Abstract
Background
Alcoholic cirrhosis represents 1% of all cause-of-deaths worldwide. Its incidence is higher in males and results from the combination of environmental and genetic factors. Among all the genetic determinants of alcoholic cirrhosis, the patatin-like phospholipase domain protein 3 (PNPLA3) rs738409 represents the most widely validated determinant. Recent cross-sectional studies on alcohol abusers identified transmembrane-6 superfamily member 2 (TM6SF2) rs58542926, membrane bound O-acyltransferase domain containing 7 (MBOAT7) rs641738, and cluster of differentiation 14 (CD14) rs2569190 as new genetic risk factors for alcoholic cirrhosis. We aimed to develop a gene-based risk score to predict the incidence of alcoholic cirrhosis in males with at-risk alcohol consumption.
Materials and methods
A total of 416 male at-risk alcohol drinkers were retrospectively examined. The association between alcoholic cirrhosis incidence and PNPLA3, CD14, TM6SF2, and MBOAT7 variants was tested. Age at onset of at-risk alcohol consumption, age, and body mass index (BMI) were included as covariates to determine the prediction score for alcoholic cirrhosis incidence by evaluating time-dependent receiver operating characteristic curves.
Results
We found that PNPLA3, CD14, and TM6SF2 were associated with alcoholic cirrhosis prevalence. PNPLA3 and CD14 were also associated with its incidence. The best predictive score formula was (age at onset of at-risk alcohol consumption × 0.1) + (number of CD14 allele T) + (number of PNPLA3 allele M) + (BMI × 0.1). A threshold of 7.27 was identified as cutoff for the predictive risk of alcoholic cirrhosis development in 36 years from the onset of at-risk alcohol consumption with 70.1% sensitivity and 78.7% specificity.
Conclusion
We developed the first score for alcoholic cirrhosis prediction that combines clinical and genetic factors.
Keywords: alcoholic cirrhosis, PNPLA3, CD14, predictive score
Introduction
Alcoholic chronic liver disease, with a prevalence of ~12% in the European and North-American populations, is characterized by a broad spectrum of conditions ranging from simple steatosis to alcoholic steatohepatitis, cirrhosis, and hepatocellular carcinoma.1,2 The alcoholic etiology represents the cause of almost half of all deaths due to cirrhosis and of 1% of all-cause deaths worldwide3 and its specific clinical sequelae and comorbidities are associated with high hospital costs.4 Environmental factors, aging, genetic predisposition, and gender play a pivotal role in alcoholic cirrhosis development and progression.5,6 The most established environmental factors influencing the susceptibility to chronic alcohol-related liver damage are represented by dose and pattern of alcohol intake, diet, obesity, diabetes, and smoking.5 Older age during chronic ethanol at-risk consumption is also considered a risk factor for more pronounced liver damage.6 Indeed, we have shown that despite a lower daily alcohol intake and a shorter duration of at-risk alcohol consumption, the time to cirrhosis development is reduced in alcoholics with a relatively older age at onset of at-risk alcohol consumption, compared with those who started drinking at a younger age.7 Although the underlying mechanisms are not completely clear, many putative factors that can increase liver sensitivity to alcohol with age have been described including aging-related increased gut permeability which results in bacterial and pathogen-associated molecular patterns translocation and leads to Kupffer cells activation.6
Predisposition to progressive alcoholic chronic liver disease is strongly influenced by genetic heritability.1 Among all the genetic determinants of liver disease, the patatin-like phospholipase domain protein 3 (PNPLA3) rs738409 (I148M) variant has been strongly linked to hepatic damage at different levels (steatosis, necroinflammation, and fibrosis) in both nonalcoholic8–13 and alcoholic liver disease in several cross-sectional studies and meta-analyses.14–20 Additionally, we previously demonstrated, for the first time in a retrospective analysis of time-dependent data, that the PNPLA3 I148M variant is independently associated with a shorter time of cirrhosis development in heavy drinkers.7 PNPLA3 is a lipase that promotes the release of triglycerides from intracellular lipid droplets21,22 and of retinol from hepatic stellate cells.23 PNPLA3 I148M is a loss-of-function variant resulting in accumulation of triglycerides in hepatocytes, accumulation of retinyl palmitate in hepatic stellate cells,21–23 and in increased secretion of profibrotic proteins from hepatic stellate cells.24 Recently, a genome-wide association study performed on alcohol abusers with and without cirrhosis identified rs641738 C>T in the membrane bound O-acyltransferase domain containing 7 (MBOAT7) and rs58542926 (E167K) in the transmembrane-6 super-family member 2 (TM6SF2) as new loci associated with alcohol-related cirrhosis.25 MBOAT7 is an enzyme involved in the phospholipid acyl chain remodeling that transfers polyunsaturated fatty acids to lysophosphatidylinositol and other lysophospholipids.26 TM6SF2 encodes for a protein of unknown function involved in the lipid secretion from the liver.27,28 These two loci of susceptibility for alcoholic liver disease have been associated also with the entire spectrum of nonalcoholic fatty liver diseases.25,29–33 To date, the role of PNPLA3, TM6SF2, and MBOAT7 in liver disease onset and progression has been largely replicated making them the most widely recognized genetic determinant of both alcoholic and nonalcoholic liver disease.
Among other possible genetic predispositions to alcoholic cirrhosis, it has been shown that the rs2569190 (C-159T) variant in the promoter region of the cluster of differentiation 14 (CD14) gene is an independent risk factor for alcoholic cirrhosis34–38 and decompensated alcoholic cirrhosis,39 when compared to alcoholics, but the role of this polymorphism in the early stages of alcoholic liver disease is not clear.38 Gut-derived endotoxin/lipopolysaccharide intestinal absorption is increased by ethanol overuse and is a powerful Kupffer cell and inflammation activator in the liver via the CD14 endotoxin receptor.40
To date, no data are available on whether the TM6SF2, MBOAT7, and CD14 variants are associated with a time-dependent risk of alcoholic cirrhosis development.
Alcoholic cirrhosis development and progression is, additionally, strongly influenced by gender. Specifically, females are more susceptible to liver alcohol damage (ie, more damage for the same amount of ethanol consumed) than males.5,41,42 This is probably due also to the higher number of Kupffer cells per volume unit in female compared to male liver.43
Although females are more susceptible to liver alcohol damage, males have a higher prevalence of alcoholic cirrhosis, probably due to the higher amount of alcohol they drink compared to females.44,45 Thus, since alcoholic cirrhosis is far more prevalent in males and considering the gender-related differences of Kupffer cell density in the liver, we decided to perform our study only in males.
Our aim was to develop, for the first time, a risk score for alcoholic cirrhosis development over time based on combined clinical and genetic factors. We first investigated how genetic factors can interplay with constitutional and environmental features in a population of male at-risk drinkers with or without cirrhosis. Then, for the same population, we developed a gene-based risk score to predict the prevalence over time of alcoholic cirrhosis with at-risk alcohol consumption.
Materials and methods
Study population
A total of 988 consecutive Caucasian male individuals with alcohol disorder and at-risk alcohol consumption from the Department of Clinical Medicine, Policlinico Umberto I, Rome, Italy were recruited between 2005 and 2014. Specifically, 565 male patients from the initial study cohort previously described, which included both genders,7 and 423 additional patients recruited during a 4-year extension of the enrollment period were retrospectively examined. At admittance, patients underwent a detailed clinical examination and interview in order to collect accurate, reliable, and objective medical history about past and current health status and alcohol consumption as previously described.7
Briefly, at-risk alcohol consumption was defined as three or more alcohol units per day for at least 5 years and one unit of alcohol was defined as 12 g of ethanol. Lifetime drinking history (LDH) was used to calculate quantitative indices of alcohol consumption patterns, throughout the life span.46 In particular, the duration (in years) of at-risk alcohol consumption was calculated from the reported age at onset of at-risk drinking to the end of at-risk alcohol consumption assessed at the first visit or during subsequent follow-up in subjects with still active alcohol intake at first visit. The amount of daily alcohol intake was extrapolated from the LDH and expressed as the average number of alcohol units per day.
The possible presence of cirrhosis was also assessed and the time of cirrhosis diagnosis was reported using data from hospital discharge papers, medical records, physical examination, blood tests, imaging, and endoscopy. Specifically, cirrhosis diagnosis and its timing were based on liver biopsy or on the first occurrence of at least two of the following features: current or past cirrhosis complications (eg, ascites, variceal gastrointestinal bleeding, hepatic encephalopathy), the presence of hyperbilirubinemia, hypoalbuminemia, prolonged international normalized ratio, low platelet count, irregular liver surface at ultrasound/computed tomography, reduced portal vein flow at ultrasound, liver elastometry, gastroesophageal varices at endoscopy. In the absence of a pathological diagnosis or overt cirrhosis, individuals with positivity of only one of the above-mentioned parameters were excluded from the current analyses.
The dry weight to calculate body mass index (BMI) and the presence of diabetes at diagnosis of cirrhosis, or for those patients without a diagnosis of cirrhosis, at first visit were also reported. Patients were asked whether their body weight was stable (changes <5 kg) compared to that at age 25, at the first visit for noncirrhotic patients, and before the diagnosis of cirrhosis in cirrhotic patients. Blood samples were collected after an overnight fast and analyzed for biochemical parameters. DNA was collected from all the individuals.
Individuals with other etiological factors (n=234) of liver disease (ie, hepatitis B and C infection, autoimmune hepatitis, primary biliary cholangitis, primary sclerosing cholangitis, Wilson’s disease, hemochromatosis, Budd–Chiari syndrome), with incomplete LDH data (n=130), incomplete clinical/biochemical data to achieve a diagnosis of alcoholic cirrhosis and/or a defined age at cirrhosis diagnosis (n=132), lack of body weight stability (n=41), or with poor DNA quality (n=35) were excluded from the analyses. Thus, a total of 416 males with at-risk alcohol consumption were analyzed in the current report.
The study was conducted in accordance with the Declaration of Helsinki and approval was obtained from the local ethical committee “Azienda Umberto I.” Every patient provided written informed consent for participation.
Genotyping of PNPLA3 rs738409, CD14 rs2569190, TM6SF2 rs2569190, and MBOAT rs641738 variants
Buffy coat fraction was recovered from whole blood EDTA and DNA extraction was performed using QIAamp® DNA Blood Kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s instructions. DNA quantification and suitability for genotyping analysis were checked using the ΔΔCT method coamplifying a sample of known concentration with unknown samples for the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase.
PNPLA3 rs738409, CD14 rs2569190, TM6SF2 rs58542926, and MBOAT7 rs641738 variants were genotyped by TaqMan® genotyping assay (Thermo Fisher Scientific, Waltham, MA, USA). Primers and probes (FAM and VIC labeled) were directly supplied by Thermo Fisher Scientific (TaqMan). Post-PCR allelic discrimination was performed on a CFX384 Real-Time System (Bio-Rad Laboratories Inc, Hercules, CA, USA) by measuring allele-specific fluorescence. For all variants, genotyping was performed in triplicate with 100% concordance between replicates. After quality control, a total of 416 individuals were included in the study. The genotyping success rate for PNPLA3 rs738409, CD14 rs2569190, and TM6SF2 rs58542926 was ≥98%. Due to the small amount of DNA available for 34 samples, MBOAT7 rs641738 genotyping was performed in 382 out of 416 individuals (92%).
Statistical analyses
Variables are shown as medians and interquartile ranges. To evaluate the association of alcoholic cirrhosis with the different genetic variants in a cross-sectional fashion, genotype, allele, and categorical variable distributions were compared by chi-squared test. Group differences for continuous variables were evaluated by the Student’s t-test or the Mann–Whitney U test as appropriate, after assessment of normality by the Kolmogorov–Smirnov test. Multivariate binary logistic analyses were performed by introducing as covariates age, BMI, and the presence of diabetes at first visit as known cirrhosis cofactors. Time of progression to cirrhosis was examined by Kaplan–Meier estimates of cumulative incidence rates. The start of follow-up corresponded to onset of at-risk alcohol consumption. Follow-up time refers therefore to years of exposure to at-risk alcohol consumption. It corresponds to the number of years from age at onset of at-risk alcohol consumption to age at cirrhosis onset or final followup without cirrhosis, measuring hence the period of exposure to the pathogenic agent (ie, at-risk alcohol consumption). Both recessive and dominant models were assessed for each gene, and the most informative selected through minimization of the Akaike information criterion (AIC). Missing data were handled using pairwise complete case analysis.
Log-rank test was used to compare differences in cumulative incidence across PNPLA3 I148M and CD14 C159T genotypes. Univariate and multivariable Cox regression analyses were used to evaluate the risk of developing cirrhosis. Cox proportional hazards models were used to evaluate the effect of PNPLA3 I148M and CD14 C159T genotypes on cirrhosis incidence after adjusting for confounding factors such as age and those present since the beginning of at-risk alcohol consumption (ie, BMI, amount of daily alcohol consumption, and age at onset of at-risk alcohol consumption). The final multivariate Cox regression model was chosen via forward-stepwise selection targeting the AIC. A score to predict alcoholic cirrhosis incidence in 36 years from the onset of at-risk alcohol consumption was calculated by rounding logarithms of HRs at the multivariate analysis, and evaluated using time-dependent receiver operating characteristic (ROC) curves. Also, a score based on the four gene variants under consideration was evaluated, and then discarded since it did not give any improvement in terms of area under the curve (AUC) over the more parsimonious score (which is based on only two of the variants considered).
The score was internally validated by bootstrapping the sample 1,000 times in order to check that the estimated AUC was within the nonparametric bootstrap CI. Analyses were carried out using R version 3.3.3 (R Development Core Team, Vienna, Austria) and P-values <0.05 were considered as statistically significant.
Results
Demographic, clinical, and alcohol consumption characteristics of the study population
A total of 416 male individuals with at-risk alcohol consumption were included in the current analyses. Individuals were adults (median age of 47 years), with median age at onset of at-risk alcohol consumption of 22 years, median daily alcohol consumption of 14 alcohol units, and median duration of at-risk alcohol consumption of 21 years.
The study population was stratified based on the presence or absence of cirrhosis into two groups: alcoholic cirrhosis (n=90) and noncirrhotic alcohol consumers (NCAC; n=326). Individuals from the NCAC group were younger (45 vs 55 years) and, as expected, healthier with lower BMI, lower diabetes prevalence, and lower aspartate aminotransferase level compared to individuals from the alcoholic cirrhosis group (Table 1). There were no differences between the two groups in the age at onset of at-risk alcohol consumption and the duration of at-risk alcohol consumption, whereas individuals from the NCAC group had higher daily alcohol consumption compared to those with cirrhosis (15 vs 12 alcohol units per day, P=0.018).
Table 1.
Demographic and clinical characteristics according to the presence or absence of cirrhosis
| AC | NCAC | P-value | |
|---|---|---|---|
|
| |||
| n | 90 | 326 | |
| Agea (years) | 55 (49–61) | 45 (37–52) | <0.001 |
| BMI (kg/m2) | 26.9 (24.3–30.1) | 25.0 (23.1–27.8) | <0.001 |
| Diabetes,a n (%) | 28 (31.1) | 17 (5.7) | <0.001 |
| Age at onset of at-risk alcohol consumption (years) | 23 (18–40) | 22 (17–29) | 0.092 |
| Daily alcohol consumption (unit) | 12 (8–16) | 15 (10–20) | 0.018 |
| Duration of at-risk alcohol consumption (years) | 24 (13–32) | 20 (12–30) | 0.364 |
| INRa | 1.36 (1.20–1.60) | 0.98 (0.92–1.02) | <0.001 |
| Serum bilirubin (mg/dL)a | 2.01 (1.17–4.11) | 0.56 (0.39–0.75) | <0.001 |
| Serum creatinine (mg/dL)a | 0.83 (0.70–1.00) | 0.80 (0.70–0.90) | 0.226 |
| Serum AST (U/L)a | 44 (30–55) | 32 (20–55) | <0.001 |
| Serum ALT (U/L)a | 29 (22–46) | 31 (21–52) | 0.326 |
| Age at diagnosis of cirrhosis (years) | 48 (44–59) | – | – |
| Child–Pugh class, n (%)b | |||
| A | 31 (34.40) | – | – |
| B | 40 (44.40) | – | – |
| C | 19 (21.10) | – | – |
| MELD scoreb | 13.94 (8.95–17.61) | – | – |
| PNPLA3, n (%) | |||
| II | 20 (22.20) | 150 (46.00) | |
| IM | 47 (52.20) | 143 (43.90) | |
| MM | 23 (25.60) | 33 (10.10) | <0.001 |
| CD14, n (%) | |||
| CC | 12 (13.30) | 82 (25.20) | |
| CT | 45 (50.00) | 170 (52.10) | |
| TT | 33 (36.70) | 74 (22.70) | 0.008 |
| TM6SF2, n (%)c | |||
| EE | 71 (83.53) | 277 (86.00) | |
| EK | 11 (12.94) | 44 (13.70) | |
| KK | 3 (3.53) | 1 (0.30) | 0.028 |
| MBOAT7, n (%)d | |||
| CC | 17 (22.10) | 86 (28.20) | |
| CT | 45 (58.40) | 163 (53.40) | |
| TT | 15 (19.50) | 56 (18.40) | 0.555 |
Notes: Data are expressed as median and interquartile range or as proportions. Genotype and allele frequencies were in Hardy–Weinberg equilibrium (P>0.05).
Data refer to the first visit.
Child–Pugh classes and MELD score were assessed only in individuals with cirrhosis.
407 patients (AC, n=85 and NCAC, n=322).
382 patients (AC, n=77 and NCAC, n=305). P-values <0.05 shown in bold were considered statistically significant.
Abbreviations: AC, alcoholic cirrhosis; ALT, alanine transferase; AST, aspartate transferase; BMI, body mass index; CD14, cluster of differentiation 14; INR, international normalized ratio; MBOAT7, membrane bound O-acyltransferase domain containing 7; MELD, model for end-stage liver disease; NCAC, noncirrhotic alcohol consumer; PNPLA3, patatin-like phospholipase domain containing 3; TM6SF2, transmembrane-6 superfamily member 2; n, number.
PNPLA3 I148M, CD14 C159T, and TM6SF2 E167K frequency is higher in individuals with cirrhosis
As shown in Table 1, the frequency of PNPLA3 M allele, CD14 T allele, and TM6SF2 K allele was higher in the alcoholic cirrhosis group than in the NCAC group without cirrhosis (P<0.001; P=0.008 and P=0.028, respectively). No differences between the two groups were observed for the MBOAT7 T allele frequency.
To assess the cross-sectional independent association between PNPLA3 M allele, CD14 T allele, or TM6SF2 K allele and alcoholic cirrhosis, we performed separate multivariate binary logistic analyses for each variant. For all the multivariate analyses, age at first visit, BMI, and the presence of diabetes were included as covariates. PNPLA3, CD14, and TM6SF2 were all identified as independent factors associated with alcoholic cirrhosis (Table 2). Specifically, the risk of alcoholic cirrhosis was increased >2-fold for each PNPLA3 M allele, and ~2-fold increased for each CD14 T allele. For TM6SF2, the risk of cirrhosis was increased only for homozygosity (KK). TM6SF2 was not associated with alcoholic cirrhosis when a dominant model was run (ie, wild-type vs heterozygous + homozygous).
Table 2.
Genetic polymorphism associations with alcoholic cirrhosis at multivariate binary logistic regression analyses
| Multivariate analysis | |||
|---|---|---|---|
| OR | 95% CI | P-value | |
| PNPLA3 allele M | 2.153 | 1.466–3.162 | <0.001 |
| IM vs II | 2.004 | 1.082–3.710 | 0.027 |
| MM vs II | 4.708 | 2.178–10.177 | <0.001 |
| CD14 allele T | 1.706 | 1.160–2.508 | 0.007 |
| CT vs CC | 1.620 | 0.795–3.530 | 0.175 |
| TT vs CC | 2.894 | 1.308–6.403 | 0.009 |
| TM6SF2 allele K | 1.465 | 0.761–2.819 | 0.253 |
| EK vs EE | 1.005 | 0.456–2.217 | 0.990 |
| KK vs EE | 21.284 | 1.294–349.985 | 0.032 |
Notes: In all multivariate binary logistic regression models, one for each genetic polymorphism, the following variables were included as covariates: age at first visit, body mass index, and diabetes. P-values <0.05 shown in bold were considered statistically significant.
Abbreviations: CD14, cluster of differentiation 14; PNPLA3, patatin-like phospholipase domain containing 3; TM6SF2, transmembrane-6 superfamily member 2.
PNPLA3 I148M and CD14 C159T genetic polymorphisms are independent risk factors for alcoholic cirrhosis development
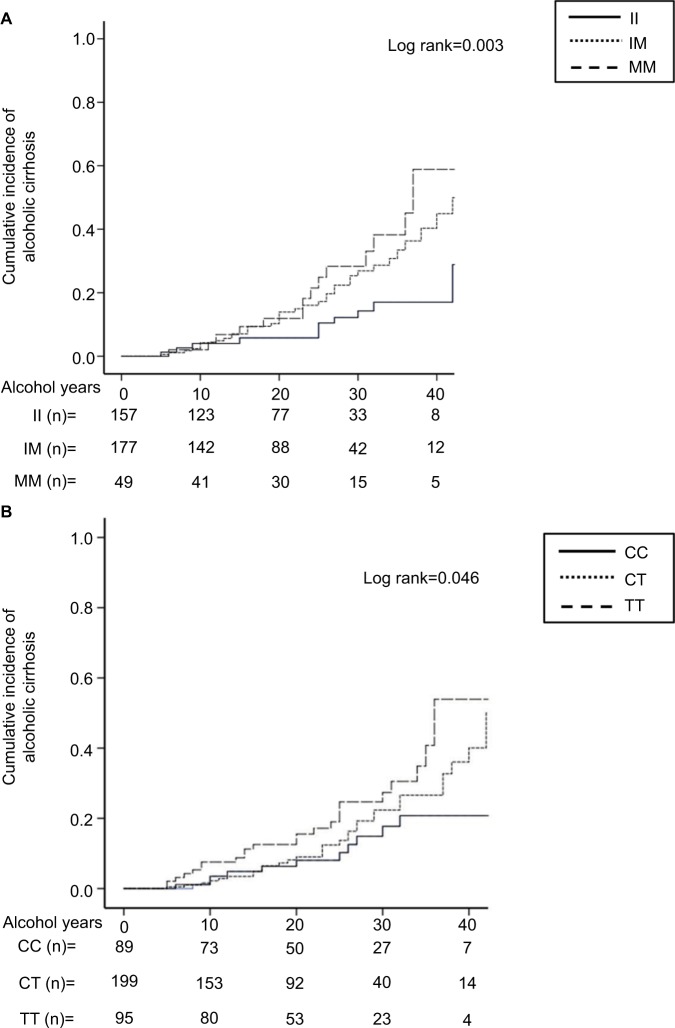
As shown in Figure 1A and B, carriers of PNPLA3 or CD14 mutant alleles showed an increased incidence of cirrhosis compared to wild-type individuals (log-rank 0.003 and 0.046, respectively). No association between the development of alcoholic cirrhosis and TM6SF2 or MBOAT7 was detected (log-rank P-value: 0.296 and 0.276, respectively).
Figure 1.
Cumulative incidence of alcoholic cirrhosis in individuals according to (A) PNPLA3 I148M genotype and (B) CD14 C159T genotype. Alcohol years indicate the duration of at-risk alcohol consumption (daily intake of ≥3 alcohol units). It corresponds to the number of years from age at onset of at-risk alcohol consumption to age at cirrhosis onset or final follow-up without cirrhosis, measuring hence the period of exposure to the pathogenic agent. The number at the bottom indicates the number of individuals at risk at the following time points: 0, 10, 20, 30, and 40 years.
Abbreviations: PNPLA3 II, individuals with two 148I alleles; MM, individuals with two 148M alleles; IM, heterozygotes; CD14 CC, individuals with two 159C alleles; TT, individuals with two 159T alleles; CT, heterozygotes.
To test the time-dependent risk of developing alcoholic cirrhosis, we performed univariate and multivariate Cox regression analyses.
At the univariate level, BMI, age at onset of at-risk alcohol consumption, PNPLA3 M allele heterozygosity, homozygosity or carriage, and CD14 T allele homozygosity or carriage were found to be risk factors for alcoholic cirrhosis development (Table 3). No association with alcoholic cirrhosis development was detected for the TM6SF2 and the MBOAT7 variants. In particular, TM6SF2 was not associated even under a dominant genetic model (ie, wild-type vs heterozygous + homozygous). When all the four variants were considered together, the risk to develop alcoholic cirrhosis was higher in carriers of the four mutant alleles. However, the risk conferred by having all the four variants was not higher than considering only PNPLA3 M and CD14 T alleles in the same model.
Table 3.
Risk of developing alcoholic cirrhosis: univariate Cox regression
| HR | 95% CI | P-value | |
|---|---|---|---|
| Agea | 0.988 | 0.961–1.016 | 0.395 |
| BMI (kg/m2) | 1.072 | 1.026–1.120 | 0.002 |
| Daily alcohol consumption (unit) | 0.983 | 0.955–1.012 | 0.251 |
| Age at onset of at-risk alcohol consumption (years) | 1.114 | 1.087–1.142 | <0.001 |
| PNPLA3 allele M | 1.737 | 1.250–2.414 | 0.001 |
| IM vs II | 2.177 | 1.202–3.945 | 0.010 |
| MM vs II | 3.052 | 1.520–6.127 | 0.002 |
| CD14 allele T | 1.503 | 1.080–2.092 | 0.016 |
| CT vs CC | 1.839 | 0.937–3.607 | 0.076 |
| TT vs CC | 2.406 | 1.176–4.923 | 0.016 |
| TM6SF2 allele K | 1.257 | 0.795–1.990 | 0.328 |
| EK vs EE | 1.075 | 0.571–2.024 | 0.822 |
| KK vs EE | 2.204 | 0.685–7.093 | 0.185 |
| MBOAT7 allele T | 1.271 | 0.937–1.723 | 0.123 |
| CT vs CC | 1.261 | 0.749–2.121 | 0.383 |
| TT vs CC | 1.615 | 0.880–2.966 | 0.122 |
| PNPLA3 allele M + CD14 allele T | 1.537 | 1.225–1.926 | <0.001 |
| PNPLA3 allele M + CD14 allele T + TM6SF2 allele K + MBOAT7 allele T | 1.428 | 1.192–1.711 | <0.001 |
Notes: Numbers of individuals (and proportion) in each of the gene allele groups, stratified by AC/NCAC: PNPLA3 II 20/150 (22/46), IM 47/143 (52/44), MM 23/33 (26/10); CD14 CC 12/82 (13/25), CT 45/170 (50/52), TT 33/74 (37/23); TM6SF2 EE 71/277 (84/86), EK 11/44 (13/14), KK 3/1 (3.5/0.3); MBOAT7 CC 17/86 (22/28), CT 45/163 (58/53), TT 15/56 (20/18).
Data refer to the first visit. P-values <0.05 shown in bold were considered statistically significant.
Abbreviations: AC, alcoholic cirrhosis; BMI, body mass index; CD14, cluster of differentiation 14; MBOAT7, membrane bound O-acyltransferase domain containing 7; NCAC, non-cirrhotic alcohol consumer; PNPLA3, patatin-like phospholipase domain containing 3; TM6SF2, transmembrane-6 superfamily member 2.
Next, to test whether the association between risk alleles and alcoholic cirrhosis development was independent from other risk factors, we performed two separate multivariate Cox regression analyses using the sum of PNPLA3 M allele and CD14 T allele, or the sum of PNPLA3 M, CD14 T, TM6SF2 K, and MBOAT7 T alleles.
For each Cox regression multivariate analysis, the following variables were included as covariates: age at first visit, BMI, daily alcohol consumption, and age at onset of at-risk alcohol consumption (Table 4). The model with the sum of PNPLA3 M allele and CD14 T allele showed a significant HR of 1.599 that did not improve in the model with the sum of PNPLA3 M, CD14 T, TM6SF2 K, and MBOAT7 T alleles (HR=1.405). Younger age at first visit, higher BMI, and older age at onset of at-risk alcohol consumption were also independent risk factors for alcoholic cirrhosis development in both models.
Table 4.
Risk of developing alcoholic cirrhosis: multivariate Cox regression
| PNPLA3 + CD14 model | PNPLA3 + CD14 + TM6SF2 + MBOAT7 model | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Agea | 0.659 | 0.589–0.738 | <0.001 | 0.679 | 0.604–0.763 | <0.001 |
| BMI (kg/m2) | 1.077 | 1.028–1.128 | 0.002 | 1.075 | 1.024–1.127 | 0.003 |
| Daily alcohol consumption (unit) | 0.990 | 0.962–1.018 | 0.480 | 0.995 | 0.969–1.023 | 0.735 |
| Age at onset of at-risk alcohol consumption (years) | 1.596 | 1.428–1.785 | <0.001 | 1.543 | 1.375–1.731 | <0.001 |
| PNPLA3 allele M + CD14 allele T | 1.599 | 1.254–2.039 | <0.001 | – | – | – |
| PNPLA3 allele M + CD14 allele T + TM6SF2 allele K + MBOAT7 allele T | – | – | – | 1.405 | 1.159–1.702 | 0.001 |
Notes:
Data refer to the first visit. In both multivariate Cox regression models the following variables, other than the genetic variants, were included as covariates: age at first visit, BMI, daily alcohol consumption, and age at onset of at-risk alcohol consumption. P-values <0.05 shown in bold were considered statistically significant.
Abbreviations: BMI, body mass index; CD14, cluster of differentiation 14; MBOAT7, membrane bound O-acyltransferase domain containing 7; PNPLA3, patatin-like phospholipase domain containing 3; TM6SF2, transmembrane-6 superfamily member 2.
Based on these results, time-dependent ROC curves for alcoholic cirrhosis incidence were calculated using PNPLA3 and CD14 allele variants, age at onset of at-risk alcohol consumption, and BMI in order to develop a predictive score for alcoholic cirrhosis development within 36 years from the onset of at-risk alcohol consumption.
The best predictive score was:
(age at onset of at-risk alcohol consumption × 0.1) + (number of CD14 T allele*1) + (number of PNPLA3 M allele*1) + (BMI × 0.1).
The threshold of 7.27 performed well in predicting the risk of alcoholic cirrhosis development, with a sensitivity of 70.1% and a specificity of 78.7%, whereas the AUC was 75%. The negative predictive value and the positive predictive value were 90.2% and 48.6%, respectively. Calibration was assessed by means of Grønnesby and Borgan test, which confirmed a good calibration (P=0.182). The reported AUC was validated internally via the bootstrap. The bootstrap-estimated 95% CI was 64%–85%, and the reported AUC was well within the CI. Fictitious examples of the use of the score are shown in Table 5 for the purpose of illustration.
Table 5.
Development of a prediction risk score for alcoholic cirrhosis in 10 fictitious patients
| Patient A | Patient B | Patient C | Patient D | Patient E | Patient F | Patient G | Patient H | Patient I | Patient J | |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of PNPLA3 and/or CD14 risk alleles | 0 | 0 | 0 | 1 | 1 | 2 | 2 | 3 | 3 | 4 |
| Age at onset of at-risk alcohol consumption | 15 | 30 | 45 | 15 | 30 | 15 | 30 | 15 | 30 | 15 |
| BMI | 28 | 35 | 28 | 35 | 35 | 35 | 24 | 25 | 18 | 18 |
| Score | 4.3 | 6.5 | 7.3 | 6 | 7.5 | 7 | 7.4 | 7 | 7.8 | 7.3 |
| Predicted AC | No | No | Yes | No | Yes | No | Yes | No | Yes | Yes |
Notes: The AC prediction score is calculated for 10 fictitious subjects. When the subjects do not have any PNPLA3 or CD14 risk allele, the AC prediction score is below the cutoff (>7.27) for cirrhosis development if either the age at onset of at-risk alcohol consumption is relatively young or the BMI is in the normal range (<25). When one or two risk alleles are present, the AC prediction score is below the cutoff for cirrhosis development if the age at onset of at-risk alcohol consumption is young and the BMI is below the overweight cutoff. In the case of three or four risk alleles, the subjects are always at-risk of cirrhosis development.
Abbreviations: AC, alcoholic cirrhosis; BMI, body mass index; CD14, cluster of differentiation 14; PNPLA3, patatin-like phospholipase domain containing 3.
Discussion
Chronic alcohol-related liver disease is complex, multifactorial, and multistage with interindividual variations in the progression and outcomes. Alcoholic cirrhosis development depends on environmental and genetic factors and the interplay between them is under investigation.47
In the present study, PNPLA3 I148M and CD14 C159T variants have been identified as two genetic components independently associated with the time-dependent risk of developing alcoholic cirrhosis in an Italian (Caucasian) population of male at-risk drinkers.
The association between the PNPLA3 I148M variant and alcoholic cirrhosis has been extensively reported in cross-sectional studies.14,17,19,20 In a previous retrospective study, we demonstrated for the first time that the PNPLA3 I148M variant is also independently associated with the incidence of alcoholic cirrhosis over time.7 In the present study, we confirmed the time-dependent association between PNPLA3 I148M variant and the incidence of alcoholic cirrhosis analyzing a larger population of male alcoholics.
CD14 endotoxin receptor is an essential mediator in inflammatory signaling and Kupffer cell activation in the liver.40 The CD14 C159T variant has been associated with a gain of function, an increased Kupffer cell activation, and inflammatory response.41 The cross-sectional association between the CD14 C159T variant and alcoholic cirrhosis has been previously described.34–36 In the present study, in agreement with previous studies, using a cross-sectional analysis in male heavy drinkers, we confirmed that the CD14 C159T variant is independently associated with alcoholic cirrhosis prevalence. Additionally, here we demonstrated, for the first time, that the CD14 C159T variant is independently associated with the development of alcoholic cirrhosis over time.
A recent genome-wide association study on alcoholic liver disease identified new loci of susceptibility to alcoholic cirrhosis on the TM6SF2 and MBOAT7 genes.25 The TM6SF2 (E167K) variant has been widely associated with liver disease prevalence.25,29,30,48 In the present study, we confirmed that homozygosity for the minor allele is independently associated with alcoholic cirrhosis using a cross-sectional analysis. However, in the Cox regression analysis, the TM6SF2 variant was not associated with the time-dependent risk of cirrhosis development. The lack of association in the time-dependent analysis could be due to the low prevalence of homozygosity and insufficient sample size.
The MBOAT7 variant has been previously associated with both alcoholic and nonalcoholic liver disease.25,32,33 However, we did not find any association between the MBOAT7 variant and alcoholic cirrhosis in both the cross-sectional and longitudinal analyses. This could be due to a lack of power for this variant. The MBOAT7 variant was the latest polymorphism described to be relevant in this research field and we did not have enough DNA to analyze it in 8% of our study population. This reduced the number of individuals successfully genotyped from 416 to 382. Additionally, the effect size of the MBOAT7 variant on liver disease has been previously shown to be lower compared to PNPLA3 and TM6SF2 variants,32 meaning that a larger number of individuals for each group is needed to detect the association.
On the basis of the genetic associations we found with the time-dependent development of alcoholic cirrhosis, in the present study we developed a score with good accuracy (AUC 75%) to predict the risk of alcoholic cirrhosis onset in 36 years from the beginning of at-risk alcohol consumption, encompassing either genetic or environmental factors. These include the CD14 C159T and PNPLA3 I148M variants, BMI, and age at onset of at-risk alcohol consumption. We did not add the other genetic variants in the score because a similar performance was obtained with an extended score. The final score we proposed is parsimonious, therefore, both in the statistical sense (being based on a minimal set of predictors) and in the practical sense (since it does not require assessment of variants of two additional candidate genes).
Concerning environmental factors that can increase the risk of alcoholic cirrhosis, in the present study, we found that BMI also is an independent predictor of alcoholic cirrhosis subsequent development. Higher BMI is a risk factor for hepatic steatosis and steatohepatitis and is considered as a cofactor favoring alcoholic cirrhosis onset.5,49–51
Finally, our results confirmed, as in our previous report,7 that a relatively older age at onset of at-risk alcohol consumption is independently associated with alcoholic cirrhosis susceptibility and helps to predict disease development. It is probably referable to the lower resilience of the liver to handle the same levels and patterns of alcohol consumption as in previous life years.6,34,35
Our study is limited by its retrospective design. However, prospective studies on this topic would be too long and unfeasible. A second limitation is that, since in most cases it would have been impossible to know the time of diabetes occurrence, in the present study we did not include the presence of diabetes in the multivariate analyses aimed to investigate associations with the incidence of alcoholic cirrhosis over time and in the prediction score for alcoholic cirrhosis development. As in other genetic studies performed in at-risk drinker populations,34,52 our score was developed for males and does not permit generalization of our findings to the entire population of at-risk drinkers. However, in our previous study, 76% of the entire population of at-risk drinkers consecutively enrolled were males, and this makes our score useful in most of the cases. Although we performed an internal validation, the absence of an independent verification cohort is another limitation of the study and the applicability and reproducibility of the study should be validated in a multicenter cohort. Additionally, very recently a loss-of-function variant in HSD17B13 (rs72613567) was associated with a reduced risk of chronic liver disease and of progression from steatosis to steatohepatitis.53 The inclusion of this protective variant in the risk score could improve the performance of similar risk scores in the future.
In conclusion, identifying genetic and environmental candidate factors that confer susceptibility to alcoholic cirrhosis development could potentially inform the clinician on patient management. Our study not only evaluated the weight of the single independent risk factors in alcoholic cirrhosis onset, but also, for the first time, these factors were combined to build a practical and an easy-to-use score. It would be useful, for clinicians, to predict, since the first visit, the risk over time of alcoholic cirrhosis in male patients with at-risk alcohol consumption and to select a population with a high risk to develop cirrhosis in order to plan a patient-tailored therapy (ie, pharmacotherapy for alcohol dependence, dietician counseling, etc).
Future aims are to expand the population study patients to female gender and to different ethnic groups to better delineate how the above-mentioned factors variate and to eventually adjust consistently the alcoholic cirrhosis predicting score.
Acknowledgments
The abstract of this paper was presented at the EASL Annual Meeting in 2017 as a poster with interim findings. The poster’s abstract was published in “Poster Abstracts” in the Journal of Hepatology (2017, volume 66, issue 1, supplement, page S348; doi.org/10.1016/S0168-8278(17)31030-9). This research was supported by grants of Sapienza, University of Rome and by “Wilhelm and Martina Lundgren Science” Fund.
Footnotes
Author contributions
AF contributed to the statistical analysis. FF, MM, and SGC contributed to the analysis and interpretation of data and drafted the manuscript. RMM, AMo, and MAB performed sample processing and sequencing. FF, AMa, EP, LP, ADS, FA, CR, MR, and MLA collected sample and clinical information. FF, RMM, AF, and SGC contributed to the study concept and design. MLA, MC, and SGC supervised and oversaw the study. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work
Disclosure
The authors reports no conflicts of interest in this work.
References
- 1.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141(5):1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcellin P, Kutala BK. Liver diseases: a major, neglected global public health problem requiring urgent actions and large-scale screening. Liver Int. 2018;38(Suppl 1):2–6. doi: 10.1111/liv.13682. [DOI] [PubMed] [Google Scholar]
- 3.Masarone M, Rosato V, Dallio M, et al. Epidemiology and natural history of alcoholic liver disease. Rev Recent Clin Trials. 2016;11(3):167–174. doi: 10.2174/1574887111666160810101202. [DOI] [PubMed] [Google Scholar]
- 4.Heslin KC, Elixhauser A, Steiner CA. Identifying in-patient costs attributable to the clinical sequelae and comorbidities of alcoholic liver disease in a national hospital database. Addiction. 2017;112(5):782–791. doi: 10.1111/add.13702. [DOI] [PubMed] [Google Scholar]
- 5.Day CP. Genes or environment to determine alcoholic liver disease and non-alcoholic fatty liver disease. Liver Int. 2006;26(9):1021–1028. doi: 10.1111/j.1478-3231.2006.01323.x. [DOI] [PubMed] [Google Scholar]
- 6.Magdaleno F, Blajszczak C, Nieto N. Key events participating in the pathogenesis of alcoholic liver disease. Biomolecules. 2017;7(4):E9–E23. [Google Scholar]
- 7.Burza MA, Molinaro A, Attilia ML, et al. PNPLA3 I148M (rs738409) genetic variant and age at onset of at-risk alcohol consumption are independent risk factors for alcoholic cirrhosis. Liver Int. 2014;34(4):514–520. doi: 10.1111/liv.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan X, Waterworth D, Perry JR, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83(4):520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotronen A, Johansson LE, Johansson LM, et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52(6):1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- 11.Hotta K, Yoneda M, Hyogo H, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:172. doi: 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51(4):1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 13.Romeo S, Sentinelli F, Cambuli VM, et al. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. J Hepatol. 2010;53(2):335–338. doi: 10.1016/j.jhep.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 14.Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42(1):21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 15.Stickel F, Hampe J, Trépo E, Datz C, Romeo S. PNPLA3 genetic variation in alcoholic steatosis and liver disease progression. Hepatobiliary Surg Nutr. 2015;4(3):152–160. doi: 10.3978/j.issn.2304-3881.2014.11.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chamorro AJ, Torres JL, Mirón-Canelo JA, González-Sarmiento R, Laso FJ, Marcos M. Systematic review with meta-analysis: the I148M variant of patatin-like phospholipase domain-containing 3 gene (PNPLA3) is significantly associated with alcoholic liver cirrhosis. Aliment Pharmacol Ther. 2014;40(6):571–581. doi: 10.1111/apt.12890. [DOI] [PubMed] [Google Scholar]
- 17.Salameh H, Raff E, Erwin A, et al. PNPLA3 gene polymorphism is associated with predisposition to and severity of alcoholic liver disease. Am J Gastroenterol. 2015;110(6):846–856. doi: 10.1038/ajg.2015.137. [DOI] [PubMed] [Google Scholar]
- 18.Shen JH, Li YL, Li D, Wang NN, Jing L, Huang YH. The rs738409 (I148M) variant of the PNPLA3 gene and cirrhosis: a meta-analysis. J Lipid Res. 2015;56(1):167–175. doi: 10.1194/jlr.M048777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausch V, Peccerella T, Lackner C, et al. Primary liver injury and delayed resolution of liver stiffness after alcohol detoxification in heavy drinkers with the PNPLA3 variant I148M. World J Hepatol. 2016;8(35):1547–1556. doi: 10.4254/wjh.v8.i35.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stickel F, Buch S, Nischalke HD, et al. Genetic variants in PNPLA3 and TM6SF2 predispose to the development of hepatocellular carcinoma in individuals with alcohol-related cirrhosis. Am J Gastroenterol. 2018 doi: 10.1038/s41395-018-0041-8. [DOI] [PubMed] [Google Scholar]
- 21.Pirazzi C, Adiels M, Burza MA, et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol. 2012;57(6):1276–1282. doi: 10.1016/j.jhep.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 22.Pingitore P, Pirazzi C, Mancina RM. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim Biophys Acta. 2014;18414(4):574–580. doi: 10.1016/j.bbalip.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Pirazzi C, Valenti L, Motta BM, et al. PNPLA3 has retinylpalmitate lipase activity in human hepatic stellate cells. Hum Mol Genet. 2014;23(15):4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pingitore P, Dongiovanni P, Motta BM, et al. PNPLA3 overexpression results in reduction of proteins predisposing to fibrosis. Hum Mol Genet. 2016;25(23):5212–5222. doi: 10.1093/hmg/ddw341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buch S, Stickel F, Trépo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47(12):1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 26.Gijón MA, Riekhof WR, Zarini S, Murphy RC, Voelker DR. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283(44):30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smagris E, Gilyard S, Basuray S, Cohen JC, Hobbs HH. Inactivation of Tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J Biol Chem. 2016;291(20):10659–10676. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Hare EA, Yang R, Yerges-Armstrong LM, et al. TM6SF2 rs58542926 impacts lipid processing in liver and small intestine. Hepatology. 2017;65(5):1526–1542. doi: 10.1002/hep.29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dongiovanni P, Petta S, Maglio C, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61(2):506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 30.Kozlitina J, Smagris E, Stender S, et al. Exomewide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46(4):352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mancina RM, Sentinelli F, Incani M, et al. Transmembrane-6 super-family member 2 (TM6SF2) E167K variant increases susceptibility to hepatic steatosis in obese children. Dig Liver Dis. 2016;48(1):100–101. doi: 10.1016/j.dld.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European Descent. Gastroenterology. 2016;150(5):1219–1230. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donati B, Dongiovanni P, Romeo S, et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. 2017;7(1):4492. doi: 10.1038/s41598-017-04991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Järveläinen HA, Orpana A, Perola M, Savolainen VT, Karhunen PJ, Lindros KO. Promoter polymorphism of the CD14 endotoxin receptor gene as a risk factor for alcoholic liver disease. Hepatology. 2001;33(5):1148–1153. doi: 10.1053/jhep.2001.24236. [DOI] [PubMed] [Google Scholar]
- 35.Martins A, Cortez-Pinto H, Machado M, et al. Are genetic polymorphisms of tumour necrosis factor alpha. interleukin-10, CD14 endotoxin receptor or manganese superoxide dismutase associated with alcoholic liver disease? Eur J Gastroenterol Hepatol. 2005;17:1099–1104. doi: 10.1097/00042737-200510000-00014. [DOI] [PubMed] [Google Scholar]
- 36.Meiler C, Muhlbauer M, Johann M, et al. Different effects of a CD14 gene polymorphism on disease outcome in patients with alcoholic liver disease and chronic hepatitis C infection. World J Gastroenterol. 2005;11(38):6031–6037. doi: 10.3748/wjg.v11.i38.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nezi V, Deutsch M, Gazouli M, et al. Polymorphisms of the CD14 genes are associated with susceptibility to alcoholic liver disease in Greek patients. Alcohol Clin Exp Res. 2013;37(2):244–251. doi: 10.1111/j.1530-0277.2012.01925.x. [DOI] [PubMed] [Google Scholar]
- 38.Zeng T, Zhang C-L, Han X-Y, Zhao S, Xie K-Q. Association between CD14−159C>T polymorphisms and the risk for alcoholic liver disease. Eur J Gastroenterol Hepatol. 2013;25(10):1–1189. doi: 10.1097/MEG.0b013e3283612ff1. [DOI] [PubMed] [Google Scholar]
- 39.Campos J, Gonzalez-Quintela A, Quinteiro C, et al. The -159C/T polymorphism in the promoter region of the CD14 gene is associated with advanced liver disease and higher serum levels of acute-phase proteins in heavy drinkers. Alcohol Clin Exp Res. 2005;29(7):1206–1213. doi: 10.1097/01.alc.0000171977.25531.7a. [DOI] [PubMed] [Google Scholar]
- 40.Zeng T, Zhang CL, Xiao M, Yang R, Xie KQ. Critical roles of Kupffer cells in the pathogenesis of alcoholic liver disease: from basic science to clinical trials. Front Immunol. 2016;7:538–552. doi: 10.3389/fimmu.2016.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eagon PK. Alcoholic liver injury: influence of gender and hormones. World J Gastroenterol. 2010;16(11):1377–1384. doi: 10.3748/wjg.v16.i11.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirpich IA, Mcclain CJ, Vatsalya V, et al. Liver injury and endotoxemia in male and female alcohol-dependent individuals admitted to an alcohol treatment program. Alcohol Clin Exp Res. 2017;41(4):747–757. doi: 10.1111/acer.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcos R, Correia-Gomes C, Miranda H, Carneiro F. Liver gender dimorphism: insights from quantitative morphology. Histol Histopathol. 2015;30(12):1431–1437. doi: 10.14670/HH-11-648. [DOI] [PubMed] [Google Scholar]
- 44.Wilsnack RW, Wilsnack SC, Kristjanson AF, Vogeltanz-Holm ND, Gmel G. Gender and alcohol consumption: patterns from the multinational GENACIS project. Addiction. 2009;104(9):1487–1500. doi: 10.1111/j.1360-0443.2009.02696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellinger JL, Shedden K, Winder GS, et al. The high burden of alcoholic cirrhosis in privately insured persons in the United States. Hepatology. 2018;68(3):872–882. doi: 10.1002/hep.29887. [DOI] [PubMed] [Google Scholar]
- 46.Skinner HA, Sheu WJ. Reliability of alcohol use indices. The lifetime drinking history and the MAST. J Stud Alcohol. 1982;43(11):1157–1170. doi: 10.15288/jsa.1982.43.1157. [DOI] [PubMed] [Google Scholar]
- 47.Stickel F, Moreno C, Hampe J, Morgan MY. The genetics of alcohol dependence and alcohol-related liver disease. J Hepatol. 2017;66(1):195–211. doi: 10.1016/j.jhep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 48.Sookoian S, Castaño GO, Scian R, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61(2):515–525. doi: 10.1002/hep.27556. [DOI] [PubMed] [Google Scholar]
- 49.Yen YH, Chang KC, Tsai MC, et al. Elevated body mass index is a risk factor associated with possible liver cirrhosis across different etiologies of chronic liver disease. J Formos Med Assoc. 2018;117(4):268–275. doi: 10.1016/j.jfma.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 50.Romeo S, Sentinelli F, Dash S, et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes. 2010;34(1):190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 51.Naveau S, Giraud V, Borotto E, et al. Excess weight risk factor for alcoholic liver disease. Hepatology. 1997;25(1):108–111. doi: 10.1002/hep.510250120. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Guo T, Yang F, et al. Single-nucleotide rs738409 polymorphisms in the PNPLA3 gene are strongly associated with alcoholic liver disease in Han Chinese males. Hepatol Int. 2018;12(5):429–437. doi: 10.1007/s12072-018-9889-3. [DOI] [PubMed] [Google Scholar]
- 53.Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378(12):1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]