Abstract
Imbalance of protein homeostasis (proteostasis) is known to cause cellular malfunction, cell death, and diseases. Elaborate regulation of protein synthesis and degradation is one of the important processes in maintaining normal cellular functions. Protein degradation pathways in eukaryotes are largely divided into proteasome-mediated degradation and lysosome-mediated degradation. Proteasome is a multisubunit complex that selectively degrades 80% to 90% of cellular proteins. Proteasome-mediated degradation can be divided into 26S proteasome (20S proteasome + 19S regulatory particle) and free 20S proteasome degradation. In 1980, it was discovered that during ubiquitination process, wherein ubiquitin binds to a substrate protein in an ATP-dependent manner, ubiquitin acts as a degrading signal to degrade the substrate protein via proteasome. Conversely, 20S proteasome degrades the substrate protein without using ATP or ubiquitin because it recognizes the oxidized and structurally modified hydrophobic patch of the substrate protein. To date, most studies have focused on protein degradation via 26S proteasome. This review describes the 26S/20S proteasomal pathway of protein degradation and discusses the potential of proteasome as therapeutic targets for cancer treatment as well as against diseases caused by abnormalities in the proteolytic system.
Keywords: Proteasome, Ubiquitination, Oxidative stress, Protein degradation, Cancer
INTRODUCTION
Protein quality control refers to the regulation of intracellular misfolded proteins that are produced through refolding, degradation, or aggregation because of various stress conditions.1 Adequate balance between normal protein synthesis and abnormal protein degradation is essential to maintain cellular proteostasis and ensure normal cellular functions.2 Various stress factors, such as diseases, aging, and UV exposure, increase the misfolding and mutation of proteins.3,4 This misfolded protein is refolded through holdase and foldase activity of the chaperone protein to ensure its normal function as a native protein.4,5 However, when misfolded proteins are accumulated beyond repair, they may not only cause cytotoxicity but also lead to abnormal cellular status. Therefore, rapid degradation of these proteins via protein degradation pathways is essential.6
The regulatory systems for intracellular protein degradation can be largely divided into the ubiquitin (Ub)–proteasome system and autophagy–lysosome system.7 Proteasome is a highly conserved multicatalytic protease found in eukaryotes (from yeast to human) and present in both nucleus and cytoplasm. It is a multisubunit complex that cleaves and selectively degrades the peptide bonds of abnormal or short-lived protein.8 According to the presence of Ub/ATP, proteasome may be classified into 26S proteasome and 20S proteasome.9 p53, a tumor suppressor called “guardian of the genome”, is a labile protein and is known to be a typical substrate protein that is degraded via two types of proteasomes.10 When ubiquitination occurs in p53 through E3 Ub ligase MDM2, 26S proteasome recognizes Ub to initiate degradation. p53 degradation through 20S proteasome occurs by default.11 In normal cells, p53 is rapidly degraded via 26S proteasome and 20S proteasome, thereby maintaining low expression. However, p53 is mostly mutated in various cancer cells, such as lung cancer, colon cancer, breast cancer, and liver cancer.12 Because mutant p53 does not form MDM2-p53 feedback loop by inhibiting MDM2-dependent ubiquitination or inhibiting the transcriptional activity of MDM2, it exhibits high expression status by interfering with proteasome-mediated degradation of p53.13,14 Mutant p53 eventually induces migration, proliferation, and drug-resistant of cancer cells through the gain of oncogenic functions.12
In addition, many proteins, such as p27, NF-κB or TGF-β receptor, are degraded through proteasomes similar to p53 and are known to regulate tumorigenesis in a variety of cancers. In breast cancer, the degradation of p27 by Ub-proteasome pathway promotes the proliferation and migration of cancer cells by regulating the G1/S transition via activation of cyclin E/Cdk2.15–17 Proteasomal degradation of IκB, which inhibits NF-κB, activates NF-κB, thereby increasing the proliferation and drug resistance of cancer cells in multiple myeloma (MM). Bortezomib, a proteasome inhibitor (PI), inhibits growth and increases apoptosis of MM by inactivating NF-κB through inhibition of IκB degradation.18,19 The E3 Ub ligase Smurf1, which is known to inhibit cell growth and migration of lung cancer, suppresses TGF-β signaling through Smad7-dependent TGFβ-RI ubiquitination.20,21
There are several previous as well as ongoing studies on proteasome-mediated degradation pathway from the discovery of Ub-proteasome degradation in late-1970 to the awarding of Nobel Prize in Chemistry for the discovery of Ub-dependent protein degradation in 2004. This review discusses the types of protein degradation systems, the types and functions of proteasomes, and the importance of regulation of protein degradation in cancer.
PROTEIN DEGRADATION SYSTEM: LYSOSOME VERSUS PROTEASOME
1. Lysosome
Lysosome mediates autophagy-lysosomal degradation via acidic hydrolase that degrades intra- and intercellular cytoplasmic material or organelles. Autophagy (self-eating, generally referred to as macroautophagy) is evolutionarily conserved in eukaryotes to deliver extra-/intracellular protein (which should be sequentially degraded) or cell organelle to lysosome.22 Unlike Ub-proteasome degradation, the cellular components and organelle transported through autophagy are randomly degraded upon binding to lysosomes. Autophagy is largely divided into the following types: 1) macroautophagy, 2) microautophagy, and 3) chaperone-mediated autophagy (CMA).23 Firstly, macroautophagy involves the formation of double-membraned autophagosome (AP) that covers cytosolic components and transports it to lysosome to degrade components. Among the three types of autophagy processes, only macroautophagy forms AP to degrade intracellular matter.24 Eighteen of the 31 autophagy-related (ATG) proteins (ATG1–10, ATG12–14, ATG16–18, ATG29, and ATG31) are proteins involved in AP formation, generally through vesicle nucleation, elongation, AP maturation, lysosome fusion, and degradation in a sequential manner.25–27 Secondly, microautophagy is directly decomposed into lysosome through the invagination of cytosolic components by the lysosome.28 Thirdly, as for CMA, the proteins damaged by oxidative stress exposes KFERQ motif; HSC70, the chaperone protein, recognizes this motif and transports it to lysosome for degradation.29 Resultantly, autophagy-lysosome protein degradation is a non-selective protein degradation process, whereas Ub-proteasome protein degradation involves selective protein degradation.30
2. Proteasome
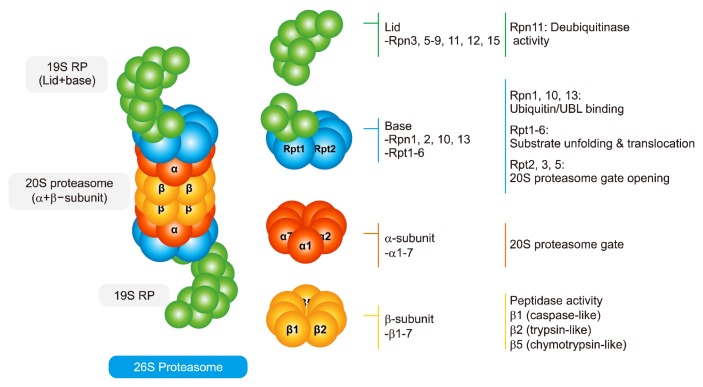
Proteasome present in both nucleus and cytoplasm degrades 80%–90% of intracellular proteins.31 26S proteasome (approximately 2,000 kDa) is a major proteasome, consisting of 20S proteasome core particle (approximately 700 kDa) with peptidase activity and two 19S regulatory particles (RP, PA700; approximately 900 kDa) (Fig. 1).32,33 Two groups of 7 α-subunits (α1–α7) and two groups of 7 β-subunits (β1–β7) at the outer of 20S proteasome form a cylindrical structure. The α-subunit acts as a gate to 20S proteasome, and β-subunit has three peptidase activities, namely, caspase-like (β1, acidic amino acids), trypsin-like (β2, basic amino acids), and chymotrypsin-like (β5, hydrophobic amino acids) activities, to directly degrade proteins.9,34,35 19S RP comprises a lid (nine Rpn3, 5–9, 11, 12, and 15) and base (four Rpn 1, 2, 10, 13, and six Rpt 1–6). Ub binds to damaged protein, and recognizes Rpn1, 10 and 13 as the Ub binding site of proteasome to bind with proteasome (recruitment). The recognized damaged protein is unfolded by Rpt 1–6, the heterohexameric AAA-ATPase. When the α-subunit gate of 20S proteasome is opened by Rpt2, 3, and 5, the damaged protein is translocated and degraded toward 20S proteasome (commitment).31,36 Rpn 11 separates Ub from the damaged protein being degraded so that Ub can be reused (deubi-qutination). Proteins are degraded into 3–25 peptides through 20S proteasome, and finally degraded into amino acids by peptidase to be reused where needed.
Figure 1.
Schematic diagram of proteasome structure and ubiquitination process. The 26S proteasome is a complex of multiple subunits consisting of 20S proteasomes (two α-subunits and two β-subunits) and two 19S regulatory particles (RP). Seven a-subunits act as a gate to 20S proteasome, and three subunits (β1, β2, and β5) of the seven β-subunits have protease activity to degrade substrate proteins. Rpn1, 10, and 13 in the lid region recognize and bind with ubiquitin bound to substrate protein, resulting in unfolding and translocation of substrate protein by Rpt1–6 with ATPase activity. Rpt2, 3, and 5 open 20S proteasome gates to allow the degradation of substrate protein via 20S proteasome.
PROTEASOME DIVERSITY
1. Hybrid proteasome
The 20S proteasome is known as the proteolytic machinery involved in the degradation of oxidized protein by oxidative stress (free 20S “core” proteasome, to be discussed later). Because there are two α-subunits in 20S proteasome, two RP can bind to them. In addition to 19S RP, other 20S proteasome regulators are 11S (called PA28 or REG) and PA200 (Blm10 in yeast).37 Depending on the type of the RP that binds, it can form a variety of hybrid proteasomes (i.e., 19S-20S-11S or 19S-20S-PA200) different from 26S or 20S. Through this, it intervenes in not only protein degradation but also immune response and DNA repair.38,39
2. Immunoproteasome
When there is a viral, bacterial, or fungal infection, host cells secrete interferon, a type of cytokine. Interferon acts as an “alarm signal” to protect surrounding cells; in particular, IFN-γ substitutes three β-subunits (β1, β2, and β5) that have peptidase activities of 20S proteasome with β1i, β2i, and β5i, respectively. The substituted 20S proteasome binds with two 11S RP (PA28), referred to as immunoproteasome (11S-20S-11S), which degrades antigen present inside the cell into antigenic peptides. The degraded peptide binds with major histocompatibility complex (MHC) class I via endoplasmic reticulum and Golgi, and is presented on the cell surface. The T cell receptor of CD8+ cytotoxic T cells recognizes the peptide bound to MHC class I and initiates an immune response.40,41
26S PROTEASOME PATHWAY (UBIQUITIN/ATP-DEPENDENT)
1. Ubiquitin-proteasome system
If proteasome degrades all proteins, along with damaged proteins, without discrimination, it will cause another problem in the cells. To resolve such issues, Ub (8.5kDa), a type of small and highly conserved protein composed 76 amino acids, is expressed in eukaryotes.42 Glycine 76 present in carboxyl terminal of Ub binds with the lysine residue of substrate proteins through isopeptide bond, a type of covalent bond.43 Ub bound to the substrate protein also attaches one or multiple Ub with methionine (Met, M), the first residue of Ub, or seven lysine residues (Lysine, K / K6, K11, K27, K29, K33, K48, or K63) of Ub.42 Eventually, it forms mono-, multi-, or poly-Ub chains (also known as ubiquitination or ubiquitylation). Furthermore, it has been recently reported to cause heterogeneous polyubiquitination (single-branch or multiple-branch chain), linear polyubiquitination; ubiquitination occurs in the residues of serine (Ser, S), cysteine (Cys, C), or threonine (Thr, T).44 Depending on the Lys residue of Ub involved in ubiquitination, it is known to mediate various functions (such as DNA repair, NF-κB activation, and gene expression). Among them, ubiquitination through Lys 48 of Ub is known to be essential for protein degradation process.45 Finally, Ub selectively binds to damaged proteins, thereby acting as a degrading signal for 26S proteasome to degrade substrate protein.
2. Ubiquitination
Ubiquitination is a sequential ATP-dependent process mediated by three enzymes (E1–E3) as post-translational modification occurring in eukaryotic cells.46 When Ub is activated by ATP, the C-terminal carboxyl group of Ub binds to Cys residue of E1 enzyme (Ub-activating enzyme, 2). Activated Ub that binds to E1 is transferred to the E2 enzyme (Ub-conjugating enzymes, approximately 40 in number). The Ub attached to E2 binds with E3 enzyme (Ub ligase enzyme, approximately 600), which in turn binds with abnormal or unnecessary substrate protein to eventually transport Ub to damaged protein. Particularly, E3 has specificity for substrate proteins.46 Recently, a new ubiquitination enzyme, E4, has been reported to be involved in the ubiquitination of proteins as an Ub chain assembly factor. p300 is involved in p53 polyubiquitination as E4 enzyme, and Ube4b, another E4 enzyme, is known to be involved in p53 degradation by increasing the ubiquitination chain of p53 through MDM2.47,48 Finally, ubiquitinated proteins are degraded through 26S proteasome.
It has been previously reported that only one E1 exists in cells as UBE1 (Ub-activating enzyme E1). In 2007, another E1 enzyme, UBE1L2 (Ub-activating enzyme E1-like 2, also known UBA6), was discovered; thus, it is currently known that two E1 enzymes exist. When comparing the specificity of transferring Ub to E2 using two E1 enzymes (UBE1 and UBA6), the two E1 enzymes showed the same efficiency, but the specificity of E2 was different.49
E3 may be divided into two types: a type with Really Interesting New Gene (RING) finger domain and another type with Homologous to the E6-AP Carboxyl Terminus (HECT) domain. The RING type serves as a bridge between E2 attached with Ub and substrate proteins and increase the activity of E2. Finally, Ub bound to E2 directly binds with substrate protein, allowing the substrates to degrade through proteasome. Unlike the RING type, the HECT type binds to Ub attached to E2 and forms thioester intermediate with Cys residue of E3. Subsequently, E3 ensures that Ub is directly bound to the substrate protein to be degraded.50 E3 protein expression is low because it self-degrades through auto-ubiquitination.51,52 p53 is a well-known substrate protein that is degraded through the 26S proteasome, which is ubiquitinated by the RING type E3 Ub ligase, MDM2, and degraded through the 26S proteasome.53
20S PROTEASOME PATHWAY (UBIQUITIN/ATP-INDEPENDENT)
1. Changes in the protein degradation system by oxidative stress
At low concentration, reactive oxygen species (ROS) acts as signaling molecule in cells to mediate cell proliferation, enzyme activation, protein synthesis, immune system, or differentiation.54 However, at high concentration, ROS oxidizes proteins and causes cytotoxicity by increasing intracellular oxidative stress. To overcome oxidative stress, cells eliminate ROS using antioxidant enzymes, such as peroxiredoxins, catalase, and glutathione peroxidase, which are the primary defensive system. However, when such stress is excessive, it causes dysfunction of antioxidant enzyme, resulting in oxidation and structural changes of normal proteins.54 In addition, under oxidative stress, the adapter protein extracellular mutant 29 (Ecm29) of 26S proteasome separates 26S proteasome into 20S proteasome and 19S RP, thereby inhibiting 26S proteasome activity as well as protein degradation through ubiquitination. Finally, oxidized proteins are accumulated in the cell without being normally degraded.55
2. Degradation of oxidized protein
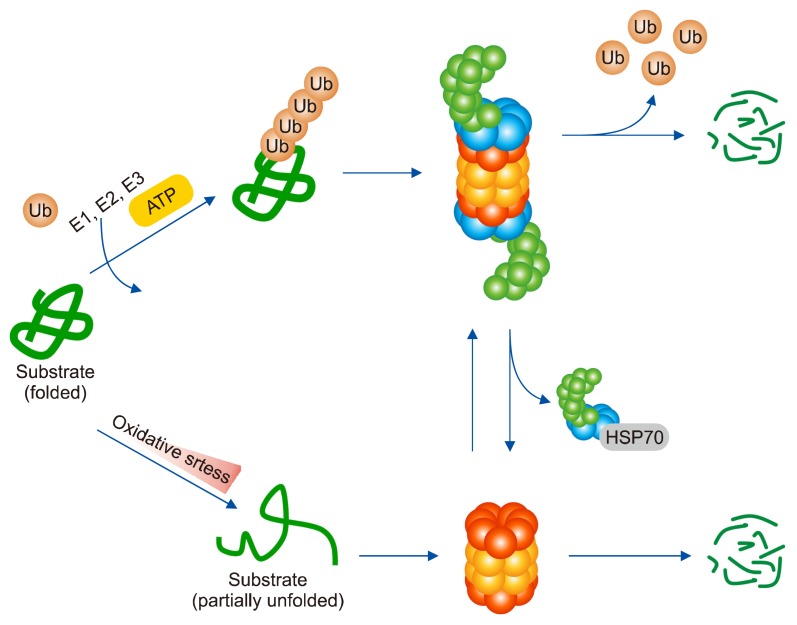
Previous studies have shown that free 20S proteasome is more effective in eliminating oxidized substrate protein caused by ROS than 26S proteasome.56 The 19S RP separated from 20S proteasome by oxidative stress is held in the chaperone HSC70 and can be assembled into 26S proteasome by binding with 20S proteasome again after approximately 3 to 5 hours (Fig. 2).57 Oxidized protein degradation through 20S proteasome is a Ub/ATP-independent degradation process that does not involve ATP and Ub, because there is no bound 19S RP.58 Unlike 26S proteasome that recognizes Ub, 20S proteasome recognizes the hydrophobic region of the protein whose structure has changed by oxidative stress as the degrading signal and degrades the oxidized protein.37
Figure 2.
Cross-talk between 26S and 20S proteasome under oxidative stress. The 26S proteasome degrades not only abnormally folded or damaged proteins but also fully folded proteins through ubiquitination. Under oxidative stress, however, the 19S regulatory particle that constitutes the 26S proteasome is dissociated from the 20S proteasome. The dissociated 20S proteasome mediates oxidized protein degradation. 19S regulatory particle is held by chaperone HSP70, and over time it binds to the 20S proteasome and is reassembled into 26S proteasome. Ub, ubiquitin.
REGULATION OF PROTEIN DEGRADATION IN CANCER
If an abnormality occurs in the regulation of protein degradation, normal proteins will be degraded and/or abnormal proteins will not be degraded, resulting in proteasome-related diseases, such as neurodegenerative diseases, cardiac dysfunction, and cancer.59–61 Some examples of the regulations of degradation of key proteins that are associated with cancer are given below (Fig. 3).
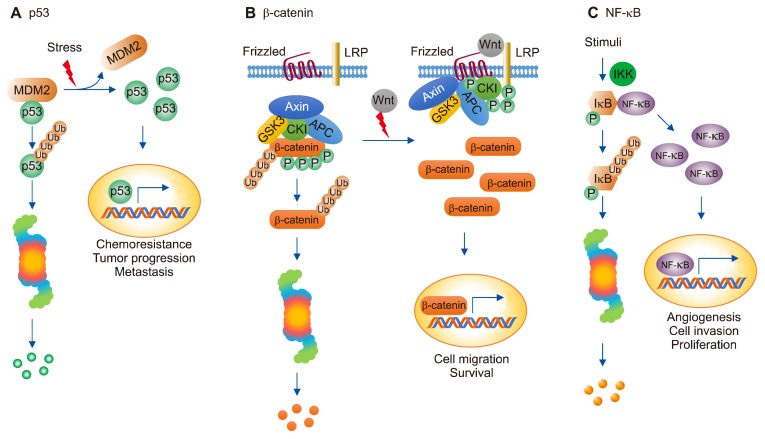
Figure 3.
Regulation of protein degradation in cancer. p53 (A) β-catenin (B) and NF-κB (C) play an important role in regulating signal transduction pathways through proteasome-mediated protein degradation of itself or its interacting partners in cancer. LRP, lipoprotein receptor related protein.
1. p53
The tumor suppressor protein p53, called the “guardian of the genome” in 1980, regulates cell cycle arrest, DNA repair, angiogenesis, and apoptosis.62 p53 increases the expression of E3 Ub ligase MDM2, and MDM2 maintains the autoregulatory feedback loop that ubiquitinates p53.63 Consequently, p53 exists in low levels in the cell after being degraded via 26S proteasome (Fig. 3A). However, it is known that about 50% to 70% of p53 is mutated in cancer cells.64 As a result, the binding between MDM2 and p53 is reduced, and the mutated p53 cannot be degraded through 26S proteasome, resulting in a high level of the mutant p53 protein. Thus, the cell cycle of the cancer cells continues, and chemoresistance appears to promote tumor progression and metastasis.12
2. β-catenin
In the absence of Wnt, β-catenin binds with Axin complex (Axin; scaffolding protein, APC; tumor suppressor, CK1; casein kinase 1 and GSK3; glycogen synthase kinase 3), a type of destruction complex, and is constantly phosphorylated by CK1 and GSK3. Phosphorylated β-catenin is ubiquitinated by β-Trcp, the E3 Ub ligase, and degraded via proteasome. Conversely, in the presence of Wnt, the Fz (Frizzled) family receptor forms receptor complex with low-density lipoprotein receptor related protein 5/6 (LRP5/6) and recruits Dvl, which inhibits the activity of Axin complex through LRP5/6 phosphorylation and Axin recruitment. Next, β-catenin cannot be degraded; it goes into the nucleus, binds with the transcription factor, and mediates cell migration and survival (Fig. 3B).65,66 Axin degrades APC through the Ub-proteasomal degradation pathway. In some colon cancer cells, the role of Axin as a tumor suppressor disappears and is known to play a role as oncogene because APC is absent or truncated.67,68
3. NF-κB
The NF-κB, composed of five families of transcription factors (p50, p52, p65/RelA, c-Rel, and RelB) plays an important role in cell growth, inflammation, immunity, and apoptosis.69,70 NF-κB binds with IκB and exists in the cytoplasm in a stable form. However, when the N-terminal serine residues of IκB are phosphorylated by IκB kinase, the NF-κB-IκB binding is dissociated, and IκB is ubiquitinated.71 Finally, IκB is degraded through 26S proteasome, and NF-κB separated from IκB is translocated to the nucleus and increases the expression of anti-apoptotic gene to increase angiogenesis, cell invasion, oncogenesis, and proliferation of cancer cells, or increase the transcription of the genes that mediate pro-inflammatory response to secrete cytokines for regulating inflammation-associated cancer (Fig. 3C).72 In the absence of stimulation, NF-κB in the nucleus activates the transcription of NFKBIA gene encoding IκB and is regulated through NF-κB/IκB negative feedback loop.73
PROTEASOME INHIBITORS AS CANCER TREATMENT TARGET
Because proteasome is a multisubunit multicatalytic proteinase complex that degrades many proteins in the cells, several studies in the past two decades have been conducted on the inhibitors that target proteasome. PI inhibit proteasome activity by reversibly or irreversibly binding to the catalytic threonine residue of β5 (chymotrypsin-like) or β1 (caspase-like) subunit with 20S proteasome activity to prevent substrate protein degradation, thus indicating a potential for anti-cancer drugs (Table 1).74 For example, PI stabilize IκB to prevent dissociation of NF-κB and IκB and the translocation of NF-κB into the nucleus, thereby reducing cancer cell proliferation and increasing apoptosis. Moreover, PI inhibit p21 and p27 degradation, thereby increasing the expressions of the two proteins; leading to cell cycle arrest or apoptosis; and increasing the expression of pro-apoptotic factors p53, Bax, and NOXA, which results in DNA repair or apoptosis of cancer cells. Finally, PI reportedly activates JNK through phosphorylation to increase apoptosis of cancer cells.74
Table 1.
Properties of proteasome inhibitors
| Name | Target | Type | Route | Disease | Phase | Ref. No. |
|---|---|---|---|---|---|---|
| Bortezomib | β5 > β1 | Reversible | IV, SQ | MM | Approved | 74, 75, 77 |
| Carfilzomib | β5 | Irreversible | IV | MM | Approved | 74, 75, 77 |
| Ixazomib | β5 > β1 | Reversible | Oral, IV | MM | Approved | 74, 75, 76, 77 |
| Marizomib | β5 > β2 > β1 | Irreversible | Oral, IV | MM, leukemia | Not approved | 74, 75, 77 |
| Delanzomib | β5 > β1 | Reversible | Oral, IV | MM, solid tumors | Not approved | 74, 75, 77 |
| Oprozomib | β5 | Irreversible | Oral | MM, solid tumors | Not approved | 74, 75, 77 |
IV, intravenous; SQ, subcutaneous; MM, multiple myeloma.
To date, three PIs have been approved by the USA Food and Drug Administration (FDA) for use in clinical cancer therapy. Bortezomib (intravenous or subcutaneous injections), a first-generation PI, is a reversible drug that binds to β5 and β1 (proteasome dissociation half-life: 110 minutes). First approved by the FDA in 2003, it is used in patients with newly diagnosed MM, relapsed/refractory MM (RRMM), mantle cell lymphoma, and advanced solid tumors. The second-generation PI carfilzomib (intravenous injections) was approved by the FDA in 2012; it irreversibly binds with β5 of 20S proteasome. It is used to treat RRMM. Finally, ixazomib (oral administration) is a reversible PI (proteasome dissociation half-life: 8 minutes), approved by the FDA as the first approved drug for oral administration in 2015. Ixazomib is also used for RRMM owing to its high absorptiveness and long terminal phase half-life (9.5 days).75,76 However, bortezomib is known to cause general side effects, such as nausea, fatigue, diarrhea, and rashes, as well as severe dose-limiting toxicity, such as peripheral neuropathy. In addition, it is known to activate mitochondrial apoptosis pathway.77,78 Carfilzomib is more effective than bortezomib; however, 18% of patients with MM treated with this drug demonstrated cardiovascular adverse events, such as arrhythmia, hypertension, and heart attacks.79 Currently, PI has various side effects and cytotoxicity, which limits its use in treatments.
CONCLUSION
Stress cause from various factors not only impairs protein degradation mechanism but also causes protein damages, modification, and misfolded protein accumulation, which eventually accumulate as intracellular inclusion bodies in the cells. Malfunctioning proteins ultimately lead to cancer, neurological disorders, and viral infection, etc. therefore, it is important to degrade abnormal proteins as quickly as possible for the cells to survive. Proteasome, one of the regulatory systems for intracellular protein degradation, is directly involved in the regulation of protein degradation, and more studies should be conducted on this in the future. Starting from bortezomib, which was first approved in 2003 by the FDA as the therapeutic PI, many inhibitors have been studied and approved; however, the major problem is that they exhibit various side effects because they directly inhibit the activity of the proteasome. Several studies are ongoing to resolve this issue.
Proteasome is generated and assembled with subunits of RP by transcription factors nuclear respiratory factors 1/2 (Nrf1/2) and Rpn4.80 The threonine residue at the N-terminal of proteasome β-subunit acts as a nucleophile to attack the substrate, forming the acyl-enzyme intermediate and hydrolyzing the substrate protein.81 Consequently, substrate protein is cleaved into two products (peptides), and the sequential reaction degrades substrate proteins into peptides. When Ub binds to the terminated proteasome, p62 binds to form AP, which gets degraded via lysosome. In this review, the proteasome pathway has been discussed separately from the lysosome pathway; however, the two pathways are mutually supplementary because inhibition of proteasome activity leads to lysosome positioning in the aggresome, resulting in cross-talk that promotes protein degradation.
Ub-dependent degradation through 26S proteasome is commonly known, but it is not always the case. Ornithine decarboxylase is known to be Ub-independent/26S proteasome-dependent degradation with antizyme as the mediator. Recently, it has been reported that 26S proteasome recognizes an adaptor protein having a UBA or UBL domain linked to Ub instead of directly recognizing the Ub chain, resulting in 26S proteasome-dependent degradation.58,82 p53 is regulated by both Ub/ATP-dependent or Ub/ATP-independent degradation processes. The 20S proteasome-mediated degradation of p21 is promoted by regulators such as PCNA or 14–3–3τ.53,83,84 In conclusion, it is expected that new therapeutic targets will be developed through a variety of studies of regulatory factors for many other substrate proteins that undergo proteasome-mediated degradation in cancer.
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1A2 B6009042).
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 2011;3: a004374. 10.1101/cshperspect.a004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta 2014;1843:182–96. 10.1016/j.bbamcr.2013.06.031 [DOI] [PubMed] [Google Scholar]
- 3.Kikis EA, Gidalevitz T, Morimoto RI. Protein homeostasis in models of aging and age-related conformational disease. Adv Exp Med Biol 2010;694:138–59. 10.1007/978-1-4419-7002-2_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta 2004;1739:5–25. 10.1016/j.bbadis.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 5.Herrmann J, Soares SM, Lerman LO, Lerman A. Potential role of the ubiquitin-proteasome system in atherosclerosis: aspects of a protein quality disease. J Am Coll Cardiol 2008;51:2003–10. 10.1016/j.jacc.2008.02.047 [DOI] [PubMed] [Google Scholar]
- 6.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med (Berl) 2003;81:678–99. 10.1007/s00109-003-0464-5 [DOI] [PubMed] [Google Scholar]
- 7.Nedelsky NB, Todd PK, Taylor JP. Autophagy and the ubiquitin-proteasome system: collaborators in neuroprotection. Biochim Biophys Acta 2008;1782:691–9. 10.1016/j.bbadis.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanayama H, Tanaka K, Aki M, Kagawa S, Miyaji H, Satoh M, et al. Changes in expressions of proteasome and ubiquitin genes in human renal cancer cells. Cancer Res 1991;51:6677–85. [PubMed] [Google Scholar]
- 9.Ben-Nissan G, Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014;4: 862–84. 10.3390/biom4030862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asher G, Shaul Y. p53 proteasomal degradation: poly-ubiquitination is not the whole story. Cell Cycle 2005;4:1015–8. 10.4161/cc.4.8.1900 [DOI] [PubMed] [Google Scholar]
- 11.Asher G, Reuven N, Shaul Y. 20S proteasomes and protein degradation “by default”. Bioessays 2006;28:844–9. 10.1002/bies.20447 [DOI] [PubMed] [Google Scholar]
- 12.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25:304–17. 10.1016/j.ccr.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol 2007;27:8284–95. 10.1128/MCB.00050-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res 2003;1:1001–8. [PubMed] [Google Scholar]
- 15.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, et al. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 1995;269:682–5. 10.1126/science.7624798 [DOI] [PubMed] [Google Scholar]
- 16.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev 1997;11: 1464–78. 10.1101/gad.11.11.1464 [DOI] [PubMed] [Google Scholar]
- 17.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin Cancer Biol 2003;13: 41–7. 10.1016/S1044-579X(02)00098-6 [DOI] [PubMed] [Google Scholar]
- 18.Sánchez-Serrano I. Success in translational research: lessons from the development of bortezomib. Nat Rev Drug Discov 2006;5:107–14. 10.1038/nrd1959 [DOI] [PubMed] [Google Scholar]
- 19.Chauhan D, Hideshima T, Mitsiades C, Richardson P, Anderson KC. Proteasome inhibitor therapy in multiple myeloma. Mol Cancer Ther 2005;4:686–92. 10.1158/1535-7163.MCT-04-0338 [DOI] [PubMed] [Google Scholar]
- 20.Li H, Xiao N, Wang Y, Wang R, Chen Y, Pan W, et al. Smurf1 regulates lung cancer cell growth and migration through interaction with and ubiquitination of PIPKIγ. Oncogene 2017;36:5668–80. 10.1038/onc.2017.166 [DOI] [PubMed] [Google Scholar]
- 21.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem 2001;276:12477–80. 10.1074/jbc.C100008200 [DOI] [PubMed] [Google Scholar]
- 22.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ 2005;12 Suppl 2:1542–52. 10.1038/sj.cdd.4401765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation path-way with a central role in health and disease. Biochim Biophys Acta 2009;1793:664–73. 10.1016/j.bbamcr.2008.07.014 [DOI] [PubMed] [Google Scholar]
- 24.Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists: mechanisms and experimental approaches. Redox Biol 2015;4:242–59. 10.1016/j.redox.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007;9:1102–9. 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- 26.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 2010;30:1049–58. 10.1128/MCB.01344-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura S, Yoshimori T. New insights into autophagosome-lysosome fusion. J Cell Sci 2017;130:1209–16. 10.1242/jcs.196352 [DOI] [PubMed] [Google Scholar]
- 28.Uttenweiler A, Schwarz H, Mayer A. Microautophagic vacuole invagination requires calmodulin in a Ca2+-independent function. J Biol Chem 2005;280:33289–97. 10.1074/jbc.M506086200 [DOI] [PubMed] [Google Scholar]
- 29.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 2004;15:4829–40. 10.1091/mbc.e04-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 2008;4:141–50. 10.4161/auto.5190 [DOI] [PubMed] [Google Scholar]
- 31.Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell 2017;169:792–806. 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams J. The development of proteasome inhibitors as anti-cancer drugs. Cancer Cell 2004;5:417–21. 10.1016/S1535-6108(04)00120-5 [DOI] [PubMed] [Google Scholar]
- 33.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol 2006;4:e267. 10.1371/journal.pbio.0040267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 2009;85:12–36. 10.2183/pjab.85.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackburn C, Gigstad KM, Hales P, Garcia K, Jones M, Bruzzese FJ, et al. Characterization of a new series of non-covalent protea-some inhibitors with exquisite potency and selectivity for the 20S beta5-subunit. Biochem J 2010;430:461–76. 10.1042/BJ20100383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grice GL, Nathan JA. The recognition of ubiquitinated proteins by the proteasome. Cell Mol Life Sci 2016;73:3497–506. 10.1007/s00018-016-2255-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raynes R, Pomatto LC, Davies KJ. Degradation of oxidized proteins by the proteasome: distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol Aspects Med 2016;50:41–55. 10.1016/j.mam.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci 2008;9:826–38. 10.1038/nrn2499 [DOI] [PubMed] [Google Scholar]
- 39.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol 2005;15:27–33. 10.1016/j.tcb.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat Rev Immunol 2012;12:813–20. 10.1038/nri3339 [DOI] [PubMed] [Google Scholar]
- 41.Groettrup M, Kirk CJ, Basler M. Proteasomes in immune cells: more than peptide producers? Nat Rev Immunol 2010;10:73–8. 10.1038/nri2687 [DOI] [PubMed] [Google Scholar]
- 42.Callis J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 2014;12:e0174. 10.1199/tab.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park CW, Ryu KY. Cellular ubiquitin pool dynamics and homeostasis. BMB Rep 2014;47:475–82. 10.5483/BMBRep.2014.47.9.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiq-uitin-based signals for proteasomal degradation. J Cell Sci 2012; 125:539–48. 10.1242/jcs.093567 [DOI] [PubMed] [Google Scholar]
- 45.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 2009;10:755–64. 10.1038/nrm2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolch W, Pitt A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat Rev Cancer 2010;10:618–29. 10.1038/nrc2900 [DOI] [PubMed] [Google Scholar]
- 47.Wu H, Leng RP. UBE4B, a ubiquitin chain assembly factor, is required for MDM2-mediated p53 polyubiquitination and degradation. Cell Cycle 2011;10:1912–5. 10.4161/cc.10.12.15882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 2003;300:342–4. 10.1126/science.1080386 [DOI] [PubMed] [Google Scholar]
- 49.Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 2007;447:1135–8. 10.1038/nature05902 [DOI] [PubMed] [Google Scholar]
- 50.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci 2012;125: 531–7. 10.1242/jcs.091777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Bie P, Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ 2011;18:1393–402. 10.1038/cdd.2011.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranaweera RS, Yang X. Auto-ubiquitination of Mdm2 enhances its substrate ubiquitin ligase activity. J Biol Chem 2013;288: 18939–46. 10.1074/jbc.M113.454470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulikov R, Letienne J, Kaur M, Grossman SR, Arts J, Blattner C. Mdm2 facilitates the association of p53 with the proteasome. Proc Natl Acad Sci U S A 2010;107:10038–43. 10.1073/pnas.0911716107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol 2014;24:R453–62. 10.1016/j.cub.2014.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Chemmama IE, Yu C, Huszagh A, Xu Y, Viner R, et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J Biol Chem 2017; 292:16310–20. 10.1074/jbc.M117.803619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shringarpure R, Grune T, Davies KJ. Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell Mol Life Sci 2001;58:1442–50. 10.1007/PL00000787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grune T, Catalgol B, Licht A, Ermak G, Pickering AM, Ngo JK, et al. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic Biol Med 2011;51:1355–64. 10.1016/j.freeradbiomed.2011.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orlowski M, Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch Biochem Biophys 2003;415:1–5. 10.1016/S0003-9861(03)00197-8 [DOI] [PubMed] [Google Scholar]
- 59.Dantuma NP, Bott LC. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front Mol Neurosci 2014;7:70. 10.3389/fnmol.2014.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mearini G, Schlossarek S, Willis MS, Carrier L. The ubiquitin-proteasome system in cardiac dysfunction. Biochim Biophys Acta 2008;1782:749–63. 10.1016/j.bbadis.2008.06.009 [DOI] [PubMed] [Google Scholar]
- 61.Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: therapeutic implications. Eur J Cancer 2004; 40:2217–29. 10.1016/j.ejca.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 62.Meng X, Franklin DA, Dong J, Zhang Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res 2014;74:7161–7. 10.1158/0008-5472.CAN-14-1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993;7:1126–32. 10.1101/gad.7.7a.1126 [DOI] [PubMed] [Google Scholar]
- 64.Ferraiuolo M, Verduci L, Blandino G, Strano S. Mutant p53 protein and the hippo transducers YAP and TAZ: a critical oncogenic node in human cancers. Int J Mol Sci 2017;18:E961. 10.3390/ijms18050961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 2012;149:1245–56. 10.1016/j.cell.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 66.Pohl SG, Brook N, Agostino M, Arfuso F, Kumar AP, Dharmarajan A. Wnt signaling in triple-negative breast cancer. Oncogenesis 2017;6:e310. 10.1038/oncsis.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci 2007;120:3327–35. 10.1242/jcs.03485 [DOI] [PubMed] [Google Scholar]
- 68.Choi J, Park SY, Costantini F, Jho EH, Joo CK. Adenomatous polyposis coli is down-regulated by the ubiquitin-proteasome pathway in a process facilitated by Axin. J Biol Chem 2004;279: 49188–98. 10.1074/jbc.M404655200 [DOI] [PubMed] [Google Scholar]
- 69.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer 2013;12:86. 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen F, Castranova V, Shi X. New insights into the role of nuclear factor-kappaB in cell growth regulation. Am J Pathol 2001; 159:387–97. 10.1016/S0002-9440(10)61708-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verma IM. Nuclear factor (NF)-kappaB proteins: therapeutic targets. Ann Rheum Dis 2004;63 Suppl 2:ii57–61. 10.1136/ard.2004.028266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gambhir S, Vyas D, Hollis M, Aekka A, Vyas A. Nuclear factor kappa B role in inflammation associated gastrointestinal malignancies. World J Gastroenterol 2015;21:3174–83. 10.3748/wjg.v21.i11.3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer 2012;12:121–32. 10.1038/nrc3204 [DOI] [PubMed] [Google Scholar]
- 74.Dou QP, Zonder JA. Overview of proteasome inhibitor-based anti-cancer therapies: perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr Cancer Drug Targets 2014;14:517–36. 10.2174/1568009614666140804154511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kubiczkova L, Pour L, Sedlarikova L, Hajek R, Sevcikova S. Proteasome inhibitors - molecular basis and current perspectives in multiple myeloma. J Cell Mol Med 2014;18:947–61. 10.1111/jcmm.12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta N, Hanley MJ, Xia C, Labotka R, Harvey RD, Venkatakrishnan K. Clinical pharmacology of Ixazomib: the first Oral proteasome inhibitor. [published online ahead of print August 17, 2018]. Clin Pharmacokinet. 10.1007/s40262-018-0702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Merin NM, Kelly KR. Clinical use of proteasome inhibitors in the treatment of multiple myeloma. Pharmaceuticals (Basel) 2014; 8:1–20. 10.3390/ph8010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bu R, Hussain AR, Al-Obaisi KA, Ahmed M, Uddin S, Al-Kuraya KS. Bortezomib inhibits proteasomal degradation of IκBα and induces mitochondrial dependent apoptosis in activated B-cell diffuse large B-cell lymphoma. Leuk Lymphoma 2014;55:415–24. 10.3109/10428194.2013.806799 [DOI] [PubMed] [Google Scholar]
- 79.Waxman AJ, Clasen S, Hwang WT, Garfall A, Vogl DT, Carver J, et al. Carfilzomib-associated cardiovascular adverse events: a systematic review and meta-analysis. JAMA Oncol 2018;4:e174519. 10.1001/jamaoncol.2017.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol 2018;19: 697–712. 10.1038/s41580-018-0040-z [DOI] [PubMed] [Google Scholar]
- 81.Dahlmann B. Proteasomes. Essays Biochem 2005;41:31–48. 10.1042/bse0410031 [DOI] [PubMed] [Google Scholar]
- 82.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 2008;9: 679–90. 10.1038/nrm2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang B, Liu K, Lin HY, Bellam N, Ling S, Lin WC. 14–3–3Tau regulates ubiquitin-independent proteasomal degradation of p21, a novel mechanism of p21 downregulation in breast cancer. Mol Cell Biol 2010;30:1508–27. 10.1128/MCB.01335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 2008;22:2496–506. 10.1101/gad.1676108 [DOI] [PMC free article] [PubMed] [Google Scholar]