Abstract
The avian pineal gland, an independent circadian oscillator, receives external photic cues and translates them for the rhythmical synthesis of melatonin. Our previous study found that monochromatic green light could increase the secretion of melatonin and expression of CLOCK and BMAL1 in chick pinealocytes. This study further investigated the role of BMAL1 and CLOCK in monochromatic green light-induced melatonin secretion in chick pinealocytes using siRNAs interference and overexpression techniques. The results showed that si-BMAL1 destroyed the circadian rhythms of AANAT and melatonin, along with the disruption of the expression of all the seven clock genes, except CRY1. Furthermore, overexpression of BMAL1 also disturbed the circadian rhythms of AANAT and melatonin, in addition to causing arrhythmic expression of BMAL1 and CRY1/2, but had no effect on the circadian rhythms of CLOCK, BMAL2 and PER2/3. The knockdown or overexpression of CLOCK had no impact on the circadian rhythms of AANAT, melatonin, BMAL1 and PER2, but it significantly deregulated the circadian rhythms of CLOCK, BMAL2, CRY1/2 and PER3. These results suggested that BMAL1 rather than CLOCK plays a critical role in the regulation of monochromatic green light-induced melatonin rhythm synthesis in chicken pinealocytes. Moreover, both knockdown and overexpression of BMAL1 could change the expression levels of CRY2, it indicated CRY2 may be involved in the BMAL1 pathway by modulating the circadian rhythms of AANAT and melatonin.
Keywords: pinealocyte, circadian rhythm, melatonin, siRNA interference, monochromatic light
Introduction
Avian circadian system is a complex of mutually coupled pacemakers residing in the pineal gland, the retina and the hypothalamic suprachiasmatic nucleus (SCN), which is synchronized to environmental light cycles (1). As one of the biological clock oscillators, the pineal gland secretes melatonin into the blood with high level during the subjective night and low level during the subjective day (2) and is a typical model system for the study of circadian rhythm. Melatonin plays an important role in the circadian and seasonal rhythms, immunomodulation and neurotransmission (3). In birds, pinealectomy abolished the circadian patterns of locomotor activity and body temperature (4, 5), which was attributed to the absence of the rhythmic secretion of melatonin, as cyclic melatonin administration restored the circadian rhythmicity of the pinealectomised birds (6, 7, 8). The change in the duration of melatonin production serves as a biological signal for the organisation of light-dependent functions, such as reproduction and behaviour (9, 10, 11). Light seems to play a definitive role in the rhythmic secretion of melatonin (12, 13). Our previous studies have shown that monochromatic green light (GL) could increase the amount of plasma melatonin in chick with rhythmic secretion (14, 15). However, the mechanism underlying the regulation of melatonin secretion by GL remains unclear.
The circadian oscillator in vertebrates is driven by the core circadian-feedback loop that consists of positive and negative components (16). In the nucleus, the positive component brain and muscle ARNT-like-1/2 (BMAL1/2) and its paralog circadian locomotor output cycles kaput (CLOCK) form a heterodimer via the Per-Arnt-Sim domain. The BMAL-CLOCK heterodimer binds to E-box enhancer elements presented in the promoter of target genes, thereby activating the expression of negative genes, such as Cryptochrome (Crys) and Period (Pers) (17, 18, 19). The BMAL1-CLOCK heterodimer also activates the transcription of arylalkylamine N-acetyltransferase (Aanat), which is a rate-limiting enzyme for melatonin synthesis (20, 21). Moreover, there was an interesting phenomenon in vivo, wherein GL illumination not only promoted the circadian rhythm secretion of melatonin in chick plasma but also enhanced the circadian rhythm expression of BMAL1 and CLOCK in three circadian pacemakers, including the pineal gland, the retina and the SCN (15, 22, 23). This phenomenon suggested that BMAL1 and CLOCK might play critical roles in GL-induced melatonin rhythmic secretion, but further in vitro studies were needed to investigate the role of BMAL1 and CLOCK in rhythmic melatonin synthesis in chick pinealocytes exposed to GL.
Because the avian pineal gland can directly receive external photic information and respond to light entrainment, it is often chosen as a prominent model even for cellular analyses (24). Therefore, siRNA interference and overexpression experiments were used in the present study to investigate the role of BMAL1 and CLOCK in monochromatic GL-induced rhythmic synthesis of melatonin in chick pinealocytes.
Materials and methods
Animals
Newly hatched Arbor Acre male broilers (Beijing Huadu Breeding, P. R. China) were housed under 12 h white light: 12 h dark cycles (lights on at 08:30; zeitgeber time (ZT) 0). Food and water were available ad libitum. All experimental procedures for this research were approved by the China Agricultural University Animal Care and Use Committee.
Pineal gland cell culture
The 7-day-old chicks were killed by decapitation. The pineal glands were removed and separated from extra tissue in cold D-Hank’s solution under sterile conditions, followed by digestion with 0.1% collagenase IV (Worthington, USA) for 30 min at 37°C and incubation with 0.25% trypsin (Gibco) for 10 min at 37°C. After the tissue was filtered through a 200 mesh metal filter, the filtrates were collected and plated onto collagen-coated six-well plastic culture plates (Corning) at a density of 106 cells/well in DMEM supplemented with 10% foetal bovine serum (25, 26). The cells were cultured under 12 h GL (560 nm): 12 h dark cycles (lights on at 08:30; zeitgeber time (ZT) 0) supplied by cold light-emitting diode (LED) bulbs. This LED system was placed 15 cm above the cell plates at the top inside a CO2 incubator. The LED energy output was tuned by changing the lamp’s voltage and current by a transformer. The voltage was 13.89 V in GL with an intensity of 0.16 W/m2.
Interference experiments
The knockdown of endogenous BMAL1 and CLOCK was performed using the synthesised siRNA (Shanghai GenePharma Co, Ltd). The siRNA sequences used to target chicken BMAL1 were sense 5ʹ-GGACUACGCAGACCAACAATT-3ʹ and antisense 5ʹ-UUGUUGGUCUGCGUAGUCCTT-3ʹ and those used to target chicken CLOCK were sense 5ʹ-GCUCGGAAGAUGGAUAAAUTT-3ʹ and antisense 5ʹ-AUUUAUCCAUCUUCCGAGCTT-3ʹ. The negative control siRNA were sense 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ and antisense 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ. After cultivation for 2 days, the pinealocytes were transiently transfected with si-BMAL1, si-CLOCK and negative control siRNA at ZT0 using a Lipofectamine 2000 transfection reagent according to the manufacturer’s protocol (11668019, Invitrogen). The cells were continuously cultured for 2 days, and the harvested cells and supernatants were then subjected to dim red light at six separate time points at 4-h intervals (ZT0, ZT4, ZT8, ZT12, ZT16 and ZT20).
Overexpression experiments
Full-length BMAL1 (NM_001001463.1) and CLOCK (NM_204174.2) segments in chicken with the homologous arm of the pcDNA3.1 vector were amplified by PCR using chicken pineal gland cDNA. The PCR products were cloned into plasmid pcDNA3.1 with EcoR I and Xho I using the seamless assembly cloning kit (C5891, CloneSmarter, USA). After cultivation for 2 days, the pinealocytes were transiently transfected with pcDNA3.1-BMAL1, pcDNA3.1-CLOCK or pcDNA3.1 (negative control). The cells were cultured and harvested in accordance with the interference experiment methods. The blank control group was the chick pinealocytes cultured under GL, we only retained the negative control group in order to make the figures clear and concise since the statistical analysis showed that expressions of all genes in blank and negative control groups were no significant difference.
Quantitative real-time PCR
Total RNA was isolated from pinealocytes using TRIzol reagents (CW0580A, CWBIO Company, China). The first-strand cDNA was synthesised according to the RevertAid first strand cDNA synthesis kit (K1622, Thermo, USA). PCR amplification was performed using the AceQ qPCR SYBR green master mix (Q111-02, Vazyme Biotech, China). PCR cycle primers are shown in Table 1. Each sample was assayed in triplicate, and relative mRNA levels were normalised to the expression levels of the housekeeping gene GAPDH.
Table 1.
The primers of clock genes.
| Genes | Primer sequences (5ʹ-3ʹ) | Product size (bp) | Accession no. |
|---|---|---|---|
| BMAL1 | F:GTAGACCAGAGGGCGACAG | 215 | NM_001001463.1 |
| R:ATGAAACTGAACCAGCGACTC | |||
| BMAL2 | F:CGGCGTTCCTTCTTCTGTC | 156 | NM_204133.1 |
| R:TTCCTCTTCCACTCCACC | |||
| CLOCK | F:GATCACAGGGCACCTCCAATA | 301 | NM_204174.2 |
| R:CTAGTTCTCGCCGCCTTTCT | |||
| CRY1 | F:GATGTGGCTATCCTGTAGTTCCT | 281 | NM_204245.1 |
| R:GCTGCTGGTAGATTTGTTTCAT | |||
| CRY2 | F:GCACGGCTGGATAAACACT | 141 | NM_204244.1 |
| R:AAATAAGC GGCAGGACAAA | |||
| PER2 | F:ATGAAACGAGCCATCCCG | 206 | NM_204262.1 |
| R:CAGTTGTCGTGATTTTGCCTA | |||
| PER3 | F:CAGTGCCTTTGTTGGGTTAC | 217 | XM_417528.2 |
| R:GATGGATTCACAAAACTGGAC | |||
| AANAT | F:GGACCAGGACAGGCTCAG | 224 | NM_205158.1 |
| R:CGAAACCACACTTCTCGTAG | |||
| GAPDH | F:ATCACAGCCACACAGAAGACG | 124 | NM_204305.1 |
| R:TGACTTTCCCACAGCCTTA |
Western immunoblot analysis
Protein concentrations were measured by the bicinchoninic acid assay (CW0014, CWBIO Company) after the total protein extracts were collected from pinealocytes using the radioimmunoprecipitation lysis buffer (CW2333, CWBIO Company). Equal amounts of protein from each sample were separated using SDS-PAGE and transferred onto PVDF membranes using electroblotting. The membranes were incubated in a solution containing 5% skim milk in phosphate buffer saline at room temperature for 1 h. The membranes were incubated with a CLOCK rabbit polyclonal antibody (1:1000, ab461, Abcam) and BMAL1 rabbit polyclonal antibody (1:1000, ab93806, Abcam) overnight at 4°C. After washing the membranes in Tris-buffered saline-Tween, they were incubated with goat anti-rabbit horseradish peroxidase-conjugated immunoglobulin (Ig) G (1:8000, CW0103, CWBIO Company) for 1 h at room temperature. The blot bands were detected using the Ecl Western blot kit (1627003, Millipore) after washing. The intensity of the signals for CLOCK and BMAL1 were quantified using Image-Pro plus software and were normalised relative to the values obtained for β-ACTIN (1:4000, CW0096, CWBIO Company).
Measurement of melatonin levels
The melatonin content in cell supernatants was measured using the competitive inhibition ELISA kit (CEA908Ge, USCN Life Science INC, China) according to the manufacturer’s protocol. First, 50 μL of each sample dilution was added into the wells and incubated with 50 μL of detection reagent A for 1 h. Then, 100 μL of detection reagent B was added and incubated for 30 min at 37°C. Next, 90 μL of the substrate solution was added and incubated for 15–25 min at 37°C. The reaction was stopped with 50 μL of stop solution, and optical density (OD) values were immediately measured using an ELISA analyser at 450 nm (Bio-Rad, Model 680).
Statistical analysis
Significant difference analysis
Data are presented as the mean ± standard error of the mean (s.e.m.).To test the significance of the differences in the expression at the same time point between experimental and negative control groups, and the differences in the expression at each time point between the groups, one-way ANOVA followed by Tukey test using SPSS version 22 (IBM Corp., Armonk) was used.
Unimodal cosinor regression analysis
The 24-h rhythm of clock genes was analysed by MATLAB 7.0 software (MathWorks Inc., USA). The cosinor formula y = a + b × cos (x × pi/12–c × pi/12) was used, where a, b and c denote the mesor, amplitude and acrophase levels of the rhythm, respectively and R2 is the fitting degree. Statistically significant differences in gene rhythms were indicated by a P value of <0.05, which was calculated using the website http://www.danielsoper.com/statcalc3/calc.aspx?id=15 (27).
Results
Efficiency of interference and overexpression of BMAL1 and CLOCK
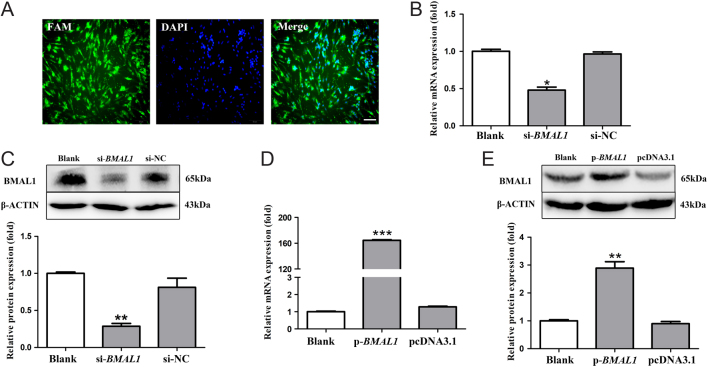
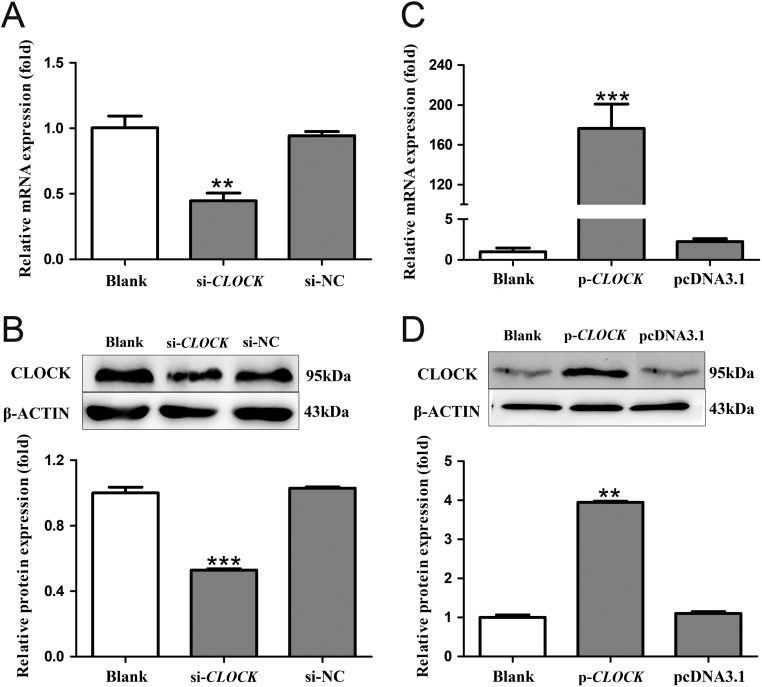
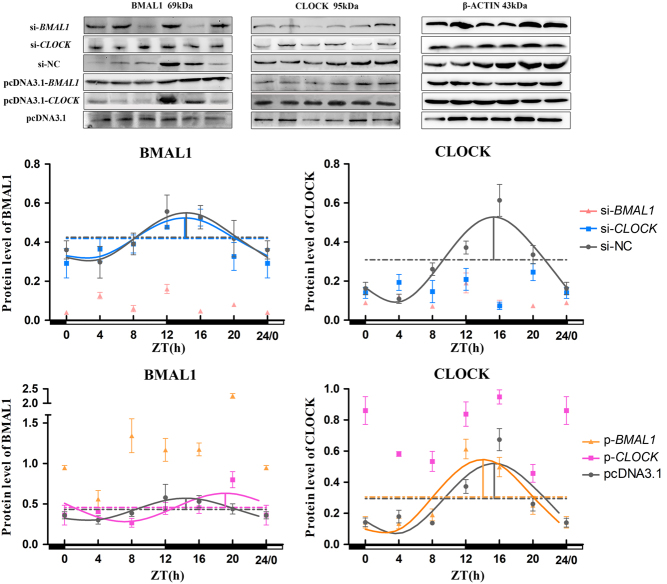
The 85–87% chick pinealocytes were successfully transfected, which clearly observed after 100 pmol/well FAM-siRNA was transfected (Fig. 1A). The mRNA and protein levels of BMAL1 and CLOCK in chick pinealocytes after transfection with siRNAs and overexpression plasmids were examined to evaluate the interference and overexpression efficiencies. Compared with the blank control group, the mRNA and protein levels of BMAL1 in cells transfected with si-BMAL1 were decreased by 52.18 and 71.29%, respectively (P < 0.05, one-way ANOVA) (Fig. 1B and C). pcDNA3.1-BMAL1 significantly increased the mRNA and protein levels of BMAL1 by 162.47 and 1.89 fold compared with those of the blank control group, respectively (P < 0.05, one-way ANOVA) (Fig. 1D and E). Furthermore, compared with the blank control group, si-CLOCK suppressed the mRNA and protein levels of CLOCK by 55.49 and 47.19%, respectively (P < 0.05, one-way ANOVA) (Fig. 2A and B). On the contrary, CLOCK overexpression increased the mRNA and protein levels of CLOCK by 175.45 and 2.95 fold compared with those of blank control group, respectively (P < 0.05, one-way ANOVA) (Fig. 2C and D). These results demonstrated the BMAL1 and CLOCK knockdown and overexpression were successfully made.
Figure 1.
Efficiency of interference and overexpression of BMAL1 in pinealocytes under green light. (A) Fluorescent dye-labelled siRNA (100 pmol/well; FAM fluorescent) was transfected into chick pinealocytes to evaluate the transfection efficiency. (B and C) The expression of BMAL1 mRNA and cBMAL1 protein after transfecting si-BMAL1 and the negative control siRNA. (D and E) The expression of BMAL1 mRNA and cBMAL1 protein after transfecting pcDNA3.1-BMAL1 and pcDNA3.1. The results are presented as the mean ± s.e.m. N = 3 per time point. Significant differences between different treatment groups are shown with asterisks, where *P < 0.05, **P < 0.01 and ***P < 0.001. (A–C) Scale bar = 50 μm.
Figure 2.
Efficiency of interference and overexpression of CLOCK in pinealocytes under green light. (A and B) The expression of CLOCK mRNA and cCLOCK protein after transfecting si-CLOCK and a negative control siRNA. (C and D) The expression of CLOCK mRNA and CLOCK protein after transfecting pcDNA3.1-CLOCK and pcDNA3.1. The results are presented as the mean ± s.e.m. N = 3 per time point. Significant differences between different treatment groups are shown with asterisks, where *P < 0.05, **P < 0.01 and ***P < 0.001.
The analysis of AANAT transcription level and melatonin secretion
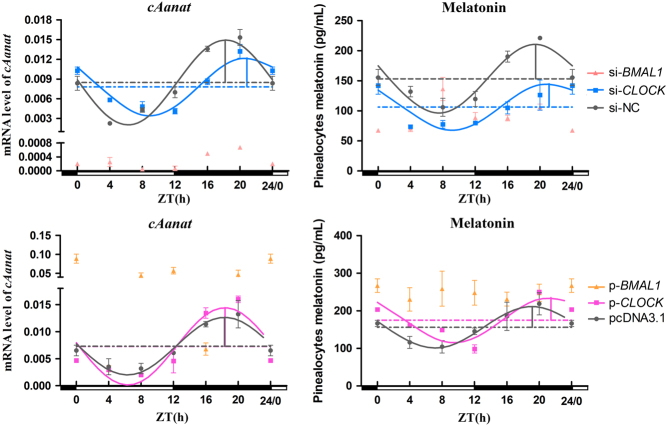
Significant daily variations in AANAT transcription level and melatonin secretion were observed in the different interference groups (P < 0.05, one-way ANOVA). The cosinor analysis showed that si-BMAL1 not only decreased the levels of AANAT mRNA and melatonin (except at ZT12 for melatonin; P < 0.05, one-way ANOVA) but also destroyed their circadian rhythms (Fig. 3). However, si-CLOCK significantly reduced melatonin secretion for the whole day, except ZT0, as well as the mRNA expression of AANAT during the night (P < 0.05, one-way ANOVA); however, it did not abolish the rhythms of AANAT mRNA and melatonin (Fig. 3). si-CLOCK decreased the amplitude of AANAT mRNA and mesor of melatonin and delayed the acrophases of AANAT mRNA and melatonin by 2.68 and 1.65 h, respectively (Table 2).
Figure 3.
Temporal changes in AANAT mRNA and melatonin secretion after BMAL1 and CLOCK interference and overexpression in pinealocytes under green light. Pink points and lines represent the BMAL1 interference group, blue points and lines represent the CLOCK interference group, yellow points and lines represent the BMAL1 overexpression group and purple points and lines represent the CLOCK overexpression group. The horizontal white bar on each figure represents the day, and the horizontal black bar represents the night. The solid points indicate the relative mRNA levels or the plasma melatonin levels. The curved lines indicate the rhythmic levels of genes and melatonin by the cosinor analysis. The horizontal dotted line represents the mesor. The vertical solid line represents acrophase and amplitude (the distance between maximum and mesor). The results are presented as the mean ± s.e.m. N = 3 per time point.
Table 2.
Circadian rhythm parameters (mean ± s.e.m.) of all clock genes, AANAT and melatonin in interference treatments, as determined by cosinor analyses.
| Group | BMAL1 | BMAL2 | CLOCK | BMAL1 | CLOCK | CRY1 | CRY2 | PER2 | PER3 | AANAT | Melatonin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mesor (×10−3) (for gene) | si-BMAL1 | ND | ND | ND | ND | ND | 22.10 ± 1.24 | ND | ND | ND | ND | ND |
| si-CLOCK | 4.06 ± 0.21 | ND | ND | 0.42 ± 0.09 | ND | 35.93 ± 0.86 | ND | 3.86 ± 0.01 | ND | 7.81 ± 0.34 | 106.41 ± 11.48* | |
| si-NC | 3.70 ± 0.53 | 4.40 ± 0.90 | 4.65 ± 0.09 | 0.43 ± 0.14 | 0.31 ± 0.06 | 32.06 ± 8.90 | 3.49 ± 0.72 | 2.93 ± 0.52 | 2.09+0.48 | 8.47 ± 0.38 | 153.33 ± 12.53 | |
| Amplitude (×10−3) (for gene) | si-BMAL1 | ND | ND | ND | ND | ND | 8.38 ± 0.33 | ND | ND | ND | ND | ND |
| si-CLOCK | 1.38 ± 0.12 | ND | ND | 0.10 ± 0.06 | ND | 13.94 ± 0.11 | ND | 1.13 ± 0.04* | ND | 4.40 ± 0.12* | 38.65 ± 10.69 | |
| si-NC | 1.66 ± 0.04 | 2.22 ± 0.07 | 3.84 ± 0.18 | 0.13 ± 0.03 | 0.22 ± 0.01 | 12.96 ± 3.90 | 2.09 ± 0.47 | 0.96 ± 0.04 | 1.26+0.46 | 6.47 ± 3.83 | 57.12 ± 3.93 | |
| Acrophase (h) | si-BMAL1 | ND | ND | ND | ND | ND | 9.39 ± 0.13* | ND | ND | ND | ND | ND |
| si-CLOCK | 12.00 ± 0.76 | ND | ND | 14.12 ± 1.73 | ND | 10.77 ± 0.03* | ND | 23.64 ± 0.23 | ND | 20.86 ± 0.02* | 21.12 ± 0.38* | |
| si-NC | 12.77 ± 0.34 | 15.88 ± 0.10 | 15.07 ± 0.03 | 14.30 ± 0.39 | 15.29 ± 0.18 | 21.77 ± 0.73 | 16.48 ± 0.25 | 0.22 ± 0.00 | 23.64 ± 0.53 | 18.18 ± 0.06 | 19.47 ± 0.05 | |
| R2 | si-BMAL1 | 0.26 | 0.34 | 0.35 | 0.22 | 0.25 | 0.59 | 0.28 | 0.00 | 0.26 | 0.05 | 0.22 |
| si-CLOCK | 0.52 | 0.21 | 0.03 | 0.80 | 0.04 | 0.88 | 0.21 | 0.90 | 0.21 | 0.93 | 0.80 | |
| si-NC | 0.82 | 0.93 | 0.84 | 0.90 | 0.87 | 0.96 | 0.96 | 0.76 | 0.89 | 0.96 | 0.91 | |
| P | si-BMAL1 | 0.22 | 0.19 | 0.10 | 0.34 | 0.54 | 0.02 | 0.20 | – | 0.23 | 0.92 | 0.38 |
| si-CLOCK | 0.02 | 0.33 | 0.92 | 0.00 | 0.90 | 0.00 | 0.34 | 0.00 | 0.32 | 0.00 | 0.00 | |
| si-NC | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F | si-BMAL1 | 1.66 | 1.91 | 2.56 | 1.21 | 0.78 | 5.33 | 1.79 | – | 1.64 | 0.16 | 1.13 |
| si-CLOCK | 4.99 | 1.24 | 0.16 | 11.77 | 0.19 | 34.62 | 1.23 | 39.00 | 1.27 | 60.60 | 16.26 | |
| si-NC | 43.51 | 127.63 | 51.67 | 46.20 | 25.63 | 206.59 | 226.90 | 15.03 | 99.97 | 357.03 | 52.83 |
Note: ND represents not determined as there was no circadian rhythm. Significant differences (P < 0.05) between the treatment and si-NC group are shown by an asterisk (*).
BMAL1 overexpression caused the loss of day–night variations in melatonin secretion (P = 0.871, one-way ANOVA), although significant daily variations in the level of AANAT mRNA and melatonin were observed in other overexpression groups (P < 0.05, one-way ANOVA). The cosinor analysis showed that the overexpression of BMAL1 increased the AANAT mRNA level and secretion of melatonin, except at ZT16 for AANAT and melatonin and ZT 20 for melatonin (P < 0.05, one-way ANOVA) but abolished the circadian rhythms of AANAT mRNA and melatonin (Fig. 3). However, AANAT mRNA and melatonin still maintained a remarkable circadian rhythm during the overexpression of CLOCK (Fig. 3). Simultaneously, the amplitude in AANAT expression was increased by the overexpression of CLOCK, and the acrophases of melatonin were delayed by 2.32 h (Table 3).
Table 3.
Circadian rhythm parameters (mean ± s.e.m.) of all clock genes, AANAT, and melatonin in overexpression treatments, as determined by cosinor analyses.
| Group | BMAL1 | BMAL2 | CLOCK | BMAL1 | CLOCK | CRY1 | CRY2 | PER2 | PER3 | AANAT | Melatonin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mesor (×10−3) | p-BMAL1 | ND | 70.64 ± 6.62* | 2.36 ± 0.14* | ND | 0.31 ± 0.07 | ND | ND | 1.22 ± 0.05(×103)* | 23.27 ± 4.18* | ND | ND |
| p-CLOCK | 3.44 ± 0.18 | 3.52 ± 0.56 | ND | 0.46 ± 0.07 | ND | ND | ND | 4.21 ± 0.00* | 2.55 ± 0.11* | 7.27 ± 0.66 | 175.17 ± 3.51 | |
| pcDNA3.1 | 3.49 ± 0.15 | 3.43 ± 0.30 | 4.04 ± 0.02 | 0.43 ± 0.15 | 0.29 ± 0.09 | 30.39 ± 1.65 | 2.93 ± 0.29 | 2.62 ± 0.20 | 1.98 ± 0.13 | 7.32 ± 1.10 | 156.13 ± 19.14 | |
| Amplitude (×10−3) | p-BMAL1 | ND | 38.21 ± 0.87* | 1.83 ± 0.09* | ND | 0.24 ± 0.03 | ND | ND | 0.16 ± 0.02(×103)* | 22.56 ± 3.92* | ND | ND |
| p-BMAL1 | 1.03 ± 0.05* | 1.74 ± 0.61 | ND | 0.18 ± 0.03 | ND | ND | ND | 1.12 ± 0.00 | 0.98 ± 0.10 | 7.17 ± 0.32* | 60.08 ± 1.97 | |
| pcDNA3.1 | 1.53 ± 0.06 | 1.75 ± 0.02 | 3.88 ± 0.17 | 0.14 ± 0.05 | 0.23 ± 0.04 | 10.41 ± 1.8 | 1.19 ± 0.10 | 1.13 ± 0.05 | 1.01 ± 0.08 | 5.33 ± 0.24 | 55.31 ± 9.65 | |
| Acrophase (h) | p-BMAL1 | ND | 9.12 ± 0.08* | 12.70 ± 0.32* | ND | 14.04 ± 0.12* | ND | ND | 2.74 ± 0.09* | 5.67 ± 0.23* | ND | ND |
| p-CLOCK | 15.31 ± 0.72* | 15.95 ± 0.77 | ND | 19.14 ± 0.20* | ND | ND | ND | 19.86 ± 1.01* | 22.16 ± 0.09 | 18.36 ± 0.42 | 21.45 ± 0.09* | |
| pcDNA3.1 | 13.51 ± 0.17 | 15.35 ± 0.25 | 15.05 ± 0.18 | 14.45 ± 1.05 | 15.39 ± 0.23 | 21.71 ± 0.27 | 16.54 ± 0.46 | 22.18 ± 0.59 | 22.34 ± 0.36 | 18.31 ± 0.50 | 19.13 ± 0.31 | |
| R2 | p-BMAL1 | 0.37 | 0.80 | 0.63 | 0.37 | 0.85 | 0.28 | 0.27 | 0.65 | 0.77 | 0.06 | 0.08 |
| p-CLOCK | 0.62 | 0.57 | 0.34 | 0.56 | 0.17 | 0.38 | 0.15 | 0.53 | 0.52 | 0.86 | 0.81 | |
| pcDNA3.1 | 0.80 | 0.97 | 0.86 | 0.83 | 0.72 | 0.86 | 0.68 | 0.85 | 0.83 | 0.95 | 0.99 | |
| P | p-BMAL1 | 0.13 | 0.00 | 0.00 | 0.48 | 0.01 | 0.21 | 0.24 | 0.00 | 0.00 | 0.86 | 0.80 |
| p-CLOCK | 0.00 | 0.01 | 0.13 | 0.05 | 0.52 | 0.07 | 0.51 | 0.01 | 0.01 | 0.00 | 0.00 | |
| pcDNA3.1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F | p-BMAL1 | 2.31 | 17.32 | 7.88 | 0.96 | 11.71 | 1.72 | 1.61 | 7.58 | 15.30 | 0.25 | 0.34 |
| p-CLOCK | 7.72 | 6.18 | 2.26 | 3.80 | 0.80 | 2.86 | 0.80 | 5.21 | 5.15 | 29.75 | 15.67 | |
| pcDNA3.1 | 41.67 | 371.68 | 42.12 | 35.60 | 21.49 | 39.79 | 12.18 | 43.48 | 36.08 | 194.50 | 254.87 |
Note: p-BMAL1 represents the overexpression BMAL1 group, p-CLOCK represents the overexpression CLOCK group. ND represents not determined as there was no circadian rhythm. Significant differences (P < 0.05) between the treatment and pcDNA3.1 groups are shown by an asterisk (*).
Transcription level analysis of positive clock genes
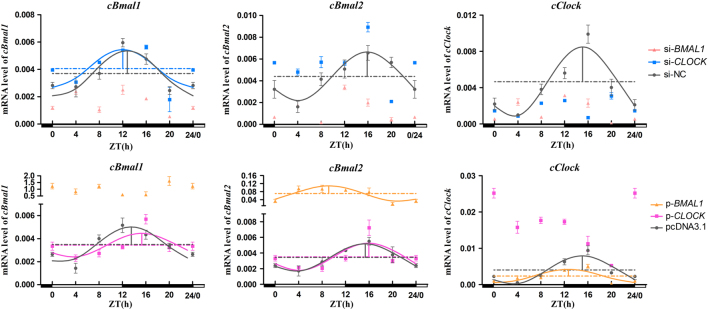
Transcripts of all the positive clock genes (BMAL1, BMAL2 and CLOCK) in the interference groups displayed day–night variations (P < 0.05, one-way ANOVA). Compared with si-NC, si-BMAL1 reduced the mRNA levels of positive genes throughout the day, except at ZT4 (P < 0.05, one-way ANOVA), whereas si-CLOCK only significantly declined its own expression throughout the day (P < 0.05, one-way ANOVA) (Fig. 4). Thecosinor analysis showed that the knockdown of BMAL1 and CLOCK resulted in a circadian arrhythmy of all positive genes, except BMAL1 mRNA in si-CLOCK-transfected cells that still presented a significant circadian rhythm (Table 2).
Figure 4.
Temporal changes in positive clock genes after BMAL1 and CLOCK interference and overexpression in pinealocytes under green light. The figure is marked the same as Fig. 3.
The mRNA expression of all positive clock genes in pinealocytes showed a significant diurnal variation after BMAL1 and CLOCK overexpression (P < 0.05, one-way ANOVA). Moreover, the overexpression of BMAL1 significantly promoted the mRNA expression of BMAL1/2 throughout the day (P < 0.05, one-way ANOVA), whereas the overexpression of CLOCK only significantly enhanced its own expression throughout the day except ZT16 (P < 0.05, one-way ANOVA) (Fig. 4). The cosinor analysis showed that the overexpression of BMAL1 and CLOCK only abolished their own rhythm (Fig. 4). Moreover, BMAL1 overexpression significantly increased the mesors and amplitudes of BMAL2 and CLOCK. Compared with the pcDNA3.1 group, the acrophases of BMAL2 and CLOCK in the BMAL1 overexpression group were increased by 6.23 and 2.35 h, respectively. However, the acrophase of BMAL1 in the CLOCK overexpression group was delayed by 1.80 h (Table 3).
Protein level analysis of positive clock genes
The protein expression of BMAL1 displayed evident diurnal oscillations in the BMAL1 interference group (si-BMAL1) and CLOCK interference group (si-CLOCK) (P < 0.05, one-way ANOVA), but the daily variation of CLOCK was destroyed by si-CLOCK (P = 0.124, one-way ANOVA). The protein expression of both CLOCK and BMAL1 throughout the day were decreased by si-BMAL1 except ZT4 for CLOCK (P < 0.05, one-way ANOVA), while si-CLOCK only decreased its own protein levels (P < 0.05, t-test) (Fig. 5). The cosinor analysis showed that the circadian rhythms of BMAL1 and CLOCK protein were disturbed by si-BMAL1, whereas si-CLOCK had no impact on the rhythmicity of the BMAL1 protein (Fig. 5 and Table 2).
Figure 5.
Temporal changes in positive clock proteins after BMAL1 and CLOCK interference and overexpression in pinealocytes under green light. The figure is marked the same as Fig. 3.
The BMAL1 and CLOCK protein levels also displayed a remarkable diurnal oscillation after BMAL1 and CLOCK overexpression (P <0.05, one-way ANOVA). The overexpression of BMAL1 and CLOCK remarkably increased their own protein levels (P < 0.05, one-way ANOVA). The cosinor analysis showed that the overexpression of BMAL1 and CLOCK disrupted their own circadian rhythms of protein levels (Fig. 5). The acrophase of BMAL1 protein was delayed by 4.69 h after the overexpression of CLOCK, while the acrophase of CLOCK protein was advanced by 1.35 h after the overexpression of BMAL1 (Table 3).
Transcription level analysis of negative clock genes
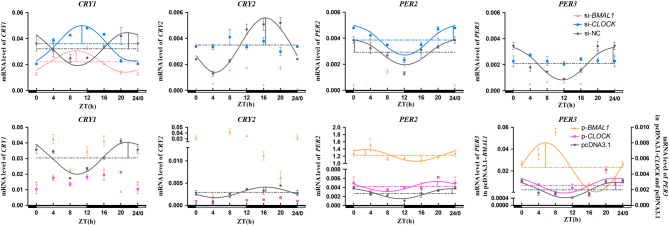
The results showed that there were significant diurnal variations at 24 h for the mRNA level of negative genes in the interference groups (P < 0.05, one-way ANOVA). si-BMAL1 significantly decreased the levels of CRY2 mRNA and PER3 mRNA in one entire day, except at ZT20 for PER3 (P < 0.05, one-way ANOVA) (Fig. 6). The circadian rhythms of CRY2 mRNA and PER3 mRNA were destroyed by si-BMAL1 and si-CLOCK, and the cosinor analysis showed that the mRNA level of CRY1 still retained a robust circadian rhythm in si-BMAL1 and si-CLOCK accompanying an opposite phase change with a negative control group (Fig. 6). si-BMAL1 caused rhythm disturbances of PER2 mRNA, whereas si-CLOCK had no effect on the circadian rhythm of PER2 mRNA (Fig. 6). si-BMAL1 and si-CLOCK advanced the acrophase of CRY1 mRNA by 12.38 and 11.00 h, respectively (Table 2).
Figure 6.
Temporal changes in negative clock genes after BMAL1 and CLOCK interference and overexpression in pinealocytes under green light. The figure is marked the same as Fig. 3.
The mRNA level of all negative clock genes in pcDNA3.1-BMAL1- and pcDNA3.1-CLOCK-transfected cells showed circadian oscillations (P < 0.05, one-way ANOVA), but only the overexpression of BMAL1 significantly promoted the transcription levels of CRY2, PER2 and PER3 (P < 0.05, one-way ANOVA) (Fig. 6E). According to the cosinor analysis, the overexpression of BMAL1 and CLOCK abolished the circadian rhythms of CRY1/2 mRNA, did not disturb the circadian rhythms of PER2/3 mRNA and increased the mesors and amplitudes of PER2/3 mRNA (Fig. 6 and Table 3).
Discussion
The circadian release of avian melatonin can be controlled by the optical environment (13). Previous studies showed that monochromatic GL could enhance melatonin synthesis and expression of positive clock genes, including CLOCK and BMAL1/2, in the pineal gland of chicks (14, 15, 28). These studies suggested that the positive clock genes were involved in the modulation of circadian release of melatonin by the stimulation of monochromatic light. The present study further clarified the roles of positive clock genes in the monochromatic GL-induced melatonin secretion in chick pinealocytes. Both the knockdown and overexpression of BMAL1 resulted in arrhythmicity in the circadian levels of AANAT mRNA and melatonin in the pinealocytes exposed to GL. Our results corroborated previous study results in which the overexpression of BMAL1 in chick pineal cells caused deregulation of the melatonin rhythm in a continuous dark condition (29). Rhythm disturbances caused by the deletion of BMAL1 have also been reported in mammals. The loss of mBmal1 in mice resulted in an immediate and complete loss of rhythmic behaviour under free-running conditions (30). On the contrary, the knockdown and overexpression of CLOCK had no effect on the circadian rhythms and release levels of melatonin. A similar finding was reported in mice that mClock-deficient mice continued to exhibit robust behavioural and molecular rhythms (31). However, AANAT also maintained an obvious circadian rhythm, but its amplitude was changed by the knockdown and overexpression of CLOCK. This was consistent with previous studies in which the knockdown of CLOCK significantly reduced the circadian expression of AANAT in chicken cone photoreceptors (32). Consequently, our data demonstrated that BMAL1 rather than CLOCK plays a critical role in the regulation of monochromatic GL-induced melatonin synthesis in chicken pinealocytes. However, an additional study as to whether the predominant role of BMAL1 in regulation of light-induced melatonin synthesis is green light specific or not is still required.
Our qPCR and Western blot results showed that si-BMAL1 broke the circadian rhythm of CLOCK in chick pinealocytes exposed to monochromatic GL, while si-CLOCK had no effect on the circadian rhythm of BMAL1 mRNA and BMAL1 protein. These results were consistent with the changes in the circadian expression of AANAT and the release of melatonin. The stability of BMAL1 rhythm in the si-CLOCK group may be due to the effect of orphan nuclear receptors RORs and REV-ERBα, which directly activate and repress the transcription of BMAL1, respectively, by binding with an Rev-erb/Ror-binding element in the BMAL1 promoter (33, 34, 35, 36). Therefore, our results demonstrated that compared with CLOCK, BMAL1 had a more predominant role in maintaining the circadian rhythms of melatonin synthesis and secretion in chick pinealocytes when stimulated by monochromatic GL, although some reports have suggested that AANAT transcription could be promoted by Bmal1/Clock heterodimers (20, 21).
The circadian rhythms of chicks were regulated by the transcription–translation feedback loop, which consists of a highly conserved set of genes including positive (BMAL1/2, CLOCK) and negative (CRY1/2, PER2/3) genes (18, 37). The deletion of mBmal1 caused rhythm disturbances and a decrease in the expression of mBmal2 in mice (30), whereas the overexpression of BMAL2 deregulated the melatonin rhythm in chick pineal cells (29). However, our study showed that both si-BMAL1 and si-CLOCK disrupted the circadian rhythm of BMAL2, but the overexpression of BMAL1 and CLOCK had no such effect. Compared with the changes in the circadian rhythms of BMAL2, AANAT and melatonin, BMAL2 was not synchronised with the rhythms disturbance of AANAT and melatonin in both si-CLOCK (BMAL2 with arrhythmicity vs AANAT and melatonin with rhythmicity) and pcDNA3.1-BMAL1 (BMAL2 with rhythmicity vs AANAT and melatonin with arrhythmicity) treatments. It indicated that BMAL2 was not an essential requirement for the modulation of circadian rhythm of melatonin synthesis and secretion in chick pinealocytes stimulated by monochromatic GL.
In mammals, mBmal1-deficient mice showed extremely low expression levels and loss of rhythmicity of mPer1/2 in SCN; however, the expression of mCry1 and mPer1/2 continued to oscillate in mClock-deficient mice (30, 31). In the present study, the knockdown of BMAL1 destroyed the circadian rhythms of negative clock genes, except CRY1, while the overexpression of BMAL1 abolished the circadian rhythms of CRY1/2. For negative clock genes, however, we found that the circadian rhythm of CRY2 mRNA was always consistent with the changes of AANAT and melatonin when BMAL1 was downregulated or upregulated. Previous studies have reported that mCry2 plays an important role in maintaining the biological rhythms of mammals because mice lacking mCry2 showed a delay in the free-running periodicity of locomotor activity (38, 39, 40). These data suggested that CRY2 is involved in BMAL1-mediated modulation of the circadian levels of AANAT and melatonin in chick pinealocytes stimulated by monochromatic GL. Consistent with the changes in AANAT and melatonin rhythmicity, both knockdown and overexpression of CLOCK had no effect on the circadian rhythm of PER2 mRNA, which was contrary to that in CRY1/2 and PER3. Different regulation mechanisms for circadian expression among Period family genes under light stimulation have also been reported in mammals (41, 42, 43). Therefore, combined with previous findings, our results indicated that PER2 may have a role in the regulation of AANAT expression when CLOCK expression is altered.
Taken together, our study showed that BMAL1 and CLOCK have different output pathways to regulate the cellular mechanism of chick pinealocytes cultured under GL. Moreover, BMAL1 rather than CLOCK plays a predominant role in the regulation of diurnal rhythms of melatonin secretion, and CRY2 may be involved in this regulatory pathway.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by the Chinese National Natural Science Foundation (31472157, 31672501 and 31873000), the Beijing Natural Science Foundation (6182018) and Beijing Advanced Innovation Center for Food Nutrition and Human Health.
Acknowledgements
The authors would like to thank all the members of neurobiology laboratory.
References
- 1.Cassone VM, Westneat DF.The bird of time: cognition and the avian biological clock. Frontiers in Molecular Neuroscience 2012. 5 1–8. ( 10.3389/fnmol.2012.00032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zawilska JB, Skene DJ, Arendt J.Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacological Reports 2009. 61 383–410. ( 10.1016/S1734-1140(09)70081-7) [DOI] [PubMed] [Google Scholar]
- 3.Hardeland R, Pandi-Perumal SR, Cardinali DP.Melatonin. International Journal of Biochemistry and Cell Biology 2006. 38 313–316. ( 10.1016/j.biocel.2005.08.020) [DOI] [PubMed] [Google Scholar]
- 4.Gaston S, Menaker M.Pineal function: the biological clock in the sparrow? Science 1968. 160 1125–1127. ( 10.1126/science.160.3832.1125) [DOI] [PubMed] [Google Scholar]
- 5.Binkley S, Kluth E, Menaker M.Pineal function in sparrows: circadian rhythms and body temperature. Science 1971. 174 311–314. ( 10.1126/science.174.4006.311) [DOI] [PubMed] [Google Scholar]
- 6.Chabot CC, Menaker M.Effects of physiological cycles of infused melatonin on circadian rhythmicity in pigeons. Journal of Comparative Physiology A Sensory 1992. 170 615–622. ( 10.1007/BF00199337) [DOI] [PubMed] [Google Scholar]
- 7.Cassone VM, Bartell PA, Earnest BJ, Vinod K.Duration of melatonin regulates seasonal changes in song control nuclei of the house sparrow, Passer domesticus: independence from gonads and circadian entrainment. Journal of Biological Rhythms 2008. 23 49–58. ( 10.1177/0748730407311110) [DOI] [PubMed] [Google Scholar]
- 8.Surbhi, Kumari Y, Rani S, Tsutsui K, Kumar V.Duration of melatonin regulates seasonal plasticity in subtropical Indian weaver bird (Ploceus philippinus). General and Comparative Endocrinology 2015. 220 46–54. ( 10.1016/j.ygcen.2014.06.004) [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Harpole CE, Trivedi AK, Cassone VM.Circadian regulation of bird song, call, and locomotor behavior by pineal melatonin in the zebra finch. Journal of Biological Rhythms 2012. 27 145–155. ( 10.1177/0748730411435965) [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Di KQ, Hao EY, Ye M, Zha QC, Li LH, Bai K, Huang RL.Effects of exogenous melatonin and photoperiod on sexual maturation in pullets. Journal of Animal Physiology and Animal Nutrition 2016. 100 46–52. ( 10.1111/jpn.12337) [DOI] [PubMed] [Google Scholar]
- 11.Sinkalu VO, Ayo JO, Adelaiye AB, Hambolu JO.Melatonin modulates tonic immobility and vigilance behavioural responses of broiler chickens to lighting regimens during the hot-dry season. Physiology and Behavior 2016. 165 195–201. ( 10.1016/j.physbeh.2016.07.019) [DOI] [PubMed] [Google Scholar]
- 12.Deep A, Schwean-Lardner K, Crowe TG, Fancher BI, Classen HL.Effect of light intensity on broiler behaviour and diurnal rhythms. Applied Animal Behaviour Science 2012. 136 50–56. ( 10.1016/j.applanim.2011.11.002) [DOI] [Google Scholar]
- 13.Surbhi, Kumar V.Avian photoreceptors and their role in the regulation of daily and seasonal physiology. General and Comparative Endocrinology 2015. 220 13–22. ( 10.1016/j.ygcen.2014.06.001) [DOI] [PubMed] [Google Scholar]
- 14.Jin E, Jia L, Li J, Yang G, Wang Z, Cao J, Chen Y.Effect of monochromatic light on melatonin secretion and arylalkylamine N-acetyltransferase mRNA expression in the retina and pineal gland of broilers. Anatomical Record Advances in Integrative Anatomy and Evolutionary Biology 2011. 294 1233–1241. ( 10.1002/ar.21408) [DOI] [PubMed] [Google Scholar]
- 15.Jiang N, Wang Z, Cao J, Dong Y, Chen Y.Role of monochromatic light on daily variation of clock gene expression in the pineal gland of chick. Journal of Photochemistry and Photobiology B-Biology 2016. 164 57–64. ( 10.1016/j.jphotobiol.2016.09.020) [DOI] [PubMed] [Google Scholar]
- 16.Dunlap JC.1996. Molecular bases for circadian clocks. Cell 96 271–290. ( 10.1016/S0092-8674(00)80566-8) [DOI] [PubMed] [Google Scholar]
- 17.Buhr ED, Takahashi JS.Molecular components of the mammalian circadian clock. Handbook of Experimental Pharmacology 2013. 2 3–27. ( 10.1007/978-3-642-25950-0_1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cassone VM.Avian circadian organization: a chorus of clocks. Frontiers in Neuroendocrinology 2014. 35 76–88. ( 10.1016/j.yfrne.2013.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isorna E, De Pedro N, Valenciano AI, Alonso-Gómez AL, Delgado MJ.Interplay between the endocrine and circadian systems in fishes. Journal of Endocrinology 2017. 232 R141–R159. ( 10.1530/JOE-16-0330) [DOI] [PubMed] [Google Scholar]
- 20.Chong NW, Bernard M, Klein DC.Characterization of the chicken serotonin N-acetyltransferase gene. Activation via clock gene heterodimer/E-box interaction. Journal of Biological Chemistry 2000. 275 32991–32998. ( 10.1074/jbc.M005671200) [DOI] [PubMed] [Google Scholar]
- 21.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ.Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nature Reviews Genetics 2005. 6 544–556. ( 10.1038/nrg1633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang N, Wang Z, Cao J, Dong Y, Chen Y.Effect of monochromatic light on circadian rhythmic expression of clock genes in the hypothalamus of chick. Journal of Photochemistry and Photobiology B-Biology 2017. 173 476–484. ( 10.1016/j.jphotobiol.2017.06.027) [DOI] [PubMed] [Google Scholar]
- 23.Cao J, Bian J, Wang Z, Dong Y, Chen Y.Effect of monochromatic light on circadian rhythmic expression of clock genes and arylalkylamine N-acetyltransferase in chick retina. Chronobiology International 2017. 34 1149–1157. ( 10.1080/07420528.2017.1354013) [DOI] [PubMed] [Google Scholar]
- 24.Earnest DJ, Cassone VM.Cell culture models for oscillator and pacemaker function: recipes for dishes with circadian clocks? Methods in Enzymology 2005. 393 558–578. ( 10.1016/S0076-6879(05)93029-2) [DOI] [PubMed] [Google Scholar]
- 25.Haldar C, Araki M.Morphometric analysis of photoreceptive, neuronal and endocrinal cell differentiation of avian pineal cells: an in vitro immunohistochemical study on the developmental transition from neuronal to photo-endocrinal property. Zoological Science 2002. 19 781–787. ( 10.2108/zsj.19.781) [DOI] [PubMed] [Google Scholar]
- 26.Horvath G, Reglodi D, Opper B, Brubel R, Tamas A, Kiss P.Effects of PACAP on the oxidative stress-induced cell death in chicken pinealocytes is influenced by the phase of the circadian clock. Neuroscience Letters 2010. 484 148–152. ( 10.1016/j.neulet.2010.08.039) [DOI] [PubMed] [Google Scholar]
- 27.Singh D, Rani S, Kumar V.Daily expression of six clock genes in central and peripheral tissues of a night-migratory songbird: evidence for tissue-specific circadian timing. Chronobiology International 2013. 30 1208–1217. ( 10.3109/07420528.2013.810632) [DOI] [PubMed] [Google Scholar]
- 28.Ma S, Wang Z, Cao J, Dong Y, Chen Y.Effect of monochromatic light on circadian rhythm of clock genes in chick pinealocytes. Photochemistry and Photobiology 2018. 94 1263–1272. ( 10.1111/php.12963) [DOI] [PubMed] [Google Scholar]
- 29.Okano T, Yamamoto K, Okano K, Hirota T, Kasahara T, Sasaki M, Takanaka Y, Fukada Y.Chicken pineal clock genes: implication of BMAL2 as a bidirectional regulator in circadian clock oscillation. Genes to Cells 2011. 6 825–836. ( 10.1046/j.1365-2443.2001.00462.x) [DOI] [PubMed] [Google Scholar]
- 30.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA.Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 2000. 103 1009–1017. ( 10.1016/S0092-8674(00)00205-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeBruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM.A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 2006. 50 465–477. ( 10.1016/j.neuron.2006.03.041) [DOI] [PubMed] [Google Scholar]
- 32.Haque R, Ali FG, Biscoglia R, Abey J, Weller J, Klein D.CLOCK and NPAS2 have overlapping roles in the circadian oscillation of arylalkylamine N-acetyltransferase mRNA in chicken cone photoreceptors. Journal of Neurochemistry 2010. 113 1296–1306. ( 10.1111/j.1471-4159.2010.06698.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U.The orphan nuclear receptor REV-ERB alpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002. 110 251–260. ( 10.1016/S0092-8674(02)00825-5) [DOI] [PubMed] [Google Scholar]
- 34.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB.A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004. 43 527–537. ( 10.1016/j.neuron.2004.07.018) [DOI] [PubMed] [Google Scholar]
- 35.Akashi M, Takumi T.The orphan nuclear receptor ROR regulates circadian transcription of the mammalian core-clock Bmal1. Nature Structural and Molecular Biology 2005. 12 441–448. ( 10.1038/nsmb925) [DOI] [PubMed] [Google Scholar]
- 36.Guillaumond F, Dardente H, Giguère V, Cermakian N.Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. Journal of Biological Rhythms 2005. 20 391–403. ( 10.1177/0748730405277232) [DOI] [PubMed] [Google Scholar]
- 37.Fukada Y, Okano T.Circadian clock system in the pineal gland. Molecular Neurobiology 2002. 25 19–30. ( 10.1385/MN:25:1:019) [DOI] [PubMed] [Google Scholar]
- 38.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM.mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999 98 193–205. ( 10.1016/S0092-8674(00)81014-4) [DOI] [PubMed] [Google Scholar]
- 39.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J. et al Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. PNAS 1999. 96 12114–12119. ( 10.1073/pnas.96.21.12114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D. et al Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999. 398 627–630. ( 10.1038/19323) [DOI] [PubMed] [Google Scholar]
- 41.Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, et al A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO Journal 1998. 17 4753–4759. ( 10.1093/emboj/17.16.4753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan L, Takekida S, Shigeyoshi Y, Okamura H.Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience 1999. 94 141–150. ( 10.1016/S0306-4522(99)00223-7) [DOI] [PubMed] [Google Scholar]
- 43.Takekida S, Yan L, Maywood ES, Hastings MH, Okamura H.Differential adrenergic regulation of the circadian expression of the clock genes Period1 and Period2 in the rat pineal gland. European Journal of Neuroscience 2000. 12 4557–4561. ( 10.1046/j.0953-816X.2000.01324.x) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a