Abstract
Estrogens are traditionally considered to be female sex steroid hormones and most of the studies examining estrogen regulation of metabolic function in the liver have been conducted in females. However, the liver expresses high levels of estrogen receptor alpha (ESR1) in both males and females, which mediates the hepatic response to estrogens. In this data article, we investigated whether metabolic disorders in Esr1 knockout (Esr1-/-) male rats were linked with loss of transcriptional regulation by ESR1 in liver. To identify the ESR1 regulated genes in the mutant liver, RNA-sequencing was performed on liver RNAs purified from young male rats. The raw data were analyzed using the CLC Genomics Workbench and high-quality RNA-sequencing reads were aligned to the Rattus norvegicus genome. Transcriptome data obtained from Esr1-/- liver RNAs were compared to that of wild type rats. Based on an absolute fold change of 2 with a p-value ≤ 0.05, a total of 618 differentially expressed genes were identified in the Esr1-/- male liver. Pathway analyses demonstrated that the majority of differentially expressed genes are regulators of carbohydrate and lipid metabolism in the liver. These differentially expressed genes and their potential roles were further examined in a companion manuscript, “Disruption of ESR1 alters the expression of genes regulating hepatic lipid and carbohydrate metabolism in male rats” (Khristi et al., 2018).
Specifications table
| Subject area | Biology, Endocrinology |
| More specific subject area | Metabolic regulation in the liver |
| Type of data | RNA-seq data tables and figures |
| How data were acquired | RNA-Sequencing, Ingenuity Pathway Analysis |
| Data format | Normalized, filtered and analyzed data; Bioinformatic prediction |
| Experimental factors | Liver transcriptome profile in Esr1 knockout (Esr1-/-) male rats |
| Experimental features | Liver tissues were collected from 10-week-old wild type and Esr1-/- male rats. Total RNA was isolated, and cDNA-libraries were prepared for RNA-sequencing. RNA-seq raw data reads were analyzed using CLC Genomics Workbench. Differentially expressed genes were further analyzed for their involvement in carbohydrate and lipid metabolism by IPA. |
| Data source location | A basic science laboratory at the University of Kansas Medical Center, Kansas City, KS, USA. |
| Data accessibility | Raw data have not yet been submitted to any public repository. |
| Related research article | V. Khristi, A. Ratri, S. Ghosh, S. Borosha, E. Dai, R. Roy, et al., Disruption of ESR1 alters the expression of genes regulating hepatic lipid and carbohydrate metabolism in male rats, Endocrinology (2018), Under review[1]. |
Value of the data
-
•
This data article provides liver transcriptomic analyses of Esr1-/- male rats.
-
•
Pathway analyses of the differentially expressed genes in the Esr1-/- liver show their involvement in carbohydrate and lipid metabolism.
-
•
Differentially expressed genes are also linked to development of obesity, hepatic steatosis, and other liver diseases.
1. Data
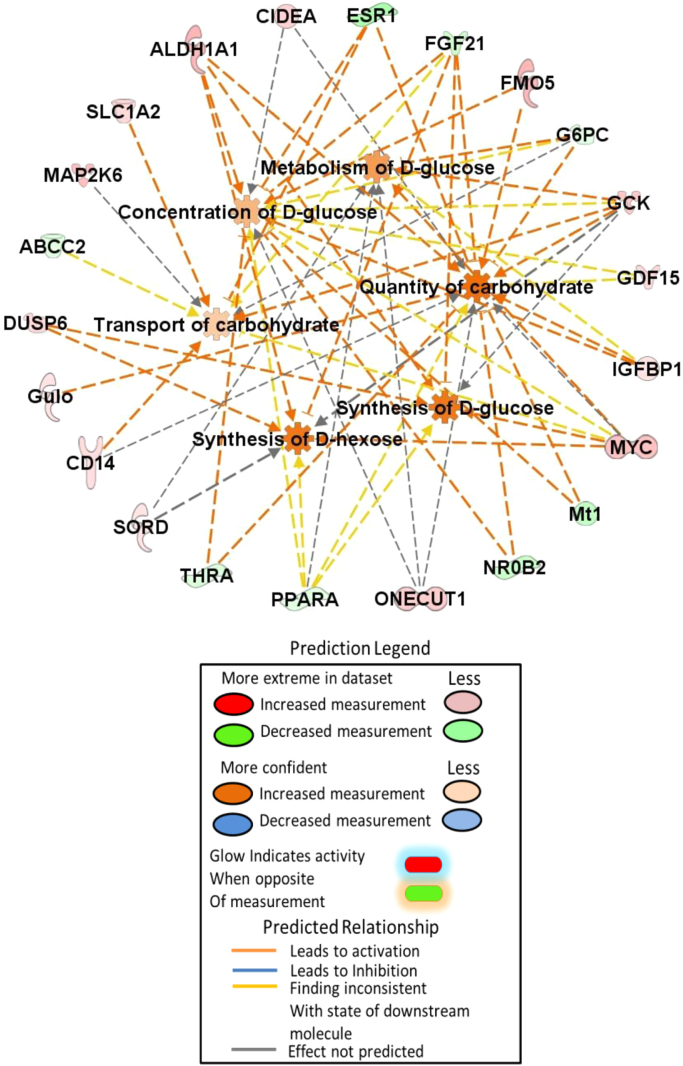
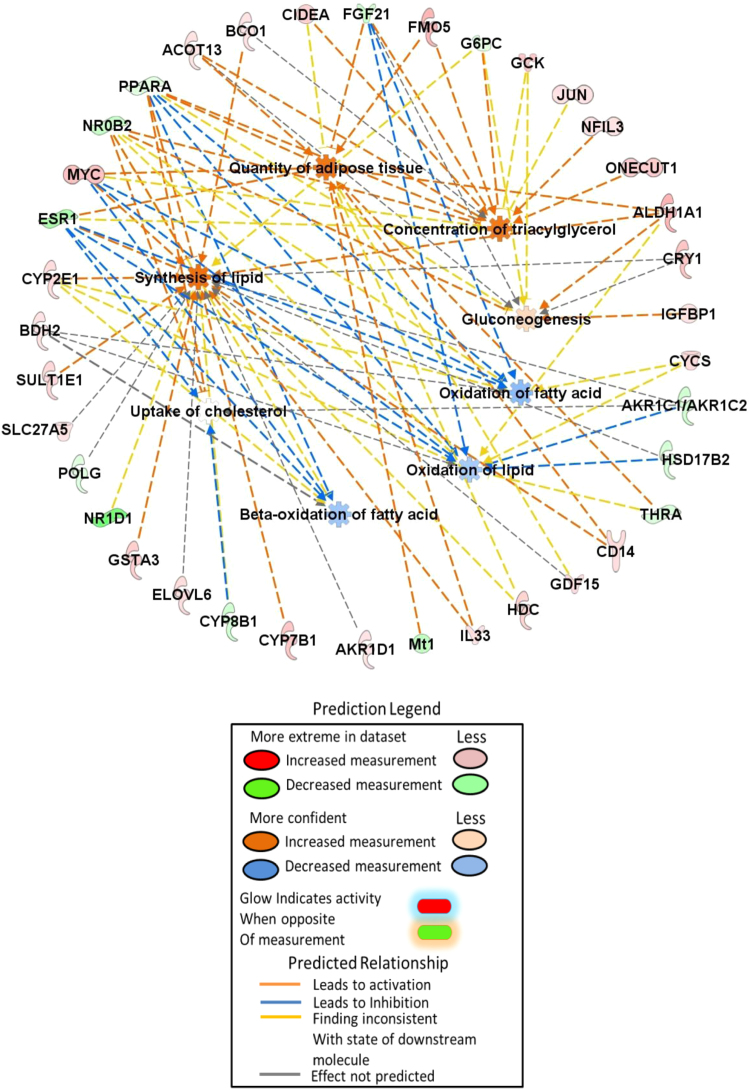
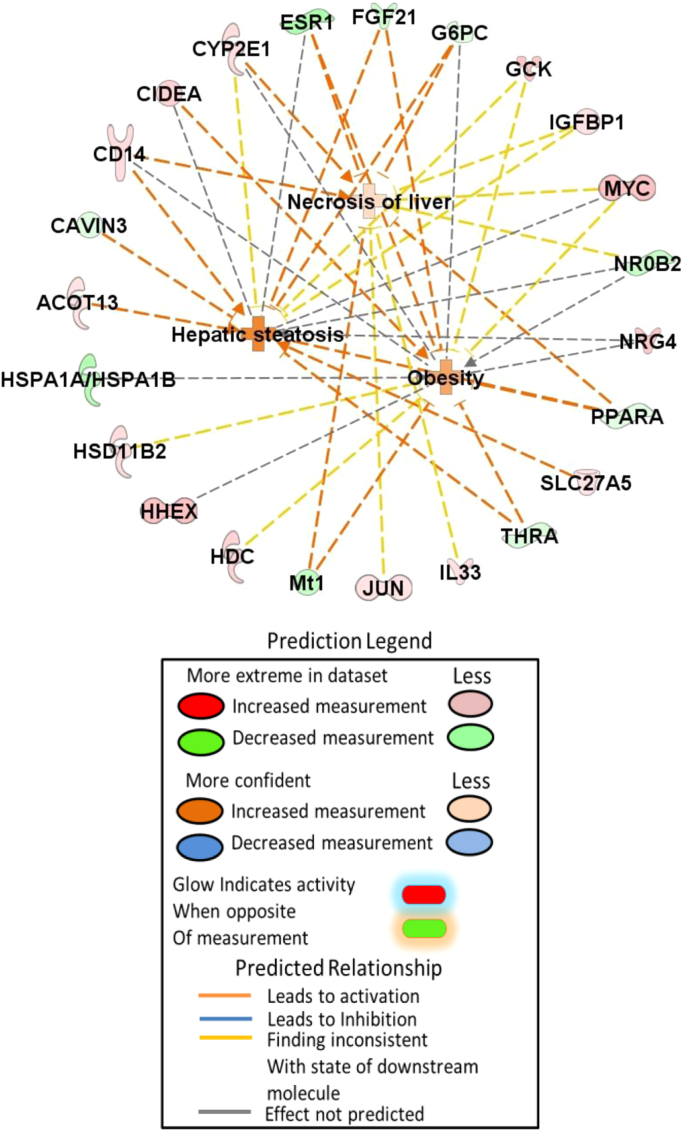
In this data article, we present analyzed RNA-seq data showing the differentially expressed genes in the Esr1-/- male liver (Table S1). Bioinformatic analyses show that these differentially expressed genes are linked to pathways of carbohydrate metabolism (Table 1, Fig. 1), lipid metabolism (Table 2, Fig. 2) and hepatic diseases including hepatic steatosis, necrosis of the liver, and obesity (Fig. 3).
Table 1.
List of pathways involved in ‘Carbohydrate metabolism’.
| Diseases or functions annotation | p-value | Predicted activation state | Activation z-score | Molecules | # Molecules |
|---|---|---|---|---|---|
| Gluconeogenesis | 0.0000000 | 0.343 | ACOT13, ALDH1A1, CRY1, FGF21, G6PC, GCK, IGFBP1, NR0B2, PPARA | 9 | |
| Concentration of D-glucose | 0.0000001 | 0.69 | ALDH1A1, CIDEA, ESR1, FGF21, FMO5, G6PC, GCK, GDF15, IGFBP1, Mt1, MYC, NR0B2, ONECUT1, PPARA, THRA | 15 | |
| Metabolism of D-glucose | 0.0000001 | 1 | G6PC, GCK, IGFBP1, MYC, ONECUT1, PPARA, SORD | 7 | |
| Synthesis of D-hexose | 0.0000023 | 1.735 | ALDH1A1, DUSP6, FGF21, GCK, MYC, PPARA, SORD | 7 | |
| Quantity of carbohydrate | 0.0000024 | Increased | 2.257 | ALDH1A1, CD14, CIDEA, ESR1, FGF21, FMO5, G6PC, GCK, GDF15, Gulo, IGFBP1, Mt1, MYC, NR0B2, ONECUT1, PPARA, THRA | 17 |
| Homeostasis of D-glucose | 0.0000017 | ACOT13, CIDEA, CRY1, FGF21, FMO5, G6PC, GCK, PPARA, SLC27A5, THRA | 10 | ||
| Transport of carbohydrate | 0.000010 | 0.478 | ABCC2, CD14, ESR1, FGF21, G6PC, GCK, MAP2K6, MYC, SLC1A2 | 9 | |
| Synthesis of D-glucose | 0.0000023 | 1.735 | ALDH1A1, DUSP6, FGF21, GCK, MYC, PPARA | 6 | |
| Quantity of glucose-6-phosphate | 0.000032 | G6PC, GCK, MYC | 3 | ||
| Regulation of D-glucose | 0.0000024 | FGF21, MYC, PPARA | 3 | ||
| Utilization of D-glucose | 0.000075 | GCK, MYC, PPARA, THRA | 4 | ||
| Transport of monosaccharide | 0.000119 | 0.555 | ESR1, FGF21, G6PC, GCK, MAP2K6, MYC, SLC1A2 | 7 | |
| Phosphorylation of D-glucose | 0.00021 | G6PC, GCK | 2 | ||
| Metabolism of glucose-6-phosphate | 0.0000057 | G6PC, GCK | 2 | ||
| Transport of D-glucose | 0.00044 | 0.555 | ESR1, FGF21, GCK, MAP2K6, MYC, SLC1A2 | 6 | |
| Quantity of glycogen | 0.0000100 | 1.172 | FGF21, G6PC, GCK, MYC, PPARA | 5 | |
| Synthesis of carbohydrate | 0.000583 | 1.809 | ALDH1A1, DUSP6, FGF2, G6PC, GCK, Gulo, IGFBP1, MYC, NR1D1, PPARA, SORD | 11 | |
| Gluconeogenesis of hepatocytes | 0.0000268 | ALDH1A1, NR0B2 | 2 | ||
| Import of D-glucose | 0.0014 | 0 | ESR1, FGF21, MYC, SLC1A2 | 4 | |
| Metabolism of carbohydrate | 0.0000320 | 1.483 | ALDH1A1, CYP2E1, DUSP6, FGF21, G6PC, GCK, Gulo, IGFBP1, MYC, NR1D1, ONECUT1, PPARA, SORD | 13 | |
| Uptake of carbohydrate | 0.00205 | 0.306 | ABCC2, CD14, FGF21, G6PC, GCK, MYC, PPARA, SLC1A2 | 8 | |
| Synthesis of glycogen | 0.0000560 | G6PC, GCK, IGFBP1, NR1D1 | 4 | ||
| Quantity of lactic acid | 0.00214 | GCK, MYC, PPARA | 3 | ||
| Production of lactic acid | 0.0000750 | GCK, MYC, PPARA | 3 |
Fig. 1.
Mechanistic diagram of selected pathways involved in the carbohydrates metabolism.
Table 2.
List of pathways involved in ‘Lipid metabolism’.
| Diseases or functions annotation | p-value | Predicted activation state | Activation z-score | Molecules | # Molecules |
|---|---|---|---|---|---|
| Synthesis of terpenoid | 0.0000000 | 1.169 | AKR1C1/AKR1C2, AKR1D1, ALDH1A1, BCO1, CRY1, CYP7B1, CYP8B1, ESR1, G6PC, GDF15, GSTA3, HDC, HSD17B2, NR1D1, POLG, PPARA, SLC27A5, SULT1E1 | 18 | |
| Metabolism of terpenoid | 0.0000000 | 0.342 | ADH7, AKR1C1/AKR1C2, AKR1D1, ALDH1A1, BCO1, CYP2E1, CYP7B1, CYP8B1, ESR1, G6PC, GSTA3, HDC, HSD11B2, HSD17B2, NR0B2, SLC27A5, SULT1E1, UGT2B11 | 18 | |
| Quantity of steroid | 0.0000000 | 0.547 | ABCC2, ACOT13, BCO1, CRY1, CYP8B1, ESR1, FGF21, FMO5, G6PC, GCK, Gulo, HSD11B2, IL33, JUN, NFIL3, NR0B2, POLG, PPARA, SLC1A2, SULT1E1, THRA, ZBTB16 | 22 | |
| Concentration of lipid | 0.0000000 | 1.111 | ABCC2, ACOT13, ALDH1A1, BCO1, CD14, CIDEA, CRY1, CYP2E1, CYP8B1, EFNA5, ESR1, FGF21, FMO5, G6PC, GCK, Gulo, HSD11B2, IL33, JUN, MYC, NFIL3, NR0B2, ONECUT1, POLG, PPARA, SLC1A2, SULT1E1, THRA, ZBTB16 | 29 | |
| Synthesis of steroid | 0.0000000 | 0.601 | AKR1C1/AKR1C2, AKR1D1, CRY1, CYP7B1, CYP8B1, ESR1, G6PC, GDF15, GSTA3, HDC, HSD17B2, NR1D1, POLG, PPARA, SLC27A5, SULT1E1 | 16 | |
| Steroid metabolism | 0.0000000 | −0.594 | AKR1C1/AKR1C2, AKR1D1, CYP2E1, CYP7B1, CYP8B1, ESR1, G6PC, GSTA3, HDC, HSD11B2, HSD17B2, NR0B2, SLC27A5, SULT1E1, UGT2B11 | 15 | |
| Concentration of cholesterol | 0.0000000 | 1.352 | ACOT13, BCO1, CYP8B1, ESR1, FGF21, FMO5, G6PC, GCK, Gulo, IL33, JUN, NFIL3, NR0B2, PPARA, SLC1A2, THRA | 16 | |
| Synthesis of lipid | 0.0000000 | Increased | 2.092 | AKR1C1/AKR1C2, AKR1D1, ALDH1A1, BCO1, CD14, CRY1, CYP2E1, CYP7B1, CYP8B1, ELOVL6, ESR1, G6PC, GDF15, GSTA3, HDC, HSD17B2, IL33, MYC, NR0B2, NR1D1, POLG, PPARA, SLC27A5, SULT1E1 | 24 |
| Homeostasis of lipid | 0.0000000 | AKR1C1/AKR1C2, CIDEA, CYP7B1, CYP8B1, FGF21, G6PC, GCK, Mt1, NR0B2, NR1D1, NR1D2, PPARA | 12 | ||
| Oxidation of lipid | 0.0000000 | −0.47 | AKR1C1/AKR1C2, ALDH1A1, BDH2, CYCS, CYP2E1, ESR1, FGF21, HSD17B2, MYC, NR0B2, PPARA, THRA | 12 | |
| Synthesis of bile acid | 0.0000000 | −0.132 | AKR1D1, CYP7B1, CYP8B1, NR1D1, PPARA, SLC27A5 | 6 | |
| Concentration of acyl glycerol | 0.00000 | 1.388 | ACOT13, BCO1, CIDEA, ESR1, FGF21, FMO5, G6PC, GCK, JUN, MYC, NFIL3, NR0B2, ONECUT1, PPARA, THRA | 15 | |
| Concentration of triacylglycerol | 0.00000 | 1.651 | ACOT13, BCO1, CIDEA, ESR1, FGF21, FMO5, G6PC, GCK, JUN, MYC, NFIL3, NR0B2, ONECUT1, PPARA | 14 | |
| Uptake of cholesterol | 0.0000158 | 0 | AKR1C1/AKR1C2, CYP8B1, ESR1, NR0B2, PPARA | 5 | |
| Concentration of bile acid | 0.000019 | ABCC2, CYP8B1, ESR1, NR0B2 | 4 | ||
| Quantity of ketone body | 0.0000334 | ACOT13, FGF21, GCK, PPARA | 4 | ||
| Inactivation of glucocorticoid | 0.0000352 | AKR1D1, HSD11B2 | 2 | ||
| Inactivation of lipid | 0.0000566 | AKR1D1, HSD11B2, SULT1E1 | 3 | ||
| Secretion of lipid | 0.0000772 | 0.927 | ABCC2, CIDEA, ESR1, HSD17B2, MAP2K6, NFIL3, PPARA, SULT1E1 | 8 | |
| Absorption of cholesterol | 0.0000842 | AKR1C1/AKR1C2, CYP8B1, NR0B2, PPARA | 4 | ||
| Uptake of lipid | 0.000206 | −0.7 | AKR1C1/AKR1C2, CD14, CYP8B1, ESR1, NR0B2, PPARA, SLC27A5 | 7 | |
| Production of ketone body | 0.000348 | FGF21, GCK | 2 | ||
| Metabolism of retinoid | 0.000453 | ADH7, ALDH1A1, BCO1, CYP2E1 | 4 | ||
| Concentration of fatty acid | 0.000463 | 1.773 | ACOT13, CIDEA, CYP2E1, EFNA5, FGF21, G6PC, GCK, MYC, PPARA | 9 | |
| Synthesis of ketone body | 0.00052 | PPARA, SLC27A5 | 2 | ||
| Metabolism of triacylglycerol | 0.000537 | ALDH1A1, CYP2E1, G6PC, NR0B2, PPARA | 5 | ||
| Abnormal quantity of lipid | 0.00055 | ACOT13, CYP8B1, ESR1, FGF21, GCK, NR0B2 | 6 | ||
| Hydroxylation of lipid | 0.000607 | CYP2E1, CYP4A22, CYP7B1 | 3 | ||
| Conversion of lipid | 0.000622 | 0.905 | AKR1C1/AKR1C2, CYP2E1, HSD11B2, HSD17B2, Mt1, PPARA | 6 | |
| Metabolism of sterol | 0.000856 | AKR1D1, CYP7B1, CYP8B1, HSD17B2, NR0B2 | 5 | ||
| Regulation of lipid | 0.00138 | FGF21, HSD11B2, PPARA | 3 | ||
| Modification of long-chain acyl-coenzyme A | 0.00154 | ELOVL6, PPARA | 2 | ||
| Activation of lipid | 0.00161 | FGF21, HSD11B2, SLC27A5 | 3 | ||
| Synthesis of sterol | 0.00183 | 1 | CYP7B1, CYP8B1, HDC, PPARA | 4 | |
| Hydroxylation of fatty acid | 0.00187 | CYP2E1, CYP4A22 | 2 | ||
| Transport of lipid | 0.00209 | 0.386 | ABCC2, AKR1C1/AKR1C2, CD14, GDF15, IL33, PPARA, SLC27A5 | 7 | |
| Fatty acid metabolism | 0.00277 | 0.767 | ABCC2, AKR1C1/AKR1C2, CD14, CYP2E1, CYP4A22, DBP, ELOVL6, GDF15, IL33, MYC, PPARA, SLC27A5 | 12 | |
| Abnormal quantity of bile salt | 0.00306 | CYP8B1, NR0B2 | 2 | ||
| Uptake of bile salt | 0.00306 | NR0B2, PPARA | 2 | ||
| Metabolism of bile acid | 0.00352 | AKR1C1/AKR1C2, SLC27A5 | 2 | ||
| Regulation of steroid | 0.00352 | HSD11B2, PPARA | 2 | ||
| Homeostasis of bile salt | 0.00452 | CYP8B1, NR0B2 | 2 | ||
| Homeostasis of cholesterol | 0.005 | AKR1C1/AKR1C2, CYP7B1, G6PC, NR1D1 | 4 | ||
| Conjugation of lipid | 0.00506 | SLC27A5, UGT2B11 | 2 | ||
| Transport of steroid | 0.00529 | −0.106 | ABCC2, AKR1C1/AKR1C2, GDF15, IL33, PPARA | 5 | |
| Metabolism of cholesterol | 0.00531 | AKR1D1, CYP7B1, CYP8B1, NR0B2 | 4 |
Fig. 2.
Mechanistic diagram of selected pathways involved in the synthesis and oxidation of lipids.
Fig. 3.
Mechanistic diagram of genes involved in hepatic steatosis, necrosis of the liver, and obesity.
2. Experimental design, materials, and methods
2.1. Esr1 knockout rats
The Holtzman Sprague-Dawley (HSD) Esr1-mutant rat model was generated by targeted deletion of exon 3 in the Esr1 gene [2]. Deletion of exon 3 caused a frameshift and null mutation in the ESR1 coding sequence [2]. All animals were screened for the presence of the mutation by PCR using tail-tip DNA samples (REDExtract-N-Amp Tissue PCR Kit, Sigma-Aldrich) and primers targeting the flanking intron sequences [2]. All procedures were performed in accordance with the protocols approved by the University of Kansas Medical Center Animal Care and Use Committee.
2.2. Sample collection from wild type and Esr1-/- rats
Liver tissues were collected from 10 to 12-week-old Esr1-/- and age matched wild type male rats. The tissue samples were collected immediately after euthanization, cut into small species, snap frozen in liquid nitrogen, and stored at −80 °C until they were processed for RNA extraction. Total RNA from liver tissues was extracted using TRI Reagent (Millipore-Sigma) following the manufacturer׳s instructions. RNA quality was assessed using the Agilent Bioanalyzer and samples with a RIN score ≥ 9 were included in the RNA-seq library preparation.
2.3. Library preparation and RNA-sequencing
The library preparation and sequencing of RNA was performed at the Genome Sequencing facility of the University of Kansas Medical Center. Five hundred nanogram of liver total RNA was used for the RNA-seq library preparation. Libraries were prepared using a TruSeq Stranded mRNA kit (Illumina) following the manufacturer׳s instructions. Briefly, mRNA was enriched from total RNA by oligo-dT magnetic beads, purified, and chemically fragmented. The first strand of cDNA was synthesized using random hexamer primers and reverse transcriptase. Then, double stranded (ds) cDNA was generated by removing the RNA template and synthesizing a replacement strand, incorporating dUTP in place of dTTP. ds cDNA was purified from the second strand reaction mix by AMPure XP beads (Beckman Coulter). The cDNA ends were blunted and poly (A) tails were added to the 3’ ends. Finally, after ligation of indexing adaptors (Illumina), the suitable DNA fragments were selected for PCR amplification for 15 cycles. Three replicate cDNA libraries were prepared for each of the wild type and Esr1-/- groups and sequenced on an Illumina HiSeq. 2500 platform.
2.4. Analyses of the RNA-sequencing data
RNA-seq data were analyzed using CLC Genomics Workbench (Qiagen Bioinformatics). Raw reads of RNA-seq were analyzed as described in a previous publication [3]. Clean reads were obtained by removing low quality reads through trimming. High quality reads of liver RNA-seq were aligned to the Rattus norvegicus genome (Rn6, downloaded from NCBI database). RNA-seq data were mapped with the following parameters: (a) maximum number of allowed mismatches was set at 2; (b) minimum length and similarity fraction were set at 0.8; and (c) minimum number of hits per read was set at 10. Gene expression values were reported as RPKM (Reads Per Kilobase of transcript per Million mapped reads) [4]. In this study, gene expression values showing an absolute fold change of 2 with p-value ≤ 0.05 were considered differentially expressed. A total of 618 genes were differentially expressed in Esr1-/- liver, 410 downregulated and 208 upregulated (Table S1).
2.5. Pathway analysis of differentially expressed genes in Esr1-/- liver
Differentially expressed genes in the Esr1-/- liver were subjected to Ingenuity Pathway Analysis (IPA; Qiagen Bioinformatics). The pathways and genes involved in carbohydrate and lipid metabolism are listed in Tables 1 and 2. The selected pathways from carbohydrate metabolism shown elevated glucose level in Esr1-/- rats shown in Fig. 1. In lipid metabolism, synthesis of lipid and concentration of triglyceride were increased in Esr1-/- rats. In contrast, oxidation of lipid and fatty acid was lower in Esr1-/- rats (Fig. 2). The genes involved in hepatic steatosis and obesity are shown in Fig. 3.
3. Statistical analysis
RNA-sequencing included three cDNA libraries in each group. Each library was prepared from pooled total RNA of two individual rats. Differentially expressed genes were identified by using CLC Genomics Workbench as described in a previous publication [5].
Acknowledgments
This project was partially supported by funding from the KUMC-KIDDRC Genomics Core and an NIH grant (HD072100). We thank the University of Kansas Medical Center– Genomics Core for generating the sequence data sets. The Genomics Core is supported by the School of Medicine, University of Kansas, the Kansas Intellectual and Developmental Disability Research Center (NIH U54 HD090216) and the Molecular Regulation of Cell Development and Differentiation - COBRE (5P20GM104936-10).
Footnotes
Transparency document associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.12.089.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.12.089.
Contributor Information
Vincentaben Khristi, Email: vkhristi@kumc.edu.
Anamika Ratri, Email: aratri@kumc.edu.
Subhra Ghosh, Email: sghosh3@kumc.edu.
Shaon Borosha, Email: shaon.borosha@vanderbilt.edu.
Eddie Dai, Email: eddiedai02@gmail.com.
V. Praveen Chakravarthi, Email: praghavulu@kumc.edu.
M.A. Karim Rumi, Email: mrumi@kumc.edu.
Michael W. Wolfe, Email: mwolfe2@kumc.edu.
Transparency document. Supplementary material
Supplementary material
.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Khristi V., Ratri A., Ghosh S., Borosha S., Dai E., Roy R. Disruption of ESR1 alters the expression of genes regulating hepatic lipid and carbohydrate metabolism in male rats. Endocrinology. 2018 doi: 10.1016/j.mce.2019.04.005. (Under review) [DOI] [PubMed] [Google Scholar]
- 2.Rumi M.A., Dhakal P., Kubota K., Chakraborty D., Lei T., Larson M.A. Generation of Esr1-knockout rats using zinc finger nuclease-mediated genome editing. Endocrinology. 2014;155:1991–1999. doi: 10.1210/en.2013-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khristi V., Chakravarthi V.P., Singh P., Ghosh S., Pramanik A., Ratri A. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell Endocrinol. 2018;474:214–226. doi: 10.1016/j.mce.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 5.Khristi V., Chakravarthi V.P., Singh P., Ghosh S., Pramanik A., Ratri A. Differentially regulated genes in Esr2-mutant rat granulosa cells. Data Brief. 2018;19:1008–1011. doi: 10.1016/j.dib.2018.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material