Summary
Th17 cells contribute to the pathogenesis of inflammatory and autoimmune diseases and cancer. To reveal the Th17 cell-specific proteomic signature regulating Th17 cell differentiation and function in humans, we used a label-free mass spectrometry-based approach. Furthermore, a comprehensive analysis of the proteome and transcriptome of cells during human Th17 differentiation revealed a high degree of overlap between the datasets. However, when compared with corresponding published mouse data, we found very limited overlap between the proteins differentially regulated in response to Th17 differentiation. Validations were made for a panel of selected proteins with known and unknown functions. Finally, using RNA interference, we showed that SATB1 negatively regulates human Th17 cell differentiation. Overall, the current study illustrates a comprehensive picture of the global protein landscape during early human Th17 cell differentiation. Poor overlap with mouse data underlines the importance of human studies for translational research.
Subject Areas: Immunology, Components of the Immune System, Omics, Proteomics
Graphical Abstract
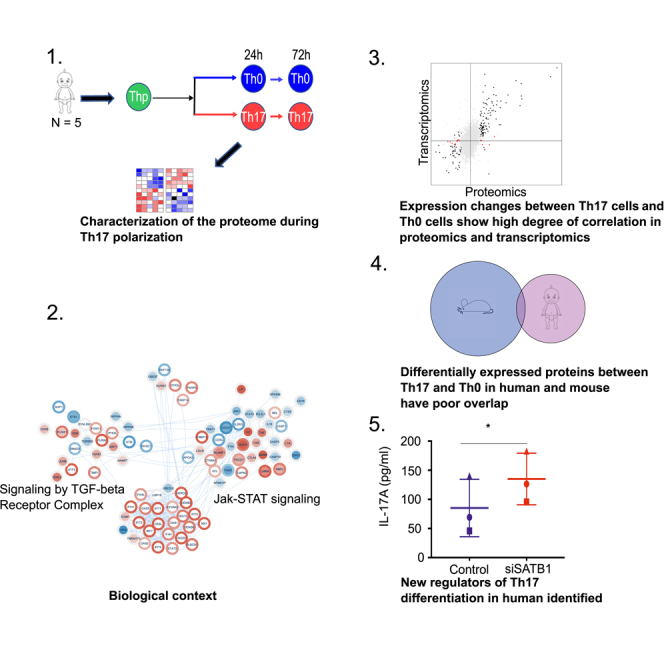
Highlights
-
•
Quantitative proteomics analysis of early human Th17 cell polarization
-
•
The proteome and transcriptome highly correlate during early Th17 polarization
-
•
Poor overlap of proteome profiles of human and mouse during early Th17 polarization
-
•
The results underline the importance of human studies for translational research
Immunology; Components of the Immune System; Omics; Proteomics
Introduction
CD4+ T cells are key players of the adaptive immune system. Upon antigenic stimulation, the naive CD4+ T (T helper precursor [Thp]) cells polarize into distinct T helper ([Th], i.e., Th1, Th2, Th9, and Th17) and regulatory T (Treg) cells (O'Shea and Paul, 2010). Although their primary function is to provide protective immunity against various intracellular and extracellular pathogens, they can also exhibit inappropriate responses in inflammatory and autoimmune diseases (McKinstry et al., 2010, Ghoreschi et al., 2011, Cosmi et al., 2014). Th17 cells are a subset of Th cells that are characterized by the expression of their key transcription factors, STAT3 and RORC and the chemokine receptor CCR6 as well as by the secretion of signature cytokines interleukin (IL)-17A and IL-17F. They are critical in combating fungal infections and contribute to the pathogenesis of several inflammatory and autoimmune diseases and various cancers (Hernández-Santos and Gaffen, 2012, Bailey et al., 2014, Burkett et al., 2015). The characterization of the molecular mechanisms regulating the differentiation and function of the Th17 cells is therefore of great interest for research into the etiology and treatment of these diseases.
Until now, our understanding of the proteins that are most important in Th17 differentiation and function in both human and mouse has originated from transcriptional profiling analyses (Ciofani et al., 2012, Tuomela et al., 2012, Yosef et al., 2013, Gaublomme et al., 2015). These analyses have identified a number of genes that now serve as key markers for Th17 cells. However, this approach is limited as some transcriptional changes are not necessarily reflected at the proteome level (Vogel and Marcotte, 2012). For example, several post-transcriptional and post-translational mechanisms modulate the stability and activity of many of the proteins involved (Liu et al., 2016). Characterization of the cellular proteome of Th17 cells enables the discovery of unique protein signatures and regulated cellular pathways that drive Th17 cell development and function. A better understanding of the proteome during the differentiation process facilitates the rational design of drugs targeting Th17-mediated inflammatory and autoimmune diseases.
Mass-spectrometry (MS)-based proteomic analysis is a powerful tool for comprehensively profiling the proteome in different cellular systems, including T cells (Cox and Mann, 2011, Howden et al., 2013). Previously, MS-based proteomic analysis of differentiating T cells focused mainly on classical Th1 and Th2 cells using in vitro differentiation systems (Loyet et al., 2005, Rautajoki et al., 2007). In addition, addressing disease-related traits, the proteomic profiles were compared for in vivo differentiated Th1 and Th1/Th17 cell clones isolated from biopsies of gut samples from patients with Crohn disease (Riaz et al., 2016). Recently, a number of studies identified a distinct set of differentially regulated proteins when comparing the proteomes of CD4+CD25+ Foxp3 expressing natural Treg cells and induced Treg (iTreg) with CD4+ conventional T cells both in human and mouse (Kubach et al., 2007, Duguet et al., 2017, Cuadrado et al., 2018, Schmidt et al., 2018). Most recently, a study reported Th17 proteome profiles in mouse (Mohammad et al., 2018). Although studies of the molecular profiles and mechanisms governing different Th and Treg cell differentiation have been mostly performed in mouse, previous reports that have compared the transcriptomic profiles of human and mouse have revealed significant differences between the two species (Schwanhüusser et al., 2011, Vogel and Marcotte, 2012). As the findings from studies based on mouse disease models often cannot be replicated in humans, studies in humans are critical (Mestas and Hughes, 2004, Mak et al., 2014).
In the current study, we utilized a label-free MS-based approach to build a quantitative dataset on the cellular proteome of naive CD4+ human T cells, CD3/CD28 activated T (Th0) cells, and Th17 cells at 24 and 72 hr after the initiation of polarization. Statistical analysis of the data revealed a Th17-cell-specific proteome signature with a number of proteins regulated during Th17 cell differentiation already at the early stage of the differentiation process. Moreover, selected proteins with previously known and unknown Th17-related functions were validated in additional samples by distinct methods to confirm the results. Furthermore, the proteomics and transcriptomics data generated in this study were compared to determine the degree of concordance between these two. Notably, a comparison of our human Th17-regulated proteome with the mouse Th17 proteome demonstrated poor overlap between the two species. Last, using the RNA interference (RNAi) approach, we demonstrated SATB1 as a negative regulator of human Th17 cell differentiation process in contrast to mouse, where it positively regulates Th17 cell differentiation (Ciofani et al., 2012). This study illustrates the global protein landscape and the mRNA-protein associations during early human Th17 cell differentiation. This dataset provides a valuable resource of candidate proteins potentially regulating the differentiation and functions of Th17 cells in humans. Further investigation on these candidate proteins might lead to the rational design of targets with therapeutic potential for modulating Th17-mediated immune responses in humans.
Results
Quantitative Proteomic Analysis during Initiation of Human Th17 Differentiation
We investigated the quantitative changes in the cellular proteome of human naive Th cells during the early stages of human Th17 cell differentiation using shotgun label-free quantitative (LFQ) proteomics. Naive CD4+ T cells isolated from the human umbilical cord blood were either activated by T cell receptor (TCR) cross-linking with CD3 and CD28 antibodies (Th0 cells) or polarized with a cytokine cocktail of IL-1β, TGF-β, and IL-6 in combination with TCR/CD28 cross-linking to initiate Th17 cell differentiation. Polarization was monitored by the expression of Th17 cell marker genes, including CCR6, IL-17A, and IL-17F, and the master transcription factor RORC (Figures S1A–S1D). The Thp cell sample and the Th0 and Th17 cell samples at 24- and 72-hr time points post-initiation of polarization were collected from five individual donors. To rule out the possibility of polarization toward interferon (IFN)-γ-expressing pathogenic Th17 cells, expression of the IFN-γ cytokine was monitored in three separate Th17 cultures prepared in a similar manner (Figures S1E and S1F). The samples were prepared using the filter-aided sample preparation protocol and analyzed in triplicate by liquid chromatography-tandem MS (LC-MS/MS) (Figure S1G).
Using the label-free MS approach, we identified more than 5,600 unique proteins among the Thp, Th0, and Th17 cell subsets (Table S1). Samples in the normalized data clustered according to the biological replicates and cell lineages indicating successful normalization, good reproducibility, and general good quality of the data (Figure S2). The proportion of missing values in the samples was low in general (<7%), being largest in the Thp cells (∼4.4%–6.1%).
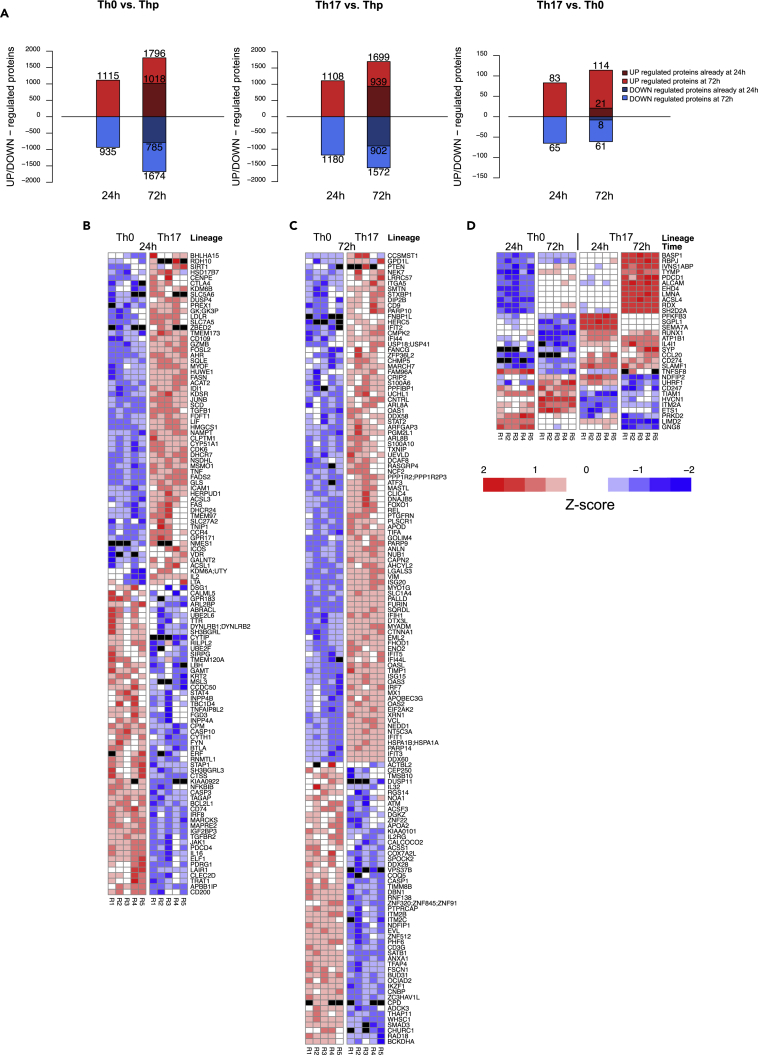
Furthermore, a considerable number of proteins were detected as differentially expressed (DE) between the different cell subsets. On comparing Th0 and Thp cells, 2,050 and 3,470 proteins were detected as DE (false discovery rate [FDR] ≤ 0.05) at 24 and 72 hr after the onset of the polarization, respectively (Figure 1A; Tables S1 and S2). Similarly, on comparing Th17 and Thp cells, 2,288 proteins were DE at 24 hr and 3,271 at 72 hr, respectively. Among the proteins DE in response to TCR activation (Th0 versus Thp), 935 and 1,674 proteins were downregulated and 1,115 and 1,796 proteins were upregulated in Th0 at 24 and 72 hr, respectively. Likewise, in response to combined TCR activation and Th17-polarizing cytokines (Th17 versus Thp), 1,180 and 1,572 proteins were downregulated and 1,108 and 1,699 proteins were upregulated in Th17 cells at 24 and 72 hr, respectively. As expected, owing to the similarity of the cells at the early stage of Th17 polarization, the differential expression analysis between Th17 and Th0 cells revealed only 148 and 175 proteins differentially regulated at 24 and 72 hr, respectively (Figure 1A; Tables S1 and S2). Among the proteins that were DE in the Th17-polarizing conditions in a time-point-specific fashion (i.e., detected as DE only at either time points), 60 and 92 proteins were upregulated and 56 and 51 proteins were downregulated at 24 and 72 hr, respectively (Figures 1B and 1C). Interestingly, only a small number of DE proteins were found to be common at both time points (Figure 1D), suggesting that the differentially regulated proteins have a stage-specific function in driving Th17 differentiation.
Figure 1.
The Differentially Expressed (DE) Proteins in the Proteomics Data
(A–D) (A) The number of up- and downregulated proteins between Th0 and Thp cells, Th17 and Thp cells, and Th17 and Th0 cells at 24 and 72 hr. At 72 hr, the number of proteins detected up- or downregulated already at 24 hr is shown in darker color. The Z score standardized expressions of the DE proteins between Th17 and Th0 unique to 24 hr (B), unique to 72 hr (C), and common to both time points (24 and 72 hr) (D). Black color in the heatmaps stands for undetected/missing value.
Among the proteins detected as DE in this data, several are previously known to be preferentially expressed and regulated during Th17 differentiation. These included AHR (Veldhoen et al., 2008), JUNB (Carr et al., 2017), FOSL2 (Ciofani et al., 2012), RBPJ (Meyer zu Horste et al., 2016), SIRT1 (Lim et al., 2015), REL (Chen et al., 2011), CCL20 (Eriksson et al., 2015), and TNFSF8 (Sun et al., 2010). Notably, our analysis also identified several other proteins (e.g., UHRF1, GK, ICAM1, KDSR, ATF3, IL4IT, APOD, VIM, PALLD, PTGFRN, and KDM6A) differentially upregulated with unexplored/unknown functions in Th17 cells (Figures 1B and 1C). Interestingly, proteins related to antiviral pathways (e.g., IRF7, MX1, OAS, OAS31, and OASL) were upregulated, suggesting the role of IFN signaling in driving Th17 differentiation. We observed the upregulation of the ADP ribosyl transferase poly (ADP-ribose) polymerase (PARP) superfamily proteins, including PARP-9, PARP-10, and PAPR-14, together with the RNA helicase DEAD-box proteins, such as DDX58 and DDX60. PARP-14 is highly expressed and positively regulated during the development of Th17 and T helper follicular cells in mouse by modulating the phosphorylation of STAT3, a key transcriptional regulator of both (Mehrotra et al., 2015). Several negative regulators of Th17 cell development (Ciofani et al., 2012, Yosef et al., 2013), including IRF8, ETS1, STAT4, FYN, CASP1, and SMAD3, were significantly downregulated in differentiating Th17 cells. Interestingly, among the downregulated proteins, we also observed several proteins with unexplored function in Th17 biology including ELF1, IL16, BTLA, LAIR1, UHRF1, ANXA1, NDFIP1, and NDFIP2.
Functional Analysis of the Th17 DE Proteins
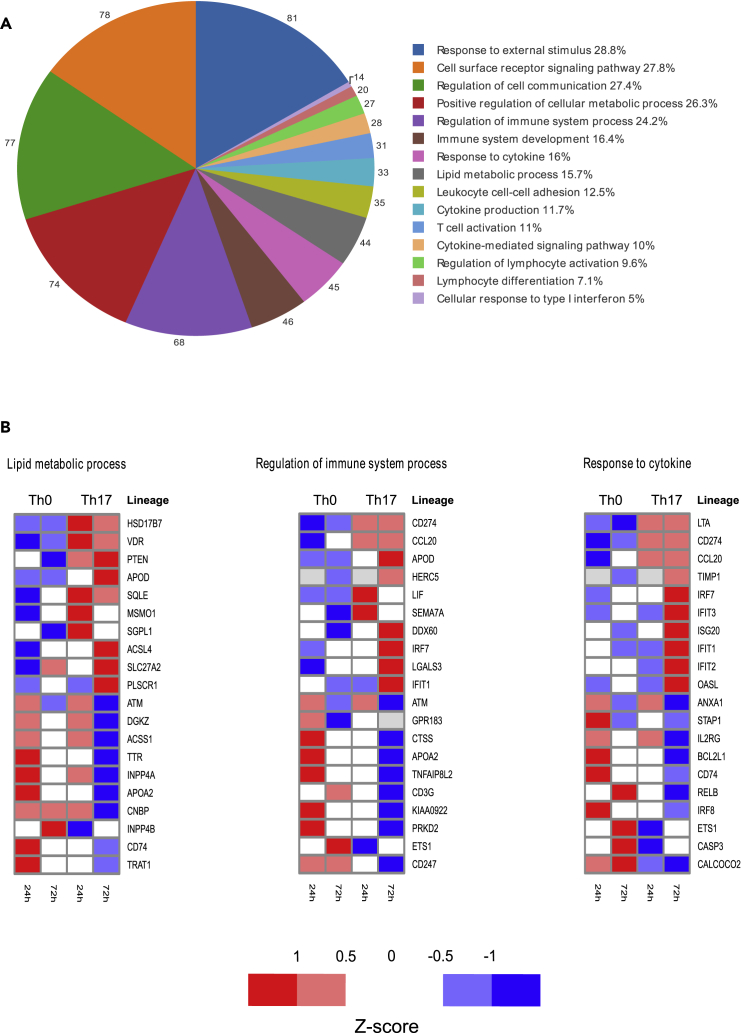
We examined the enrichment of Gene Ontology (GO) annotations for biological processes among the DE proteins over both time points. The GO analysis revealed significant (FDR ≤0.05) enrichment of approximately 300 biological processes (Table S3). The enriched processes included a number of important immunological processes, such as regulation of immune system development, immune system processes, response to external stimulus, cell surface receptor signaling pathways, cytokine production, cytokine-mediated signaling pathways, positive regulation of cellular metabolic processes, T cell activation, and lymphocyte differentiation (Figure 2A; Table S3).
Figure 2.
Enriched GO Biological Processes for the Proteins Differentially Expressed between Th17 and Th0 in the Proteomics Data
(A) The proportions of DE proteins for the 15 chosen representative GO biological processes out of all the annotated DE proteins.
(B) The averaged standardized expressions of 10 of the most upregulated and 10 of the most downregulated proteins in each of the three chosen representative GO biological processes. The GO analysis was performed using DAVID over both time points. The detected proteome was used as a reference background.
We observed that proteins related to lipid metabolic process were highly regulated in Th17 cells, which is in agreement with the recent studies showing the role of fatty acid metabolism in regulating Th17 cell differentiation and function (Figure 2B) (Berod et al., 2014, Santori et al., 2015). Known proteins of interest, such as VDR (Chang et al., 2010), are involved in lipid metabolism and have previously been linked to Th17 differentiation. MSMO1 and CYP51A1 are both also involved in lipid metabolism and have been reported to regulate RORC expression and the pathogenic activity of Th17 cells (Santori et al., 2015). Our analysis additionally identified a number of other proteins related to lipid metabolism signaling whose functions in regulating Th17 differentiation have not yet been defined (Table S3). As expected, we observed a number of proteins that were common to the immune system process and response to cytokines, including CD274 (PD-L1), CCL20, IFIT1, IRF7, and ETS1 (Figure 2B). Notably, proteins induced in response to IFN and antiviral pathways were highly enriched for the GO term “response to cytokine,” including OASL, ISG20, IRF7, and IFN-induced protein with tetratricopeptide repeat (IFIT) family proteins.
The Ingenuity Pathways Analysis (IPA; https://www.qiagenbioinformatics.com) was used to assign the molecular functional types and cellular locations of the proteins DE between Th17 and Th0 cells (Figures S3A and S3B; Table S3). The IPA analysis revealed distinct molecular function types for the Th17-regulated proteins, including enzymes (∼23%), transcription regulators (∼12%), transporters and kinases (5% each), transmembrane receptors (4.6%), peptidases and cytokines (3.5% each), phosphatases (2.5%), and G-protein receptors (1.1%). In addition, a small fraction of the Th17 DE proteins was recognized as growth factors, ligand-dependent nuclear receptors, ion channels, and translation regulators (<1% each). A major fraction of the Th17 DE proteins (>37%) were classified as others owing to their undefined functions (Figure S3A). Moreover, the IPA analysis for cellular compartment revealed four major cell locations for the proteins DE between Th17 and Th0 cells, including the cytoplasm (45.4%), nucleus (23.8%), plasma membrane (19.9%), and extracellular space (8.2%), with only 2.8% of the DE proteins belonging to an undefined (other) cell location (Figure S3B; Table S3). Thus the enrichment of proteins assigned to various molecular functional types and their location at different cellular compartments indicate the involvement of distinct signaling pathways and transcriptional mechanisms during differentiation of Th17 cells. In conclusion, our analysis identified proteins from almost all the functional types and cellular compartments, providing a valuable resource for further investigation of candidate proteins potentially regulating the differentiation and function of Th17 cells in humans.
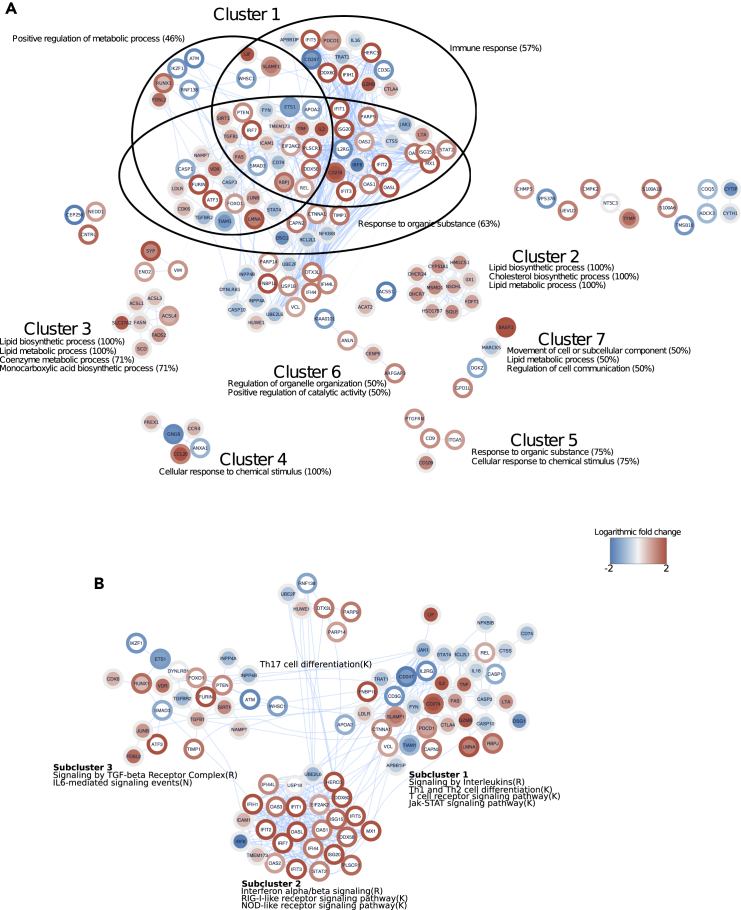
An Interaction Network for the DE Proteins during Early Th17 Cell Polarization
To understand the relationships between proteins identified in our data, we performed a protein-protein interaction network analysis for the proteins DE in Th17 cells at both time points by mining the STRING database (Szklarczyk et al., 2017). Of the 291 unique proteins detected as DE between Th17 and Th0, 146 were found to have known experimental or predicted interactions (Figure 3). Altogether 443 interactions between the 146 proteins were detected. To further examine the possible functional elements among the interacting proteins, we identified clusters based on the interactions in the resulting network (Table S4). This yielded altogether 15 clusters, with seven of the clusters having four or more interacting proteins. The average interaction score between the cluster members in the identified clusters was high (0.77–0.93, range 0–1). To infer the functional relevance of the identified clusters, we examined the prevalence of the enriched GO biological processes within the seven clusters (Table S3). To further complement our network analysis, we included the changes in expression between Th17 and Th0 for the interacting proteins. The functional interaction analysis identified cluster 1 as the largest cluster with altogether 78 interconnected proteins related to distinct GO biological processes, including immune responses, positive regulation of cellular metabolic processes, and response to organic substances as the top three representative GO biological processes (Figure 3A). A large proportion of the proteins included in cluster 1 were either upregulated at 24 or 72 hr or downregulated at 24 hr, whereas only a small portion seemed to be downregulated at 72 hr. In addition, all the proteins of clusters 2 and 3 were related to lipid metabolic processes (Figure 3A). Furthermore, all proteins included in clusters 2 and 3 were found to be differentially regulated only at 24 hr (except for ACSL4, which was upregulated also at 72 hr).
Figure 3.
The Interaction Network among the DE Proteins between Th17 and Th0
The network is based on all high-confidence experimental and predicted interactions from the STRING database.
(A) The network of protein-protein interactions from the DE proteins between Th17 and Th0 over both time points. The representative significant GO biological processes and cluster identities are shown only for clusters 1–7 (clusters sized >3).
(B) Cluster 1 divided into sub-clusters and the enriched pathways identified using the ReactomeFIViz Cytoscape plug-in. The letter in parentheses after each pathway name denotes the source of the annotation: R, Reactome; K, KEGG; N, NCI PID. The edges in the networks were bundled for a clearer representation. The logarithmic fold change between Th17 and Th0 at 24 hr (node inner circles) and at 72 hr (node outer circles) was implemented as continuous color mapping. For a clearer representation, the logarithmic fold changes are truncated to a range of −2 to 2.
Furthermore, to gain insight into the biological implication of expression changes in cluster 1, we analyzed the proteins in the context of biological pathways. A close examination identified three distinct functional sub-clusters within cluster 1 that were further called for functional enrichment of biological pathways (Figure 3B). The proteins in sub-clusters 1 and 3 were enriched for biological pathways involving signaling by TGF-β receptor complex, IL-6-mediated signaling events, signaling by interleukins, Th1 and Th2 cell differentiation, and the JAK-STAT signaling pathway. Interestingly, we observed a number of proteins in these two sub-clusters that are known modulators of Th17 cell differentiation, including CDK6, RUNX1, JUNB, SIRT1, RBPJ, TNF, SLAMF1, and FAS (positive regulators of Th17 cell differentiation) and FOSL2, SMAD3, ETS1, STAT4, and FYN (negative regulators of Th17 cell differentiation). Noticeably, the majority of proteins in sub-cluster 2 were upregulated in Th17 condition at 72 hr and enriched for antiviral immune pathways including IFN-α/β signaling, RIG-I-like receptor signaling pathway, and NOD-like receptor pathway (Figure 3B). The enrichment of antiviral proteins in the network at this early stage of differentiation is interesting because they are known to negatively modulate Th17 differentiation (Ramgolam et al., 2009, Ye et al., 2017). However, the expression of IRF-8 was downregulated in Th17 cells at 24 hr. IRF-8 is a known suppressor of Th17 cell differentiation in mouse (Ouyang et al., 2011). Further studies are needed to characterize the role of proteins related to antiviral pathway in human Th17 cell differentiation. To summarize, we identified protein-protein interaction networks of the proteins related to distinct biological processes and cellular pathways indicating their coordinated regulation during early Th17 cell differentiation program.
Validation of Mass-Spectrometry-Identified Th17 DE Proteins
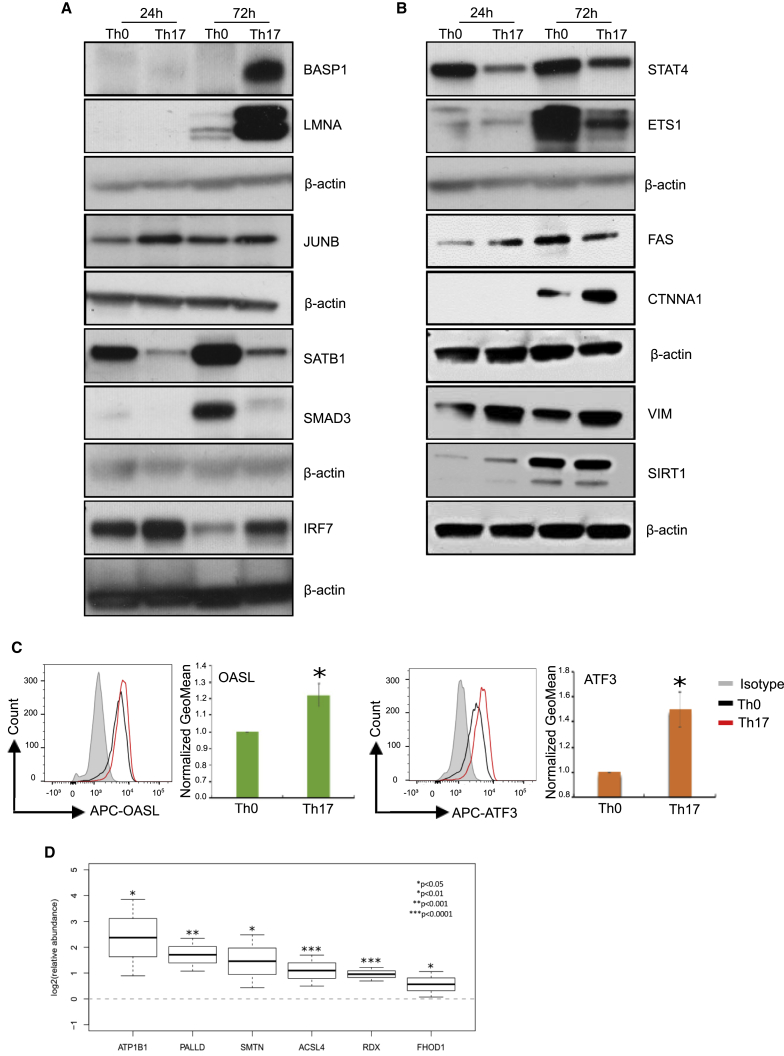
MS-based global proteomics approaches for protein identification rely on probability algorithms and benefit from further validation by targeted MS or other methods, such as immunoblotting or flow cytometry. We used all these three means to validate the expression profiles of selected candidate proteins. Results using western blot and flow cytometry analysis are shown in Figures 4A, 4B, and S4). We have previously shown the changes in protein expression of BASP1 (Figure 4A), LMNA (Figure 4A), and ATP1B1 (in Figure 4D) in Th17 cells, and these were used here as marker genes that further confirmed consistency between our current and previous results (Tuomela et al., 2012). Western blot analysis for the selected Th17 upregulated proteins (JUNB, SIRT1, CTNNA1, FAS, VIM, and IRF7) and downregulated proteins (SATB1, SMAD3, STAT4, and ETS1) showed consistent results with the proteomic analysis (Figures 4A, 4B, and S4). Using flow cytometry, we also validated the expression of OASL and ATF3 at 72 hr after the initiation of Th17 cell polarization. Again, the results were in agreement with the proteome data showing Th17-specific increased expression of OASL and ATF3 (Figure 4C). The function of BASP1, LMNA, CTNNA1, OASL, ATF3, and VIM in modulating Th17 cell differentiation and functions remains to be studied.
Figure 4.
Validation of the Mass-Spectrometry (MS)-Identified Proteins Using Immunoblotting, Flow Cytometry, and Targeted Mass-Spectrometry-Based Selected Reaction Monitoring (SRM) Analysis
(A and B) Immunoblot validations of MS identified up- or downregulated proteins with known and unknown Th17-related function. Blots show the protein extracts from the Th0 and Th17 cells at 24 and 72 hr. A representative blot from three biological replicates is shown.
(C) Flow cytometry validations of OASL and ATF3 detected as differentially upregulated in Th17 cells by MS analysis. A representative histogram image from Th17 (red), Th0 (black), and isotype control (gray) as well as the mean of four biological replicates are shown. Error bars represent standard deviation values across biological replicates (n=4; *p<0.05 for Th0 versus Th17; two-tailed t-test analysis).
(D) Confirmation of the protein expression of six proteomics candidates, ATP1B1, PALLD, ACSL4, FHOD1, SMTN, and RDX, using targeted SRM analysis. The SRM data is from four biological replicates and represented as the mean values +/- 95% confidence interval. (n = 4; *p < 0.05, **p < 0.01, ***p < 0.0001, for Th17 versus Th0).
In addition, we confirmed the expression of five additional candidates with unknown function in Th17 biology, namely, PALLD, ACSL4, SMTN, RDX, and FHOD1 in addition to ATP1B1 using a targeted MS-based selected reaction monitoring analysis (Figure 4D). Long-chain acyl-CoA synthetase 4 (ACSL4) belongs to the class of enzymes that catalyses addition of a coenzyme-A (CoA) group to free long-chain fatty acids, specifically arachidonic acid and eicosapentaenoic acid, and converts them into acyl-CoA forms (Kang et al., 1997). As recent studies have advocated the role of lipid metabolism in regulating Th17 cell differentiation and function (Berod et al., 2014, Wang et al., 2015, Sun et al., 2017, Young et al., 2017), it will be of interest to investigate how ACSL4 regulates Th17 cell function. The formin-homology-domain-containing protein FHOD, radixin (RDX), and smoothelin (SMTN) are cytoskeleton proteins that regulate cell migration and contraction (Niessen et al., 2005, Schulze et al., 2014). Radixin is a part of membrane-associated ERM (ezrin-radixin-moesin) family proteins that plays a key role in activation and T cell homeostasis by regulating cell adhesion, migration, and mobility and promotes T cell-antigen-presenting cell interaction (Faure et al., 2004, Müller et al., 2013). The role of these proteins in regulating Th17 cell functions remains to be established. To summarize, we confirmed the protein expression changes for the selected proteins detected by proteomics analysis by alternative methods in separate, independent biological replicates. These candidates provide interesting targets for further functional studies to reveal unknown mechanisms regulating Th17 cell differentiation and function.
Comparison of the Transcriptome and Proteome of the Differentiating Th17 Cells
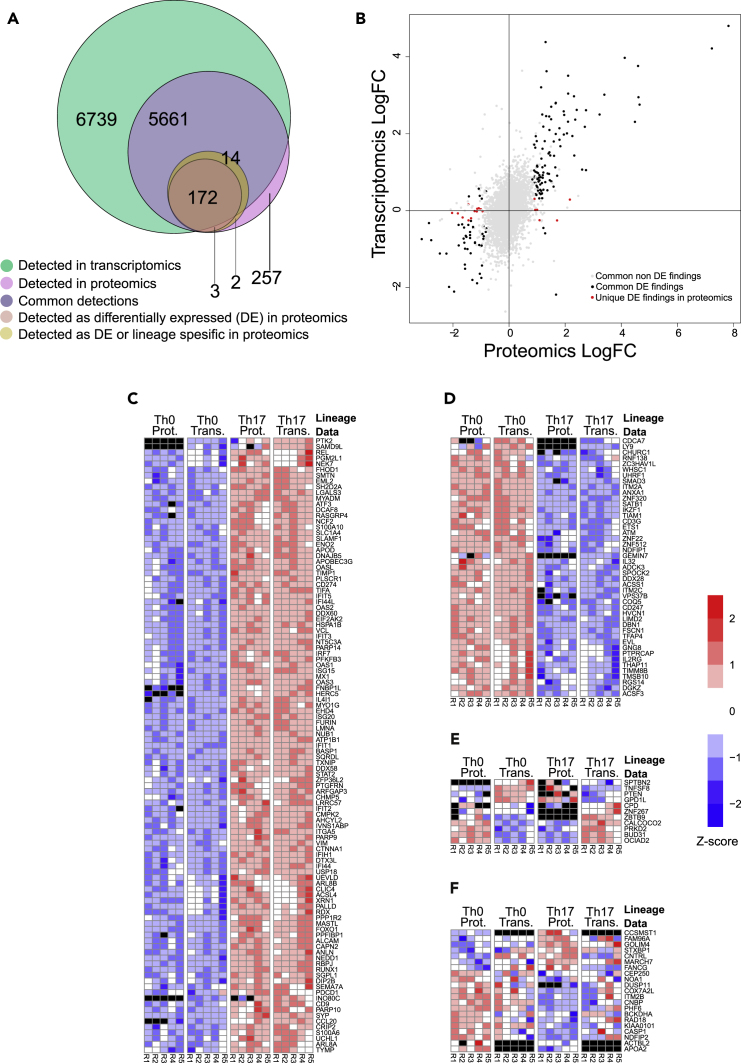
To find out the degree of concordance between protein and corresponding mRNA expression, we compared proteome and RNA sequencing (RNA-seq) datasets from the same human Th0 and Th17 cells at the 72-hr time point. Altogether 12,400 transcripts were detected from RNA-seq data (Figure 5A; Table S5). Proteomics and transcriptomics showed high degree of overlap as there were transcripts corresponding to 95.5% (5,661) of the 5,923 proteins detected by LC-MS (Table S5). Approximately half (54.3%, 6,739) of the protein-coding genes detected in RNA-seq data was not detected in the corresponding proteomics data. This is consistent with recent studies on other types of human T cells showing that approximately 35%–60% of the protein-coding genes detected by RNA-seq data were not detected in the corresponding proteomic data (Cuadrado et al., 2018, Schmidt et al., 2018). To gain further insight into the Th17-cell-specific protein expression changes and their relative mRNA expression changes, we examined the overlap of differentially regulated proteins with the mRNA transcriptome data. Of total 191 differentially regulated proteins, 186 were detected in transcriptomics data, among which 172 were DE (Th17 versus Th0). Altogether 262 proteins were uniquely detected in proteomics data, of which 257 proteins were not differentially regulated between Th0 and Th17 cells. Three proteins were DE in proteomics but not detected in transcriptomics data, and two proteins were detected only in one cell type in proteomics data, but not detected in transcriptomics data (Figure 5A; Table S5). The Pearson correlation coefficient analysis of the logarithmic fold changes showed a good correlation for all the common detections between proteomics and transcriptomics (0.519, p value < 0.001, n = 5,661). Importantly, an improved correlation was observed for the common DE proteins and DE transcripts (0.825, p < 0.001, n = 154). As anticipated the DE findings unique to proteomics among the common detection showed a poor correlation (0.233, p = 0.35, n = 18) (Figure 5B, Table S5).
Figure 5.
Correspondence of the Differentially Regulated Proteins between Th17 and Th0 and the Transcriptomics Data at 72 hr
(A) Venn diagram demonstrating the total number of common and unique detections and the total number of differentially regulated and differentially expressed (DE) proteins detected uniquely in the proteomics and commonly with the transcriptomics data. Differentially regulated proteins refer to those proteins detected as differentially expressed or detected only in one condition.
(B–E) (B) The logarithmic fold changes (logFC) of the all the common detections in proteomics and transcriptomics. The differentially expressed (DE) proteins detected as DE also in transcriptomics are marked with black and those uniquely DE in proteomics are marked with red. The Pearson correlation coefficient over the logFCs over all the common detections was 0.519 (p value < 0.001, n = 5,661), over the common DE findings 0.825 (p<0.001, n = 154), and over the DE findings unique to proteomics among the common detections 0.233 (p = 0.35, n = 18). The Z score standardized expression levels of the DE proteins and the corresponding DE transcripts between Th17 and Th0 (C) upregulated in Th17 in both datasets, (D) downregulated in Th17 in both datasets, and (E) divergently regulated between datasets.
(F) The Z score standardized expression levels of the DE proteins not detected as DE in transcriptomics. Black color in the heatmaps stands for undetected/missing value.
For a more comprehensive comparison of the proteomics and transcriptomics data, we focused our analyses on the common differentially regulated proteins (163 proteins) either detected only in one condition (nine proteins; detected either in Th0 or Th17 condition) or DE proteins (154 proteins) having corresponding DE transcripts between Th17 and Th0 (Figures 5C–5E). Noticeably, among the 163 differentially regulated proteins with corresponding detected transcripts, a large majority (93.2%) was changed in a similar fashion between mRNA and protein levels (Figures 5C and 5D; Table S5). Among these, 107 proteins and their corresponding mRNAs were consistently upregulated, including RBPJ, RUNX1, CCL20, BASP1, ATP1B1, and LMNA, and 45 proteins and their corresponding mRNAs were consistently downregulated, such as SMAD3, SATB1, ETS1, IL2RG, and Nedd4-family-interacting protein 1 (Ndfip1). However, 11 molecules showed opposite expression changes between mRNA and protein expression for both up- and downregulated genes at both time points (Figure 5E). This anti-correlation profile of proteins and mRNA observed in our data was also seen in another study comparing human iTreg transcriptome and proteome (Schmidt et al., 2018). Of note, 21 proteins DE between Th17 and Th0 cells were not detected as DE in the transcriptome data (Figure 5F and Table S5), suggesting that they may be post-transcriptionally regulated. Furthermore, the transcripts corresponding to CCSMST1, ACTBL2, and APOA2 proteins were not detected in transcriptomics data across all replicates. Further validation and evaluation of the differences in the expression profiles of these proteins with available good-quality antibodies such as GOLIM4, CNTRL, CASP1, RAD18, CNBP, NDFIP2, and ITM2B are required to understand the mechanisms responsible for their regulation. Thus our comparative analysis of the transcriptome and the proteome showed mostly similar, and also opposite expression, changes in the mRNA-protein levels, consistent with dynamic regulation of mRNA and protein expression. Moreover, our analysis highlighted the importance of proteomic analysis in complementing measurements on the transcriptome level.
Comparison of the Human and Mouse Proteomes during Early Th17 Cell Polarization
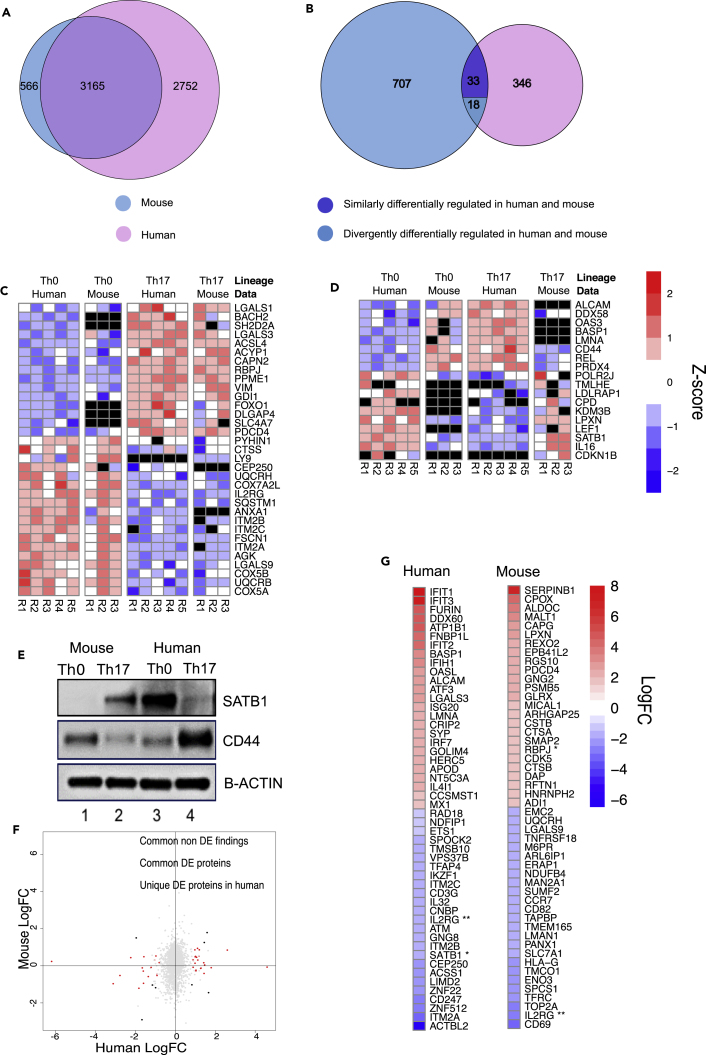
Mouse models have served as a useful tool for characterizing the biochemical and in vivo functions of genes. However, when used to model human diseases, the extent to which findings from mouse genetic models translate to humans has gained limited success. Importantly, using a systems biology approach, previous studies have revealed differences between mouse and human (Mestas and Hughes, 2004, Shay et al., 2013, Pishesha et al., 2014). Our recent study identified conserved and diversified gene signatures from the comparison of human and mouse transcriptomes during Th17 cell polarization (Tuomela et al., 2016). To further establish the level of dissimilarity, we made direct comparison with data from a recently published proteomics dataset from mouse Th17 cells cultured for 72 hr (Mohammad et al., 2018). The proteome profiles of CD4+ T cells 72 hr after TCR activation and Th17 polarization were compared using the aforementioned mouse data and the human data from the current study.
Comparison of the proteins detected in human (5,917 proteins) and mouse (3,731 proteins) revealed high degree of overall overlap as homologs for almost 85% of the mouse proteins were also detected in human (Figure 6A, Table S6). To make the comparison of the differentially regulated proteins more comprehensive, we used a threshold of FDR 0.1 (used in Mohammad et al., 2018) in both datasets to define the DE proteins. In agreement with our previous study (Tuomela et al., 2016), there was very limited overlap between the proteins differentially regulated between Th17 and Th0 cells, respectively, in human and mouse. Of the 758 and 397 proteins differentially regulated at 72 hr between Th17 and Th0 in mouse and in human, respectively, only 33 proteins were detected as differentially regulated in a similar fashion in human and mouse (Figure 6B; Table S6). Moreover, 18 proteins showed regulation in expression the opposite direction in human and mouse.
Figure 6.
Comparison of Differential Regulation between Th17 and Th0 at 72 hr in Human and Mouse
(A–D) Venn diagrams demonstrating (A) the total number of common and unique detections in human and mouse proteomics data and (B) the number of common and unique differentially regulated proteins detected. The numbers of differentially regulated proteins behaving in the similar or opposite fashion is shown in the overlap. Differentially regulated proteins refer to those proteins detected as DE or only in one condition. The Z score standardized expression levels of the commonly detected differentially regulated proteins regulated (C) similarly or (D) in an opposite fashion in human and mouse.
(E) Immunoblot validations of SATB1 and CD44 expression in human and mouse during Th17 cell polarization. Blots show the protein extracts from the Th0 and Th17 cells at 72 hr. A representative blot from three biological replicates is shown.
(F) The logarithmic fold changes (logFC) of all the common detections in human and mouse. The DE proteins detected as DE also in mouse are marked with black, and those uniquely DE in humans are marked with red. The Pearson correlation coefficient for the logFCs of all the common detections was 0.102 (p value < 0.001, n = 3,143), 0.304 (p value = 0.46, n = 8) with the common DE proteins detected, and 0.34 (p = 0.04, n = 73) for the DE findings unique to humans among the common detections.
(G) The logFCs and identities of the 25 most up- and downregulated DE proteins in humans and mouse. The common detection is marked with **. Proteins detected as DE in the other species but not among the 25 most up- and downregulated are marked with *. Mouse genes corresponding to mouse proteins are transformed as orthologous human genes for comparison using Ensembl BioMart. Black color in the heatmaps stands for undetected/missing value.
Of the 33 proteins whose expression was regulated similarly in human and mouse (Figure 6B), 15 and 18 proteins were upregulated and downregulated, respectively, in Th17 conditions at 72 hr (Figure 6C). Further analysis of the 18 proteins that showed opposite expression patterns between Th17 and Th0 in the two species at 72 hr revealed eight proteins (including DDX58, CD44, REL, and PRDX4) with upregulation in human and downregulation in mouse. The 10 proteins differentially upregulated in mouse and downregulated in human Th17 conditions included IL16, SATB1, LEF1, and LPXN (Figure 6D). The relative changes in expression of CD44 and SATB1 were validated both in human and mouse with western blot. RORyt and BATF proteins were used as polarization markers for mouse Th17 cell differentiation (Figure S5A). The immunoblot results demonstrated that the expression of these proteins was consistent with comparative analysis of quantitative proteomic data (Figures 6E and S5B).
SATB1, a chromatin organizer and transcription factor, is known to control the expression of a large number of genes involved in T cell development, activation, and differentiation both in mouse and human (Alvarez et al., 2000, Ahlfors et al., 2010, Notani et al., 2010, Burute et al., 2012). Both in mouse and human, SATB1 regulates Th2 cell differentiation by directly controlling the expression of its target genes and mediates topological looping of the transcriptionally active chromatin to the Th2 cytokine locus (Cai et al., 2006, Notani et al., 2010). FOXP3-mediated inhibition of SATB1 expression in human Treg cells is critical for their suppressive function as well as repression of genes for effector T cell differentiation (Beyer et al., 2011). Although the expression of SATB1 is conserved between Th2 and Treg cells, its opposite expression profile in human and mouse in Th17 cells suggests a different role in driving Th17 cell differentiation.
CD44 is a transmembrane glycoprotein with diverse functions in a variety of cell types (Baaten et al., 2010b). In T cells, CD44 deficiency in Th1 cells leads to compromised cell survival and abrogated generation of cellular memory to viral infection (Baaten et al., 2010a). CD44 is also required for the suppressive function of Treg cells by promoting persistent expression of FOXP3 and suppressive cytokines TGF-α1 and IL-10 (Bollyky et al., 2009). Mice lacking Cd44 gene have an increased secretion of IL-17 and severe experimental autoimmune encephalomyelitis (EAE) disease score (Flynn et al., 2013). However, a contrasting study revealed CD44 deletion to downregulate Th1/Th17 differentiation, promote Th2 differentiation, and ameliorate clinical EAE disease score (Guan et al., 2011). Because, the majority of the understanding on the regulation and functions of CD44 in T cells have come from studies in mouse, the opposite expression profile of CD44 in human and mouse Th17 cells deserves further studies to clarify its function in humans. Our results suggest these proteins to have different roles in polarizing Th17 cells in human and mouse. Moreover, the vast majority of the proteins detected as DE between Th17 and Th0 at 72 hr after the initiation of Th17 polarization in human were not detected as DE in mouse, and vice versa. Furthermore, we detected a number of proteins in one species, but not in the other. For instance, the proteins CEP250, ANXA1, ALCAM, OAS3, and BASP1 were not detected at all in Th17 cells in mouse, whereas LY9 and CDKN1B were not detected in human Th17 cells. Moreover, the proteins, BACH2, SH2D2A, FOXO1, DLGAP4, SLC4A7, TMLHE, LDLRAP1, CPD, and KDM3B were not detected in mouse Th0 condition.
To address the level of conservation (similarity) of protein signatures between human and mouse, Pearson correlation coefficients were calculated for logarithmic fold changes (Th17 versus Th0) of the proteins detected in human and mouse. Overall, the analysis revealed very little correlation (R2 = 0.102, p value < 0.001, n = 3,143), although it was somewhat better for the proteins that were DE in both human and mouse (R2 = 0.304, p value = 0.46, n = 8) and for the DE proteins unique to human among the common detections (R2 = 0.34, p = 0.04, n = 73) (Figure 6F, Table S6). Upon examination of 25 most up- and downregulated DE proteins in human and mouse, we found only IL2RG detected in both species. In addition, SATB1 (human) and RBPJ (mouse) were also detected as DE in both species, but not among the 25 most up- and downregulated in one of the species (Figure 6G, Table S6). Overall, our analysis identified a very low degree of similarities in DE protein signatures between human and mouse, as well as a number of proteins even with opposite expression patterns, underscoring why studies using mouse genetic models have limited success in recapitulating the results in humans.
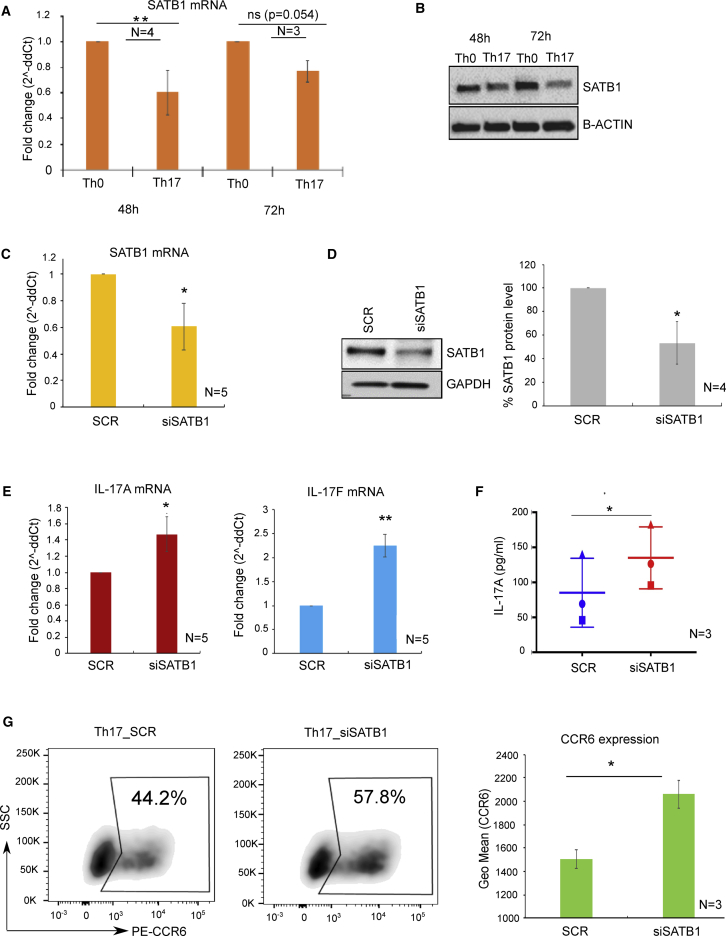
Repression of SATB1 Expression Contributes to Human Th17 Cell Differentiation
The expression of SATB1 in distinct Th and Treg cells is shown to be conserved between human and mouse (Ribeiro de Almeida et al., 2009, Ahlfors et al., 2010, Beyer et al., 2011). SATB1 expression has been previously reported to be upregulated in mouse Th17 cells (Ciofani et al., 2012, Mohammad et al., 2018). However, we found SATB1 transcription to be downregulated during human Th17 cell differentiation (Tuomela et al., 2012, Tuomela et al., 2016). This observation was further complemented by the findings, both at transcript and protein levels from our current transcriptomics and proteomics data and further validated by real-time quantitative PCR analysis and western blot analysis (Figures 4A, 6D, 6E, 7A, and 7B; Table S4). To examine the role of SATB1 in human Th17 cells, RNAi was used to silence its expression. Introduction of SATB1-specific small interfering RNA (siRNA) (siSATB1) in differentiating Th17 cells resulted in a significant decrease in the expression of SATB1 both at mRNA and protein levels when compared with non-targeting scramble siRNA (Figures 7C, 7D, and S5C). SATB1 silencing in Th17 cells led to increased transcription of marker cytokines IL-17A and IL-17F genes as well as the secretion of IL-17A cytokine (Figures 7E and 7F). In addition, depletion of SATB1 resulted in the increased expression of Th17-specific chemokine receptor, CCR6 (Figure 7G). Overall, depletion of SATB1 results in increased expression of the Th17 cell signature genes suggesting it acts as a negative regulator of Th17 cell differentiation in humans.
Figure 7.
SATB1 Is a Negative Regulator of Human Th17 Cell Differentiation
The mRNA expression profile of SATB1 in Th0 and Th17 cells at 48 and 72 hr after cell activation (A). Western blot analysis of SATB1 expression in Th0 and Th17 cells at 48 and 72hr after cell activation (B). The RNA interference (RNAi)-mediated knockdown of SATB1 at 24 hr post-Th17 cell polarization using specific siRNAs. Scramble siRNA (SCR) was used as a negative control. Silencing of SATB1 at mRNA (C) and protein (D) level was measured. Effect of SATB1 knockdown on IL-17A and IL-17F mRNA levels (E) by real-time TaqMan PCR analysis and on IL-17A protein secretion (F) was quantitated by ELISA assay from SATB1 siRNA nucleofected Th17 cells relative to NT-siRNA at 72 hr post-initiation of Th17 polarization. CCR6 surface expression detected by flow cytometry was measured (G) from SATB1 siRNA nucleofected Th17 cells relative to NT-siRNA at 72 hr post-initiation of Th17 polarization. Data are representative of 3–5 independent experiments. Error bars represent standard deviation values across biological replicates (n = 3–5; *p < 0.05, **p < 0.01 for Th0 versus Th17 and SCR versus siSATB1; two-tailed t-test analysis).
Discussion
The role of Th17 cells has been established in the pathogenesis of inflammatory and autoimmune diseases and cancer (Ghoreschi et al., 2011, Bailey et al., 2014). Since their discovery a decade ago, most studies have used mouse model to identify key regulators and molecular mechanisms driving Th17 cell development and function. However, the molecular mechanisms and signaling pathways that regulate human Th17 cell differentiation remain largely unclear. Our understanding of the human Th17 cell development and functions rely on studies of individual gene function or studies utilizing genome-wide transcriptional profiling to gain a holistic systems view (Tuomela et al., 2012, Tuomela et al., 2016, Gaublomme et al., 2015). However, genome-wide transcriptome analysis only describes the general characteristics of molecular changes (Stubbington et al., 2015) and does not necessarily reflect the specific phenotypic changes, which are controlled at the protein level.
In the recent years, global proteomic approaches have been adapted for the global analysis of complex proteomes in several distinct human immune cell types, including B and plasma cells (Salonen et al., 2013), macrophages and dendritic cells (Becker et al., 2012, Eligini et al., 2015, Worah et al., 2016), neutrophils (Tomazella et al., 2009), resting T cells (Howden et al., 2013) cytotoxic T cells (CTL) (Hukelmann et al., 2016, van Aalderen et al., 2017), natural killer T cells (Scheiter et al., 2013), Th1 and Th2 cells (Filén et al., 2009), and Treg cells (Kubach et al., 2007, Duguet et al., 2017, Cuadrado et al., 2018, Schmidt et al., 2018). In addition, a recent study describing the proteomes of Th1/Th17 clones derived from the gut biopsies of the patient with Crohn disease identified protein signatures of “mixed” Th1/Th17 phenotypes using MS-based protein identification and quantification (Riaz et al., 2016).
In the present study, we have used a label-free MS approach to reveal qualitative and quantitative proteomic differences between Th0 and Th17 cells during early stages of cellular stimulation and differentiation from naive CD4+ T cell subsets isolated from the umbilical cord blood. To understand the dynamics of protein expression changes during early stages of Th17 cell differentiation, we characterized proteome changes at 24 and 72 hr after initiation of the Th17 cell polarization. Despite the fact that the vast majority of the proteome is shared between TCR-activated cells (Th0) and cells primed for Th17 cell polarization at this early stage, our analyses demonstrated significant temporal and lineage-specific changes in the proteome. Overall, we observed clear differences in protein abundance associated with Th17 cell polarization.
Our analysis of the cellular proteome during early Th17 cell differentiation identified the differential expression of 291 proteins. The detected DE proteins included key protein markers used to characterize early differentiating human Th17 cells, such as cell surface markers (CCR4, CTLA-4, ICOS, and PDCD1), cytokines (CCL20 and TNF-α), and nuclear proteins (RUNX1, SIRT1, AHR, RBPJ, JUNB, REL, and FOSL2). These findings support the overall strength of the proteomic data generated and the MS-based approach implemented in this study. In addition, we discovered a number of additional proteins with significant differential expression between Th17 and Th0 cells that might have functional effects on Th17 development and function. Such examples are two glycosylphosphatidylinositol (GPI)-anchored cell surface glycoproteins, namely, semaphorin 7A (SEMA7A; also known as CD108) and CD109. SEMA7A is a negative regulator of T cell responses (Czopik et al., 2006). It has important roles in inflammatory and autoimmune disorders, including rheumatoid arthritis, multiple sclerosis, and colitis (Suzuki et al., 2007, Jongbloets et al., 2013, Gutiérrez-Franco et al., 2016). Yet, the molecular mechanism of how SEMA7A regulates differentiation and functioning of Th17 cells is not known. Likewise, CD109 is a co-receptor for TGF-β that negatively regulates TGF-β signaling. CD109 does this by coupling with caveolin-1, which mediates internalization and degradation of TGF-β receptors via receptor-mediated endocytosis (Bizet et al., 2011) or via regulating SMAD7/Smurf2-mediated degradation of TGFBR1 (Bizet et al., 2012). The regulation of TGF-β signaling contributes to the development of Th17 cells. At lower concentrations, TGF-β supports Th17 differentiation, and at higher concentrations, it promotes the differentiation of Treg cells. Besides its role in controlling TGF-β signaling, in human keratinocytes CD109 amplifies the activation of STAT3 (Litvinov et al., 2011), a transcription factor also required for Th17 development.
Interestingly, FURIN, found to be upregulated in Th17 cells in our analysis, is a pro-protein convertase enzyme involved in the processing of CD109 into the 180-kDa secreted form and 25-kDa GPI-anchored form, suggesting that FURIN participates in regulating CD109-mediated molecular functions. In mouse, the deficiency of FURIN in T cells causes T cell expansion/activation, impaired production of TGF-β1, and breakdown of the peripheral tolerance (Pesu et al., 2008). However, the molecular mechanisms by which FURIN regulates Th17 cell development are still unclear. Consistent with these findings, Th17-specific upregulation of FURIN and CD109 in our analysis suggests that they may participate in controlling the development and function of Th17 cells via an overlapping pathway.
A number of proteins involved in the regulation of nuclear structure organization or function were found to be differentially upregulated (e.g., LMNA, MYOF, BASP1, IRF7, BHLHA15, PARP9, PARP10, PARP14, and CDK6) or downregulated (e.g., ELF1, ACTBL2, CHURC, CNBP, SATB1, IRF8, and IKZF1) in Th17 cells. These observations suggest an important role of these proteins in influencing nuclear architecture and function, which are active parts of regulating chromatin structure or transcription. In mouse, IRF8 has been shown to inhibit Th17 differentiation where IRF8 physically interacts with RORγt, and suppresses Il-17a transcription (Ouyang et al., 2011). The reduced expression of IRF8 in human Th17 cells when compared with Th0 cells further supports its role as a negative regulator of Th17 cell differentiation. The expression of IKZF1 (Ikaros) was downregulated in Th17 cells across all replicates. However, a study of Ikaros knockout mouse has shown that IKZF1 positively regulates the development and function of Th17 cells (Wong et al., 2013). The role of IKZF1 in the human system has not been reported. Further studies are needed to uncover the mechanisms and functions of these proteins in the regulation of Th17 cell development.
The mapping of protein-protein interactions is important in understanding the molecular mechanisms regulating cellular processes. The STRING analysis showed a highly cohesive protein-protein network and revealed abundant functional interactions between the proteins involving distinct GO biological processes. The majority of the protein-protein interactions in the network were related to the regulation of cellular metabolic processes with a number of the interactions involving proteins of lipid metabolic process. The role of a few proteins (e.g., FADS2 and FASN, ACC1, AHR, and HSD17B7) of the lipid metabolism pathway from our network has been recently reported to regulate Th17 function (Berod et al., 2014, Hu et al., 2015, Santori et al., 2015, Wang et al., 2015, Sun et al., 2017).
Among the validated Th17-regulated proteins, JUNB, SATB1, SMAD3, STAT4, ATF3, and ETS1 have previously been shown to have functional roles either in the context of Th17 cells or other Th cell subsets. In addition, RUNX1, ATP1B1, KDSR, LMNA, VDR, and BASP1 were validated at the protein level also in our previous report (Tuomela et al., 2012). The STAT3-dependent expression of JUNB and its role in positively regulating Th17 cell differentiation have been recently described in mouse model (Carr et al., 2017). Furthermore, SMAD3 (Martinez et al., 2009) and ETS1 (Moisan et al., 2007) negatively regulate the murine Th17 cell function. STAT4 is a major regulator of Th1 cell differentiation and function, and is a critical component in maintaining the balance between Th17 and Treg cells under disease states (Xu et al., 2011). Importantly, some of the validated proteins, including BASP1, LMNA, IRF7, ATF3, ACSL4, FHOD1, SMTN, ATP1B1, RDX, PALLD, and OASL have previously been poorly characterized or not reported to regulate Th17 cell differentiation and function and are hence candidates for functional follow-up studies. For example, we have previously reported that ATF3 directly regulates the human IFN-γ expression and promotes Th1 differentiation (Filén et al., 2010). However, the role of ATF3 in regulating the differentiation and function of human Th17 cells remains to be studied. In a mouse model, IRF7 silencing in resting CD4+ cells resulted in the increased secretion of Th17-associated cytokines (e.g., IL-17A, IL-17F, IL-21, IL-22, and IL-9), suggesting that IRF7 is a negative regulator of the Th17 response (Tao et al., 2014). However, the molecular mechanisms responsible for the expression of IRF7 and how it regulates human Th17 cell polarization are still unclear.
Recent studies have addressed the role of lipid metabolism in regulating Th17 cell differentiation and function (Berod et al., 2014, Wang et al., 2015, Sun et al., 2017, Young et al., 2017). As an enzyme involved in lipid metabolic pathways, ACSL4 is an interesting candidate for further studies on regulation of Th17 cell differentiation. Other candidates of interest, although not related to lipid metabolism, are FHOD1, RDX, and SMTN. These structural proteins regulate cell migration and contraction (Niessen et al., 2005, Schulze et al., 2014). However, their role in Th17 cell differentiation and function remains undetermined.
A comparison between the proteomic and transcriptomic profiles can provide valuable insight into the relationship between mRNA and protein expression that may not be feasible to achieve by separate analysis of either dataset alone (Nagaraj et al., 2011, Worah et al., 2016). To that end, we compared the current proteomics data with RNA-seq data from the same samples processed for MS-based proteome analysis. Almost 95% of the proteins detected by LFQ analysis had transcripts for the corresponding protein-coding genes and displayed good correlation in their relative abundance versus Th0 (0.519) (Figure 5B) consistent with the recent studies on other cells including human Treg cells (Weekes et al., 2014, Cuadrado et al., 2018, Schmidt et al., 2018). The DE proteins showed even improved correlation (0.819). However, we detected 21 proteins DE in a Th17-specific manner that were not observed at the transcriptome level. However, of these 21 DE proteins, only two proteins ITM2B and COX7A2L were also found to be DE and downregulated in Th17 condition in our reanalysis of the mouse proteomics data from Mohammad et al., 2018. Moreover, they reported that the expression of ITM2B was DE in mouse Th17 transcriptomics analysis with opposite expression profile compared with their protein data (i.e., downregulated at the RNA level and upregulated at the protein level when Th17 polarized cells were compared with activated cells). COX7A2L was found to be downregulated in mouse both at the transcript and protein levels. The expression of CEP250 was observed as DE and downregulated in human proteomics data, whereas it was detected only in the Th0 condition in mouse proteomics data and not detected as DE in mouse transcriptomics data. Thus based on our analysis it appears that the majority of these 21 DE proteins are uniquely post-transcriptionally regulated in humans. To conclude, our study provides several candidate proteins whose roles in the regulation of Th17 differentiation and function are yet to be determined.
Comparative analysis of transcriptional changes in human and mouse during immune cell differentiation, including Th17 cell specification, revealed a conserved signature between these species (Shay et al., 2013, Tuomela et al., 2016). This includes a panel of potential therapeutic targets that are regulated in a similar fashion in human and mouse and thus suitable for follow-up studies in experimental in vivo mouse models. In this study, we complemented our previous study by comparing human and mouse proteomes, which demonstrated remarkable differences in the regulation of protein expression between human and mouse during Th17 cell priming. Interestingly, only 33 proteins were differentially regulated in a similar fashion, whereas 18 showed regulation to the opposite direction in human and mouse. Although consistent with our previous results comparing DE transcripts from human and mouse during Th17 cell differentiation (Tuomela et al., 2016), the lack of overlap at the protein level is even more striking. Moreover, our results are in line with other recent reports, including those from the ImmGen consortium, suggesting inter-species differences in the cellular machinery due to the operations of conservation and diversification of the trans- and cis-regulatory programs (Mestas and Hughes, 2004, Shay et al., 2013). Although the proteins regulated in a similar manner in human and mouse are good candidates for follow-up functional in vivo studies in experimental mouse models, further investigation on the proteins regulated in opposite manner among common DE proteins in human and mouse along with human-specific DE proteins is critical for understanding their role in regulating Th17 cell differentiation and function in humans.
The expression of SATB1 was downregulated in human Th17 cells when compared with Th0 cells, suggesting its role as a negative regulator of human Th17 cell differentiation. However, a previous mouse study reported that SATB1 positively regulates Th17 function by controlling the expression of key Th17-related genes including Il-22, Il-17A and Il-17F, Il21, Il1R1, and Ccl20 (Ciofani et al., 2012). Opposite regulation of this key chromatin organizer (i.e., SATB1) during early specification of human and mouse Th17 cells suggests fundamental differences in the process between the two species. Moreover, recently, conditional deletion of SATB1 in mouse showed impaired production of IL-17- and IFN-γ in pathogenic T cells, and the mouse were resistant to the induction of EAE, suggesting a role of SATB1 for the generation of pathogenic Th cells (Akiba et al., 2018). Using the RNAi approach, we showed that SATB1 knockdown induced an elevated expression of marker cytokines IL-17A and IL-17F and CCR6, a chemokine expressed on the surface of Th17 cells. This suggests SATB1 to be a negative regulator of Th17 cell priming. Further studies are required to gain insight into the molecular mechanisms through which SATB1 controls Th17 differentiation and function in humans.
In summary, our results revealed differences in the proteome of the cell types and time points studied and shed light on various perspectives of the early regulation of human Th17 cell priming. Beyond previous transcriptomics applications describing the regulatory programs that govern Th17 cell development and function in human and mouse, our study reports comprehensive proteomics data describing the initiation of human Th17 cell priming of naive CD4+ T cells derived from the umbilical cord blood. Hence, this study complements previous investigations to understand human Th17 cell priming. This study has resulted in the identification of a panel of proteins with unknown or poorly characterized functions in the context of Th17 development and function. We provide several previously unknown candidate proteins for further studies to reveal their influence on human Th17 specification and immunomodulatory and therapeutic potential for Th17-mediated immune diseases.
Limitations of the Study
This study describes the global cellular proteomic landscape during early human Th17 cell differentiation. However, the current data only represents unfractionated cellular lysates. The fractionation of the cellular compartments could have provided further insight into the differentiation process. Furthermore, mechanistic follow-up studies on key findings reported here are required and are interesting targets for future studies. Our comparison of human to mouse Th17 proteomics data showed very limited overlap. It would be important to consider the kinetics of the process in the mouse—here we compared our data with data from a reported parallel study in mouse, where only one time point had been analyzed. It will be important to functionally validate in the mouse system the few findings with consistent results in human. Understanding the molecular mechanisms of proteins regulated in a similar fashion in human and mouse is crucial for translational research.
Ethical Aspects
The usage of blood of unknown donors was approved by the Ethics Committee of the Hospital District of Southwest Finland. Experiments using animals' cells were in accordance with the relevant University Animal Welfare guidelines.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank all voluntary blood donors and personnel of Turku University Hospital, Department of Obstetrics and Gynecology, Maternity Ward (Hospital District of Southwest Finland), for cord blood collection; the staff of the Turku Proteomics Facility, Biocenter Finland; and Marjo Hakkarinen and Sarita Heinonen for excellent technical assistance. S.K.T. is supported by the Juvenile Diabetes Research Foundation Ltd (JDRF; grant 3-PDF-2018-574-A-N); T.V. is supported by the Doctoral Programme in Mathematics and Computer Sciences (MATTI) of the University of Turku; M.M.K. is supported by the University of Turku Graduate School (UTUGS) Turku Doctoral Programme of Molecular Medicine (TuDMM) and a central grant from Finnish Cultural Foundation; R.L. has been supported by the Academy of Finland, AoF, Centre of Excellence in Molecular Systems Immunology and Physiology Research (2012-2017) grant 250114; by the AoF grants 292335, 294337, 292482, and 31444; and by grants from the JDRF, the Sigrid Jusélius Foundation (SJF), the Finnish Cancer Foundation, and the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 115797 (INNODIA). This Joint Undertaking receives support from the Union's Horizon 2020 research and innovation programme and“EFPIA,” “JDRF,” and “The Leona M. and Harry B. Helmsley Charitable Trust.” L.L.E. has received grants from the European Research Council (ERC; grant 677943), the European Union's Horizon 2020 research and innovation programme (grant 675395), the AoF (grants 296801, 304995, 310561, 313343), the Juvenile Diabetes Research Foundation (JDRF; grant 2-2013-32), the Finnish Funding Agency for Innovation (TEKES; grant 1877/31/2016), and SJF.
Author Contributions
S.K.T. designed and performed the experiments, analyzed and interpreted data, prepared figures, and wrote the manuscript. T.V. performed computational analysis of the data and interpreted the results, prepared figures, and wrote the manuscript. A.S. designed and performed experiments, analyzed data, and prepared figures; M.K. designed and performed the experiments, analyzed data, and wrote part of methods; S.D.B. processed and analyzed the samples for the label-free proteomics, analyzed data, prepared figures, and wrote part of methods; R.M. implemented the label-free and targeted proteomics analyses, analyzed data, and edited the manuscript; E.K. performed the experiments and analyzed data; V.S. performed experiments; R.S.D.A. performed experiments; O.R. provided expertise and edited the manuscript; S.G. provided expertise and supervision and edited the manuscript; L.L.E. provided expertise, participated in the interpretation of the results, provided guidance and supervision, and wrote the manuscript; and R.L. designed the study setup, provided expertise, participated in the interpretation of the results, provided guidance and supervision, and wrote the manuscript. All authors have contributed to the manuscript.
Declaration of Interests
The authors declare that they have no conflict of interest.
Published: January 25, 2019
Footnotes
Supplemental Information includes Transparent Methods, five figures, and seven tables and can be found with this article online at https://doi.org/10.1016/j.isci.2018.12.020.
Contributor Information
Laura L. Elo, Email: laura.elo@utu.fi.
Riitta Lahesmaa, Email: rilahes@utu.fi.
Data and Software Availability
The PRIDE accession number (Vizcaíno et al., 2016) for the mass spectrometry proteomic profiling data presented in this paper is PXD008973. The selected reaction monitoring (SRM) validation data are available through PASSEL with the dataset identifier PASS01204. The RNAseq data from this study is submitted to the Gene Expression Omnibus (GEO) with identifier GSE118974.
Supplemental Information
4
References
- van Aalderen M.C., van den Biggelaar M., Remmerswaal E.B.M., van Alphen F.P.J., Meijer A.B., Ten Berge I.J.M., van Lier R.A.W. Label-free analysis of CD8+T cell subset proteomes supports a progressive differentiation model of human-virus-specific T cells. Cell Rep. 2017;19:1068–1079. doi: 10.1016/j.celrep.2017.04.014. [DOI] [PubMed] [Google Scholar]
- Ahlfors H., Limaye A., Elo L.L., Tuomela S., Burute M., Gottimukkala K.V., Notani D., Rasool O., Galande S., Lahesmaa R. SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood. 2010;116:1443–1453. doi: 10.1182/blood-2009-11-252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba Y., Kuwabara T., Mukozu T., Mikami T., Kondo M. SATB1 is required for the development of experimental autoimmune encephalomyelitis to maintain T cell receptor responsiveness. Microbiol. Immunol. 2018;62:255–268. doi: 10.1111/1348-0421.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J.D., Yasui D.H., Niida H., Joh T., Loh D.Y., Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Baaten B.J.G., Li C.R., Deiro M.F., Lin M.M., Linton P.J., Bradley L.M. CD44 regulates survival and memory development in Th1 cells. Immunity. 2010;32:104–115. doi: 10.1016/j.immuni.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baaten B.J.G., Li C.R., Bradley L.M. Multifaceted regulation of T cells by CD44. Commun. Integr. Biol. 2010;3:508–512. doi: 10.4161/cib.3.6.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.R., Nelson M.H., Himes R.A., Li Z., Mehrotra S., Paulos C.M. Th17 cells in cancer: the ultimate identity crisis. Front. Immunol. 2014;5:276. doi: 10.3389/fimmu.2014.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L., Liu N.C., Averill M.M., Yuan W., Pamir N., Peng Y., Irwin A.D., Fu X., Bornfeldt K.E., Heinecke J.W. Unique proteomic signatures distinguish macrophages and dendritic cells. PLoS One. 2012;7:e33297. doi: 10.1371/journal.pone.0033297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C.N., Bähre H. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Beyer M., Thabet Y., Müller R.U., Sadlon T., Classen S., Lahl K., Basu S., Zhou X., Bailey-Bucktrout S.L., Krebs W. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizet A.A., Liu K., Tran-Khanh N., Saksena A., Vorstenbosch J., Finnson K.W., Buschmann M.D., Philip A. The TGF-β co-receptor, CD109, promotes internalization and degradation of TGF-β receptors. Biochim. Biophys. Acta. 2011;1813:742–753. doi: 10.1016/j.bbamcr.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Bizet A.A., Tran-Khanh N., Saksena A., Liu K., Buschmann M.D., Philip A. CD109-mediated degradation of TGF-?? receptors and inhibition of TGF-?? responses involve regulation of SMAD7 and Smurf2 localization and function. J. Cell Biochem. 2012;113:238–246. doi: 10.1002/jcb.23349. [DOI] [PubMed] [Google Scholar]
- Bollyky P.L., Falk B.A., Long S.A., Preisinger A., Braun K.R., Wu R.P., Evanko S.P., Buckner J.H., Wight T.N., Nepom G.T. CD44 costimulation promotes FoxP3+ regulatory T cell persistence and function via production of IL-2, IL-10, and TGF-beta. J. Immunol. 2009;183:2232–2241. doi: 10.4049/jimmunol.0900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett P.R., Zu Horste G.M., Kuchroo V.K. Pouring fuel on the fire: Th17 cells, the environment, and autoimmunity. J. Clin. Invest. 2015;125:2211–2219. doi: 10.1172/JCI78085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burute M., Gottimukkala K., Galande S. Chromatin organizer SATB1 is an important determinant of T-cell differentiation. Immunol. Cell Biol. 2012;90:852–859. doi: 10.1038/icb.2012.28. [DOI] [PubMed] [Google Scholar]
- Cai S., Lee C.C., Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Carr T.M., Wheaton J.D., Houtz G.M., Ciofani M. JunB promotes Th17 cell identity and restrains alternative CD4+T-cell programs during inflammation. Nat. Commun. 2017;8:301. doi: 10.1038/s41467-017-00380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.H., Chung Y., Dong C. Vitamin D suppresses Th17 cytokine production by inducing C/EBP Homologous Protein (CHOP) expression. J. Biol. Chem. 2010;285:38751–38755. doi: 10.1074/jbc.C110.185777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Hardy K., Pagler E., Ma L., Lee S., Gerondakis S., Daley S., Shannon M.F. The NF-κB transcription factor c-Rel is required for Th17 effector cell development in experimental autoimmune encephalomyelitis. J. Immunol. 2011;187:4483–4491. doi: 10.4049/jimmunol.1101757. [DOI] [PubMed] [Google Scholar]
- Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmi L., Maggi L., Santarlasci V., Liotta F., Annunziato F. T helper cells plasticity in inflammation. Cytometry A. 2014;85:36–42. doi: 10.1002/cyto.a.22348. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- Cuadrado E., van den Biggelaar M., de Kivit S., Chen Y.-Y., Slot M., Doubal I., Meijer A., van Lier R.A.W., Borst J., Amsen D. Proteomic analyses of human regulatory T cells reveal adaptations in signaling pathways that protect cellular identity. Immunity. 2018;48:1046–1059.e6. doi: 10.1016/j.immuni.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Czopik A.K., Bynoe M.S., Palm N., Raine C.S., Medzhitov R. Semaphorin 7A is a negative regulator of T cell responses. Immunity. 2006;24:591–600. doi: 10.1016/j.immuni.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Duguet F., Locard-Paulet M., Marcellin M., Chaoui K., Bernard I., Andreoletti O., Lesourne R., Burlet-Schiltz O., Gonzalez de Peredo A., Saoudi A. Proteomic analysis of regulatory T cells reveals the importance of Themis1 in the control of their suppressive function. Mol. Cell. Proteomics. 2017;16:1416–1432. doi: 10.1074/mcp.M116.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eligini S., Brioschi M., Fiorelli S., Tremoli E., Banfi C., Colli S. Human monocyte-derived macrophages are heterogenous: proteomic profile of different phenotypes. J. Proteomics. 2015;124:112–123. doi: 10.1016/j.jprot.2015.03.026. [DOI] [PubMed] [Google Scholar]
- Eriksson P., Andersson C., Cassel P., Nyström S., Ernerudh J. Increase in Th17-associated CCL20 and decrease in Th2-associated CCL22 plasma chemokines in active ANCA-associated vasculitis. Scand. J. Rheumatol. 2015;44:80–83. doi: 10.3109/03009742.2014.952332. [DOI] [PubMed] [Google Scholar]
- Faure S., Salazar-Fontana L.I., Semichon M., Tybulewicz V.L., Bismuth G., Trautmann A., Germain R.N., Delon J. ERM proteins regulate cytoskeleton relaxation promoting T cell-APC conjugation. Nat. Immunol. 2004;5:272–279. doi: 10.1038/ni1039. [DOI] [PubMed] [Google Scholar]
- Filén J.-J., Filén S., Moulder R., Tuomela S., Ahlfors H., West A., Kouvonen P., Kantola S., Björkman M., Katajamaa M. Quantitative proteomics reveals GIMAP family proteins 1 and 4 to be differentially regulated during human T helper cell differentiation. Mol. Cell. Proteomics. 2009;8:32–44. doi: 10.1074/mcp.M800139-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filén S., Ylikoski E., Tripathi S., West A., Björkman M., Nyström J., Ahlfors H., Coffey E., Rao K.V., Rasool O., Lahesmaa R. Activating transcription factor 3 is a positive regulator of human IFNG gene expression. J. Immunol. 2010;184:4990–4999. doi: 10.4049/jimmunol.0903106. [DOI] [PubMed] [Google Scholar]
- Flynn K.M., Michaud M., Madri J.A. CD44 deficiency contributes to enhanced experimental autoimmune encephalomyelitis: a role in immune cells and vascular cells of the blood-brain barrier. Am. J. Pathol. 2013;182:1322–1336. doi: 10.1016/j.ajpath.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaublomme J.T., Yosef N., Lee Y., Gertner R.S., Yang L.V., Wu C., Pandolfi P.P., Mak T., Satija R., Shalek A.K. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Hirahara K., O'Shea J.J. T helper 17 cell heterogeneity and pathogenicity in autoimmune disease. Trends Immunol. 2011;32:395–401. doi: 10.1016/j.it.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H., Nagarkatti P.S., Nagarkatti M. CD44 reciprocally regulates the differentiation of encephalitogenic Th1/Th17 and Th2/regulatory T cells through epigenetic modulation involving DNA methylation of cytokine gene promoters, thereby controlling the development of experimental autoimmune encephalomyelitis. J. Immunol. 2011;186:6955–6964. doi: 10.4049/jimmunol.1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Franco A., Costa C., Eixarch H., Castillo M., Medina-Rodríguez E.M., Bribián A., de Castro F., Montalban X., Espejo C. Differential expression of sema3A and sema7A in a murine model of multiple sclerosis: Implications for a therapeutic design. Clin. Immunol. 2016;163:22–33. doi: 10.1016/j.clim.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Hernández-Santos N., Gaffen S.L. Th17 cells in immunity to Candida albicans. Cell Host Microbe. 2012;11:425–435. doi: 10.1016/j.chom.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden A.J.M., Geoghegan V., Katsch K., Efstathiou G., Bhushan B., Boutureira O., Thomas B., Trudgian D.C., Kessler B.M., Dieterich D.C. QuaNCAT: Quantitating proteome dynamics in primary cells. Nat. Methods. 2013;10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Wang Y., Hao L.Y., Liu X., Lesch C.A., Sanchez B.M., Wendling J.M., Morgan R.W., Aicher T.D., Carter L.L. Sterol metabolism controls TH17 differentiation by generating endogenous RORγ agonists. Nat. Chem. Biol. 2015;11:141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- Hukelmann J.L., Anderson K.E., Sinclair L.V., Grzes K.M., Murillo A.B., Hawkins P.T., Lamond A.I., Cantrell D.A. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 2016;17:104–112. doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloets B.C., Ramakers G.M.J., Pasterkamp R.J. Semaphorin7A and its receptors: Pleiotropic regulators of immune cell function, bone homeostasis, and neural development. Semin. Cell Dev. Biol. 2013;24:129–138. doi: 10.1016/j.semcdb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Kang M.J., Fujino T., Sasano H., Minekura H., Yabuki N., Nagura H., Iijima H., Yamamoto T.T. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc. Natl. Acad. Sci. U S A. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubach J., Lutter P., Bopp T., Stoll S., Becker C., Huter E., Richter C., Weingarten P., Warger T., Knop J. Human CD4+CD25+ regulatory T cells: proteome analysis identifies galectin-10 as a novel marker essential for their anergy and suppressive function. Blood. 2007;110:1550–1558. doi: 10.1182/blood-2007-01-069229. [DOI] [PubMed] [Google Scholar]
- Lim H.W., Kang S.G., Ryu J.K., Schilling B., Fei M., Lee I.S., Kehasse A., Shirakawa K., Yokoyama M., Schnölzer M. SIRT1 deacetylates ROR t and enhances Th17 cell generation. J. Exp. Med. 2015;212:607–617. doi: 10.1084/jem.20132378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov I.V., Bizet A.A., Binamer Y., Jones D.A., Sasseville D., Philip A. CD109 release from the cell surface in human keratinocytes regulates TGF-beta receptor expression, TGF-beta signalling and STAT3 activation: relevance to psoriasis. Exp. Dermatol. 2011;20:627–632. doi: 10.1111/j.1600-0625.2011.01288.x. [DOI] [PubMed] [Google Scholar]
- Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Loyet K.M., Ouyang W., Eaton D.L., Stults J.T. Proteomic profiling of surface proteins on Th1 and Th2 cells. J. Proteome Res. 2005;4:400–409. doi: 10.1021/pr049810q. [DOI] [PubMed] [Google Scholar]
- Mak I.W.Y., Evaniew N., Ghert M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- Martinez G.J., Zhang Z., Chung Y., Reynolds J.M., Lin X., Jetten A.M., Feng X.H., Dong C. Smad3 differentially regulates the induction of regulatory and inflammatory T cell differentiation. J. Biol. Chem. 2009;284:35283–35286. doi: 10.1074/jbc.C109.078238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinstry K.K., Strutt T.M., Swain S.L. Regulation of CD4+ T-cell contraction during pathogen challenge. Immunol. Rev. 2010;236:110–124. doi: 10.1111/j.1600-065X.2010.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra P., Krishnamurthy P., Sun J., Goenka S., Kaplan M.H. Poly-ADP-ribosyl polymerase-14 promotes T helper 17 and follicular T helper development. Immunology. 2015;146:537–546. doi: 10.1111/imm.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J., Hughes C.C.W. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Meyer zu Horste G., Wu C., Wang C., Cong L., Pawlak M., Lee Y., Elyaman W., Xiao S., Regev A., Kuchroo V.K. RBPJ controls development of pathogenic Th17 cells by regulating IL-23 receptor expression. Cell Rep. 2016;16:392–404. doi: 10.1016/j.celrep.2016.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad I., Nousiainen K., Bhosale S.D., Starskaia I., Moulder R., Rokka A., Cheng F., Mohanasundaram P., Eriksson J.E., Goodlett D.R. Quantitative proteomic characterization and comparison of T helper 17 and induced regulatory T cells. PLoS Biol. 2018;16:e2004194. doi: 10.1371/journal.pbio.2004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan J., Grenningloh R., Bettelli E., Oukka M., Ho I.C. Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med. 2007;204:2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N., Fischer H.J., Tischner D., van den Brandt J., Reichardt H.M. Glucocorticoids induce effector T cell depolarization via ERM proteins, thereby impeding migration and APC conjugation. J. Immunol. 2013;190:4360–4370. doi: 10.4049/jimmunol.1201520. [DOI] [PubMed] [Google Scholar]
- Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen P., Rensen S., van Deursen J., De Man J., De Laet A., Vanderwinden J.M., Wedel T., Baker D., Doevendans P., Hofker M. Smoothelin-A is essential for functional intestinal smooth muscle contractility in mice. Gastroenterology. 2005;129:1592–1601. doi: 10.1053/j.gastro.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Notani D., Gottimukkala K.P., Jayani R.S., Limaye A.S., Damle M.V., Mehta S., Purbey P.K., Joseph J., Galande S. Global regulator SATB1 recruits β-catenin and regulates TH2 differentiation in Wnt-dependent manner. PLoS Biol. 2010;8:e1000296. doi: 10.1371/journal.pbio.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J., Paul W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4 + T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X., Zhang R., Yang J., Li Q., Qin L., Zhu C., Liu J., Ning H., Shin M.S., Gupta M. Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat. Commun. 2011;2:314. doi: 10.1038/ncomms1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesu M., Watford W.T., Wei L., Xu L., Fuss I., Strober W., Andersson J., Shevach E.M., Quezado M., Bouladoux N. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008;455:246–250. doi: 10.1038/nature07210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishesha N., Thiru P., Shi J., Eng J.C., Sankaran V.G., Lodish H.F. Transcriptional divergence and conservation of human and mouse erythropoiesis. Proc. Natl. Acad. Sci. U S A. 2014;111:4103–4108. doi: 10.1073/pnas.1401598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramgolam V.S., Sha Y., Jin J., Zhng X., Markovic-Plese S. IFN-beta inhibits human Th17 cell differentiation. J. Immunol. 2009;183:5418–5427. doi: 10.4049/jimmunol.0803227. [DOI] [PubMed] [Google Scholar]
- Rautajoki K.J., Marttila E.M., Nyman T.A., Lahesmaa R. Interleukin-4 inhibits caspase-3 by regulating several proteins in the Fas pathway during initial stages of human T helper 2 cell differentiation. Mol. Cell. Proteomics. 2007;6:238–251. doi: 10.1074/mcp.M600290-MCP200. [DOI] [PubMed] [Google Scholar]
- Riaz T., Sollid L.M., Olsen I., de Souza G.A. Quantitative proteomics of gut-derived Th1 and Th1/Th17 clones reveal the presence of CD28+ NKG2D- Th1 cytotoxic CD4+ T cells. Mol. Cell. Proteomics. 2016;15:1007–1016. doi: 10.1074/mcp.M115.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C., Heath H., Krpic S., Dingjan G.M., van Hamburg J.P., Bergen I., van de Nobelen S., Sleutels F., Grosveld F., Galjart N., Hendriks R.W. Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J. Immunol. 2009;182:999–1010. doi: 10.4049/jimmunol.182.2.999. [DOI] [PubMed] [Google Scholar]
- Salonen J., Rönnholm G., Kalkkinen N., Vihinen M. Proteomic changes during B cell maturation: 2D-DIGE approach. PLoS One. 2013;8:e77894. doi: 10.1371/journal.pone.0077894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santori F.R., Huang P., van de Pavert S.A., Douglass E.F., Jr., Leaver D.J., Haubrich B.A., Keber R., Lorbek G., Konijn T., Rosales B.N. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. 2015;21:286–297. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiter M., Lau U., van Ham M., Bulitta B., Gröbe L., Garritsen H., Klawonn F., König S., Jänsch L. Proteome analysis of distinct developmental stages of human natural killer (NK) cells. Mol. Cell. Proteomics. 2013;12:1099–1114. doi: 10.1074/mcp.M112.024596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Marabita F., Kiani N.A., Gross C.C., Johansson H.J., Éliás S., Rautio S., Eriksson M., Fernandes S.J., Silberberg G. Time-resolved transcriptome and proteome landscape of human regulatory T cell (Treg) differentiation reveals novel regulators of FOXP3. BMC Biol. 2018;16:47. doi: 10.1186/s12915-018-0518-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze N., Graessl M., Blancke Soares A., Geyer M., Dehmelt L., Nalbant P. FHOD1 regulates stress fiber organization by controlling the dynamics of transverse arcs and dorsal fibers. J. Cell Sci. 2014;127:1379–1393. doi: 10.1242/jcs.134627. [DOI] [PubMed] [Google Scholar]
- Schwanhüusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shay T., Jojic V., Zuk O., Rothamel K., Puyraimond-Zemmour D., Feng T., Wakamatsu E., Benoist C., Koller D., Regev A., ImmGen Consortium Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl. Acad. Sci. U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington M.J.T., Mahata B., Svensson V., Deonarine A., Nissen J.K., Betz A.G., Teichmann S.A. An atlas of mouse CD4+ T cell transcriptomes. Biol. Direct. 2015;10:14. doi: 10.1186/s13062-015-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Fu J., Zhou Y. Metabolism controls the balance of Th17/T-regulatory cells. Front. Immunol. 2017;8:1632. doi: 10.3389/fimmu.2017.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Yamada H., Shibata K., Muta H., Tani K., Podack E.R., Yoshikai Y. CD30 ligand/CD30 plays a critical role in Th17 differentiation in mice. J. Immunol. 2010;185:2222–2230. doi: 10.4049/jimmunol.1000024. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Okuno T., Yamamoto M., Pasterkamp R.J., Takegahara N., Takamatsu H., Kitao T., Takagi J., Rennert P.D., Kolodkin A.L. Semaphorin 7A initiates T-cell-mediated inflammatory responses through α1β1 integrin. Nature. 2007;446:680–684. doi: 10.1038/nature05652. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Zhang X., Chopra M., Kim M.J., Buch K.R., Kong D., Jin J., Tang Y., Zhu H., Jewells V., Markovic-Plese S. The role of endogenous IFN-beta in the regulation of Th17 responses in patients with relapsing-remitting multiple sclerosis. J. Immunol. 2014;192:5610–5617. doi: 10.4049/jimmunol.1302580. [DOI] [PubMed] [Google Scholar]
- Tomazella G.G., da Silva I., Laure H.J., Rosa J.C., Chammas R., Wiker H.G., de Souza G.A., Greene L.J. Proteomic analysis of total cellular proteins of human neutrophils. Proteome Sci. 2009;7:32. doi: 10.1186/1477-5956-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomela S., Salo V., Tripathi S.K., Chen Z., Laurila K., Gupta B., Äijö T., Oikari L., Stockinger B., Lähdesmäki H., Lahesmaa R. Identification of early gene expression changes during human Th17 cell differentiation. Blood. 2012;119:e151–e160. doi: 10.1182/blood-2012-01-407528. [DOI] [PubMed] [Google Scholar]
- Tuomela S., Rautio S., Ahlfors H., Öling V., Salo V., Ullah U., Chen Z., Hämälistö S., Tripathi S.K., Äijö T. Comparative analysis of human and mouse transcriptomes of Th17 cell priming. Oncotarget. 2016;7:13416–13428. doi: 10.18632/oncotarget.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M., Hirota K., Westendorf A.M., Buer J., Dumoutier L., Renauld J.C., Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yosef N., Gaublomme J., Wu C., Lee Y., Clish C.B., Kaminski J., Xiao S., Meyer Zu Horste G., Pawlak M. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell. 2015;163:1413–1427. doi: 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes M.P., Tomasec P., Huttlin E.L., Fielding C.A., Nusinow D., Stanton R.J., Wang E.C., Aicheler R., Murrell I., Wilkinson G.W. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.Y., Hatfield J.K., Brown M.A. Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T Cell Development. J. Biol. Chem. 2013;288:35170–35179. doi: 10.1074/jbc.M113.481440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worah K., Mathan T.S.M., Vu Manh T.P., Keerthikumar S., Schreibelt G., Tel J., Duiveman-de Boer T., Sköld A.E., van Spriel A.B., de Vries I.J.M. Proteomics of human dendritic cell subsets reveals subset-specific surface markers and differential inflammasome function. Cell Rep. 2016;16:2953–2966. doi: 10.1016/j.celrep.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang Y., Qiu G., Lal G., Yin N., Wu Z., Bromberg J.S., Ding Y. Stat4 is critical for the balance between Th17 cells and regulatory T cells in colitis. J. Immunol. 2011;186:6597–6606. doi: 10.4049/jimmunol.1004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Wang Y., Liu X., Li L., Opejin A., Hsueh E.C., Luo H., Wang T., Hawiger D., Peng G. TLR7 signaling regulates Th17 cells and autoimmunity: novel potential for autoimmune therapy. J. Immunol. 2017;199:941–954. doi: 10.4049/jimmunol.1601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N., Shalek A.K., Gaublomme J.T., Jin H., Lee Y., Awasthi A., Wu C., Karwacz K., Xiao S., Jorgolli M. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.E., Flaherty S., Woodman K.M., Sharma-Walia N., Reynolds J.M. Fatty acid synthase regulates the pathogenicity of Th17 cells. J. Leukoc. Biol. 2017;102:1229–1235. doi: 10.1189/jlb.3AB0417-159RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
4