kim YH, Mook-Jung I. RAGE: the beneficial and deleterious effects by diverse mechanisms of actions. Mol Cells. 2011;31(2):91–97. doi: 10.1007/s10059-011-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, Stern DM, Nawroth PP. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83(11):876–886. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 5.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63(10):2649–2657. [PubMed] [Google Scholar]
- 6.Arumugam T, Simeone DM, Van Golen K, Logsdon CD. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res. 2005;11(15):5356–5364. doi: 10.1158/1078-0432.CCR-05-0092. [DOI] [PubMed] [Google Scholar]
- 7.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279(7):5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 8.Arumugam T, Ramachandran V, Logsdon CD. Effect of cromolyn on S100P interactions with RAGE and pancreatic cancer growth and invasion in mouse models. Journal of the National Cancer Institute. 2006;98(24):1806–1818. doi: 10.1093/jnci/djj498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparvero LJ, Asafu-Adjei D, Kang R, Tang D, Amin N, Im J, Rutledge R, Lin B, Amoscato AA, Zeh HJ, Lotze MT. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takada M, Hirata K, Ajiki T, Suzuki Y, Kuroda Y. Expression of receptor for advanced glycation end products (RAGE) and MMP-9 in human pancreatic cancer cells. Hepato-gastroenterology. 2004;51(58):928–930. [PubMed] [Google Scholar]
- 11.Takada M, Koizumi T, Toyama H, Suzuki Y, Kuroda Y. Differential expression of RAGE in human pancreatic carcinoma cells. Hepato-gastroenterology. 2001;48(42):1577–1578. [PubMed] [Google Scholar]
- 12.Kang R, Tang D, Schapiro NE, Livesey KM, Farkas A, Loughran P, Bierhaus A, Lotze MT, Zeh HJ. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell death and differentiation. 2010;17(4):666–676. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao L, Weinstein SJ, Albanes D, Taylor PR, Graubard BI, Virtamo J, Stolzenberg-Solomon RZ. Evidence That Serum Levels of the Soluble Receptor for Advanced Glycation End Products Are Inversely Associated with Pancreatic Cancer Risk: A Prospective Study. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-10-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krechler T, Jachymova M, Mestek O, Zak A, Zima T, Kalousova M. Soluble receptor for advanced glycation end-products (sRAGE) and polymorphisms of RAGE and glyoxalase I genes in patients with pancreas cancer. Clin Biochem. 2010;43(10–11):882–886. doi: 10.1016/j.clinbiochem.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Kang R, Tang D, Loze MT, Zeh HJ. Apoptosis to autophagy switch triggered by the MHC class III-encoded receptor for advanced glycation endproducts (RAGE) Autophagy. 2011;7(1):91–93. doi: 10.1038/cdd.2009.149. [DOI] [PubMed] [Google Scholar]
- 16.Kang R, Tang D, Lotze MT, Zeh HJ., 3rd RAGE regulates autophagy and apoptosis following oxidative injury. Autophagy. 2011;7(4) doi: 10.4161/auto.7.4.14681. [DOI] [PubMed] [Google Scholar]
- 17.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6(3):322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25(7):717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii S, Mitsunaga S, Yamazaki M, Hasebe T, Ishii G, Kojima M, Kinoshita T, Ueno T, Esumi H, Ochiai A. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008;99(9):1813–1819. doi: 10.1111/j.1349-7006.2008.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer AMS. 16. Marine Pharmacology and the Late 2011 Marine Pharmaceuticals Pipeline. Toxicon. 2012;60(2):104. doi: 10.1016/j.toxicon.2012.04.017. [DOI] [Google Scholar]
- 21.Cimino G, De Stefano S, Minale L, Trivellone E. 12-epi-scalarin and 12-epi-deoxoscalarin, Sesterterpenes from the sponge Spongia nitens. Journal of the Chemical Society Perkin transactions 1. 1977;13:1587–1593. [PubMed] [Google Scholar]
- 22.Kallifatidis G, Hoepfner D, Jaeg T, Guzman EA, Wright AE. The marine natural product manzamine A targets vacuolar ATPases and inhibits autophagy in pancreatic cancer cells. Marine drugs. 2013;11(9):3500–3516. doi: 10.3390/md11093500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, Tang Y, Kang Q, Feng Y, Chen A. Curcumin inhibits gene expression of receptor for advanced glycation end-products (RAGE) in hepatic stellate cells in vitro by elevating PPARgamma activity and attenuating oxidative stress. British journal of pharmacology. 2012;166(8):2212–2227. doi: 10.1111/j.1476-5381.2012.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fattorusso E, Magno S, Santacroce C, Sica D. Scalarin, a new pentacyclic C-25 terpenoid from the sponge Cacospongia scalaris. Tetrahedron. 1972;28:5993–5997. [Google Scholar]
- 25.Pettit GR, Cichacz A, Tan R, Hoard MS, Melody N, Pettit R. Antineoplastic agents. 386. Isolation of sesterstatins 1–3 from the marine sponge Hyrtios erecta. Journal of natural products. 1998;61(1):13–16. doi: 10.1021/np970203+. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y-J, Lee J-W, Lee D-G, Lee H-S, Kang JS, Yun J. Cytotoxic sesterterpenoids isolated from the marine sponge Scalarispongia sp. International journal of molecular sciences. 2014;15(11):20045–20053. doi: 10.3390/ijms151120045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukamoto S, Miura S, van Soest RW, Ohta T. Three new cytotoxic sesterterpenes from a marine sponge Spongia sp. Journal of natural products. 2003;66(3):438–440. doi: 10.1021/np020497l. [DOI] [PubMed] [Google Scholar]
- 28.Guzman EA, Johnson JD, Linley PA, Gunasekera SE, Wright AE. A novel activity from an old compound: Manzamine A reduces the metastatic potential of AsPC-1 pancreatic cancer cells and sensitizes them to TRAIL-induced apoptosis. Investigational new drugs. 2011;29(5):777–785. doi: 10.1007/s10637-010-9422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang A, Herter-Sprie G, Zhang H, Lin EY, Biancur D, Wang X, Deng J, Hai J, Yang S, Wong KK, Kimmelman AC. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer discovery. 2018;8(3):276–287. doi: 10.1158/2159-8290.CD-17-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guzman EA, Harmody D, Pitts TP, Vera-Diaz B, Winder PL, Yu Y, Wright AE. Inhibition of IL-8 secretion on BxPC-3 and MIA PaCa-2 cells and induction of cytotoxicity in pancreatic cancer cells with marine natural products. Anti-cancer drugs. 2017;28(2):153–160. doi: 10.1097/CAD.0000000000000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.