Summary
Sucrose non‐fermenting‐1‐related protein kinase‐1 (SnRK1) is an essential energy‐sensing regulator and plays a key role in the global control of carbohydrate metabolism. The SnRK1 gene has been found to increase starch accumulation in several plant species. However, its roles in improving starch quality have not been reported to date. In this study, we found that the IbSnRK1 gene was highly expressed in the storage roots of sweet potato and strongly induced by exogenous sucrose. Its expression followed the circandian rhythm. Its overexpression not only increased starch content, but also decreased proportion of amylose, enlarged granule size and improved degree of crystallinity and gelatinization in transgenic sweet potato, which revealed, for the first time, the important roles of SnRK1 in improving starch quality of plants. The genes involved in starch biosynthesis pathway were systematically up‐regulated, and the content of ADP‐glucose as an important precursor for starch biosynthesis and the activities of key enzymes were significantly increased in transgenic sweet potato. These findings indicate that IbSnRK1 improves starch content and quality through systematical up‐regulation of the genes and the increase in key enzyme activities involved in starch biosynthesis pathway in transgenic sweet potato. This gene has the potential to improve starch content and quality in sweet potato and other plants.
Keywords: IbSnRK1, starch content, starch quality, sweet potato
Introduction
In plants, starch is the most important storage carbohydrate, which is widely used in our life and industries (Horrer et al., 2016; Thalmann et al., 2016; Zeeman et al., 2010). Starch usually exists in crop plants storage tissues such as endosperms and storage tubers/roots. Its production is critical to both yield and quality of crops, and high content and quality of starch in crops have become the main targets of breeders (Burrell, 2003; Volpicella et al., 2016).
Starch in plants has two forms of glucan polymer, amylose (10%–25%) and amylopectin (75%–90%) (Tester et al., 2004). Amylose is a linear or infrequently branched molecule which is composed of α‐1, 4‐linked D‐glucosyl units and <1% α‐1,‐6 linkages. Amylopectin is a highly branched glucan which consists of short α‐1,‐4‐linked D‐glucosyl chains with 5%–6% α‐1, 6 linkages (Ball and Morell, 2003; James et al., 2003; Tetlow et al., 2004). Starch biosynthesis is a complicated process, and a series of enzymes are generally considered to be linked with its biosynthesis, including invertase (acid invertase and neutral invertase), sucrose synthase (SuSy), ADP‐glucose pyrophosphorylase (AGPase), granule‐bound starch synthase (GBSS, also called Waxy), soluble starch synthase (SS), starch‐branching enzyme (SBE) and starch‐debranching enzyme (DBE) (Abe et al., 2013; James et al., 2003).
Sucrose non‐fermenting‐1‐related protein kinase‐1 (SnRK1) is an essential energy‐sensing regulator and plays a key role in the global control of carbohydrate metabolism (Halford and Hrdie, 1998). The genes encoding SnRK1 have been cloned from several plant species such as rye (Alderson et al., 1991), potato (Lakatos et al., 1999), Arabidopsis (Kleinow et al., 2000), maize (Lumbreras et al., 2001), sorghum (Jain et al., 2008) and Malus hupehensis Rehd. (Li et al., 2010). Antisense inhibition of SnRK1 in developing pollen grains caused an almost complete loss of starch accumulation in transgenic barley (Zhang et al., 2001). Kanegae et al. (2005) found that SnRK1 had a role in starch accumulation of rice. Starch level was increased by 30% in the tubers of the SnRK1‐overexpressing potato plants (McKibbin et al., 2006). The soluble sugar and starch contents were significantly increased in transgenic tomato overexpressing a heterologous SnRK1 gene from M. hupehensis Rehd. (Wang et al., 2012). However, roles of SnRK1 in improving starch quality have not been reported to date.
Sweet potato, Ipomoea batatas (L.) Lam., is an important starch crop. Its potential as food and carbohydrate source has been widely recognized (Jarret et al., 1992; Jiang et al., 2013). Improving starch content and quality of this crop remains an urgent demand, especially in the field of biotechnology. Several genes encoding the starch biosynthesis enzymes, including GBSSI, SSI, SBEI and SBEII, have been isolated from sweet potato and their overexpression or down‐regulation by RNAi impacts starch content, composition and physicochemical properties of this crop (Hamada et al., 2006; Kimura et al., 2001; Otani et al., 2007; Shimada et al., 2006; Wang et al., 2017b; Zhou et al., 2015). Tanaka et al. (2009) found that the storage root dry matter content and starch content were significantly increased in transgenic sweet potato overexpressing SRF1 encoding a Dof zinc finger transcription factor. Overexpression of IbAATP encoding a plastidic ATP/ADP transporter protein significantly increased starch and amylose contents, enlarged starch granules and altered fine structure of amylopectin in transgenic sweet potato (Wang et al., 2016b).
In our previous study, one IbSnRK1 gene was isolated from sweet potato cv. Lushu 3 (Jiang et al., 2013), and its overexpression increased the nitrate N content in roots and soluble protein and starch contents in leaves of transgenic sweet potato (Ren et al., 2018). In this study, we found that its overexpression not only increased starch content, but also decreased proportion of amylose, enlarged granule size and improved degree of crystallinity and gelatinization in the storage roots of transgenic sweet potato.
Results
Expression of IbSnRK1 in sweet potato
Real‐time quantitative PCR (qRT‐PCR) analysis showed that the expression levels of IbSnRK1 in sweet potato cultivars Lizixiang, Xushu 18, Shangshu 19 and Lushu 3 with different starch content were positively correlated with their starch contents (Figure 1a). The highest expression level of IbSnRK1 was found in the storage roots among various tissues of Lushu 3 (Figure 1b). The expression of IbSnRK1 was strongly induced by exogenous sucrose and reached the highest level at 12 h after 175 mm sucrose treatment in the leaves of Lushu 3 (Figure 1c). Its expression in Lushu 3 followed the circandian rhythm and peaked at 10 am (P2, P5, P8 and P11) in each cycle, which displayed a sine‐wave pattern with the circadian rhythm treatment (Figure 1d).
Figure 1.

Expression analysis of IbSnRK1 in sweet potato. (a) Starch content and IbSnRK1 expression in the storage roots of different cultivars. (b) Expression of IbSnRK1 in different tissues of Lushu 3. L, leaf; P, petiole; S, stem; SR, storage root; FR, fibrous root. (c) Expression of IbSnRK1 in response to 175 mm sucrose treatment. The treatment with H2O was used as control. (d) Time settings used to examine the circadian rhythm during 16‐h light/8‐h dark photoperiods with light on at 5 am and off at 9 pm. Total RNA was extracted from the in vitro grown plants sampled at 10 pm (P1, P4, P7 and P10), 10 am (P2, P5, P8 and P11) and 4 pm (P3, P6, P9 and P12), respectively. The results are expressed as relative values with respect to P1 (set to 1.0). Data are presented as the mean ± SD (n = 3). * and different lowercase letters indicate a significant difference at P < 0.05; ** and different capital letters indicate a significant difference at P < 0.01 by Student's t‐test.
Production of the IbSnRK1‐overexpressing sweet potato plants
A total of 285 putatively transgenic plants were produced from the 1800 cell aggregates of sweet potato cv. Lizixing cocultivated with Agrobacterium tumefaciens (Figure S1a–e). GUS assay showed that 18 of them, named L1, L2, L3, …, L18, respectively, had visible GUS activity in leaf, stem and root tissues (Figure S1f–h). PCR analysis of genomic DNA confirmed the presence of IbSnRK1 in the 18 GUS‐positive plants and the absence of IbSnRK1 in the wild type (WT) and empty vector control (VC) (Figure S1i). The transgenic plants, WT and VC, were transferred to soils in a greenhouse and then in a field, and no morphological variations were observed among them (Figure S1j–m).
Starch and amylose contents
qRT‐PCR analysis showed that the expression levels of IbSnRK1 were significantly higher in the transgenic sweet potato plants than in WT and VC (Figure 2a). The starch content in the transgenic plant storage roots was increased by 1.7%–31.3% compared with WT, while the proportion of amylose (except for L2, L3 and L6) was decreased to 16.1%–17.8% from 18.0% of WT (decreased by 1.4%–10.7%) (Figure 2b,c). No differences in starch and amylose contents were found between WT and VC (Figure 2b,c).
Figure 2.

Expression analysis of IbSnRK1 (a), starch content (b) and amylose proportion (c) in the storage roots of the transgenic sweet potato plants, WT and VC. The sweet potato β‐actin gene was used as an internal control. Data are presented as the mean ± SD (n = 3). * and ** indicate a significant difference compared with WT at P < 0.05 and P < 0.01, respectively, by Student's t‐test.
Starch granule morphology and size
Four transgenic sweet potato plants with the highest starch content, L13, L14, L17 and L18, were selected for their starch quality evaluation. The number of starch granules in the amyloplasts of freshly harvested storage roots of the four transgenic plants was obviously increased compared with WT and VC (Figure 3a), but no variations in morphology of starch granules were observed among them (Figure 3b). Starch granule diameters of WT and VC were 5.2–76.0 μm and 5.2–66.9 μm, with a mean volume diameter (MV) of 22.1 and 22.3 μm, respectively, and their distribution displayed a unimodal pattern with a single peak at approximately 18 μm (Figure 3c). Most of the starch granules in WT and VC were <50 μm in diameter, and the proportions of <25 μm, 25–50 μm and >50 μm in WT were 51.5%, 42.1% and 6.4%, respectively, and in VC 50.5%, 43.5% and 6.0%, respectively (Figure 3d). In the transgenic plants, starch granule sizes ranged from 4.0 to 400.0 μm, with the MV of 25.4 to 29.4 μm, and their distributions exhibited two peaks at approximately 16 and 130 μm (Figure 3c). There were no differences in the proportion of <25 μm between the transgenic plants and WT/VC, but the proportions of 25–50 μm were decreased to 11.2%–17.5% and the proportions of >50 μm were increased to 41.6%–46.6% (Figure 3d).
Figure 3.
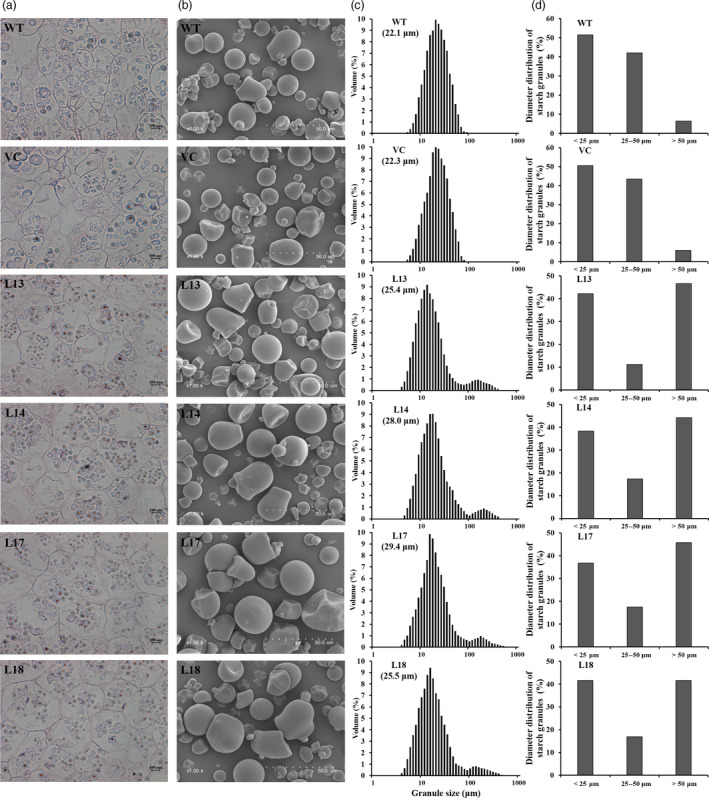
Morphology and size distribution of starch granules from the storage roots of the transgenic sweet potato plants, WT and VC. (a) Starch granules in amyloplasts. The bar indicates a length of 100 μm. (b) Scanning electron micrographs of the starch granules. The dotted line indicates a length of 50 μm. (c) Size distribution and mean volume diameter (MV) of starch granules. (d) Comparison of the starch granule diameter.
Chain length distribution
Chain length distribution (CLD) of amylopectin exhibited a similar pattern in the four transgenic plants and WT/VC, and four peaks were present at approximately degree of polymerization (DP) 7, DP 12, DP 17 and DP 46 in L13, L14, WT and VC, but only three peaks (DP 7, DP 12 and DP 46) were found in L17 and L18 (Figure 4a). The value for the amylopectin glucan chain of WT was subtracted from the corresponding value of the transgenic plants to generate the CLD difference models (Figure 4b). The results indicated that the number of chains with DP 5–8 was markedly increased, but that of DP 9–19 was decreased, followed by the increase in DP 20–48 (Figure 4b).
Figure 4.

Chain length distribution (CLD) of amylopectin from the transgenic sweet potato plants, WT and VC. (a) CLD of the amylopectin after normalization to the total peak area. (b) Differences of CLD between the transgenic plants and WT were calculated as follow: the normalized CLD value for each transgenic plant and VC minus the value obtained for WT. DP, degree of polymerization.
Analysis of X‐ray diffraction
X‐ray diffractograms showed that starch from the transgenic sweet potato plants, WT and VC, exhibited a similar pattern with three strong reflections at 2θ of about 15°, 17° and 23° (Figure 5a), which indicated their crystal types were all A‐type (Table S1). Degree of crystallinity of starch from WT and VC was 42.1% and 41.8%, respectively, but that of the transgenic plants was increased to 44.9%–47.4% (Table S1).
Figure 5.

Wide‐angle X‐ray powder diffraction spectra (a) and differential scanning calorimeter thermograms (b) of starches from the storage roots of the transgenic sweet potato plants, WT and VC.
Thermal characterization
The transgenic sweet potato plants exhibited different patterns of starch gelatinization from WT and VC (Figure 5b). The starch of WT gelatinized at a temperature range of 60.2 °C (onset tempertature, T o) to 82.4 °C (conclusion temperature, T c) with an enthalpy (ΔH) of 11.1 J/g. However, T o, T c and ΔH of starch from the transgenic plants were significantly decreased, indicating their starch was earlier to gelatinize, compared with WT (Table S2).
Expression of starch biosynthesis genes
The genes involved in starch biosynthesis pathway, including VacINV, NI, SuSy, HXK, UGPase, PGM, AGP‐L1, GBSSI, SSI, SSII, SSIII and SBEII (except for GBSSII, SSIV and SBEI) encoding acid invertase, neutral invertase, sucrose synthase, hexokinase, UDP‐glucose pyrophosphorylase, phosphoglucomutase, AGPase, GBSSI, SSI, SSII, SSIII and SBEII, respectively, were systematically up‐regulated in the storage roots of transgenic sweet potato plants compared with WT and VC (Figure 6). The transcript level of PUL encoding starch‐debranching pullulanase was significantly decreased, but IsaI encoding starch‐debranching isoamylase showed no change in expression level (Figure 6).
Figure 6.

Expression of the genes involved in starch biosynthesis pathway in the storage roots of the transgenic sweet potato plants, WT and VC. Data are presented as the mean ± SD (n = 3). * and ** indicate a significant difference compared with WT at P < 0.05 and P < 0.01, respectively, by Student's t‐test.
The activities of starch biosynthesis key enzymes
The activities of key enzymes, acid invertase, neutral invertase, SuSy, AGPase, SS, GBSS and SBE for starch biosynthesis were significantly increased to 7.3–12.2, 2.58–4.26, 1.12–1.61, 2.0–4.1, 2.0–3.1, 2.9–5.4 and 1.6–2.8 folds in the storage roots of the transgenic sweet potato plants compared with WT, which were consistent with the expression of the corresponding genes (Figures 6 and 7). No differences were found in the activities between WT and VC.
Figure 7.

Enzyme activities of acid invertase, neutral invertase, SuSy, AGPase, GBSS, SS and SBE in the storage roots of the transgenic sweet potato plants, WT and VC. Data are presented as the mean ± SD (n = 3). * and ** indicate a significant difference compared with WT at P < 0.05 and P < 0.01, respectively, by Student's t‐test.
The contents of components related to starch biosynthesis
The contents of sucrose, glucose, fructose, glucose 6‐phosphate (glc‐6‐P) and glucose‐1‐phosphate (Glc‐1‐P) were significantly decreased, but those of uridine diphosphate glucose (UDP‐glc), adenosine triphosphate (ATP), adenosine diphosphate glucose (ADP‐glc) and ADP were significantly increased in transgenic sweet potato plants compared with WT (Figure 8). No differences in the content of these components were found between WT and VC (Figure 8).
Figure 8.

The content of components related to starch biosynthesis in the storage roots of the transgenic sweet potato plants, WT and VC. Data are presented as the mean ± SD (n = 3). * and ** indicate a significant difference compared with WT at P < 0.05 and P < 0.01, respectively, by Student's t‐test.
Discussion
The SnRK1 gene has been shown to increase starch accumulation in several plant species such as barley (Zhang et al., 2001), rice (Kanegae et al., 2005) and potato (McKibbin et al., 2006), but its roles in improving starch quality of plants have not been reported to date. In the present study, the expression of IbSnRK1 was strongly induced by exogenous sucrose (Figure 1c) and followed the circandian rhythm (Figure 1d). Endogenous circadian clock has been shown to affect starch accumulation in several plant species (Wang et al., 2001). Its overexpression not only increased starch content, but also decreased proportion of amylose, increased number of starch granules, enlarged mean granule size and improved degree of crystallinity and gelatinization in transgenic sweet potato. These findings reveal, for the first time, the important roles of SnRK1 in improving starch quality of plants (Figures 3, 4, 5; Tables S1 and S2).
The content and composition of starch directly impact the starch yield and quality of crops (Bahaji et al., 2014; Burrell, 2003; Volpicella et al., 2016). The amylose–amylopectin ratio is a key factor that affects starch properties and is also important for food and bio‐industry applications of starches (Zeeman et al., 2010; Zhou et al., 2015). Low‐amylose starch is very important for optimizing industrial applications and allowing consumers to select crop varieties for health benefits (BeMiller and Whistler, 2009; Chung et al., 2011). Sweet potato starch is normally composed of 20%–30% amylose and 70%–80% amylopectin (Zhou et al., 2015). Its starch granules normally range from 4 to 40 μm, with an average size of 19 μm (Hoover, 2001). In this study, the content of starch and the number and size of starch granules were significantly increased, while the proportion of amylose was significantly decreased in the transgenic sweet potato lines (Figures 2 and 3). The increased number and size of starch granules might contribute to more accumulation of starch in the transgenic lines. By practical experience, the enlarged starch granules are beneficial to the extraction and separation of starch from the storage roots of sweet potato and its industrial applications.
There is a negative correlation between degree of crystallinity and amylose content, but a positive correlation between gelatinization and amylose content in plants (Cheetham and Tao, 1998; Copeland et al., 2009; Zhou et al., 2015). Our study showed that the four transgenic lines exhibited altered starch chain lengths, increased degree of crystallinity and decreased gelatinization temperature and enthalpy (Figure 4 and 5; Tables S1 and S2). These results indicated that the alternation of starch chain lengths led to the low proportion of amylose, which resulted in the increased degree of crystallinity and decreased gelatinization temperature and enthalpy in the transgenic lines.
As is well known, the starch biosynthesis is a sophisticated and systematic process (Ohdan et al., 2005). Sucrose can switch on the expression of starch biosynthesis‐related genes to enhance starch accumulation (Ahn et al., 2010; Hattori et al., 1991). Invertase (acid invertase and neutral invertase) and SuSy are the key enzymes for the conversion of sucrose to starch in starch biosynthesis pathway and catalyse the conversion of sucrose to glucose and fructose and the cleavage of sucrose and UDP to UDP‐glucose and fructose, respectively (Keeling and Myers, 2010; Koch, 2004; Roitsch and González, 2004). Hajirezaei et al. (2000) found that both invertase and SuSy exhibited significantly higher activities at different developmental stages of potato tubers. AGPase plays a critical role in the first committed step of starch biosynthesis and converts Glc‐1‐P into ADP‐glc, and ADP‐glc is the precursor for the biosynthesis of amylose and amylopectin (Yang et al., 2017). GBSS is mainly involved in the elongation of glucan chains in the amylose production, and Waxy mutants almost completely lack amylose (Sano et al., 1985). In addition to its roles in amylose biosynthesis, GBSSI is also found to be responsible for the extension of long glucans within the amylopectin fraction (Delrue et al., 1992; Maddelein et al., 1994; Van de Wal et al., 1998).
A series of enzymes, such as SS, SBE and DBE (isoamylase, Isa and pullulanase, PUL), work together to catalyse the biosynthesis of amylopectin (Abe et al., 2013; Facon et al., 2013; McMaugh et al., 2014; Pei et al., 2015). It is reported that SSI, SSII, SSIII, SBEI, SBEII, Isa and PUL were closely linked to the amylopectin chains. SSI increased the proportion of chains with DP 5–8 and DP ≥ 18 and reduced the proportion of chains with DP 9–17 in transgenic sweet potato (Wang et al., 2016b). SSII mainly synthesized amylopectin chains with DP 8–12 and DP 13–25 in sweet potato (Takahata et al., 2010). The deficiency of OsSSIIIa in the rice duI mutant led to the decrease in the chains with DP > 30 (Ryoo et al., 2007). Overexpression of SBEII increased the proportion with DP 6–12, particularly with DP 6, and decreased the gelatinization temperature in potato (Brummell et al., 2015).
Purcell et al. (1998) reported that SnRK1 was involved in the control of SuSy in potato. McKibbin et al. (2006) found that overexpression of StSnRK1 decreased glucose level and increased starch content by enhancing the activities of SuSy and AGPase in transgenic potato. SbSnRK1α from the wild potato species Solanum berthaultii had significant effects on StvacINV1‐associated sucrose degradation in transgenic potato (Lin et al., 2015). The expression levels of SuSy and AGPase were increased and the activities of both enzymes were also enhanced in the IbSnRK1‐overexpressing tobacco plants (Jiang et al., 2013). Overexpression of StSnRK1 from potato up‐regulated SuSy, AGPase and SS III and enhanced the activities of the corresponding enzymes in transgenic tobacco (Wang et al., 2017b).
In this study, most of the genes involved in starch biosynthesis pathway were systematically up‐regulated, the activities of key enzymes were increased, and the related components were also altered in the IbSnRK1‐overexpressing sweet potato plants (Figures 6, 7, 8). Our results support that SnRK1 increases flux through the starch biosynthesis pathway by systematically up‐regulating the starch biosynthesis genes, which leads to increased starch content (Figure 9), as reported by Halford and Hey (2009). Furthermore, the significant up‐regulation of SSI, SSII, SSIII and SBEII responsible for amylopectin chains resulted in the increase in amylopectin proportion, which led to the increase in degree of crystallinity and the decrease in gelatinization temperature and enthalpy in the transgenic plants (Figure 5) (Brummell et al., 2015; Ryoo et al., 2007; Takahata et al., 2010; Wang et al., 2016b).
Figure 9.

Diagram showing the regulation of starch biosynthesis in the storage roots of the IbSnRK1‐overexpressing sweet potato plants. Biosynthesis pathways are shown with solid arrows and regulatory interactions are shown with broken arrows. Fold changes (the mean of the four transgenic lines L13, L14, L17 and L18) are shown in colour, red boxes, white boxes and blue boxes indicate up‐regulation, no obvious change and down‐regulation of expression of genes encoding these enzymes (proteins), respectively.
In conclusion, this study reveals, for the first time, the important roles of SnRK1 in improving starch quality of plants. Its overexpression increased starch content, decreased proportion of amylose, enlarged granule size and improved degree of crystallinity and gelatinization by increasing the expression levels of genes and the activities of key enzymes involved in starch biosynthesis pathway in transgenic sweet potato. This gene has the potential to improve starch content and quality in sweet potato and other plants.
Experimental procedures
Plant materials
Sweet potato cultivars Lizixiang, Xushu 18, Shangshu 19 and Lushu 3 grown in a field were employed for analysing the expression of SnRK1. Lizixiang was used for characterizing the function of IbSnRK1.
Expression analysis of IbSnRK1 in sweet potato
Total RNA was isolated from the storage roots of four sweet potato cultivars and five tissues (leaf, petiole, stem, storage root and fibrous root) of Luhsu 3 using Trozol Kit (Transgen, Beijing, China). The first‐strand cDNA was synthesized using PrimeScript™ RT reagent Kit (TaKaRa, Dalian, China). qRT‐PCR was conducted to determine the transcript levels of IbSnRK1 using SYBR Premix Ex Taq (TaKaRa, Dalian, China) with a 7500 Real‐Time PCR system (Applied Biosystems, Foster, CA). Specific primers of IbSnRK1 and the sweet potato Actin (AY905538) gene as internal control were listed in Table S3. The gene expression was quantified using comparative C T method (Schmittgen and Livak, 2008).
The response of IbSnRK1 to exogenous sucrose was examined according to the method of Wang et al. (2016b). Leaf petioles from Lushu 3 plants grown in a field were treated for 1 day with water followed by treatments of either 175 mm sucrose or water (control) in the dark at 28 °C, and sampled at 0, 3, 6, 12, 24 and 48 h after treatment to analyse the expression of IbSnRK1.
The response of IbSnRK1 to the circadian rhythm was investigated as described by Leterrier et al. (2008). In vitro grown plants of Lushu 3 were subjected to a 16‐h light/8‐h dark regimen for 1 month in a growth chamber at 28 °C prior to modifying the light conditions. Total RNA was extracted from each whole plant sampled at different time points (Figure 1d) for detecting the expression of IbSnRK1 by qRT‐PCR.
Production of the transgenic sweet potato plants
The expression vector pCAMBIA3301‐IbSnRK1 contained IbSnRK1 under the control of CaMV 35S promoter and NOS terminator and glucuronidase (gusA) and bar genes driven by a CaMV 35S promoter, respectively. Embryogenic suspension cultures of Lizixiang were prepared as described by Liu et al. (2001). Transgenic plants were produced using 0.5 mg/L phosphinothricin (PPT) as selection pressure according to the method of Yu et al. (2007) and identified by GUS assay and PCR analysis as described by Wang et al. (2016a). The transgenic plants, WT and VC, were transplanted in pots with a mixture of soil, vermiculite and humus (1:1:1, v/v/v) in a greenhouse and then grown in a field for further evaluation. The expression of IbSnRK1 in the freshly harvested storage roots of the transgenic plants was analysed by qRT‐PCR as described above.
Quantification of starch and amylose
The freshly harvested storage roots were employed to quantify starch content (Smith and Zeeman, 2006). Amylose content was determined using the colorimetric assay (Wang et al., 2016b). Standard curves were established with the standard samples of potato amylose and amylopectin (Sigma‐Aldrich, Shanghai, China).
Analysis of starch granule morphology and size
Central pieces of the freshly harvested storage roots were used to prepare the paraffin sections (Li et al., 2017) and observed with a microscope (BX51 plus DP70, Olympus, Kyoto, Japan). The starch samples from the freshly harvested storage roots (Wang et al., 2017b) were spread on an aluminium stub using double‐sided adhesive tape, coated with gold and then observed with variable pressure scanning electron microscope (Hitachi S3400N, Tokyo, Japan) to examine the shape of starch granules. The starch granule size was measured with a Mastersize 2000 laser diffraction instrument (Malvern Instruments Ltd., Worcestershire, UK) according to the method of Zhou et al. (2015).
Measurement of CLD
Starch samples were digested with isoamylase Pseudomonas amyloderamosa (Sigma‐Aldrich, Shanghai, China) as described by Nishi et al. (2001). CLD of amylopectin was analysed with high‐performance anion‐exchange chromatography with pulsed amperometric detection (HPAEC‐PAD, Dionex‐ICS 3000, Dionex Co., Sunnyvale, CA).
Analysis of X‐ray diffraction
Starch samples were scanned through the 2θ range of 5–40° using a D8 Advance Bruker X‐ray diffractometer (Bruker AXS, Karlsruhe, Germany) to determine X‐ray diffraction. The crystal type and degree of crystallinity were calculated with Jade 5.0 software (Materials Data Inc., Livermore, CA).
Analysis of thermal characteristics
Starch samples and distilled water were mixed (1:3, w/w), and the suspension was sealed in an aluminium pan with an empty aluminium pan as control for 24 h under room temperature. The samples were heated over a temperature range of 30–95 °C (raising the temperature at 10 °C/min) with a differential scanning calorimeter (DSC Q2000, TA Instruments Ltd., Crawley, UK), and the T o, peak temperature (T p), T c and ΔH were measured with Universal Analysis 2000 (TA Instruments Ltd., Crawley, UK).
Expression analysis of starch biosynthesis genes
Expression of starch biosynthesis‐related genes in the freshly harvested storage roots was analysed by qRT‐PCR as described above. The specific primers were listed in Table S3.
Activity analysis of starch biosynthesis enzymes
The freshly harvested storage roots were grounded into power in liquid nitrogen. Activities of acid invertase, neutral invertase, SuSy, AGPase, GBSS, SS and SBE were measured according to the methods of Vargas et al. (2007), Baroja‐Fernández et al. (2012), Zhang et al. (2012) and Wang et al. (2016b), respectively.
Quantification of components related to starch biosynthesis
The freshly harvested storage roots were used to quantify the contents of sucrose, glucose and fructose as described by Li et al. (2017). The contents of glc‐6‐P and glc‐1‐P were determined according to the method of Baroja‐Fernández et al. (2012). UDP‐glc, ADP‐glc, ATP and ADP were measured with the method of Gámez‐Arjona et al. (2011).
Statistical analysis
All experiments were independently performed three times, and the data were presented as the mean ± SE. Results were analysed by Student's t‐test in a two‐tailed analysis using SPSS 20.0 Statistic Program. Significance was defined as P < 0.05 (* or lowercase letters) and P < 0.01 (** or capital letters), respectively.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Production of transgenic sweet potato plants overexpressing the IbSnRK1 gene.
Table S1 X‐ray diffraction patterns of starches from the storage roots of the transgenic sweet potato plants, WT and VC.
Table S2 The thermal characteristics of starches from the transgenic sweet potato plants, WT and VC.
Table S3 Primers used in this study.
Acknowledgements
This work was supported by China Agriculture Research System (CARS‐10) and National Natural Science Foundation of China (31461143017, 31271777).
Contributor Information
Hong Zhai, Email: zhaihong@cau.edu.cn.
Qingchang Liu, Email: liuqc@cau.edu.cn.
References
- Abe, N. , Nakamura, Y. and Fujita, N. (2013) Thermal properties, morphology of starch granules and crystallinity of endosperm starch in SSI and BE isozymes double mutant lines. J. Appl. glycosci. 60, 171–176. [Google Scholar]
- Ahn, Y.O. , Kim, S.H. , Kim, C.Y. , Lee, J.S. , Kwak, S.S. and Lee, H.S. (2010) Exogenous sucrose utilization and starch biosynthesis among sweetpotato cultivars. Carbohyd. Res. 345, 55–60. [DOI] [PubMed] [Google Scholar]
- Alderson, A. , Sabelli, P.A. , Dickinson, J.R. , Cole, D. , Richardson, M. , Kreis, M. , Shewry, P.R. et al. (1991) Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc. Natl Acad. Sci. 88, 8602–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahaji, A. , Li, J. , Sánchez‐López, Á.M. , Baroja‐Fernández, E. , Muñoz, F.J. , Ovecka, M. , Almagro, G. et al. (2014) Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 32, 87–106. [DOI] [PubMed] [Google Scholar]
- Ball, S.G. and Morell, M.K. (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233. [DOI] [PubMed] [Google Scholar]
- Baroja‐Fernández, E. , Muñoz, F.J. , Li, J. , Bahaji, A. , Almagro, G. , Montero, M. , Etxeberria, E. et al. (2012) Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl Acad. Sci. 109, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BeMiller, J.N. and Whistler, R. (2009) Starch: Chemistry and Technology. Salt Lake City, UT: Academic Press. [Google Scholar]
- Brummell, D.A. , Watson, L.M. , Zhou, J. , McKenzie, M.J. , Hallett, I.C. , Simmons, L. , Carpenter, M. et al. (2015) Overexpression of starch branching enzyme II increases short‐chain branching of amylopectin and alters the physicochemical properties of starch from potato tuber. BMC Biotechnol. 15, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell, M.M. (2003) Starch: the need for improved quality or quantity‐an overview. J. Exp. Bot. 54, 451–456. [DOI] [PubMed] [Google Scholar]
- Cheetham, N.W.H. and Tao, L.P. (1998) Variation in crystalline type with amylose content in maize starch granules: an X‐ray powder diffraction study. Carbohyd. Polym. 36, 277–284. [Google Scholar]
- Chung, H.J. , Liu, Q. , Lee, L. and Wei, D.Z. (2011) Relationship between the structure, physicochemical properties and in vitro digestibility of rice starches with different amylose contents. Food Hydrocolloid, 25, 968–975. [Google Scholar]
- Copeland, L. , Blazek, J. , Salman, H. and Tang, C. (2009) Form and functionality of starch. Food Hydrocolloid, 23, 1527–15934. [Google Scholar]
- Delrue, B. , Fontaine, T. , Routier, F. , Decq, A. , Wieruszeski, J.M. , Koornhuyse, N.V.D. , Maddelein, M.L. et al. (1992) Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule‐bound starch synthase activity accumulate a structurally modified amylopectin. J. Bacteriol. 174, 3612–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facon, M. , Lin, Q.H. , Azzaz, A.M. , Hennen‐Bierwagen, T.A. , Myers, A.M. , Putaux, J.L. , Roussel, X. et al. (2013) Distinct functional properties of isoamylase‐type starch debranching enzymes in monocot and dicot leaves. Plant Physiol. 163, 1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámez‐Arjona, F.M. , Li, J. , Raynaud, S. , Baroja‐Fernández, E. , Muñoz, F.J. , Ovecka, M. , Ragel, P. et al. (2011) Enhancing the expression of starch synthase class IV results in increased levels of both transitory and long‐term storage starch. Plant Biotechnol. J. 9, 1049–1060. [DOI] [PubMed] [Google Scholar]
- Hajirezaei, M.R. , Takahata, Y. , Trethewey, R.N. , Willmitzer, L. and Sonnewald, U. (2000) Impact of elevated cytosolic and apoplastic invertase activity on carbon metabolism during potato tuber development. J. Exp. Bot. 51, 439–445. [DOI] [PubMed] [Google Scholar]
- Halford, N.G. and Hey, S.J. (2009) Snf1‐related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 419, 247–259. [DOI] [PubMed] [Google Scholar]
- Halford, N.G. and Hrdie, D.G. (1998) SNF1‐related protein kinases: global regulators of carbon metabolism in plants?. Plant Mol. Biol. 37, 735–748. [DOI] [PubMed] [Google Scholar]
- Hamada, T. , Kim, S.H. and Shimada, T. (2006) Starch‐branching enzyme I gene (IbSBEI) from sweet potato (Ipomoea batatas); molecular cloning and expression analysis. Biotechnol. Lett. 28, 1255–1261. [DOI] [PubMed] [Google Scholar]
- Hattori, T. , Fukumoto, H. , Nakagawa, S. and Nakamura, K. (1991) Sucrose‐induced expression of genes coding for storage root storage protein, sporamin, of sweet potato in leaves and petioles. Plant Cell Physiol. 32, 79–86. [Google Scholar]
- Hoover, R. (2001) Composition, molecular structure, and physicochemical properties of tuber and root starches: a review. Carbohyd. Polym. 45, 253–267. [Google Scholar]
- Horrer, D. , Flütsch, S. , Pazmino, D. , Matthews, J.S.A. , Thalmann, M. , Nigro, A. , Leonhardt, N. et al. (2016) Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr. Biol. 26, 362–370. [DOI] [PubMed] [Google Scholar]
- Jain, M. , Li, Q.B. and Chourey, P.S. (2008) Cloning and expression analyses of sucrose non‐fermenting‐1‐related kinase 1 (SnRK1b) gene during development of sorghum and maize endosperm and its implicated role in sugar‐to‐starch metabolic transition. Physiol. Plantarum, 134, 161–173. [DOI] [PubMed] [Google Scholar]
- James, M.G. , Denyer, K. and Myers, A.M. (2003) Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 6, 215–222. [DOI] [PubMed] [Google Scholar]
- Jarret, R.L. , Gawel, N. and Whittemore, A. (1992) Phylogenetic relationships of the sweetpotato [Ipomoea batatas (L.) Lam.]. J. Amer. Soc. Hort. Sci. 117, 633–637. [Google Scholar]
- Jiang, T. , Zhai, H. , Wang, F.B. , Yang, N.K. , Wang, B. , He, S.Z. and Liu, Q.C. (2013) Cloning and characterization of a carbohydrate metabolism‐associated gene IbSnRK1 from sweetpotato. Hortic. Sci. 158, 22–32. [Google Scholar]
- Kanegae, H. , Miyoshi, K. , Hirose, T. , Tsuchimoto, S. , Mori, M. , Nagato, Y. and Takano, M. (2005) Expressions of rice sucrose non‐fermenting‐1 related protein kinase 1 genes are differently regulated during the caryopsis development. Plant Physiol. Biochem. 43, 669–679. [DOI] [PubMed] [Google Scholar]
- Keeling, P.L. and Myers, A.M. (2010) Biochemistry and genetics of starch synthesis. Annu. Rev. Food Sci. Technol. 1, 271–303. [DOI] [PubMed] [Google Scholar]
- Kimura, T. , Otani, M. , Noda, T. , Ideta, O. , Shimada, T. and Saito, A. (2001) Absence of amylose in sweet potato [Ipomoea batatas (L.) Lam.] following the introduction of granule‐bound starch synthase I cDNA. Plant Cell Rep. 20, 663–666. [Google Scholar]
- Kleinow, T. , Bhalerao, R. , Breuer, F. , Umeda, M. , Salchert, K. and Koncz, C. (2000) Functional identification of an Arabidopsis Snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 23, 115–122. [DOI] [PubMed] [Google Scholar]
- Koch, K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7, 235–246. [DOI] [PubMed] [Google Scholar]
- Lakatos, L. , Klein, M. , Höfgen, R. and Bánfalvi, Z. (1999) Potato StubSNF1 interacts with StubGAL83: a plant protein kinase complex with yeast and mammalian counterparts. Plant J. 17, 569–574. [DOI] [PubMed] [Google Scholar]
- Leterrier, M. , Holappa, L.D. , Broglie, K.E. and Beckles, D.M. (2008) Cloning, characterization and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol. 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G.J. , Peng, F.T. , Zhang, L. , Shi, X.Z. and Wang, Z.Y. (2010) Cloning and characterization of a SnRK1‐encoding gene from Malus hupehensis Rehd. and heterologous expression in tomato. Mol. Biol. Rep. 37, 947–954. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, Y.N. , Zhang, H. , Zhang, Q. , Zhai, H. , Liu, Q.Z. and He, S.Z. (2017) The plasma membrane‐localized sucrose transporter IbSWEET10 contributes to the resistance of sweet potato to fusarium oxysporum. Front. Plant Sci. 8, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. , Liu, T.F. , Liu, J. , Xun, L. , Ou, Y.B. , Zhang, H.L. , Li, M. et al. (2015) Subtle regulation of potato acid invertase activity by a protein complex of invertase, invertase inhibitor, and sucrose nonfermenting1‐related protein kinase. Plant Physiol. 168, 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q.C. , Zhai, H. , Wang, Y. and Zhang, D.P. (2001) Efficient plant regeneration from embryogenic suspension cultures of sweetpotato. In Vitro Cell Dev‐An. 37, 564–567. [Google Scholar]
- Lumbreras, V. , Albà, M.M. , Kleinow, T. , Koncz, C. and Pagès, M. (2001) Domain fusion between SNF1‐related kinase subunits during plant evolution. EMBO Rep. 2, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddelein, M.L. , Libessart, N. , Bellanger, F. , Delrue, B. , D'Hulst, C. , Koornhuyse, N.V.D. , Fontaine, T. et al. (1994) Toward an understanding of the biogenesis of the starch granule: determination of granule‐bound and soluble starch synthase functions in amylopectin synthesis. J. Biol. Chem. 269, 25150–25157. [PubMed] [Google Scholar]
- McKibbin, R.S. , Muttucumaru, N. , Paul, M.J. , Powers, S.J. , Burrell, M.M. , Coates, S. , Purcell, P.C. et al. (2006) Production of high‐starch, low‐glucose potatoes through over‐expression of the metabolic regulator SnRK1. Plant Biotechnol. J. 4, 409–418. [DOI] [PubMed] [Google Scholar]
- McMaugh, S.J. , Thistleton, J.L. , Anschaw, E. , Luo, J.X. , Konik‐Rose, C. , Wang, H. , Huang, M. et al. (2014) Suppression of starch synthase I expression affects the granule morphology and granule size and fine structure of starch in wheat endosperm. J. Exp. Bot. 65, 2189–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, A. , Nakamura, Y. , Tanaka, N. and Satoh, H. (2001) Biochemical and genetic analysis of the effects of amylose‐etender mutation in rice endosperm. Plant Physiol. 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- Ohdan, T. , Francisco, P.B. Jr , Sawada, T. , Hirose, T. , Terao, T. , Satoh, H. and Nakamura, Y. (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56, 3229–3244. [DOI] [PubMed] [Google Scholar]
- Otani, M. , Hamada, T. , Katayama, K. , Kitahara, K. , Kim, S.H. , Takahata, Y. , Suganum, T. et al. (2007) Inhibition of the gene expression for granule‐bound starch synthase I by RNA interference in sweet potato plants. Plant Cell Rep. 26, 1801–1807. [DOI] [PubMed] [Google Scholar]
- Pei, J.L. , Wang, H.J. , Xia, Z.Q. , Liu, C. , Chen, X. , Ma, P.G. , Lu, C. et al. (2015) Phylogeny and expression pattern of starch branching enzyme family genes in cassava (Manihot esculenta Crantz) under diverse environments. Mol. Cell. Biochem. 406, 273–284. [DOI] [PubMed] [Google Scholar]
- Purcell, P.C. , Smith, A.M. and Halford, N.G. (1998) Antisense expression of a sucrose nonfermenting‐1‐related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose‐inducibility of sucrose synthase transcripts in leaves. Plant J. 14, 195–202. [Google Scholar]
- Ren, Z.T. , Zhao, H.Y. , He, S.Z. , Zhai, H. , Zhao, N. and Liu, Q.C. (2018) Overexpression of IbSnRK1 enhances nitrogen uptake and carbon assimilation in transgenic sweetpotato. J. Integr. Agric. 17, 296–305. [Google Scholar]
- Roitsch, T. and González, M.C. (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 9, 606–613. [DOI] [PubMed] [Google Scholar]
- Ryoo, N. , Yu, C. , Park, C.S. , Baik, M.Y. , Park, I.M. , Cho, M.H. , Bhoo, S.H. et al. (2007) Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white‐core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 26, 1083–1095. [DOI] [PubMed] [Google Scholar]
- Sano, Y. , Maekawa, M. and Kikuchl, H. (1985) Temperature effects on the Wx protein level and amylose content in the endosperm of rice. J. Hered. 76, 221–222. [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative C T method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shimada, T. , Otani, M. , Hamada, T. and Kim, S.H. (2006) Increase of amylose content of sweetpotato starch by RNA interference of the starch branching enzyme II gene (IbSBEII). Plant Biotechnol. 23, 85–90. [Google Scholar]
- Smith, A.M. and Zeeman, S.C. (2006) Quantification of starch in plant tissues. Nat. Protoc. 1, 1342–1345. [DOI] [PubMed] [Google Scholar]
- Takahata, Y. , Tanaka, M. , Otani, M. , Katayama, K. , Kitahara, K. , Nakayachi, O. , Nakayama, H. et al. (2010) Inhibition of the expression of the starch synthase II gene leads to lower pasting temperature in sweetpotato starch. Plant Cell Rep. 29, 535–543. [DOI] [PubMed] [Google Scholar]
- Tanaka, M. , Takahata, Y. , Nakayama, H. , Nakatani, M. and Tahara, M. (2009) Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta, 230, 737–746. [DOI] [PubMed] [Google Scholar]
- Tester, R.F. , Karkalas, J. and Qi, X. (2004) Starch—composition, fine structure and architecture. J. Cereal Sci. 39, 151–165. [Google Scholar]
- Tetlow, I.J. , Morell, M.K. and Emes, M.J. (2004) Recent developments in understanding the regulation of starch metabolism in higher plants. J. Exp. Bot. 55, 2131–2145. [DOI] [PubMed] [Google Scholar]
- Thalmann, M.R. , Pazmino, D. , Seung, D. , Horrer, D. , Nigro, A. , Meier, T. , Kölling, K. et al. (2016) Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell, 28, 1860–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wal, M. , D'Hulst, C. , Vincken, J.P. , Bule′on, A. , Visser, R. and Ball, S. (1998) Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J. Biol. Chem. 273, 22232–22240. [DOI] [PubMed] [Google Scholar]
- Vargas, W.A. , Pontis, H.G. and Salerno, G.L. (2007) Differential expression of alkaline and neutral invertases in response to environmental stresses: characterization of an alkaline isoform as a stress‐response enzyme in wheat leaves. Planta, 226, 1535–1545. [DOI] [PubMed] [Google Scholar]
- Volpicella, M. , Fanizza, I. , Leoni, C. , Gadaleta, A. , Nigro, D. , Gattulli, B. , Mangini, G. et al. (2016) Identification and characterization of the sucrose synthase 2 gene (Sus2) in durum wheat. Front. Plant Sci. 7, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.J. , Yeh, K.W. and Tsai, C.Y. (2001) Regulation of starch granule‐bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 161, 635–644. [Google Scholar]
- Wang, X.L. , Peng, F.T. , Li, M.J. , Yang, L. and Li, G.J. (2012) Expression of a heterologous SnRK1 in tomato increases carbon assimilation, nitrogen uptake and modifies fruit development. J. Plant Physiol. 169, 1173–1182. [DOI] [PubMed] [Google Scholar]
- Wang, B. , Zhai, H. , He, S.Z. , Zhang, H. , Ren, Z.T. , Zhang, D.D. and Liu, Q.C. (2016a) A vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Hortic. Sci. 201, 153–166. [Google Scholar]
- Wang, Y.N. , Li, Y. , Zhang, H. , Zhai, H. , Liu, Q.C. and He, S.Z. (2016b) A plastidic ATP/ADP transporter gene, IbAATP, increases starch and amylose content and alters starch structure in transgenic sweetpotato. J. Integr. Agric. 15, 1968–1982. [Google Scholar]
- Wang, F.B. , Ye, Y.X. , Chen, X.H. , Wang, J.Z. , Chen, Z.Y. and Zhou, Q. (2017a) A sucrose non‐fermenting‐1‐related protein kinase 1 gene from potato, StSnRK1, regulates carbohydrate metabolism in transgenic tobacco. Physiol. Mol. Biol. Plants, 23, 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.N. , Li, Y. , Zhang, H. , Zhai, H. , Liu, Q.C. and He, S.Z. (2017b) A soluble starch synthase I gene, IbSSI, alters the content, composition, granule size and structure of starch in transgenic sweet potato. Sci. Rep. 7, 2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Gao, T. , Xu, M.J. , Dong, J. , Li, H.X. , Wang, P.F. , Li, G.Z. et al. (2017) Functional analysis of a wheat AGPase plastidial small subunit with a truncated transit peptide. Molecules, 22, 386–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B. , Zhai, H. , Wang, Y.P. , Zang, N. , He, S.Z. and Liu, Q.C. (2007) Efficient Agrobacterium tumefaciens‐mediated transformation using embryogenic suspension cultures in sweetpotato, Ipomoea batatas (L.) Lam. Plant Cell Tissue Organ Cult. 90, 265–273. [Google Scholar]
- Zeeman, S.C. , Kossmann, J. and Smith, A.M. (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Shewry, P.R. , Jones, H. , Barcelo, P. , Lazzeri, P.A. and Halford, N.G. (2001) Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 28, 431–442. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Li, H.W. , Yuan, L.M. , Wang, Z.Q. , Yang, J.C. and Zhang, J.H. (2012) Post‐anthesis alternate wetting and moderate soil drying enhances activities of key enzymes in sucrose‐to‐starch conversion in inferior spikelets of rice. J. Exp. Bot. 63, 215–227. [DOI] [PubMed] [Google Scholar]
- Zhou, W.Z. , Yang, J. , Hong, Y. , Liu, G.L. , Zheng, J.L. , Gu, Z.B. and Zhang, P. (2015) Impact of amylose content on starch physicochemical properties in transgenic sweet potato. Carbohyd. Polym. 122, 417–427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Production of transgenic sweet potato plants overexpressing the IbSnRK1 gene.
Table S1 X‐ray diffraction patterns of starches from the storage roots of the transgenic sweet potato plants, WT and VC.
Table S2 The thermal characteristics of starches from the transgenic sweet potato plants, WT and VC.
Table S3 Primers used in this study.


