Summary
Auxin is as an efficient initiator and regulator of cell fate during somatic embryogenesis (SE), but the molecular mechanisms and regulating networks of this process are not well understood. In this report, we analysed SE process induced by Leafy cotyledon1‐like 1 (GhL1L1), a NF‐YB subfamily gene specifically expressed in embryonic tissues in cotton. We also identified the target gene of GhL1L1, and its role in auxin distribution and cell fate specification during embryonic development was analysed. Overexpression of GhL1L1 accelerated embryonic cell formation, associated with an increased concentration of IAA in embryogenic calluses (ECs) and in the shoot apical meristem, corresponding to altered expression of the auxin transport gene GhPIN1. By contrast, GhL1L1‐deficient explants showed retarded embryonic cell formation, and the concentration of IAA was decreased in GhL1L1‐deficient ECs. Disruption of auxin distribution accelerated the specification of embryonic cell fate together with regulation of GhPIN1. Furthermore, we showed that PHOSPHATASE 2AA2 (GhPP2AA2) was activated by GhL1L1 through targeting the G‐box of its promoter, hence regulating the activity of GhPIN1 protein. Our results indicate that GhL1L1 functions as a key regulator in auxin distribution to regulate cell fate specification in cotton and contribute to the understanding of the complex process of SE in plant species.
Keywords: cotton, auxin, cell fate, GhPIN1, GhPP2AA2, GhL1L1
Introduction
Somatic embryogenesis (SE) is a process of asexual reproduction in plants in which somatic cells undergo differentiation and redifferentiation to form embryos. It resembles zygotic embryogenesis, whereby globular‐, torpedo‐ and cotyledonary‐stage embryos are formed. SE consists of direct SE and indirect SE. In former, embryoids are produced from the explant directly during in vitro culture, while indirect SE is more complex whereby somatic cell dedifferentiation leads to callus formation and then redifferentiation into embryogenic calluses (ECs) and somatic embryos. Cotton undergoes indirect SE, and the process can be divided into several stages, including the dedifferentiation of cotton somatic cells and transition from nonembryogenic calluses (NEC) to ECs, followed by the development of somatic embryos (Yang et al., 2012).
As an excellent natural provider of fibre, cotton needs a reproducible and highly efficient plant regeneration scheme for transgenic research and genetic engineering. The morphological and molecular mechanisms of SE have been studied in cotton in our laboratory (Jin et al., 2014; Yang et al., 2012; Zhou et al., 2016). GhHmgB3‐deficient hypocotyls dedifferentiate more rapidly but fail to differentiate into ECs (Hu et al., 2011). By contrast, overexpression of GhCKI prevents EC and plant regeneration by blocking the transition from NECs to ECs (Min et al., 2015). However, the precise mechanisms of gene regulation during cotton SE have not been elucidated.
Transcription factors are considered to play important roles during the process of SE, and the Leafy cotyledon (LEC) genes are major regulators of embryo development and cellular differentiation (Kwong et al., 2003; Lee et al., 2003; Lotan et al., 1998). Mutations of LEC1 result in defective embryo maturation (Lotan et al., 1998; Meinke, 1992; Meinke et al., 1994). LEC2, FUS3 and ABI3 have been considered as marker genes during embryogenic cell formation (Braybrook and Harada, 2008; Gazzarrini et al., 2004; Wang and Perry, 2013). LEC genes are found to regulate auxin homeostasis during embryogenesis (Braybrook et al., 2006; Gazzarrini et al., 2004; Kagaya et al., 2005; Stone et al., 2008), and LEC1 and LEC1‐like (L1L) are partially functionally redundant (Kwong et al., 2003; Yamamoto et al., 2009). LEC1 acts as a coactivator of PIF4 to co‐regulate etiolation‐related genes during postembryonic growth in the dark (Huang et al., 2015). LEC2 is considered to regulate YUCCA4, which encodes an auxin biosynthetic enzyme, required for somatic embryo formation (Braybrook et al., 2006; Stone et al., 2001, 2008). FUS3 interacts with LEC2 to promote auxin biosynthesis (Tang et al., 2017). Disruption of auxin homeostasis by GhCKI overexpression, which might act downstream of GhLEC1, leads to defective embryogenesis (Min et al., 2015).
Auxin regulation during plant SE has been well documented in model systems (Kim et al., 2000; Komamine et al., 2005; Quint and Gray, 2006). Auxin gradients modulate the response and transduction of the auxin signal to regulate the expression of genes during phase changes between cell division and cell differentiation during SE (Chugh and Khurana, 2002; Jimenez, 2005; Rose and Nolan, 2006; Yang and Zhang, 2010; Yang et al., 2012). Auxin plays an important role in WUS expression, which is essential for embryonic stem cell fate determination during SE in Arabidopsis (Mayer et al., 1998; Su et al., 2009). Research has also demonstrated that the interactions between auxin, ethylene, gibberellic acid and stress response regulate SE (Wang et al., 2018; Zheng et al., 2013, 2016; Zhou et al., 2016).
Auxin gradients are established by local auxin biosynthesis, degradation or polar auxin transport (Vanneste and Friml, 2009; Wabnik et al., 2013). DR5, a highly active synthetic auxin response element (AuxRE) reporter gene, reveals the distribution of auxin in tissues and cells (Ulmasov et al., 1997; Zhang et al., 2016). Auxin biosynthesis spatially and temporally regulated by YUCs is an essential source of auxin during embryogenesis, floral development and vascular patterning (Cheng et al., 2006; Zhao, 2010). It has been demonstrated that PIN‐dependent auxin transport and the auxin gradient play important roles during the formation of the apical‐basal embryo axis (Friml et al., 2003). The Arabidopsis PIN gene family consists of eight members, and auxin efflux in the embryo is mediated by PIN1, PIN4 and PIN7 (Friml et al., 2003; Paponov et al., 2005). PIN1 is expressed in proembryogenic cells in a nonpolar manner during the early developmental stages and then becomes polarized to the basal side of provascular cells during the early globular stage (Friml et al., 2003; Steinmann et al., 1999). The localization of PIN1 changes from the apical cells in 16‐cell stage embryos to the basal side in early heart stage embryos (Grunewald and Friml, 2010; Guenot et al., 2012). Directional auxin movement depends on the phosphorylation status of the PINs, which affects their polar subcellular localization. PINOID kinase and PP2A phosphatase are important regulators of PIN targeting and so of auxin distribution (Friml et al., 2004; Michniewicz et al., 2007; Zhang et al., 2010). Moreover, some auxin polar transport inhibitors, such as 2,3,5‐triiodobenzoic acid (TIBA), can inhibit auxin transport by affecting the localization of PIN proteins and can be used to investigate the accumulation of auxin (Geldner et al., 2001; Klima et al., 2015).
In this study, a LEC1‐type gene, GhL1L1, was identified by RNA‐Seq as being expressed during cotton SE (Yang et al., 2012), and is specifically expressed in cotton embryonic tissues. Overexpression of GhL1L1 reorganized cell patterning during cell dedifferentiation and accelerated cell fate specification during embryonic development, with a change in auxin homeostasis. Disruption of GhL1L1 expression resulted in the opposite phenotype. We propose that GhL1L1 mediates auxin distribution to regulate cell fate specification by initiating the interaction between GhPP2AA2 and GhPIN1 proteins through binding to the cis‐element within the promoter of GhPP2AA2.
Results
GhL1L1 is specifically expressed in cotton embryonic tissues
GhL1L1 was identified during a RNA‐Seq profiling analysis during cotton SE (Yang et al., 2012). The 651 bp cDNA encodes a NF‐YB subfamily protein of 216 amino acids. GhL1L1 protein has a highly conserved central B domain (Figure S1). The transcript of GhL1L1 specifically accumulates in ECs, somatic embryos [somatic globular embryos (SGEs), somatic torpedo embryos (STEs), somatic cotyledon embryos (SCEs)] and zygotic embryos [zygotic globular embryos (ZGEs), zygotic torpedo embryos (ZTEs), zygotic cotyledon embryos (ZCEs)], with the greatest abundance in SGEs. Expression of GhL1L1 was very low in root, stem, leaf and other nonembryonic tissues, as well as in cells in the dedifferentiation stage (0, 6, 24, 48 h, 5 days and NECs) (Figure 1a). The expression of GhL1L1 was also detected during the dedifferentiation and redifferentiation stages in four cotton varieties with different regeneration abilities. The highest expression levels were observed in YZ1, the genotype with the highest regeneration efficiency, followed by relatively high levels in Jin668, a genotype that regenerates with moderate efficiency. By contrast, expression levels were low in Simian3 and H7124, which are recalcitrant to regeneration in both Gossypium hirsutum and Gossypium barbadense (Figure 1b).
Figure 1.
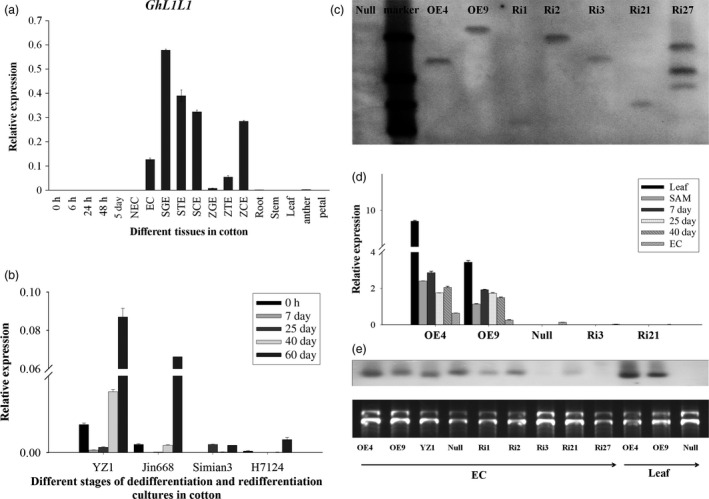
Expression analysis of GhL1L1. (a) qRT‐PCR analysis of GhL1L1 (0, 6, 24, 48 h and 5 days explants; NEC, nonembryogenic callus; EC, embryogenic callus; SGE, somatic globular embryo; STE, somatic torpedo embryo; SCE, somatic cotyledon embryo; ZGE, zygotic globular embryo; ZTE, zygotic torpedo embryos; ZCE, zygotic cotyledon embryo). (b) qRT‐PCR analysis of GhL1L1 in YZ1, Jin668, Simian3 and H7124. (c) Southern blotting of transgenic cotton plants, OE4 and OE9 represent the overexpression lines, Ri1, Ri2 and Ri3 represent RNA interference of 3′ untranslated region lines, Ri21and Ri27 represent RNA interference of coding region lines. (d) qRT–PCR analysis of GhL1L1 in different transgenic and null lines in leaf, shoot apical meristem (SAM), 7, 25, 40 days explants and ECs. (e) Northern blot analysis of GhL1L1 in different transgenic and null lines in ECs and leaf. The data (in a, b and d) are shown as the mean ± SE (n = 3).
A total of 41 NF‐YB subfamily genes were identified genome‐wide in G. hirsutum (TM‐1) (Figure S2a). Phylogenetic analysis revealed six LEC1‐type genes in G. hirsutum, GhL1L1A (Gh_A05G1515), GhL1L1D (Gh_D05G1686), GhL1L2A (Gh_A13G1116), GhL1L2D (Gh_D13G1387), GhL1L3A (Gh_A08G0216) and GhL1L3D (Gh_D08G0296), among which A and D represent different copies that are distributed to the A and D subgenomes, respectively, and which encode similar proteins, with several nucleotide polymorphisms in the CDS regions of the A and D subgenomes.
The expression patterns of the 41 NF‐YB genes were analysed in G. hirsutum (TM‐1) using public data sets. The six LEC1‐type genes were specifically expressed in 20‐day seeds, with the most abundant expression observed for GhL1L1D (Gh_D05G1686). Some of the other NF‐YB genes were also expressed in other tissues (Figure S2b). Three B3 domain genes [GhLEC2 (Gh_A09G0695), GhFUS3 (Gh_A07G2123), GhABI3 (Gh_D07G1550)] were also identified in upland cotton. All the LEC1‐type genes and B3 domain genes showed expression patterns specifically during cotton embryogenesis (Figure S3).
GhL1L1 positively regulates cell fate specification during cotton SE
To understand the function of GhL1L1 in cotton, one overexpression and two RNAi vectors including the coding region and 3′ untranslated region of GhL1L1 were constructed and transformed into G. hirsutum YZ1. Several single T‐DNA insertion lines were identified by Southern blot analysis and selected for further analysis (Figures 1c and S4). qRT‐PCR and northern blotting revealed that GhL1L1 transcript accumulated in leaf, shoot apical meristem (SAM) and dedifferentiation stage explants, and was also high in ECs of the overexpression lines, but low levels accumulated in the RNAi lines (Figure 1d,e). We selected two overexpression lines (OE4, OE9, with higher expression levels in OE4 than in OE9), one 3′ untranslated region RNAi line (Ri3), one coding region RNAi line (Ri21) and a null line (a negative plant line isolated from the offspring of OE4) for further study.
Calluses were induced from each transgenic lines (OE4, OE9, Ri3, Ri21) and the null on MSB medium in vitro. Seven days postinduction, calluses could be observed at the ends of null and RNAi line explants, while only a little expansion with adventitious roots was observed at the ends of OE4 and OE9 explants. Histological observation showed reorganized cell patterning from the cambium areas in OE4 and OE9, while vascular cells overproliferated in Ri3 and Ri21. During the development of SE, the polar growth was evident for Ri3 and Ri21 on 15 and 25 days postinduction. However, the explant ends of the overexpression lines (OE4 and OE9) showed little difference (Figure 2a). The callus proliferation rate (CPR) was then measured among those lines during the dedifferentiation stage. The results showed that overexpression of GhL1L1 inhibited callus proliferation at both ends of the hypocotyls, while increased CPR was observed only in the RNAi line (Ri21) at all tested time points, compared with the null (Figure 2b). Therefore, we conclude that the expression of GhL1L1 leads to a reorganization of the patterning of cambium cells and restricts uncontrolled callus proliferation.
Figure 2.
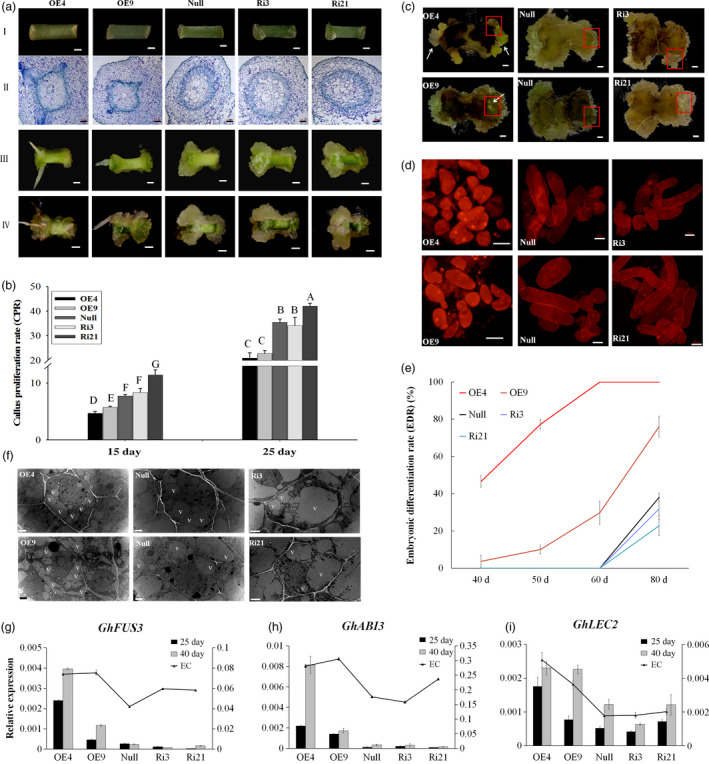
GhL1L1 positively regulates cell fate specification during cotton SE. (a) The phenotypes of different GhL1L1 transgenic and null lines at 7 (I, scale bars = 1 mm; II, scale bars = 200 μm), 15 (III, scale bars = 5 mm) and 25 days postinduction (IV, scale bars = 5 mm). (b) The callus proliferation rate (CPR) of different transgenic and null lines at 15 and 25 days post‐induction. Different capital letters denote significant differences by multiple comparisons using Statistix 8.0 software. (c) The ECs or embryos were observed for the overexpression line (OE4 and OE9) explants (white arrows) at 40 days postinduction, scale bars = 2.5 mm. (d) Cellular features of the calluses from red boxes of (c), scale bars = 50 μm. (e) The embryonic differentiation rate (EDR) of different transgenic lines and null at 40, 50, 60 and 80 days post‐induction. (f) The ECs of different transgenic lines and null scanned by transmission electron microscopy. v, vacuole. Scale bars = 2 μm. qRT–PCR analysis shows GhFUS3 (g), GhABI3 (h) and GhLEC2 (i) in GhL1L1 transgenic and null lines at 25 and 40 days post‐induction and ECs (embryogenic calluses from the corresponding transgenic lines and null). The data (in b, e, g, h and i) are shown as the mean ± SE (n = 3).
Overexpression of GhL1L1 promoted embryonic cell differentiation during cell culture, which accounted for an embryonic differentiation rate (EDR) of 46.6% in OE4 and 3.8% in OE9 with observable ECs at 40 days postinduction. Round small cells and a large nucleus were observed in cells of overexpression lines (Figure 2c,d and e). However, in null and RNAi lines no ECs were present, with large long cells and an unclear nucleus (Figure 2c,d). Some ECs were observed in OE4 at 25 days postinduction, while the null and RNAi lines produced none until at least 60 days postinduction, and were clearly visible at approximately 80 days postinduction. An EDR of less than 40% (null, 38%; Ri3, 31.9%; Ri21, 22.8%) were seen in null and RNAi lines, with more than 70% (OE4, 100%; OE9, 76%) in overexpression lines at the same time point (Figure 2e). Compared with null, many small vacuoles were observed in ECs of OE4 and OE9, while large vacuoles were present in ECs of Ri3 and Ri21 (Figure 2f).
ABI3, FUS3 and LEC2 are considered as marker genes during embryonic stem cell fate determination (Braybrook and Harada, 2008; Gazzarrini et al., 2004; Wang and Perry, 2013). GhABI3 and GhFUS3 were specifically expressed in embryonic cells (ECs and somatic embryos) (Figure S3d,e). The expression of GhABI3 and GhFUS3 could not be detected at 7 days postinduction, but the expression levels of the three genes were significantly up‐regulated in OE4 and OE9 compared with the RNAi lines and null at 25 and 40 days postinduction (Figure 2g–i). Some embryonic cells were present when overexpressing GhL1L1 even at 25 days postinduction. These results suggest that overexpression of GhL1L1 accelerates cell fate specification, while repression of GhL1L1 retards embryonic cell differentiation.
GhL1L1 affects auxin accumulation and auxin distribution in cotton
Given the importance of auxin during cotton SE, the concentration of IAA in the different transgenic lines at 25 and 40 days postinduction and in ECs was measured by HPLC‐MS. After 25 and 40 days of induction, the IAA concentration of explants in the overexpression lines was higher than in the null and RNAi explants. Additionally, the concentration of endogenous IAA was increased in the GhL1L1 overexpression ECs. By contrast, it was decreased in the ECs of RNAi lines compared with null (Figure 3a), and the transcription of two auxin response genes, ARF19 and IAA33, was increased after overexpressing GhL1L1 in ECs (Figure 3b,c).
Figure 3.
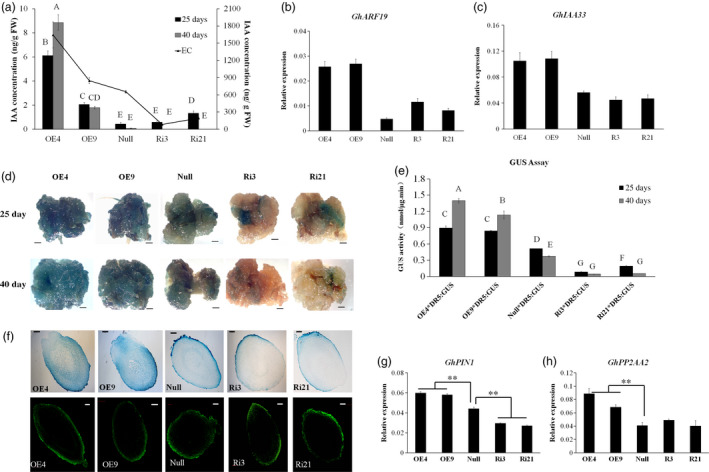
Auxin accumulation and distribution in GhL1L1 transgenic and null lines during SE. (a) The IAA concentration in different transgenic and null lines at 25 and 40 days postinduction and ECs (embryogenic calluses from the corresponding transgenic and null lines). Expression analysis of the auxin response genes GhARF19 (b) and GhIAA33 (c) by qRT‐PCR. GUS expression (d) and GUS activity (e) of explants from five F1 hybrids ( OE4/DR5::GUS, OE9/DR5::GUS, Ri3/DR5::GUS, Ri21/DR5::GUS, null/DR5::GUS ) at 25 and 40 days postinduction, scale bars = 2.5 mm. Different capital letters denote significant differences by multiple comparisons using Statistix 8.0 software. (f) GUS staining (upper) and IAA immunofluorescence (below) of longitudinal section of torpedo embryos from GhL1L1 transgenic and null lines, scale bars = 100 μm. Expression analysis of GhPIN1 (g) and GhPP2AA2 (h) in ECs of different transgenic and null lines. The data (in a, b, c, e, g and h) are shown as the mean + SE (n = 3). Statistical analyses were performed using Student's t‐test. **p < 0.01.
To investigate the function of GhL1L1 in auxin accumulation and distribution, GhL1L1 transgenic lines containing DR5::GUS were generated to monitor auxin distribution. Explants of the hybrids were sampled for GUS staining and GUS activity at 25 and 40 days postinduction, respectively. Clear GUS staining was observed in the calluses of explants from the hybrids of OE‐GhL1L1/DR5::GUS, with only a little GUS staining in the hybrid of null/DR5::GUS and almost no GUS staining in RNAi‐GhL1L1/DR5::GUS (Figure 3d). As expected, GUS activity corresponded to GUS staining. GUS activity was much stronger in the calluses of explants from hybrids of OE‐GhL1L1/DR5::GUS and was lower in RNAi‐GhL1L1/DR5::GUS lines than the hybrid of null/DR5::GUS (Figure 3e). GUS staining showed that auxin was uniformly distributed in the apical and basal parts of DR5::GUS torpedo embryos, but it was asymmetrically distributed in the GhL1L1‐deficient or overexpressor torpedo embryos (Figure 3f). To detail the IAA accumulation in embryos, immunolocalization in torpedo embryos with a monoclonal antibody against IAA was performed. The results were similar to the GUS staining results (Figure 3f). In addition, GUS expression was markedly increased in the SAM of the OE‐GhL1L1/DR5::GUS hybrids compared with the null/DR5::GUS and RNAi‐GhL1L1/DR5::GUS hybrids (Figure S5). We conclude that GhL1L1 affects auxin accumulation and distribution in cotton embryonic tissues.
Polar auxin transport and local auxin biosynthesis determine auxin distribution. To investigate the relationship between GhL1L1 and auxin distribution, the expression of some auxin biosynthetic genes and polar auxin transport genes were analysed in ECs from GhL1L1 transgenic and null lines. GhYUC2, GhYUC4, GhYUC8 and GhYUC10, which are key auxin biosynthesis genes, showed irregular changes (Figure S6a–d), indicating that the auxin accumulation was not due to the activation of local auxin biosynthesis. PIN1 and PIN4 mediate auxin efflux and distribution during Arabidopsis embryogenesis (Friml et al., 2003). The expression level of GhPIN1 was up‐regulated in ECs from GhL1L1 overexpression lines but decreased in RNAi lines (Figure 3g), while the transcript of GhPIN4 showed irregular changes in all lines (Figure S6e). Moreover, the expression of PHOSPHATASE 2A (GhPP2AA2, Gh_A11G0044) was increased in GhL1L1 overexpression lines (Figure 3h). GhPP2AA2 is homologous to AtPP2AA2 (AT3G25800), which functions in the dephosphorylation of PIN auxin efflux carriers (Michniewicz et al., 2007; Skottke et al., 2011). Thus, we speculated that GhL1L1 might regulate the expression of GhPIN1 and GhPP2AA2 to mediate auxin distribution.
GhL1L1 binds to the promoter of GhPP2AA2 to activate its expression
The promoter of GhPP2AA2 has been cloned and shown to contain two CCAAT‐box motifs and a G‐box motif, which are candidate binding sites for LEC1‐type genes (Dorn et al., 1987; Li et al., 1992; Mendes et al., 2013). They were designated A, B and C regions (Figure 4a). Yeast one‐hybrid (Y1H) bait vectors were constructed, named proGhPP2AA2‐F, proGhPP2AA2‐ΔC, proGhPP2AA2‐ΔG and proGhPP2AA2‐mG (Figure 4b) and experiments were conducted to study protein‐promoter interactions. Positive results were observed following co‐transformation of GhL1L1 either with proGhPP2AA2‐F or proGhPP2AA2‐ΔC, but not with either deficiency or mutation of the G‐box element (Figure 4c).
Figure 4.
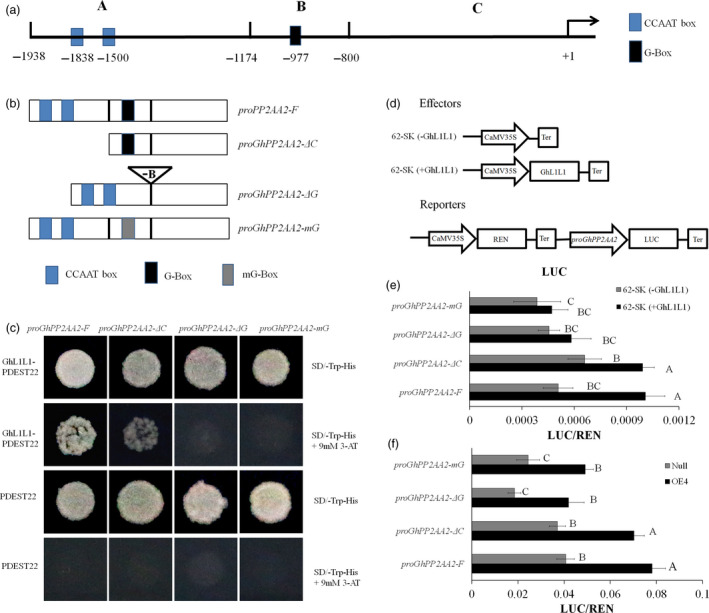
GhL1L1 binds to the G‐box in the promoter to activate the expression of GhPP2AA2. (a) Schematic diagram of different regions of the promoter of GhPP2AA2, named A, B, C. (b) proPP2AA2‐F represents the full length of the promoter, proGhPP2AA2‐ΔC represents deletion of the A region, proGhPP2AA2‐ΔG represents deletion of the B region and proGhPP2AA2‐mG represents mutation of the B region (G‐box, CACGTT mutated to CAAGGT). (c) Transformant yeast colonies visible on the medium (SD‐Trp‐His + 3‐AT) show that GhL1L1 was able to bind to proPP2AA2‐F and proGhPP2AA2‐ΔC. No visible yeasts were observed when co‐transforming GhL1L1 either with proGhPP2AA2‐ΔG or proGhPP2AA2‐mG . Empty pDEST22 vector was used as a negative control. 3AT, 3‐Amino‐1,2,4‐triazole. (d) Schematic diagram of effectors and reporter. GhL1L1 activates gene expression by binding to the G‐box element in the promoter of GhPP2AA2 in tobacco leaf protoplasts (e) and OE4 and null EC protoplasts (f) in vivo. 62‐SK (‐GhL1L1) served as negative controls. Data are shown as the mean ± SE (n = 3). Different capital letters denote significant differences by multiple comparisons using Statistix 8.0 software.
The dual‐luciferase reporter system was also applied to quantify the interaction between GhL1L1 and ProGhPP2AA2 in vivo. The effectors and reporters were transformed into protoplasts from tobacco leaves to exclude the effects of background genes. Simultaneously, the Renilla luciferase (REN) gene driven by the 35S promoter was co‐expressed as an internal control (Figure 4d). Compared with the negative control, GhL1L1 enhanced the activity of the LUC reporter under the control of proGhPP2AA2‐F and proGhPP2AA2‐ΔC (Figure 4e). ProGhPP2AA2‐F, proGhPP2AA2‐ΔC, proGhPP2AA2‐ΔG and proGhPP2AA2‐mG were also used to transform protoplasts from OE4 and null. The activation of LUC expression driven by proGhPP2AA2‐F and proGhPP2AA2‐ΔC was also observed in OE4 (Figure 4f). The results showed that GhL1L1 is able to bind to the G‐box element of the GhPP2AA2 promoter to activate GhPP2AA2 expression.
GhPP2AA2 interacts with GhPIN1 in vitro and in vivo
PP2A phosphatase was identified as an important regulator of PIN activity and auxin distribution (Michniewicz et al., 2007). To understand the relationship between GhPP2AA2 and GhPIN1, both in vitro and in vivo experiments were performed. To obtain GhPIN1 protein, recombinant protein GhPIN1‐HL‐GST was produced by removing the transmembrane domain from the hydrophilic loop of GhPIN1 (GhPIN1‐HL). GhPIN1‐HL was fused to glutathione S‐transferase (GST) and GhPP2AA2 was fused to maltose‐binding protein (MBP). In vitro pull‐down assays showed that recombinant GhPIN1 and GhPP2AA2 interacted with each other (Figure 5a). These interactions were further confirmed by bimolecular fluorescence complementation (BiFC) assays in vivo. Strong YFP fluorescence signals indicated that GhPP2AA2 interacted with GhPIN1‐HL (Figure 5b,c). Moreover, tobacco epidermal cells were transformed with the FRET 2in1 vectors, GhPP2AA2 fused to GFP and GhPIN1 or GhPIN1‐HL fused to mCherry, which showed that GhPP2AA2 and GhPIN1 colocalized at the cell membrane (Figure 5d). These results indicated that GhPIN1 directly interacts with GhPP2AA2.
Figure 5.
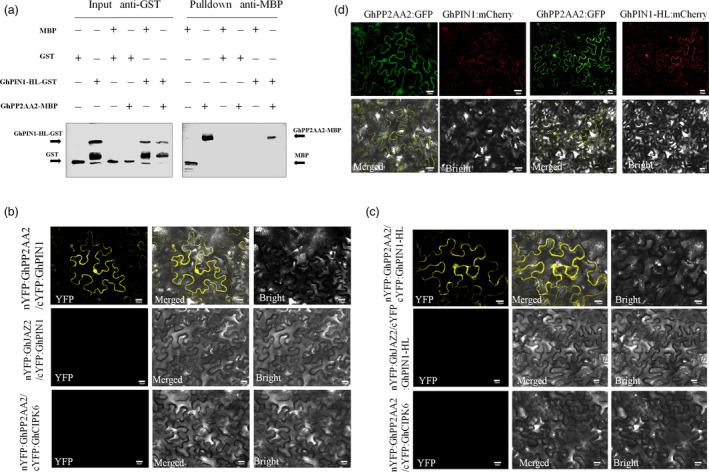
GhPP2AA2 interacts with GhPIN1 protein in vitro and in vivo. (a) GhPP2AA2 interacts with the GhPIN1 hydrophilic loop (GhPIN1‐HL) in the pull‐down assay in vitro. GST and GST‐GhPIN1‐HL proteins were used to pull down interacting proteins. MBP or MBP‐GhPP2AA2 proteins were detected by Western blotting with anti‐MBP antibodies and anti‐GST as input. BiFC assays revealed the interaction of GhPP2AA1 with GhPIN1 (b) and GhPIN1‐HL (c) in vivo. Yellow fluorescence (YFP) indicated a positive interaction. nYFP:GhJAZ2 and cYFP:GhCIPK6 fusion proteins served as negative controls. (d) GhPP2AA2 and GhPIN1 (GhPIN1‐HL) colocalized as indicated by GFP and mCherry. Scale bars = 20 μm in b, c and d.
Disrupted trafficking of GhPIN1 inhibits auxin polar transport and accelerates cell fate specification during embryo formation
To confirm whether disrupted PIN1 protein affects cotton cell fate, a synthetic inhibitor of PIN1, TIBA, was added to MSB medium to culture GhL1L1 transgenic and null explants. CPR was measured at 15 and 25 days postinduction. As shown previously, CPR of OE4 and OE9 was less than null, while CPR of Ri21 increased on MSB medium without TIBA (Figure 2c). However, callus proliferation was suppressed by TIBA in all the lines, especially in RNAi lines, and the difference in CPR among different lines could not be clearly observed at 15 days postinduction. However, callus growth between the apical and basal parts of the explants in RNAi lines was poorly differentiated (Figure 6a,b). TIBA treatment accelerated the presence of ECs in all lines, but especially in RNAi lines, where they were observed at least 10 days earlier than in other lines. EDR was increased to 86.4% in OE4 and 32.3% in OE9 at 40 days post‐induction, and it was increased to at least 90% in null and RNAi lines at approximately 80 days postinduction (Figures 2e and 6c,d). Moreover, the expression level of GhABI3 and GhFUS3 increased to a high level at 40 days postinduction in overexpression line OE4, in accordance with the phenotype (Figure 6e–h).
Figure 6.
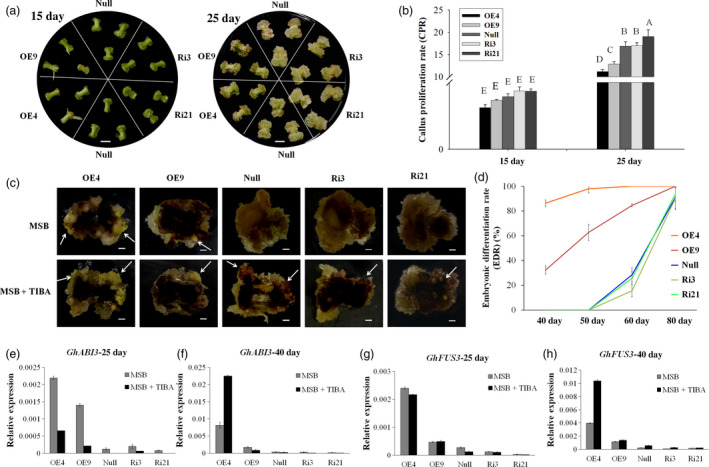
TIBA treatment affects callus proliferation and auxin distribution during cotton SE. (a) The phenotypes of different GhL1L1 transgenic and null lines at 15 and 25 days postinduction treated with TIBA, scale bars = 5 mm. (b) The CPR of different transgenic and null lines at 15 and 25 days postinduction treated with TIBA. Different capital letters denote significant differences by multiple comparisons using Statistix 8.0 software. (c) ECs or embryos were observed from the transgenic and null lines (white arrows) at 50 days postinduction treated with TIBA, scale bars = 2 mm. (d) The embryonic differentiation rate (EDR) of different transgenic and null lines at 40, 50, 60 and 80 days postinduction treated by TIBA. qRT–PCR analysis of GhABI3 (e, f) and GhFUS3 (g, h) in GhL1L1 transgenic and null explants induced on MSB medium or supplemented with TIBA at 25, 40 days postinduction. The data (in b, d, e, f, g and h) are shown as the mean ± SE (n = 3).
TIBA treatments were also applied to DR5::GUS transgenic seedlings during cotton SE. GUS staining was uniformly distributed at both ends of the explants before induction, while it was accumulated to the morphological basal parts of explants at 1, 3, 7, 15 and 40 days postinduction without TIBA treatment (Figure S7a), demonstrating that the auxin distribution polarized to the morphological basal region of the explants. When TIBA was added to the medium, the polar distribution of auxin was disturbed, with most of the GUS staining accumulating in the morphological basal parts of the explants, with little diffusion to the morphological apical part (Figure S7b), similar to the overexpression explants of GhL1L1 (Figure 2a) and the OE4/DR5::GUS explants induced on MSB medium (Figure S7c). These results suggest that GhL1L1 overexpression may disturb auxin polar distribution, to accelerate cell fate specification during embryo formation.
Discussion
GhL1L1 displays a conserved B domain and specific expression during embryogenesis
LEC1 encodes a CCAAT‐binding transcription factor of the HAP3 subunit. LEC1 and L1L are LEC1‐type subunits with a conserved B domain (Kwong et al., 2003). The B domain of LEC1 and L1L is necessary for its activity in embryogenesis (Lee et al., 2003). We demonstrate that GhL1L1 and other LEC1‐type proteins are highly conserved in the B domain (Figure S1). LEC1 is required during seed maturation, and embryos of the lec1 mutant are intolerant to seed desiccation (Lotan et al., 1998). The expression pattern of L1L is similar to that of LEC1 (Kwong et al., 2003). Additionally, genes such as LEC2, FUS3 and ABI3, closely related B3 domain transcription factors, have been reported to play major roles in embryo maturation (Braybrook et al., 2006; Stone et al., 2001). We found that the expression of GhL1L1 was only detected in seeds (Figure S2b) or embryonic cells (Figure 1a). These results show that GhL1L1 is highly conserved and specifically expressed in cotton. Additionally, GhL1L1 is an important player in embryogenesis.
Somatic embryogenesis accompanied by complex auxin dynamics
Somatic embryogenesis is a process whereby somatic cells regenerate into embryos, then to new plants by in vitro culture without fertilization. Callus induction is fundamental to cotton SE, which is the foundation of producing transgenic cotton via Agrobacterium‐mediated transformation. The molecular mechanisms underlying callus induction are complex. The molecular mechanisms underlying callus induction have been documented, including auxin induction, cytokinin induction, wound induction and formation via the reacquisition of embryonic or meristematic fates (Ikeuchi et al., 2013). Callus formation is associated with a variety of hormones. An intermediate ratio of auxin and cytokinin can increase callus induction and proliferation (Skoog and Miller, 1957). Brassinosteroid, abscisic acid or ethylene also can induce callus formation in some plants (Goren et al., 1979; Hu et al., 2000; Wang et al., 2018). TIBA inhibits auxin distribution and PIN1 localization, which are important for embryogenesis (Forestan et al., 2010; Geldner et al., 2001). Auxin levels change dynamically during cotton SE (Yang et al., 2012). The strictly polar auxin distribution can be observed in the explants of DR5::GUS induced on MSB medium (Figure S7a). However, polar auxin transport was disrupted after treatment with TIBA (Figure S7b), the proliferation of NECs was repressed by TIBA (Figure 6c), while the embryonic cell formation was advanced by TIBA treatment (Figure 6c,d). We propose that cell fate specification accompanied by complex auxin dynamics, encompassing not only auxin levels but also auxin distribution.
GhL1L1 affects auxin polar distribution
GhL1L1 is an important regulator for cell fate specification. Overexpression of GhL1L1 accelerated embryonic cell formation and restricted callus proliferation, with altered auxin distribution. By contrast, GhL1L1‐deficient explants dedifferentiated vigorously but showed retarded embryonic cell formation (Figure 2). A new idea presented here is that GhL1L1 regulates auxin distribution during cell fate specification. In this study, the concentration of free IAA increased in ECs and SAM after overexpressing GhL1L1 (Figures 3 and S5). Additionally, the GUS staining increased in GhL1L1 overexpression lines but decreased in GhL1L1 RNAi lines (Figure 3), paralleled with the polar growth of explants in transgenic lines. This indicates that GhL1L1 participates in cell fate specification by regulating auxin distribution. Moreover, the postinduction phenotype of explants from GhL1L1 overexpression lines was similar to the postinduction phenotype of the wild type treated with TIBA. Furthermore, the polarized growth of callus on the both ends of explants was decreased. The calluses of GhL1L1 overexpression lines were more dramatically suppressed (Figure 6a,b).
Gradients of auxin are mediated by its efflux via asymmetrically localized PIN proteins (Benková et al., 2003; Paponov et al., 2005). Polar auxin transport and correct apical–basal axis formation of the embryo require PIN1, PIN4 and PIN7 (Friml et al., 2003; Guenot et al., 2012). The expression of GhPIN1 was altered in ECs of the GhL1L1 overexpression lines (Figure 3g), and auxin accumulated. Hence, we suppose that auxin efflux might be decreased because of the effect on GhPIN1 activity, resulting in an increased auxin concentration. PIN polarity is related to the phosphorylation status of PIN proteins. The absence of PP2A, in particular PP2AA1, PP2AA2 or PP2AA3, increases PIN1 phosphorylation in embryos. PP2A regulates the dephosphorylation of PIN proteins (Friml et al., 2004; Michniewicz et al., 2007). Our data confirm that GhL1L1 positively regulates GhPP2AA2 to affect the activity of GhPIN1 (Figure 4). Thus, overexpression of GhL1L1 or treatment with TIBA affected the auxin polar transport by affecting the activity of GhPIN1.
Conclusion
Based on an integration of the relationships between different morphological and biochemical changes in cotton, GhL1L1 repressed the initial cell dedifferentiation and callus proliferation, but it played a positive role in embryonic cell formation. GhL1L1 activated the expression of GhPP2AA2 by binding to the cis‐element G‐box in the promoter of GhPP2AA2 to interact with GhPIN1 protein and affect the activity of GhPIN1, which was also affected by TIBA treatment (Figure 7).
Figure 7.
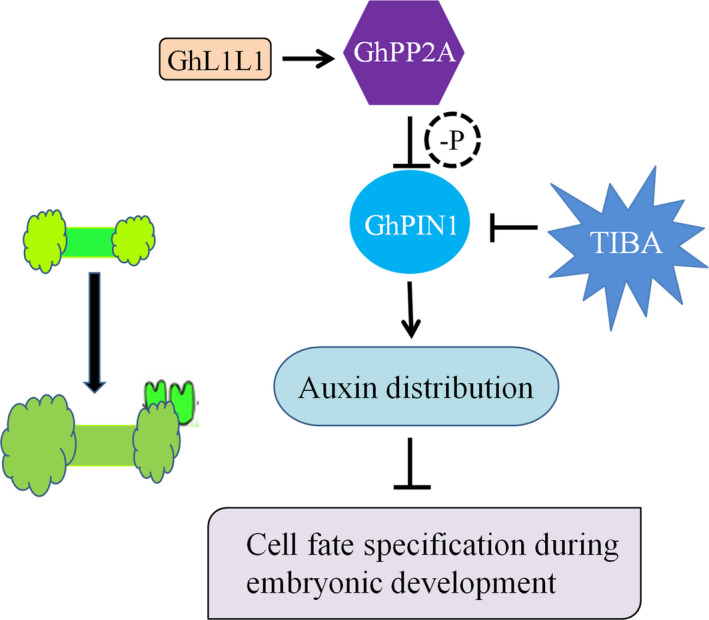
Schematic showing the role of GhL1L1 during cotton SE. GhL1L1 activates the expression of GhPP2AA2, which might dephosphorylate GhPIN1, which was also affected by TIBA treatment, to affect the auxin distribution, and then affects cell fate specification during embryonic development.
Experimental procedures
GhL1L1 sequencing analysis, vectors construction and transformation
The full‐length GhL1L1 was obtained from cDNA amplification of cotton ECs. The amino acid sequence alignments and phylogenetic relationship were analysed with the ClustalX and MEGA6 software respectively. The coding sequence was inserted into the vector PK2GW7 to construct the vector 35S::GhL1L1 by Gateway Technology (Invitrogen, Carlsbad, CA) for overexpression. The 3′ untranslated region and coding region were cloned into the RNAi vector pHellsgate4 by recombination reaction. The overexpression and RNAi vectors were transformed into G. hirsutum ‘YZ1’ plants via Agrobacterium tumefaciens (EHA105) as described previously (Jin et al., 2005, 2006). All primers used in the vectors construction are listed in Table S1.
Plant materials, callus induction and TIBA treatment
Cotton materials, YZ1 [cotton (G. hirsutum)], Jin668 [cotton (G. hirsutum)], Simian3 [cotton (G. hirsutum)], H7124 [cotton (G. barbadense)], 3–79 [cotton (G. barbadense)], DR5::GUS transgenic plants, GhL1L1 transgenic plants and F1 hybrids of DR5::GUS transgenic plants with GhL1L1 transgenic plants were used in this study. The DR5::GUS and GhL1L1 transgenic plants were in the YZ1 background. The callus induction procedure was performed as follows. Hypocotyls of etiolated seedlings sampled at 0 h or cut into 5–7‐mm sections as explants were cultured on MSB medium at different time points or subcultured for EC and somatic embryos as described previously (Yang et al., 2012). The MSB medium was supplemented with 5 μm TIBA (Sigma, St. Louis, MO) to monitor the disruption of the auxin distribution.
Southern and northern blotting, RT–PCR and qRT–PCR
To determine the copy number of T‐DNA inserted in transgenic cotton, Southern blotting was performed as previously described (Li et al., 2010). Genomic DNA was extracted from leaves of transgenic cotton using the Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China). Approximately, 15 μg of DNA was hybridized with the probe of an NPTII fragment using a DIG‐High Prime DNA Labeling and Detection Starter Kit II (Roche, Mannheim, Germany).
To determine the expression level of GhL1L1 in wild‐type and transgenic plants, total RNA was isolated from ECs and leaves using a PureLink RNA Mini Kit (Invitrogen). Northern blotting was performed as previously described (Tu et al., 2007). Approximately, 15 μg of RNA was hybridized with a GhL1L1 probe fragment labelling with the DIG‐High Prime DNA Labeling and Detection Starter Kit II. RT‐PCR and qRT‐PCR were performed as previously described (Hao et al., 2012). The expression level of GhUBQ7 (DQ116441) was used as the internal control (Tu et al., 2007). The primers used for Southern and northern blotting, RT–PCR and qRT–PCR are listed in Table S1.
Callus proliferation rate and EDR calculation
The hypocotyl of GhL1L1 transgenic plants and DR5::GUS were cut into approximately 7 mm pieces and induced on MSB medium or supplemented with 5 μm TIBA in the culture room. The CPR was calculated as the fold change in weight gained of explants at 15 and 25 days postinduction as described (Wang et al., 2018). The EDR (the explants with ECs or embryos/the total explants) was calculated by the rate of ECs or embryos occurred on the explants at 40, 50, 60 and 80 days postinduction on MSB medium or supplemented with 5 μm TIBA.
The CPR and EDR experiments were conducted with three biological replicates, and each replicate represented at least four culture dishes with at least 10 explants each dish.
GUS assay and histochemical analysis
The hypocotyl of GhL1L1 transgenic plants induced on MSB medium or supplemented with 5 μm TIBA were photographed using a Nikon D40 camera (Nikon corporation, Tokyo, Japan) at 15 and 25 days postinduction. Calluses were removed and stained with propidium iodide to visualize cellular structure at 40 days postinduction, and the features were photographed under a microscope (Zeiss, Oberkochen, Germany). The hypocotyls of five F1 hybrids (OE4/DR5::GUS, OE9/DR5::GUS, Ri3/DR5::GUS, Ri21/DR5::GUS, null/DR5::GUS) were induced on MSB medium. Histochemical localization and quantitative analyses of GUS activity was performed as described previously (Cai et al., 2008; Deng et al., 2012). The features of GUS staining were observed and photographed under a microscope (Leica, Wetzlar, Germany). To evaluate the microstructure of torpedo embryos after GUS staining and the structure of hypocotyls, samples were fixed in 50% FAA and cut into 8‐μm‐thick sections as in a previous study (Yang et al., 2012). GUS‐stained sections were directly observed after being deparaffinized with xylene. To observe the structure of the hypocotyl, the deparaffinized hypocotyl sections were stained with 1% toluidine blue. The photographs were obtained under a microscope (Zeiss). To observe the structure of transgenic EC cells, transmission electron microscope analysis was performed as previously described (Sun et al., 2014). The experiments were conducted with three biological replicates.
Endogenous IAA extraction, quantification and immunofluorescence localization
To estimate the concentration of endogenous IAA, samples were immediately ground in liquid nitrogen and extracted in 1 mL 80% cold methanol, which contains 10 ng/mL 2H5‐IAA (Olchemlm, Olomouc, Czech Republic), as an internal standard. Further extraction and quantitative analyses of IAA were performed as described previously (Liu et al., 2012). The experiments were conducted with three biological replicates.
Samples of torpedo embryos were fixed in 50% FAA as described previously (Hou and Huang, 2005). Sections were incubated with anti‐rabbit Dylight 488 secondary antibody (Thermo Scientific, Waltham, MA) for immunofluorescence. Fluorescence was assayed using a confocal laser‐scanning microscope (Olympus, Tokyo, Japan).
Yeast one‐hybrid assay
To characterize the interaction between GhL1L1 and the promoter of GhPP2AA2 in yeast, the promoter sequence of GhPP2AA2 (−1 to −1939) was amplified by PCR using YZ1 genomic DNA and cloned into the pHis‐1 bait vector to generate pHis‐1‐proGhPP2AA2. It was then divided to three regions: −1174 to −1953‐bp region containing two CCAAT boxes (at approximately −1838, −1500 bp) named A, −800 to −1173‐bp region containing a G‐box motif (at approximately −977 bp) named B and −1 to −799‐bp region containing basic promoter elements without the CCAAT‐box motif and G‐box named C. Three vectors of the promoters were constructed in the pHis‐1 bait vector, with proGhPP2AA2‐ΔC representing deletion of the A region, proGhPP2AA2‐ΔG representing deletion of the B region and ProGhPP2AA2‐mG representing mutation of the B region (G‐box, CACGTT mutated to CAAGGT). The Y1H assay was performed as previously described (Min et al., 2015). The primers used in the Y1H assay are listed in Table S1.
Dual‐luciferase reporter assay system
The transient dual‐luciferase reporter assays were performed as described previously (Hellens et al., 2005). The full‐length and three‐variant promoters (proGhPP2AA‐F, proGhPP2AA2‐ΔC, proGhPP2AA2‐ΔG and proGhPP2AA2‐mG) were cloned into pGreenII 0800‐LUC at the PstI and BamHI sites. Moreover, GhL1L1 was cloned into vector 62‐SK to obtain 62‐SK (+GhL1L1). These plasmids and 62‐SK (+GhL1L1) or negative 62‐SK (−GhL1L1) were co‐transformed into protoplasts from tobacco leaves. These plasmids were also transformed into protoplasts from ECs of OE4 and null respectively. Firefly luciferase and Renilla spp. luciferase activities were then quantified using the dual‐luciferase assay reagents (Promega, Madison, WI) with a Multimode Plate Reader (PerkinElmer). The primers used in the LUC assays are listed in Table S1.
In vitro pull‐down assay
PIN1 is a membrane protein and is difficult to express in prokaryotes (Michniewicz et al., 2007). CDS deletion of the transmembrane domain of GhPIN1 (GhPIN1‐HL) was cloned and constructed into pGEX‐4T‐1 to obtain GST‐GhPIN1‐HL recombinant proteins. For MBP‐GhPP2AA2 recombinant proteins, the CDS of GhPP2AA2 was cloned into pMAL‐c4x. The GST fusion proteins and MBP fusion proteins were purified using glutathione beads (Promega) and amylose resin (NEB, Ipswich, MA). The pull‐down assay was performed as described previously (Yang et al., 2017). The primers used in the pull‐down assay are listed in Table S1.
Bimolecular fluorescence complementation assays and colocalization
Bimolecular fluorescence complementation assays were performed as described previously (Grefen and Blatt, 2012). The sequences of GhPP2AA2 and GhPIN1 (or GhPIN1‐HL) were constructed in pDONR221 via recombination reactions. The pBIFCt‐2in1‐NN vectors were constructed via attL and attR site (LR) recombination (Invitrogen) for BiFC. nYFP:GhJAZ2 (nuclear‐localized protein) and cYFP:GhCIPK6 (membrane‐localized protein) fusion proteins were used as negative control. The pFRETgc‐2in1‐NN vector was constructed by LR recombination for colocalization (Hecker et al., 2015). All the vectors were transformed into Agrobacterium tumefaciens GV3101 and used to infect tobacco epidermal cells. The YFP, GFP and mCherry fluorescence were assayed using a confocal laser‐scanning microscope (Olympus). The primers used in the BiFC assays and colocalization were listed in Table S1.
Statistical analysis
All experiments were conducted with at least three biological replicates, and the values are displayed as the mean ± SD. Statistical significance was determined using Student's t‐test, *P < 0.05; **P < 0.01 were considered statistically significant, and multiple comparisons were performed using Statistix 8.0 software (Analytical Software, Tallahassee, FL ).
Conflicts of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Alignment analysis of LEC1‐type subunit.
Figure S2 NF‐YB subfamily in cotton.
Figure S3 Expression analysis by qRT‐PCR.
Figure S4 Southern blotting of transgenic cotton plants.
Figure S5 GUS staining of the shoot apical meristem (SAM).
Figure S6 qRT‐PCR analysis of the genes expression.
Figure S7 GUS staining of DR5::GUS explants.
Table S1 The primers used in this study.
Acknowledgements
We thank Prof. Sheng Luan and Dr. Congcong Hou (University of California,Berkeley) for providing plasmids pDONR221‐P1P4, pDONR221‐P3P2, pBIFCt‐2in1‐NN and pFRETgc‐2in1‐NN; Dongqin Li and Hongbo Liu (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for their technical assistance with the liquid chromatography/mass spectrometry; Huazhi Song (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for assistance with the confocal laser‐scanning microscope; and Prof. Keith Lindsey (Department of Biosciences, Durham University) for modifying the manuscript. This work was supported by funding from the National Key Project of Research and Development Plan (2016YFD0101006) and National R&D Project of Transgenic Crops of Ministry of Science and Technology of China (2016ZX08005‐004‐002).
Contributor Information
Xiyan Yang, Email: yxy@mail.hzau.edu.cn.
Xianlong Zhang, Email: xlzhang@mail.hzau.edu.cn.
References
- Benková, E. , Michniewicz, M. , Sauer, M. , Teichmann, T. , Seifertová, D. , Jürgens, G. and Friml, J. (2003) Local, efflux‐dependent auxin gradients as a common module for plant organ formation. Cell, 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Braybrook, S.A. and Harada, J.J. (2008) LECs go crazy in embryo development. Trends Plant Sci. 13, 624–630. [DOI] [PubMed] [Google Scholar]
- Braybrook, S.A. , Stone, S.L. , Park, S. , Bui, A.Q. , Le, B.H. , Fischer, R.L. , Goldberg, R.B. et al. (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl Acad. Sci. USA, 103, 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, M. , Qiu, D. , Yuan, T. , Ding, X. , Li, H. , Duan, L. , Xu, C. et al. (2008) Identification of novel pathogen‐responsive cis‐elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant, Cell Environ. 31, 86–96. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Dai, X. and Zhao, Y. (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis . Genes Dev. 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh, A. and Khurana, P. (2002) Gene expression during somatic embryogenesis—recent advances. Curr. Sci. 83, 715–730. [Google Scholar]
- Deng, F. , Tu, L. , Tan, J. , Li, Y. , Nie, Y. and Zhang, X. (2012) GbPDF1 is involved in cotton fiber initiation via the core cis‐element HDZIP2ATATHB2. Plant Physiol. 158, 890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn, A. , Bollekens, J. , Staub, A. , Benoist, C. and Mathis, D. (1987) A multiplicity of CCAAT box‐binding proteins. Cell, 50, 863–872. [DOI] [PubMed] [Google Scholar]
- Forestan, C. , Meda, S. and Varotto, S. (2010) ZmPIN1‐mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 152, 1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J. , Vieten, A. , Sauer, M. , Weijers, D. , Schwarz, H. , Hamann, T. , Offringa, R. et al. (2003) Efflux‐dependent auxin gradients establish the apical‐basal axis of Arabidopsis . Nature, 426, 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J. , Yang, X. , Michniewicz, M. , Weijers, D. , Quint, A. , Tietz, O. , Benjamins, R. et al. (2004) A PINOID‐dependent binary switch in apical‐basal PIN polar targeting directs auxin efflux. Science, 306, 862–865. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S. , Tsuchiya, Y. , Lumba, S. , Okamoto, M. and McCourt, P. (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell, 7, 373–385. [DOI] [PubMed] [Google Scholar]
- Geldner, N. , Friml, J. , Stierhof, Y.D. , Jürgens, G. and Palme, K. (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature, 413, 425–428. [DOI] [PubMed] [Google Scholar]
- Goren, R. , Altman, A. and Giladi, I. (1979) Role of ethylene in abscisic acid‐induced callus formation in citrus bud cultures. Plant Physiol. 63, 280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen, C. and Blatt, M.R. (2012) A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques, 53, 311–314. [DOI] [PubMed] [Google Scholar]
- Grunewald, W. and Friml, J. (2010) The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29, 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenot, B. , Bayer, E. , Kierzkowski, D. , Smith, R.S. , Mandel, T. , Zadnikova, P. , Benkova, E. et al. (2012) Pin1‐independent leaf initiation in Arabidopsis . Plant Physiol. 159, 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, J. , Tu, L. , Hu, H. , Tan, J. , Deng, F. , Tang, W. , Nie, Y. et al. (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker, A. , Wallmeroth, N. , Peter, S. , Blatt, M.R. , Harter, K. and Grefen, C. (2015) Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiol. 168, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P. , Allan, A.C. , Friel, E.N. , Bolitho, K. , Grafton, K. , Templeton, M.D. , Karunairetnam, S. et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Z.X. and Huang, W.D. (2005) Immunohistochemical localization of IAA and ABP1 in strawberry shoot apexes during floral induction. Planta, 222, 678–687. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Bao, F. and Li, J. (2000) Promotive effect of brassinosteroids on cell division involves a distinct CycD3‐induction pathway in Arabidopsis . Plant J. 24, 693–701. [DOI] [PubMed] [Google Scholar]
- Hu, L. , Yang, X. , Yuan, D. , Zeng, F. and Zhang, X. (2011) GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton. Plant Biotechnol. J. 9, 1038–1048. [DOI] [PubMed] [Google Scholar]
- Huang, M. , Hu, Y. , Liu, X. , Li, Y. and Hou, X. (2015) Arabidopsis LEAFY COTYLEDON1 mediates postembryonic development via interacting with PHYTOCHROME‐INTERACTING FACTOR4[OPEN]. Plant Cell, 27, 3099–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi, M. , Sugimoto, K. and Iwase, A. (2013) Plant callus: mechanisms of induction and repression. Plant Cell, 25, 3159–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, V. (2005) Involvement of plant hormones and plant growth regulators on in vitro somatic embryogenesis. Plant Growth Regul. 47, 91–110. [Google Scholar]
- Jin, S. , Zhang, X. , Liang, S. , Nie, Y. , Guo, X. and Huang, C. (2005) Factors affecting transformation efficiency of embryogenic callus of Upland cotton (Gossypium hirsutum) with Agrobacterium tumefaciens . Plant Cell Tissue Organ Cult. 81, 229–237. [Google Scholar]
- Jin, S. , Zhang, X. , Nie, Y. , Guo, X. , Liang, S. and Zhu, H. (2006) Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plantarum, 50, 519–524. [Google Scholar]
- Jin, F. , Hu, L. , Yuan, D. , Xu, J. , Gao, W. , He, L. , Yang, X. et al. (2014) Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: evidence for stress response functions in SE development. Plant Biotechnol. J. 12, 161–173. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y. , Toyoshima, R. , Okuda, R. , Usui, H. , Yamamoto, A. and Hattori, T. (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46, 399–406. [DOI] [PubMed] [Google Scholar]
- Kim, E.Y. , Lee, E.K. , Cho, D.Y. and Soh, W.Y. (2000) Relationship between auxin‐induced cell proliferation and somatic embryogenesis in culture of carrot cotyledons. J. Plant Biol. 43, 115–120. [Google Scholar]
- Klima, P. , Lankova, M. and Zazimalova, E. (2015) Inhibitors of plant hormone transport. Protoplasma, 253, 1–14. [DOI] [PubMed] [Google Scholar]
- Komamine, A. , Murata, N. and Nomura, K. (2005) 2004 SIVB Congress Symposium Proceeding: mechanisms of somatic embryogenesis in carrot suspension cultures—morphology, physiology, biochemistry, and molecular biology. In Vitro Cell. Dev. Biol. Plant, 41, 6–10. [Google Scholar]
- Kwong, R.W. , Bui, A.Q. , Lee, H. , Kwong, L.W. , Fischer, R.L. , Goldberg, R.B. and Harada, J.J. (2003) LEAFY COTYLEDON1‐LIKE defines a class of regulators essential for embryo development. Plant Cell, 15, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. , Fischer, R.L. , Goldberg, R.B. and Harada, J.J. (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl Acad. Sci. USA, 100, 2152–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.Y. , Mantovani, R. , Hooft van Huijsduijnen, R. , Andre, I. , Benoist, C. and Mathis, D. (1992) Evolutionary variation of the CCAAT‐binding transcription factor NF‐Y. Nucleic Acids Res. 20, 1087–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Liu, D. , Tu, L. , Zhang, X. , Li, W. , Zhu, L. , Tan, J. et al. (2010) Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep. 29, 193–202. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Li, X. , Xiao, J. and Wang, S. (2012) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice‐bacterium interaction. Plant Methods, 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan, T. , Ohto, M.‐A. , Yee, K.M. , West, M.A. , Lo, R. , Kwong, R.W. , Yamagishi, K. et al. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell, 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F. , Schoof, H. , Haecker, A. , Lenhard, M. , Jürgens, G. and Laux, T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell, 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. (1992) A homoeotic mutant of arabidopsis thaliana with leafy cotyledons. Science, 258, 1647–1650. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. , Franzmann, L.H. , Nickle, T.C. and Yeung, E.C. (1994) Leafy cotyledon mutants of Arabidopsis . Plant Cell, 6, 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes, A. , Kelly, A.A. , van Erp, H. , Shaw, E. , Powers, S.J. , Kurup, S. and Eastmond, P.J. (2013) bZIP67 regulates the omega‐3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell, 25, 3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz, M. , Zago, M.K. , Abas, L. , Weijers, D. , Schweighofer, A. , Meskiene, I. , Heisler, M.G. et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell, 130, 1044–1056. [DOI] [PubMed] [Google Scholar]
- Min, L. , Hu, Q. , Li, Y. , Xu, J. , Ma, Y. , Zhu, L. , Yang, X. et al. (2015) LEAFY COTYLEDON1‐CASEIN KINASE I‐TCP15‐PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant Physiol. 169, 2805–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov, I.A. , Teale, W.D. , Trebar, M. , Blilou, I. and Palme, K. (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 10, 170–177. [DOI] [PubMed] [Google Scholar]
- Quint, M. and Gray, W.M. (2006) Auxin signaling. Curr. Opin. Plant Biol. 9, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, R.J. and Nolan, K.E. (2006) Invited review: genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and Medicago truncatula . In Vitro Cell. Dev. Biol. Plant, 42, 473–481. [Google Scholar]
- Skoog, F. and Miller, C.O. (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro . Symp. Soc. Exp. Biol. 11, 118–130. [PubMed] [Google Scholar]
- Skottke, K.R. , Yoon, G.M. , Kieber, J.J. and DeLong, A. (2011) Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 7, e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T. , Geldner, N. , Grebe, M. , Mangold, S. , Jackson, C.L. , Paris, S. , Gälweiler, L. et al. (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science, 286, 316–318. [DOI] [PubMed] [Google Scholar]
- Stone, S.L. , Kwong, L.W. , Yee, K.M. , Pelletier, J. , Lepiniec, L. , Fischer, R.L. , Goldberg, R.B. et al. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl Acad. Sci. USA, 98, 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L. , Braybrook, S.A. , Paula, S.L. , Kwong, L.W. , Meuser, J. , Pelletier, J. , Hsieh, T.F. et al. (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl Acad. Sci. USA, 105, 3151–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y.H. , Zhao, X.Y. , Liu, Y.B. , Zhang, C.L. , O'Neill, S.D. and Zhang, X.S. (2009) Auxin‐induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis . Plant J. 59, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Zhu, L. , Xu, L. , Yuan, D. , Min, L. and Zhang, X. (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5, 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, L.P. , Zhou, C. , Wang, S.S. , Yuan, J. , Zhang, X.S. and Su, Y.H. (2017) FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana . New Phytol. 213, 1740–1754. [DOI] [PubMed] [Google Scholar]
- Tu, L.L. , Zhang, X.L. , Liang, S.G. , Liu, D.Q. , Zhu, L.F. , Zeng, F.C. , Nie, Y.C. et al. (2007) Genes expression analyses of sea‐island cotton (Gossypium barbadense L.) during fiber development. Plant Cell Rep. 26, 1309–1320. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T. , Murfett, J. , Hagen, G. and Guilfoyle, T.J. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell, 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste, S. and Friml, J. (2009) Auxin: a trigger for change in plant development. Cell, 136, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Wabnik, K. , Robert, H.S. , Smith, R.S. and Friml, J. (2013) Modeling framework for the establishment of the apical‐basal embryonic axis in plants. Curr. Biol. 23, 2513–2518. [DOI] [PubMed] [Google Scholar]
- Wang, F. and Perry, S.E. (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Liu, N. , Wang, T. , Li, J. , Wen, T. , Yang, X. , Lindsey, K. et al. (2018) The GhmiR157a/GhSPL10 regulatory module controls initial cellular dedifferentiation and callus proliferation in cotton by modulating ethylene‐mediated flavonoid biosynthesis. J. Exp. Bot. 69, 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A. , Kagaya, Y. , Toyoshima, R. , Kagaya, M. , Takeda, S. and Hattori, T. (2009) Arabidopsis NF‐YB subunits LEC1 and LEC1‐LIKE activate transcription by interacting with seed‐specific ABRE‐binding factors. Plant J. 58, 843–856. [DOI] [PubMed] [Google Scholar]
- Yang, X.Y. and Zhang, X.L. (2010) Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 29, 36–57. [Google Scholar]
- Yang, X. , Zhang, X. , Yuan, D. , Jin, F. , Zhang, Y. and Xu, J. (2012) Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 12, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Li, C. , Cai, Z. , Hu, Y. , Nolan, T. , Yu, F. , Yin, Y. et al. (2017) SINAT E3 ligases control the light‐mediated stability of the brassinosteroid‐activated transcription factor BES1 in Arabidopsis . Dev. Cell, 41, 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Nodzynski, T. , Pencík, A. , Rolcík, J. and Friml, J. (2010) PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl Acad. Sci. USA, 107, 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Zeng, J.Y. , Long, H. , Xiao, Y.H. , Yan, X.Y. and Pei, Y. (2016) Auxin regulates cotton fiber initiation via GhPIN‐mediated auxin transport. Plant Cell Physiol., 58, 385. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. (2010) Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Q. , Zheng, Y. and Perry, S.E. (2013) AGAMOUS‐Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 161, 2113–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Q. , Zheng, Y. , Ji, H. , Burnie, W. and Perry, S.E. (2016) Gene regulation by the AGL15 transcription factor reveals hormone interactions in somatic embryogenesis. Plant Physiol. 172, 2374–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T. , Yang, X. , Guo, K. , Deng, J. , Xu, J. , Gao, W. , Lindsey, K. et al. (2016) ROS homeostasis regulates somatic embryogenesis via the regulation of auxin signaling in cotton. Mol. Cell Proteomics, 15, 2108–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignment analysis of LEC1‐type subunit.
Figure S2 NF‐YB subfamily in cotton.
Figure S3 Expression analysis by qRT‐PCR.
Figure S4 Southern blotting of transgenic cotton plants.
Figure S5 GUS staining of the shoot apical meristem (SAM).
Figure S6 qRT‐PCR analysis of the genes expression.
Figure S7 GUS staining of DR5::GUS explants.
Table S1 The primers used in this study.


