Abstract
Targeted genome editing is an advanced technique that enables precise modification of the nucleic acid sequences in a genome. Genome editing is typically performed using tools, such as molecular scissors, to cut a defined location in a specific gene. Genome editing has impacted various fields of biotechnology, such as agriculture; biopharmaceutical production; studies on the structure, regulation, and function of the genome; and the creation of transgenic organisms and cell lines. Although genome editing is used frequently, it has several limitations. Here, we provide an overview of well-studied genome-editing nucleases, including single-stranded oligodeoxynucleotides (ssODNs), transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and CRISPR-Cas9 RNA-guided nucleases (CRISPR-Cas9). To this end, we describe the progress toward editable nuclease-based therapies and discuss the minimization of off-target mutagenesis. Future prospects of this challenging scientific field are also discussed.
Keywords: genome editing, nucleases, DSB, NHEJ, HDR, ZFNs, TALENs, CRISPR-Cas9, ssODNs, off-target mutagenesis
Main Text
In modern biotechnological and medical research, tools for precise and predetermined genome modification are used to identify genes with diverse functionality. To study the functions of genes, small interfering RNAs (siRNAs) are widely used to suppress gene expression. However, the use of siRNA-mediated knockdown is questionable because its threshold success rate is below 70%.1 This method is hindered by several issues, including low effectiveness of engineered constructs at the chromosomal target, time-consuming processing, labor-intensive selection criteria, and possible undesirable mutagenic effects.2 Although siRNA is useful to study gene function, the knockdown deficiency often occurs. siRNA-mediated knockdown offers temporary inhibition of gene function, which limits our ability to correlate phenotype and genotype. In the early stages of molecular biotechnology, genes were categorized by features of their mutant phenotype. Subsequent development of genome editing is more advantageous than the siRNA technology for assessing the function of a gene or genotype.2, 3
Currently, in addition to siRNA technology, modified DNA-binding proteins, such as zinc-finger proteins and transcription activator-like effectors (TALEs), have been widely used for sequencing target DNA and for gene regulation.3, 4, 5 In modern biotechnological research, the methods for gene modification primarily use three types of endonucleases: zinc-finger nucleases (ZFNs), TALENs, and CRISPR-Cas9. Recently, the use of CRISPR-Cas9 has increased tremendously compared with that of other endonucleases (e.g., ZFNs and TALENs); the latter are not used as frequently because of several drawbacks, such as single-site targeting, occurrence of nonspecific mutations, and low efficiency.6, 7, 8, 9, 10 Single-stranded nucleotide sequences with a complementary sequence of approximately 20 pairs have been designed to target a desired region in the genome.11, 12 These three types of endonucleases are based on similar mechanisms in modification of the genome, cleaving chromosomal DNA in a specific location for targeted alteration of the genome.13
Site-specific programmable nucleases can generate DNA double-strand breaks (DSBs),14 which can lead to 2-fold increases in homologous recombination.15 Therefore, site-specific programmable nucleases can be employed in targeted mutagenesis.16 There are two main mechanisms underlying DSB repair: non-homologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ, the principal DNA repair mechanism found in eukaryotes, repairs DSBs by ligating the broken ends of DNA. This ligation is mediated by specific protein factors that re-connect the strand without a homologous DNA template.17, 18 There are two major pathways in NHEJ: (1) direct ligation of two ends of the targeted DNA, or (2) end-joining via deletion or insertion of nucleic acids. In both of these events, the occurrence of NHEJ-mediated deletions and mutations are well documented.19, 20 In contrast, HDR can also be implemented to repair DSBs, but requires a homologous DNA template (Figure 1).21, 22, 23, 24 These mechanisms, however, cannot be used to implement a gene knockin into non-dividing cells.25 Hence, a CRISPR-Cas9-based method, known as homology-independent targeted integration (HITI), was developed for this purpose. Because HITI does not require a homologous arm, it has a recognition site for flanking Cas9 so that Cas9 can cut both target and donor sequences. The donor then integrates in the genome and repairs DSBs in an independent pathway similar to that of NHEJ. This technique has allowed integration of a gene into non-dividing cells with only 2.1% insertion in the opposite direction.26
Figure 1.
Genome Engineering Using Programmable Nucleases
Zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR-Cas9 are used to induce targeted double-strand breaks (DSBs) at the desired chromosomal locus. Non-homologous end joining (NHEJ) or homology-directed repair (HDR), one of the two cellular repair pathways, is then used to repair the DSB. NHEJ can be used to knock out genes, whereas HDR can be used either for gene correction or to introduce precise alterations into the genome; this is directed by a homologous DNA template. Adapted from Chandrasegaran and Carroll22 and Ramalingam et al.,23 copyright (2015) Creative Commons Attribution 4.0 International.
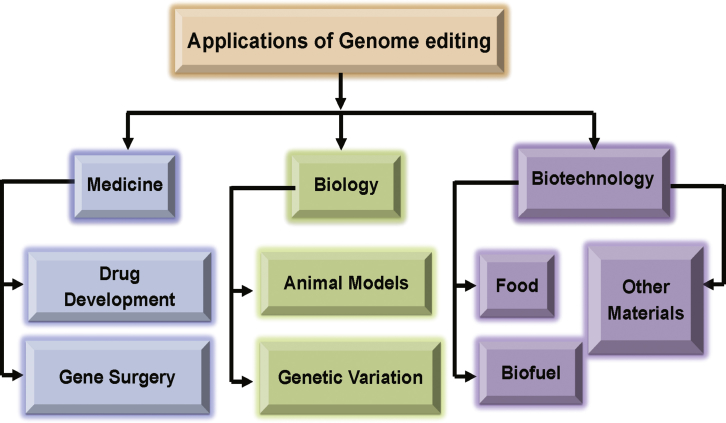
In early 2012, CRISPR-Cas9 nucleases were developed as a tool for modifying the genomes of various cell lines and those of several living organisms.27 For DNA modification, CRISPR-Cas9 systems outperform the TALEN and ZFN systems.10, 28, 29, 30 In addition to its editing capability, the CRISPR-Cas9 system possesses curative potential31 and can be used to correct gene mutations in various genetic diseases such as β-thalassemia, cystic fibrosis, hemophilia, Duchenne muscular dystrophy, and hereditary tyrosinemia type I.32, 33, 34, 35, 36, 37, 38, 39 Systems based on ZFNs, TALENs, and CRISPR-Cas9 have been used to alter the genomes of plants, animals, and even humans. These tools are now mainly used to alter the desired sequences in specific genes and have impacted numerous fields, such as biopharmaceutical development; gene surgery; alteration, regulation, and function of genome structure; and production of biofuel, food, and transgenic cell lines and animals (Figure 2). Although commonly used for genome alteration, these tools still have numerous issues.
Figure 2.
An Overview of Applications of Genome Editing
Genome editing can be used in various fields of biotechnology, including biopharmaceutical development and gene therapy; genome structure alteration, regulation, and function; and production of biofuel, food, and transgenic cell lines and animals.
In this review, we provide an overview of common genome-editing nucleases including ZFNs, TALENs, CRISPR-Cas9, and single-stranded oligonucleotides (ssODNs). These tools are used for genome editing in various fields of cellular and molecular biotechnology, including genome alteration in cells and embryos, development of biocompatible drugs, targeting of genetic diseases, and development of traits in plants or farm animals. Finally, we discuss the prospect of using genome editing for editable nuclease-based therapies and for limiting off-target mutagenesis. We also discuss the future prospects of these technologies, including their use in genome editing and modeling specific diseases.
Origin of Programmable Nucleases
Although double-strand breaks in the DNA cause severe fatal effects in cells, DNA breaks can be repaired by NHEJ and HDR.15 Gene targeting by HDR is not an efficient mechanism in higher eukaryotes. One in a million treated cells may undergo this kind of genome modification.21 Molecular scissors, such as restriction endonucleases, cannot induce a DSB at a targeted chromosomal location. This is because they can only identify very short DNA templates of 4–8 bp. Inducing a targeted DSB is a major problem when employing HDR technology for genome engineering of eukaryotic cells.22 This led to the development of a generalized delivery system to target genomic DSB at a distinctive chromosomal locus and to help stimulate HDR.22
Numerous options, such as programmable nucleases, ZFNs, TALENs, and RNA-guided CRISPR-Cas9, have been used to induce a targeted DSB. These programmable nucleases possess several common features, which can be used in the editing of eukaryotic genomes. Using a rare-cutting endonuclease, DNA DSBs can be repaired via homologous recombination, which can improve the repair by 2- to 3-fold efficiency in the targeted region. Conversely, DSBs can also be repaired by NHEJ in the absence of a gene-marking vector or homologous donor DNA.16, 40 Programmable nucleases for NHEJ- or HDR-mediated repair of site-specific DSBs are commonly used for targeted genetic modifications, gene disruption, gene insertion, gene correction with point mutagenesis, and chromosomal rearrangements.13, 41
Designed Nucleases
ZNFs
Zinc-finger (ZF) motifs were first described by Klug et al.42 as zinc-binding domains in transcription factor IIIA in Xenopus oocytes; ZFs occur more frequently in eukaryotes than in prokaryotes.42 Every ZF is composed of approximately 30 amino acids in a conserved ββα configuration, where the zinc atom is attached to two pairs of cysteine and histidine residues. Furthermore, each ZF is combined with DNA via an α-helix introduced into the main channel of the DNA double helix, and by a recognized 3- to 4-bp sequence. Generally, most ZFs are attached to a 3-bp target template and then change the recognition site to a 4-bp cross-strand. This mechanism influences the specificity of neighboring ZFs, thereby complicating the generation of ZF proteins (ZFPs). ZFP generation is mediated by a simple modular design because all ZFs identify a triplet sequence (Figure 3A).22 To generate DNA-binding ZFPs, three to six ZFs are linked together in a tandem sequence (Figure 3A).22 ZFPs are generally composed of tandem arrays of C2H2 zinc fingers, which are mainly responsible for sequence specificity of ZFNs.43 Altering DNA-binding specificities of ZFNs is an important feature in construction of a programmable nuclease; this is due to mutagenesis that can occur in ZFNs.44, 45
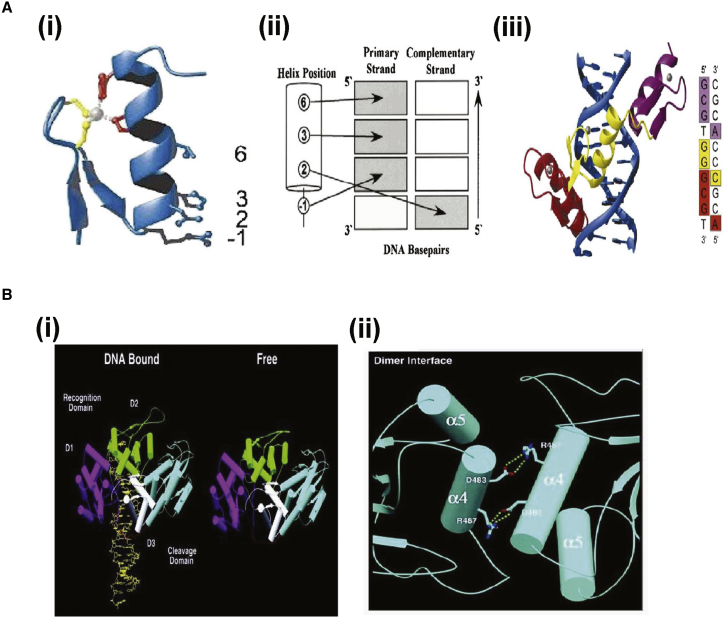
Figure 3.
Illustration of DNA Recognition by ZFPs and Crystal Structures of FokI and How FokI Bound to DNA
(A) DNA recognition by ZFPs. (Ai) Structure of a single ZFN, (Aii) DNA recognition by ZFNs, and (Aiii) structure of three-finger Zif268 bound to its cognate site. (B) Crystal structures of FokI and FokI bound to DNA. (Bi) Structures of FokI-DNA complex and of FokI enzyme alone. In both structures, the FokI cleavage domain piggybacks on the recognition domain. (Bii) Native FokI crystallizes as a dimer. The dimer interface is at the FokI nuclease domain, which is formed by two salt bridges between arginine (R) and aspartic acid (D) residues of the FokI monomers. Reproduced from Chandrasegaran and Carroll,22 Wah et al.,48 Miller et al.,300 and Pavletich et al.,301 copyright (2015) Creative Commons Attribution 4.0 International.
ZFNs are generally composed of two important domains: the DNA-binding ZF protein (ZFP) domain and the nuclease domain derivative from the FokI restriction enzyme.46 A type IIS restriction enzyme, FokI is usually used to identify non-palindromic pentadeoxy-ribonucleotide in the DNA double helix. FokI cleaves the nucleotides downstream of the recognition site. The functionality of FokI is regulated by two distinguishable protein domains: (1) domain FR and (2) domain FN (Figure 3B).22 The former is attached to a specific recognition site in the DNA to help propagate a signal to the latter via allosteric interaction. Upon receiving the signal, the latter activates the endonuclease activity of FokI after which the final cleavage is induced (Figure 3B).22 The modular characteristics of FokI endonuclease were confirmed by crystal structures of local FokI and DNA-binding FokI; the cleavage domain was isolated from the recognition domain to prevent any contact with DNA.47, 48 Nevertheless, ZFNs produced by traditional methods often showed limited DNA targeting activity and cytotoxicity when off target.49, 50 Studies on ZFNs have confirmed that, on average, a single functional ZFN pair can be generated per 100-bp DNA sequence,51 implying that ZFNs may allow efficient genome editing.
Different types of computer-based programs are currently available for searching possible ZFN target sites in a certain DNA sequence. Among such programs, PROGNOS (http://bao.rice.edu/Research/BioinformaticTools/prognos.html) and ZiFiT (http://zifit.partners.org/ZiFiT/) are most commonly used.52 In addition, EENdb (http://eendb.cbi.pku.edu.cn/) can be used along with other databases.52
TALENs
TALENs are simple modular codes for DNA recognition processed by the TALE proteins. TALEs are naturally occurring proteins commonly found in Xanthomonas, a pathogenic bacterium that infects plants.53, 54 TALENs can act as a versatile platform for programmable DNA-binding proteins. There are several similarities between TALENs and ZFNs. For example, similar to ZFNs, a FokI nuclease domain is also found in TALENs. Moreover, TALENs utilize different types of DNA-binding domains, mostly isolated from Xanthomonas spp.13 The central domain of a TALE consists of a repeating unit composed of approximately 34 amino acids. Each of these units is recognized as a single base pair. The specificity of TALEs mainly depends on the two hypervariable amino acids at the 12th and 13th positions, which are called repeat variable diresidues (RVDs).55, 56
Four different RVD amino acid diresidues (i.e., NI, HD, NG/HG, and NN) are mainly used to identify adenine (A), cytosine (C), thymine (T), and guanine (G)/adenine (A), respectively. This recognition process of TALE modules appears to function independently from neighboring modules, unlike the mechanism used by ZFPs. The presence of a DNA recognition code mainly provides close interaction between the array of amino acid repeats and the nucleotide sequence of a genome; therefore, new TALENs can be blueprinted with desired sequence specificities.57, 58 The modular character of TALE patterns combined with the simple DNA detection code makes TALENs suitable for constructing common nucleases.59 Similar to programs used to design ZFNs, web-based computer programs, such as E-TALEN (http://www.e-talen.org/E-TALEN/),60 are also available for designing TALENs.
In mammalian genomes, many types of complex TALENs can also target distinctive loci. TALENs are generally simpler to construct than are ZFNs. However, the designed sequences for encoding TALENs are much longer than those for ZFNs. The TALE and ZFN motifs are similar in size; however, TALE motifs recognize only one base, whereas ZFNs recognize 3–4 bp sequences. These consensus series of TALE motifs with highly repetitive sequences increase the complexity of TALEN-encoding genes in E. coli,22 presenting a challenge in increasing the numbers of these genes. These TALEN-encoding genes also complicate virus-mediated delivery into mammalian cells. To overcome those challenges, different types of effector domains can be combined with TALE repeats for targeted genetic alterations.
Cloning DNA segments that encode TALE arrays involves certain technical challenges posed by large similar repeat sequences, requirement of approximately 20 RVDs, and increased consumption of cost.61 Numerous techniques for rapid assembly of custom TALE arrays include: Golden Gate cloning systems,62 solid-phase assembly,59, 63 and ligation-independent cloning.64 All of these systems are currently used for high-throughput sequencing. In brief, Golden Gate cloning systems utilize a type IIS restriction enzyme for producing four-base extensions at individual DNA fragments encoding RVD modules. DNA fragments with complementary extensions can be linked to generate numerous RVD elements in a planned array.65, 66 Any DNA sequences can be targeted by TALENs, which is the main advantage of this type of nuclease over other types. Small DNA sequences can be mutated using TALENs, which cannot be accomplished by ZFNs or RNA-guided engineered nucleases (RGENs). The one limitation for the synthesis of TALENs is that the 5′ ends of binding sites, which start with thymine (T), are recognized by two amino acid terminal folds.55
To overcome this thymine-specific recognition, extensive research has focused on TALE variants. Such efforts have made it possible to help recognize bases other than thymine at the 5′ end. This reorganization process has been useful for increasing the range of sites targetable by TALENs.67, 68 The production of customized TALENs or genome-modified cell lines has been pursued actively by commercial organizations such as Cellectis Bioresearch (Paris, France), Transposagen Biopharmaceuticals (Lexington, KY, USA), and Life Technologies (Grand Island, NY, USA).3
CRISPR-Cas9
A number of in-depth reviews have covered adaptive immunity in bacteria, which involves the mechanism of adaptive resistance based on the CRISPR-Cas system.69, 70, 71, 72, 73, 74, 75, 76, 77 Bacteria and archaea commonly use this mechanism to degrade complementary sequences that are present in viral and plasmid DNA.78 These organisms generally capture DNA fragments of approximately 20 bp from viruses and plasmids, which facilitates the formation of a CRISPR sequence. These sequences, referred to as protospacers, can be inserted into their own genome. Pre-CRISPR RNA is transcribed from CRISPR regions of type II CRISPR systems belonging to two major classes. Pre-CRISPR RNA is then processed to form a short CRISPR RNA (crRNA). This process is facilitated by the transcription of trans-activating crRNAs (tracrRNAs), which are combined with crRNAs.79 The crRNA-tracrRNA complex ultimately associates with Cas9 to form an active endonuclease for the degradation of foreign DNA; this active endonuclease is called dual RNA-Cas9.78 RNA-Cas9 targets a 23-bp sequence that consists of a guide sequence (20-bp) in crRNA and the 5′-NGG-3′sequence identified as the protospacer-adjacent motif (PAM).80
The crystal structure of Cas9 proteins, derived from different species, can exist alone as an inactive form, although it can become active through the binding of a single-chain guide RNA (sgRNA).81 A new RGEN-encoding plasmid can be easily prepared by cloning DNA sequences that encode either crRNA or sgRNA into a suitable vector. This process is much simpler than complicated protein-engineering methods for the synthesis of RGENs.82, 83, 84 Due to a basic design and simple preparation methods, RGENs are more suitable for targeting endogenous loci in human cells than are ZFNs and TALENs. In the case of RGENs, only RNA needs to be designed, rather than protein-engineering two nucleases to recognize the target sequence. The CRISPR-Cas9 system is more advantageous for new targets because it can target many objects simultaneously; this system has been widely used in research laboratories worldwide. Moreover, the CRISPR-Cas9 system is economical for small laboratories, making it a reasonable choice for genome engineering procedures.22 In addition to its utility in editing genomic sequences, the CRISPR-Cas9 system is a non-mutagenic gene regulation tool that is completely sequence specific. Several companies, such as Sigma-Aldrich, System Biosciences, ToolGen, and Transposagen Biopharmaceuticals, commercially offer RGENs for laboratory use.
ssODNs
Engineering the mammalian genome is a powerful genetic approach for developing novel therapeutics for treating hereditary diseases. In addition to complete gene knockout, there is also single-nucleotide exchange for re-engineering of mammalian genomes via ssODNs. This method has been used to generate nucleotide changes and often works in combination with other genome-editing tools.85, 86, 87, 88 The lower efficiency of ssODNs makes them incompatible for therapeutic application. However, adjuvants can be used to enhance the frequency of ssODNs. The overall activity of genome editing by ssODNs can be improved approximately 5- to 10-fold by inducing double-strand DNA breaks before introducing ssODNs.89, 90, 91, 92 TALENs have also been used in combination with ssODNs to exchange single nucleotides at specific sites in a gene.66, 93, 94, 95 To repair single-point mutations, combining TALENs with ssODNs is used to generate substantial changes at both genotypic and phenotypic levels.95, 96, 97
The core mechanism of single-point mutation repair involves reduction of ssODNs by TALENs, which are required for gene editing of a specific entry site provided for the oligonucleotide. An oligonucleotide with an adjacent stretch of RNA and DNA has been developed for correction of a single mutation in episomal and chromosomal targets in mammalian cells.98, 99 This type of oligonucleotide is capable of correcting a site-specific mutation in tissue-cultured cells when administered in vivo. This stable and steady process of gene correction, mediated by an RNA-DNA oligonucleotide, was established by clonal analysis at the sequence level.100 This RNA-DNA oligonucleotide may be a promising curative approach for genetic diseases. To inhibit the expression of a desired gene, ODNs can be used as antisense oligonucleotides that hybridize with target mRNAs complementary to the sequence of the antisense oligonucleotide.101
In mammalian cells, a 40-nt ssODN is sufficient for genetic modification.102, 103 In yeast, however, 20–70 bases are needed to induce modifications. A plasmid vector, bearing a mutant neomycin phosphotransferase (NPT) gene, can be co-transfected with a 40-base single-stranded oligomer that comprises the sequence of wild-type NPT in mammalian cells. Cells containing NTP are selected by G418 antibiotic screening. These cells contain wild-type DNA molecules that result from recombination between two DNA molecules. This method is advantageous in altering or introducing point mutations into the genomes of human cells.102 In yeast, sense oligonucleotides generate nearly 20-fold more transformants than do antisense oligonucleotides. This discrepancy was confirmed with oligonucleotides designed to create modifications at six different locations along the gene. The contrary result was likely caused by the length and sequence of the oligonucleotide, number of alterations, and host strain.103 This implies that ssODNs can be used in various gene-targeting approaches including the development of transgenic animals, improvement of isogenic cell lines, site-specific mutagenesis, and gene therapy.
Applications of Genome Editing in Mammals
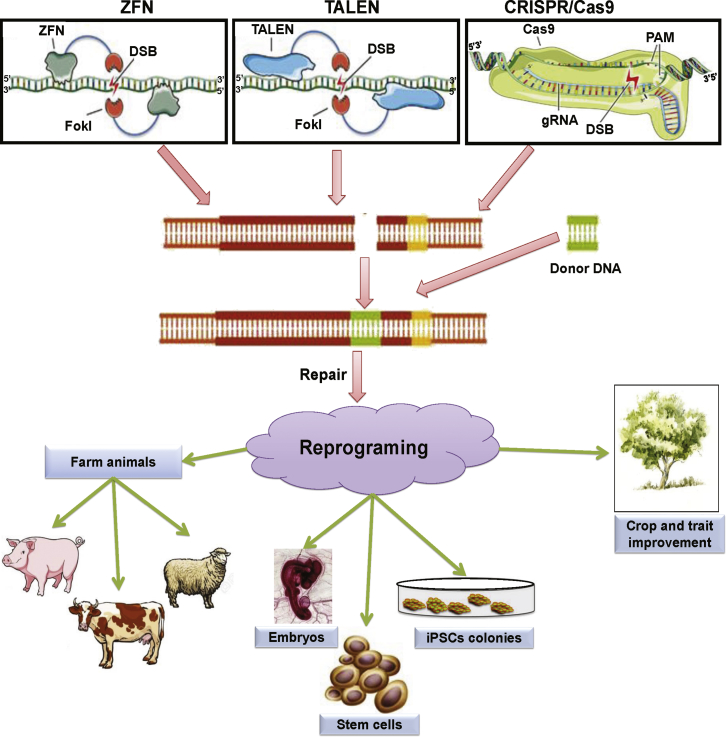
In life sciences research, biotechnologies that enable deletion, insertion, and modification of DNA sequences in cells or organisms allow us to evaluate specific gene functions. These genome-modification technologies have also enabled the examination and large-scale manipulation of genes and protein networks. In addition to enabling manipulation of transcriptional regulation at a specific locus, genome editing can reveal the mechanisms regulating genetic systems. In modern biotechnology, the underlying regulatory mechanisms of genetic building blocks are helpful for reverse engineering of useful biological systems. The concurrent modifications of multiple genes can have adverse effects that trigger complex polygenic disorders. Conversely, genome-editing technologies can be used to directly correct harmful mutations in humans.104 Potential applications of programmable nucleases in humans are discussed below and overviewed in Figure 4.
Figure 4.
An Overview of Potential Applications of Programmable Nucleases
Programmable nucleases can edit the genome and reprogram genetic information, which consequently affects genome structure and function. This technology can be used to produce farm animals, transgenic cell lines (i.e., embryos, stem cells, and induced pluripotent stem cells [iPSCs]), and transgenic plants.
Genome Editing in Farm Animals
With the rapidly growing population, there is an urgent need to develop biotechnological approaches that enhance animal production, while reducing the risks associated with climate change and/or environmental pollution. In the recent past, genotyping and whole-genome sequencing improved our ability to analyze the genetics of farm animals. Production of genetically edited pigs was a milestone in genome manipulation of farm animals.105 In recent years, genome editing has been performed via deletion, addition, and modification of base pairs at targeted loci. All of these genomic alterations are permanent and heritable in the following generations of livestock. Similar to the process occurring in plant species, ZFNs, TALENs, and CRISPR-associated endonuclease Cas9 (CRISPR-Cas9) are universally used to modify gene functions via NHEJ.106, 107, 108 However, insertions and additions via NHEJ can vary in size and sequence, making screening of nonfunctional clones more difficult.109 Specific genes of interest in the genomes of cattle, sheep, and pigs have been edited to yield viable zygotes and living animals. MSTN, the gene encoding myostatin, can generate more extensive muscling in cattle, sheep, and pigs, whereas the POLLED allele in cattle is commonly targeted for editing along with protein-encoding genes such as RELA.110, 111, 112, 113, 114, 115 Editing of the CD163 gene for resistance to the porcine reproductive and respiratory syndrome (PRRS) in pigs is a recent achievement and has been used extensively in the production of PRRS-resistant pigs.116, 117
Using ZFN for genome editing begins with the introduction of plasmid DNA or mRNA encoding ZFN into the target cells or embryos via microinjection or transfection.118 Then the translated ZFN protein binds to its specific target sequence and cleaves the target DNA by activating FokI nucleases at 30°C. The binding of ZFN to targeted loci is slow at a lower temperature, which affects cell-cycle progression.119 Insertion of ZFN-encoding plasmid DNA or mRNA can potentially cause constant transcription, leading to nonspecific DNA cleavage. In the case of ZFN-encoding plasmid DNA, a temporary transfection protocol can be used to dilute the plasmid DNA; this is a major advantage of using ZFN. Alternatively, microinjection of ZFN-encoding mRNA is more accurate than transfecting with ZFN-encoding plasmid DNA.120 Thus, microinjection of ZFN-encoding mRNA can reduce the risk for permanent assimilation of ZFNs, making ZFNs more efficient genome-editing tools than the conventional approach.
Recently, transgenic pigs have been used as alternatives to mouse models for evaluating human diseases and developing therapies; this is because symptoms of classical diseases in mice do not fully mimic those in humans. Pigs are more suitable models for investigating human diseases, such as cystic fibrosis, diabetes, and cancer, because pigs possess human-like genetic, anatomical, and physiological characteristics.121, 122 Pigs are also vital organ donors in generating xenografts of human organs.123 Knockout pigs, engineered using ZFNs, generally carry a hemizygous transgenic EGFP reporter allele. The endogenous porcine gene (such as peroxisome proliferators activated receptor-γ [PPAR-γ]) was the first gene successfully targeted by ZFN. These types of knockout pigs are useful in research on cardiovascular diseases.124 Biallelic knockout live pigs are produced by using ZFNs to target an endogenous gene.125 Furthermore, cattle with knocked out β-lactoglobulin (BLG), a major milk serum protein and allergen, have been produced via gene targeting with ZFN.126, 127
For knockout of the BLG gene, bovine fetal fibroblasts are transfected with ZFN mRNAs. Successful cell transfection is confirmed using the T7 endonuclease PCR to show that approximately 15% of the cells carry a mutated variant, whereas 3% are positive for the biallelic BLG gene knockout. The mutated BLG gene is also verified to detect the existence of any off-target mutagenesis. These studies indicate that specific genome editing with ZFNs in higher domestic animals results in less off-target mutagenesis than do other similar approaches.126
The Gram-negative bacterium Xanthomonas is a plant pathogen that naturally produces TALEs. Xanthomonas infects numerous plant species such as rice, citrus, cotton, pepper, tomato, and soybeans.128 In cattle, two important genes in fibroblasts, viz. ACAN and GDF8, can be modified using TALENs. ACAN is responsible for congenital achondroplasia, whereas GDF8 (growth differentiation factor 8/MSTN) acts as a controller of muscular growth. Bovine fibroblasts bearing the GDF8 gene showed approximately 29% modifications when treated with TALENs.101 Conversely, the ACAN gene showed 77% modification after treatment with TALENs.101 To produce live progeny with desirable genetic modifications, customized cells are used for somatic cell nuclear transfer.110 Recently, TALENs were used to generate a porcine model of hypercholesterolemia by modifying the LDL receptor gene. TALEN-based alteration of the porcine DMD gene was also developed for the model of Duchenne muscular dystrophy (DMD).110 To improve disease resistance in pigs, 20 ng/μL TALEN mRNA was recently introduced into porcine zygotes to target the RELA gene. Sixteen out of 56 porcine embryos were successfully transformed. The RELA gene is involved in tolerance to infection with the African swine fever virus.129 Successful transformation was confirmed by DNA sequence analysis. Among the mutants, one-third was either homozygous or heterozygous. The genome sequences of the MSTN locus in both cattle and sheep show a high level of similarity; hence the same TALENs can be used in sheep and cattle.130 To generate live progeny, TALEN mRNA was microinjected into ovine zygotes and then transferred into recipient ewes. After successful pregnancy and delivery, fewer offspring were found to have the heterozygous gene.130
The specificity and efficacy of genome editing via CRISPR-Cas are similar to those achieved using ZFNs and TALENs. The von Willebrand factor (vWF) gene in pigs has been targeted by injecting Cas9 mRNA and sgRNA into zygotes. However, premature embryonic development was not affected by this process. After a successful pregnancy and delivery, the survival rate of piglets was approximately 88%. In-vivo piglet fetal development was not hampered by this genome modification.131 Other genes, such as the p65 and the adenomatous polyposis coli (APC) gene loci, have also been targeted in porcine fetal fibroblasts by the CRISPR-Cas9 system.132 All of these endonucleases act as valuable gene-editing tools that are revolutionizing biological research and molecular medicine. To further expand this technology, we need to investigate model organisms and farm animals, develop biomedical models, and alter genes for the treatment of genetic diseases.
Genome Editing in Embryos
All the discussed programmable nucleases can also be used as genome-editing tools for human embryos. Altering genes in somatic cells at various clinical phases can be helpful in developing potential therapeutic applications. In recent years, CRISPR-Cas9 was used as a therapeutic tool to prevent the onset of β-thalassemia (a fatal blood disorder) by eliminating mutated human β-globulin (HBB) gene, which is responsible for the onset of β-thalassemia.133 Nonetheless, this approach was not successful because of the ethical issues raised by the scientific community. To escape the ethical issues, it was decided to use “non-viable” embryos, which were tripronuclear zygotes generated by fertilization with two sperms. These non-viable embryos are commonly discarded in clinics.134
In vitro fertilization may generate approximately 2%–5% polyspermic zygotes, which can produce blastocysts; however, this does not manifest in the in-vivo process.135 Polyspermic zygotes are a model for assessing the target efficacy and off-target effects of CRISPR-Cas9.136, 137 A subunit of adult human hemoglobin is coded by the β-globin (HBB) gene, which is mutated in β-thalassemia.138 The location of the HBB gene on chromosome 11, within the β-globin gene group, consists of four other globin genes: HBE, HBG2, HBG1, and HBD.139 Three designed guide RNAs (gRNAs; G1, G2, and G3) can be transfected into human cells to target various sections of the HBB gene; in 29 independent clones, specific editing of the HBB gene has been observed.134 Genome editing in germline cells or early embryos can offer an opportunity for the treatment of genetic diseases. Recently, the naturally occurring C-C chemokine receptor type 5 (CCR5Δ32) allele was successfully introduced into early human three pro-nuclei (3PN) embryos using the CRISPR-Cas system. However, the efficiency of HDR was low in the CCR5Δ32 allele.140 The CCR5Δ32 gene was chosen because this gene encodes the major co-receptor used by the HIV-1 virus in targeting human immune cells.141 People with this allele show slow development of resistance to HIV infections.142, 143 Subsequent procedures with premature human 3PN embryos generated 3PN embryos with the altered CCR5Δ32 allele. Conversely, ZFNs have been used without a specific rat gene for the production of knockout rats.144 In this process, the GFP, immunoglobin M, and RAB38 genes were targeted successfully to achieve the complete knockout of GFP, immunoglobin M, and Rab38 transgenes without any cleavage at off-target sites.145
Modifications of a genome, if introduced via its editing, can be heritable. Heritable alteration of the genome has been reported in rats after administering ZFN into one-celled embryos.146 Several studies have generated multiple germline mutations in mice that turned out to aid in the development of genetically engineered animals. In mice, two loci, such as TET1 and TET2, can be targeted through the modifications of CRISPR-Cas9. Treated parent mice have successfully transmitted this modification to the next generation when confirmed by DNA sequencing.147, 148
The genome-editing methods used in mammalian cells are not completely similar to those used in model organisms such as zebrafish. The Cas9/gRNA system has been successfully used in embryos to execute site-specific cleavage in zebrafish. The successful exogenous insertion of a foreign DNA fragment was then achieved as a donor DNA was provided during the experiment. DSBs induced by the Cas9 nuclease generate biallelic conversion of the ETSRP or GATA5 gene in treated tissues. Microinjecting Cas9 mRNA and gRNA into zebrafish embryos leads to insertion of site-specific 1–24 bp or deletion of 7–32 bp. Cas9/gRNA has produced biallelic conversion of ETSRP, GATA4, and GATA5 genes in targeted somatic cells.149 In another study on zebrafish, the CRISPR-Cas system was used to target one hemizygous EGFP reporter gene and four endogenous loci (TYR, GOL, MITFA, and DDX19).150 The resulting mutation rate of 75%–99% indicates that in most cells, the biallelic gene was disrupted. The five genomic loci in embryos were efficiently modified by the biallelic gene to show diverse phenotypes. CRISPR-Cas-induced mutations were highly specific in somatic tissues; additionally, germline transmission of these mutations has also been confirmed in the first generation.150
Genome Editing in Stem Cells and iPSCs
Research on human induced pluripotent stem cells (iPSCs) and human embryonic stem cells (ESCs) has advanced the study of human genetics and cell-based therapies. Pluripotent stem cells can self-renew and undergo unlimited division.151 These properties make them suitable for cloning and genotyping. Genome-modification tools have been successfully used to introduce highly specific alterations into iPSCs, rendering them distinguishable from background noise; this allows assessment of numerous developmental and disease-associated characteristics. Genome editing in human iPSCs aids in the production of synthetic organs and regenerative medicine, as well as in gene therapies.152 Therefore, interest in modified human iPSCs has increased because of their potential use in adapted cell therapy.151, 153
To study the gene functions in mice, researchers generally target ESCs via homologous recombination (HR); however, similar to HDR, HR was also unsuccessful in human ESCs.154 ESCs differ between humans and mice, likely reflecting differences in DNA repair processes. Although both ZFNs and TALENs have been shown to be successful in editing iPSC genomes, cloning using CRISPR-Cas remains the most straightforward technique. Despite the successful achievement in targeting and modification of genes, isolation of edited iPSC clones was difficult and expensive. However, genome modification in treated human iPSCs is currently easily demonstrated by microscopy or flow cytometry. After modified iPSCs have been identified and genotyped, assessment should be made further for their pluripotency (extracellular and intracellular) and any chromosomal abnormalities.153 Both human ESCs and iPSCs will likely play important roles in developing new technologies for modifications of the human genome.151 These technologies can also aid in engineering dendritic-cell-directed cancer vaccines, T cell immunotherapy, and experimental biology. Moreover, modifiable human ESCs and iPSCs can be used to create human cell lines for increased production of biomolecules, which have numerous industrial applications.155, 156
Human monogenic diseases (e.g., sickle cell anemia, spinal muscular atrophy, and X-linked adrenoleukodystrophy) can be easily treated by gene complementation or alteration. Recently, several studies on human monogenic diseases have successfully used genome-editing tools such as ZFNs, TALENs, and CRISPR-Cas9.155, 156, 157, 158, 159 Several gene therapy approaches have been developed and used for effective treatment of diseases such as X-linked severe combined immunodeficiency (X-SCID), Wiskott-Aldrich syndrome (WAS), and adenosine deaminase deficiency (ADA).153 Ex-vivo gene complementation of retroviral-mediated gene therapy, which targets hematopoietic stem cells, has been used to permanently correct X-SCID-related abnormalities.160 In an infection with the HIV-1 virus, CCR5 acts as a chemokine receptor that facilitates viral entrance into the host cell. Deletion of 32 bp at the coding region of this receptor renders it nonfunctional and consequently resistant to HIV-1 infectivity.143
In other studies on HIV-1 infection in T lymphocytes or bone marrow hematopoietic stem cells, modification of the CCR5 receptor was achieved using ZFNs in CD4+ T cells.157 This modification eventually activates NHEJ machinery, resulting in random mutations. Enforced expression of the four transcription factors, OCT4, SOX2, KLF4, and c-MYC, in patient-derived fibroblasts led to the development of disease-specific human iPSC lines. Specific demarcation of such iPSCs enables the analysis of diseases such as Down syndrome, also called trisomy 21.153 Modeling of Huntington’s disease (HD) has also been performed using the CRISPR-based genome-editing technology and patient-derived iPSCs.158 To create a Parkinson’s disease (PD) model, ZFNs were used to introduce two-point mutations into a particular type of α-synuclein gene of human ESCs known as A53T. This is a dominant mutation in α-synuclein as commonly observed in a PD patient. As such, it is crucial evidence to acknowledge the sporadic form of the PD or less familial pattern of molecular pathogenesis.161 However, successful alteration of 1 bp restored the A53T mutation in patient-derived iPSCs without affecting the rest of the genome.161 Several similar genome-editing studies have been performed in iPSCs.153 In one such study, a genome-editing tool was used to correct or disrupt a gene via point mutation.153 These studies have also generated novel models for diseases such as Barth syndrome, HIV, and β-thalassemia.153 Moreover, innovative research has been conducted with artificial nucleases. Most of the prior work has been aimed at increasing the efficiency of gene editing and developing a highly modified iPSC/ESC line.153 The revolutionary research on this approach formed a TALEN-mediated Cas9-inducible human ESC line. Six different genes in this cell line were targeted using CRISPR, ultimately generating a double- or triple-knockout line.162 For therapeutic applications in patients with genetically linked diseases, genome editing is promising for the correction of disease-linked mutations in iPSC-derived progenitor cells. However, this approach is currently limited by the lack of well-established protocols.
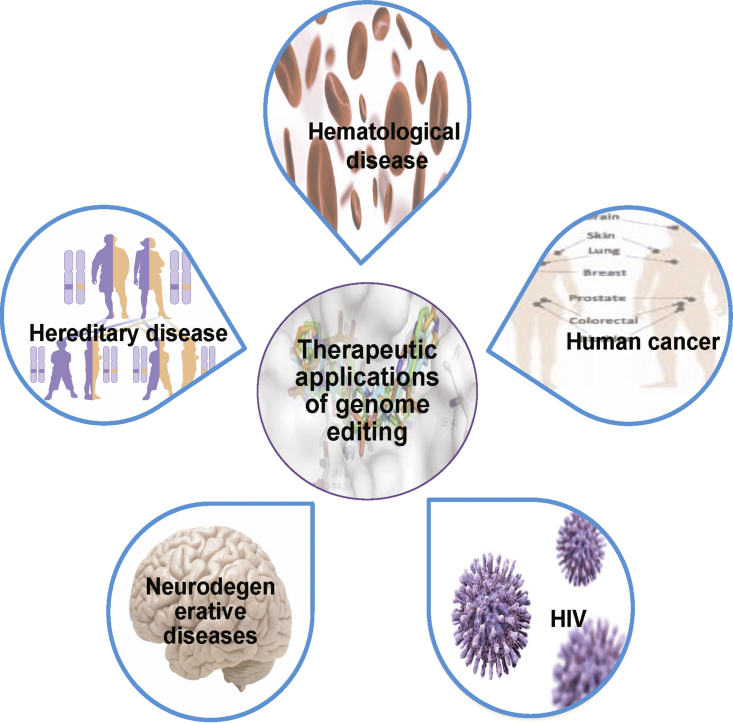
Therapeutic Applications
The development of genome editing for therapeutic applications is challenging. Consistent alterations in multiple genes can have severe effects that induce complex polygenic disorders, which can be treated using advanced genome engineering tools such as ZFNs, TALENs, and CRISPR-Cas9. The therapeutic applications, possibilities, and challenges associated with programmable nucleases are illustrated in Figure 5.
Figure 5.
Schematic Diagram of Potential Therapeutic Applications of Genome Editing
Treatment of Hematological Disease
Inherited Hematological Disorders
Gene therapy has been used for various human diseases, including hematological disease, cancer, AIDS, diabetes, heart failure, and numerous neurodegenerative diseases. More than 2,000 clinical trials using gene therapy have been carried out worldwide.163 Several gene therapy products have been approved for marketing; these include Gendicine in China, Cerepro in Europe,163 Luxterna,164 and chimeric antigen receptor-modified T cells (CAR-T cells) in the United States.165 Hematological diseases include genetic diseases and different types of malignancy. In order to develop a therapy for a genetic disease, it is necessary to identify a mechanism that can be used to overcome the effects of the causative mutation. The generation of iPSCs from patients with β-thalassemia has been performed using a non-viral method and a TALEN-based modification.166 Modification of the mutation in these iPSCs can correct the function of the HBB gene. After sequencing, the HBB gene of targeted iPSCs did not show any additional mutations. This approach can be used in the development of effective therapy against β-thalassemia.166
Similarly, the genome-editing mechanism of CRISPR-Cas9 was also used to correct the β-thalassemia mutation.167 In that study, the targeted cells showed a normal karyotype with proper pluripotency and no off-target mutations. The corrected human iPSC lines retain their expression of HBB, which suggests potential clinical application in gene therapy for β-thalassemia.167 Sickle cell disease (SCD), another common genetic condition, is caused by a homozygous mutation in the sixth codon of the HBB gene. In this mutation, glutamic acid is replaced with a valine in the amino acid sequence. The consequence is abnormal production of β-globin and faulty red blood cells.168 Recently, a ZFN-based strategy was used to repair two mutated β-globin alleles in iPSCs derived from a patient with SCD.169 However, transcription of the repaired alleles was suppressed by the co-integration of a gene cassette in the first intron.169 This issue can be resolved by using a piggyback transposon, which can facilitate the removal of this gene cassette in iPSCs without leaving unnecessary sequences.168 Two TALENs were also designed to target the HBB gene mutation in SCD.170 To correct the HBB gene, CRISPR-Cas9 technology was used in iPSCs derived from patients with SCD. This genome-editing technology relies on a donor template that includes the wild-type of HBB DNA, which shows normal activity.171
A common blood-related genetic disease, hemophilia A, is caused by several mutations in the blood coagulation factor VIII (F8) gene. Two types of chromosomal inversions that cover a fraction of the F8 gene normally result in approximately 50% of hemophilia A.172 A particular TALEN pair is used to convert a 140-kbp chromosomal segment that passes through the F8 gene in human iPSCs; this segment is responsible for hemophilia A. The modified segment can be returned to the original state using the same TALEN pair. The expression of F8 mRNA was the same as that of wild-type in modified iPSCs cell lines. The results of this study indicated that a TALEN-based genome-editing mechanism can help correct gene rearrangements that cause genetic disorders such as hemophilia A.173 Another genetic disorder, hemophilia B, is caused by the scarcity of blood coagulation factor IX, which is encoded by the F9 gene. Mutations in this gene generally occur in exons 2–8.174 Successful repair of the mutated F9 gene was attained using ZFN technology in vivo combined with a targeting vector consisting of wild-type F9 exons 2–8. The resulting gene targeting successfully repaired the hemophilia B phenotype. The repair mechanism was consistent, and neither failure nor restoration of mutation had been reported.175
The phagocyte-NADPH oxidase enzyme complex includes a subunit called phagocyte oxidase (phox), which can produce reactive oxygen species (ROS). Mutations in phox cause the immunodeficiency chronic granulomatous disease (CGD).176 The correction of X-linked CGD (X-CGD) in iPSCs was accomplished using ZFNs. In this case, ZFNs targeted a single copy of the therapeutic minigene GP91phox located at the adeno-associated virus integration site 1 (AAVS1) locus. Further studies have shown that the inserted AAVS1 alleles had no off-target insertions, and that these clones were converted to mature neutrophils with proper ROS production.177 The heterozygous mutations in the RUNX1 gene can lead to an unusual autosomal dominant disease called familial platelet disorder (FPD) and, further, to acute myeloid leukemia (AML). This gene encodes an important transcription factor that is involved in leukemogenesis. iPSCs of patients with FPD and/or AML having a RUNX1 non-sense mutation Y260X showed deficient megakaryopoietic differentiation.178 To correct the Y260X mutation, the donor vector was comprised of a cDNA sequence of exons 5–8 and two ZFNs specific for the RUNX1 gene. This mechanism can correct FPD in the iPSC line to produce a wild-type variant of the gene and restored megakaryopoietic differentiation.178
Fanconi’s anemia (FA), a genetic disorder related to the failure of the bone marrow, causes hematological and solid malignancies.179 FA is caused by a point mutation in the FA complementation group C (FANCC). This mutation can cause aberrant splicing that removes exon 4 from the FANCC gene.180 An in situ study of this gene mutation was carried out using fibroblasts derived from FA patients.181 These authors applied CRISPR-Cas technology to amend the FANCC mutation for the intervention of the FA disorder. The results of this study were satisfactory, generating a high rate of corrected clones via HDR mechanism.181 Overall, the results of these studies indicate that genome-editing technologies can potentially alleviate inborn hematological disorders.
Acquired Hematological Disorders
In acquired hematological disease, genome-editing tools can be used to examine gene function and to develop disease models. AML is a common myeloid disorder. AML can be treated using CRISPR-Cas9 to remove the C4BPB gene, which encodes the primary protein responsible for AML.28 Similarly, ZFN genome-editing technology has been used to disrupt the Tet2 catalytic domain of the TET2 gene, which is responsible for myelodysplastic syndrome in zebrafish.182 Conversely, myeloid malignant cells with multiple gene mutations can be modeled using the CRISPR-Cas9 genome-editing technology. This technology allows the production of animals with mutations in various genes. The majority of these mutations are found in cases of myeloid malignancies.172 Genome-editing technology together with transcriptomic analysis can be used as a novel approach to identify and confirm genes responsible for drug resistance in AML.183 Genome-editing technology can be used in lymphoid malignancies as a new therapeutic approach.
Adult T cell leukemia (ATL) is enormously destructive to mature human T cells. A virus known as human T cell lymphotropic virus type 1 (HTLV1) is responsible for ATL.184 Currently, there is no vaccine against HTLV1. A promising result was nonetheless obtained using ZFNs to target the HTLV1 provirus and destroy virally infected cells. The major advantage of this technology is the formation of two long terminal repeats (LTRs) for each provirus; LTRs possess enhanced binding affinity for their targets.185 ZFN technology has proven to be a promising tool to combat ATL. Burkitt’s lymphoma (BL) is a latent viral infectious disease. The causal organism of BL is a herpesvirus known as the Epstein-Barr virus (EBV). Recently, the CRISPR-Cas9 system was used to disrupt the EBV genome in a BL cell line. The results of that study indicated that CRISPR-Cas9 suppressed the activity of EBV and restored the pathway of cellular apoptosis.186 BL cell lines require MCL-1, an anti-apoptotic protein, for their functioning and survival. The deletion of this protein by CRISPR-Cas9 technology induces apoptosis in the BL cell lines.187 Genome-editing technologies can be used to achieve a better understanding of the molecular mechanisms involved in inherited and acquired hematological disorders. However, in using these technologies, we need to optimize gene delivery and avoid destructive off-target side effects.
Treatment of Hereditary Disease
Targeting a specific sequence is challenging when correcting defective genes in patients with inherited disorders. Currently, gene therapy and RNAi have shown the greatest precision in targeting specific sequences.188 This ability currently renders gene therapy and RNAi as the two most powerful therapeutic tools for the treatment of hereditary diseases. Gene therapy involves the reinstallation of omitted gene function via viral transgene expression. Optimized oligonucleotide designs showed a localized distribution with improved safety profiles. RNAi thus enabled the targeting of numerous genes in one tissue simultaneously. RNAi suppressed the expression of faulty genes by knocking down target mRNA. Gene therapy and RNAi are thus currently used to treat severe combined immunodeficiency (SCID), transthyretin-related hereditary amyloidosis, WAS, cancer, and age-related macular degeneration.189, 190, 191, 192, 193, 194, 195 However, these technologies still have limited efficacy in treating certain genetic diseases.188 To overcome these limitations, several nuclease-based genome-editing approaches have been developed and used for the potential treatment of hereditary diseases. Naturally occurring mutations can confer resistance in non-monogenic diseases such as cardiovascular disease, Alzheimer’s disease (AD), and hemoglobinopathies.34
Genome-editing technologies are efficient tools for introducing protective mutations into affected individuals. NHEJ with programmable nucleases was successfully used to inactivate the HTT gene in HD and the FGFR3 gene in achondroplasia. In both cases, natural allele integrity was maintained. Cell-culture models, currently used to study HD and AD, and to develop suitable therapies against neurodegeneration, are discussed later in this review. NHEJ-based technology was also successfully used to treat nucleotide expansion disorders such as spinocerebellar and Friedreich’s ataxia.188 HDR has also been used to convert a mutant sequence to wild-type sequence in CCR5 in HIV and PCSK9 in hypercholesterolemia.196, 197 Mouse models are used to assess the efficacy of CRISPR-Cas9-mediated in-vivo genome editing in adult animals. Tyrosinemia type I (HTI) is a lethal genetic disease caused by a mutation in the enzyme fumarylacetoacetate hydrolase (FAH).34 A point mutation of G to A in the last nucleotide of exon 8, which leads to the exclusion of exon 8 during splicing, causes the abnormality in mice and humans. The instability of the FAH protein causes the accumulation of toxic metabolites, resulting in severe liver damage.198 To correct the G-to-A splicing mutation of Fah protein in the mouse model, single-strand DNA (ssDNA) was used along with unguided Cas9 technology for the treatment of hereditary tyrosinemia.34 Another example is primate DMD. DMD is caused by three different mutations in the dystrophin gene. These mutations result in the loss of dystrophin and degeneration of primate muscle, similar to the phenotype of DMD patients.199 CRISPR-Cas9 was delivered directly into the muscle cells of mice with DMD using adeno-associated viral delivery. Once delivered, CRISPR-Cas9 excised the mutated exon, generating an excised dystrophin. DMD mice that underwent this treatment showed better muscle strength without off-target effects.200, 201, 202 In summary, the CRISPR-Cas9 system can be used as an efficient tool to correct or mitigate genetic diseases.
Treatment of Cancer
Cancer is a polygenic disease caused by defects in multiple genes associated with oncogenes and tumor suppressor genes. The genomes of normal cells and cancer cells are important for modeling cancer and uncovering genes that are responsible for the overall process. Genetically engineered mouse models (GEMMs), with specific mutations in genes, oncogenes, and tumor suppressors, are used to study cancer biology.203 A phenomenon called passenger mutations, occurring directly or indirectly, aids in the conversion of normal cells into cancer cells by activating oncogenes or inactivating tumor suppressor genes. CDNA-based overexpression and RNAi-based inactivation can cause off-target effects. However, CRISPR-Cas9 produces less off-target integration and, therefore, is frequently used in cancer research. Different types of cell lines with one or more targeted mutations are suitable for exploring the effects of mutations associated with cancer phenotypes.
CRISPR-Cas9 technology also enables analysis of oncogenic signaling pathways via sequential or multiplex gene editing.203 Using GEMMs and non-germline GEMMs, researchers have uncovered the fundamental features of tumor instigation, maintenance, and evolution. Moreover, these models can be used to screen anticancer agents and identify drug-resistance mechanisms (Figure 6).204, 205 Genome-editing technology based on CRISPR allows us to generate large stockpiles of ESC lines with numerous arrangements of basic or restrictive mutations in oncogenes and tumor suppressor genes. The main advantage of the CRISPR-Cas9 system in mouse cancer models is the systematic generation of models with multiple oncogenic alleles. This property aids in assessing allele-specific tumor development and therapeutic responses. The CRISPR-Cas9 method is also used for improving existing cancer models. These next-generation models will help us to better understand and discover functional remedies against cancer.203
Figure 6.
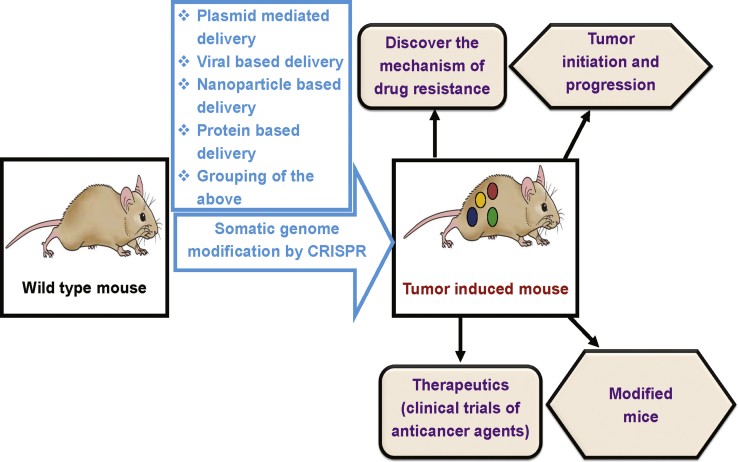
A Schematic Diagram for Production of Genetically Engineered Mouse Models Using Programmable Nuclease (CRISPR-Cas9)
Genetically engineered mouse models (GEMMs) can be used to discover mechanisms of drug resistance and those of tumor initiation and progression, and to develop new anticancer drugs.
In recent years, this system has also been used to edit the somatic genome both ex vivo and in vivo. Ex-vivo editing of the TP53 tumor suppressor gene has been performed in Eμ-Myc transgenic mice.206 A similar approach was used for ex-vivo disruption of the mixed heredity leukemia 3 tumor suppressor gene in AML.207 In another study, CRISPR-Cas9 was used to rapidly generate mouse models of AML by ex-vivo modification of single or multiple genes via lentivirus.208 These studies showed that CRISPR-Cas9 ex-vivo somatic genome editing can be used to rapidly produce mouse models of several human malignancies. Similarly, CRISPR-Cas9 was used in hepatocytes of living animals to deliver the plasmids of Cas9 and sgRNAs, respectively, targeting the PTEN and TP53 tumor suppressor genes in vivo.109 These studies show that somatic modification of cancer genes by CRISPR technology in wild-type mice can proficiently onset certain types of cancer.
For curative modification of single or multiple mutations, CRISPR-based technology offers efficient delivery and editing via viral or non-viral methods. CRISPR-Cas9 technology can be used to modify T cells and the immune response.209 Additionally, this technology may be used for specific ex-vivo manufacturing of immune cells for potential immunotherapy. For example, new CAR-T cells possess an introduced chimeric antigen receptor.210, 211 In summary, the use of CRISPR-Cas9 technology in cancer biology has allowed us to shift from fundamental research to experimental and translational applications.
Several viral infections are involved in carcinogenesis; these include: (1) hepatitis B and C viruses in liver cancer, (2) EBV in nasopharyngeal carcinoma, and (3) human papillomavirus (HPV) in cervical cancer. Inactivation of these oncogenic viruses may alleviate tumorigenesis.163 The CRISPR-Cas9 genome-editing system can be used to alleviate such carcinogenesis via defense against oncogenic viral infections. Antiviral and anti-proliferative effects were obtained using the CRISPR-Cas9 system in HPV-mediated cervical carcinoma and EBV-mediated BL cells.186, 212 In both cases, treatment with CRISPR-Cas9 inhibited proliferation of tumor cells while reducing the overall viral load.
If cancer is treated as a genetic disease, CRISPR-Cas9 can be used as a genome-editing tool to correct oncogenic abnormalities in the genome. Collaboration between genetic mutations and epigenetic alterations may cause the initiation and progression of cancer. CRISPR-Cas9 can be used to correct genetic mutations in monogenic diseases and transform the epigenetic states of a cell.163 With the help of sgRNAs, dead Cas9 (dCas9; a Cas9 should lose endonuclease activity due to point mutations at the endonuclease domains) and epigenetic modifiers can enter target sites for epigenetic regulation; this is a prospective tool in anticancer therapeutics. Recently, there has been increased interest in improving anticancer immune responses, and resistance to chemotoxicity and radiotoxicity. Cancer is polygenic and heterogeneous in nature; this presents the main obstacle in CRISPR-Cas9-mediated cancer therapy.163
Bladder cancer is a common urologic cancer that can be mitigated by traditional chemotherapy and radiation treatment. However, the limitations of these traditional treatments include: (1) mass cell killing, (2) nonspecific targeting, (3) serious side effects, and (4) numerous abnormalities of a genome.213 Recently, the CRISPR-Cas9 system was used in combination with modular AND gate circuits, human cancer-specific promoter of the telomerase reverse transcriptase gene, and a human bladder-specific promoter gene. This circuit can identify bladder cancer cells and is used to effectively treat bladder cancer.214 The CRISPR-Cas9 genome-editing system can be used to produce logic circuits. The Cas9 protein of the CRISPR-Cas9 system can be joined with sgRNA to generate an effector complex that leads to a double-strand DNA cleft.215 These circuits were shown to robustly and precisely inhibit the proliferation of carcinoma cells in the bladder; these circuits also restored apoptosis in these cells while lowering their motility.216 Chromosomal relocation plays a vital role in the expression of genes that show therapeutic activity against cancer.
In human non-small-cell lung cancer (NSCLC), the combination of two oncogenes (echinoderm microtubule-associated protein-like 4 [EML4] and anaplastic lymphoma kinase [ALK]) was shown to play an important role.216 Using CRISPR-Cas9 technology with a viral delivery method in somatic cells, researchers successfully rearranged the chromosomes of adult mammals in vivo. This process produced a mouse model of EML4-ALK-mediated lung cancer.217 These mouse models provide information about the molecular mechanisms underlying tumor formation; this can be used to assess drug resistance and the efficacy of targeted therapeutics in vivo. Conversely, CRISPR-Cas9-mediated technology for chromosomal rearrangement is limited by spaces between the excised sites and their affinity for Cas9.218 Colorectal cancer (CRC), another commonly occurring human carcinoma, is caused by several mutations in normal colon epithelium. The loss of APC gene and successive mutations in KRAS (encoding Kirsten rat sarcoma viral oncogene homolog), SMAD4 (encoding the SMAD protein), and TP53 (encoding the p53 protein) genes are mainly responsible for colorectal carcinogenesis. In a rodent model, mutations in these genes were found to induce intestinal adenocarcinoma similar to human CRC.219 Using CRISPR-Cas9 genome-editing methods, genetic alterations can be introduced into human intestinal cells.82, 215 sgRNAs and CRISPR-Cas9 can be used to target the APC, SMAD4, and TP53 tumor repressor genes. This technology has also been used to isolate single cells from human organoids and to generate mutations that recapitulate the adenoma-carcinoma sequence.219
Treatment of Neurodegenerative Diseases
The most common neurodegenerative diseases are mainly differentiated as either age-reliant or selective neurodegeneration. The pathogenesis of common neurodegenerative diseases, such as PD, AD, HD, amyotrophic lateral sclerosis (ALS), and spinal muscular atrophy (SMA), remains unclear.220 To date, there are no effective therapies for these diseases. However, the generation of cellular models will help acquire information about the pathogenesis of these conditions and will lead to facilitated drug screening. Using stem cells for cell replacement was recently explored as a possible therapy for these diseases. The prevalence of various neurodegenerative diseases is higher in developed counties such as the United States (around 7 million occurrences).221 As mentioned earlier, genome-editing tools are used to prevent the inheritance of AD. In future applications, it may be possible to use these tools to develop curative therapies against neurodegeneration. Genome-editing technologies, such as TALENs, ZFNs, and CRISPR-Cas9, have already been used to alter or generate genetic mutations and model particular neurodegenerative diseases (Figure 7).222, 223 In higher animals, it is difficult to produce large-animal disease models, necessitating the use of cell models. Additionally, the lack of ESC lines, derived from large animals, is a major obstacle in gene manipulation.220
Figure 7.
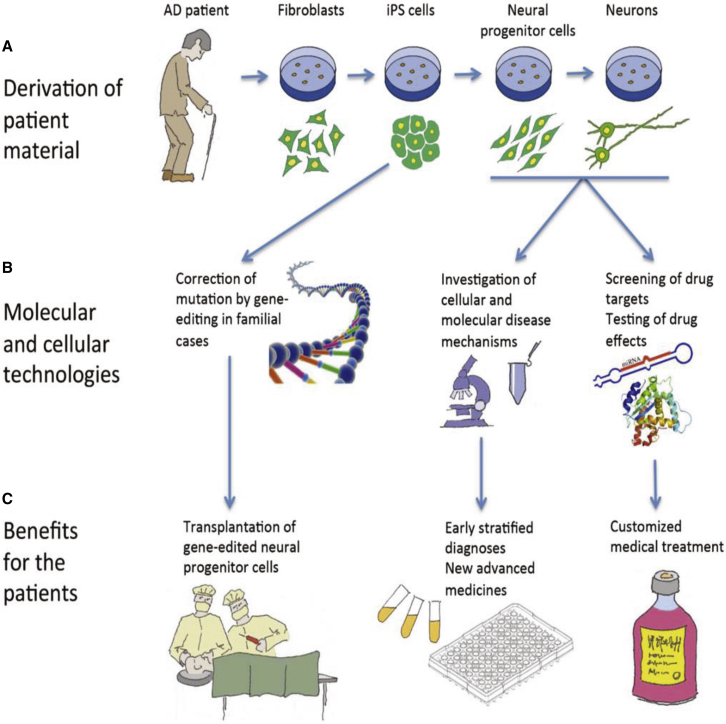
Schematic Illustration of the Use of Induced Pluripotent Stem Cells in Relation to Alzheimer’s Disease
(A) Induced pluripotent stem cells (iPSCs), derived from a skin biopsy acquired from a patient with Alzheimer’s disease (AD), are differentiated into neural progenitor cells and neurons. (B) In familial cases, the disease-causing mutation can be corrected by gene editing of the iPSCs; the neural progenitor cells and neurons can be used for research and drug screening. (C) Patients can benefit from cell therapy, better diagnostic procedures, customized treatments, and novel medical approaches. Reproduced from Freude et al.,223 copyright (2014) Creative Commons Attribution 4.0 International.
Recent progress in genome-editing technologies has enabled the production of large-animal models for exploring neurodegenerative diseases.220 The CRISPR-Cas9 system can be used to create mutations that impede the open reading frame and inactivate the gene. Hence neurodegenerative diseases, caused by malfunction of specific genes in animals, can be easily studied using CRISPR-Cas9.220 CRISPR-Cas9 genome-editing technology can also be used to assess mosaic mutations in various types of cells. CRISPR-Cas9 can target each gene in an embryo, which is advantageous in generating animal models of neurodegenerative diseases. This technique is particularly useful in large-animal models. Neurodegenerative diseases, such as PD, can originate from mutations in the PARKIN and PINK1 genes.
In animal models, CRISPR-Cas9 can be used for targeted silencing of candidate genes.220 This may enable us to mimic mutations observed in patients with PD. Neurodegenerative diseases can also originate from mutant cytotoxic proteins. PD and HD are partially caused by mutations in α-synuclein and polyglutamine extended huntingtin, respectively.220 Using CRISPR-Cas9 technology in mammalian cells can efficiently generate these proteins, while other tools, such as NHEJ, can be used to increase their production.224 Promising results in human immunotherapy are also being reported. Human ALS is frequently caused by the C9ORF72 gene, which encodes dipeptide repeat (DPR) proteins. CRISPR-Cas9-based knockout screening of the human genome was thus used to identify suppressors and enhancers of C9ORF72-encoded DPRs. Using this technology, numerous modifiers were identified; one of these modifiers, TMX2, ameliorated endoplasmic reticular stress in a patient with C9ORF72 ALS, resulting in dramatic survival of neurons.225
Treatment of HIV
The first case of HIV infection was reported in 1981.226 Since then, HIV has become a major public health concern, affecting more than 35 million people globally.226 Although antiretroviral therapy (ART) can reduce the symptoms of HIV-1, it is not possible to achieve full recovery. The main obstacle for curative therapy is perseverance of HIV reservoirs, which cannot be eliminated by antiretroviral therapy.227 HIV viral DNA assimilates into host genomes, forming organized viral reservoirs. It is possible that deleting or neutralizing viral DNA would eliminate HIV persistence.228 Some limitations of current antiviral therapies include drug toxicity, resistance to antiretroviral therapy, and failure to eliminate a dormant viral infection. In addition, the medications and expenses of lifelong treatment, as well as large numbers of patients with HIV/AIDS, render treatment grueling and costly. To eliminate dormant viral reservoirs, it is important to focus on individualized therapies that can block viral DNA and show minimal drug toxicity.229
Nuclease-mediated genome editing is an encouraging approach in therapeutic applications against HIV-1.229, 230 The engineered nucleases mainly include ZFNs, TALENs, and CRISPR-Cas9; these can disrupt HIV-1 proviral DNA that has converted into active DNA and integrated into the host genome. These technologies can also interrupt the entry of HIV-1, which occurs via co-receptors C-C chemokine receptor 5 (CCR5) or C-C-C chemokine receptor 4 (CXCR4). CCR5 can be targeted by ZFNs; this treatment for patients with HIV-1 is in phase II medical trials.229, 230 Because the entry of HIV-1 requires co-receptors, such as CD4 and either CCR5 or CXCR4, these co-receptors are potential targets for therapeutic applications. A deletion of 32 bp in the CCR5 gene of a patient with HIV-1 eliminated the existing infection and rendered the patient resistant to infections with R5 type HIV-1.143, 231
As discussed previously, antiretroviral therapy cannot eliminate HIV from dormant reservoirs. Dormant viral reservoirs are mostly found within memory CD4+ T cells and can persist for approximately 60 years after receiving antiretroviral therapy.232 The cleavage of HIV-1 proviral DNA, induced via CRISPR-Cas9, is dose dependent and does not show adverse effects in patients. The use of ZFNs has also been successful in eliminating the HIV-1 provirus by targeting the transactivation response element of HIV-1 LTR. However, using ZFNs to eliminate proviral DNA needs to be further optimized by protein engineering.155 The delivery of ZFNs into human CD4+ T and CD34+ hematopoietic stem progenitor cells (HSPCs) to target the CCR5 gene was assessed using a mouse model of HIV infection.
Using an adenoviral vector in patients with HIV showed promising results. This vector expresses a ZFN and was used to target the CCR5 gene in CD4 T cells isolated from 12 patients with HIV.104 The CRISPR-Cas9 gene-editing system can also remove proviral DNA by a sgRNA-guided method.233 This technology of proviral DNA removal by CRISPR-Cas9 also helps to inactivate viral genes in other diseases. CRISPR-Cas9-mediated removal of HIV proviral DNA was achieved using sgRNAs, which can target preserved sites within the U3 region of viral LTR in dormant infections.230 In addition to inactivation of proviral DNA and prevention of replication in infected cells, CRISPR-Cas9 can provide resistance against recurrent HIV infections and can target multiple spots inside the HIV genome.234 The Cas9 protein of CRISPR-Cas9 can persist in human HSPCs. HSPCs can then differentiate into monocytes and macrophages without undesirable side effects. TALENs and CRISPR-Cas9 can also be used for natural deletion of CCR5Δ32 in iPSCs with a piggy-Bac transposon donor sequence.235
An HIV-GFP Jurkat cell line (JLat10.6) has also been used to test the efficacy of the CRISPR-Cas9 system in silencing HIV-1 DNA.236 The results of that study indicated that CRISPR-Cas9 effectively targeted and silenced HIV-1 proviral DNA in the JLat10.6 cell line. This spurred the notion to match gRNA with a targeted viral DNA sequence.236 Recently, internal antisense long non-coding RNAs (lncRNAs) were tested as novel tools for inhibiting the HIV-1 provirus, which was expressed from the NEF gene.229 High expression of NEF lncRNA can cause viral inhibition, whereas low expression shows the opposite result. The 5′ LTR region may be a suitable target for the CRISPR-Cas9 genome-editing system.229 Several antiviral therapies can be used to control viral infections. However, the virus is untraceable in the blood, which poses a major problem. Recent studies show that the CRISPR-Cas9 genome-editing system can act as a novel antiviral tool by targeting viral coding regions. The system can be used to block assimilation and progression of HIV-1 infection, to eliminate hidden viral reservoirs, and to confer lasting immunization against HIV-1.
Application of Genome Editing in Plants
Crop Improvement
Traditional approaches to improving crop species have generally depended on conventional and transgenic breeding methods. However, these methods suffer from several limitations. The former is limited by a declining genetic base that depends on germplasm collections. In addition, randomly occurring variants are generated by mutagenic effects. The latter generates genetically modified (GM) crops, which is concerning with respect to health and environmental safety. The addition of foreign genes into a genome can limit commercialization and will possibly require a complex regulatory system. To overcome these difficulties, newly introduced techniques based on site-specific nucleases (SSNs) may be suitable tools for successful genome editing.
Genome modification in plants was first conducted in the early 1990s; however, due to the low efficiency of genome engineering, this research did not advance. In recent years, precise genome modification in model plants and other important crops has been revolutionized with the help of SSNs.237, 238 To date, research on SSNs has identified three common programmable nucleases: ZFNs, TALENs, and CRISPR-Cas-mediated RNA-guided DNA endonucleases, which are used for editing plant genomes (Table 1).3 Among these, the RNA-based CRISPR-Cas system is the most suitable for use in various organisms, especially in plants.239 The overall editing mechanism depends on the introduction of DSBs at target sites commonly repaired by either HR or NHEJ process.240 NHEJ can generate gene knockouts by producing frameshift mutations in coding sequences; alternatively, HR results in gene substitution, combination, or accumulation. To date, most genome modification procedures have mainly focused on model plant species. As such, enormous efforts have been invested into crop plant research. In the tobacco plant, resistance against one or more herbicides was introduced by a missense mutation in donor templates, which was then inserted into protoplasts along with ZFNs. Similarly, the same ALS gene in a tobacco protoplast was replaced using TALEN-mediated genome modification.241
Table 1.
Practical Applications of Nucleases in Crop Plants
| Crop Plants | Treated Tissue | Used Nucleases | References | Practical Applications |
|---|---|---|---|---|
| Rice | embryo | TALENs | 250 | resistance to bacterial blight |
| 251 | ||||
| Cas9/sgRNA | 283 | |||
| 284 | ||||
| 285 | ||||
| 286 | ||||
| 287 | ||||
| Maize | maize cells | TALEN and CRISPR-Cas9 | 288 | reduction of phytate (inositol phosphate), creation of male sterile plants |
| meganucleases | 289 | |||
| 290 | ||||
| 291 | ||||
| ZFN | 242 | |||
| Soya bean | hairy root and somatic embryo | ZFN | 292 | production of seeds with high monosaturated oleic acid and low polyunsaturated fatty acid, linoleic acid |
| Cas9/sgRNA | 293 | |||
| TALENs | 294 | |||
| Cotton | embryogenic callus cells | meganucleases | 295 | resistance to herbicide |
| Canola | immature seed | ZFN | 296 | decreased levels of palmitic acid and increased total levels of C18 fatty acids |
| Wheat | genomic DNA | CRISPR-Cas9 | 285 | resistance to powdery mildew |
| TALEN | 248 | |||
| sgRNAs | 297 | |||
| Cas9/sgRNA | 248 | |||
| Sorghum | immature embryos | CRISPR-Cas9 | 284 | expression of clover fluorescence protein |
| Barley | grains of winter barley | TALENs | 298 | creation of homozygous mutants |
| 299 |
Continuous expression of ZFN and a simple donor molecule add the PAT herbicide tolerance gene at the IPK1 locus. This genome modification imparts herbicide resistance and leads to high accumulation of phytate and low levels of inorganic phosphate caused by IPK1 expression.242 GM organism (GMO) crops have been commercially used in the recent past. However, these crops are still not accepted in the developed countries because of serious health and environmental considerations. Conversely, specific manipulation of genomes by SSN can be used to overcome problems associated with classical transgenic breeding and can prevent the introduction of foreign genes and proteins, reducing the probable risks associated with transgenic procedures.243, 244 The US Department of Agriculture (USDA; Washington, DC, USA) has provided public authorization to use ZFN techniques for production of GM corn.245 The regulatory agenda for GM crops in European Union (EU) countries is mainly focused on genome modification procedures rather than on final products. Therefore, crops modified by genome-editing tools, such as CRISPR-Cas9 and others, may not be classified as GMOs.244, 246 Finally, site-specific techniques can offer more accurate methods for crop improvement and may be promising biotechnological tools for plant breeding.
Trait Improvement
Without the domestication of plants, we could not maintain the balance between supply and demand or provide food, medicine, chemicals, renewable materials, animal feed, and biofuels. The process of domestication includes improvement of crop performance and crop properties that are directly related to human welfare. Genome editing can accelerate the overall plant breeding process by inserting or modifying genes that can directly improve the characteristics of crops.
Among genome-editing methods, CRISPR-Cas9 is more advantageous because multiple traits can be customized simultaneously using this procedure. The simplest form of gene modification is NHEJ-mediated gene knockout. This technique was successfully used to eliminate genes that negatively affected food quality, decreased resistance to pathogens or diseases, and decreased the value of end products.247 TALEN and CRISPR-Cas9 can be used to target genes located at the mildew resistance locus (MLO) in the wheat plant. The MLO homoalleles can be effectively knocked out to promote resistance to powdery mildew in the wheat plant.248 SSNs enable the addition of numerous genes near existing transgenic loci to facilitate stacking of targeted molecular traits. This mechanism can introduce multiple traits into crop plants with minimal risk of segregation, which is the main obstacle in classical plant breeding and in modern genetic engineering methods.
After stacking, the complete group of transgenes can be moved to another plant because such a group acts as a solitary locus. A maize line, containing an herbicide resistance marker and an artificial ZFN target site, was produced using this process.249 CRISPR-Cas9 techniques are accessible and straightforward to use in both forward and reverse genetics, aiding in basic research conducted with model plant species. This is helpful for generating genomic data, and for more rapid gene discovery and trait improvement in various plant species.247 A modification TALEN technique was developed by engineering HAX3 from the Brassicaceae pathogen Xanthomonas campestris. These techniques have been widely used in many crop plants including rice, barley, and maize. The genes that are responsible for disease vulnerability in rice can be easily mutated by TALEN to produce disease-resistant rice with normal phenotypes.250 In rice, numerous mutants have been produced using TALENs to knock out eight Brachypodium genes with high efficiency.251
Genome-editing methods, such as CRISPR-Cas9, play a vital role in identifying novel traits in common profitable crop plants. The delivery and expression of engineered nucleases in plant cells are critical because some plant species do not show positive results. For generating novel traits, it is important to establish a balance between the specific plant tissue and the method used for transformation; this is a major issue that needs to be addressed. To overcome these difficulties, Gemini virus-mediated replicons can be used to transmit DNA for genome engineering in various plant species.252 New approaches to plant breeding, along with in-depth understanding of the whole-plant genome, will facilitate the development of important traits in plants.
Minimizing Off-Target Mutagenesis Risks of Nucleases
Genome-editing nucleases are the multipurpose tools that are extensively applied for studying the potential of genetic materials, producing GMOs, and preclinical investigation of genetic diseases. However, the frequency of off-targeted activities (more than 50%) is a major concern in RGEN target sites, particularly in therapeutic and clinical applications.253 To minimize the off-target mutations, a specific proofreading approach needs to be elucidated. However, CRISPR-Cas9 is reported as an editing approach with proofreading capability. The mechanism of CRISPR-Cas9 was first discovered in bacteria, where it is used as an immune response against invading pathogens such as bacteriophages.254, 255, 256 In the early stages of this discovery, the CRISPR-Cas9 system was mainly used as a tool for genome editing in various types of cell lines. Later, living organisms, such as C. elegans,255, 257 zebrafish,149 mice,258, 259 rats,147, 148 rabbits,260 and monkeys,261 were more easily modified with CRISPR-Cas9 than with TALENs or ZFNs. In humans, the CRISPR-Cas9 system can also play an important role in correcting genetic mutations such as those involved in β-thalassemia,32 cystic fibrosis,262 hemophilia A,33 cataracts,263 hereditary HTI,34 and DMD.35
Numerous studies have shown that in human cells, nucleases are involved in host defense mechanisms to fight against viruses such as hepatitis B264, 265 and HIV.233, 234, 235, 236 Frequent use of nucleases has raised concerns about nonspecific activity at off-target sites, especially when nucleases are used for clinical purposes. Nonspecific activity at off-target sites may cause stable and injurious cytotoxicity or even tumorigenicity, impeding normal cellular function in humans. Nonspecific activity at off-target sequences is less frequent in the case of CRISPR-Cas9 than in cases of ZFNs and TALENs.50, 266, 267, 268, 269 Off-target cleavage activity of CRISPR-Cas9 can be decreased by mutagenesis-related approaches or by eradicating the nuclease function of CRISPR-Cas9. For deactivation of the RuvC nuclease domain, an aspartate-to-alanine mutation is highly effective and attenuates the activity of CRISPR-Cas9.254 In this procedure, just one strand is cleaved off dsDNA, producing a simple gap rather than an entire blunt dsDNA break, which is less efficient in promoting HR.
Several groups have shown that the double-nick or DSBs strategy increases cleavage at on-target sites; this can be used in combination with wild-type (WT) Cas9, which effectively reduces off-target mutations.270, 271, 272 This nicking activity may cause point mutations; hence this process is also risky.273 The fusion of FokI nuclease with an inactive Cas9 protein can control the genome-editing process. In the gene-editing process, both Streptococcus pyogenes Cas9 (SpCas9) and Staphylococcus aureus (Sa) Cas9 have been used extensively. However, the specificity and cleavage activity of SaCas9 is higher than that of SpCas9; this is because the former can be guided to a 21- to 23-nt site, whereas the latter can only recognize a 17-nt sequence.274, 275
Combining the Cas9 protein and sgRNA-mediated on- and off-target cleavage has been used for DNA plasmid transfections in different cell lines. This process results in overexpression of the Cas9 protein and sgRNA, which may cause off-target mutagenesis because larger amounts of DNA are transfected.29 As the number of interactions increases, the chances of off-target site cleavage by Cas9 binding also increase. Direct delivery of Cas9 into cells has been used to reduce the off-target effects. The Cas9 protein is unstable and has a moderately short half-life, making its presence transient. Using the Cas9 protein instead of the Cas9 plasmid produces 13 times more on-target cleavage compared with off-target cleavage.276 In a 72-hr time-course experiment, the Cas9 protein was no longer detected after 24 hr, whereas a noticeable increase in Cas9 plasmid transfection persisted for up to 72 hr.276 Chemical modification at the 3′ end of sgRNAs improves their cleavage activity and specificity during genome editing.277
Future Prospects of Genome Editing
As we have shown, genome-editing technologies have the potential to significantly advance cell-based therapy and improve our understanding of many genetic diseases. Efforts made over the last decade are leading to the generation of specific and permanent multiple-hit pluripotent cell (PC) lines for reprogramming into disease-linked cell types. However, well-developed protocols for these methods are still lacking. It is important to optimize this methodology and to achieve competent gene alteration and biallelic gene editing before translating these approaches into clinical applications. In a few studies, the relative effectiveness of gene editing was compared among the available options such as ZFNs, TALENs, and CRISPR/Cas9. Recently, it was shown in human pluripotent stem cells (hPSCs) that the CRISPR-Cas9 system is more effective than TALEN-mediated gene editing.83
In some cases, a combination of techniques is preferable to using traditional approaches to improve efficiency; examples of such combinations are integrase-defective lentiviral vectors (IDLVs)/ZFN,278, 279 transposon/CRISPR,32 and TALEN/CRISPR (iCRISPR).162 For patient therapy, genome-editing approaches must precisely target disease-associated mutations in either iPSC-derived progenitor cells or in mature cells for autologous transplant; these procedures must be considered safe and not disturb other loci in the genome. Interestingly, a complete genome analysis by Veres et al.280 confirmed that a slight risk of off-target mutations is posed by both CRISPR-Cas9 and TALEN targeted to the sortilin 1 (SORT1) gene in human ESCs (hESCs). Presently, there are promising advances in gene alteration for a variety of diseases. Schwank et al.262 recently corrected the function of cystic fibrosis transmembrane conductance regulator (CFTR) gene using CRISPR-Cas9 in adult stem cells (SCs) derived from patients with cystic fibrosis (Figure 8). In that study, treatment with CRISPR-Cas9 generated few off-target effects. The results of the forskolin-response assay, performed in that study, confirmed that the activity of disease-linked transmembrane receptor was recovered. However, cystic fibrosis affects multiple organs (Figure 8),262 making it difficult to treat cystic fibrosis using the CRISPR-Cas9 system.
Figure 8.
Schematic Illustration Showing Functional Repair of CFTR by CRISPR-Cas9 in Intestinal Stem Cell Organoids Acquired from Patients with Cystic Fibrosis
Reproduced from Schwank et al.262 Copyright 2013. Reproduced with permission from Elsevier.
Recently, Sánchez-Rivera and Jacks203 predicted that nuclease-based genome editing will be a vital link, connecting the bench and clinical applications in cancer treatment. Successful expansion of genome profiling for extensive characterization of patient-derived tumors is creating comprehensive roadmaps for developing cell- and whole-animal-based empirical approaches. Single or multiplex nuclease- and/or small-molecule-based technologies will advance the development of personalized genome editing, which can be used to rapidly recognize genotype-specific weaknesses and harmful synthetic interactions.281 Such personalized techniques can potentially enable rapid identification of lethal mechanisms and development of respective therapeutic approaches.282
Several technical restrictions limit the usage of CRISPR-Cas9 for therapeutic targeting of oncogenes in human patients, and prospects of this gene therapy are currently controversial. Recently, a study used this technology to achieve accurate in-vivo genetic alterations in the hepatic tissue of adult mice. This study successfully corrected an inherited genetic disorder via HDR in a mouse model.34 Future developments in this technology will permit therapeutic modifications of single or various driver mutations. Such developments will also increase the effectiveness of editing and delivery via CRISPR-Cas9 using both non-viral and viral delivery vectors. In addition to modifying cancer-linked mutations, CRISPR-Cas9 components can be employed for specific ex-vivo editing of immune cells for immunotherapeutic applications. For example, CRISPR-Cas9 components can be used to improve T cells modified with the chimeric antigen receptor (CAR-T cells);210 in these cells, CAR is incorporated into a safe harbor locus.
Currently, CRISPR-Cas9 is a leading RNA-programmed gene-editing platform. The field of genome alteration has undergone a rapid scientific revolution that will transform basic biological and biomedical research.254 Genome editing can be used in various types of iPSC-based therapy and cancer therapy, offering exciting prospects for better assessment of disease progression and development of effective treatments.
Concluding Remarks
There is tremendous promise in using the CRISPR-Cas9 system for targeted editing of the genome in cells and whole organisms. This technique will advance human therapies, agricultural biotechnology, and microbial gene engineering. The revolutionary discovery of the CRISPR-Cas9 genome-editing system will impact the fields of modified medicine, human genetic alteration, and drug development. Nonetheless, various obstacles to genome-editing technology still need to be overcome. The major concerns associated with genome editing in higher organisms are off-target mutations in the genome and precision in inserting Cas9 into a cellular system. Overcoming these limitations will advance the field of human gene therapy. Both off-target mutations and imprecise delivery of Cas9 into the cell can be lethal because they can cause cellular alterations and death. Several studies have shown that Cas9-guided RNA complexes can alleviate genetic disease when injected into an adult mouse model. Traditional methods, such as nucleic acid and protein delivery, can be helpful in identifying suitable delivery methods for genome-editing nucleases. In addition to nucleases, supplementary methods for genome editing have been considered. Recently, another technology, called triplex-forming peptide nucleic acids, was successfully used for genome modifications. These technologies will revolutionize the fields of medical and pharmaceutical sciences by helping us identify biochemical pathways in infections and by simplifying drug delivery methods. The limiting factors of genome editing in animal and plant species need to be addressed before application in humans because of the potential impact on human welfare. Thus, detailed molecular studies and suitable disease models are required before using genome editing in the treatment of human diseases.
Author Contributions
S.K.S., M.S.R., and K.-H.K. designed the study; F.K.S. and S.M.K.R. drafted the manuscript; S.K.S. extracted all figures; M.A.H.M.J. and S.M.R.I. partially wrote the manuscript; S.K.S., M.S.R., and K.-H.K. wrote and edited the manuscript. All authors reviewed and approved the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the KU-Research Professor Program of Konkuk University. This study was partially supported by a grant from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning, South Korea (grant 2016R1E1A1A01940995).
Contributor Information
Subbroto Kumar Saha, Email: subbroto@konkuk.ac.kr.
Ki-Hyun Kim, Email: kkim61@hanyang.ac.kr.
References
- 1.Krueger U., Bergauer T., Kaufmann B., Wolter I., Pilk S., Heider-Fabian M., Kirch S., Artz-Oppitz C., Isselhorst M., Konrad J. Insights into effective RNAi gained from large-scale siRNA validation screening. Oligonucleotides. 2007;17:237–250. doi: 10.1089/oli.2006.0065. [DOI] [PubMed] [Google Scholar]
- 2.McManus M.T., Sharp P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 3.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malankhanova T.B., Malakhova A.A., Medvedev S.P., Zakian S.M. Modern genome editing technologies in Huntington’s disease research. J. Huntingtons Dis. 2017;6:19–31. doi: 10.3233/JHD-160222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith A.J.P., Deloukas P., Munroe P.B. Emerging applications of genome-editing technology to examine functionality of GWAS-associated variants for complex traits. Physiol. Genomics. 2018;50:510–522. doi: 10.1152/physiolgenomics.00028.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 9.Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waryah C.B., Moses C., Arooj M., Blancafort P. Zinc fingers, TALEs, and CRISPR systems: a comparison of tools for epigenome editing. Methods Mol. Biol. 2018;1767:19–63. doi: 10.1007/978-1-4939-7774-1_2. [DOI] [PubMed] [Google Scholar]
- 11.Ishida K., Gee P., Hotta A. Minimizing off-target mutagenesis risks caused by programmable nucleases. Int. J. Mol. Sci. 2015;16:24751–24771. doi: 10.3390/ijms161024751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thurtle-Schmidt D.M., Lo T.W. Molecular biology at the cutting edge: a review on CRISPR/CAS9 gene editing for undergraduates. Biochem. Mol. Biol. Educ. 2018;46:195–205. doi: 10.1002/bmb.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 14.Ranjha L., Howard S.M., Cejka P. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma. 2018;127:187–214. doi: 10.1007/s00412-017-0658-1. [DOI] [PubMed] [Google Scholar]
- 15.Choulika A., Perrin A., Dujon B., Nicolas J.-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bibikova M., Golic M., Golic K.G., Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 18.Hefferin M.L., Tomkinson A.E. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst.) 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 19.van Gent D.C., van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 20.Miyaoka Y., Mayerl S.J., Chan A.H., Conklin B.R. Detection and quantification of HDR and NHEJ induced by genome editing at endogenous gene loci using droplet digital PCR. Methods Mol. Biol. 2018;1768:349–362. doi: 10.1007/978-1-4939-7778-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capecchi M.R. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 22.Chandrasegaran S., Carroll D. Origins of programmable nucleases for genome engineering. J. Mol. Biol. 2016;428(5 Pt B):963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramalingam S., Annaluru N., Chandrasegaran S. A CRISPR way to engineer the human genome. Genome Biol. 2013;14:107. doi: 10.1186/gb-2013-14-2-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roche P.J., Gytz H., Hussain F., Cameron C.J., Paquette D., Blanchette M. Efficient homology directed repair by Cas9: DNA localization and cationic polymeric transfection in mammalian cells. bioRxiv. 2018 [Google Scholar]
- 25.Nami F., Basiri M., Satarian L., Curtiss C., Baharvand H., Verfaillie C. Strategies for in vivo genome editing in nondividing cells. Trends Biotechnol. 2018;36:770–786. doi: 10.1016/j.tibtech.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K., Izpisua Belmonte J.C. In vivo genome editing via the HITI method as a tool for gene therapy. J. Hum. Genet. 2018;63:157–164. doi: 10.1038/s10038-017-0352-4. [DOI] [PubMed] [Google Scholar]
- 27.Rouet R., Thuma B.A., Roy M.D., Lintner N.G., Rubitski D.M., Finley J.E., Wisniewska H.M., Mendonsa R., Hirsh A., de Oñate L. Receptor-mediated delivery of CRISPR-Cas9 endonuclease for cell-type-specific gene editing. J. Am. Chem. Soc. 2018;140:6596–6603. doi: 10.1021/jacs.8b01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 29.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotta A., Yamanaka S. From genomics to gene therapy: induced pluripotent stem cells meet genome editing. Annu. Rev. Genet. 2015;49:47–70. doi: 10.1146/annurev-genet-112414-054926. [DOI] [PubMed] [Google Scholar]
- 31.Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie F., Ye L., Chang J.C., Beyer A.I., Wang J., Muench M.O., Kan Y.W. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park C.-Y., Kim D.H., Son J.S., Sung J.J., Lee J., Bae S., Kim J.H., Kim D.W., Kim J.S. Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sürün D., Schwäble J., Tomasovic A., Ehling R., Stein S., Kurrle N., von Melchner H., Schnütgen F. High efficiency gene correction in hematopoietic cells by donor-template-free CRISPR/Cas9 genome editing. Mol. Ther. Nucleic Acids. 2018;10:1–8. doi: 10.1016/j.omtn.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marangi M., Pistritto G. Innovative therapeutic strategies for cystic fibrosis: moving forward to CRISPR technique. Front. Pharmacol. 2018;9:396. doi: 10.3389/fphar.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.March J., Dickson G., Popplewell L. Development of CRISPR/dCas9 systems to address muscle fibrosis in Duchenne muscular dystrophy. Neuromuscul. Disord. 2018;28(Suppl 1):S7. [Google Scholar]
- 39.Shao Y., Wang L., Guo N., Wang S., Yang L., Li Y., Wang M., Yin S., Han H., Zeng L. Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats. J. Biol. Chem. 2018;293:6883–6892. doi: 10.1074/jbc.RA117.000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouet P., Smih F., Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guha T.K., Wai A., Hausner G. Programmable genome editing tools and their regulation for efficient genome engineering. Comput. Struct. Biotechnol. J. 2017;15:146–160. doi: 10.1016/j.csbj.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J., McLachlan A.D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tupler R., Perini G., Green M.R. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 44.Desjarlais J.R., Berg J.M. Redesigning the DNA-binding specificity of a zinc finger protein: a data base-guided approach. Proteins. 1992;12:101–104. doi: 10.1002/prot.340120202. [DOI] [PubMed] [Google Scholar]
- 45.Rebar E.J., Pabo C.O. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 46.Kim Y.-G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wah D.A., Hirsch J.A., Dorner L.F., Schildkraut I., Aggarwal A.K. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 48.Wah D.A., Bitinaite J., Schildkraut I., Aggarwal A.K. Structure of FokI has implications for DNA cleavage. Proc. Natl. Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramirez C.L., Foley J.E., Wright D.A., Müller-Lerch F., Rahman S.H., Cornu T.I., Winfrey R.J., Sander J.D., Fu F., Townsend J.A. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cornu T.I., Thibodeau-Beganny S., Guhl E., Alwin S., Eichtinger M., Joung J., Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger Nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 51.Kim H.J., Lee H.J., Kim H., Cho S.W., Kim J.-S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee C.M., Cradick T.J., Fine E.J., Bao G. Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol. Ther. 2016;24:475–487. doi: 10.1038/mt.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 54.Moscou M.J., Bogdanove A.J. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 55.Mak A.N.-S., Bradley P., Cernadas R.A., Bogdanove A.J., Stoddard B.L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.-K., Shi Y., Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maeder M.L., Thibodeau-Beganny S., Osiak A., Wright D.A., Anthony R.M., Eichtinger M., Jiang T., Foley J.E., Winfrey R.J., Townsend J.A. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sander J.D., Dahlborg E.J., Goodwin M.J., Cade L., Zhang F., Cifuentes D., Curtin S.J., Blackburn J.S., Thibodeau-Beganny S., Qi Y. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heigwer F., Kerr G., Walther N., Glaeser K., Pelz O., Breinig M., Boutros M. E-TALEN: a web tool to design TALENs for genome engineering. Nucleic Acids Res. 2013;41:e190. doi: 10.1093/nar/gkt789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holkers M., Maggio I., Liu J., Janssen J.M., Miselli F., Mussolino C., Recchia A., Cathomen T., Gonçalves M.A. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Briggs A.W., Rios X., Chari R., Yang L., Zhang F., Mali P., Church G.M. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmid-Burgk J.L., Schmidt T., Kaiser V., Höning K., Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat. Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim Y., Kweon J., Kim A., Chon J.K., Yoo J.Y., Kim H.J., Kim S., Lee C., Jeong E., Chung E. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 66.Ding Q., Lee Y.-K., Schaefer E.A., Peters D.T., Veres A., Kim K., Kuperwasser N., Motola D.L., Meissner T.B., Hendriks W.T. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doyle E.L., Hummel A.W., Demorest Z.L., Starker C.G., Voytas D.F., Bradley P., Bogdanove A.J. TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat -1. PLoS ONE. 2013;8:e82120. doi: 10.1371/journal.pone.0082120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamb B.M., Mercer A.C., Barbas C.F., 3rd Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res. 2013;41:9779–9785. doi: 10.1093/nar/gkt754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Terns M.P., Terns R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koonin E.V., Wolf Y.I. Evolution of the CRISPR-Cas adaptive immunity systems in prokaryotes: models and observations on virus-host coevolution. Mol. Biosyst. 2015;11:20–27. doi: 10.1039/c4mb00438h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Belkum A., Soriaga L.B., LaFave M.C., Akella S., Veyrieras J.-B., Barbu E.M., Shortridge D., Blanc B., Hannum G., Zambardi G. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. MBio. 2015;6 doi: 10.1128/mBio.01796-15. e01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westra E.R., Dowling A.J., Broniewski J.M., van Houte S. Evolution and ecology of CRISPR. Annu. Rev. Ecol. Evol. Syst. 2016;47:307–331. [Google Scholar]
- 74.Mojica F.J.M., Montoliu L. On the origin of CRISPR-Cas technology: from prokaryotes to mammals. Trends Microbiol. 2016;24:811–820. doi: 10.1016/j.tim.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 75.Li R., Fang L., Tan S., Yu M., Li X., He S., Wei Y., Li G., Jiang J., Wu M. Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 2016;26:1273–1287. doi: 10.1038/cr.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mansoor M., Melendez A.J. Advances in antisense oligonucleotide development for target identification, validation, and as novel therapeutics. Gene Regul. Syst. Bio. 2008;2:275–295. doi: 10.4137/grsb.s418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patterson A.G., Yevstigneyeva M.S., Fineran P.C. Regulation of CRISPR-Cas adaptive immune systems. Curr. Opin. Microbiol. 2017;37:1–7. doi: 10.1016/j.mib.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 78.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mojica F.J., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 81.Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ding Q., Regan S.N., Xia Y., Oostrom L.A., Cowan C.A., Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.-Y., Song D.W., Lee K.J., Jung M.H., Kim S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Engstrom J.U., Kmiec E.B. DNA replication, cell cycle progression and the targeted gene repair reaction. Cell Cycle. 2008;7:1402–1414. doi: 10.4161/cc.7.10.5826. [DOI] [PubMed] [Google Scholar]
- 86.Parekh-Olmedo H., Kmiec E.B. Progress and prospects: targeted gene alteration (TGA) Gene Ther. 2007;14:1675–1680. doi: 10.1038/sj.gt.3303053. [DOI] [PubMed] [Google Scholar]
- 87.Aarts M., te Riele H. Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Ther. 2011;18:213–219. doi: 10.1038/gt.2010.161. [DOI] [PubMed] [Google Scholar]
- 88.Rivera-Torres N., Banas K., Bialk P., Bloh K.M., Kmiec E.B. Insertional mutagenesis by crispr/cas9 ribonucleoprotein gene editing in cells targeted for point mutation repair directed by short single-stranded DNA oligonucleotides. PLoS ONE. 2017;12:e0169350. doi: 10.1371/journal.pone.0169350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferrara L., Kmiec E.B. Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004;32:5239–5248. doi: 10.1093/nar/gkh822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ferrara L., Kmiec E.B. Targeted gene repair activates Chk1 and Chk2 and stalls replication in corrected cells. DNA Repair (Amst.) 2006;5:422–431. doi: 10.1016/j.dnarep.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 91.Radecke F., Peter I., Radecke S., Gellhaus K., Schwarz K., Cathomen T. Targeted chromosomal gene modification in human cells by single-stranded oligodeoxynucleotides in the presence of a DNA double-strand break. Mol. Ther. 2006;14:798–808. doi: 10.1016/j.ymthe.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 92.Radecke F., Radecke S., Schwarz K. Unmodified oligodeoxynucleotides require single-strandedness to induce targeted repair of a chromosomal EGFP gene. J. Gene Med. 2004;6:1257–1271. doi: 10.1002/jgm.613. [DOI] [PubMed] [Google Scholar]
- 93.Doyle E.L., Booher N.J., Standage D.S., Voytas D.F., Brendel V.P., Vandyk J.K., Bogdanove A.J. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G., 2nd, Tan W., Penheiter S.G., Ma A.C., Leung A.Y. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merkert S., Bednarski C., Göhring G., Cathomen T., Martin U. Generation of a gene-corrected isogenic control iPSC line from cystic fibrosis patient-specific iPSCs homozygous for p.Phe508del mutation mediated by TALENs and ssODN. Stem Cell Res. (Amst.) 2017;23:95–97. doi: 10.1016/j.scr.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 96.Rivera-Torres N., Strouse B., Bialk P., Niamat R.A., Kmiec E.B. The position of DNA cleavage by TALENs and cell synchronization influences the frequency of gene editing directed by single-stranded oligonucleotides. PLoS ONE. 2014;9:e96483. doi: 10.1371/journal.pone.0096483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strouse B., Bialk P., Niamat R.A., Rivera-Torres N., Kmiec E.B. Combinatorial gene editing in mammalian cells using ssODNs and TALENs. Sci. Rep. 2014;4:3791. doi: 10.1038/srep03791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoon K., Cole-Strauss A., Kmiec E.B. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proc. Natl. Acad. Sci. USA. 1996;93:2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cole-Strauss A., Yoon K., Xiang Y., Byrne B.C., Rice M.C., Gryn J., Holloman W.K., Kmiec E.B. Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science. 1996;273:1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- 100.Alexeev V., Yoon K. Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA-DNA oligonucleotide. Nat. Biotechnol. 1998;16:1343–1346. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- 101.Xu L., Zhao P., Mariano A., Han R. Targeted myostatin gene editing in multiple mammalian species directed by a single pair of TALE nucleases. Mol. Ther. Nucleic Acids. 2013;2:e112. doi: 10.1038/mtna.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Campbell C.R., Keown W., Lowe L., Kirschling D., Kucherlapati R. Homologous recombination involving small single-stranded oligonucleotides in human cells. New Biol. 1989;1:223–227. [PubMed] [Google Scholar]
- 103.Yamamoto T., Moerschell R.P., Wakem L.P., Komar-Panicucci S., Sherman F. Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics. 1992;131:811–819. doi: 10.1093/genetics/131.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tan W.S., Carlson D.F., Walton M.W., Fahrenkrug S.C., Hackett P.B. Precision editing of large animal genomes. Adv. Genet. 2012;80:37–97. doi: 10.1016/B978-0-12-404742-6.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Porteus M.H., Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 107.Bogdanove A.J., Voytas D.F. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 108.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N.S., Cai W., Yang G., Bronson R., Crowley D.G. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carlson D.F., Tan W., Lillico S.G., Stverakova D., Proudfoot C., Christian M., Voytas D.F., Long C.R., Whitelaw C.B., Fahrenkrug S.C. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lillico S.G., Proudfoot C., Carlson D.F., Stverakova D., Neil C., Blain C., King T.J., Ritchie W.A., Tan W., Mileham A.J. Live pigs produced from genome edited zygotes. Sci. Rep. 2013;3:2847. doi: 10.1038/srep02847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan W., Carlson D.F., Lancto C.A., Garbe J.R., Webster D.A., Hackett P.B., Fahrenkrug S.C. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc. Natl. Acad. Sci. USA. 2013;110:16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qian L., Tang M., Yang J., Wang Q., Cai C., Jiang S., Li H., Jiang K., Gao P., Ma D. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci. Rep. 2015;5:14435. doi: 10.1038/srep14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dowidar Y.A., El-Sayed M.A., Elrefy A.M., Shoura H.E. Detection of myostatin gene MSTN in some goat breeds (Capra hircus) J. Genet. Eng. Biotechnol. 2018 doi: 10.1016/j.jgeb.2018.04.002. Published online April 26, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kang J.-D., Kim S., Zhu H.-Y., Jin L., Guo Q., Li X.-C., Zhanga Y.-C., Xinga X.-X., Xuana M.-F., Zhanga G.-L. Generation of cloned adult muscular pigs with myostatin gene mutation by genetic engineering. RSC Adv. 2017;7:12541–12549. [Google Scholar]
- 116.Whitworth K.M., Rowland R.R., Ewen C.L., Trible B.R., Kerrigan M.A., Cino-Ozuna A.G., Samuel M.S., Lightner J.E., McLaren D.G., Mileham A.J. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016;34:20–22. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- 117.Telugu B.P., Park K.-E., Park C.-H. Genome editing and genetic engineering in livestock for advancing agricultural and biomedical applications. Mamm. Genome. 2017;28:338–347. doi: 10.1007/s00335-017-9709-4. [DOI] [PubMed] [Google Scholar]
- 118.Hauschild-Quintern J., Petersen B., Cost G.J., Niemann H. Gene knockout and knockin by zinc-finger nucleases: current status and perspectives. Cell. Mol. Life Sci. 2013;70:2969–2983. doi: 10.1007/s00018-012-1204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Doyon Y., Choi V.M., Xia D.F., Vo T.D., Gregory P.D., Holmes M.C. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat. Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 120.Watanabe M., Nakano K., Matsunari H., Matsuda T., Maehara M., Kanai T., Kobayashi M., Matsumura Y., Sakai R., Kuramoto M. Generation of interleukin-2 receptor gamma gene knockout pigs from somatic cells genetically modified by zinc finger nuclease-encoding mRNA. PLoS ONE. 2013;8:e76478. doi: 10.1371/journal.pone.0076478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Flisikowska T., Merkl C., Landmann M., Eser S., Rezaei N., Cui X., Kurome M., Zakhartchenko V., Kessler B., Wieland H. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143:1173–1175.e7. doi: 10.1053/j.gastro.2012.07.110. [DOI] [PubMed] [Google Scholar]
- 122.Luo Y., Bolund L., Sørensen C.B. Pig gene knockout by rAAV-mediated homologous recombination: comparison of BRCA1 gene knockout efficiency in Yucatan and Göttingen fibroblasts with slightly different target sequences. Transgenic Res. 2012;21:671–676. doi: 10.1007/s11248-011-9563-1. [DOI] [PubMed] [Google Scholar]
- 123.Cooper D.K., Ayares D. The immense potential of xenotransplantation in surgery. Int. J. Surg. 2011;9:122–129. doi: 10.1016/j.ijsu.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 124.Yang D., Yang H., Li W., Zhao B., Ouyang Z., Liu Z., Zhao Y., Fan N., Song J., Tian J. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hauschild J., Petersen B., Santiago Y., Queisser A.-L., Carnwath J.W., Lucas-Hahn A., Zhang L., Meng X., Gregory P.D., Schwinzer R. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 2011;108:12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu S., Luo J., Song Z., Ding F., Dai Y., Li N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 2011;21:1638–1640. doi: 10.1038/cr.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wei J., Wagner S., Maclean P., Brophy B., Cole S., Smolenski G., Carlson D.F., Fahrenkrug S.C., Wells D.N., Laible G. Cattle with a precise, zygote-mediated deletion safely eliminate the major milk allergen beta-lactoglobulin. Sci. Rep. 2018;8:7661. doi: 10.1038/s41598-018-25654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Boch J., Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 129.Palgrave C.J., Gilmour L., Lowden C.S., Lillico S.G., Mellencamp M.A., Whitelaw C.B.A. Species-specific variation in RELA underlies differences in NF-κB activity: a potential role in African swine fever pathogenesis. J. Virol. 2011;85:6008–6014. doi: 10.1128/JVI.00331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Proudfoot C., Carlson D.F., Huddart R., Long C.R., Pryor J.H., King T.J., Lillico S.G., Mileham A.J., McLaren D.G., Whitelaw C.B., Fahrenkrug S.C. Genome edited sheep and cattle. Transgenic Res. 2015;24:147–153. doi: 10.1007/s11248-014-9832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hai T., Teng F., Guo R., Li W., Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Petersen B., Niemann H. Molecular scissors and their application in genetically modified farm animals. Transgenic Res. 2015;24:381–396. doi: 10.1007/s11248-015-9862-z. [DOI] [PubMed] [Google Scholar]
- 133.Otieno M. CRISPR-Cas9 human genome editing: challenges, ethical concerns and implications. J. Clin. Res. Bioeth. 2015;6:253–255. [Google Scholar]
- 134.Liang P., Xu Y., Zhang X., Ding C., Huang R., Zhang Z., Lv J., Xie X., Chen Y., Li Y. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Munné S., Cohen J. Chromosome abnormalities in human embryos. Hum. Reprod. Update. 1998;4:842–855. doi: 10.1093/humupd/4.6.842. [DOI] [PubMed] [Google Scholar]
- 136.Bredenoord A.L., Pennings G., de Wert G. Ooplasmic and nuclear transfer to prevent mitochondrial DNA disorders: conceptual and normative issues. Hum. Reprod. Update. 2008;14:669–678. doi: 10.1093/humupd/dmn035. [DOI] [PubMed] [Google Scholar]
- 137.Sathananthan A.H., Tarin J.J., Gianaroli L., Ng S.C., Dharmawardena V., Magli M.C., Fernando R., Trounson A.O. Development of the human dispermic embryo. Hum. Reprod. Update. 1999;5:553–560. doi: 10.1093/humupd/5.5.553. [DOI] [PubMed] [Google Scholar]
- 138.Hill R.J., Konigsberg W., Guidotti G., Craig L.C. The structure of human hemoglobin. I. The separation of the α and β chains and their amino acid composition. J. Biol. Chem. 1962;237:1549–1554. [PubMed] [Google Scholar]
- 139.Schechter A.N. Hemoglobin research and the origins of molecular medicine. Blood. 2008;112:3927–3938. doi: 10.1182/blood-2008-04-078188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kang X., He W., Huang Y., Yu Q., Chen Y., Gao X., Sun X., Fan Y. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J. Assist. Reprod. Genet. 2016;33:581–588. doi: 10.1007/s10815-016-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Martinson J.J., Chapman N.H., Rees D.C., Liu Y.-T., Clegg J.B. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet. 1997;16:100–103. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- 142.Marmor M., Sheppard H.W., Donnell D., Bozeman S., Celum C., Buchbinder S., Koblin B., Seage G.R., 3rd, HIV Network for Prevention Trials Vaccine Preparedness Protocol Team Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J. Acquir. Immune Defic. Syndr. 2001;27:472–481. doi: 10.1097/00126334-200108150-00009. [DOI] [PubMed] [Google Scholar]
- 143.Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.-M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 144.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Geurts A.M., Cost G.J., Freyvert Y., Zeitler B., Miller J.C., Choi V.M., Jenkins S.S., Wood A., Cui X., Meng X. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gupta R.M., Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Invest. 2014;124:4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li W., Teng F., Li T., Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 148.Li D., Qiu Z., Shao Y., Chen Y., Guan Y., Liu M., Li Y., Gao N., Wang L., Lu X. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 149.Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.W., Xi J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jao L.-E., Wente S.R., Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li M., Suzuki K., Kim N.Y., Liu G.-H., Izpisua Belmonte J.C. A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J. Biol. Chem. 2014;289:4594–4599. doi: 10.1074/jbc.R113.488247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhu Z., Huangfu D. Human pluripotent stem cells: an emerging model in developmental biology. Development. 2013;140:705–717. doi: 10.1242/dev.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Byrne S.M., Mali P., Church G.M. Genome editing in human stem cells. Methods Enzymol. 2014;546:119–138. doi: 10.1016/B978-0-12-801185-0.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sancho-Martinez I., Li M., Izpisua Belmonte J.C. Disease correction the iPSC way: advances in iPSC-based therapy. Clin. Pharmacol. Ther. 2011;89:746–749. doi: 10.1038/clpt.2010.341. [DOI] [PubMed] [Google Scholar]
- 155.Qu X., Wang P., Ding D., Li L., Wang H., Ma L., Zhou X., Liu S., Lin S., Wang X. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41:7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P.-Q., Reik A., Chu V., Paschon D.E., Zhang L., Kuball J. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 2012;18:807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Perez E.E., Wang J., Miller J.C., Jouvenot Y., Kim K.A., Liu O., Wang N., Lee G., Bartsevich V.V., Lee Y.L. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.An M.C., O’Brien R.N., Zhang N., Patra B.N., De La Cruz M., Ray A., Ellerby L.M. Polyglutamine disease modeling: epitope based screen for homologous recombination using CRISPR/Cas9 system. PLoS Curr. 2014;6 doi: 10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ott de Bruin L.M., Volpi S., Musunuru K. Novel genome-editing tools to model and correct primary immunodeficiencies. Front. Immunol. 2015;6:250. doi: 10.3389/fimmu.2015.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hacein-Bey-Abina S., Le Deist F., Carlier F., Bouneaud C., Hue C., De Villartay J.-P., Thrasher A.J., Wulffraat N., Sorensen R., Dupuis-Girod S. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 161.Soldner F., Laganière J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L.I., Myers R.H., Lindquist S. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.González F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xiao-Jie L., Hui-Ying X., Zun-Ping K., Jin-Lian C., Li-Juan J. CRISPR-Cas9: a new and promising player in gene therapy. J. Med. Genet. 2015;52:289–296. doi: 10.1136/jmedgenet-2014-102968. [DOI] [PubMed] [Google Scholar]
- 164.Dias M.F., Joo K., Kemp J.A., Fialho S.L., da Silva Cunha A., Jr., Woo S.J., Kwon Y.J. Molecular genetics and emerging therapies for retinitis pigmentosa: basic research and clinical perspectives. Prog. Retin. Eye Res. 2018;63:107–131. doi: 10.1016/j.preteyeres.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 165.Salmikangas P., Kinsella N., Chamberlain P. Chimeric antigen receptor T-cells (CAR T-Cells) for cancer immunotherapy—moving target for industry? Pharm. Res. 2018;35:152. doi: 10.1007/s11095-018-2436-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ma N., Liao B., Zhang H., Wang L., Shan Y., Xue Y., Huang K., Chen S., Zhou X., Chen Y. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J. Biol. Chem. 2013;288:34671–34679. doi: 10.1074/jbc.M113.496174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Song B., Fan Y., He W., Zhu D., Niu X., Wang D., Ou Z., Luo M., Sun X. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 2015;24:1053–1065. doi: 10.1089/scd.2014.0347. [DOI] [PubMed] [Google Scholar]
- 168.Sun N., Zhao H. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol. Bioeng. 2014;111:1048–1053. doi: 10.1002/bit.25018. [DOI] [PubMed] [Google Scholar]
- 169.Zou J., Mali P., Huang X., Dowey S.N., Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sun N., Liang J., Abil Z., Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol. Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 171.Huang X., Wang Y., Yan W., Smith C., Ye Z., Wang J., Gao Y., Mendelsohn L., Cheng L. Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells. 2015;33:1470–1479. doi: 10.1002/stem.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pellagatti A., Dolatshad H., Yip B.H., Valletta S., Boultwood J. Application of genome editing technologies to the study and treatment of hematological disease. Adv. Biol. Regul. 2016;60:122–134. doi: 10.1016/j.jbior.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 173.Park C.Y., Kim J., Kweon J., Son J.S., Lee J.S., Yoo J.E., Cho S.R., Kim J.H., Kim J.S., Kim D.W. Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc. Natl. Acad. Sci. USA. 2014;111:9253–9258. doi: 10.1073/pnas.1323941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bolton-Maggs P.H.B., Pasi K.J. Haemophilias A and B. Lancet. 2003;361:1801–1809. doi: 10.1016/S0140-6736(03)13405-8. [DOI] [PubMed] [Google Scholar]
- 175.Li H., Haurigot V., Doyon Y., Li T., Wong S.Y., Bhagwat A.S., Malani N., Anguela X.M., Sharma R., Ivanciu L. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Merling R.K., Sweeney C.L., Chu J., Bodansky A., Choi U., Priel D.L., Kuhns D.B., Wang H., Vasilevsky S., De Ravin S.S. An AAVS1-targeted minigene platform for correction of iPSCs from all five types of chronic granulomatous disease. Mol. Ther. 2015;23:147–157. doi: 10.1038/mt.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zou J., Sweeney C.L., Chou B.K., Choi U., Pan J., Wang H., Dowey S.N., Cheng L., Malech H.L. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117:5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Connelly J.P., Kwon E.M., Gao Y., Trivedi N.S., Elkahloun A.G., Horwitz M.S., Cheng L., Liu P.P. Targeted correction of RUNX1 mutation in FPD patient-specific induced pluripotent stem cells rescues megakaryopoietic defects. Blood. 2014;124:1926–1930. doi: 10.1182/blood-2014-01-550525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schifferli A., Kühne T. Fanconi anemia: overview of the disease and the role of hematopoietic transplantation. J. Pediatr. Hematol. Oncol. 2015;37:335–343. doi: 10.1097/MPH.0000000000000374. [DOI] [PubMed] [Google Scholar]
- 180.Whitney M.A., Saito H., Jakobs P.M., Gibson R.A., Moses R.E., Grompe M. A common mutation in the FACC gene causes Fanconi anaemia in Ashkenazi Jews. Nat. Genet. 1993;4:202–205. doi: 10.1038/ng0693-202. [DOI] [PubMed] [Google Scholar]
- 181.Osborn M.J., Gabriel R., Webber B.R., DeFeo A.P., McElroy A.N., Jarjour J., Starker C.G., Wagner J.E., Joung J.K., Voytas D.F. Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum. Gene Ther. 2015;26:114–126. doi: 10.1089/hum.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gjini E., Mansour M.R., Sander J.D., Moritz N., Nguyen A.T., Kesarsing M., Gans E., He S., Chen S., Ko M. A zebrafish model of myelodysplastic syndrome produced through tet2 genomic editing. Mol. Cell. Biol. 2015;35:789–804. doi: 10.1128/MCB.00971-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rathe S.K., Moriarity B.S., Stoltenberg C.B., Kurata M., Aumann N.K., Rahrmann E.P., Bailey N.J., Melrose E.G., Beckmann D.A., Liska C.R., Largaespada D.A. Using RNA-seq and targeted nucleases to identify mechanisms of drug resistance in acute myeloid leukemia. Sci. Rep. 2014;4:6048. doi: 10.1038/srep06048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nagai M., Osame M. Human T-cell lymphotropic virus type I and neurological diseases. J. Neurovirol. 2003;9:228–235. doi: 10.1080/13550280390194028. [DOI] [PubMed] [Google Scholar]
- 185.Tanaka A., Takeda S., Kariya R., Matsuda K., Urano E., Okada S., Komano J. A novel therapeutic molecule against HTLV-1 infection targeting provirus. Leukemia. 2013;27:1621–1627. doi: 10.1038/leu.2013.46. [DOI] [PubMed] [Google Scholar]
- 186.Wang J., Quake S.R. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc. Natl. Acad. Sci. USA. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O’Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 188.Cox D.B., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gaspar H.B., Cooray S., Gilmour K.C., Parsley K.L., Adams S., Howe S.J., Al Ghonaium A., Bayford J., Brown L., Davies E.G. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 190.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Morris E.C., Fox T., Chakraverty R., Tendeiro R., Snell K., Rivat C., Grace S., Gilmour K., Workman S., Buckland K. Gene therapy for Wiskott-Aldrich syndrome in a severely affected adult. Blood. 2017;130:1327–1335. doi: 10.1182/blood-2017-04-777136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Guzman-Aranguez A., Loma P., Pintor J. Small-interfering RNAs (siRNAs) as a promising tool for ocular therapy. Br. J. Pharmacol. 2013;170:730–747. doi: 10.1111/bph.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mansoori B., Sandoghchian Shotorbani S., Baradaran B. RNA interference and its role in cancer therapy. Adv. Pharm. Bull. 2014;4:313–321. doi: 10.5681/apb.2014.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rizk M., Tüzmen Ş. Update on the clinical utility of an RNA interference-based treatment: focus on Patisiran. Pharm. Genomics Pers. Med. 2017;10:267–278. doi: 10.2147/PGPM.S87945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liu R., Paxton W.A., Choe S., Ceradini D., Martin S.R., Horuk R., MacDonald M.E., Stuhlmann H., Koup R.A., Landau N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 197.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 198.Paulk N.K., Wursthorn K., Wang Z., Finegold M.J., Kay M.A., Grompe M. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51:1200–1208. doi: 10.1002/hep.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chen Y., Zheng Y., Kang Y., Yang W., Niu Y., Guo X., Tu Z., Si C., Wang H., Xing R. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Amoasii L., Long C., Li H., Mireault A.A., Shelton J.M., Sanchez-Ortiz E., McAnally J.R., Bhattacharyya S., Schmidt F., Grimm D. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 2017;9:eaan8081. doi: 10.1126/scitranslmed.aan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tabebordbar M., Zhu K., Cheng J.K.W., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sánchez-Rivera F.J., Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Engelman J.A., Chen L., Tan X., Crosby K., Guimaraes A.R., Upadhyay R., Maira M., McNamara K., Perera S.A., Song Y. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen Z., Cheng K., Walton Z., Wang Y., Ebi H., Shimamura T., Liu Y., Tupper T., Ouyang J., Li J. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–617. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Malina A., Mills J.R., Cencic R., Yan Y., Fraser J., Schippers L.M., Paquet M., Dostie J., Pelletier J. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 2013;27:2602–2614. doi: 10.1101/gad.227132.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen C., Liu Y., Rappaport A.R., Kitzing T., Schultz N., Zhao Z., Shroff A.S., Dickins R.A., Vakoc C.R., Bradner J.E. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 210.Sadelain M., Brentjens R., Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 212.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 213.Amit D., Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J. Transl. Med. 2010;8:134. doi: 10.1186/1479-5876-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu Y., Zeng Y., Liu L., Zhuang C., Fu X., Huang W., Cai Z. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun. 2014;5:5393. doi: 10.1038/ncomms6393. [DOI] [PubMed] [Google Scholar]
- 215.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 217.Blasco R.B., Karaca E., Ambrogio C., Cheong T.C., Karayol E., Minero V.G., Voena C., Chiarle R. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 218.Maddalo D., Manchado E., Concepcion C.P., Bonetti C., Vidigal J.A., Han Y.C., Ogrodowski P., Crippa A., Rekhtman N., de Stanchina E. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 220.Tu Z., Yang W., Yan S., Guo X., Li X.J. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol. Neurodegener. 2015;10:35. doi: 10.1186/s13024-015-0031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Jung Y.W., Hysolli E., Kim K.Y., Tanaka Y., Park I.H. Human induced pluripotent stem cells and neurodegenerative disease: prospects for novel therapies. Curr. Opin. Neurol. 2012;25:125–130. doi: 10.1097/WCO.0b013e3283518226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ross C.A., Akimov S.S. Human-induced pluripotent stem cells: potential for neurodegenerative diseases. Hum. Mol. Genet. 2014;23(R1):R17–R26. doi: 10.1093/hmg/ddu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Freude K., Pires C., Hyttel P., Hall V.J. Induced pluripotent stem cells derived from Alzheimer’s disease patients: the promise, the hope and the path ahead. J. Clin. Med. 2014;3:1402–1436. doi: 10.3390/jcm3041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Corrigendum: increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2016;34:210. doi: 10.1038/nbt0216-210c. [DOI] [PubMed] [Google Scholar]
- 225.Kramer N.J., Haney M.S., Morgens D.W., Jovičić A., Couthouis J., Li A., Ousey J., Ma R., Bieri G., Tsui C.K. CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet. 2018;50:603–612. doi: 10.1038/s41588-018-0070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.UNAIDS . Joint United Nations Programme on HIV/AIDS; 2012. Global Report: UNAIDS Report on the Global AIDS Epidemic 2012. [Google Scholar]
- 227.Siliciano R.F., Greene W.C. HIV latency. Cold Spring Harb. Perspect. Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Donahue D.A., Wainberg M.A. Cellular and molecular mechanisms involved in the establishment of HIV-1 latency. Retrovirology. 2013;10:11. doi: 10.1186/1742-4690-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Saayman S., Ali S.A., Morris K.V., Weinberg M.S. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin. Biol. Ther. 2015;15:819–830. doi: 10.1517/14712598.2015.1036736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., Luo B., Alvarez-Carbonell D., Garcia-Mesa Y., Karn J. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Biti R., Ffrench R., Young J., Bennetts B., Stewart G., Liang T. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat. Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 232.Siliciano J.D., Kajdas J., Finzi D., Quinn T.C., Chadwick K., Margolick J.B., Kovacs C., Gange S.J., Siliciano R.F. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 233.Zhu W., Lei R., Le Duff Y., Li J., Guo F., Wainberg M.A., Liang C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Liao H.K., Gu Y., Diaz A., Marlett J., Takahashi Y., Li M., Suzuki K., Xu R., Hishida T., Chang C.J. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 237.Curtin S.J., Voytas D.F., Stupar R.M. Genome engineering of crops with designer nucleases. Plant Genome. 2012;5:42–50. [Google Scholar]
- 238.Voytas D.F. Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 2013;64:327–350. doi: 10.1146/annurev-arplant-042811-105552. [DOI] [PubMed] [Google Scholar]
- 239.Pennisi E. The CRISPR craze. Science. 2013;341:833–836. doi: 10.1126/science.341.6148.833. [DOI] [PubMed] [Google Scholar]
- 240.Wyman C., Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 241.Zhang Y., Zhang F., Li X., Baller J.A., Qi Y., Starker C.G., Bogdanove A.J., Voytas D.F. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013;161:20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Shukla V.K., Doyon Y., Miller J.C., DeKelver R.C., Moehle E.A., Worden S.E., Mitchell J.C., Arnold N.L., Gopalan S., Meng X. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 243.Lusser M., Parisi C., Plan D., Rodríguez-Cerezo E. Deployment of new biotechnologies in plant breeding. Nat. Biotechnol. 2012;30:231–239. doi: 10.1038/nbt.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Podevin N., Davies H.V., Hartung F., Nogué F., Casacuberta J.M. Site-directed nucleases: a paradigm shift in predictable, knowledge-based plant breeding. Trends Biotechnol. 2013;31:375–383. doi: 10.1016/j.tibtech.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 245.Waltz E. Tiptoeing around transgenics. Nat. Biotechnol. 2012;30:215–217. doi: 10.1038/nbt.2143. [DOI] [PubMed] [Google Scholar]
- 246.Hartung F., Schiemann J. Precise plant breeding using new genome editing techniques: opportunities, safety and regulation in the EU. Plant J. 2014;78:742–752. doi: 10.1111/tpj.12413. [DOI] [PubMed] [Google Scholar]
- 247.Bortesi L., Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 248.Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 249.Ainley W.M., Sastry-Dent L., Welter M.E., Murray M.G., Zeitler B., Amora R., Corbin D.R., Miles R.R., Arnold N.L., Strange T.L. Trait stacking via targeted genome editing. Plant Biotechnol. J. 2013;11:1126–1134. doi: 10.1111/pbi.12107. [DOI] [PubMed] [Google Scholar]
- 250.Li T., Liu B., Spalding M.H., Weeks D.P., Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 251.Shan Q., Wang Y., Chen K., Liang Z., Li J., Zhang Y., Zhang K., Liu J., Voytas D.F., Zheng X. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant. 2013;6:1365–1368. doi: 10.1093/mp/sss162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Baltes N.J., Gil-Humanes J., Cermak T., Atkins P.A., Voytas D.F. DNA replicons for plant genome engineering. Plant Cell. 2014;26:151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhang X.-H., Tee L.Y., Wang X.-G., Huang Q.-S., Yang S.-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 256.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 257.Tzur Y.B., Friedland A.E., Nadarajan S., Church G.M., Calarco J.A., Colaiácovo M.P. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics. 2013;195:1181–1185. doi: 10.1534/genetics.113.156075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yang D., Xu J., Zhu T., Fan J., Lai L., Zhang J., Chen Y.E. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J. Mol. Cell Biol. 2014;6:97–99. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 262.Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 263.Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 264.Ramanan V., Shlomai A., Cox D.B., Schwartz R.E., Michailidis E., Bhatta A., Scott D.A., Zhang F., Rice C.M., Bhatia S.N. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Seeger C., Sohn J.A. Targeting hepatitis B virus with CRISPR/Cas9. Mol. Ther. Nucleic Acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Porteus M.H., Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 267.Guilinger J.P., Pattanayak V., Reyon D., Tsai S.Q., Sander J.D., Joung J.K., Liu D.R. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods. 2014;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Li T., Huang S., Zhao X., Wright D.A., Carpenter S., Spalding M.H., Weeks D.P., Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X., Skarnes W.C. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 273.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 279.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C.A., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Khaled W.T., Liu P. Cancer mouse models: past, present and future. Semin. Cell Dev. Biol. 2014;27:54–60. doi: 10.1016/j.semcdb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 282.Crystal A.S., Shaw A.T., Sequist L.V., Friboulet L., Niederst M.J., Lockerman E.L., Frias R.L., Gainor J.F., Amzallag A., Greninger P. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Feng Z., Zhang B., Ding W., Liu X., Yang D.L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., Zhu J.K. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., Zhang K., Liu J., Xi J.J., Qiu J.L., Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 286.Xu R., Li H., Qin R., Wang L., Li L., Wei P., Yang J. Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice (N. Y.) 2014;7:5. doi: 10.1186/s12284-014-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zhou H., Liu B., Weeks D.P., Spalding M.H., Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Liang Z., Zhang K., Chen K., Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics. 2014;41:63–68. doi: 10.1016/j.jgg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 289.Gao H., Smith J., Yang M., Jones S., Djukanovic V., Nicholson M.G., West A., Bidney D., Falco S.C., Jantz D., Lyznik L.A. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61:176–187. doi: 10.1111/j.1365-313X.2009.04041.x. [DOI] [PubMed] [Google Scholar]
- 290.Marton I., Honig A., Omid A., De Costa N., Marhevka E., Cohen B., Zuker A., Vainstein A. From Agrobacterium to viral vectors: genome modification of plant cells by rare cutting restriction enzymes. Int. J. Dev. Biol. 2013;57:639–650. doi: 10.1387/ijdb.130205av. [DOI] [PubMed] [Google Scholar]
- 291.Roth N., Klimesch J., Dukowic-Schulze S., Pacher M., Mannuss A., Puchta H. The requirement for recombination factors differs considerably between different pathways of homologous double-strand break repair in somatic plant cells. Plant J. 2012;72:781–790. doi: 10.1111/j.1365-313X.2012.05119.x. [DOI] [PubMed] [Google Scholar]
- 292.Curtin S.J., Zhang F., Sander J.D., Haun W.J., Starker C., Baltes N.J., Reyon D., Dahlborg E.J., Goodwin M.J., Coffman A.P. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 2011;156:466–473. doi: 10.1104/pp.111.172981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Jacobs T.B., LaFayette P.R., Schmitz R.J., Parrott W.A. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015;15:16. doi: 10.1186/s12896-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Haun W., Coffman A., Clasen B.M., Demorest Z.L., Lowy A., Ray E., Retterath A., Stoddard T., Juillerat A., Cedrone F. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 2014;12:934–940. doi: 10.1111/pbi.12201. [DOI] [PubMed] [Google Scholar]
- 295.D’Halluin K., Vanderstraeten C., Van Hulle J., Rosolowska J., Van Den Brande I., Pennewaert A., D’Hont K., Bossut M., Jantz D., Ruiter R., Broadhvest J. Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol. J. 2013;11:933–941. doi: 10.1111/pbi.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gupta M., DeKelver R.C., Palta A., Clifford C., Gopalan S., Miller J.C., Novak S., Desloover D., Gachotte D., Connell J. Transcriptional activation of Brassica napus β-ketoacyl-ACP synthase II with an engineered zinc finger protein transcription factor. Plant Biotechnol. J. 2012;10:783–791. doi: 10.1111/j.1467-7652.2012.00695.x. [DOI] [PubMed] [Google Scholar]
- 297.Upadhyay S.K., Kumar J., Alok A., Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 2013;3:2233–2238. doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Wendt T., Holm P.B., Starker C.G., Christian M., Voytas D.F., Brinch-Pedersen H., Holme I.B. TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol. Biol. 2013;83:279–285. doi: 10.1007/s11103-013-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Gurushidze M., Hensel G., Hiekel S., Schedel S., Valkov V., Kumlehn J. True-breeding targeted gene knock-out in barley using designer TALE-nuclease in haploid cells. PLoS ONE. 2014;9:e92046. doi: 10.1371/journal.pone.0092046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Miller J.C., Pabo C.O. Rearrangement of side-chains in a zif268 mutant highlights the complexities of zinc finger-DNA recognition. J. Mol. Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- 301.Pavletich N.P., Pabo C.O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]