Cancer is a major health burden, however advances in early diagnosis and improved surgery and therapeutic options have improved outcomes over the past several decades. Nevertheless, many radiological and chemotherapeutic treatments yield severe side effects, while relapse and subsequent outgrowth of treatment resistant tumours is common. The majority of cancer treatments rely on inhibition of cancer cells pro-growth signalling, blocking proliferation or inducing DNA damage. Our cells generate single and double strand DNA breaks as an undesirable consequence of normal metabolism, as well as in a planned manner in order to diversify the genomes of immune and reproductive cells. It is critical to understand the functions of the DNA damage repair (DDR) machinery and the consequences of DNA damage in normal and cancerous cells so that potent, specific and minimally toxic cancer treatments can be developed.
Cancer frequently develops due to loss of normal DDR regulation, which drives genomic instability and acquisition of further mutations. Often, families with mutations in central mediators of DDR have a strong predisposition to cancer (for example Ataxia telangiectasia and xeroderma pigmentosum syndromes). The genetic causes of sporadic cancers are often less obvious and only detectable by genomic sequencing of large cohorts of cancer and control cases. Recent work by Chen et al. tested mutations in a panel of 62 cancer susceptibility genes in over 15,000 breast cancer patients and healthy individuals (1). They found that 0.38% of wild-type BRCA breast cancer patients had a loss of function mutation in the DDR gene RAD51D, while such mutations were detected in 0.1% of the healthy population. For example, RAD51D mutation carriers had higher grade cancers and early relapse compared to wild types, implicating RAD51D loss as rare but penetrant breast cancer mutation associated with more aggressive disease.
During cancer progression the disruption of normal DDR drives accumulation of additional genetic defects that facilitate more rapid evolution of the tumour cell population and promote intrinsic resistance to DNA damaging chemotherapeutic agents. Herein we present a discussion of our current understanding of cancer-associated DDR defects and treatment options, with a focus on the RAD51 paralog family of repair proteins.
Types of DDR
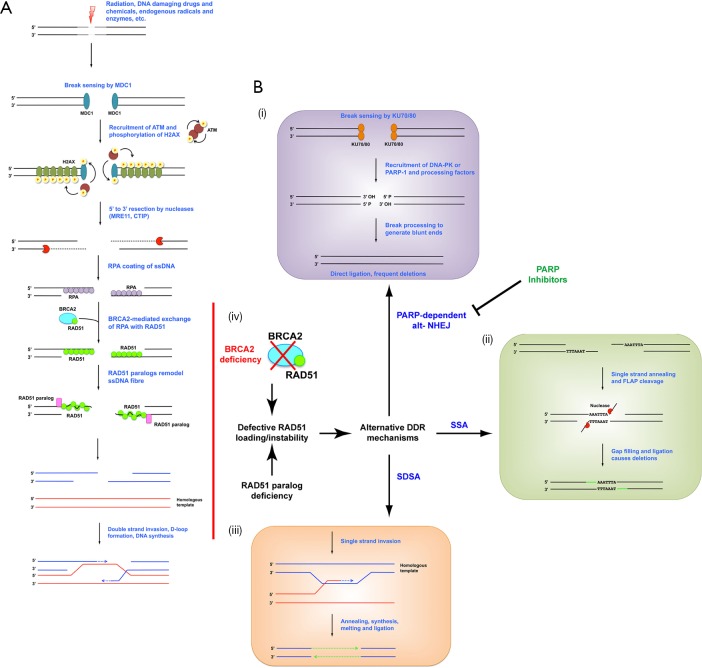
DDR can be mechanistically divided into two major categories based on the type of damage: single strand break repair (SSBR) and double strand break repair (DSBR). SSBR can be further sub-classified based on the type of base, nucleotide or structural damage of the initial lesion, reviewed elsewhere (2). DSBR consists of two major mechanisms which differ based on template dependence. Homology dependent repair requires alignment of damaged DNA fibres with a homologous template, while homology independent does not require a template. Homologous recombination (HR) is an error-free pathway that uses a homologous undamaged copy as a template for re-synthesis of both ends of a double strand break (DSB), which are then correctly aligned, gap filled and ligated (Figure 1A). Alternatively, non-homologous end joining (NHEJ) is template-independent and ligates enzymatically blunted DSBs resulting in deletions and rearrangements (Figure 1B: i). Single strand annealing (SSA) is a deletion-generating process that aligns microhomologous regions of broken single-stranded DNA (ssDNA) ends, clips unannealed flaps, and gap fills and ligates the strands (Figure 1B: ii). Finally, synthesis dependent strand annealing (SDSA) is an insertion-prone mechanism that utilizes regions of microhomology to extend one end of an ssDNA fibre, which is then annealed and ligated to the other broken end (Figure 1B: iii). Between all of these pathways, only HR maintains sequence fidelity.
Figure 1.
DSBR pathways and the effect of RAD51 paralog loss on pathway choice. (A) Schematic of homologous recombination. Double strand breaks are detected by the sensor protein MDC1, which recruits the ATM dimer. ATM autophosphorylates, becomes a monomer and then phosphorylates H2A.X, which serves as a recruitment signal for additional complexes including exonucleases. Exonucleases resect the broken ends in a 5' to 3' direction to generate 3' overhanging ssDNA fibres, which are then coated with RPA. BRCA2 loads RAD51 onto the fibres by exchanging it with RPA. The RAD51-ssDNA fibres are remodelled and stabilized by RAD51 paralogs, and filaments are engaged in homology searching the genome. RAD51 catalyses the annealing of the ssDNA fibres to their homologous template for resynthesis. The junction structure is then resolved and ligated to generate intact duplex. (B) Schematic of alternative DSBR mechanisms: (i) alt-NHEJ. Binding of the KU70/80 heterodimer to the break recruits DNA-PK or PARP-1 and their associated end processing factors. Once 3' hydroxyl and 5' phosphate termini are restored ends are blunt-ligated; (ii) SSA. Regions of microhomology between ssDNA fibres are annealed, generating 3' overhanging flaps. These are cleaved by structure-specific endonucleases and the remaining gaps are filled and ligated; (iii) SDSA. Single strand invasion occurs as in HR, but only one end of a break aligns with a homologous template for polymerase extension. The newly synthesized DNA is then melted from its template and annealed to the existing 3' ssDNA overhang on the other break end; (iv) effect of RAD51 loading defects on pathway choice. RAD51 paralog or BRCA2 deficiency lead to instability or loss of RAD51 ssDNA fibres. Without RAD51 fibres HR cannot be completed and alternative error prone repair mechanisms are engaged. In these circumstances PARP-dependent alt-NHEJ is a critical repair mechanism. Chemical inhibition of PARP causes an overuse of classical DNA-PK driven NHEJ with subsequent cytotoxic deletions and chromosome structural aberrations. NHEJ, non-homologous end joining; ssDNA, single-stranded DNA; SSBR, single strand break repair; DSBR, double strand break repair; RPA, replication protein A; SSA, single strand annealing; SDSA, synthesis dependent strand annealing; HR, homologous recombination; DDR, DNA damage repair.
NHEJ can be carried out at any stage of the cell cycle, but is repressed in late S and G2 so that the more accurate HR mechanism can predominate. Cancer cells often have loss-of-function or over-activation mutations in critical components of DDR which cause repair deficiencies and dependence on compensatory mechanisms or inappropriate overuse of an activated pathway. The balance between NHEJ and HR is controlled by a range of factors including cell cycle-dependent expression, complex formation, phosphorylation and ubiquitination of repair proteins (3,4).
The ordered process of DNA transactions that comprise HR require sequential assembly of protein complexes at the DNA break [reviewed in (5) and outlined in Figure 1A]. At DSBs, the phosphorylation at serine 139 of the histone variant H2A.X (commonly referred to as γH2A.X) serves as a recruitment signal for MDC1, which in turn is phosphorylated by ATM. Phosphorylated MDC1 recruits the ubiquitin E3 ligase RNF8 catalysing Lys 63 linked ubiquitination of H2A.X and subsequent ubiquitin-dependent recruitment of BRCA1 along with its associated interacting proteins. Extended fibres of single stranded DNA are generated by nuclease resection of the double stranded broken ends. These ssDNA fibres are initially coated with replication protein A (RPA), which is then displaced by the ATP-dependent recombinase RAD51A (the literature commonly refers to RAD51A simply as RAD51 and its close relatives as RAD51 paralogs and we will utilize this convention for consistency). RAD51 is an ATP and magnesium-dependent ssDNA binding protein. BRCA2 loads RAD51 ring-like oligomers along ssDNA to form a nucleoprotein filament that is then engaged in a homology search throughout the genome to identify the corresponding intact sequence. The homologous intact DNA is then melted and the RAD51-loaded ssDNA is annealed to the complementary strand to generate a displacement loop (D-loop) bubble-like structure. The 3' end of the original broken strand is thus annealed to a complete copy which is used as a template for DNA polymerase to extend the 3' broken end past the original break point. Completion of this resynthesis process for both ends of a DSB yields two complementary 3' extended ssDNA filaments that can be resolved and ligated to reform intact dsDNA, thus maintaining sequence fidelity and repairing the initial DSB.
The RAD51 paralog family
Loading of Rad51 onto ssDNA fibres is critical for homology searching and formation of the D-loop. Rad51 knockout mice are not viable (6) and the work of our lab and others showed that it is frequently overexpressed in cancers, where it promotes efficient repair of chemotherapy-induced DNA breaks, resistance to chemotherapy and supports metastasis (7). The paralogs RAD51B, C, D, XRCC2, XRCC3 and DMC1 are divergent proteins with limited sequence homology to each other or to RAD51 except for their critical ATP-binding domains, Walker A and Walker B, which are well conserved. DMC1 has meiosis-specific functions and will not be discussed further here. The remaining mitotic paralogs form two major multi-protein complexes: the RAD51B/C/D/XRCC2 (BCDX2) and the RAD51C/XRCC3 (CX3) complexes (8) (RAD51C is common to both complexes). The BCDX2 complex is formed of two smaller subcomplexes, RAD51B/C and RAD51D/XRCC2 (9,10). The RAD51D N-terminal region binds XRCC2 while its C-terminal region binds the C-terminus of RAD51C in an ATP dependent manner (9,10). This makes RAD51D a linker between RAD51B/C and XRCC2 in the BCDX2 complex.
The Rad51D/XRCC2 complex binds ssDNA and like RAD51 is able to catalyse in vitro D-loop formation in an ATP and magnesium dependent manner (11). The ATPase activity of the BCDX2 complex is stimulated by ssDNA but is only minimally enhanced by dsDNA (8). While this RAD51-like in vitro behaviour of RAD51D and the BCDX2 complex would indicate that perhaps the paralogs carry out similar functions to RAD51, this is not the case. In fact, deletion of any of the mitotic paralogs inhibits the localization of RAD51 to radiation-induced DSBs and causes a subsequent reduction in the frequency of sister chromatid exchange events (12,13), indicative of a HR defect.
RAD51C is a component of both paralog complexes and deficient cells display exquisite sensitivity to a range of DNA damaging agents including radiation, alkylating and cross-linking agents. While the DNA damage sensitivity of paralog deficient cells has been widely demonstrated, direct comparisons of the behaviour of paralog mutants are less common. Takata et al. found that all mitotic paralog knockouts were very sensitive to a range of agents, but interestingly RAD51C or RAD51D knockouts have a slightly more serious RAD51 recruitment defect than XRCC2/3 knockouts (12). Consistent with its central role in both complexes, Rad51C knockout mice are not viable, and p53+/− Rad51C+/− double heterozygous mice are particularly tumour prone (14). Mutation of RAD51C is well established risk factor for breast and ovarian cancer with mutation rates of approximately 0.5% [for example (15) and many others]. RAD51D is less frequently mutated in cancers than RAD51C; given that RAD51D participates in one paralog complex and RAD51C in both, defects in RAD51D most likely have less impact on HR efficiency.
Two recent biophysical studies on the nematode RAD51 paralog RFS-1/RIP-1 have highlighted a critical role for the paralogs in regulating the structure and stability of RAD51 coated ssDNA filaments (16,17). These studies measured the stability of RAD51-ssDNA binding along with the flexibility of the resulting fibres. They found that upon nucleotide binding the nematode paralog RFS-1/RIP-1 caps the 5' end of a RAD51-ssDNA filament and propagates a conformational change in RAD51 that increases fibre flexibility while decreasing the rate at which RAD51 unbinds from DNA (Figure 1A). The increased stability of RAD51-ssDNA filaments and flex would favour DNA repair by HR by making the homology search more efficient and allowing a longer time for location of a homologous template. The literature consistently demonstrates that RAD51 paralog deficient cells fail to localize RAD51 to DNA damage. This poor RAD51 fibre loading or stability phenocopies the failure of RAD51 loading in BRCA2 deficient cells (Figure 1B: iv).
Genetics of RAD51 paralog mutation in breast and ovarian cancer
Many studies have examined the potential relationships between mutation of RAD51 paralogs and cancer [for example (15,18,19) and others]. Mutations in XRCC2 and XRCC3 are rare and sometimes have uncertain pathogenicity (20-22). RAD51B, C and D mutations are uncommon but have been clearly demonstrated in breast and ovarian cancers [(1,15,19,23-26) and others].
There have been very few previous reports of RAD51D mutation in breast cancer (19). The recent study by Chen et al. identified germline mutations of RAD51D in 0.38% of unselected breast cancer patients (1). Sixty percent of the mutations identified in this study were K91fs, an early frameshift mutation that codes for a protein lacking both Walker domains, with other frameshifts and premature stop codons making the balance. The early frameshifts and stop codons in these mutants would produce proteins lacking one or both Walker domains and it seems likely that such proteins would not be functional. The intrinsic genetic instability of RAD51D deficient cells would likely facilitate further acquisition of cancer driving-mutations. In support of this idea the RAD51D mutant cases reported in this study were diagnosed younger, tended to be from more aggressive subtypes, and relapsed earlier than wild type cases (1).
Potential therapeutic options for RAD51 paralog mutated cancer
Available evidence supports a role for RAD51 paralogs downstream of BRCA2 in HR. In vitro experiments indicate that RAD51 paralog loss largely phenocopies BRCA2 loss. BRCA2 deficient cancers are HR defective and reliant on homology-independent alternative NHEJ and SSA for DSBR. Their sensitivity to PARP inhibitors relies on overuse of DNA-PK driven error prone classical NHEJ once PARP activity is lost (27). Restoration of functional HR would make such cancers less sensitive to PARP inhibitors and DNA damaging chemotherapy, and this is in fact a well-established resistance mechanism. Given the HR defects, overuse of other error-prone pathways, large scale genomic deletions and telomere dysfunction displayed by RAD51 paralog deficient cells in various model systems, it appears likely that RAD51 paralog deficient cancers should respond well to PARP inhibition (Figure 1B: iv).
A recent ovarian cancer relapse study highlighted this effect (28). In this study matched pre-treatment and post-progression samples (12 samples) collected as part of a PARP inhibitor trial were sequenced for mutations in core HR pathway genes. Truncating mutations were found in BRCA1, RAD51C or RAD51D in 6 out of 12 pre-treatment biopsies. Secondary mutations restoring the reading frame were detected in 5 of these 6 PARP inhibitor resistant, relapsed tumours. One particularly interesting case had two simultaneous metastases with RAD51D mutations- one that was responding to PARP inhibition and the other resistant. The original pre-treatment tumour and the PARP inhibitor sensitive metastasis had a RAD51D frameshift mutation, and the simultaneous PARP inhibitor resistant metastasis acquired a second frameshift mutation that restores the reading frame to produce a functional RAD51D protein. Overall the evidence indicates that RAD51 paralogs participate in HR by promoting normal RAD51 function and that paralog deficiency causes a HR defect which is synthetically lethal with PARP inhibition.
While HR-restoring reversion mutations do occur in cancers which have relapsed on PARP inhibitor treatment, other modes of acquired resistance contribute significantly to the burden of disease. PARP inhibitor-treated BRCA2 deficient cells can accumulate cytotoxic levels of replication stress, which may contribute to the synthetic lethality independently of the HR defect (29,30). These recent works show that replication stress and BRCA2-null synthetic lethality is mediated by MRE11 and MUS81 driven excessive resection of stalled replication forks. BRCA2 restricts binding of MRE11 and MUS81 to stalled replication forks, thus limiting resection and preventing replication fork collapse, although the precise mechanism differs in each case.
Biophysical analyses of paralog-induced RAD51-ssDNA fibre structural changes support a model where the paralogs promote HR and chemoresistance by increasing the efficiency of the homology search. The role of RAD51 paralogs in HR and phenotypic similarity between paralog and BRCA1/2 loss would tend to indicate that paralog mutant and BRCA1/2 mutant cancers could be successfully treated with a similar strategy. As discussed above BRCA1/2 have replicative roles, which are independent of HR, and some of the synthetic lethality of the BRCA2/PARP inhibitor phenotype can be attributed to replication fork protection. While it seems likely that cancer-associated RAD51 paralog loss would cause PARP inhibition synthetic lethality, it may be to a lesser extent than BRCA2 deficiency due to the board scope of BRCA2 functions. It will be interesting to determine if the paralogs also remodel replication stress-associated RAD51 fibres. Recent studies have highlighted the low frequency but moderate penetrance of RAD51 paralog loss in breast and ovarian cancer, and demonstrated that regaining normal RAD51 paralog function can promote chemoresistance. We expect that further studies will consider if paralog mutation status can be incorporated with BRCA mutation testing to predict breast and ovarian cancer responses to chemotherapy and PARP inhibition.
Acknowledgements
None.
Provenance: This is an invited Editorial commissioned by Section Editor Yazhou He, MD (Institute of Genetics and Molecular Medicine, Western General Hospital/Usher Institute of Population Health Sciences, University of Edinburgh, Edinburgh, UK).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Chen X, Li Y, Ouyang T, et al. Associations between RAD51D germline mutations and breast cancer risk and survival in BRCA1/2-negative breast cancers. Ann Oncol 2018;29:2046-51. 10.1093/annonc/mdy338 [DOI] [PubMed] [Google Scholar]
- 2.Abbotts R, Wilson DM. Coordination of DNA single strand break repair. Free Radic Biol Med 2017;107:228-44 10.1016/j.freeradbiomed.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibata A, Conrad S, Birraux J, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J 2011;30:1079-92. 10.1038/emboj.2011.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heyer WD, Ehmsen KT, Liu J. Regulation of Homologous Recombination in Eukaryotes. Annu Rev Genet 2010;44:113-39. 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavin MF, Kozlov S, Gatei M, et al. ATM-dependent phosphorylation of all three members of the MRN complex: From sensor to adaptor. Biomolecules 2015;5:2877-902. 10.3390/biom5042877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim DS, Hasty P. A Mutation in Mouse rad51 Results in an Early Embryonic Lethal That Is Suppressed by a Mutation in p53. Mol Cell Biol 1996;16:7133-43. 10.1128/MCB.16.12.7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiegmans AP, Al-Ejeh F, Chee N, et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget 2014;5:3261-72. 10.18632/oncotarget.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masson JY, Tarsounas MC, Stasiak AZ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 2001;15:3296-307. 10.1101/gad.947001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller KA, Sawicka D, Barsky D, et al. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res 2004;32:169-78. 10.1093/nar/gkg925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruver AM, Miller KA, Rajesh C, et al. The ATPase motif in RAD51D is required for resistance to DNA interstrand crosslinking agents and interaction with RAD51C. Mutagenesis 2005;20:433-40. 10.1093/mutage/gei059 [DOI] [PubMed] [Google Scholar]
- 11.Kurumizaka H, Ikawa S, Nakada M, et al. Homologous Pairing and Ring and Filament Structure Formation Activities of the Human Xrcc2 Rad51D Complex. J Biol Chem 2002;277:14315-20. 10.1074/jbc.M105719200 [DOI] [PubMed] [Google Scholar]
- 12.Takata M, Sasaki MS, Tachiiri S, et al. Chromosome Instability and Defective Recombinational Repair in Knockout Mutants of the Five Rad51 Paralogs. Mol Cell Biol 2001;21:2858-66. 10.1128/MCB.21.8.2858-2866.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun J, Buechelmaier ES, Powell SN. Rad51 Paralog Complexes BCDX2 and CX3 Act at Different Stages in the BRCA1-BRCA2-Dependent Homologous Recombination Pathway. Mol Cell Biol 2013;33:387-95. 10.1128/MCB.00465-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuznetsov SG, Haines DC, Martin BK, et al. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res 2009;69:863-72. 10.1158/0008-5472.CAN-08-3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song H, Dicks E, Ramus SJ, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol 2015;33:2901-7. 10.1200/JCO.2015.61.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor MRG, Špírek M, Chaurasiya KR, et al. Rad51 Paralogs Remodel Pre-synaptic Rad51 Filaments to Stimulate Homologous Recombination. Cell 2015;162:271-86. 10.1016/j.cell.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor MRG, Špírek M, Jian Ma C, et al. A Polar and Nucleotide-Dependent Mechanism of Action for RAD51 Paralogs in RAD51 Filament Remodeling. Mol Cell 2016;64:926-39. 10.1016/j.molcel.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golmard L, Caux-Moncoutier V, Davy G, et al. Germline mutation in the RAD51B gene confers predisposition to breast cancer. BMC Cancer 2013;13:484. 10.1186/1471-2407-13-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buys SS, Sandbach JF, Gammon A, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 2017;123:1721-30. 10.1002/cncr.30498 [DOI] [PubMed] [Google Scholar]
- 20.Hilbers FS, Luijsterburg MS, Wiegant WW, et al. Functional Analysis of Missense Variants in the Putative Breast Cancer Susceptibility Gene XRCC2. Hum Mutat 2016;37:914-25. 10.1002/humu.23019 [DOI] [PubMed] [Google Scholar]
- 21.Al Zoubi MS, Zavaglia K, Mazanti C, et al. Polymorphisms and mutations in GSTP1, RAD51, XRCC1 and XRCC3 genes in breast cancer patients. Int J Biol Markers 2017;32:e337-43. 10.5301/ijbm.5000258 [DOI] [PubMed] [Google Scholar]
- 22.Manuguerra M, Saletta F, Karagas MR, et al. XRCC3 and XPD/ERCC2 Single Nucleotide Polymorphisms and the Risk of Cancer: A HuGE Review. Am J Epidemiol 2006;164:297-302. 10.1093/aje/kwj189 [DOI] [PubMed] [Google Scholar]
- 23.Couch FJ, Hart SN, Sharma P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 2015;33:304-11. 10.1200/JCO.2014.57.1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera B, Di Iorio M, Frankum J, et al. Functionally null RAD51D missense mutation associates strongly with ovarian carcinoma. Cancer Res 2017;77:4517-29. 10.1158/0008-5472.CAN-17-0190 [DOI] [PubMed] [Google Scholar]
- 25.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010;42:410-4. 10.1038/ng.569 [DOI] [PubMed] [Google Scholar]
- 26.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol 2016;34:1460-8. 10.1200/JCO.2015.65.0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci 2011;108:3406-11. 10.1073/pnas.1013715108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondrashova O, Nguyen M, Shield-Artin K, et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov 2017;7:984-98. 10.1158/2159-8290.CD-17-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rondinelli B, Gogola E, Yücel H, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol 2017;19:1371-8. 10.1038/ncb3626 [DOI] [PubMed] [Google Scholar]
- 30.Ray Chaudhuri A, Callen E, Ding X, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016;535:382-7. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]