Summary
The optimization of plant architecture in order to breed high‐yielding soya bean cultivars is a goal of researchers. Tall plants bearing many long branches are desired, but only modest success in reaching these goals has been achieved. MicroRNA156 (miR156)‐SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE ( SPL ) gene modules play pivotal roles in controlling shoot architecture and other traits in crops like rice and wheat. However, the effects of miR156‐ SPL modules on soya bean architecture and yield, and the molecular mechanisms underlying these effects, remain largely unknown. In this study, we achieved substantial improvements in soya bean architecture and yield by overexpressing GmmiR156b. Transgenic plants produced significantly increased numbers of long branches, nodes and pods, and they exhibited an increased 100‐seed weight, resulting in a 46%–63% increase in yield per plant. Intriguingly, GmmiR156b overexpression had no significant impact on plant height in a growth room or under field conditions; however, it increased stem thickness significantly. Our data indicate that GmmiR156b modulates these traits mainly via the direct cleavage of SPL transcripts. Moreover, we found that GmSPL9d is expressed in the shoot apical meristem and axillary meristems (AMs) of soya bean, and that GmSPL9d may regulate axillary bud formation and branching by physically interacting with the homeobox gene WUSCHEL ( WUS ), a central regulator of AM formation. Together, our results identify GmmiR156b as a promising target for the improvement of soya bean plant architecture and yields, and they reveal a new and conserved regulatory cascade involving miR156‐ SPL ‐ WUS that will help researchers decipher the genetic basis of plant architecture.
Keywords: soya bean, GmmiR156b, SPL, shoot branching, plant architecture, yield
Introduction
Soya bean [Glycine max (L.) Merr.] is a major source of plant proteins and oils. As the demand for soya bean grows, the genetic improvement of soya bean cultivars to increase yields will become increasingly important (Teng et al., 2015). Shoot architecture is the most important trait of high‐yielding crops, and shoot branching is a major component of shoot architecture (Barbier et al., 2017; Mathan et al., 2016). Since the 1990s, a major goal of plant scientists has been to improve soya bean yields by breeding cultivars with optimal branching and an ideal shoot architecture. Three models are considered to define the ideal soya bean architecture (ISA): (i) tall plants with a large stature and at least five branches, (ii) intermediate plants with a moderate stature and two or three branches and (iii) compact plants with a small stature and one or two branches (Chavarria et al., 2017). Despite extensive effort, only modest improvement in soya bean architecture has been achieved. Further, no single gene has been identified to promote the ISA and high yields.
Soya bean has a unique plant architecture, with leaves, inflorescences and pods at each node. Therefore, the production of high‐yielding soya bean plants with an ISA requires coordination between branching (branch numbers, lengths and angles) and vertical growth (main stem‐containing nodes) (Pedersen and Lauer, 2004). Achieving the ISA has been an important research topic for decades, but the mechanism has remained elusive. Genomewide analyses using a homology‐finding approach have identified 406 genes that are potentially associated with branching in soya bean, and 57 of these genes are colocalized with quantitative trait loci for soya bean branching (Tan et al., 2013). However, no functional validation of the roles of these genes in soya bean architecture has been reported (Arite et al., 2007; Booker et al., 2004; Finlayson, 2007; Morris et al., 2001; Takeda et al., 2003; Zou et al., 2006).
Plant architecture is determined by axillary meristem (AM) activity and bud outgrowth (Wai and An, 2017; Wang and Jiao, 2018). As the activity of axillary buds is normally suppressed by the activity of the shoot apical meristem (SAM) (Domagalska and Leyser, 2011), plant architecture is controlled by the balance between the activities of the SAM and AM and is strictly regulated by genetic regulatory networks. To date, our knowledge of plant architecture comes mainly from studies of the model plants Arabidopsis thaliana (hereafter, Arabidopsis) and rice (Oryza sativa). In Arabidopsis, many genes involved in AM initiation (e.g. LATERAL ORGAN BOUNDARIES1) and lateral bud outgrowth (e.g. BRANCHED1) have been identified (Bell et al., 2012; Finlayson, 2007; Teichmann and Muhr, 2015). Notably, the homeodomain transcription factor WUSCHEL (WUS), which defines the shoot stem cell niche, is also involved in AM initiation (Wang et al., 2017). In rice, several genes, including MONOCULM 1 and two WUS orthologs (TILLERS ABSENT1 and MONOCUM 3), regulate tillering (Lu et al., 2015; Tanaka et al., 2015). Notably, the SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE (SPL) family gene OsSPL14, also known as IDEAL PLANT ARCHITECTURE1 (IPA1), is a key regulator of ideal rice architecture and high rice yields (Jiao et al., 2010; Miura et al., 2010; Zhang et al., 2017). IPA1 is targeted by the microRNA (miRNA) OsmiR156, which is an upstream master regulator of ideal plant architecture (IPA) in rice.
MiRNAs are small noncoding RNAs (19–23 nt in length) that play crucial roles in plant development (Ha and Kim, 2014). MiR156s were originally identified as key regulators of the juvenile‐to‐adult phase transition in plants (Wang et al., 2009; Wu et al., 2009). Since then, their effects on plant architecture have been recognized in various species, including Arabidopsis and rice (Aung et al., 2014; Schwarz et al., 2008; Wang et al., 2015b). MiR156s affect shoot branching and plant architecture by cleaving SPL transcripts (Du et al., 2017; Liu et al., 2017; Wang et al., 2015b). Overexpression of miR156s promotes axillary bud initiation and outgrowth while suppressing SAM activity, causing a dwarf plant phenotype (Schwarz et al., 2008; Wang et al., 2015a). MiR156 overexpression also modifies other traits, including flowering, root length and grain yields (Wang and Wang, 2015; Wang and Zhang, 2017; Yu et al., 2015). Thus, miR156‐SPL modules coordinately regulate yield‐related traits in crops. Soya bean has 28 miR156s in its genome; however, the roles of those miR156s in determining plant architecture and yield‐related traits are unknown.
Previously, we generated transgenic soya bean lines overexpressing GmmiR156b and confirmed the roles of GmmiR156b in flowering (Cao et al., 2015). Here, we report that miR156b is a master regulator of ISA and that overexpressing GmmiR156b promotes the first type of ISA, with a substantial yield increase per plant of up to 63%. GmSPL9d is the main target of GmmiR156b, and its encoded protein interacts directly with GmWUSs. We also found new high yield‐related roles for GmmiR156b, including in the development of soya bean leaves and stems and the determination of seed size. Our findings provide a promising avenue for the genetic improvement of soya bean architecture and novel insight into the molecular mechanisms underlying establishment of the ideal architecture and high yields in soya bean.
Results
GmmiR156b overexpression affects shoot branching but not plant height
To investigate whether GmmiR156b affects other traits in soya bean, we performed phenotypic studies using two transgenic lines with substantially increased levels of GmmiR156b (Figure S1) in a growth room and under natural field conditions (with a distance of 60 cm between rows and between the plants in each row). T4 transgenic plants of these GmmiR156b‐overexpressing (miR156bOE) lines showing varied levels of GmmiR156b expression exhibited a dramatically different shoot architecture compared with wild‐type controls. As shown in Figure 1, lines miR156bOE‐5 and miR156bOE‐11 grew bigger and had significantly more branches than nontransformed control plants at harvest time (Figure 1a,b). The average number of branches per transgenic plant was 12–15, while the average number of branches per control plant was only 8–9 (Figures 1c and S2). It is well known that planting density has a strong impact on branching and plant architecture. To assess whether the increased number of branches in the miR156bOE plants was due to the large interplant distance, we planted T5 plants in the same field with two different interplant distances (row and plant spacing: 40 cm or 50 cm). The miR156bOE plants were bigger and exhibited greater branching activity than wild‐type control plants regardless of the plant/row spacing (Figure S3). Our results suggest a pivotal role for GmmiR156b in the genetic control of branching in soya bean.
Figure 1.

Characteristics of miR156b OE transgenic soya bean plants at the harvest stage. (a) Architecture of miR156b OE and wild‐type plants at the harvest stage. Scale bar, 20 cm. (b) Branching ability of the miR156b OE lines and wild type after harvest. Scale bar, 20 cm. (c) The mean total numbers of branches per miR156b OE plant and wild‐type plant (n = 20). (d) Heights of the miR156b OE lines and wild‐type controls (n = 20). (e) Stem morphology of miR156b OE and wild‐type plants at the harvest stage in the field. Yellow arrows show the sites of cotyledons at the postgermination stage. Red lines indicate the sites used to measure stem circumference in the transgenic and nontransgenic plants. Scale bar, 5 cm. (f) Statistical comparison of the stem circumference values of miR156b OE and wild‐type plants (n = 10). The data in the graphs represent means ± SEs. Statistical significance was estimated by Student's t‐tests. **P < 0.01; ***P < 0.001; ns, no significance.
Interestingly, the heights of both transgenic lines were comparable to that of the wild‐type control plants, even though the lines showed different levels of GmmiR156b overexpression (Figure 1d). This is different from the dwarf phenotypes observed in other plants (Wang et al., 2015a). In addition, the miR156bOE transgenic plants had significantly sturdier stems than did the control plants (Figure 1e,f). Notably, the severity of the phenotypes of miR156bOE‐5 transgenic line was stronger than that of the miR156bOE‐11, indicating that the level of GmmiR156b expression may determine the phenotypes.
GmmiR156b overexpression increased the yield per plant substantially
To determine the effects of GmmiR156b overexpression on yield, we characterized the pod number, seed number and 100‐seed weight per plant, which are major factors affecting soya bean yields. As shown in Figure 2, the total number of pods per transgenic plant was significantly higher than in the wild‐type control (Figure 2a). Accordingly, the average seed number per transgenic plant was increased by approximately 28% (Figure 2b). Furthermore, GmmiR156b overexpression had a large impact on seed size (including the length, width and thickness), resulting in bigger seeds compared with those of wild‐type control plants (Figure 2c–e) and a significant increase in the 100‐seed weight per plant (Figure 2f). Consequently, the highest seed yields per plant were obtained from the transgenic plants, and the average grain yield per plant among the miR156bOE plants was increased by 46% (miR156bOE‐11) to 63% (miR156bOE‐5) (Figure 2g). Intriguingly, the protein and lipid contents of the transgenic soya bean seeds were not significantly different compared with wild type (Figure 2h,i), suggesting a major role for GmmiR156b in determining soya bean yields.
Figure 2.

GmmiR156b overexpression can increase the grain yield per plant but does not alter seed quality. Total pod number (a) and seed number (b) per transgenic plant and wild‐type plant (n = 20) at the harvest stage. (c) Seed morphology of miR156b OE lines and wild type after harvest. Scale bar, 5 mm. (d) Soya bean seed characters and (e) size parameters of the miR156b OE lines and wild‐type control plants after harvest (n > 40). T, L and W are the abbreviations for seed thickness, length and width, respectively. Scale bar, 5 mm. (f) The 100‐seed weights of the miR156b OE lines and wild‐type control plants (n = 20). (g) Yield per plant in miR156b OE lines and wild type (n = 15). (h) The protein and lipid contents of seeds from miR156b OE and wild‐type plants. The data in the graphs represent means ± SEs. Statistical significance was estimated by Student's t‐tests. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance.
GmmiR156b overexpression shortens the plastochron of trifoliolate leaves
The plastochron was shorter in miR156bOE plants than in wild‐type plants. At 70 days after emergence (DAE), the number of trifoliolate leaves per miR156bOE plant was significantly higher than in wild type under field conditions (Figure 3a). The total number of trifoliolate leaves, including on the main stem and branches, per miR156bOE plant increased about 120%. In particular, the number of leaves on the branches was greatly increased compared with wild type because of the increased branch number (Figure 3b,c). Both miR156bOE lines grew faster than wild type in terms of trifoliolate leaf initiation (Figure 3d). Accordingly, the node number per miR156bOE plant was markedly increased (Figure 3e). Decreased plastochron lengths were also observed when the miR156bOE plants were grown indoors (Figure S4). The total number of trifoliolate leaves on the primary stem was significantly different between the miR156bOE and wild‐type control plants at 30 DAE (Figure S3). This observation is consistent with previous results obtained for Arabidopsis miR156f (Wang et al., 2008), suggesting that GmmiR156 has a conserved function in leaf plastochron determination.
Figure 3.
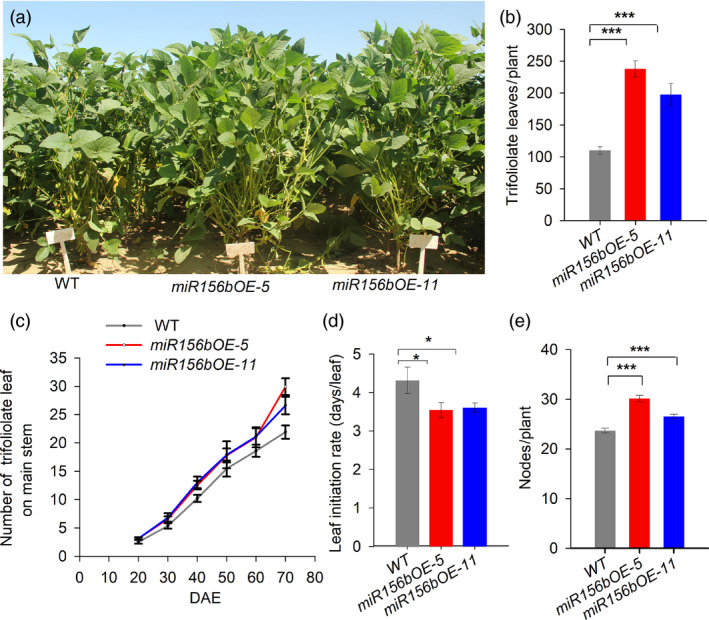
GmmiR156b overexpression shortens the length of the plastochron to increase the number of trifoliolate leaves and nodes per plant. (a) Transgenic miR156b OE‐5, miR156b OE‐11 and wild‐type control plants were grown in a field at 70 days after emergence (DAE). Scale bar, 20 cm. (b) The total trifoliolate leaf number per transgenic and wild‐type plant (n = 20). (c) Dynamic changes in the trifoliolate leaf number on the main stem in wild type and miR156b OE transgenic lines (n = 10). (d) The leaf initiation rates were significantly increased in the miR156b OE lines (n = 10). (e) Average node number per transgenic and wild‐type plant (n = 20). All data in the graphs represent means ± SEs. Statistical significance was estimated by Student's t‐tests. *P < 0.05; ***P < 0.001.
GmmiR156b overexpression stimulates SAM and AM activity
To investigate the cytological basis for the regulatory effects of GmmiR156 on the leaf plastochron and branching in soya bean, we performed the following experiments. First, we harvested shoot growth tips from potted miR156bOE and wild‐type plants (which exhibited a clear difference in trifoliolate leaf growth at 15 DAE; Figure 4a). As miR156OE‐5 line showed stronger shoot phenotypes, we used line miR156bOE‐5 for further morphological observation of the shoot tips [including leaf primordia (LP), the SAM and AMs]. As shown in Figure 4b, newly emerged trifoliolate leaves in the miR156bOE shoot apices were much bigger than those in wild type, and there was one more young trifoliolate leaf visible at the miR156bOE‐5 shoot apex than in wild type (Figure 4c,g). Although no significant differences were noted between the shoot apices of miR156OE‐5 and the control (Figure 4d,e,h,i), scanning electron micrographs showed different developmental characteristics in these shoot apices, including the number of LP and morphology of the SAM (Figure 4f,j). Longitudinal (paraffin) sections of the shoot apices confirmed that the overexpression of GmmiR156b promoted shoot development (Figure 4k,l). Notably, GmmiR156b overexpression also accelerated the initiation and growth of LP and axillary bud primordia (Figure 4m,n). There were three axillary bud primordia in the axil of the LP at the miR156bOE‐5 apex, compared to only two axillary bud primordia in the axil of the LP at the apex of the wild‐type plants. Meanwhile, LP were initiated in the SAM of the miR156bOE plants but not in the wild‐type control.
Figure 4.

GmmiR156b overexpression enhanced the meristematic activity in transgenic plants at the vegetative growth stage. (a) Leaf phenotypes of miR156b OE lines grown in a growth room for 15 days after emerging from soil. Yellow arrows indicate trifoliolate leaves. Scale bar, 2 cm. (b) Shoot apical tissues from miR156b OE lines after removing two visible and mature leaves under a stereoscopic microscope. Scale bar, 5 mm. Dissected immature leaves from wild‐type (c–e) and miR156b OE‐5 (g–i) soya bean shoot apical tissues. (f and j) Scanning electron microscopic images of the shoot apical meristem (SAM) in wild‐type (f) and miR156 OE‐5 (j) plants. Yellow arrows indicate leaf primordia (LP). The numbers represent LP at different developmental stages. Blue arrows show initiated stipule primordia. At least ten individuals were examined for each genotype; representative images are shown. (k–n) Morphological structure of a wild‐type (k and m) and miR156b OE‐5 SAM (l and n). Scale bars in k and l, 200 μm. Scale bars in m and n, 50 μm. (o and p) In situ hybridization analyses of Histone H4 in wild‐type (o) and miR156b OE‐5 (p) shoot apices at 15 days after emergence.
To determine whether GmmiR156b overexpression raised the meristematic activity of the SAM and AMs, we performed in situ hybridization for Histone H4 mRNA to detect proliferating cells in S phase of the cell cycle (Gaudin et al., 2000). As expected, the level of Histone H4 expression in the miR156bOE‐5 apex was dramatically higher than that in wild type (Figure 4o,p). These results demonstrate that GmmiR156b is a positive regulator of SAM and AM formation and development.
GmSPL9d may play a major role in GmmiR156b‐mediated SAM and AM development
MiR156s mediate various biological processes through their SPL gene targets. In soya bean, 17 SPL genes, including two GmSPL2s, five GmSPL6s, four GmSPL9s and six GmSPL13s, were found (Table S1 and Figure S5). Among them, two GmSPL13 genes (GmSPL13Ba and GmSPL13Bb) were predicted to be regulated by GmmiR156b at the translational level; the rest of the GmSPL mRNAs are likely cleaved by GmmiR156b because they contain sequences that are complementary to the miRNA (Figure 5a). Next, we performed a 5′ Rapid Amplification of cDNA Ends (RACE) assay and found that all of the 15 GmSPL mRNAs were efficiently cleaved between base pairs 10 and 11 of the GmmiR156b target sites (Figure 5a), suggesting that these SPLs are targeted by GmmiR156b.
Figure 5.

5′ RACE validation of the target genes of GmmiR156b and expression analyses. (a) Experimental validation of miR156b target genes and cleavage sites using 5′ RACE. Vertical arrows indicate the 5′ termini of the miRNA‐guided cleavage products, as identified by 5′ RACE, with the frequency of clones shown. (b and c) qRT‐PCR analysis of GmSPL expression in the shoot apex (a) and axillary buds (b) of miR156b OE‐5 and wild‐type plants. GmELF1b was used as an endogenous control for gene expression. Asterisks represent significantly decreased expression of the GmSPL genes in miR156 OE‐5 plants (Student's t‐test; **P < 0.01; ***P < 0.001).
To identify specific SPL genes targeted by GmmiR156b during SAM and AM development, we analysed the expression of 15 GmSPL genes in the shoot apex and axillary buds of miR156bOE‐5 and wild‐type plants at 15 DAE. Our qRT‐PCR results show that GmSPL2a, GmSPL9a and (especially) GmSPL9d were significantly down‐regulated in the shoot apex and axillary buds of miR156bOE‐5 plants, while the expression of other target genes was slightly down‐regulated or not affected (Figure 5b,c). This result indicates that GmSPL2a, GmSPL9a and GmSPL9d may be targets of GmmiR156b in the SAM and AM, and that SPL9d is likely the main target of GmmiR156b. Meanwhile, we also analysed the expression of 15 predicted targets in other organs and found that expression of these target genes showed different patterns of organ specificity. Among them, GmSPL2a was the most abundant target gene in flower; GmSPL6a was most abundant in stem, leaf and root, while GmSPL6d was most abundance in seed. Interestingly, GmSPL9d also the highest level of expression in nodule (Figures S6). Together, these results indicate that miR156b may regulate the IPA and high yield trait through different target genes in soya bean.
GmSPL9d overexpression suppresses branching in Arabidopsis
To further test whether GmSPL9d functions in SAM and AM development, we ectopically expressed GmSPL9d under the control of the 35S promoter in Arabidopsis ecotype Columbia 0 (Col‐0) plants. Only high levels of GmSPL9d expression significantly reduced the branching and height of Col‐0 plants (Figure 6a–c). This is likely due to the fact that GmSPL9d mRNAs are cleaved by Arabidopsis miR156s because GmSPL9d is phylogenetically similar to AtSPL9, and both genes possess the same miR156b target sequence (Figure S7).
Figure 6.

GmSPL9d has a conserved function in branching control. (a) Overexpression of GmSPL9d in Col‐0. Scale bar, 5 cm. (b) qRT‐PCR analysis of GmSPL9d expression in three transgenic Arabidopsis lines. (c) Branch number in Col‐0 and transgenic Arabidopsis lines overexpressing SPL9d. (d) Overexpression of the mutated gene (7mGmSPL9d) inhibited branching and the height of the spl9‐2 mutant. Scale bar, 5 cm. (e) qRT‐PCR analysis of GmSPL9d expression in spl9‐2 mutant plants ectopically expressing GmSPL9d. (f) Quantitative analysis of branch number in wild‐type plants (Col‐0) and spl9‐2 mutant plants overexpressing 7mGmSPL9d (n > 10). The data were surveyed at 30 days after emergence from soil. ACTIN8 was used as an internal control for gene expression. All data are represented as means ± SEs. Asterisks represent a significantly decreased branch number in spl9‐2 mutants overexpressing 7mGmSPL9d compared with that in Col‐0 (Student's t‐test; *P < 0.05; ***P < 0.001).
Previously, it was shown that a loss of function in AtSPL9 resulted in a dramatically increased number of branches and a bushy phenotype in Arabidopsis (Schwarz et al., 2008). To confirm the effect of GmSPL9d on shoot architecture, we overexpressed a mutated version of GmSPL9d (7mGmSPL9d) with seven‐point mutations at the miR156b cleavage site (mismatches) without affecting any amino acids in the spl9‐2 mutant. As shown in Figure 6d–f, GmSPL9d was highly expressed in spl9‐2‐expressing 7mGmSPL9d plants, and the 7mGmSPL9d‐OE spl9‐2 plants were significantly shorter than spl9‐2 plants; moreover, the number of branches was significantly lower than in spl9‐2 mutant and Col‐0 plants. Taken together, these results demonstrate that GmSPL9d has a conserved function in shoot branching in plants, but that it may have a distinct role in apical dominance in soya bean. Our data also indicate that the expression level of GmSPL9d is closely related to the activity of apical growth and branching.
GmSPL9d interacts with GmWUSa/b
We next explored the molecular mechanism by which GmSPLs affect soya bean plant architecture. Because WUSs are key regulators of both the SAM and AM in plants and GmWUSa is expressed in the SAM and AM and affects Arabidopsis shoot architecture (Tanaka et al., 2015; Xin et al., 2017), we speculated that two transcription factors (GmSPL9d and GmWUSa) work together to regulate soya bean shoot architecture. To test this, we first performed in situ hybridization. Our results show that GmSPL9d was coexpressed with GmWUSa in the SAM and AMs (Figure 7a–j). To further address the relationship between GmSPL9d and GmWUSa/b, we performed several protein interaction assays. Yeast two‐hybrid (Y2H) assays showed that GmSPL9d can physically interact with GmWUSa/b, but that the interaction with GmWUSa is stronger (Figure 7k). The physical interaction between GmSPL9d and GmWUSa/b was further confirmed by a pull‐down assay (Figure 7l). The interaction of GmSPL9d with GmWUSa/b was then confirmed using a bimolecular fluorescence complementation (BiFC) system (Figure 7m). To validate the GmSPL9d‐GmWUSa/b interaction, we performed co‐immunoprecipitation (Co‐IP) experiments using a transient expression system in Nicotiana benthamiana leaves. Constructs expressing GmSPL9d‐GFP and GmWUSa/b‐flag fusion proteins were cotransformed into tobacco. As shown in Figure 7n, the GmSPL9d‐GFP fusion protein was detected in cell extracts when it was coexpressed with GmWUS‐flag; by contrast, no protein interaction was detected when GmSPL9d‐GFP was coexpressed with myc‐flag. Our in vitro and in vivo results demonstrate that GmSPL9d can directly interact with GmWUSa/b.
Figure 7.
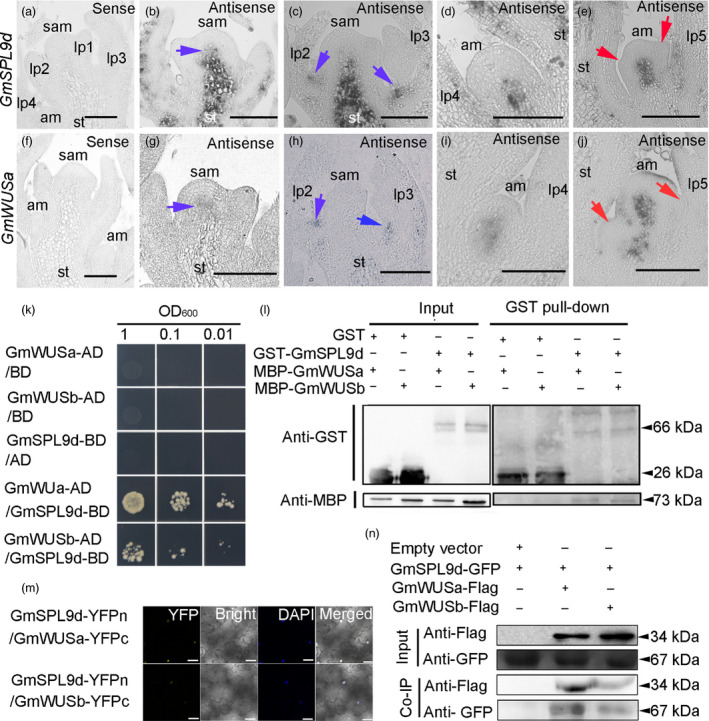
GmSPL9d interacts with GmWUS. (a–e) In situ expression patterns of GmSPL9d in the shoot apical meristem (SAM) and axillary meristems (AMs). (a) Vegetative shoot apices at 15 days after emergence (DAE) were hybridized with a GmSPL9d sense strand control. (b) The GmSPL9d antisense probe signal appeared in the SAM (blue arrow). (c) The GmSPL9d signal appeared in the leaf axillary region (blue arrows) at the pre‐AM formation stage. (d) The GmSPL9d signal was detected in AM bumps. (e) GmSPL9d was expressed in established axillary buds. Red arrows show leaf primordia (LP). (f–j) In situ expression of GmWUSa in the SAM and at different stages of axillary bud formation in wild type. (f) The vegetative shoot apex of plants at 15 DAE was hybridized with a GmWUSa sense strand control. (g) The GmWUSa antisense signal appeared in the SAM (blue arrow). (h) A GmWUSa signal also appeared in the leaf axillary region (blue arrows) at the pre‐AM formation stage. (i) The GmWUSa signal detected in the AM bumps where GmSPL9d was expressed. (j) GmWUSa expression in an established axillary bud. Red arrows indicate LP. Note: lp1–5 represent LP at different developmental stages. st, stem. Scale bar, 50 μm. (k) Y2H assay validating the interaction of GmSPL9d with GmWUSa/b. (l) A GST pull‐down assay showing the interaction between GmSPL9d and GmWUSa/b. The sizes of GST tag, GST‐GmSPL9d fusion protein, MBP tag and MBP‐GmWUSa/b fusion proteins were indicated. (m) BiFC in tobacco leaves. Visible light indicates the interaction between GmSPL9d and GmWUSa/b in the nucleus. DAPI staining was used as a nuclear marker. Panels (left to right): YFP; bright; DAPI staining; merged channels. Scale bar, 25 μm. (n) Co‐IP of GmSPL9d‐GFP and GmWUSa/b‐flag. Transgenic plants expressing GFP were used as a negative control. The sizes of Flag tag, GmWUSa/b‐Flag fusion protein, GFP tag and GmSPL9d‐GFP fusion protein were indicated.
To examine which domain of GmWUS is involved in its interaction with GmSPL9d, we fused the full‐length or truncated GmWUSa coding sequence to the Gal4‐DNA‐binding domain to produce bait vectors and fused the full‐length GmSPL9d coding sequence to the GAL4 activation domain (AD; GAL4‐AD‐GmSPL9d) to produce the prey vector. Y2H assays revealed that the acidic domain (amino acids 220–234) of GmWUSa is essential for the interaction between GmWUSa and GmSPL9d (Figure S8a), and the results were confirmed by BiFC assays (Figure S8b).
As SPL9 and WUS are markers of plant growth and architecture (Mayer et al., 1998; Wang et al., 2009), we questioned whether the SPL9‐WUS protein interaction is a conserved regulatory mode during plant growth and development. In Y2H and BiFC assays, SPL9 interacted strongly with WUS in Arabidopsis (Figure S9). To determine whether other SPLs interact with WUS, we included SPL2 in our assays. The results confirmed an interaction between SPL2 and WUS (Figure S10), indicating that SPL‐WUS interactions might be a conserved mechanism of SAM and AM regulation in plants.
Discussion
The roles of miR156 in branching have been shown in monocots (e.g. rice and wheat) and dicots (e.g. Arabidopsis and alfalfa). Overexpression of miR156 increases branching, resulting in bushy plants (Chuck et al., 2007; Fu et al., 2012; Liu et al., 2017; Luo et al., 2012; Schwab et al., 2005; Xie et al., 2012). In monocots, tiller number is related to the number of spikes or panicles per plant. The fact that miR156 overexpression causes an increase in tiller number and reduction in spike number (Liu et al., 2017; Wang et al., 2015a,b) has hindered the application of miR156 in IPA breeding. In soya bean, branch number is directly correlated with the pod number per plant, and long branches with effective nodes are a key requirement for the first ISA model. Our finding that GmmiR156b overexpression dramatically increased the number of branches in soya bean (Figure 1a–c) supports the notion that GmmiR156 is a central regulator of shoot branching in higher plants. Intriguingly, overexpression of GmmiR156b enabled soya bean plants to produce an increased number of long branches with effective nodes (Figure 1b), thereby resulting in increased numbers of pods and seeds, and a high yield per plant (Figure 2a,b). Previously, we reported that overexpression of GmmiR156b delayed flowering of soya bean for about 20 days in the controlled growth chamber (Cao et al., 2015). Such a long time delay in flowering will greatly affect maturation of plants and eventual yield in soya bean. Unexpectedly, these GmmiR156bOE plants only showed slight delay in flowering time under natural field conditions. The flowering time difference between the GmmiR156bOE plants grown in the controlled and natural conditions may be caused by the environmental factors, such as light, diurnal temperature variation and so on. Thus, GmmiR156b is an ideal target for the genetic improvement of soya bean architecture to obtain increased yields. Previous studies have shown that the expression levels of miR156s and their target genes are correlated with agronomic traits in crops, such as rice, switchgrass and alfalfa (Aung et al., 2014; Fu et al., 2012; Wang et al., 2015a). In this study, we also found a positive correlation between GmmiR156b expression level and ISA. The transgenic line miR156bOE‐5 with higher level of miR156b expression showed bigger statue, more branches and higher yield (Figure 1). Therefore, genetic control of miR156 expression levels is crucial for achieving ISA and high yield in soya bean.
MiR156s exert their functions by negatively regulating SPL genes. In rice and wheat, IPA1 (OsSPL14) and its wheat ortholog TaSPL17 are targets of miR156 in the regulation of tillering and plant architecture (Jiao et al., 2010; Luo et al., 2012; Miura et al., 2010; Wang and Zhang, 2017). In Arabidopsis, miR156 regulates branching through SPL9 and SPL15 (Schwarz et al., 2008). In soya bean, we obtained expression and preliminary genetic evidence in Arabidopsis showing that GmSPL9d may be the main target of GmmiR156b in branching (Figures 5b,c, 7c–e, and 6c). Because of the uniqueness of soya bean growth and development, further analysis of GmSPL9d loss of function mutations will confirm its major role in ISA of soya bean. Because several other GmSPL genes were also down‐regulated in the axillary buds of GmmiR156bOE soya bean plants (Figure 5c), we cannot exclude the possibility that GmmiR156b accomplishes its role in shoot architecture through several SPL genes. In plants, the SPL genes exist as a large gene family. In soya bean, we found that these GmSPL genes showed organ‐specific expression patterns and were differentially regulated by miR156b (Figure S6). It is conceivable that GmmiR156s control complex traits through a regulatory network composed of GmSPL and their downstream genes. Therefore, the GmmiR156‐GmSPL module is a key regulatory hub essential for soya bean ISA and high yield.
Symbiotic nitrogen fixation is a unique trait in soya bean, and number of nitrogen‐fixing nodule is one of the most important factors that determine the nitrogen fixation efficiency which plays an important role in grain yield and seed quality. It was previously reported that overexpression of GmmiR156 using rhizogenesis‐mediated hairy root transformation significantly reduced nodule number in soya bean in a controlled growth condition (Yan et al., 2013). In this study, we found that several GmSPL genes including GmSPL9d were also highly expressed in mature nodules in soya bean (Figure S6), supporting the notion that the GmmiR156‐GmSPL module may also modulate nodulation and symbiotic nitrogen fixation of soya bean. It will be interesting to further investigate whether the overexpression of GmmiR156b affects nodulation and symbiotic nitrogen fixation in stable transgenic plants under controlled and natural environments, and how miR156b coordinately regulates symbiotic nitrogen fixation and ISA. It is also important to identify which target gene(s) are required for regulation of symbiotic nitrogen fixation.
Branching is suppressed by apical dormancy in most plants. Therefore, an optimal balance between apical growth and branching is critical to achieve an IPA and high yields. In rice and other plants, overexpression of GmmiR156 results in increased tillers but stunted plants (Liu et al., 2017; Wang et al., 2015a). By contrast, GmmiR156b overexpression did not affect the activity of the soya bean apical meristem and primary shoot growth (Figures 1a,d and 3a). This finding implies that GmmiR156b does not mediate the activity of the apical meristem in soya bean; moreover, it is possible that the regulation of apical meristems and AMs is not tightly linked in soya bean. Genetic control of apical dominance is a fundamental question in plant biology, and extensive progress has been made using model plants such as Arabidopsis in past decades. Because of unique architecture of miR156bOE plants, it will be very interesting to dissect how GmmiR156 regulate SAM and AM and uncover the molecular mechanisms underlying the ideal architectures of soya bean plants.
In this study, we produced several lines of evidence showing that GmmiR156b plays additional novel roles that contribute to the ideal architecture and high yields in soya bean. Firstly, overexpression of GmmiR156b resulted in plants with a sturdier stem (Figure 1e,f), which may protect plants against lodging. Secondly, GmmiR156bOE plants produced significantly more and bigger seeds (Figure 2c–e), resulting in substantially increased yields per plant (Figure 2f,g). Thirdly, GmmiR156b overexpression shortened the plastochron to increase trifoliolate leaves (Figure 3a–d), which might enhance photosynthetic activity to secure extra consumption by additional vegetative and reproductive organs. Finally, GmmiR156b did not affect the quality of soya bean seeds (Figure 2h,i). To our knowledge, we have identified novel roles for GmmiR156b in coordinating branching and plant apical growth that contribute to an ISA and regulate stem thickness, which in turn affects lodging resistance. This is also the first work to show that an ISA and high yields can be achieved by manipulating GmmiR156b. Our findings indicate a potential avenue for breeding high‐yielding soya bean plants with an ideal architecture. Our data also indicate the existence of a novel and conserved molecular mechanism by which SPLs complex with WUSs to determine plant architecture. Finally, our findings provide novel insight into the molecular mechanisms underlying species‐specific shoot architecture and high yield‐related traits, and they provide a strategy for the deliberate breeding of crops with an IPA and high yields.
Experimental procedures
Soya bean materials and growth conditions
Soya bean cultivar Williams 82 and two T4 transgenic 35S:miR156b lines, miR156bOE‐5 and miR156bOE‐11 (Cao et al., 2015), were planted at the Experimental Station of Hebei Academy of Agriculture and Forestry Science (37°56′N, 114°43′E) in the summer of 2016 and 2017. Seeds were sown in fields with a proper soil moisture content (15%–20%) in three‐row plots with a row length of 6 m, adopting a completely random block design with three biological replicates. After 3 weeks, the seedlings were manually thinned to achieve interrow and interplant distances of 40, 50 or 60 cm, respectively. Plant architecture and yield‐related traits were evaluated during growth and at the harvest stage. Transgenic and wild‐type plants were grown in pots in a growth room under standard conditions (16‐h days, daytime temperature of 26 °C and night‐time temperature of 23 °C). Samples were collected at 15 DAE and used for gene expression analyses. Arabidopsis ecotype Col‐0 and the spl9‐2 mutant (SALK_006573.53.25.x) used in this study were obtained from the Arabidopsis Resource Center (ABRC, Columbus, OH). Arabidopsis plants were grown under standard conditions (16‐h days, daytime temperature of 22 °C and night‐time temperature of 18 °C).
Protein and lipid content determination
We used a MATRIX‐I FT‐NIR Spectrometer (Bruker Corp., Billerica, MA) to detect the protein and lipid contents in our plants after harvest. We used seeds from five plants of each miR156bOE line and wild type. Each plant was assessed three times.
Plasmid construction for the generation of transgenic Arabidopsis plants
For GmSPL9d overexpression, pTF101 (a plant binary vector) harbouring 35S:GmSPL9d‐GFP or 35S:7mGmSPL9d‐GFP was generated. Full‐length (1080 bp) GmSPL9d cDNA was amplified and inserted into pTF101 using SmaI and BamHI. For site‐directed mutagenesis, seven synonymous mutations in the miR156b‐binding site of GmSPL9d were designed as described by Jiao et al. (2010) and inserted into pTF101 using SmaI and BamHI. Each construct was transformed into Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation using the floral dip method (Zhang et al., 2006). As pTF101 contains the bar gene, the transformants were screened on Murashige and Skoog medium containing Basta. The seeds were stratified in darkness at 4 °C for 2 days and then transferred to a culture room at 22 °C under a 16 h of light/8 h of dark photoperiod. At 7 days after stratification, the seedlings were transplanted to soil for molecular characterization and homozygote identification. Homozygous transgenic lines were used for phenotypic analyses. The primers used are summarized in Table S2.
Y2H assays
Yeast transformation was performed following the manufacturer's instructions (Clontech Laboratories, Mountain View, CA). The full‐length coding sequences of GmSPL9d and GmWUSa/b and various deletion derivatives of GmWUSa were amplified with the listed primers (Table S2). The resulting products were cloned into pDONOR207 by the BP reaction (Invitrogen, Carlsbad, CA), and then into pGBKT7/pGADT7 by LR recombination (Invitrogen). The constructs were then cotransformed into Saccharomyces cerevisiae strain AH109. Yeast transformants were confirmed by growth on SD/‐Leu/‐Trp medium. The transformed yeast cells were incubated in liquid SD/‐Leu/‐Trp medium and grown at 28 °C to an optical density (OD600) value of 1, and the yeast cells were then collected and diluted to different ODs (0.1, 0.01, and 0.001). Protein–protein interactions were assayed in growth experiments using the suspended, transformed yeast on plates containing SD/‐Ade/‐His/‐Leu/‐Trp medium at 2–3 days after incubation at 28 °C.
BiFC
The full‐length coding sequences of GmSPL9d and GmWUSa/b and all of the deleted derivatives of GmWUSa were cloned into the N‐terminus of yellow fluorescent protein (YFP) and the C‐terminus of YFP via Gateway reactions with the pDONOR vector system (Invitrogen). The primers used are listed in Table S2. The constructs were transformed in GV3101 cells, and the transformed clones were cultured and infiltrated into N. benthamiana leaves as described previously (Song et al., 2011). The infiltrated tobacco leaves were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) to detect the nucleus of each plant cell, and fluorescence was imaged using a Leica confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) at 2 days after transformation. A 512 nm laser line was used to stimulate YFP, and an A530–585 bandpass filter (Leica Microsystems, Wetzlar, Germany) was used to collect the YFP signal.
Protein expression constructs and protein purification
To purify recombinant GST‐GmSPL9d, MBP‐GmWUSa and MBP‐GmWUSb, the open‐reading frames of GmSPL9d, MBP‐GmWUSa and MBP‐GmWUSb were cloned into pGEX‐4T‐1 and pMAL‐c2x. The GST‐GmSPL9d and MBP‐GmWUSa expression plasmids were then transformed into Escherichia coli strain BL21. Protein purification was performed using glutathione agarose (Thermo Fisher Scientific, Waltham, MA) and amylose resin (NEB, Ipswich, MA) according to the manufacturers' instructions. The primers used to construct the plasmids are listed in Table S2.
In vitro pull‐down assay
Purified GST and GST‐GmSPL9d were incubated with the same volume of GST beads in GST binding buffer for 2 h at room temperature, washed with GST binding buffer four times to remove redundant proteins and then incubated with MBP‐GmWUSa and MBP‐GmWUSb for another 2 h at room temperature. The cultures were then washed four times to remove redundant MBP‐GmWUSa or MBP‐GmWUSb. Samples were then collected, mixed with 2× SDS protein loading buffer and boiled for 10 min for Western blotting. Anti‐MBP (SAB2104172; Sigma‐Aldrich, St. Louis, MO) and anti‐GST (HT601‐01; Beijing TransGen Biotech Co., Ltd., Beijing, China) antibodies were used to detect GST‐ and MBP‐tagged proteins.
Co‐IP
GmWUSa and GmWUSb in pDONOR207 were recombined with pEarlygate100 (35S:Flag‐pEarlygate) to generate the GmWUSa‐Flag fusion protein. GmSPL9d cDNA was recombined with pTF101 (with a GFP tag) to generate a GmSPL9d‐GFP fusion expression cassette. The constructs were introduced into N. benthamiana leaves through GV3101 infiltration. The leaves were harvested at 2 days after infiltration and frozen in liquid nitrogen. The frozen leaves were then homogenized and mixed with protein extraction buffer [50 mm Na2HPO4/NaH2PO4 (pH 7.4), 150 mm NaCl, 1% Triton X‐100, 15% glycerol, 1 mm phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Roche, Basel, Switzerland)]. After protein extraction, flag beads (ANTI‐FLAG M2 Affinity Gel; Sigma‐Aldrich) were washed four times with phosphate‐buffered saline and incubated with the extracted proteins at room temperature for 1 h. The precipitated samples were washed at least four times with protein wash buffer [50 mm Na2HPO4/NaH2PO4 (pH 7.4), 150 mm NaCl, 0.1% Triton X‐100, 10% glycerol, 1 mm PMSF and protease inhibitor cocktail (Roche)] and then eluted with 2× SDS protein loading buffer and boiled for 10 min for Western blotting. The Flag‐ and GFP‐tagged proteins were detected with anti‐Flag antibodies [monoclonal ANTI‐FLAG M2‐Peroxidase (HRP) antibody; Sigma‐Aldrich] and anti‐GFP antibodies [Goat Polyclonal to GFP (HRP); Abcam, Cambridge, UK].
Prediction of GmmiR156b targets and 5′ RACE mapping of miRNA cleavage sites
Putative targets of miR156b were predicted using psRNATarget (Dai and Zhao, 2011). For the 5′ RACE mapping of miR156b cleavage sites, total RNAs were isolated from a mixture of different organs collected from 4‐week‐old Williams 82 plants using Plant RNA Reagent (Invitrogen) according to the manufacturer's recommendations. A 5′‐Full RACE Kit with TAP (Takara Bio Inc., Kusatsu, Japan) was used to process the total RNAs and to map the 5′ termini of the primary transcripts. The cDNA samples were amplified by nested PCR according to the manufacturer's protocols. Gene‐specific primers (Table S2) were designed by Invitrogen.
RNA extraction and quantitative PCR analysis
Total RNA and small RNAs were extracted using TRIzol reagent [Tiangen Biotech (Beijing) Co., Ltd., Beijing, China] and then treated with DNase I (Takara Bio Inc.) to remove contaminating genomic DNA. First‐strand cDNA was synthesized from the total RNA using a FastQuant RT Kit [Tiangen Biotech (Beijing) Co., Ltd.]. Stem‐loop‐specific reverse transcription for miR156b and miR1520d was performed as described previously (Chen et al., 2005). qRT‐PCR was performed using SuperReal PreMix Plus [SYBR Green; Tiangen Biotech (Beijing) Co., Ltd.]. The miRNA and specific primers for the genes analysed are listed in Table S2.
Histology
The shoot apices of wild‐type plants at 15 DAE were fixed in 4% paraformaldehyde, dehydrated and embedded in Paraplast (Leica Microsystems). The wax‐embedded samples were then sectioned at a thickness of 8 μm. After being deparaffinized and dehydrated in a gradient ethanol series, the samples were stained with toluidine blue and scanned by microscopy.
Scanning electron microscopy
For scanning electron microscopy, tissues were placed in 1% glutaraldehyde in 0.025 m phosphate buffer (sodium phosphate, pH 7.2–7.4) and a vacuum was applied for 10 min. The tissues were then fixed overnight at 4 °C. Next, the tissues were rinsed twice with 0.025 m phosphate buffer for 15 min, postfixed with 1% osmium tetroxide in 0.025 m phosphate buffer for 1 h at 4 °C and moved through a gradient ethanol series (20% increments), with each increment lasting 10 min and ending with two exchanges of 100% ethanol. Each tissue was then mounted to a test stub with double‐sided adhesive tape and sputter‐coated with a gold‐palladium alloy using an Eiko IB‐3 Ion Coater (Eiko Co., Ltd., Wakayama, Japan). The samples were then examined with a Hitachi S‐3500N Scanning Electron Microscope (Tokyo, Japan).
In situ hybridization
A 380‐nt fragment of the GmSPL9d 5′‐untranslated region (UTR), a 153‐nt fragment of the GmWUSa 5′‐UTR and a 753‐nt fragment of the GmHistone H4 coding sequencing were amplified from Williams 82 cDNA using KOD Polymerase (Stratagene, San Diego, CA) and cloned into pSPT 18 (Roche) using specific primers (Table S2). Digoxigenin‐labelled sense or antisense probes were synthesized with T7 or SP6 RNA Polymerase (Roche). Shoot apical tissues from wild‐type and transgenic materials at 15 DAE were dissected and fixed in 4% paraformaldehyde. Paraffin‐embedded materials were sectioned at a thickness of 8 μm. After the sections had been deparaffinized and dehydrated, hybridization and detection were performed as described previously (Long et al., 1996).
Phylogenetic analysis
Seventeen homologous protein sequences of GmSPLs and Arabidopsis SPLs were obtained from Phytozome (www.phytozome.net) and TAIR (www.Arabidopsis.org) and imported into MEGA5 for complete alignment using Alignment Explorer/CLUSTAL (Tamura et al., 2011). A phylogenetic tree was then built with MEGA5 using the neighbour‐joining method with the bootstrapping value set at 1000 replications.
Statistical analysis
All data were analysed using IBM SPSS Statistics 19 (IBM Corp., Armonk, NY). The means and standard errors of all results were calculated, and Student's t‐tests were performed to generate P‐values. SigmaPlot 10.0 (Systat Software Inc., San Jose, CA) was used to produce graphs.
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Author contributions
ZS, YT and XL conceived the study and designed the experiments. ZS, CS, JY, QJ, LW and YW performed the experiments. FZ, BL and FK generated the transgenic soya bean materials. ZS, QZ, BZ, MZ and LZ conducted the field experiment. ZS, YW and LW analysed the data. XL wrote the manuscript with input from all of the authors.
Supporting information
Figure S1 GmmiR156b was overexpressed in different tissues of miR156bOE lines.
Figure S2 Number of different type branches between WT and miR156bOE lines.
Figure S3 Number of branches per plant at different planting densities.
Figure S4 GmmiR156b overexpression decreased the length of the plastochron.
Figure S5 Phylogenetic analysis of GmSPL proteins.
Figure S6 Expression analysis of GmSPLs in different organs.
Figure S7 Alignment between soybean GmSPL9d and Arabidopsis SPL.
Figure S8 Interaction between GmSPL9d and various deleted derivatives of GmWUSa.
Figure S9 WUS interacts with SPL9 in Arabidopsis.
Figure S10 WUS interacts with SPL2 in Arabidopsis.
Table S1 Target gene prediction of miR156b in psRNATarget.
Table S2 Primers in this study.
Acknowledgements
We thank Kan Wang (Iowa State University) for providing the pTF101.1 vector. We thank Dr. Xigang Liu (Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences) for providing technical assistance with the in situ hybridization experiment. We also thank the ABRC for providing the spl9‐2 mutant seeds. This work was supported by the National Key Research and Development Program of China (2016YFA0500503), the National Natural Science Foundation of China (31230050), the Ministry of Agriculture of the People's Republic of China (2014ZX0800929B, 2018ZX0800955B) and Huazhong Agricultural University's Scientific and Technological Self‐innovation Foundation (2015RC014).
Contributor Information
Fanjiang Kong, Email: kongfj@iga.ac.cn.
Baohui Liu, Email: liubh@iga.ac.cn.
Yiping Tong, Email: yptong@genetics.ac.cn.
Xia Li, Email: xli@mail.hzau.edu.cn.
References
- Arite, T. , Iwata, H. , Ohshima, K. , Maekawa, M. , Nakajima, M. , Kojima, M. , Sakakibara, H. et al. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Aung, B. , Gruber, M.Y. , Amyot, L. , Omari, K. , Bertrand, A. and Hannoufa, A. (2014) MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13, 779–790. [DOI] [PubMed] [Google Scholar]
- Barbier, F.F. , Dun, E.A. and Beveridge, C.A. (2017) Apical dominance. Curr. Biol. 27, R864–R865. [DOI] [PubMed] [Google Scholar]
- Bell, E.M. , Lin, W.‐C. , Husbands, A.Y. , Yu, L. , Jaganatha, V. , Jablonska, B. , Mangeon, A. et al. (2012) Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl Acad. Sci. USA, 109, 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker, J. , Auldridge, M. , Wills, S. , McCarty, D. , Klee, H. and Leyser, O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Cao, D. , Li, Y. , Wang, J. , Nan, H. , Wang, Y. , Lu, S. , Jiang, Q. et al. (2015) GmmiR156b overexpression delays flowering time in soybean. Plant Mol. Biol. 89, 353–363. [DOI] [PubMed] [Google Scholar]
- Chavarria, G. , Caverzan, A. , Müller, M. and Rakocevic, M. (2017) Soybean – The Basis of Yield, Biomass and Productivity, Ch. 02. Rijeka: InTech. [Google Scholar]
- Chen, C. , Ridzon, D.A. , Broomer, A.J. , Zhou, Z. , Lee, D.H. , Nguyen, J.T. , Barbisin, M. et al. (2005) Real‐time quantification of microRNAs by stem‐loop RT‐PCR. Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G. , Cigan, A.M. , Saeteurn, K. and Hake, S. (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39, 544–549. [DOI] [PubMed] [Google Scholar]
- Dai, X. and Zhao, P.X. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska, M.A. and Leyser, O. (2011) Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Du, Y. , Liu, L. , Li, M. , Fang, S. , Shen, X. , Chu, J. and Zhang, Z. (2017) UNBRANCHED3 regulates branching by modulating cytokinin biosynthesis and signaling in maize and rice. New Phytol. 214, 721–733. [DOI] [PubMed] [Google Scholar]
- Finlayson, S.A. (2007) Arabidopsis TEOSINTE BRANCHED1‐LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol. 48, 667–677. [DOI] [PubMed] [Google Scholar]
- Fu, C. , Sunkar, R. , Zhou, C. , Shen, H. , Zhang, J.‐Y. , Matts, J. , Wolf, J. et al. (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 10, 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin, V. , Lunness, P.A. , Fobert, P.R. , Towers, M. , Riou‐Khamlichi, C. , Murray, J.A.H. , Coen, E. et al. (2000) The expression of D‐cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 122, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, M. and Kim, V.N. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509. [DOI] [PubMed] [Google Scholar]
- Jiao, Y. , Wang, Y. , Xue, D. , Wang, J. , Yan, M. , Liu, G. , Dong, G. et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Cheng, X. , Liu, P. and Sun, J. (2017) miR156‐targeted SBP‐box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 174, 1931–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A. , Moan, E.I. , Medford, J.I. and Barton, M.K. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis . Nature, 379, 66. [DOI] [PubMed] [Google Scholar]
- Lu, Z. , Shao, G. , Xiong, J. , Jiao, Y. , Wang, J. , Liu, G. , Meng, X. et al. (2015) MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genomics, 42, 71–78. [DOI] [PubMed] [Google Scholar]
- Luo, L. , Li, W. , Miura, K. , Ashikari, M. and Kyozuka, J. (2012) Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol. 53, 1793–1801. [DOI] [PubMed] [Google Scholar]
- Mathan, J. , Bhattacharya, J. and Ranjan, A. (2016) Enhancing crop yield by optimizing plant developmental features. Development, 143, 3283–3294. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F.X. , Schoof, H. , Haecker, A. , Lenhard, M. , Jürgens, G. and Laux, T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell, 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Ikeda, M. , Matsubara, A. , Song, X.J. , Ito, M. , Asano, K. , Matsuoka, M. et al. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Morris, S.E. , Turnbull, C.G.N. , Murfet, I.C. and Beveridge, C.A. (2001) Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 126, 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, P. and Lauer, J.G. (2004) Soybean growth and development in various management systems and planting dates. Crop Sci. 44, 508–515. [Google Scholar]
- Schwab, R. , Palatnik, J.F. , Riester, M. , Schommer, C. , Schmid, M. and Weigel, D. (2005) Specific effects of microRNAs on the plant transcriptome. Dev. Cell, 8, 517–527. [DOI] [PubMed] [Google Scholar]
- Schwarz, S. , Grande, A.V. , Bujdoso, N. , Saedler, H. and Huijser, P. (2008) The microRNA regulated SBP‐box genes SPL9 and SPL15 control shoot maturation in Arabidopsis . Plant Mol. Biol. 67, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. , Qi, T. , Huang, H. , Ren, Q. , Wu, D. , Chang, C. , Peng, W. et al. (2011) The Jasmonate‐ZIM domain proteins interact with the R2R3‐MYB transcription factors MYB21 and MYB24 to affect Jasmonate‐regulated stamen development in Arabidopsis . Plant Cell, 23, 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T. , Suwa, Y. , Suzuki, M. , Kitano, H. , Ueguchi‐Tanaka, M. , Ashikari, M. , Matsuoka, M. et al. (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, B. , Guo, Y. and Qiu, L.J. (2013) Whole genome discovery of genes related to branching and co‐localization with QTLs in soybean. Hereditas (Beijing), 35, 793–804. [DOI] [PubMed] [Google Scholar]
- Tanaka, W. , Ohmori, Y. , Ushijima, T. , Matsusaka, H. , Matsushita, T. , Kumamaru, T. , Kawano, S. et al. (2015) Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1 . Plant Cell, 27, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann, T. and Muhr, M. (2015) Shaping plant architecture. Front. Plant Sci. 6, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, P. , Caballero‐Anthony, M. , Lassa, J. and Nair, T. (2015) Towards Asia 2025: policy and technology imperatives. Summary of the main findings of the second international conference on Asian food security held in Singapore on 21–22 August 2014. Food Secur. 7, 159–165. [Google Scholar]
- Wai, A.H. and An, G. (2017) Axillary meristem initiation and bud growth in rice. J. Plant Biol. 60, 440–451. [Google Scholar]
- Wang, Y. and Jiao, Y. (2018) Axillary meristem initiation—a way to branch out. Curr. Opin. Plant Biol. 41, 61–66. [DOI] [PubMed] [Google Scholar]
- Wang, H. and Wang, H. (2015) The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant. 233, 553–560. [DOI] [PubMed] [Google Scholar]
- Wang, L. and Zhang, Q. (2017) Boosting rice yield by fine‐tuning SPL gene expression. Trends Plant Sci. 22, 643–646. [DOI] [PubMed] [Google Scholar]
- Wang, J.W. , Schwab, R. , Czech, B. , Mica, E. and Weigel, D. (2008) Dual effects of miR156‐targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana . Plant Cell, 20, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.W. , Czech, B. and Weigel, D. (2009) miR156‐regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana . Cell, 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Sun, S. , Jin, J. , Fu, D. , Yang, X. , Weng, X. , Xu, C. et al. (2015a) Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl Acad. Sci. USA. 112, 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Wang, Z. , Amyot, L. , Tian, L. , Xu, Z. , Gruber, M.Y. and Hannoufa, A. (2015b) Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus . Mol. Genet. Genomics, 290, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Tian, C. , Zhang, C. , Shi, B. , Cao, X. , Zhang, T.Q. , Zhao, Z. et al. (2017) Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell, 29, 1373–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. , Park, M.Y. , Conway, S.R. , Wang, J.W. , Weigel, D. and Poethig, R.S. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Shen, J. , Hou, X. , Yao, J. , Li, X. , Xiao, J. and Xiong, L. (2012) Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol. 158, 1382–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, W. , Wang, Z. , Liang, Y. , Wang, Y. and Hu, Y. (2017) Dynamic expression reveals a two‐step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis . J. Plant Physiol. 214, 1–6. [DOI] [PubMed] [Google Scholar]
- Yan, Z. , Hossain, M.S. , Wang, J. , Valdes‐Lopez, O. , Liang, Y. , Libault, M. , Qiu, L. et al. (2013) miR172 regulates soybean nodulation. Mol. Plant Microbe Interact. 26, 1371–1377. [DOI] [PubMed] [Google Scholar]
- Yu, N. , Niu, Q.W. , Ng, K.H. and Chua, N.H. (2015) The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83, 673–685. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.S. , Niu, Q.W. and Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Yu, H. , Ma, B. , Liu, G. , Wang, J. , Wang, J. , Gao, R. et al. (2017) A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 8, 14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Zhang, S. , Zhang, W. , Li, G. , Chen, Z. , Zhai, W. , Zhao, X. et al. (2006) The rice HIGH‐TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48, 687–698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 GmmiR156b was overexpressed in different tissues of miR156bOE lines.
Figure S2 Number of different type branches between WT and miR156bOE lines.
Figure S3 Number of branches per plant at different planting densities.
Figure S4 GmmiR156b overexpression decreased the length of the plastochron.
Figure S5 Phylogenetic analysis of GmSPL proteins.
Figure S6 Expression analysis of GmSPLs in different organs.
Figure S7 Alignment between soybean GmSPL9d and Arabidopsis SPL.
Figure S8 Interaction between GmSPL9d and various deleted derivatives of GmWUSa.
Figure S9 WUS interacts with SPL9 in Arabidopsis.
Figure S10 WUS interacts with SPL2 in Arabidopsis.
Table S1 Target gene prediction of miR156b in psRNATarget.
Table S2 Primers in this study.


