Abstract
Live attenuated influenza vaccines (LAIVs) are promising tools for the induction of broad protection from influenza due to their ability to stimulate cross-reactive T cells against influenza pathogens. One of the major targets for cytotoxic T-cell immunity is viral nucleoprotein (NP), which is relatively conserved among antigenically distant influenza viruses. Nevertheless, a diversity of epitope composition has been found in the NP protein of different lineages of influenza A viruses. The H2N2 master donor virus which is currently used as a backbone for the LAIV and donor of the six genomic segments encoding the internal proteins, A/Leningrad/134/ 17/57 (MDV Len/17), was isolated 60 years ago. As such, NP-specific T-cell immunity induced upon vaccination with classical LAIVs with a 6:2 genome composition containing this older NP might be suboptimal against currently circulating influenza viruses. In this study, a panel of H3N2 LAIV candidates with wild-type NP genes derived from circulating viruses were generated by reverse genetics (5:3 genome composition). These viruses displayed the cold adaptation and temperature sensitivity phenotypes of MDV Len/17 in vitro. LAIVs with both 6:2 and 5:3 genome compositions were attenuated and replicated to a similar extent in the upper respiratory tract of ferrets. LAIVs were immunogenic as high neutralizing and hemagglutination inhibition serum antibody titers were detected 21 days after infection. All vaccinated animals were protected against infection with heterologous H3N2 influenza A viruses. Thus, LAIV with a 5:3 genome composition is safe, immunogenic and can induce cross-protective immunity.
Keywords: Live attenuated influenza vaccine, Influenza A virus, Nucleoprotein, Reverse engineering, Ferrets
1. Introduction
Influenza A viruses are highly contagious respiratory pathogens that continuously threaten the human population. Influenza epidemics cause severe respiratory disease worldwide, with up to 645,000 annual influenza-associated illness deaths (Iuliano et al., 2018). The most effective strategy to prevent infection with influenza virus is vaccination. Currently, there are three types of influenza vaccines available – inactivated influenza vaccines (IIV), live attenuated influenza vaccines (LAIV) and recombinant influenza vaccines. Immunization with IIV induces a humoral immune response, typically directed to the viral surface glycoproteins – hemagglutinin (HA) and neuraminidase (NA). However, this immunity is strain-specific and offers little protection against drift variants of influenza viruses. Similarly, LAIVs induce humoral immunity against the HA and NA, but LAIVs induce cross-reactive immunity more efficiently than IIV. Infection with LAIV induces no or mild upper respiratory symptoms mimicking a subclinical influenza virus infection (Hoft et al., 2017). Viral infection activates both systemic and localized innate and adaptive immune responses, which provide protection by a variety of mechanisms including viral interference (Wang et al., 2015), antiviral NK cell activation at the site of infection (Laurie et al., 2015), influenza-specific cross-reactive T cell activation and expansion (Schultz-Cherry, 2015), generation of highavidity mucosal neutralizing sIgA antibody (Mohn et al., 2017) and induction of immunological memory to influenza virus antigens (van Riet et al., 2012).
LAIV strains typically inherit their HA and NA genes from a wildtype influenza virus (seasonal or potentially pandemic) and the remaining six genes from an attenuated master donor virus (referred to as a 6:2 genome composition) (Swain et al., 2004). The internal proteins, especially the nucleoprotein (NP), are predominant targets for the CD8+ T-cell immune response in humans (Aleksandrova, 1977). While CD4+ T cells regulate the immune response, CD8+ T cells contribute to influenza virus clearance (Grant et al., 2013).
Influenza A(H3N2) viruses have drifted continuously in humans over the past 40 years (Moskophidis and Kioussis, 1998). This has resulted in decreased effectiveness of seasonal vaccines, since the viruses easily escape vaccine-induced antibody immunity to the viral antigens (Hay et al., 2001). The internal proteins of influenza virus are highly conserved over time as compared to the HA and NA proteins, making them attractive targets to improve the efficacy/effectiveness of LAIVs (Glatman-Freedman et al., 2017). Currently, commercially available LAIVs for influenza A are based on two master donor viruses (MDV), A(H2N2) A/Leningrad/134/17/57 (Len/17) - used in Russia and India, and A(H2N2) A/Ann Arbor/6/60 - used in the USA, Canada and Europe; both strains were isolated 60 years ago. Over this time, A(H2N2) influenza viruses were displaced by A(H1N1) and A(H3N2) variants and mutations accumulated in virus proteins (Quinones-Parra et al., 2014). Indeed, only one third of the predicted the human leukocyte antigen (HLA) class I-restricted NP epitopes identified in A(H2N2) MDV Len/17 are conserved in recent A(H1N1) and A(H3N2) isolates (Machkovech et al., 2015). Additionally, we have found that at least 28 HLA class I-restricted epitopes have diverged between Len/17 MDV and recent A(H3N2) influenza virus NP (Isakova-Sivak et al., 2017).
Thus, there is a risk that the T-cell immunity induced upon vaccination with classical LAIVs (with 6:2 genome composition) may be diminished against currently circulating influenza viruses. The most straightforward way to overcome T-cell epitope mismatch is to include wild-type internal proteins in the LAIV reassortant viruses. In the case of Russian LAIV, only the matrix and NP genes can be replaced because they are not involved in attenuation of the MDV Len/17 virus (Korenkov et al., 2018). Thus, we proposed that the generation of a 5:3 genome composition by introducing a wild-type NP gene into the current 6:2 genome vaccine composition might be sufficient to enhance cross-protection (Isakova-Sivak et al., 2011). We generated a panel of reverse genetics-derived 5:3 LAIVs of A(H1N1), A(H3N2) and A(H7N9) subtypes. Comparative studies in mice of A(H1N1) and A(H7N9) 5:3 LAIVs demonstrated that 5:3 LAIVs could induce broader cytotoxic Tlymphocyte (CTL) cross-protective immunity and protection against more distant strains than their 6:2 LAIV counterparts (Isakova-Sivak et al., 2016). Ferrets are the most suitable animal model to study A(H3N2) human influenza infection, due to the inability of recent A(H3N2) viruses to replicate efficiently in mouse respiratory tissues (Isakova-Sivak et al., 2017; Rekstin et al., 2017). Unfortunately, ferrets’ immunobiology has not yet been well described to study cross-reactivity of influenza NP-specific T cells. In particular, there are no data about the specificity of influenza-reactive T cells in these animals. In addition, the information regarding the contribution of nucleoproteinspecific T cells in the total T-cell-mediated response in ferrets is lacking. In the present study we did not focus on the details of ferret’s T cell responses, however our previous findings from human T-cell in vitro studies justified the necessity of LAIV virus NP renewal (Narasaraju et al., 2009).
In this study, we compared A(H3N2) LAIV reassortants with 6:2 and 5:3 genome compositions with regard to (i) growth characteristics, (ii) safety, (iii) immunogenicity and their (iv) ability to protect animals against challenge with homologous and heterologous viruses. These ferret studies will serve as a basis for conducting clinical trials of 5:3 LAIVs in volunteers, where the impact of NP specific epitopes of recent A(H3N2) influenza viruses on improving CTL immunity can be thoroughly evaluated.
2. Materials and methods
2.1. Viruses
rgLAIV strains with 6:2 or 5:3 genome compositions were generated by reverse genetics (RG). HA, NA and NP genes of A(H3N2) viruses A/ Texas/50/2012 (TX/12), A/Switzerland/9715293/2013 (SW/13) and A/Hong Kong/4801/2014 (HK/14) viruses were cloned into dual-promoter plasmids, as previously described (Korenkov et al., 2018). Six genomic segments coding for internal and non-structural proteins of A/ Leningrad/134/17/57 (H2N2) were previously cloned into RG vectors (Isakova-Sivak et al., 2011). Viruses were rescued in MDCK/293 T cells as previously described (Isakova-Sivak et al., 2011). The 6:2 LAIV strains prepared from TX/12, SW/13 and HK/14 viruses were designated TX LAIV 6:2, SW LAIV 6:2 and HK LAIV 6:2, respectively. The 5:3 LAIVs were designated TX LAIV 5:3, SW LAIV 5:3 and HK LAIV 5:3, respectively (Table 1). Wild-type influenza A(H3N2) strains A/Hong Kong/4801/2014, A/Switzerland/9715293/2013, A/Texas/50/2012, A/Perth/16/2009 (PE/09) A/Brisbane/10/2007 (BR/07), A/Panama/ 2007/99 (PA/99), A/California/7/2009-like (H1N1) and B/Brisbane/ 60/2008-like were acquired from the WHO Collaborating Centre for Reference and Research on Influenza, Victorian Infectious Diseases Reference Laboratory (VIDRL), Melbourne, Australia. Wild-type and LAIV viruses were passaged in the allantoic cavity of embryonated hens’ eggs and stored in aliquots at −80 °C. Temperature sensitive and cold adapted (ts/ca) phenotypes of the LAIV A(H3N2) viruses were determined by titration in eggs at different temperatures: 38 °C compared to 33 °C and 26 °C compared to 33 °C for the ts and ca phenotypes, respectively. Eggs inoculated with serial 10-fold virus dilutions were incubated for 48 h (at 33 °C and 38 °C) or 6 days (at 26 °C). Virus titers were calculated by Reed and Muench method and expressed in log10 EID50/mL.
Table 1.
Phenotypic characteristics of H3N2 LAIVs and Len/17 master donor virus.
| LAIV strain | Source of HA and NA (clade) | Source of NP | Distance to Len/17 NP, as substitutions | Virus titre in eggs, lgEID50/mL |
||
|---|---|---|---|---|---|---|
| 26 °C | 33 °C | 38 °C | ||||
| TX LAIV 6:2 | TX/12 (3c.1) | Len/17 | - | 7.0 ± 1.3 | 9.2 ± 0.5 | 2.2 ± 0.7 |
| TX LAIV 5:3 | TX/12 (3c.1) | TX/12 | 27 | 6.1 ± 0.5 | 8.1 ± 0.4 | 1.8 ± 0.3 |
| SW LAIV 6:2 | SW/13 (3c3.a) | Len/17 | - | 6.1 ± 0.4 | 8.1 ± 0.7 | 1.7 ± 0.4 |
| SW LAIV 5:3 | SW/13 (3c3.a) | SW/13 | 29 | 5.2 ± 0.7 | 7.8 ± 0.5 | 1.8 ± 0.3 |
| HK LAIV 6:2 | HK/14 (3c2.a) | Len/17 | - | 5.5 ± 0.5 | 7.9 ± 0.4 | 1.8 ± 0.5 |
| HK LAIV 5:3 | HK/14 (3c2.a) | HK/14 | 30 | 5.4 ± 0.7 | 7.5 ± 0.3 | 1.5 ± 0.3 |
| Len/17 | Len/17 | Len/17 | - | 6.9 ± 0.7 | 8.9 ± 0.3 | 1.9 ± 0.3 |
| TX/12 | TX/12 (3c.1) | TX/12 | 27 | 3.5 ± 0.8 | 9.0 ± 0.6 | 9.0 ± 0.5 |
| SW/13 | SW/13 (3c3.a) | SW/13 | 29 | 3.0 ± 0.8 | 7.0 ± 0.9 | 6.3 ± 1.0 |
| HK/14 | HK/14 (3c2.a) | HK/14 | 30 | 3.5 ± 0.9 | 8.4 ± 0.6 | 8.4 ± 1.1 |
TX/12: A/Texas/50/2012 (H3N2); NP accession number EPI408574; SW/13: A/Switzerland/9715293/2013 (H3N2); NP accession number EPI540519; HK/14: A/ Hong Kong/4801/2014 (H3N2); NP accession number EPI614430; Len/17: A/Leningrad/134/17/57 (H2N2); NP accession number EPI555083; Distances were calculated as number of amino-acid substitutions between wild-type and Len/17 NP sequences aligned by the multiple alignment tool of Geneious 6.0 software.
2.2. Ferrets care and study design
Adult, outbred ferrets purchased from independent breeders were housed in the Bioresources Facility at the Peter Doherty Institute for Infection and Immunity. Ferrets studies were performed under the University of Melbourne Biochemistry & Molecular Biology, Dental Science, Medicine, Microbiology & Immunology, and Surgery Animal Ethics Committee approval, in accordance with the National Health and Medical Research Council (NHMRC) Australian code of practice for the care and use of animals for scientific purposes. Female ferrets with signs of estrus received chorulon prior to the start of the study. As per routine husbandry, all ferrets were vaccinated against canine distemper disease prior to use and temperature transponders with identification chips were subcutaneously implanted in ferret withers. All male ferrets were surgically castrated. Weight and temperature were measured daily. For each experiment, animals were allocated into test groups with equal weight and sex distribution.
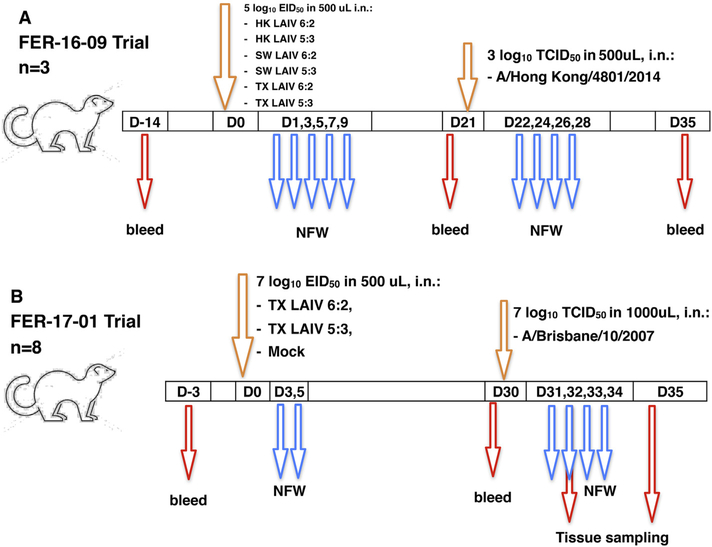
This study consisted of two consecutive experiments. In the first one (FER-16–09), groups of three ferrets were inoculated on day 0 with 5.0 log10 EID50 of each LAIV virus in a volume of 500 μL (Fig. 1A). Nasal fluid washes (NFW) were collected on days 1, 3, 5, 7 and 9 after vaccination for LAIV virus isolation. On day 21, after collecting blood samples all animals were challenged intranasally with 3.0 log10 TCID50 of HK/14 wild-type virus. NFW were collected on days 1, 3, 5 and 7 post-challenge for challenge virus isolation. Terminal bleeds were collected on day 35.
Fig. 1.
Study designs for FER-16–09 (A) and FER-17–01 (B). In FER-16–09 experiment, 7 groups of 3 ferrets were vaccinated with 105 EID50 in 500 uL with LAIVs (HK 6:2, HK 5:3, Sw SW 6:2, Sw SW 5:3, Tx TX 6:2, Tx TX 5:3) or PBS (mock) intranasally under anesthesia on day 0. On day 21 post-infection (DPI), ferrets were intranasally challenged with 103 TCID50 in 500uL of A/Hong Kong/4801/2014 (H3N2) virus. In the FER-17–01 experiment, 3 groups of 8 ferrets were vaccinated with 107 EID50 in 500 uL of LAIV (Tx 6:2 or Tx 5:3) or PBS (mock) intranasally under anesthesia on day 0. At 30 DPI, ferrets were intranasally challenged with 107 TCID50 in 1000uL of A/Brisbane/10/2007 (H3N2) virus. Animals were bled prior to LAIV vaccination, prior to wild-type virus challenge and at the end of the experiment. Nasal fluid washes were collected every second day after LAIV or wild-type virus infection as indicated. Nasal, laryngeal, tracheal, caudal left lung lobe and caudal right lung lobe tissue samples were collected from the four animals in each group on days 32 and 35.
In the second experiment (FER-17–01), groups of eight ferrets were immunized with TX LAIV 6:2 and TX LAIV 5:3 viruses at a dose of 7.0 log10 EID50 in a volume of 500 μL (Fig. 1B). NFW were collected on days 3 and 5 after inoculation. On day 30, blood samples were collected and ferrets were challenged with 7.0 log10 TCID50 wild-type A/Brisbane/ 10/2007 in a volume of 1 mL. NFW were collected on days 1, 2, 3 and 4 after challenge. Four animals in each group were euthanized on 32 or 35 days (2 and 5 days after challenge) and respiratory tract tissues were collected for further analyses.
2.3. LAIV vaccination and challenge virus inoculation procedure of ferrets
Prior to dosing, ferrets were anaesthetized by intramuscular administration of 12.5 mg/kg ketamine and 2.5 mg/kg ilium xylazil-20 in a 1:1 [v/v] mixture (Troy Laboratories). Viruses were administered by slowly dropping 0.5 or 1.0 mL Phosphate buffered saline (PBS)-diluted virus into one nostril of an anaesthetized ferret in supine position, then after approximately twenty seconds (allowing time for the inoculum move into the nasal passage) the animals were rolled on their sides. Anaesthetized ferrets were monitored and wrapped in burlap until recovery.
2.4. Nasal fluid wash collection
Ferrets were sedated for NFW collection by intramuscular administration of 5 mg/kg ilium xylazil-20 (Troy Laboratories). NFW was collected by passing 1 mL of PBS (Sigma) supplemented with 1% bovine serum albumin (BSA, Sigma) and 50 U/mL penicillin, 50 μg/mL streptomycin (SAFC Biosciences) through the nostrils of sedated ferrets. Washes were immediately mixed, 140 μL was aliquoted into 560 μL buffer AVL™ containing carrier RNA, (Qiagen, Qiamp Viral RNA Mini Kit) and the remaining NFW was placed on ice and stored at −80 °C. Viral RNA (vRNA) was extracted daily after sampling.
2.5. Tissue sample collection
For euthanasia, ferrets were anaesthetized as above and injected with 5 mL lethabarb via the intracardiac or intrahepatic routes. Euthanized ferrets were dissected aseptically. The caudal left and caudal right lung lobes were resected, halved by longitudinal sectioning and frozen at −80 °C. The larynx with upper third of the trachea were resected by longitudinal sectioning and frozen at −80 °C. For nasal tissue collection, scalps were removed from the facial skull. Mandible was removed and maxilla was resected by a coronal cut between fourth premolar and molar. Maxilla with nasal tissues was halved by midsagittal cut and frozen at −80 °C.
2.6. Serum collection
Ferrets were bled by jugular venepuncture under anesthesia. No more than 1 mL of blood was collected into serum clot activator tubes (VACUETTE® Z). Terminal bleeds were collected by cardiac puncture under anesthesia, collecting approximately 9 mL. Blood tubes were centrifuged at 1811 ×g for 10 min to separate the serum, which was aliquoted and stored at −20 °C.
2.7. Serum HI assay
Sera were treated with receptor destroying enzyme (RDE (II) SEIKEN) (1:4 v/v ratio) and adsorbed with turkey red blood cells (RBC) to remove non-specific inhibitors and quantitated against four HA units of influenza A viruses to be tested (Isakova-Sivak et al., 2011).
2.8. Extraction and quantification of NFW vRNA by RT-PCR
vRNA was extracted from NFW using the QIAamp Viral RNA Mini Kit according to the manufacturer’s instructions (Qiagen, Qiamp Viral RNA Mini Kit). vRNA was eluted in 60 uL AVE buffer and stored at −80 °C. vRNA was quantified from 4 μL vRNA by RT-PCR using the SensiFAST™ Probe Lo-ROX One-Step Kit (BIOLINE) according to manufacturer’s protocol with the CDC influenza virus real-time RT-PCR influenza A(H1/H3/H1 pdm09) subtyping panel (RUO) (LOT: 12–0134) (influenza A Matrix primers and probes), obtained through the International Reagent Resource, Influenza Division, WHO Collaborating Centre for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA). The RT-PCR consisted of 1 cycle of 45 °C for 10 min and 95 °C for 2 min and 45 cycles of 95 °C for 5 s, 60 °C for 30 s using the 7500 Fast Real-Time System (Applied Biosystems), and 7500 Fast System SDS Software version 1.4.0. The threshold was automatically set and Ct determined. Samples, no-template controls and positive plasmid controls were included in each real time RT-PCR assay. The detection limit was 10 copies of M gene as previously determined (Kendal and Skehel, 1982). Copy number was calculated using a plasmid standard curve.
2.9. Virus quantification in NFW by ViroSpot (VS) assay
Infectious virus in NFW samples was quantified by ViroSpot assay with modifications (Carolan et al., 2014). Briefly, NFW samples were serially diluted 10-fold in Maintenance Medium (MM): Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 1% Glutamax (Gibco), 1% minimum essential medium (MEM) non-essential amino acids solution (Gibco), 0.5% sodium bicarbonate solution, 0.2M HEPES, 183 U/mL penicillin/streptomycin (Gibco), and 1% amphotericin B (Gibco). Then 100 uL of the sample were transferred to Madin-Darby Canine Kidney cells with sialic acid overexpression (MDCK-SIATI) monolayer in 96 well plates and incubated at 35 °C, 5% CO2, 90 min. Next, an equal volume of MM supplemented with 3.2% carboxymethylcellulose (CMC, Sigma) and 500 AU/mL TPCK-treated trypsin was added to the virus-inoculated plates and incubated for an additional 16 h at 35 °C, 5% CO2. Supernatant was aspirated and the monolayer fixed with 10% (v/v) formalin (Sigma) solution in PBS for 10 min at room temperature (RT). Wells were washed once with 200 μL PBS and 100 μL 0.5% Triton X-100 was added to each well and incubated for 10 mins at RT. Wells were washed 3 times with 0.05% (v/v) Tween-20 in PBS (PBS-T) and stained with anti-influenza A nucleoprotein antibody (clones A1, A3 Blend; Merck) at 1:5000 dilution in 2% (w/v) instant skim milk in PBS-T for 60 mins at RT. Next, wells were washed 3 times with PBS-T and stained with goat anti-mouse IgG (H+L)-HRP conjugate (BIORAD) at 1:5000 dilution in 2% instant skim milk powder in PBS for 60 mins at RT. The plates were washed 3 times with PBS-T then virus spots were visualized by addition of 50 uL KPL TrueBlue™ peroxidase substrate (SeraCare) incubated for 10 min in the dark. The reaction was stopped by the addition of 200 ul dH2O, then plates were washed two more times with dH2O. Plates were dried at RT, and spots counted using a CTL-immunospot S6 Macro Analyzer with CTL Switchboard 2.6.0 (Cellular Technology Limited, USA). ViroSpots were calculated as the geometric mean per 1 mL NFW volume. Preliminary studies yielded similar results of the LAIV stocks quantification at 33 °C and 35 °C temperatures of incubation in ViroSpot assay (Supplementary fig. 1).
2.10. Virospot microneutralization (MN) assay
Ferret serum antibody titres were determined using the virospot MN assay (van Baalen et al., 2017). Briefly, RDE-treated serum samples were diluted 2-fold in MM and incubated 1:1 with 500 to 1000 virospot units of the LAIV viruses for 60 mins at 35 °C. The serum-virus mixture was then transferred to MDCK-SIAT-I monolayer in 96 well plates and incubated for 90 mins at 35 °C. An equal volume of MM with 3.2% CMC (final 1.6% CMC) and 500 AU/mL TPCK trypsin (Sigma) was added to the plates and incubated for an additional 16 h at 35 °C, 5% CO2. Plates were stained as above. The well with the highest serum dilution resulting in up to 50% neutralization was considered the neutralizing antibody titre.
2.11. Homogenization of respiratory tissues
All steps were performed in a 4–8 °C cold room or on ice. Tissue samples were weighed and placed on ice. MM was added to tissues to achieve 20% w/v tissue concentration. Larynx and lung lobes tissues were homogenized in gentleMACS™ M tubes (Miltenyi Biotec) using a gentleMACS Dissociator instrument (Miltenyi Biotec). Turbinates were dissociated by rotor-stator homogenization Polytron PT 2500E (ThermoFisher Scientific). Samples were clarified by centrifugation by 3000 g for 10 mins at 4 °C and supernatants were collected.
2.12. Respiratory tissue infectious virus titration
Infectious virus was quantified by 50% tissue culture infective dose (TCID50) assays as described (van Baalen et al., 2017). Tissue supernatants were titrated on MDCK-SIAT-I cell monolayers and read by hemagglutination of turkey RBC.
2.13. Statistical analysis and graphical data representation
Statisticals analysis and graphical plotting were performed with GraphPad Prism 6 software. Multiple t-test with Holm-Sidak multiple correction method (alpha 0.05) was utilized to compare data from groups of ferrets.
3. Results
3.1. Phenotypic characterization of A(H3N2) LAIVs with 6:2 and 5:3 genome compositions
We generated a panel of A(H3N2) LAIV 6:2 and 5:3 candidates from three wild-type A(H3N2) viruses for three consecutive influenza seasons (TX/12, SW/13 and HK/14). The in vitro properties of a LAIV candidate are cold adaptation (ca) and temperature sensitivity (ts). Both LAIVs 6:2 and LAIVs 5:3 displayed the ca and ts phenotypes of MDV A/Leningrad/134/17/57 (H2N2), demonstrating that the wild-type NP gene had no impact on these phenotypic characteristics of the reassortant vaccine viruses (Table 1).
3.2. A(H3N2) LAIVs 5:3 are safe and induce similar antibody immune responses as A(H3N2) LAIVs 6:2 in ferrets
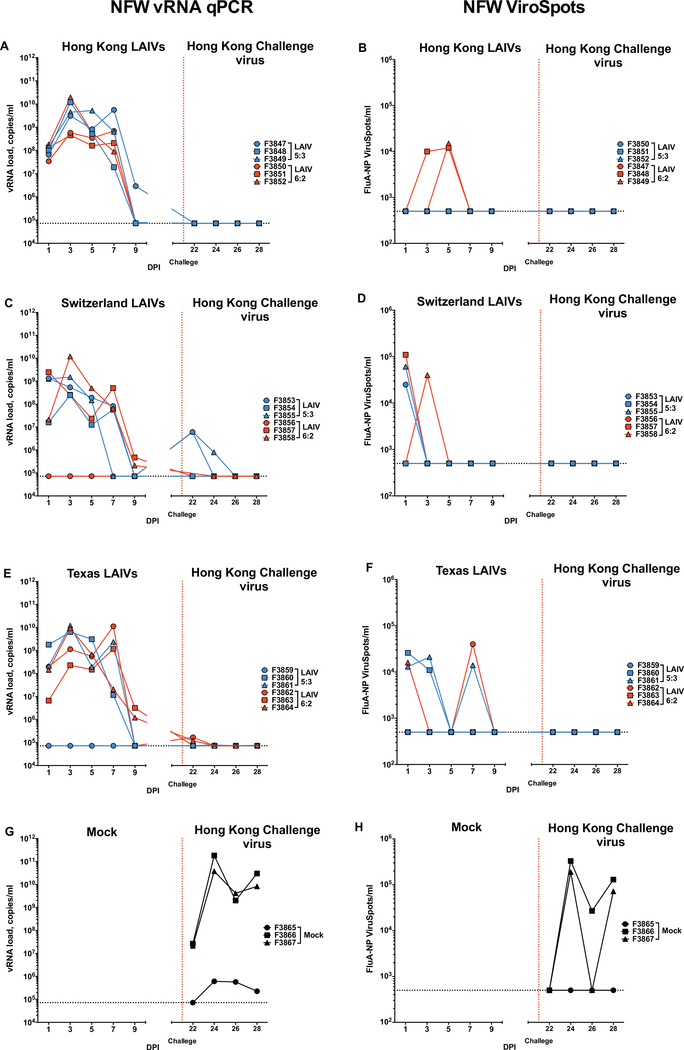
The first ferret experiment aimed to assess the impact of the wild-type NP on the safety, replication and immunogenicity of LAIVs in ferrets (Fig. 1A). Following vaccination, the majority of animals shed virus, with vRNA detected in NFW samples from day 1 to day 9 postvaccination, peaking on days 3 to 7 (Figs. 2A, C, E). Both LAIVs 6:2 and 5:3 viruses were shed in similar amounts in the upper respiratory tract. However, ferrets vaccinated with SW LAIV 6:2 and TX LAIV 6:2 shed virus for 1 day longer than ferrets vaccinated with the corresponding LAIVs 5:3 vaccine strains (Figs. 2C, E, day 7 and day 9). Infectious virus was detected only in NFW samples that had a high amount of vRNA (Fig. 2B, D, F). One ferret each from the SW LAIV 6:2 (F3656) and the TX LAIV 5:3 (F3859) groups did not shed virus as measured by vRNA or infectious virus (Fig. 2, Supplementary Table 1). These ferrets remained seronegative and were excluded from further immunological analysis (Supplementary Table 1).
Fig. 2.
LAIVs and challenge viruses shedding in ferret NFW samples in experiment FER-16–09. Animals received intranasally 105 EID50/500 uL of either LAIVs. 21 days after LAIV administration, animal were challenged with 103 TCID50 in 500uL of A/Hong Kong/4801/2014 (H3N2) virus (vertical red dotted line). The Mx gene copies was measured in NFW vRNA samples by RT-PCR (A, C, E, G) and the number of FluA-NP+ spots were estimated by ViroSpot assay (B, D, F, H). LAIVs 5:3, LAIVs 6:2 and Mock vaccinated groups data are visualized in blue, red and black colors respectively. Dotted line denotes assay limit of detection. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The majority of animals did not develop clinical signs following vaccination LAIVs with 6:2 and 5:3 genome compositions; 80% (12/15) ferrets maintained or gained weight and 93% (14/15) did not develop fever (Supplementary Fig. 2). One ferret vaccinated with HK LAIV 5:3 (F3849, female) and one ferret vaccinated with SW LAIV 6:2 (F3857, female) lost more than 10% body weight, thus were given enhanced diets (Supplementary Table 2). Both ferrets were found to be on heat (F3849 detected on day 32 and F3657 detected on day 15). One ferret from the TX LAIV 5:3 group (F3859, female) lost less than 10% body weight over the study. This animal did not shed LAIV virus after vaccination (Fig. 2E).
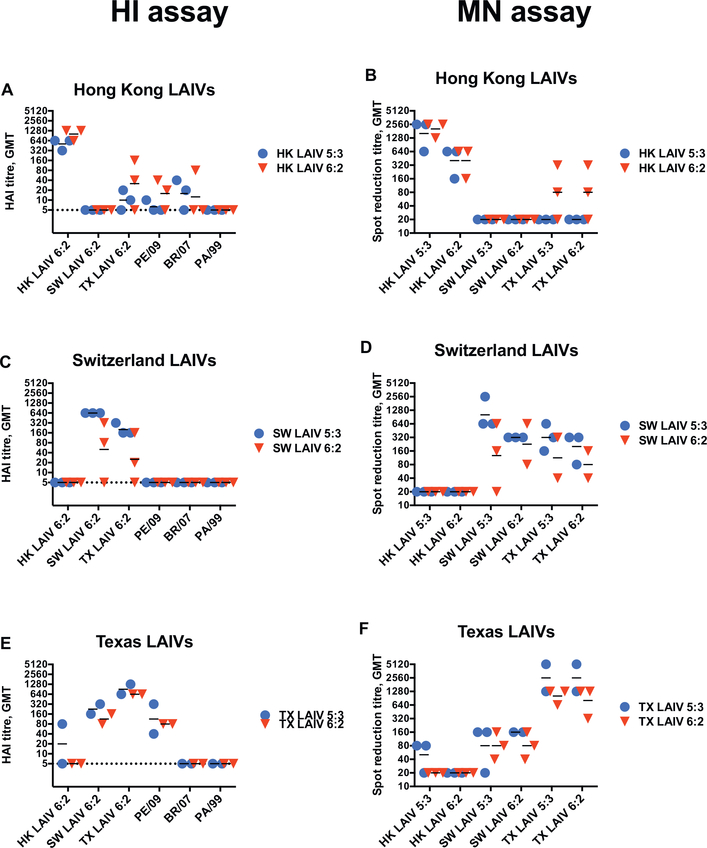
The immunogenicity of H3N2 LAIVs was measured by HI and MN assays on day 21 after LAIV administration. Assessment of sera by HI assays showed similar patterns, with high homologous antibody titres and cross-reactivity induced by vaccination with SW and TX LAIVs (Figs. 3A, C, E). There were no differences in the patterns of HI titres between animals vaccinated with LAIV 5:3 and LAIV 6:2 variants for any of the vaccines. We assessed whether the LAIVs induced cross-reactive antibodies to older A(H3N2) strains: A/Perth/16/2009, A/Brisbane/10/2007, A/Panama/2007/99. TX LAIVs generated cross-reactive HI antibodies against PE/09 virus, but HK and SW LAIVs did not. None of the animals developed cross-reactive antibodies to BR/07 or PA/99.
Fig. 3.
Hemagglutination inhibition antibody and virus neutralization titres in post-LAIV ferret serum samples in study FER-16–09. Serum samples were collected 21 days after HK LAIVs, SW LAIVs and TX LAIVs vaccination and HI titers (A, C, E respectively) or neutralizing (B, D, F respectively) titers were estimated. All Pre-LAIVs serum samples (−14 DPI) were HI-negative. HI titre in Mock samples were below the limit of detection (1/5). Serum samples from ferrets F3856 (SW LAIV 6:2) and F3859 (TX LAIV 5:3) were negative and excluded from analysis. LAIVs 5:3 and LAIVs 6:2 vaccinated groups data are visualized in blue and red colors respectively. Test antigens are shown on the x-axis. Dotted line denotes assay limit of detection. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Neutralizing serum antibodies were measured against the full panel of A(H3N2) LAIVs included in this study (Figs. 3B, D, F). Both LAIV 6:2 and LAIV 5:3 vaccination elicited high levels of neutralizing antibodies to the immunizing LAIV viruses with similar patterns of cross-reactivities to the HI antibodies. Thus, vaccination with HK LAIV 5:3 did not induce cross-reactive antibodies to the other virus strains (Fig. 3B). Vaccination with HK LAIV 6:2 induced antibodies that cross-neutralized TX LAIV, but not the SW LAIV viruses (Fig. 3B). Vaccination with SW and TX LAIVs induced cross-neutralizing antibodies to each other. At the same time, TX LAIV 5:3 vaccination induced low neutralizing titers against HK LAIV 5:3 virus (Figs. 3D, F).
Ferrets were challenged on day 21 with wild-type A/Hong Kong/ 4801/2014 virus, which has the most similar NP to all LAIVs with genome composition 5:3 and protection from challenge virus infection was assessed. Two of three ferrets from the mock-vaccinated group shed high amounts of challenge virus, whilst the third ferret shed a low amount of virus, as measured by vRNA (Fig. 2G). Infectious virus was detected in 2 of 3 animals (Fig. 2H). Neutralizing antibodies were detected on day 14 after HK/14 virus challenge only in the two mockvaccinated ferrets with a high challenge virus load. Interestingly, crossreactive antibodies to LAIV SW and TX were also induced (Supplementary Fig. 3). Shedding of challenge virus was not detected by qPCR in nasal washes of HK LAIV 6:2 and 5:3 vaccinated ferrets (Fig. 2A), though low levels of challenge virus vRNA were detected in NFWs of TX LAIV 6:2 and 5:3 and SW LAIV 5:3 vaccinated ferrets (Fig. 2C, E). This suggests that vaccination with HK LAIVs completely protect against homologous challenge and vaccination with SW LAIV 5:3 and TX LAIV 6:2 provide partial protection against challenge with the heterologous A (H3N2) A/Hong Kong/4801/2014 virus. Infectious virus was not detected in NFWs of any LAIV-vaccinated animals by ViroSpot assay on day 3 and 5 after challenge (Fig. 2B, D, F). Challenge with the wild-type HK virus did not change the distribution of virus-neutralizing antibody specificity in vaccinated ferrets by the end-point on day 35 (14 day post-challenge) (data not shown). Presumably due to the low dose of challenge virus, minimal clinical symptoms were observed in mockvaccinated animals (Supplementary Fig. 2); therefore these outcomes could not be considered for analysis of the protective efficacy of the LAIVs.
3.3. H3N2 LAIVs with 5:3 and 6:2 genome compositions show similar safety and immunogenicity profiles and protect animals from a high-dose of heterologous challenge virus
To compare the cross-protective efficacy of LAIVs 6:2 and LAIVs 5:3 more thoroughly, we vaccinated groups of eight ferrets with TX LAIV 6:2, TX LAIV 5:3 or PBS (mock), then challenged with a heterologous wild-type virus, A/Brisbane/10/2007 (Fig. 1B). Following vaccination with the TX LAIVs, cross-reactive antibodies to BR/07 were not detected by HI assay thus minimizing the contribution of antibodies to protection from challenge infection. However, there is high conservation between the NP protein sequences for TX/14 and BR/07, thus we hypothesized that there may be a difference in vaccine efficacy between 5:3 and 6:2 vaccinated groups.
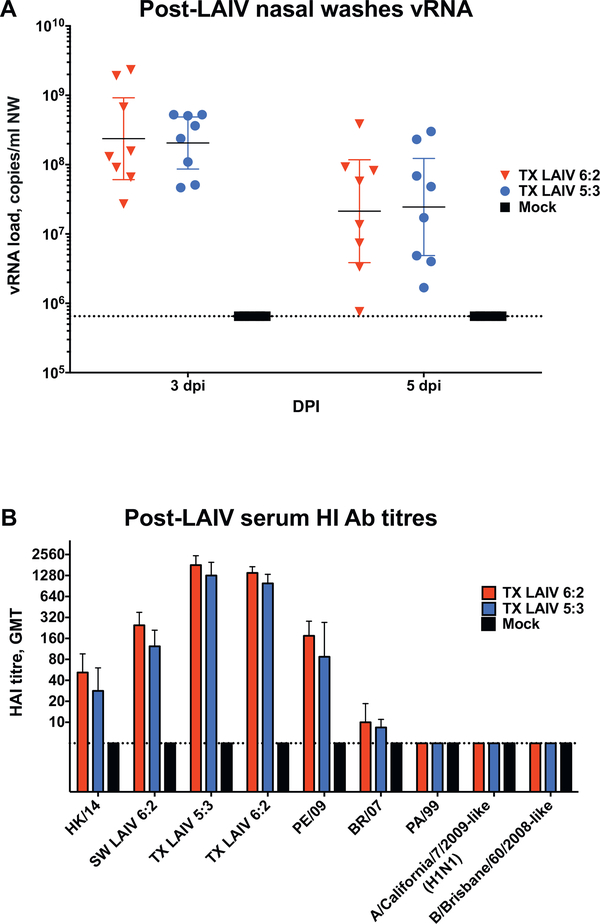
Ferrets were vaccinated with a higher vaccine dose, 7.0 Log10 EID50, instead of 5.0 Log10 EID50 in the first experiment. All immunized ferrets shed LAIV vRNA at high levels as measured in NFW samples collected on days 3 and 5 after vaccination (Fig. 4A). Importantly, virus shedding dynamics in the upper respiratory tract were similar for TX LAIV 6:2 and TX LAIV 5:3 strains. Despite the higher virus dose, none of the animals showed clinical signs of illness after vaccination: both TX LAIV 6:2 and TX LAIV 5:3 vaccinated ferrets increased body weight and no fever was detected (Supplementary Fig. 4). Young adult ferrets were used in the experiment and they matured and gained weight over the study duration (up to 25–50% body weight increases).
Fig. 4.
LAIV viruses upper respiratory shedding and Hemagglutination inhibition antibody titres after immunization with TX LAIVs in experiment FER-1701. Groups of 8 ferrets were inoculated intranasally with 107 EID50 in 500 uL of TX LAIV 6:2 (red), TX LAIV 5:3 (blue) or Mock (black) on day 0. (A) On selected days after LAIV virus inoculation vRNA levels were estimated in nasal washes by RT-PCR. Bars and whiskers denote geometric mean titre and 95% confidence interval respectively. (B) Serum samples were collected after 30 days post vaccination and assayed by HI assay. All Pre-LAIVs serum samples (3 days before vaccination) were negative. Test antigens are shown on the x-axis. Bars and whiskers denote geometric mean titre and 95% confidence interval respectively. Dotted line denotes assay limit of detection. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
High levels of homologous HI antibodies were detected on day 30 after a single vaccine dose (Fig. 4B). Both vaccine groups induced HI antibody that cross-reacted with SW/13, PE/09 and, to a lesser extent, HK/14 H3N2 viruses. There was no difference between TX LAIVs 6:2 or 5:3 variants in serum antibody response to any of the test antigens. Additionally, HK LAIVs induced serum antibodies were prone to react with distanced PA/99 and BR/07 H3N2 influenza viruses in HI assay.
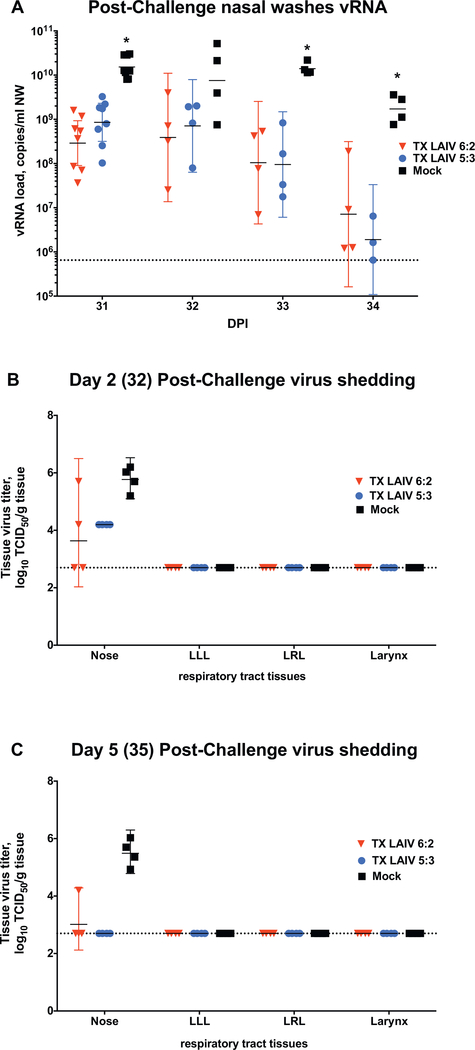
To study the cross-protective efficacy of the TX LAIV 6:2 and TX LAIV 5:3, all vaccinated animals were intranasally challenged with 7.0 Log10 TCID50 of a heterologous wild-type virus A/Brisbane/10/2007 (H3N2). A higher dose of challenge virus was used to ensure consistent respiratory infection. The challenge virus efficiently replicated in all mock-immunized animals: high copy numbers of vRNA were observed in NFWs from all ferrets (Fig. 5A) for at least 4 days after challenge. Both TX LAIV 6:2 and TX LAIV 5:3 vaccinated ferrets shed significantly lower vRNA than mock-immunized animals, and vRNA levels peaked between 1 and 3 days after challenge and mostly cleared by day 4 (Fig. 5A). Infectious challenge virus was detected only in the nasal tissues of mock-immunized ferrets on days 32 and 35, indicating the virus did not migrate down to the lower respiratory tract (Figs. 5B, C). In both LAIV 6:2 and 5:3-immunized groups much lower titers were detected in nasal tissues on day 32 (2 days after challenge), compared to the mock-vaccinated group. On day 32, all TX LAIV 5:3 vaccinated ferrets and only 2 of 4 TX LAIV 6:2 vaccinated ferrets had detectable infectious virus in the nasal tissues. By day 35 (5 days post challenge), infectious virus was not detected in TX LAIV 5:3 group, though one of four ferrets in TX LAIV 6:2 group still shed infectious virus (Fig. 5C). Therefore, these results suggest that the TX LAIV 5:3 vaccine is as efficacious as the TX LAIV 6:2 at inducing cross-protective immunity against a heterologous A(H3N2) virus. Monitoring of clinical signs upon challenge did not show any significant weight loss or temperature rise in mock-immunized animals, nor in either vaccine group (Supplementary Fig. 4).
Fig. 5.
Shedding of wild-type virus in respiratory tract after challenge of TX LAIVs immunized ferrets in experiment FER-17–01. Groups of 8 ferrets were inoculated intranasally with 107 EID50 in 500 uL of TX LAIV 6:2 (red), TX LAIV 5:3 (blue) or Mock (black) on day 0. After 30 days, ferrets were challenged with 107 TCID50 in 1000 uL of w/t A/Brisbane/10/2007 (H3N2) strain. On selected days after challenge virus inoculation (A) vRNA levels were estimated in nasal washes by RT-PCR. On days 32 (B) and 35 (C) four animals from each group were euthanized and respiratory tract tissue samples were collected for virus titre estimation. Bars and whiskers denote geometric mean titre and 95% confidence interval respectively. Asterisks denotes significant differences in values of LAIVs vs Mock vaccinated groups (Multiple t-test with Holm-Sidak multiple correction method, alpha 0,05). Dotted line denotes assay limit of detection. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Influenza is a major pathogen causing severe respiratory disease, and the main weapon against this infection is vaccination. Despite the proven clinical efficacy of currently licensed vaccines in preventing seasonal influenza in various age groups, the improvement of immunogenicity and effectiveness of these vaccines is of great interest.
Currently, the reassortant viruses used for live attenuated influenza vaccines are prepared on the backbone of the cold-adapted master donor viruses A/Leningrad/134/17/57 in Russia and A/Ann Arbor/6/ 60 in the USA. An influenza nucleoprotein, a relatively conserved protein, which is one of the major targets for CTL immunity, yet during the past 60 years, there has been significant changes in the NP of circulated influenza A virus strains. In addition, there have been years of very low vaccine efficacy against A(H3N2) viruses as the strain undergoes major antigenic change. It is worthwhile noting that the comparative evaluation of cell-mediated immunogenicity of vaccine viruses with 5:3 and 6:2 genome compositions in animal models is not easy because special humanized animals are required to detect the differences in human epitope-specific T-cell responses upon vaccination. Nevertheless, to enter a clinical trial a thorough characterization of the new vaccines in pre-clinical trials is required.
Ferrets are the most suitable animals to study A(H3N2) influenza infection in vivo. There are numerous approaches to investigate influenza vaccine safety and challenge protection in ferrets. Traditionally, for LAIV ferret studies a human dose of 7.0 Log10 of EID50 is used for vaccination (Kendal et al., 1982). In our trials, we compared 6:2 and 5:3 H3N2 LAIVs administered either at low dose (5.0 Log10) or at high dose (7.0 Log10). In both cases LAIVs 5:3 replicated in the upper respiratory tract in ferrets, comparable with 6:2 LAIVs. As measured by vRNA qPCR, the HK, SW and TX LAIV 5:3 candidates, at both dose levels, had similar infectivity and viral load dynamics in the upper respiratory tract of ferrets as their LAIV 6:2 counterparts. Both LAIV 6:2 and LAIV 5:3 variants were attenuated when administered at either dose. Although some animals lost weight after vaccination or virus infection, there was no correlation with vaccine strain or challenge group. Mature ferrets can lose up to 50% of their body weight during breeding season (Matsuoka et al., 2009). The FER-16–09 trial was performed in ferret mating season (spring), which may have contributed to the observed weight loss (Buckland et al., 2013).
Immunogenicity is a good predictor of disease protection in ferrets (Fox et al., 2014). We showed that LAIVs 5:3 induce a potent serum antibody response in ferrets. Introduction of the wild-type NP gene into Len/17-based LAIV did not influence antibody-mediated vaccine immunogenicity. Both LAIVs 6:2 and LAIVs 5:3 variants induced similar titers of virus-neutralizing and HI antibodies. Moreover, despite each LAIV inducing clade-specific serum antibodies, TX and SW LAIVs inoculation were also able to stimulate cross-reactive neutralizing antibodies to each other.
Importantly, vaccination with HK, SW or TX LAIVs prevented infection with wild type A/Hong Kong/4801/2014 (H3N2) challenge virus, as measured by shedding of the challenge virus in the upper respiratory tract. In the FER-16–09 trial, we utilized a low dose of LAIVs for priming (5 Log10 EID50) and a low-dose challenge virus (3 Log10 EID50) strategy. The challenge viruses used in this study were seasonal A(H3N2) viruses that induced mild clinical disease in ferrets and humans. There was no superiority of LAIV 6:2 or 5:3 variants in preventing challenge virus infection. In the second study, FER-17–01, where ferrets were challenged with an antigenically distant wild-type virus (A/Brisbane/10/2007); both the virus load and shedding duration were reduced dramatically in TX LAIVs 6:2 and TX LAIVs 5:3 primed ferrets.
As the influenza nucleoprotein is one of the major targets of the T cell response, we proposed that the LAIV 5:3 variant might provide an advantage in protection from wild-type influenza virus infection. However, we did not detect a difference in the efficacy against challenge virus infection with the LAIV 5:3, bearing a wild-type NP gene compared to the LAIV 6:2 in ferrets. Unfortunately, little is known about ferret T-cell immunobiology and in particular, the role of T cells in infection (Vidaña et al., 2014). This is due to the methodological limitations and the absence of detailed knowledge about MHC genes polymorphisms in ferrets. Some progress in ferrets T-cell studies was made by using human leucocyte anti-CD-antibodies (Dipiazza et al., 2016; Rutigliano et al., 2008). Recently, Dr. Reber and colleagues studied the specificity of T cells using arrays of influenza protein peptides in ferrets and demonstrated that the high cross-reactive CD4+ and CD8+ T cells were stimulated in influenza infected or vaccinated ferrets (Reber et al., 2018). Moreover, it has been shown that NP-specific T cells were the most dominated upon pH1N1 infection (about 30% of all influenza specificities of either CD4+ or CD8+ T cells) but not in the case of H3N2 infection (about 13% and 3.6% of NP-specific of all influenza specificities CD4+ and C8+ T cells, respectively). These results support the idea that in the case of H3N2 infection the NP-specific T cells are not dominating over the other specificities of T cells induced upon influenza infection. Thus, impact of the NP-specific T cells in overall T-cell response in the ferret H3N2 influenza model might be not pronounced enough to detect differences in the efficacy between LAIVs 6:2 and 5:3 variants. Also, there are cross-reactive T cells after influenza B virus infection and are likely to contribute to protection from infection with either lineage (Laurie et al., 2018), however, further work is required to understand the immunodominance of influenza proteins in ferrets. The presented data supports further investigation of T-cell reactivity and protection of LAIVs with wild-type NP in.
Using a ferret model of human influenza we have shown that reverse genetics engineered H3N2 LAIVs with wild-type nucleoproteins (5:3 genome composition) preserved in vitro growth phenotypes of Len/17 MDV. Both H3N2 LAIVs with Len/17 or wild-type nucleoproteins had similar clinical outcomes and replication dynamics in the respiratory tract of adult ferrets and both induced a pronounced humoral immune response in ferrets. Ferrets primed by H3N2 LAIVs with Len/17 or wild-type nucleoproteins were protected from challenge infection by heterologous wild type virus of the same subtype. Further studies in a humanized animal model or clinical trials in humans are needed to investigate the benefit of including a wild-type NP in LAIV strains.
Supplementary Material
Acknowledgments
We sincerely appreciate all the staff at the WHO Collaborating Centre for Reference and Research on Influenza and the Department of Microbiology & Immunology, The University of Melbourne, both located at the Peter Doherty Institute for Infection and Immunity in Melbourne for their assistance during this study.
Funding
Generation and in vitro characterization of H3N2 LAIVs was funded by Russian Scientific Foundation Grant № 14-15-00034. The Melbourne WHO Collaborating Centre for Reference and Research is supported by the Australian Government Department of Health (Agreement id. 44GFBE88).
Footnotes
Conflicts of interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2018.06.019
References
- Aleksandrova GI, 1977. Use of the genetic recombination method for obtaining vaccinal strains of the influenza virus. Vopr. Virusol 387–395. [PubMed] [Google Scholar]
- Buckland MD, Hall L, Mowlem A, 2013. A Guide to Laboratory Animal Technology. Elsevier Science. [Google Scholar]
- Carolan LA, Butler J, Rockman S, Guarnaccia T, Hurt AC, Reading P, Kelso A, Barr I, Laurie KL, 2014. TaqMan real time RT-PCR assays for detecting ferret innate and adaptive immune responses. J. Virol. Methods 205, 38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipiazza A, Richards K, Batarse F, Lockard L, Zeng H, García-Sastre A, Albrecht RA, Sant AJ, 2016. Flow cytometric and cytokine ELISpot approaches to characterize the cell-mediated immune response in ferrets following influenza virus infection. J. Virol 90, 7991–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Bell J, Broome R, 2014. Growth and Reproduction, Biology and Diseases of the Ferret. John Wiley & Sons, Inc., pp. 187–209. [Google Scholar]
- Glatman-Freedman A, Drori Y, Beni SA, Friedman N, Pando R, Sefty H, Tal I, McCauley J, Rahav G, Keller N, Shohat T, Mendelson E, Hindiyeh M, Mandelboim M, 2017. Genetic divergence of Influenza A(H3N2) amino acid substitutions mark the beginning of the 2016–2017 winter season in Israel. J. Clin. Virol 93, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant E, Wu C, Chan KF, Eckle S, Bharadwaj M, Zou QM, Kedzierska K, Chen W, 2013. Nucleoprotein of influenza A virus is a major target of immunodominant CD8+ T-cell responses. Immunol. Cell Biol 91, 184–194. [DOI] [PubMed] [Google Scholar]
- Hay AJ, Gregory V, Douglas AR, Lin YP, 2001. The evolution of human influenza viruses. Phil. Transac. R. Soc. Lond. B 356, 1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft DF, Lottenbach KR, Blazevic A, Turan A, Blevins TP, Pacatte TP, Yu Y, Mitchell MC, Hoft SG, Belshe RB, 2017. Comparisons of the humoral and cellular immune responses induced by live attenuated influenza vaccine and inactivated influenza vaccine in adults. Clin. Vaccine Immunol 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakova-Sivak I, Chen LM, Matsuoka Y, Voeten JT, Kiseleva I, Heldens JG, den Bosch H, Klimov A, Rudenko L, Cox NJ, Donis RO, 2011. Genetic bases of the temperature-sensitive phenotype of a master donor virus used in live attenuated influenza vaccines: A/Leningrad/134/17/57 (H2N2). Virology 412, 297–305. [DOI] [PubMed] [Google Scholar]
- Isakova-Sivak I, Korenkov D, Rudenko L, 2016. Reassortant viruses for influenza vaccines: is it time to reconsider their genome structures? Expert Rev. Vaccines 15, 565–567. [DOI] [PubMed] [Google Scholar]
- Isakova-Sivak I, Korenkov D, Smolonogina T, Tretiak T, Donina S, Rekstin A, Naykhin A, Shcherbik S, Pearce N, Chen LM, Bousse T, Rudenko L, 2017. Comparative studies of infectivity, immunogenicity and cross-protective efficacy of live attenuated influenza vaccines containing nucleoprotein from cold-adapted or wild-type influenza virus in a mouse model. Virology 500, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira Da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Azziz-Baumgartner E, Cheng P-Y, Dawood F, Foppa I, Olsen S, Haber M, Jeffers C, Macintyre CR, Newall AT, Wood JG, Kundi M, Popow-Kraupp T, Ahmed M, Rahman M, Marinho F, Sotomayor Proschle CV, Vergara Mallegas N, Luzhao F, Sa L, Barbosa-Ramírez J, Sanchez DM, Gomez LA, Vargas XB, Acosta Herrera A, Llanés MJ, Fischer TK, Krause TG, Mølbak K, Nielsen J, Trebbien R, Bruno A, Ojeda J, Ramos H, van der Heiden M, del Carmen Castillo Signor L, Serrano CE, Bhardwaj R, Chadha M, Narayan V, Kosen S, Bromberg M, Glatman-Freedman A, Kaufman Z, Arima Y, Oishi K, Chaves S, Nyawanda B, Al-Jarallah RA, Kuri-Morales PA, Matus CR, Corona MEJ, Burmaa A, Darmaa O, Obtel M, Cherkaoui I, van den Wijngaard CC, van der Hoek W, Baker M, Bandaranayake D, Bissielo A, Huang S, Lopez L, Newbern C, Flem E, Grøneng GM, Hauge S, de Cosío FG, de Moltó Y, Castillo LM, Cabello MA, von Horoch M, Medina Osis J, Machado A, Nunes B, Rodrigues AP, Rodrigues E, Calomfirescu C, Lupulescu E, Popescu R, Popovici O, Bogdanovic D, Kostic M, Lazarevic K, Milosevic Z, Tiodorovic B, Chen M, Cutter J, Lee V, Lin R, Ma S, Cohen AL, Treurnicht F, Kim WJ, Delgado-Sanz C, de Mateo Ontañón S, Larrauri A, León IL, Vallejo F, Born R, Junker C, Koch D, Chuang J-H, Huang W-T, Kuo H-W, Tsai Y-C, Bundhamcharoen K, Chittaganpitch M, Green HK, Pebody R, Goñi N, Chiparelli H, Brammer L, Mustaquim D, 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendal APP, Skehel JJ, 1982. Hemagglutination inhibition In: Kendal APP, Skehel JJ (Eds.), Concepts and Procedures for Laboratory-based Influenza Surveillance. Centers for Disease Control and Prevention, Atlanta, pp. 17–35. [Google Scholar]
- Kendal A, Pereira M, Skehel J, 1982. Concepts and Procedures for Laboratory-Based Influenza Surveillance. Centers for Disease Control, Atlanta. [Google Scholar]
- Korenkov D, Nguyen THO, Isakova-Sivak I, Smolonogina T, Brown LE, Kedzierska K, Rudenko L, 2018. Live attenuated influenza vaccines engineered to express the nucleoprotein of a recent isolate stimulate human influenza CD8+ T cells more relevant to current infections. Hum. Vaccines Immunother 14, 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie KL, Guarnaccia TA, Carolan LA, Yan AWC, Aban M, Petrie S, Cao P, Heffernan JM, Mcvernon J, Mosse J, Kelso A, McCaw JM, Barr IG, 2015. Interval between infections and viral hierarchy are determinants of viral interference following influenza virus infection in a ferret model. J. Infect. Dis 212, 1701–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie KL, Horman W, Carolan LA, Chan KF, Layton D, Bean A, Vijaykrishna D, Reading PC, McCaw JM, Barr IG, 2018. Evidence for viral interference and cross-reactive protective immunity between influenza B virus lineages. J. Infect. Dis 217, 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machkovech HM, Bedford T, Suchard MA, Bloom JD, 2015. Positive selection in CD8+ T-cell epitopes of influenza virus nucleoprotein revealed by a comparative analysis of human and swine viral lineages. J. Virol 89, 11275–11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Lamirande EW, Subbarao K, 2009. The ferret model for influenza. Curr. Protoc. Microbiol. 13, 1–29 (Chapter 15, Unit 15G 12). [DOI] [PubMed] [Google Scholar]
- Mohn KGI, Zhou F, Brokstad KA, Sridhar S, Cox RJ, 2017. Boosting of crossreactive and protection-associated T cells in children after live attenuated influenza vaccination. J. Infect. Dis 215, 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskophidis D, Kioussis D, 1998. Contribution ofvirus-specific CD8(+) cytotoxic T cells to virus clearance or pathologic manifestations of influenza virus infection in a T cell receptor transgenic mouse model. J. Exp. Med 188, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasaraju T, Sim MK, Ng HH, Phoon MC, Shanker N, Lal SK, Chow VT, 2009. Adaptation of human influenza H3N2 virus in a mouse pneumonitis model: insights into viral virulence, tissue tropism and host pathogenesis. Microbes Infect. 11, 2–11. [DOI] [PubMed] [Google Scholar]
- Quinones-Parra S, Grant E, Loh L, Nguyen TH, Campbell KA, Tong SY, Miller A, Doherty PC, Vijaykrishna D, Rossjohn J, Gras S, Kedzierska K, 2014. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc. Natl. Acad. Sci. U. S. A 111, 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber AJ, Music N, Kim JH, Gansebom S, Chen J, York I, 2018. Extensive T cell cross-reactivity between diverse seasonal influenza strains in the ferret model. Sci. Rep 8, 6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekstin A, Isakova-Sivak I, Petukhova G, Korenkov D, Losev I, Smolonogina T, Tretiak T, Donina S, Shcherbik S, Bousse T, Rudenko L, 2017. Immunogenicity and cross protection in mice afforded by pandemic H1N1 live attenuated influenza vaccine containing wild-type nucleoprotein. Biomed. Res. Int 2017, 9359276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutigliano JA, Doherty PC, Franks J, Morris MY, Reynolds C, Thomas PG, 2008. Screening monoclonal antibodies for cross-reactivity in the ferret model of influenza infection. J. Immunol. Methods 336, 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz-Cherry S, 2015. Role of NK cells in influenza infection. Curr. Top. Microbiol.Immunol 386, 109–120. [DOI] [PubMed] [Google Scholar]
- Swain SL, Dutton RW, Woodland DL, 2004. T cell responses to influenza virus infection: effector and memory cells. Viral Immunol. 17, 197–209. [DOI] [PubMed] [Google Scholar]
- van Baalen CA, Jeeninga RE, Penders GH, van Gent B, van Beek R, Koopmans MP, Rimmelzwaan GF, 2017. ViroSpot microneutralization assay for antigenic characterization of human influenza viruses. Vaccine 35, 46–52. [DOI] [PubMed] [Google Scholar]
- van Riet E, Ainai A, Suzuki T, Hasegawa H, 2012. Mucosal IgA responses in influenza virus infections; thoughts for vaccine design. Vaccine 30, 5893–5900. [DOI] [PubMed] [Google Scholar]
- Vidaña B, Majó N, Pérez M, Montoya M, Martorell J, Martínez J, 2014. Immune system cells in healthy ferrets: an immunohistochemical study. Vet. Pathol. Online 51, 775–786. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chua BY, Ramos JV, Parra SM, Fairmaid E, Brown LE, Jackson DC, Kedzierska K, 2015. Establishment of functional influenza virus-specific CD8(+) T cell memory pools after intramuscular immunization. Vaccine 33, 5148–5154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.