Abstract
Targeted insertion of large pieces of DNA is an important goal of genetic engineering. However, this goal has been elusive since classical methods for homology-directed repair are inefficient and often not feasible in many systems. Recent advances are described here that enable site-specific genomic insertion of relatively large DNA with much improved efficiency. Using the preferred repair pathway in the cell of nonhomologous end-joining, DNA of up to several kb could be introduced with remarkably good precision by the methods of HITI and ObLiGaRe with an efficiency up to 30-40%. Recent advances utilizing homology-directed repair (methods of PITCh; short homology arms including ssODN; 2H2OP) have significantly increased the efficiency for DNA insertion, often to 40-50% or even more depending on the method and length of DNA. The remaining challenges of integration precision and off-target site insertions are summarized. Overall, current advances provide major steps forward for site-specific insertion of large DNA into genomes from a broad range of cells and organisms.
Keywords: genome editing for site-specific insertion of large DNA; programmable nucleases of ZFN, TALENs and CRISPR-Cas9; ObLiGaRe (Obligate Ligation-Gated Recombination); HITI (Homology-Independent Targeted Integration); PITCh (Precise Integration into Target Chromosome); ssODN (single-strand oligodeoxynucleotide)
Introduction
In order to determine gene function, the initial approach in the field of genetics was to use X-irradiation or chemical mutagenesis to create mutations genome-wide (or to study spontaneous mutations), and then visualize phenotypic changes and map the mutated genes on the chromosomes. With the advent of molecular approaches, it became possible to mutate a desired stretch of DNA and then insert it into the genome for fine-scale mapping of functional regions of the gene or its regulatory elements. Initially, integration of foreign DNA into eukaryotic genomes occurred at ectopic locations beyond the control of the investigator. T-DNA was the first vector used for genetic engineering in plants [Bevan and Chilton 1982]. At about the same time, the P-element transposon was the first vector used for transformation of a multicellular animal, accomplished in Drosophila [Rubin and Spradling 1982; Spradling and Rubin 1982]. Subsequently, several other transposons were used as transformation vectors for a variety of other insects [O’Brochta and Atkinson 1996]. One of the most versatile of these is piggyBac [Lobo et al. 1999; Horn and Wimmer 2000; Handler 2002]. Generally, the size of integrated DNA was <20 Kb. Subsequently, integration of several kb DNA could be introduced at 55% efficiency (five-fold higher than with P-elements) using phiC31, but the placement of the phage att site for integration was still random with regard to the genome [Groth et al. 2004]. Depending on the site of genomic insertion, position effects influenced the extent of gene expression. To overcome this problem, many transformants at various genomic locations were analyzed to obtain their average level of gene expression. For comparative studies of several different mutations in the same gene, techniques were developed to always introduce the mutated DNA into the same ectopic location with cassette exchange methods such as Cre-Lox or FLP-FRT. However, unlike phiC31, Cre/loxP and Flp/FRT are reversible. A major advance in Drosophila was the development of P[acman], which is a BAC transgenic platform for targeted insertion of large DNA fragments over 100 kb [Venken et al. 2006; Venken et al. 2009]. Similarly, inserts of 80 kb into an att site using phiC31 was demonstrated in Drosophila [Wesolowska and Rong 2013].
Although the advances above were suitable for molecular dissection of gene function by mutation, they did not satisfy the need for site-specific insertion of DNA for use in genome editing to cure genetic diseases. Moreover, site-specific insertions into the endogenous locus overcome position effects of random integration and allow the investigator to analyze gene expression at levels dictated by its normal genomic location. This goal became a reality with the advent of programmable nucleases as molecular scissors to cleave a single desired site in the genome into which DNA could be inserted. This technology allows insertion of DNA to study its function by engineered mutations. In addition, it allows locus-specific insertion of reporter genes as well as insertion of tagged genes. Finally, these approaches can be used for gene replacement in gene therapy. The focus of this review is on recent advances for site-specific insertion of relatively large DNA.
Programmable site-specific nucleases
Zinc Finger Nucleases
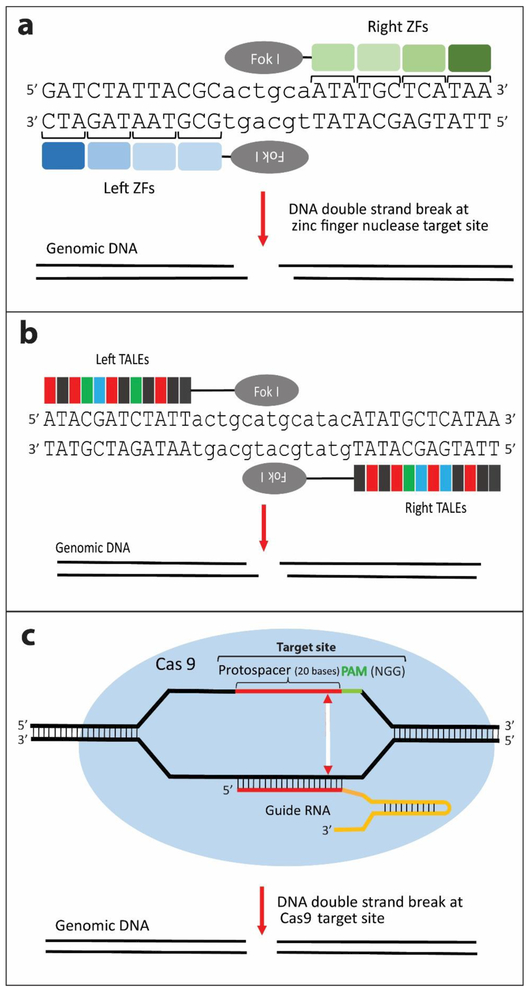
A major advance for site-specific insertions was the development of zinc finger nucleases (ZFNs) [Urnov et al. 2005; Carroll et al. 2010; Urnov et al. 2010; Carroll 2011]. Each zinc finger recognizes 3 bases and usually 3-6 fingers are used per ZFN to recognize 9-18 bases (Fig. 1a). The individual zinc fingers or two finger modules are chosen from a library so that in principle the large variety of sequence combinations makes it possible to target any sequence of interest in the genome. Because ZFNs work as a heterodimer, the number of bases recognized is doubled to 18-36, thereby enhancing target specificity. For each ZFN, the array of zinc fingers is coupled with the cleavage domain of the restriction enzyme FokI [Li et al. 1992; Kim et al. 1996]. Thus, in the ZFN chimera the zinc fingers provide targeting to the desired site in the genome and the FokI nuclease cleaves at that site, giving rise to overhanging ends (Fig. 1a). Despite the length of the target sequence, off-target cleavages were sometimes seen, often reflecting the erroneous formation of a ZFN homodimer. This problem was minimized by alteration of the amino acids at the ZFN interface so that only heterodimers were permissible, thus lowering off-target insertions and giving increased insertion site specificity [Miller et al. 2007; Szczepek et al. 2007; Söllü et al. 2010; Doyon et al. 2011; Ramalingam et al. 2011]. The introduction of ZFN methodology ushered in the era of programmable nucleases to target specific sequences for genome engineering.
Fig. 1. Programmable nucleases for site-specific cleavage in the genome.
(a) ZFNs: Each zinc finger (ZF) binds to 3 nucleotides of DNA. An array of 3-4 ZFs is coupled with the FokI nuclease for cleavage. (b) TALENs: Each TALE domain binds to a single nucleotide. An array of TALEs is coupled with the FokI nuclease. For both (a) and (b), FokI nuclease acts as an obligate heterodimer where the nuclease domain cleaves the spacer sequence to create a DSB with cohesive ends, (c) The sgRNA contains a 19–20 base sequence (red) within the CRISPR RNA (crRNA) that is complementary to the chromosomal target site, and this is combined with a transactivating CRISPR RNA (tracrRNA) (yellow) to guide the Cas9 nuclease (blue oval) to the target site where it creates DSBs with blunt ends 3 bases upstream of the PAM sequence (green).
An unanticipated problem with the design of ZFNs is that the specificity of individual zinc fingers can be influenced by the neighboring fingers [reviewed by Chandrasegaran and Carroll 2016]. This necessitated further work and validation that the final array of zinc fingers did indeed cut the desired sequence.
TALENs
Even though the problems in ZFN construction could be overcome, their high cost led to a search for alternative programmable nucleases. Transcription activator-like effector nucleases (TALENs) provided such an alternative. The TALE DNA binding module in plant virulence factors contains repeating units of 33-35 amino acids that are virtually identical except for amino acids 12 and 13 that impart specific nucleotide recognition for that TALE motif. Thus, the tandem array of amino acids recognizes the DNA sequence composed of nucleotides specified by each individual TALE motif [Boch et al. 2009; Deng et al. 2012; Mak et al. 2012] (Fig. 1b). This means that the tandem array of TALE motifs can be customized to recognize a desired DNA sequence target binding site. This is fairly straight-forward [Reyon et al. 2012], unlike the custom design of ZFNs. Moreover, the binding of each module to a single base pair is not influenced by context. Like ZFNs, the TALE that determines the target DNA binding site is coupled to FokI nuclease to direct DNA cleavage to a unique site in the genome [Christian et al. 2010; Miller et al. 2011; Mussolino et al. 2011]. However, the large size of the genes encoding TALENs and their repetitive nature introduced some difficulties into their production [reviewed by Chandrasegaran and Carroll 2016].
CRISPR-Cas
The CRISPR-Cas system used by bacteria and archaea to degrade invading DNA was harnessed as a powerful tool for genetic engineering [Jinek et al. 2012 and reviewed by Barrangou and Marraffini 2014; Doudna and Charpentier 2014; Hsu et al. 2014; Sander et al. 2014]. Unlike ZFNs and TALENs that are both protein-targeted nucleases, CRISPR-Cas9 utilizes RNA that guides the associated nuclease to its target site for cleavage. The guide RNA contains a short 19–20 base element within the CRISPR RNA (crRNA) that is complementary to the chromosomal target site, and this acts with a trans-activating CRISPR RNA (tracrRNA) to guide the Cas9 nuclease to the target site (Fig. 1c). The crRNA/tracRNA can be combined into a single guide RNA (sgRNA) that targets Cas9 to its site of cleavage [Jinek et al. 2012]. Cleavage of genomic targets with the CRISPR-Cas9 system requires a PAM sequence (eg, NGG for S. pyogenes Cas9) in the chromosome directly adjacent to the protospacer element of the target site. The guide RNA (gRNA) directs Cas9 to create a double-strand break (DSB) at the target site, 3 bp upstream of the PAM site. However, there can be some sequence-dependent variability of the efficiency of CRISR/Cas9 cleavage [Xu et al. 2015; Liu et al. 2016].
Overall, CRISPR-Cas9 has become the favored site-specific nuclease, with applications in a broad range of organisms (summarized by Chandrasegaran and Carroll 2016]. Off-target cleavage was an initial problem, but several approaches have been designed to minimize this (reviewed by [Barrangou and Doudna 2016; Chandrasegaran and Carroll 2016] and discussed further below). Creating the guide RNA is much simpler and less costly than the designer proteins to target ZFNs or TALENs to the desired genomic sequence for cleavage.
Cellular pathways for repair of DNA double-strand breaks (DSBs)
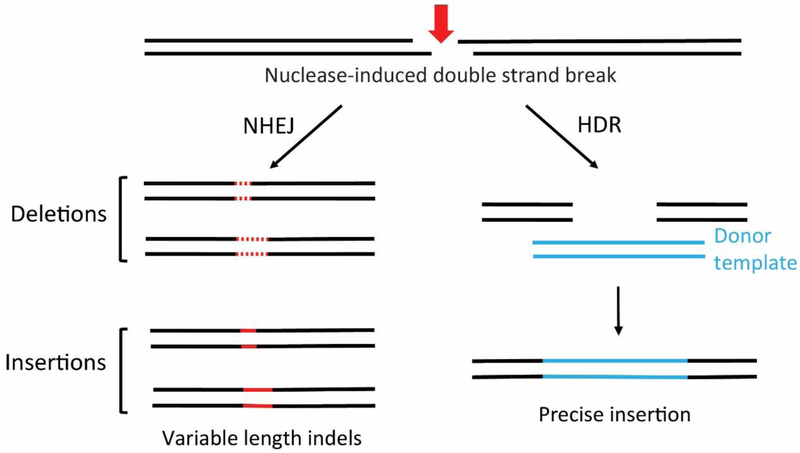
Success with programmable nucleases as site-specific molecular scissors allowed the development of genetic engineering (“genome editing”) that has now been successfully accomplished in several kinds of animals and plants [reviewed by Urnov et al. 2010; Chandrasegaran and Carroll 2016; Gaj et al. 2013; Segal and Meckler 2013; Carroll 2014; Peng et al. 2014; Salsman and Dellaire 2017]. Cells utilize two main pathways to repair DNA breaks: homology-directed repair (HDR) and classical nonhomologous end-joining (c-NHEJ) (Fig. 2). The mechanisms for the repair pathways have been summarized recently [Ceccaldi et al. 2016; Jasin and Haber 2016; Rodgers and McVey 2016; Danner et al. 2017] and are beyond the scope of this review. Briefly, HDR utilizes several hundred bases of homology on each side of the DSB in contrast to NHEJ where homology is not needed. HDR is maximal in late S phase and G2 whereas NHEJ occurs throughout the cell cycle and is the preferred pathway in G1 when resection activity is low [Sfeir and Symington 2015].
Fig. 2. The major repair pathways.
DNA DSBs are repaired by nonhomologous end-joining (NHEJ) or by homology-dependent repair (HDR). When a DNA donor is present, it can be integrated, as shown for HDR. In the absence of a DNA donor, small insertions and deletions (indels) are created by NHEJ.
Another pathway is termed alternative NHEJ (a-NHEJ, also called alt-EJ). a-NHEJ involves PARP1, the MRN complex and its partner CtIP, whereas c-NHEJ uses the Ku70/80 heterodimer and DNA-PK catalytic subunit [Dueva and Iliakis 2013]. DSB repair occurs by a-NHEJ when the c-NHEJ pathway is compromised by the absence of Ku [Fattah et al. 2010].
The DNA ends are resected in HDR but not in NHEJ. The resected 3’ single-strand DNA (ssDNA) overhangs can be repaired by any of three mechanisms: (i) HDR, (ii) alternative NHEJ (a-NHEJ) including microhomology-mediated end-joining (MMEJ)[Sfeir and Symington 2015), and (iii) single-strand annealing (SSA)[Bhargava et al. 2016]. The DNA ends at the DSB are bound by proteins: Ku70/Ku80 (in c-NHEJ) or MRN (in HDR and a-NHEJ). The other proteins downstream of MRN in the repair pathway differ for HDR and a-NHEJ. We focus on how all these various pathways have been employed for use in genome editing, especially with regard to insertion of relatively large DNA into cells from multicellular eukaryotes.
Homology-directed repair
Insertion of DNA that is several kb long is readily accomplished in yeast by homology-directed repair (HDR) [Scherer and Davis 1979; Orr-Weaver et al. 1981]. Although this is also possible in higher eukaryotes as first demonstrated in mouse cells [Smithies et al. 1985; Thomas et al. 1986; Mansour et al. 1988; Capecchi 1989], the efficiency is low, though the efficiency can be improved to one in ten thousand by increased homology [Deng and Capecchi 1992] or by induced DSBs [Rouet et al. 1994; Smih et al. 1995]. HDR occurs during S and G2 of the cell cycle reflecting the cell cycle regulated interaction of BRCA1–PALB2 that recruits BRCA2 [Orthwein et al. 2015]. Moreover, in late S and G2 a sister chromatid is present for repair utilizing hundreds of bases of homologous DNA sequences that flank the DSB [Branzei and Foiani 2008; Mladenov et al. 2016]. For genome editing, artificially introduced donor DNA with 0.5-2 kb homology arms is used instead of the sister chromatid for HDR at targeted DSBs created by site-specific molecular scissors (Fig. 2). Early experiments demonstrated that the rare-cutting endonuclease I-SceI [Rouet et al. 1994; Smih et al. 1995] or a ZFN-induced DSB [Bibikova et al. 2001] provided the entry point for HDR, thus ushering in the possibility of genome editing. In another early experiment, ZFNs were used to create site-specific cleavage in Drosophila embryos that was repaired by NHEJ in the absence of donor DNA or repaired by HDR in the presence of donor DNA [Bibikova et al. 2002; Bibikova et al. 2003]. Soon thereafter, ZFNs were used to repair a reporter insert [Porteus and Baltimore 2003] and endogenous genes in human cells [Urnov et al. 2005].
Using long stretches of homology (500-800 bp homology arms and ranging from 200 bp to several kb in other experiments), programmable nucleases coupled with the HDR pathway have been used for precise integration of transgenes from 8-15 kb in cultured cells [Moehle et al. 2007; Jiang et al. 2013]. However, the efficiency of DNA insertion is usually low, creating problems for targeted gene insertion within genomes of whole organisms, especially where large-scale screening can be difficult.
So far, targeted DNA insertion by HDR has only been reported in a few whole organisms. Instead, in most systems, DSBs are repaired by nonhomologous end-joining (NHEJ) which is the preferred pathway (e.g., by ~10-fold in Drosophila [Beumer et al. 2008; Carroll et al. 2010]). In order to increase the frequency of HDR, some investigators have disabled the NHEJ preferred pathway by knocking out DNA ligase 4 in C. elegans [Morton et al. 2006] or in Drosophila [Beumer et al. 2008; Bozas et al. 2009; Beumer et al. 2013], or by using the NHEJ inhibitor SCR7 in mammalian cells or embryos [Chu et al. 2015; Maruyama et al. 2015; Singh et al. 2015] or reducing the NHEJ pathway by other means such as gene silencing of Ku70, Ku80 or DNA ligase I [Chu et al. 2015]. Similarly, HDR efficiency can be increased by timed delivery into synchronized cells [Lin et al. 2014], since HDR is maximal in late S and G2. These and other strategies have been reviewed recently [Salsman and Dellaire 2017], but overall there is only a 5 to 10-fold increase in HDR. Thus, even with these approaches, HDR in its classical form may occur at too low a frequency to be practical for introduction of large DNA fragments into intact organisms. Promising recent advances are described later in this article for genomic insertion of DNA by homology.
Nonhomologous end-joining
The nonhomologous end-joining (NHEJ) repair pathway occurs throughout the cell cycle. Since it is preferentially used rather than HDR by most cells and organisms [Sedivy and Sharp 1989; Bozas et al. 2009; Iyama and Wilson 2013], it should be more efficient for targeted gene insertion in whole organisms. Typically the error-prone nature of NHEJ in the absence of donor DNA results in small insertions or deletions (indels) that cause gene disruption at the target site, and this has been the primary use so far of NHEJ (Fig. 2). Indeed, CRISPR-Cas9 technology has become one of the most popular ways to inactivate a gene due to disruptions created by indels.
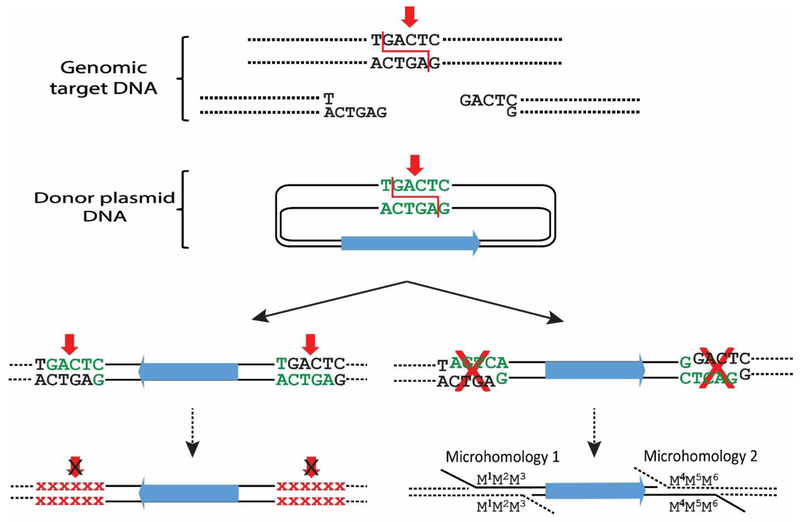
Previously, the integration of large DNA fragments via NHEJ has been inefficient as well as imprecise and not widely used. One route that has been implemented to improve efficiency is to couple NHEJ with ligation in vivo (“end-capture”) of compatible overhangs created in the donor DNA and the genomic target site by molecular scissors such as ZFNs (Fig. 3) [Orlando et al. 2010; Cristea et al. 2013; Weinthal et al. 2013]. The cohesive ends of the donor and target can pair, resulting in a product with the donor inserted at a single orientation. However, since the cleavage target sites have thus been recreated in the product, they can be re-cleaved until mutations (such as deletions by exonucleases) occur to destroy the target sites. As a result, the donor DNA is then integrated into the genomic target site by NHEJ with imprecise junctions. Potentially, exonucleolytic deletions can also occur in the donor DNA upon its initial introduction into the cell in the short window of time when the ends are not protected by the ZFNs [Cristea et al. 2013]. The donor DNA cannot be inserted readily in the opposite orientation since the cohesive ends would not be complementary between the donor and the genomic target sites (pathway at bottom right of Fig. 3). However, it is possible that exonuclease digestion could resect the ends of the donor until areas of microhomology with the genomic target region are encountered and integration could occur by microhomology-mediated end-joining (MMEJ, discussed below). Both pathways shown in Fig. 3 to produce stable products after end-capture would occur at relatively low frequency and both would result in imprecise junctions.
Fig. 3. End-capture of cohesive ends.
Diagram of “end capture” of a piece of donor DNA at the genomic site of a nuclease-induced (red arrows) DSB. Two pathways could result in stable products, but at low efficiency and with imprecise junctions. Left: The overhanging ends of the donor DNA are complementary to the overhanging ends of the genomic target site, since both the donor and genomic DNAs were cleaved by the same ZFN. However, end capture recreates the target sites that can be re-cleaved. Recutting and religation will be repeated until the target site sequence is altered by mutation and can no longer be cleaved by the programmable nuclease. Small red Xs indicate the mutated sequences. Right: Donor DNA that inserts with an opposite orientation to that shown in the left pathway will have incompatible ends to the genomic target sites, but it can be integrated via microhomology with very low efficiency.
Although the FokI nuclease of ZFNs and TALENs create cohesive ends suitable for end-capture, Cas9 creates blunt ends that are not suitable for end-capture. One recent advance has been to fuse Fok1 to an inactivated Cas9 so that overhanging ends are created by Fok1 after CRISPR direction to the target site [Guilinger et al. 2014; Tsai et al. 2014]. Alternatively, CRISPR can be used with Cpf1 that creates overhanging ends rather than the blunt ends generated by Cas9; Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system [Zetsche et al. 2015]. End-capture works best when the plasmid is cleaved after introduction into the cell, rather than prior linearization [Cristae et al. 2013; Auer et al. 2014; Bachu et al. 2015]. However, the efficiency for site-specific genomic insertion of DNA fragments by NHEJ with end-capture was not much higher than for HDR [Orlando et al. 2010; Cristea et al. 2013; Weinthal et al. 2013; Yamamoto et al. 2015].
CRISPR-Cas9 site-specific cleavage to create blunt ends has been used for DNA insertion by NHEJ without end-capture. Using this approach, insertion of ~5 kb into mammalian cells by NHEJ after CRISPR-Cas9 breaks in both the plasmid and genomic target site only had efficiencies of 0.17% in HEK293 cells and 0.45% in CHO cells, and there were indels at the insertion junctions [Bachu et al. 2015]. In contrast, ~6 kb DNA was inserted into the zebrafish genome by CRISPR-Cas9-mediated NHEJ knock-in with an efficiency of 10% for in-frame germline integrations and transmission to future generations of 31%, but the precision was low and indels were present at the integration junctions [Auer et al. 2014]. A similar approach created >10 kb (and as large as 34 kb) insertions in human cell lines and embryonic stem cells [He et al. 2016]. However, indels were often at the insertion junctions in these reports. The use of CRISPR/Cas9 and NHEJ has been optimized in the HITI methods described below.
Recent advances for site-specific insertion of relatively large DNA by NHEJ
Many shied away from use of NHEJ for site-specific insertion of DNA due to its reputation of imprecision, with indels at the junctions of the insertion. The property of imprecision is a hallmark of the alternative NHEJ pathway (a-NHEJ) and a lesser problem for the canonical NHEJ pathway (c-NHEJ). Although indels occur when the ends of the DNA DSBs are not compatible, c-NHEJ can achieve 75% or more error-free repairs according to Bétermier et al. [2014].
Two new methods are described below that utilize NHEJ with improved efficiency for site-specific insertion of large DNA. One possibility to explain the higher efficiency is that the experimental design prevents reformation of the target site after the insertion has occurred and therefore re-cutting is prevented. The examples described below and depicted in the accompanying figures show site-specific insertion of the donor DNA after creating a single cut at the target site. These methods are readily adaptable to gene replacement when two cuts are made in the genomic DNA, with each cut flanking the gene to be replaced.
Homology-Independent Targeted Integration (HITI)
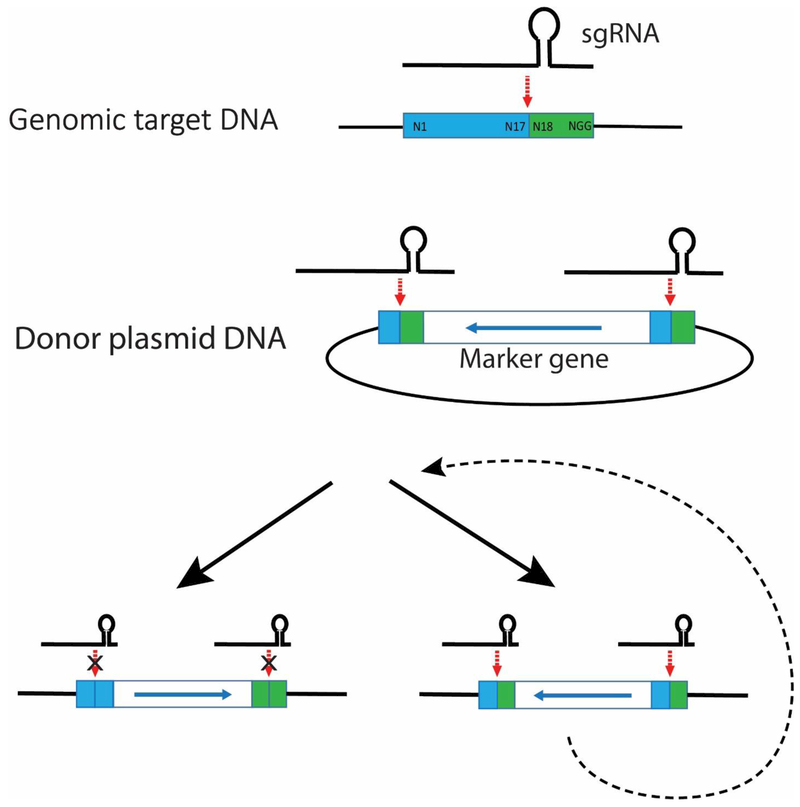
Recently, extensive optimization of CRISPR-Cas9 with NHEJ (called HITI = homology-independent targeted integration) (Fig. 4) was used for insertion of DNA [Suzuki et al. 2016]. In HITI, the sgRNA of CRISPR cleaves the same target sequence in the genome and flanking the gene of interest in the donor plasmid. Subsequently, the gene of interest is inserted into the genomic target site by NHEJ. The gene of interest can be inserted in either orientation, but if the sgRNA target site is re-created, the donor DNA is presumably excised and re-inserted until ultimately it is in the final stable orientation where the sgRNA target site is no longer present. When used for DNA site-specific integration into proliferating HEK293 cells, the efficiency was 30-40%, and even somewhat better after optimization of the nuclear localization sequence that was used for Cas9. Although the majority of the cells had precise integrations, up to a quarter exhibited imprecise integrations with indels.
Fig. 4. Homology-independent targeted integration (HITI).
The sgRNA of CRISPR cleaves the same target sequence in the genome and flanking the gene of interest in the donor plasmid (the blue box indicates the beginning of the target sequence and the green box indicates the end of the target sequence). The gene of interest is subsequently inserted into the genomic target site by NHEJ in either orientation. If the sgRNA target site is re-created, the inserted DNA is presumably excised and re-inserted until finally it is in the final stable orientation where the sgRNA target site is no longer present
The HITI approach was also successful for DNA site-specific integration in cultured post-mitotic neurons, with an efficiency of 0.58% of all cells and 56% of the transformed cells [Suzuki et al. 2016]. Moreover, the precision of integration was very high (~90%). When used for insertion into rodent neurons in whole animals where HITI with adeno-associated viral (AAV) vectors were injected into the visual cortex of adult mouse brains, the efficiency of site-specific integration was 3.5% of all cells and 10.6% of infected cells [Suzuki et al. 2016]. Intravenous injection of HITI-AAV vectors into mice resulted in nuclear GFP expression in 3-10% of cells in heart, liver, muscle and other tissues, with good precision comparable to the HEK293 experiments (Suzuki et al. 2016). Note that the HITI-mediated DNA insertions above using whole animals were somatic and not germline events.
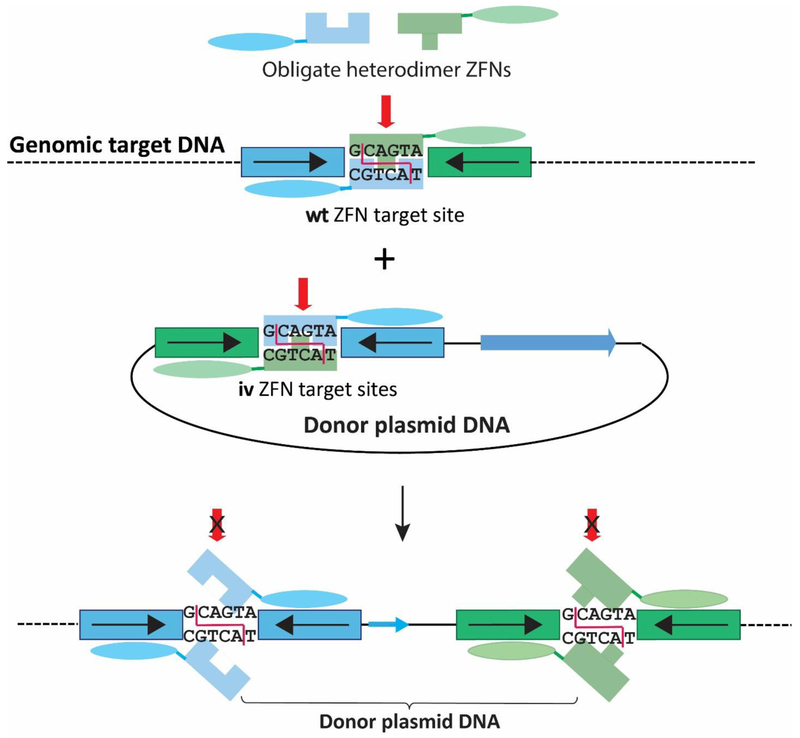
ObLiGaRe (Obligate Ligation-Gated Recombination)
Another approach using NHEJ for site-specific genomic introduction of large DNA is obligate ligation-gated recombination (ObLiGaRe). The ObLiGaRe method was initially developed for use with ZFN site-specific cleavage of genomic DNA from cultured cells [Maresca et al. 2013] and has been adapted for whole organisms [Yamamoto et al. 2015]. The pair of ZFNs binds to the target DNA sites on opposite strands and forms heterodimers through the modified FokI nuclease domain. In the ObLiGaRe approach, the donor plasmid contains the same ZFN binding sites as the genomic target site to allow end-capture of the overhanging ends after FokI digestion, but the trick is that the ZFN binding sites in the donor plasmid are inverted relative to the host target sequence (Fig. 5). The new junction formed after the integration event prevents subsequent ZFN excision since the ZFN heterodimer target sites are destroyed [Doyon et al. 2011]. This increases the efficiency of formation of the final product. By contrast, in the HITI method, only half of the products have the nuclease cleavage sites destroyed, and the other half are continually re-cleaved until the donor DNA is inserted in the opposite orientation to destroy the cleavage sites of the product (Fig. 4). An improved situation occurs with the ObLiGaRe method where the cleavage sites have been destroyed in all of the products initially (Fig. 5).
Fig. 5. Obligate ligation-gated recombination (ObLiGaRe).
A pair of ZFNs cleaves both the wild type (wt) target site of genomic DNA and the inverted target sites (iv) of the donor construct. Both the wt and iv target sites have the identical spacer sequence between the left and right ZFN binding sites, thus creating complementary overhanging ends between the donor DNA and genomic target for end-capture. Once the linearized donor has been ligated into the genomic target site, the same pair of ZFNs cannot cut (x over red arrow) the newly formed junctions since homodimer formation is prevented by the use of an obligatory heterodimeric FokI nuclease domain. Moreover, the inverted target sequence in the donor plasmid prevents adjacent placement of the green and blue sequences. Thus, the efficiency of formation of the final product of the integrated donor DNA at the genomic target site is increased as re-cutting cannot occur. The single dotted or solid lines represent dsDNA of genomic or plasmid DNA, respectively. In the example shown here, the entire donor plasmid is integrated into the target site.
Therefore, the basis of ObLiGaRe is that two separate DNA binding sites are required to make the inverted target site, thus preventing re-cleavage after genomic integration. [Maresca et al. 2013]. To achieve this, ObLiGaRe uses the FokI endonuclease domain, which has to form a dimer to be active. The heterodimeric nature of ZFNs makes possible the inversion of the target sites in the donor relative to the target. CRISPR-Cas9 in its original form is not compatible with ObLiGaRe because Cas9 is not a heterodimer. However, the newly developed dimeric CRISPR-FokI nuclease where Fok1 is fused to a catalytically inactive Cas9 (dCas9) could be very useful as molecular scissors, providing an alternative to ZFNs or TALENs for future applications [Qi et al. 2013; Guilinger et al. 2014; Tsai et al. 2014].
The transformation efficiency with ObLiGaRe of ~2% in cultured cells was about 5 times higher than using classical approaches of HDR [Maresca et al. 2013]. With ObLiGaRe, site-specific integration of 15 kb DNA into the genome of HCT116 human cells had 75-88% precision at the junctions [Maresca et al. 2013]. When ObLiGaRe was used for site-specific integration in flies, efficient insertion (~27% without any selection) of a 2.4 kb piece of DNA into somatic cell genomes was observed 3 days after injection of embryos and had 87% precision at the insertion junctions [Yamamoto et al. 2015]. Furthermore, ObLiGaRe using a 6.5 kb fragment had 6.9% efficiency for germline site-specific integration, and sequencing the insertion junctions of several fly lines revealed no sequence changes at all, nor was there any evidence for off-target insertions [Yamamoto et al. 2015]. Therefore, ObLiGaRe with NHEJ has quite good efficiency with precise integration and minimal off-target insertions.
Recent advances for site-specific insertion of relatively large DNA by homology
Although classical HDR uses several hundred bases of homology flanking the site for insertion of DNA, some recent approaches have had good success using much smaller length of homology, as described below. Although these recent approaches employ homology between donor and target sequences, they do not always use HDR.
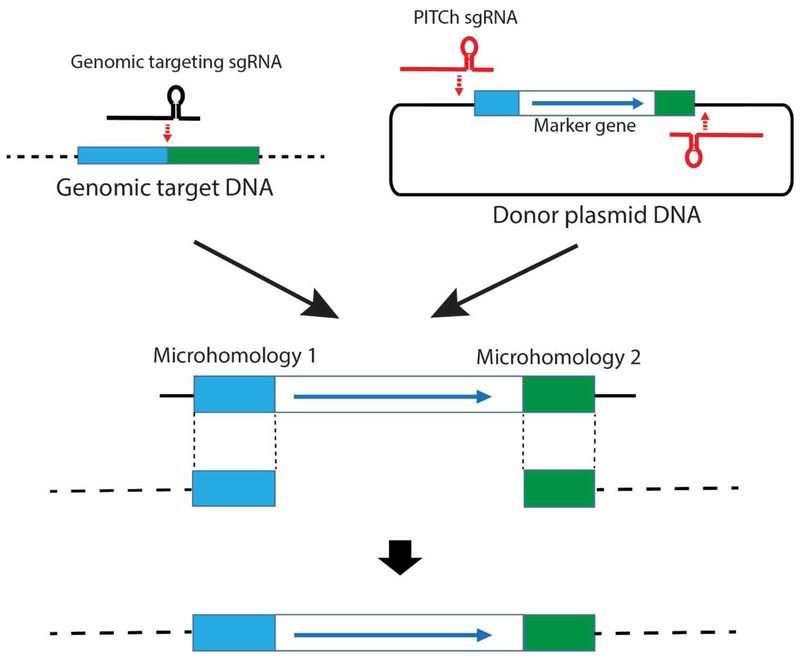
PITCh (Precise Integration into Target Chromosome)
PITCh (Precise Integration into Target Chromosome) (Fig. 6) is mediated by 5-25 bases of microhomology-mediated end-joining (MMEJ). MMEJ is a subset of the a-NHEJ pathway and occurs during M and early S and is independent of Ku and ligase 4, unlike c-NHEJ [Sfeir and Symington 2015]. PITCh has been reported for insertion of relatively large DNA into genomic sites from cultured cells, zebrafish, silkworms and frogs cleaved by TALENs or CRISPR-Cas [Nakade et al. 2014; Hisano et al. 2015; Sakuma et al. 2016]. Moreover, MMEJ appears to be even more efficient than c-NHEJ (79% MMEJ vs. 53% c-NHEJ for genomic integration in zebrafish embryos [Hisano et al. 2015]). For the PITCh system, a programmable nuclease creates a DSB in the donor DNA and the genomic target site with subsequent DNA insertion stimulated by MMEJ. When a single DSB occurs in the plasmid donor, then the plasmid carrying the DNA of interest is inserted at the genomic target site. In contrast, just the DNA of interest (without the plasmid) is inserted at the genomic target site when two DSBs flank the DNA of interest in the plasmid. In version 2 of CRIS-PITCh [Sakuma et al. 2016], targets for the CRISPR guide RNA are in the plasmid, adjacent to the 20 base microhomologous sequences flanking the DNA of interest. Using CRISPR-Cas9 for PITCh in HEK293T cells, the precision of DNA insertion was good but indels sometimes occurred at the junctions (80% perfect 5’ junction; 50% perfect 3’ junction) [Sakuma et al. 2016], as might be expected since the a-NHEJ pathway can be imprecise. The difference in precision of integration at the two junctions might be explained by MMEJ occurring at just one junction and NHEJ occurring at the other junction.
Fig. 6. Precise integration into target chromosome (PITCh).
Two regions of 5-25 base homology at the genomic target site and flanking the gene of interest in the donor plasmid are shown (blue, green) In version 2 of CRIS-PITCh, CRISPR sgRNA cuts adjacent to the microhomology regions in the plasmid, whereas a different sgRNA cuts between the two blocks of microhomology in the genome. The MMEJ pathway (a subset of a-NHEJ) uses the blocks of microhomology to insert the gene of interest into the desired genomic site. Note that the two blocks of microhomology are separated in the end product of PITCh so that the guide RNA that initially cleaved the genomic DNA cannot subsequently cleave the genomic end product. Similarly, the target sites for the PITCh guide RNAs are destroyed in the integrated product. The single dotted or solid lines represent dsDNA of genomic or plasmid DNA, respectively.
HMEJ (Homology-Mediated End-Joining)
The method of HMEJ has a similar strategy to MMEJ (Fig. 6) except that homology arms of 800 bases are used rather than 20 base microhomology arms [Yao et al. 2017; Yao et al. 2018. In an exhaustive set of experiments, HMEJ was compared to MMEJ, NHEJ and classical HDR for DNA insertion in numerous cell types in vitro and in vivo as well as mouse and monkey embryos. In almost all cases, HMEJ outperformed the other methods or in a few cases had the same efficiency as MMEJ. The efficiency of DNA insertion by HMEJ ranged from 5-50% depending on the cell type and from 15-30% in mouse embryos. It is unknown how HMEJ will perform for insertions larger than the few hundred bp of the mCherry reporter that was used. Impressively, sequencing revealed precise integrations occurred in almost all cases. Depending on the recipient cells, HMEJ seems to act by homologous recombination, NHEJ or perhaps a novel HMEJ pathway; further research is needed to clarify the mechanism. A similar approach to HMEJ with comparable results of improved efficiency of DNA insertion has also been reported by others who obtained insertion efficiency of up to 30% in stem cells [Zhang et al. 2017].
HDR with short homology arms
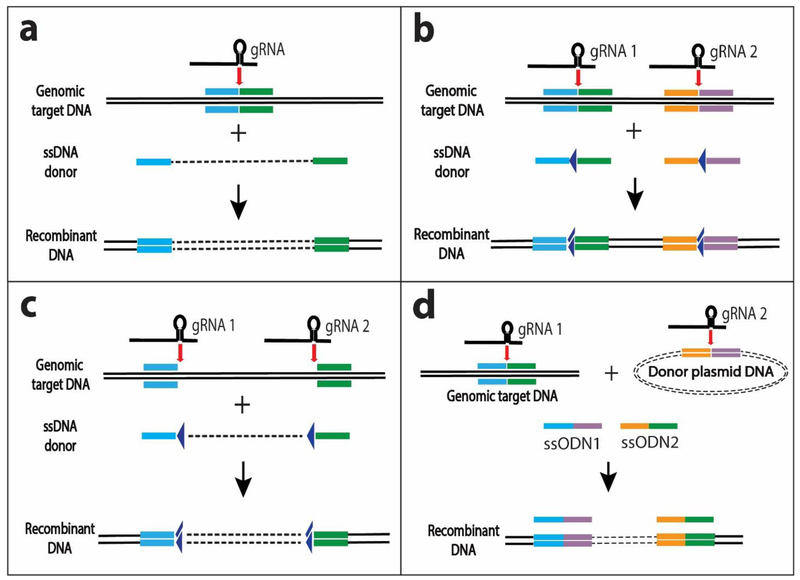
Although classical HDR uses long homology arms of 0.5-2 kb, it has been reported that HDR can work with only ca. 50 bases of homology. Specifically, donor homology length is relatively unimportant when a small piece of DNA is integrated into the target site, but the donor homology length becomes more important as the length of the donor DNA increases [Orlando et al. 2010]. HDR efficiency is enhanced when using linear DNAs as repair templates. The genomic target site is cleaved by CRISPR-Cas9 and homology arms of 30-100 bases facilitate the introduction of the donor DNA. The donor DNA can be a double-stranded PCR fragment [Paix et al. 2014 and 2016] or a long ssODN (single-strand oligodeoxynucleotide) (Fig. 7a); in both cases the donor DNA has the homology arms at its ends. It has been reported that the HDR efficiency for ssODNs can be increased to almost 60% in cultured human cells when the homology arms are asymmetric (e.g., 91 nt PAM proximal and 36 nt PAM distal) [Richardson et al. 2016]. The use of ssODN rather than double-stranded DNA (dsDNA) (as used for classical homologous recombination) can increase the insertion by as much as 500-fold, thus making genome editing possible in organisms that were previously intractable [Ferenczi et al. 2017]. Note that DNA cloning is not needed for either ssODN or double-stranded PCR fragments, thus simplifying the procedure.
Fig. 7. Genomic integration with short homology arms.
(a) A single-stranded DNA donor (long ssODN) containing homology arms 30-100 bases (blue, green) is administered together with the CRISPR guide RNA and Cas9 which cuts the desired target site in the genome. Alternatively (not shown) the donor can be dsDNA with two short ssODNs providing the bridging homology arms. (b) For future conditional knockouts (as by Cre-recombinase in mice), two guide RNAs and two short ssODNs (each with a LoxP site) are introduced to result in LoxP sites (triangles) at the genomic sites where Cas9 cleavage had occurred. The LoxP sites allow future deletion for conditional knock-out. (c) A more efficient strategy for future conditional knock-outs is to use a single long ssODN donor with LoxP sites (triangles) flanking the homology arms at the ends (blue, green). As in panel (b), the genomic DNA is cut by Cas9 that is targeted by two guide RNAs. Note that cutting the genome with two gRNAs (panel c) can also be employed for DNA replacement in the genome, in contrast to a simple DNA insertion after cutting the genome with just one gRNA (panel a). Easi-CRISPR employs the method shown here in panel (c): A long single-stranded DNA donor with short homology arms is injected with the pre-assembled Cas9 ribonucleoprotein (ctRNP) complex containing two guide RNAs to create targeted insertion. The ctRNP:complex contains crRNA + tracRNA +Cas9 protein). The dashed line indicates the inserted DNA sequence in the single-stranded donor DNA and in the double-stranded recombinant DNA. The sequence indicated by the dashed line can be deleted readily in future experiments by use of the LoxP sites that flank it. (d) The “two-hit by sgRNA and two oligos with a targeting plasmid” (2H2OP) portrayed here introduces a double-stranded donor plasmid DNA and two short bridging ssODNs (providing homology to the junctions) together with two sgRNAs to direct Cas9 cleavage of the chromosomal target site and of the donor plasmid, respectively.
HDR with these relatively short homology arms is highly efficient in many systems and also can be quite precise (e.g., >95% precision in C. elegans; Paix et al. 2016). Perhaps surprisingly, the efficiency of site-specific integration in C. elegans decreases as the length of homology increases (75% edits for 35 base homology, 47% edits for 60 base homology, 33% edits for 100 base homology) [Paix et al. 2016]. Optimal homology arms of 30-40 bases for ssODN have also been reported in mammalian cells by others [Liang et al. 2017]. Short homology arms also worked to introduce a double-stranded PCR fragment with a 739 bp mCherry insert and flanking homology arms of 36 bases into mouse zygotes with 31% editing efficiency and 100% precision [Paix et al. 2017a], similar to the efficiency found for long ssODNs [Quadros et al. 2017]. However, the efficiency decreased for DNA insertions > ~1 kb and 33 base homology arms [Paix et al. 2017a]. Interestingly, the efficiency of insertion of 714 bp in human cells was the same regardless if the homology arms were 33 bases or 518 bases [Paix et al. 2017a]. This is in contrast to the HMEJ experiments described above [Yao et al. 2017] where long homology arms perform better than short homology arms. A detailed protocol on performing edits of either DNA insertions or deletions using short homology arms has recently been published and contains useful tips [Paix et al. 2017b].
Introduction of DNA with short homology arms appears to work through DNA synthesis-dependent strand annealing (SDSA), rather than direct integration of the linear donor DNA in C. elegans [Paix et al. 2016] and in cultured human cells [Paix et al. 2017a]. Elegant experiments show that the DNA repair that is utilized is consistent with gene conversion [Paix et al. 2017a]. The accuracy of repair is asymmetric where one junction can be precise and the other junction can be imprecise, suggesting different homology requirements to initiate and resolve repair and consistent with SDSA for gene conversion [Paix et al. 2017a].
The experiments above had short homology arms to target DNA to a Cas9-cleaved single genomic site by a single guide RNA. However, a similar approach can be used with two guide RNAs to direct Cas9 cleavage to two genomic sites. This tactic has been employed to introduce two LoxP sites into mice for future conditional knock-outs by Cre recombinase. Use of Cas9 mRNA, two guide RNAs (sgRNAs) and two ssDNAs (each with a LoxP site) flanked by homologies of 40-80 bases had efficiencies of 2-5% [Bishop et al. 2016; Miano et al. 2016] with only one earlier report of 16% [Yang et al. 2013] (Fig. 7b). Presumably, the low efficiency of this approach could be explained by generation of single LoxP insertions, double LoxP insertions in trans and deletions resulting from NHEJ besides the desired two LoxP sites in cis. Although sequential injections of one sgRNA and one ssODN at the one cell stage followed by the second sgRNA and second ssODN at the two cell stage of mouse embryos helped to improve cis-insertions of the two LoxP sites [Horii et al.et al. 2017], it is cumbersome and not amenable to high throughput. The efficiency was dramatically improved to 44% when a 514 base single ssDNA donor containing two LoxP sites flanked by homology arms was introduced together with two sgRNAs and Cas9 mRNA; this approach had 86% precision [Miura et al. 2015] (Fig. 7c). Similarly, a thorough examination of many loci and testing many variables showed that the efficiency of 9% for the expected insertion using two ssODNs was greatly increased to an efficiency of 48% when using a single longer ssODN [Lanza et al. 2017]. In this case, 20-30% of the mice had an off-target insertion, but it was not observed for the same animal to have both an on-target and off-target insertion.
Easi-CRISPR (efficient additions with ssDNA inserts-CRISPR)
Optimization of this approach in a method called Easi-CRISPR (efficient additions with ssDNA inserts-CRISPR) (Fig. 7c) has had excellent results for edits flanked by LoxP sites [Quadros et al. 2017; Miura et al. 2018]. Up to almost 2 kb inserts were achieved with ssODN donor DNA flanked by 50-100 bases of homology at each end when co-injected into mouse zygotes with ctRNP (a complex of pre-assembled crRNA + tracRNA + Cas9 ribonucleoprotein) [Aida et al. 2015]. Specifically, an efficiency of 40% of the desired insert using a single ssODN donor (862 bases flanked by 91 and 93 base homologies) and ctRNP was achieved, which was better than the efficiency of 13% for the same donor and sgRNP (complex of sgRNA with Cas9). The three-fold greater efficiency when using ctRNP than sgRNP has also been noted by others [Aida 2015]. Moreover, efficiencies can be improved by using sgRNP rather than sgRNA [Chen et al. 2016]. When using Easi-CRISPR to create insertions of ssODN with ctRNP at six different loci to insert 527-893 bases, the efficiencies averaged 43%, ranging from 8.5% to an amazing 100%. Using this ssODN and ctRNP Easi-CRISPR approach for additional experiments at six other loci with ssODNs had 779-1368 base insertion frequencies of 25-67% of which 44% had the correct sequence at both junctions. Another group had 35% efficiency when using Easi-CRISPR in mice and found that the average germline transmission was 52.8% and no off-target cleavage [Ma et al. 2017].
When comparing the dsDNA donor PITCh approach [Aida et al.et al. 2016] to ssODN Easi-CRISPR [Quadros et al. 2017] for DNA insertion at the same locus in the mouse genome, PITCh had an efficiency of 33% for the correct insertion compared to 100% efficiency for Easi-CRISPR. In this case, the PITCh experiment used Exo1 nuclease that is an enhancer of targeted insertion [Aida et al. 2016]. A drawback of ssODN is that lengths over 2 kb are not commercially available [Paix et al. 2016; Quadros et al. 2017]. However, the approach of Miura et al. [2015] to create ssDNA by reverse transcription of a 4-5 kb in vitro transcribed RNA template may be promising for introduction of longer DNA, though errors may occur when producing the longer ssDNA.
2H2OP (two-hit by sgRNA and two oligos with a targeting plasmid)
A variant of the ssODN protocol succeeded in generating transformed rats with inserted DNA even as large as 200 kb for a BAC insertion with 1/15 pups (6.7%) insertion efficiency and correct sequences at the junctions [Yoshimi et al. 2016]. This method is called “two-hit by sgRNA and two oligos with a targeting plasmid” (2H2OP) (Fig. 7d). In this case, the donor vector does not have homology ends. Instead, the donor plasmid DNA and two short ssODNs as bridging oligos that provide homology are co-injected with two sgRNAs. One of the sgRNAs directs Cas9 cleavage at the target site in the chromosome and the other sgRNA directs Cas9 cleavage in the donor plasmid. The two ssODNs span the chromosomal target and inserted donor junctions at each end to increase the ligation efficiency. A related method of 3H2OP (4.3% efficiency) uses three sgRNAs for cleavage at two regions of the chromosome to replace the intervening stretch of DNA with the donor DNA that was made linear by the third sgRNA-mediated cleavage.
Despite the relatively modest insertion efficiency, 2H20P surpassed the TAL-PITCh method where no inserts were found for the same target site and the same 4.8 kb insert in contrast to 3/17 pups (17.6%) with 2H2OP. Although long inserts can be obtained with 2H2OP, a disadvantage is the lack of precision and only one of the pups with the 4.8 kb insertion had correct sequences at the insertion junctions. Another disadvantage is that the vector is also inserted at the target site, though presumably this could be overcome by using two sgRNAs to direct cleavage of the desired insert of the plasmid, in addition to a sgRNA for Cas9 cleavage of the chromosomal target site.
Conclusions and Perspectives
DNA insertion by NHEJ or by homology
Great strides are being made to develop genome editing to precisely introduce DNA at a specific location. This is necessary for experiments in basic biology and also for gene therapy to cure genetic diseases [reviewed by Pan et al. 2013]. The classical approach of HDR had excellent precision but low efficiency and is not feasible in several systems.
Table 1 summarizes the recent advances utilizing NHEJ or homology, as described in this review. NHEJ has been used in two approaches to introduce relatively long DNA into targeted genomic sites. For example, genomic insertions of 15 kb in cultured cells and 6.5 kb for fly germline transformation were achieved using the ObLiGaRe method. Although the efficiencies were modest (2-27%), the percentage of precise insertion was very good (75-100%).
Table 1.
Summary of new methods for site-specific insertion of relatively long DNA
| Method | organism | size of inserted DNA | efficiency | % of precise insertion |
|---|---|---|---|---|
| DNA insertion by NHEJ: | ||||
| HITI | HEK293 cells | 700 bp | few to 30-40% | 75-90% |
| cultured neurons | 56% of transformed cells | ~90% | ||
| mouse brain (somatic insertion) | 10.6% | 75-90% | ||
| ObLiGaRe | HCT116 cells; | 15 kb | 2% of all cells | 75-88% |
| Sciara (fungus fly) | 2.4 kb (somatic) | 27% | 87% | |
| Sciara (fungus fly) | 6.5 kb (germline) | 6.9% | 100% | |
| DNA insertion by homology: | ||||
| PITCh (5-25 bases homology) | ||||
| zebrafish embryos | 79% | 77% | ||
| HEK293T cells | 1.4 kb | ~75% | 80% perfect 5’ junction; 50% perfect 3’ junction |
|
| also silkworms (Bombyx) and frogs (Xenopus) | ||||
| HMEJ (800 bases homology) | ||||
| HEK293 cells | 700 bp | ~20% | almost always perfect | |
| stem cells | 0.7-6.1 kb | 2-8% | ||
| mouse brain | 700 bp | 10% | ||
| mouse embryos | 700 bp | 23% | 100% | |
| 35-100 base homology | <1 kb | 31-75% | 95-100% | |
| cultured human cells | <1 kb | ~60% | ||
| cultured human cells | 514 bp | 44% | 86% | |
| C. elegans (worm) | <1 kb | 33-75% | ||
| mouse embryos | 739 bp | 31% | 100% | |
| mouse embryos | <1 kb | 48% | 70-80% | |
| Easi-CRISPR (DNA replacement; ssODN donor with 50-100 bases homology) | ||||
| mouse embryos | <1 kb | 8.5-100% (average of 43%) |
||
| mouse embryos | 779-1368 bp | 25-67% | 44% | |
| 2H2OP or 3H2OP (DNA replacement by ssODN bridging oligos for 60-120 bases homology) | ||||
| rat embryos | ~1 kb | 18% | 25% | |
| rat embryos | 6.2 kb | 17% | 25% | |
| rat embryos | 200 kb | 13.3% | 50% | |
(nb – multiple entries for same cell type or organism reflect results from different experiments or from different groups)
Alternative strategies have used homology of 5-120 bases (and 800 bases homology in HMEJ) flanking the sequence to be inserted. These alternative strategies utilize a variety of pathways (not simply HDR) and sometimes the mechanism is not yet known. Usually, the inserted DNAs were ~1 kb or less with efficiencies up to 79% and even 100% and usually very good precision (often 70-100%). Larger DNA of 6.2 kb and even 200 kb was inserted with the 2H2OP method with 13-17% efficiency and 25-50% precision.
As can be seen, the wide ranges of efficiency to introduce DNA into a genomic target site vary both with the targeted locus and with the biological system. Often it is a trade-off of precision vs. efficiency, where 100% precision may have lower efficiency. The efficiency and precision are generally higher for DNA insertion (using a single guide RNA) rather than for DNA replacement (using two guide RNAs).
Off-target insertions
Beyond efficiency, yet another important consideration is that site-specific insertions should not occur elsewhere in the genome at off-target sites [Zhang 2015]. This has been minimized by using ZFNs with point mutations at the ZFN interface so that only heterodimers and not homodimers are permissible, thus lowering off-target insertions [Miller et al. 2007; Szczepek et al. 2007; Söllü et al. 2010; Doyon et al. 2011; Ramalingam et al. 2011].
Off-target insertions have been problematic with CRISPR-Cas9, but can be somewhat reduced by judicious choice of a unique target sequence [Lin et al. 2016]. Various methods have been used to estimate the frequency of cleavage at off-target sites [Stella and Montoya 2016], including the Digenome-Seq [Kim et al. 2016] and GUIDE-Seq [Tsai et al. 2015; Tsai and Joung 2016]. Off-target insertion has been minimized for CRISPR approaches such as by using a truncated sgRNA [Fu et al. 2014] or using a pair of Cas9s modified to act as nickases with staggered cuts on opposite DNA strands [Mali et al. 2013; Ran et al. 2013]. Another method paired a mutant Cas9 without nuclease activity (dCas9) with a FokI nuclease domain, which requires two sgRNAs with defined spacing and orientation before it cuts [Guilinger et al. 2014; Tsai et al. 2014]. It has been reported that use of Cas9 protein rather than Cas9 mRNA also helps to reduce off-target insertions [Wang et al. 2015]. While these techniques have improved the situation, there were still some residual off-target cleavages. Recent developments using a high fidelity Cas9 with a few engineered point mutations have further reduced off-target integrations in CRISPR approaches to barely detectable levels [Slaymaker et al. 2016; Kleinstiver et al.et al. 2016]. Even more recently, it has been reported that off-target insertions can be abolished by inactivation of Pol Θ and c-NHEJ [Saito et al. 2017; Zelensky et al. 2017].
Overall, the advances summarized here for efficient site-specific insertion of large DNA with very good precision and minimal off-target insertions hold promise to achieve gene editing in whole organisms, which are integral to and will facilitate advances in modern biology. These advances in basic science will also have translational applications in medicine and agriculture.
Acknowledgements
Research in the author’s lab is supported by NIH R01 HG008160, NIH R01 GM121455 and NSF MCB 1607411.
Abbreviations
- AAV
adeno-associated viral vector
- a-NHEJ
alternative nonhomologous end-joining
- Cas
CRISPR associated protein
- c-NHEJ
canonical nonhomologous end-joining
- CRISPR
clustered regularly interspaced short palindromic repeat
- crRNA
CRISPR RNA (the guide RNA)
- ctRNP
a complex of pre-assembled crRNA + tracRNA + Cas9 protein
- dCas9
catalytically inactive Cas9
- DSB
double-strand break
- dsDNA
double-stranded DNA
- Easi-CRISPR
efficient additions with ssDNA inserts-CRISPR
- HITI
homology-independent targeted integration
- HDR
homology-directed repair
- HMEJ
homology-mediated end-joining
- HR
homologous recombination
- Indels
small insertions and deletions
- MMEJ
microhomology-mediated end-joining
- NHEJ
nonhomologous end-joining
- ObLiGaRe
obligate ligation-gated recombination
- PAM
protospacer-adjacent motif
- PITCh
precise integration into target chromosome
- SDSA
DNA synthesis-dependent strand annealing
- sgRNA
single guide RNA containing a crRNA sequence and tracRNA (for CRISPR-Cas)
- sgRNP
complex of pre-assembled guide RNA and Cas9 protein
- SSA
single-strand annealing
- ssDNA
single-stranded DNA
- ssODN
single-strand oligodeoxynucleotide
- TALEN
transcription activator-like effector nuclease
- tracrRNA
transactivating CRISPR RNA (a structural RNA to recruit Cas9)
- 2H2OP
two-hit by sgRNA and two oligos with a targeting plasmid
- 3H2OP
three-hit by sgRNA and two oligos with a targeting plasmid
- ZFN
zinc finger nuclease
Footnotes
Conflict of interests
The authors declare no competing financial interests.
References
- Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R et al. (2015) Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 16: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida T, Nakade S, Sakuma T, Izu Y, Oishi A et al. (2016) Gene cassette knock-in in mammalian cells and zygotes by enhances MMEJ. BMC Genomics 17: 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet J-P, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24: 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachu R, Bergareche I, Chasin LA (2015) CRISPR-Cas targeted plasmid integration into mammalian cells via non-homologous end joining. Biotech Bioengineering 112: 2154–2162. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Doudna JA (2016) Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34: 933–941. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA (2014) CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell 54: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétermier M, Bertrand P, Lopez BS (2014) Is non-homologous end-joining really an inherently error-prone process? PLoS Genet 10: e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J et al. (2008) Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Nat Acad Sci 105: 19821–19826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Mukherjee K, Carroll D (2013) Donor DNA utilization during gene targeting with zinc-finger nucleases. G3: Genes/Genomes/Genetics 3: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MW, Chilton MD (1982) T-DNA of the Agrobacterium Ti and Ri plasmids. Ann Rev Genet 16: 357–384. [DOI] [PubMed] [Google Scholar]
- Bhargava R et al. (2016) Regulation of single-strand annealing and its role in genome maintenance. Trends Genet 32: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D (2003) Enhancing gene targeting using designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J et al. (2001) Stimulation of homologous recombination through targeted cleavage by a chimeric nuclease. Mol Cell Biol 21: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D (2002). Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KA et al. (2016). CRISPR/Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3: Genes/Genomes/Genetics 6: 2051–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S et al. (2009) Breaking the code of DNA binding specificity of TAL type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bozas A, Beumer KJ, Trautman JK, Carroll D (2009) Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 182: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2008) Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol Cell Biol 9: 297–308. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. (1989) Altering the genome by homologous recombination. Science 244: 1288–1292. [DOI] [PubMed] [Google Scholar]
- Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D (2014) Genome engineering with targetable nucleases. Annu Rev Biochem 83: 409–439. [DOI] [PubMed] [Google Scholar]
- Carroll D, Beumer KJ, Trautman JK (2010) High-efficiency gene targeting in Drosophila with zinc finger nucleases. Methods Mol Biol 649: 271–280. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R Rondinelli B D’Andrea AD (2016) Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26: 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasegaran S, Carroll D (2016) Origins of programmable nucleases for genome engineering. J Mol Biol 428: 963–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S et al. (2016) Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation in zygotes. J Biol Chem 291: 14457–14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermark T, Doyle EL, Schmidt C, Zhang F et al. (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S et al. (2015) Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. [DOI] [PubMed] [Google Scholar]
- Cristea S, Freyvert Y, Santiago Y, Holmes MC, Urnov FD et al. (2013) In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng 110: 871–880. [DOI] [PubMed] [Google Scholar]
- Danner E, Bashir S, Yumlu S, Wurst W, Wefers B, Kühn R (2017) Control of gene editing by manipulation of DNA repair mechanisms. Mamm Genome 287–8: 262–274. [DOI] [PubMed] [Google Scholar]
- Deng C, Capecchi MR (1992) Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol 12: 3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, Mahfouz M, Wang J et al. (2012) Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1077–1086. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J et al. (2011) Enhancing zinc-finger-nuclease activity with improved obligate heterodimer architectures. Nat Methods 8: 74–79. [DOI] [PubMed] [Google Scholar]
- Dueva D, Iliakis G (2013) Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer. Transl Cancer Res 2: 163–177. [Google Scholar]
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N et al. (2010). Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet 6; e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi A, Pyott DE, Xipnitou A, Molnar A (2017) Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc Nat Acad Sci 114: 13567–13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP (2004) Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR (2004) Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Biotechnol 32: 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM (2002) Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem Mol Biol 32: 1211–1220. [DOI] [PubMed] [Google Scholar]
- He X, Tan C, Wang F, Wang Y, Zhou R, Cui D et al. (2016) Knock-in of large reporter genes in human cells via CRISPR/Cas9 induced homology-dependent and independent DNA repair. Nucleic Acids Res 44: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano Y, Sakuma T, Nakade S, Ohga R, Ota S et al. (2015) Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci Rep 5: 8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T, Morita S, Kimura M, Terawaki N, Shibutani M, Hatada I (2017) Efficient generation of conditional knockout mice via sequential introduction of lox sites. Sci Rep 71: 7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C, Wimmer EA (2000) A versatile vector set for animal transgenesis. Dev Genes Evol 210; 630–637. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, Wilson DM (2013) DNA repair mechanisms in dividing and nondividing cells. DNA Repair 12: 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Haber JE (2016) The democratization of gene editing: insights from site-specific cleavage and double-strand break-repair. DNA Repair 44: 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ et al. (2013) Translating dosage compensation to trisomy 21. Nature 500: 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA et al. (2012) Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-G, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain. Proc Nat Acad Sci 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim S, Kim S et al. (2016) Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-sEq. Genome Res 26: 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT et al. (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza DG, Gaspero A, Lorenzo I, Liao L, Zheng P et al. (2017) Employing single-stranded DNA donors for the high throughput production of conditional knockout alleles in mice. BioRxiv doi: 10.1101/195651. [DOI] [Google Scholar]
- Li L, Wu LP, Chandrasegaran S (1992) Functional domains in FokI restriction endonuclease. Proc Nat Acad Sci 89: 4275–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Potter J, Kumar S, Ravinder N, Chesnut JD (2017) Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J Biotechnol 241: 136–146. [DOI] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA (2014) Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLIFE 3: e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Li H, Hao M, Xiong D, Luo Y et al. (2016) Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci Rep 6: 34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X et al. (2016) Sequence features associated with cleavage efficiency of CRISPR/Cas9 system. Sci Rep 6: 19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo N, Li X, Fraser MJ (1999) Transposition of the piggyBac element in embryos of Drosophila melanogaster, Aedes aegypti and Tricholposia ni. Mol Gen Genet 261: 803–810. [DOI] [PubMed] [Google Scholar]
- Ma X, Chen C, Veevers J, Zhou X, Ross RS., Feng W, Chen J (2017) CRISPR/Cas9-mediated gene manipulation to create single-amino-acid-substituted and floxed mice with a cloning-free method. Sci Rep 7: 42244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak AN, Bradley P, Cernadas RA, Bogdanov AJ, Stoddard BL (2012) The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335: 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M et al. (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotech 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR (1988) Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336: 348–352. [DOI] [PubMed] [Google Scholar]
- Maresca M, Lin V, Guo N, Yang Y (2013) Obligate Ligation-Gated Recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res 23: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR et al. (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM, Zhu QM, Lowenstein CJ. (2016) A CRISPR path to engineering new genetic mouse models for cardiovascular research. Arterioscler Thromb Vase Biol 36: 1058–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee Y-L et al. (2007) An improved zinc-finger nuclease architecture for highly specific genome cleavage. Nat Biotechnol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA., Wang J et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Miura H, Gurumurthy CB, Sato T, Sato M, Ohtsuka M (2015) CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci Rep 5: 12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M (2018) Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat Protoc 13: 195–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Magin S, Soni A, Iliakis G (2016) DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer, cell cycle and proliferation-dependent regulation. Semin Cancer Biol 37-38: 51–64. [DOI] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC et al. (2007) Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Nat Acad Sci 104 3055–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM, Carroll D (2006) Induction and repair of zinc finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Nat Acad Sci 103: 16370–16375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T et al. (2011) A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade S, Tsubota T, Sakane Y, Kume S, Sakamot N et al. (2014) Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nature Comm 5: 5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brochta DA, Atkinson PW (1996) Transposable elements and gene transformation in non-drosophilid insects. Insect Biochem Mol Biol 26: 739–753 [DOI] [PubMed] [Google Scholar]
- Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA et al. (2010) Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res 38: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ (1981) Yeast transformation: a model system for the study of recombination. Proc Nat Acad Sci 78: 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI et al. et al. (2015) Mechanism for the suppression of homologous recombination in G1 cells. Nature 528: 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak MJ, Rasoloson D, Paidemarry S, Green R, Reed RR, Seydoux G (2017a) Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Nat. Acad Sci 114: E10745–E10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Seydoux G (2017b) Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122: 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Schmidt H, Seydoux G (2016) Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res 44: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Wang Y, Smith HE, Lee CY, Calidas D et al. (2014) Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Xiao L, Li ASS, Zhang X, Sirois P et al. (2013) Biological and biomedical applications of engineered nucleases. Mol Biotechnol 55: 54–62. [DOI] [PubMed] [Google Scholar]
- Peng Y, Clark KJ, Campbell JM, Panetta M, Guo Y et al. (2014) Making designer mutants in model organisms. Development 142: 4042–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH., Baltimore D (2003) Chimeric nucleases stimulate gene targeting in human cells. Science 300: 763. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA., Doudna JA, Weissman JS. et al. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T et al. (2017) Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam S, Kandavelou K, Rajenderan R, Chandrasegaran S (2011) Creating designed zinc finger nucleases with minimal cytotoxicity. J Mol Biol 405: 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA., Hsu PD, Lin CY, Gootenberg JS, Konermann S et al. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. Erratum in Cell 155: 479-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD et al. (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nature Biotechnol 34: 339–344. [DOI] [PubMed] [Google Scholar]
- Rodgers K, McVey M (2016) Error-prone repair of DNA double-strand breaks. J Cell Physiol 231: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M (1994) Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Nat Acad Sci 91: 6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC (1982) Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–533. [DOI] [PubMed] [Google Scholar]
- Saito S, Maeda R, Adachi N (2017) Dual loss of human POLQ and LIG4 abolishes random integration. Nat Commun 8: 16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Shota Nakade S, Sakane Y, Suzuki K-IT, Yamamoto T (2016) MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nature Protocols 11: 118–133. [DOI] [PubMed] [Google Scholar]
- Salsman J, Dellaire G (2017) Precision genome editing in the CRISPR era. Biochem Cell Biol 95:. 187–201 [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S, Davis RW (1979) Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Nat Acad Sci 76: 4951–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy JM, Sharp PA (1989) Positive genetic selection for gene disruption in mammalian cells by homologous recombination. Proc Nat Acad Sci 86: 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ., Meckler JF (2013) Genome engineering at the dawn of the golden age. Annu Rev Genomics Hum Genet 14: 135–158. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Symington LS (2015) Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci 40, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E (2015) A mouse geneticist’s practical guide to CRISPR applications. Genetics 199: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX et al. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smih F, Rouet P, Romanienko PJ, Jasin M (1995) Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res 23: 5012–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS (1985) Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317: 230–234. [DOI] [PubMed] [Google Scholar]
- Söllü C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML et al. (2010) Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res 38: 8269–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM (1982) Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Stella S, Montoya G (2016) The genome editing revolution: A CRISPR-Cas TALE off-target story. Bioessays 38: S4–S13. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J et al. (2016) In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ et al. (2007) Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25: 786–793. [DOI] [PubMed] [Google Scholar]
- Thomas KR, Folger KR, Capecchi MR (1986) High frequency targeting of genes to specific sites in the mammalian genome. Cell 44: 419–428. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK (2016) Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet 17: 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V et al. (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature Biotechnol 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV et al. (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33: 187–.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM et al. (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA (2009) Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods 6: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shao Y, Guan Y, Li L, Wu L, Chen F et al. (2015) Large genomic fragment deletion and functional gene cassette knock-in via Cas9 protein mediated genome editing in once-cell rodent embryos. Sci Rep 5: 17517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal DM, Taylor RA, Tzfira T (2013) Nonhomologous end joining-mediated gene replacement in plant cells. Plant Physiol 162: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowska N, Rong YS (2013) Long-range targeted manipulation of the Drosophila genome by site-specific integration and recombinational resolution. Genetics 193: 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H et al. (2015) Sequence determinants of improved sgRNA design. Genome Res 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Bliss J, Gerbi SA (2015) Whole organism genome editing: Targeted large DNA insertion via ObLiGaRe nonhomologous end-joining in vivo capture. G3: Genes/Genomes/Genetics 5: 1843–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H et al. (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Wang X, Hu X, Liu Z, Liu J, Zhou H et al. (2017) Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res 276: 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Wang X, Liu J, Shi L, Huang P, Yang H (2018) CRISPR/Cas9-mediated targeted integration in vivo using a homology-mediated end joining-based strategy. J Vis Exp Mar 12;(133). doi: 10.3791/56844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T (2016) ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 7: 10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M (2017) Inactivation of Pol Θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F (2015) CRISPR-Cas9: Prospects and challenges. Human Gene Therapy 26: 409–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Li XL, Li GH et al. (2017) Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol 18: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]