Abstract
Background
The histone methyltransferase (HMT) family includes histone lysine methyltransferases (HKMTs) and histone/protein arginine methyltransferases (PRMTs). The role of HMT gene variants in prostate cancer remains unknown. Therefore, this study aimed to evaluate HMT gene variants in the pathogenesis and prognosis of human prostate cancer, using in vitro cell studies and bioinformatics analysis.
Material/Methods
Integrative bioinformatics analysis of the expression of 51 HMT genes in human prostate cancer was based on datasets from the Cancer Genome Atlas (TCGA). Correlation and regression analysis were used to identify critical HMTs in prostate cancer. Kaplan-Meier and the area under the receiver operating characteristics curve (AUROC) were performed to evaluate the function of the HMTs on prognosis. Gene expression and function of 22Rv1 human prostate carcinoma cells were studied.
Results
The HMT genes identified to have a role in the pathogenesis of prostate cancer included the EZH2, SETD5, PRDM12, NSD1, SETD6, SMYD1, and the WHSC1L1 gene. The EZH2, SETD5, and SMYD1 genes were selected as a prognostic panel, with the SUV420H2 HMT gene. SETD2, NSD1, and ASH1L were identified as critical genes in the development of castration-resistant prostate cancer (CRPC), similar to mixed-lineage leukemia (MLL) complex family members. Knockdown of the SETD5 gene in 22Rv1 prostate carcinoma cells in vitro inhibited cancer cell growth and migration.
Conclusions
HMT gene variants may have a role in the pathogenesis of prostate cancer. Future studies may determine the role of HMT genes as prognostic biomarkers in patients with prostate cancer.
MeSH Keywords: Epigenesis, Genetic; Histone-Lysine N-Methyltransferase; Prostatic Neoplasms
Background
Worldwide, prostate cancer (adenocarcinoma) is the fourth most common cancer and is the second most common cancer in men [1–3]. Prostate cancer is also clinically heterogeneous and can be indolent or may have a highly aggressive course, requiring different treatment approaches [4,5]. Increasing advances have been made in understanding the molecular basis for prostate cancer and distinguishing the different subtypes and the variable clinical outcomes. For example, androgen receptor-driven ETS gene fusion is an important molecular mechanism that allows the subclassification of prostate cancer, as are the activated gene fusions or mutations of RAS and RAF gene family members, a subtype of ETS-negative tumors [6]. An improved understanding of the genetic alterations that drive of prostate cancer at the molecular level may lead to new therapies that target for specific subtypes of prostate cancer.
The histone methyltransferase (HMT) family includes histone lysine methyltransferases (HKMTs) and histone/protein arginine methyltransferases (PRMTs). HMTs play crucial roles in epigenetic regulation, by controlling histone lysine methylation and demethylases. Currently, approximately 51 HMTs have been acknowledged identified in humans, with the primary functions being to catalyze the translocation of methyl moieties from S-adenosylmethionine (SAM) to histone lysine sites (Supplementary Table 1) [7,8]. Histone methylation can result in different biological effects according to the degree of histone methylated, including damage to chromatin structure, gene transcription, and alterations in cell mitosis, which may accelerate the development and progression of cancers [9]. There is now increasing evidence to validate the genetic roles of HMTs, including abnormal histone methylation, which may contribute to the genesis and progression of prostate cancer. For example, abnormal expression of the enhancer of zeste homolog 2 (EZH2) gene, resulting in an imbalance of H3K27me3 (trimethylation at lysine 27 of histone 3), has been shown to be associated with progression of prostate cancer and with castration-resistant prostate cancer (CRPC) [10,11].
Although increasing advances have been made in the understanding of epigenetic regulation of cancers, the roles of HMTs in prostate cancer remain unknown. Therefore, the aims of this preliminary study were to evaluate HMT gene variants in the pathogenesis and prognosis of human prostate cancer, using in vitro cell studies and bioinformatics analysis, including the roles the 51 known HMT genes. The study included the analysis of data available from the Cancer Genome Atlas (TCGA) database, which allowed differential mRNA expression, copy number alteration (CNA), somatic gene mutation, and DNA methylation state to be identified with clinical outcome, including progression-free survival (PFS), in patients with prostate cancer.
Material and Methods
Data sources
The dataset of gene expression, copy number alteration (CNA), DNA methylation, gene mutation and clinic data for 51 human histone methyltransferases (HMTs) of 550 prostate adenocarcinoma samples available from the Cancer Genome Atlas (TCGA) (including 52 matched tumor and adjacent normal tissues) used in this study were downloaded from The University of California, Santa Cruz (UCSC) Cancer Genomics Browser (https://genome-cancer.ucsc.edu/). A total of 399 tumor samples, with clinical data, were chosen to perform an integrated analysis of 51 HMTs. Gene expression data were log2 (x+1) transformed from the RNA-Seq by Expectation-Maximization (RSEM) count estimates and DNA methylation data, shown as the β-value, and were profiled using the Infinium HumanMethylation27 platform (Illumina, San Diego, CA, USA).
The Gleason scoring system of histological grade was used as an indicator of the degree of tumor aggression and prognosis of prostate cancer, with increasing Gleason scores reflecting worse prognosis. Two patients with a Gleason score of 10 were incorporated into the patient group with a Gleason score of 9. For CNA data, homozygous deletion (Homedel), heterozygous deletion (Hetloss), diploid, low-level amplification (Gain) and high-level amplification (Amp) were indicated as −2, −1, 0, 1, and 2 respectively. Also, a gene copy number dataset of 150 previously published metastatic prostate cancer samples was downloaded from cBioPortal for Cancer Genomics (http://www.cbioportal.org) to compare CNAs in prostate cancer of different stage further, as tumor stage is also an indicator of prognosis (Supplementary Table 2).
Statistical analysis
Statistical analysis was performed using the R software (version 3.3.4), GraphPad Prism (version 7.01) and SPSS (version 18.0). A two-tailed paired t-test was performed to compare the different expression levels of 51 HMT genes in 52 matched prostate tumor tissues and adjacent normal tissues. The correlation analysis for CNA, mRNA expression and DNA methylation, mRNA expression of 51 HMTs in 399 sequenced prostate cancer patients were performed by Spearman correlation, Kendall rank correlation, and Pearson correlation analysis. Comparison between HMT expression profiles in prostate cancer samples with Gleason score was conducted by R statistical software using one-way analysis of variance (ANOVA). Correlation analysis and clustering methods analyzed the association between androgen receptor (AR) expression and HMT gene expression. Kaplan-Meier survival curve was conducted to investigate the impact of genetic alterations of different HMTs on patient survival. Univariate and multivariate Cox regression analysis was performed to identify factors associated with patient prognosis in 399 samples of prostate cancer by using the survival package in the R statistical software package.
Generation of a prognostic HMT gene signature
The set of 399 patients with transcription, DNA methylation, CNA, mutation and clinical survival information was divided into groups for each gene that included high and low gene expression or methylation, deletion or amplification, diploid state, mutational and wild-type HMT genes, respectively. Each of the 51 gene expression features demonstrated a fit using a univariate Cox proportional-hazards analysis model without any adjustment, and 13 candidate features were generated at a p-value <0.05 using the Wald test (Wald chi-squared test). These candidate features were further analyzed using multivariate Cox proportional hazards analysis model, with adjustment for age, T-stage, Gleason score, and androgen receptor (AR) expression. Eight features with significant correlation coefficients were identified, and a panel of the top four most significant HMT genes was finally identified.
The risk score of the panel of the top four most significant HMT genes was calculated according to the gene expression values and risk factors, to divide the patients with prostate cancer into two groups according to their scores, a high-risk and a low-risk group. Prediction across this panel was then pooled and performance assessed using the area under the receiver operating characteristics curve (AUROC). Kaplan-Meier plots were generated to evaluate their impact on clinical outcome. For CNA, features were selected to fit a univariate Cox proportional hazards analysis if frequencies of each type CNA (deletion or amplification) were more than 3%. Significant genes were identified to fit a multivariate Cox proportional hazards analysis model to assess their value as prognostic cancer biomarkers. Univariate and multivariate Cox proportional hazards analysis models were performed to analyze the prognostic value of DNA methylation and mutation in prostate cancer.
Data visualization
Data visualization tools including the ggplot2 data visualization package, pheatmap package in R, progression-free survival (PFS), receiver operator characteristic (ROC) curves, and meta packages of R (vision 3.3.4). Box plots of gene expression, histograms of changes in copy number, and Kaplan-Meier plots were visualized by GraphPad Prism (version 7.01). The mutational spectrum of the KMT2C and KMT2D genes was generated by the Oncoprint and MutationMapper tools (http://www.cbioportal.org/tools.jsp) on the cBioportal website [12].
Cell culture and transfection and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The human prostate cancer cell line 22Rv1 was obtained from American Type Cell Culture (ATCC) (Catalog No. CRL-2505). Cells were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA), as recommended by the manufacturer. The target sequence of the SETD5 gene was chosen as follows: GCUUGGAGGCUGAGGAGUU. The oligonucleotides corresponding to this target sequence were annealed and cloned into the EcoR1 and AgeI sites of the pLkO.1 plasmid (Addgene, Cambridge, MA, USA). The efficiency of SETD5 interference was determined by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) using SYBR Premix Ex Taq II and SYBR Green I (Takara Biotechnology, Dalian, China). Quantitative primers of the SETD5 gene were as follows:
Forward: GGACTGCCTTATGCTACGA;
Reverse: GCCATCCTGAGAAAGACCACA.
Cell proliferation and migration assays
Cell proliferation of the human prostate cancer cell line 22Rv1 was evaluated by the MTS colorimetric cell proliferation assay (Sigma, St Louis, MO, USA) and a colony formation assay, according to the manufacturer’s instructions, with each experiment performed in triplicate. A cell migration assay was used to assess the cell invasion capacity (Corning, NY, USA) with the assay performed in duplicate.
Results
Identification of the expression profiles of histone methyltransferase (HMT) gene mRNA in prostate cancer
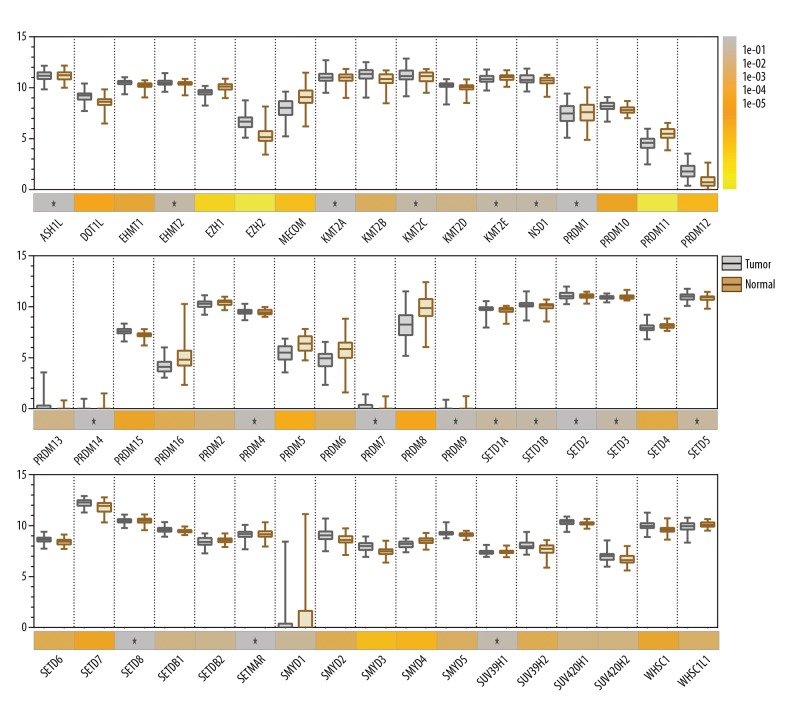
The mRNA levels of the 51 histone methyltransferase (HMT) genes in 52 matched prostate tumor tissues and adjacent normal tissues were analyzed and presented as box plots. The Student’s t-test was used to determine the significance of the gene expression levels. Compared with adjacent normal prostate tissues, mRNA levels of prostate cancer tissue of 14 HMT genes were identified that were significantly increased, including DOT1L, EZH2, EHMT1, KMT2B, PRDM10, PRDM12, PRDM15, SETD6, SETD7, SMYD2, SMYD3, SMYD5, SUV39H2, and WHSC1) (p<0.001). Compared with adjacent normal tissues, mRNA levels of prostate cancer tissue of nine HMT genes were identified that were significantly decreased, including EZH1, MECOM, PRDM11, PRDM16, PRDM5, PRDM6, PRDM8, SETD4, and SMYD4 (p<0.001) (Figure 1). Also, mRNA expression levels of the PRDM13, PRDM14, PRDM7, PRDM9, and SMYD1 genes were at very low levels in both tumor tissues and normal prostate tissues. Twenty-one significantly fold-change (FC) genes with false discovery rates (FDR) with a p-value of <0.05 were found using the Limma R software package for analyzing data from gene expression, including six 2.0-fold-change, three 1.5-fold-change, and eleven 1.2-fold-change genes, which were in accordance with 23 HMT genes previously described in the literature (Supplementary Table 3).
Figure 1.
Box plots showing histone lysine methyltransferase (HKMT) mRNA levels in 52 matched prostate cancer tissues (grey) and adjacent normal tissues (brown). Paired t-test was performed to compare the different expression profiles between tumor and adjacent normal tissues. Statistical significance is shown as the p-values by different grayscales of the lower color bars (p>0.05 are specifically shown by * inside the sub-boxes). There was reduced mRNA expression of several genes, including PRDM7, PRDM9, PRDM13, PRDM14, and SMYD1.
To compare the differences between HMT gene expression profiles between Gleason score 6, 7, 8, and 9 prostate cancers, one-way analysis of variance (ANOVA) was performed, which identified 11 HMT genes that were significantly expressed: WHSC1, EZH2, PRDM12, KMT2D, SETD5, PRDM8, EHMT1, SUV420H2, PRDM10, PRDM6, and DOT1L (p <0.001) (Supplementary Figure 1, Supplementary Table 4). Among those genes, PRDM6 and PRDM8 both showed a significantly negative correlation with the Gleason score. The EZH2, PRDM12, SUV420H2, WHSC1, PRDM10 and SETD5 genes showed a significantly positive correlation with the Gleason score (Supplementary Figure 1, Supplementary Figure 2). These findings indicated that alteration in mRNA expression levels of these genes in prostate cancer tissue samples were prognostic indicators in these cases of prostate cancer.
Changes in copy number alteration (CNA) of HMT genes in prostate carcinoma
Copy number alterations (CNAs) have previously been reported to be an important mechanism for activating oncogenes or inactivating tumor suppressor genes in the genesis and progression of human malignancy [13]. Therefore, CNAs of 51 HMT genes in 492 prostate cancer tissue samples were systematically analyzed, which identified six HMT genes: SETDB2, PRDM1, PRDM13, PRDM7, WHSC1L1, and PRDM5. These six genes showed homozygous deletion (Homedel) in more than 3% of prostate cancer tissue samples. Two HMT genes, PRDM14 and MECOM, showed high-level amplification (Amp) in more than 3% of prostate cancer samples (Table 1, Supplementary Figure 3).
Table 1.
Frequency of HMTs copy number alterations and mutations (%).
| Gene | Location | Amp | Gain | Diploid | Hetloss | Homdel | Mutation |
|---|---|---|---|---|---|---|---|
| SETDB2 | 13q14.2 | 0.00 | 0.81 | 54.88 | 27.64 | 16.67 | 0.24 |
| PRDM1 | 6q21 | 0.00 | 0.41 | 67.07 | 18.70 | 13.82 | 0.00 |
| PRDM13 | 6q16.2 | 0.00 | 1.22 | 66.06 | 19.11 | 13.62 | 0.00 |
| PRDM7 | 16q24.3 | 0.20 | 1.02 | 60.57 | 29.88 | 8.33 | 0.47 |
| WHSC1L1 | 8p11.23 | 2.64 | 11.38 | 52.44 | 27.44 | 6.10 | 0.71 |
| PRDM5 | 4q27 | 0.00 | 2.85 | 87.60 | 6.30 | 3.25 | 0.24 |
| EZH1 | 17q21.2 | 0.00 | 1.02 | 86.18 | 10.16 | 2.64 | 0.24 |
| SETD5 | 3p25.3 | 0.20 | 6.91 | 85.77 | 4.47 | 2.64 | 0.24 |
| SETD6 | 16q21 | 0.20 | 1.22 | 73.98 | 21.95 | 2.64 | 0.00 |
| PRDM11 | 11p11.2 | 0.20 | 4.47 | 88.82 | 4.27 | 2.24 | 0.00 |
| PRDM6 | 5q23.2 | 0.20 | 2.64 | 87.20 | 8.13 | 1.83 | 0.00 |
| KMT2A | 11q23.3 | 0.41 | 5.89 | 85.77 | 6.30 | 1.63 | 1.18 |
| PRDM15 | 21q22.3 | 0.00 | 4.67 | 87.40 | 6.50 | 1.42 | 0.24 |
| SETD1B | 12q24.31 | 0.81 | 4.88 | 87.20 | 5.69 | 1.42 | 0.00 |
| SETD2 | 3p21.31 | 0.00 | 7.11 | 87.80 | 3.66 | 1.42 | 0.94 |
| SETMAR | 3p26.1 | 0.20 | 7.32 | 86.79 | 4.27 | 1.42 | 0.00 |
| SMYD2 | 1q32.3 | 0.00 | 6.30 | 88.41 | 3.86 | 1.42 | 0.24 |
| KMT2C | 7q36.1 | 0.41 | 18.70 | 75.41 | 4.27 | 1.22 | 5.41 |
| SETD3 | 14q32.2 | 0.41 | 2.64 | 89.02 | 6.71 | 1.22 | 0.00 |
| PRDM10 | 11q24.3 | 0.81 | 6.10 | 87.60 | 4.47 | 1.02 | 0.24 |
| SMYD4 | 17p13.3 | 0.00 | 1.42 | 80.08 | 17.48 | 1.02 | 0.00 |
| PRDM2 | 1p36.21 | 0.20 | 0.61 | 89.23 | 9.15 | 0.81 | 0.47 |
| SETD8 | 12q24.31 | 0.61 | 4.27 | 87.40 | 6.91 | 0.81 | 0.00 |
| SMYD5 | 2p13.2 | 0.41 | 2.64 | 90.24 | 5.89 | 0.81 | 0.00 |
| DOT1L | 19p13.3 | 0.20 | 1.22 | 90.24 | 7.72 | 0.61 | 0.71 |
| PRDM9 | 5p14.2 | 0.61 | 6.30 | 89.43 | 3.05 | 0.61 | 0.71 |
| SETD1A | 16p11.2 | 0.41 | 6.10 | 86.79 | 6.10 | 0.61 | 0.47 |
| SMYD3 | 1q44 | 0.41 | 5.89 | 90.04 | 3.05 | 0.61 | 0.00 |
| SUV39H2 | 10p13 | 0.41 | 3.46 | 86.59 | 8.94 | 0.61 | 0.00 |
| SUV420H1 | 11q13.2 | 2.44 | 7.32 | 89.02 | 0.61 | 0.61 | 0.24 |
| WHSC1 | 4p16.3 | 1.02 | 2.85 | 89.43 | 6.10 | 0.61 | 0.24 |
| ASH1L | 1q22 | 1.42 | 6.10 | 90.04 | 2.03 | 0.41 | 1.88 |
| EHMT2 | 6p21.31 | 0.20 | 3.66 | 91.46 | 4.27 | 0.41 | 0.24 |
| EZH2 | 7q36.1 | 0.41 | 18.70 | 79.07 | 1.42 | 0.41 | 0.24 |
| KMT2B | 19q13.12 | 0.41 | 2.03 | 92.89 | 4.27 | 0.41 | 0.47 |
| KMT2E | 7q22.3 | 0.81 | 18.70 | 78.25 | 1.83 | 0.41 | 0.94 |
| PRDM16 | 1p36.32 | 0.41 | 1.63 | 89.43 | 8.13 | 0.41 | 0.24 |
| PRDM8 | 4q21.21 | 0.41 | 2.64 | 92.89 | 3.66 | 0.41 | 0.71 |
| SETD4 | 21q22.12 | 0.00 | 4.67 | 89.23 | 5.69 | 0.41 | 0.00 |
| SETD7 | 4q31.1 | 0.00 | 3.25 | 91.26 | 5.08 | 0.41 | 0.00 |
| SMYD1 | 2p11.2 | 0.41 | 2.24 | 91.46 | 5.49 | 0.41 | 0.00 |
| PRDM12 | 9q34.12 | 1.22 | 10.57 | 85.37 | 2.64 | 0.20 | 0.47 |
| PRDM4 | 12q23.3 | 1.02 | 5.28 | 88.82 | 4.67 | 0.20 | 0.00 |
| EHMT1 | 9q34.3 | 1.42 | 10.57 | 85.37 | 2.64 | 0.00 | 0.00 |
| KMT2D | 12q13.12 | 0.20 | 3.25 | 91.87 | 4.67 | 0.00 | 4.94 |
| MECOM | 3q26.2 | 3.66 | 13.62 | 80.69 | 2.03 | 0.00 | 1.18 |
| NSD1 | 5q35.2 | 0.81 | 4.27 | 91.67 | 3.25 | 0.00 | 0.47 |
| PRDM14 | 8q13.3 | 6.10 | 24.80 | 68.29 | 0.81 | 0.00 | 0.47 |
| SETDB1 | 1q21.3 | 1.42 | 6.71 | 91.26 | 0.61 | 0.00 | 0.47 |
| SUV39H1 | Xp11.23 | 1.02 | 2.64 | 90.85 | 5.49 | 0.00 | 0.00 |
| SUV420H2 | 19q13.42 | 0.41 | 4.07 | 91.46 | 4.07 | 0.00 | 0.24 |
Amp – high-level amplification; Gain – low-level gain; Hetless – heterozygous deletion; Homdel – homozygous deletion. Genes were ranked based on the frequency of Homdel.
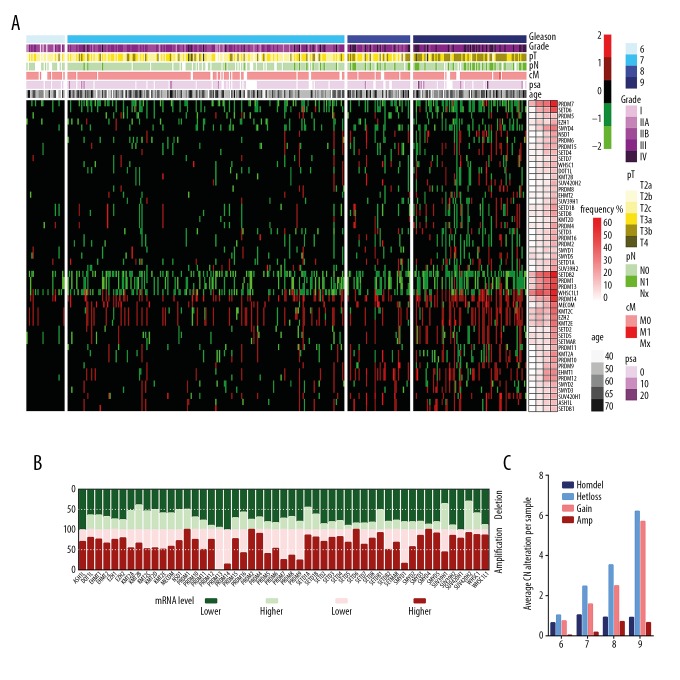
Further analysis of 51 HMT CNAs in prostate cancer tissue samples, with different Gleason scores, showed that increasing CNA events in prostate cancer were significantly correlated with a Gleason score of between 6 to 9 (Figure 2A). The CNA frequencies of most HMT genes were positively correlated with the Gleason score, for both amplification and deletion alterations (Supplementary Figure 4). Also, with increasing Gleason score, the average CNA events of HMT genes in each tumor tissue sample were all incrementally increased, except for the homozygous deletion (Homedel), which had a more stable level in prostate cancer tissue samples with different Gleason scores (Figure 2C). The frequencies of CNA events in metastatic prostate cancer tissue samples were significantly increased when compared with primary prostate cancer tissue (Supplementary Figure 5). These data indicated that, in the cases in this study, prostate cancers that had many more CNA events of the HMT genes had an increased tumor grade and worse clinical outcome.
Figure 2.
Expression profiles of altered copy numbers of histone lysine methyltransferase (HKMT) in samples of prostate cancer according to Gleason scores. (A) The pheatmap package in R shows that increasing copy number alteration (CNA) events of 51 histone methyltransferases (HMTs) in prostate cancer samples, with a Gleason score of 6 to 9, were significantly associated with the clinical stage. (B) A histogram shows the effect on HMT expression levels due to changes in gene copy numbers. The y-axis represents the proportion of genes expressed (higher or lower) associated with different types of copy number alterations. (C) A histogram shows that the average CNA events of HMTs in each sample are incrementally increased with an increase in Gleason score.
The correlations between copy number alterations and expression of 51 HMT genes were analyzed in the 399 prostate cancer tissue samples that underwent sequence analysis. To obtain a weighted ranking for the correlation coefficients, the Spearman correlation, Kendall rank correlation, and Pearson correlation analysis were performed (Table 2). As shown in Table 2, gene expression levels of almost all HMT genes were positively correlated with CNAs, including two HMT genes, WHSC1L1 and SETDB2 with correlation coefficients >0.5 (Spearman method, p<0.0001) and four HMT genes, SETD6, SMYD4, EZH2 and SETDB1 with correlation coefficients >0.3 (Spearman method, p<0.0001). To further analyze the impact caused by CNAs on gene expression levels of HMTs, the frequency of amplification and deletion with higher or lower HMT mRNA levels in tissue samples were calculated separately. The frequency of amplification with higher HMT mRNA levels and deletion with lower HMT mRNA levels, were significantly increased in prostate cancer tissue compared with the normal prostate tissue (p<0.0001, paired t-test). The difference between diploid tumor samples was non-significant (Figure 2B, Supplementary Table 5).
Table 2.
Associations between HMTs expression and CNAs or methylation state.
| Gene | CNA/mRNA Correlation | Methylation/mRNA Correlation | ||||
|---|---|---|---|---|---|---|
| Spearman | Pearson | Kendall | Spearman | Pearson | Kendall | |
| WHSC1L1 | 0.577 | 0.577 | 0.460 | −0.005 | 0.026 | −0.004 |
| SETDB2 | 0.555 | 0.535 | 0.435 | 0.122 | 0.183 | 0.083 |
| SETD6 | 0.499 | 0.530 | 0.407 | −0.232 | −0.230 | −0.157 |
| SMYD4 | 0.400 | 0.410 | 0.325 | NA | NA | NA |
| EZH2 | 0.347 | 0.363 | 0.283 | −0.232 | −0.197 | −0.160 |
| SETDB1 | 0.325 | 0.411 | 0.265 | −0.179 | −0.213 | −0.119 |
| SETD8 | 0.310 | 0.338 | 0.251 | 0.015 | −0.013 | 0.011 |
| SUV39H2 | 0.297 | 0.309 | 0.240 | −0.043 | −0.071 | −0.029 |
| SETD3 | 0.294 | 0.436 | 0.240 | −0.068 | −0.091 | −0.045 |
| EZH1 | 0.293 | 0.359 | 0.236 | −0.089 | −0.094 | −0.059 |
| EHMT1 | 0.280 | 0.355 | 0.228 | NA | NA | NA |
| SUV420H1 | 0.277 | 0.462 | 0.226 | −0.164 | −0.160 | −0.111 |
| PRDM4 | 0.275 | 0.339 | 0.223 | −0.024 | 0.033 | −0.017 |
| SETD4 | 0.265 | 0.302 | 0.216 | 0.305 | 0.259 | 0.205 |
| SETD5 | 0.243 | 0.211 | 0.196 | −0.118 | −0.078 | −0.078 |
| KMT2C | 0.243 | 0.283 | 0.195 | NA | NA | NA |
| SMYD5 | 0.241 | 0.246 | 0.196 | −0.116 | −0.101 | −0.081 |
| PRDM13 | 0.237 | 0.174 | 0.216 | 0.346 | 0.327 | 0.266 |
| PRDM12 | 0.217 | 0.205 | 0.176 | 0.356 | 0.356 | 0.244 |
| SETD2 | 0.212 | 0.176 | 0.172 | −0.161 | −0.127 | −0.112 |
| PRDM2 | 0.209 | 0.216 | 0.171 | −0.323 | −0.360 | −0.219 |
| PRDM15 | 0.207 | 0.238 | 0.168 | −0.087 | −0.069 | −0.058 |
| SETMAR | 0.196 | 0.229 | 0.158 | −0.041 | −0.053 | −0.028 |
| KMT2A | 0.193 | 0.209 | 0.156 | 0.210 | 0.247 | 0.139 |
| SMYD3 | 0.189 | 0.208 | 0.153 | 0.234 | 0.189 | 0.160 |
| WHSC1 | 0.174 | 0.270 | 0.140 | −0.133 | −0.168 | −0.088 |
| PRDM10 | 0.170 | 0.191 | 0.138 | −0.093 | −0.130 | −0.064 |
| SETD1A | 0.165 | 0.200 | 0.132 | −0.071 | −0.082 | −0.046 |
| SETD7 | 0.164 | 0.200 | 0.132 | 0.223 | 0.224 | 0.150 |
| SETD1B | 0.164 | 0.139 | 0.133 | NA | NA | NA |
| KMT2E | 0.160 | 0.169 | 0.129 | 0.129 | 0.048 | 0.087 |
| SMYD2 | 0.158 | 0.252 | 0.129 | −0.140 | −0.215 | −0.093 |
| PRDM5 | 0.155 | 0.162 | 0.125 | −0.523 | −0.551 | −0.365 |
| PRDM6 | 0.154 | 0.157 | 0.124 | −0.262 | −0.265 | −0.175 |
| PRDM14 | 0.150 | 0.132 | 0.143 | 0.046 | 0.056 | 0.036 |
| ASH1L | 0.149 | 0.145 | 0.121 | −0.298 | −0.130 | −0.205 |
| PRDM11 | 0.142 | 0.164 | 0.114 | 0.055 | 0.054 | 0.035 |
| NSD1 | 0.127 | 0.141 | 0.103 | 0.029 | 0.040 | 0.019 |
| DOT1L | 0.126 | 0.156 | 0.103 | −0.053 | −0.052 | −0.034 |
| PRDM9 | 0.122 | 0.055 | 0.118 | −0.121 | −0.013 | −0.098 |
| EHMT2 | 0.116 | 0.160 | 0.094 | −0.128 | −0.110 | −0.085 |
| SUV420H2 | 0.074 | 0.100 | 0.060 | −0.050 | −0.019 | −0.032 |
| KMT2B | 0.072 | 0.055 | 0.058 | 0.052 | 0.028 | 0.034 |
| KMT2D | 0.059 | 0.050 | 0.048 | 0.030 | 0.025 | 0.021 |
| PRDM16 | 0.058 | 0.121 | 0.047 | NA | NA | NA |
| PRDM7 | 0.056 | 0.035 | 0.050 | 0.050 | 0.037 | 0.036 |
| MECOM | 0.048 | 0.059 | 0.038 | −0.163 | −0.139 | −0.113 |
| PRDM1 | 0.034 | 0.061 | 0.028 | 0.002 | −0.080 | 0.003 |
| SMYD1 | 0.006 | 0.030 | 0.006 | 0.007 | −0.041 | 0.005 |
| PRDM8 | −0.011 | −0.044 | −0.009 | −0.529 | −0.551 | −0.372 |
| SUV39H1 | −0.050 | −0.014 | −0.040 | 0.007 | 0.049 | 0.005 |
Genes were ranked based on the Spearman correlation coefficient. Significantly associated HMTs between gene expression with CNAs or methylation were highlighted.
Methylation states of the HMT gene promoter CpG islands in prostate cancer
DNA methylation is a crucial regulator in tumor development, by altering mRNA expression. For 46 HMT genes, correlation analysis was performed to compare gene expression and promoter methylation in 399 prostate cancer tissue samples by three different statistical methods, the Spearman correlation analysis, Kendall rank correlation, and the Pearson correlation analysis. The results showed that the promoter methylation level of most HMT genes was negatively correlated with gene expression, including two HMT genes, PRDM8 and PRDM5, with an absolute value with the Spearman correlation coefficient >0.5 (P<0.0001) and one HMT gene, PRDM2 with the Spearman correlation coefficient >0.3 (P<0.0001). However, the methylation levels of the SETD4, PRDM12, and PRDM13 genes were all significantly correlated with gene expression levels, with a correlation coefficient >0.3 (P<0.0001) (Table 2).
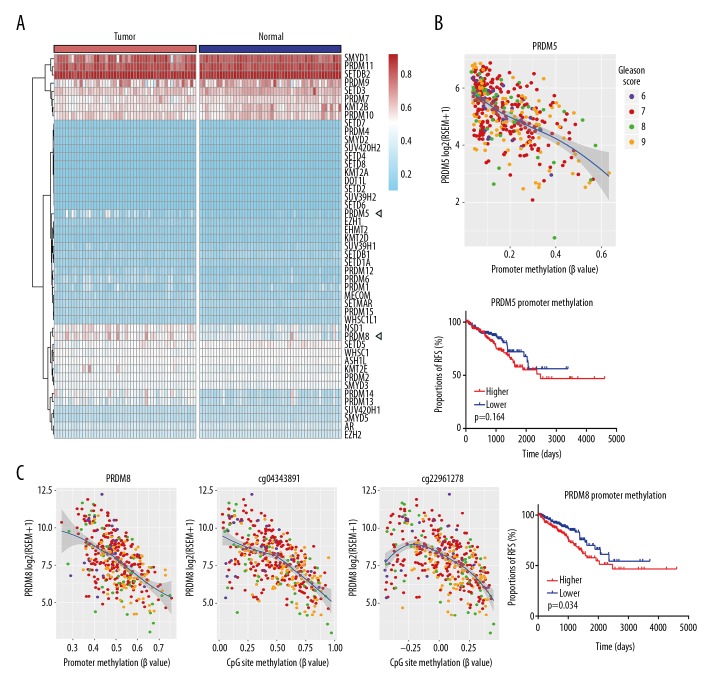
The pheatmap package in R was plotted to show the promoter methylation profiles of 46 HMT genes in 50 paired prostate tumor and adjacent normal tissue samples. Most HMT genes had a lower promoter methylation level, but some HMT genes had a higher promoter methylation level, including the three genes, PRDM7, PRDM9, and SMYD1, which had low mRNA levels (Figures 1, 3A). Further quantitative analysis of the methylation changes between matched tumor and adjacent normal prostate tissues, identified PRDM14, NSD1, and PRDM8 as the top three differentially methylated HMT genes in prostate cancer, by paired t-test and correlation analysis (Supplementary Table 6). By combining the correlation analysis of mRNA and promoter methylation and the results of differential expression and methylation, the PRDM5 and PRDM8 genes were identified and underwent further analysis (Figure 3B, 3C).
Figure 3.
DNA methylation profiles of histone methyltransferases (HMTs) in prostate cancer (adenocarcinoma). (A) The pheatmap package in R showed different promoter methylation levels in 50 paired tumor tissues and corresponding adjacent tissues. Promoter CpG sites of most histone methyltransferases (HMTs) are expected at low methylation level and the difference between tumor and normal tissues are unexpected plain. PRDM5 and PRDM8 were indicated by triangle because their mRNA levels are both 1.2-fold-change from normal to tumor tissues and have a high correlation coefficient to the corresponding gene expression level. (B, C) Scatter diagrams showed the relationship between gene expression and its promoter methylation state (β value) in 399 prostate cancers. Survival analysis show samples with lower PRDM5 promoter methylation as well as PRDM8 have a better prognosis in some extent than higher ones. (C) Two CpG sites in the PRDM8 promoter which are both 1.2-fold-change between normal and tumor tissues have an increased correlation with PRDM8 mRNA levels compared with other CpG sites.
Analysis of the PRDM8 and PRDM5 differentially methylated HMT genes in prostate cancer
Promoter methylation of the PRDM8 gene was increased, even in the normal prostate tissues. However, the 1.5-fold-change was significantly increased in the tumor tissues resulting in significant alterations with gene expression (p<0.0001, using a t-test). Different methylation levels of the PRDM8 gene were also correlated with clinical outcome in prostate tumor samples from 399 patients. Also, two CpG sites (cg04343891 and cg22961278) of the PRDM8 promoter were both differently methylated and significantly associated with PRDM8 gene expression (Figure 3C).
Promoter methylation of the PRDM5 gene was decreased in the normal prostate tissues but was significantly increased in matched prostate cancer tissues. For PRDM5 gene expression, there were six CpG sites out of eight with a lower methylation level (β ≤0.1). In more than 90% of adjacent normal prostate tissues, and in more than 5% of tumor tissues, a higher methylation level was found (β ≥0.3). The average PRDM5 mRNA levels of the two groups (β ≥0.3 and β ≤0.1) in the 399 prostate cancer samples were compared for each CpG site. The results showed that PRDM5 was differently expressed between two groups for six CpG sites (p<0.001 using a t-test) (Table 3). These data support that PRDM5 was a methylation silenced gene in prostate cancer in the cases in this study, which highlights the need for further studies to determine the potential treatment approaches for patients with different PRDM5 promoter methylation states [14].
Table 3.
Summary of PRDM5 CpG sites.
| CpG ID | Genetic location | Epigenetic location | β >0.3 in tumor(%) | β <0.1 in normal(%) | PRDM5 mRNA level in 399 PCa samples | ||
|---|---|---|---|---|---|---|---|
| β >0.3 | β <0.1 | p-Value | |||||
| cg07070341 | TSS200 | Island | 16 | 94 | 4.418 | 5.461 | 1.34E-14 |
| cg18181323 | TSS200 | Island | 8 | 98 | 3.996 | 5.418 | 2.34E-08 |
| cg19294653 | TSS200 | Island | 8 | 98 | 4.043 | 5.444 | 3.55E-11 |
| cg22135105 | TSS200 | Island | 22 | 98 | 4.438 | 5.506 | 8.41E-19 |
| cg11171429 | TSS1500 | S_Shore | 20 | 94 | 4.358 | 5.542 | 2.31E-18 |
| cg13155001 | 1st Exon; 5′UTR | Island | 10 | 100 | 4.661 | 5.201 | 6.39E-03 |
| cg06329345 | TSS1500 | S_Shore | 30 | 88 | NA | ||
| cg10416963 | TSS1500 | S_Shore | 78 | 4 | NA | ||
Methylation level of CpG sites of PRDM5 in 50 paired tumor and adjacent tissues were listed. Average PRDM5 mRNA level in the two groups (β >0.3 and β <0.1) were calculated and T.test was used to value the significance of difference.
Mutations of HMT genes in prostate cancer
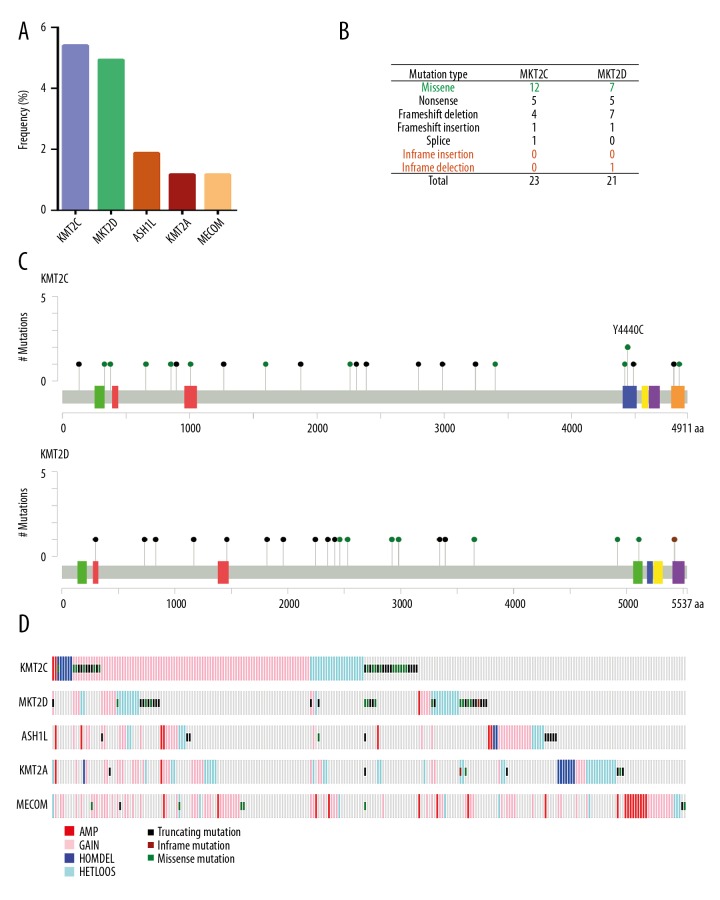
The KMT2C, KMT2D, ASH1L, KMT2A, and MECOM genes were the top five most frequently mutated HMT genes in prostate cancer in this study (Table 1, Figure 4A). Integrated analysis of mutation profiles of KMT2C and KMT2D were performed in 425 primary prostate cancer samples. The results showed that a total of 23 KMT2C gene mutations and 21 KMT2D gene mutations were identified (Figure 4B). A mutation map was performed to display the distribution and frequency of KMT2C and KMT2D mutations in 425 prostate cancers (Figure 4C). A mutualy exclusivity relationship for copy number alterations and mutations of the top five mutated HMTs in prostate cancers are shown in Figure 4D.
Figure 4.
The KMT2C and KMT2D gene mutation spectrum in prostate carcinoma. (A) Bar graphs show the top five mutated histone methyltransferases (HMTs). (B) The frequency of each mutation type for KMT2C and KMT2D from 429 primary prostate adenocarcinomas. The data were obtained from The Cancer Genome Atlas (TCGA) database. (C) The images show protein domains and the positions of specific mutations of the KMT2C and KMT2D genes. A green dot indicates a missense mutation. The black dot indicates a truncating mutation include nonsense mutation, frameshift deletion, insertion, or splice. A brown dot indicates an in-frame insertion or deletion mutation. (D) Mutual exclusivity relationship for copy number alterations and mutation of the top five mutated HMTs in prostate cancers.
Co-expression of HMT genes with androgen receptors
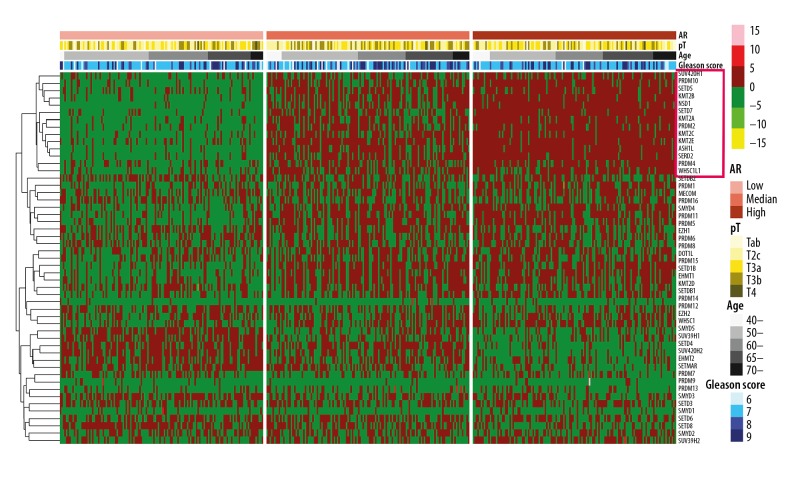
Cluster analysis showed that high expression levels of several HMT genes, including ASH1L, SETD2, KMT2B, NSD1, KMT2C, SETD7, KMT2E, KMT2A, PRDM10, SETD5, PRDM2, PRDM4, and WHSC1L1 were significantly clustered in tumor tissue samples with high expression of androgen receptors when compared with tumor tissue samples with low expression of androgen receptors, which was determined by correlation analysis between androgen receptor and HMT mRNA levels (Figure 5, Supplementary Table 5). Notably, KMT2B, KMT2C, and KMT2E have been validated previously as having a role in the progression of castration-resistant prostate cancer (CRPC) [15]. These results showed that these genes, identified in the cases in this study, may be involved in the androgen receptor response pathway in prostate cancer, and their genetic alterations may function in the development of CRPC.
Figure 5.
Co-expression of histone methyltransferases (HMTs) and androgen receptors. The pheatmap package in R shows that several histone methyltransferases (HMTs) (ASH1L, SETD2, KMT2B, NSD1, KMT2C, SETD7, KMT2E, KMT2A, PRDM10, SETD5, PRDM2, PRDM4, and WHSC1L1) were significantly clustering in the higher androgen receptor (AR) expression cluster.
Alterations in HMT gene expression were associated with clinical outcome in patients with prostate cancer
To study the clinical effects HMT gene variations on clinical outcome in patients with prostate cancer, the association between progression-free survival (PFS), CNA, mRNA levels, mutation and promoter methylation levels were examined in the 399 prostate cancer samples. Univariate Cox proportional hazards analysis was performed primarily to find potential HMT genes that were most relevant for further multivariate Cox proportional-hazards analysis, with p<0.05. The results showed that amplification and deletion of most the most significant HMT genes were associated with poor clinical outcome (HR >1). Nine HMT genes with increased levels of promoter methylation, and sixteen HMT genes with increased mRNA levels were identified as having a significant association with clinical outcome in the cases of prostate cancer included in this study (Supplementary Table 7).
Patient age, the T-stage, Gleason score, and the expression level of the androgen receptor (AR) for 28 genetic alterations were identified by multivariate analysis (p<0.1) (Supplementary Table 8). The results showed that amplification of EHMT1, KMT2B, PRDM1, PRDM12, PRDM16, PRDM4, SETD1B, SETD8, SETDB1, SMYD1, SMYD2, SMYD5, and WHSC1L1 genes, and the deletion of EHMT2, SETD4, SETD6, SMYD1, SUV39H2, and WHSC1L1 genes, were significantly associated with reduced clinical outcome and PFS. Increased mRNA levels of EZH2, SETD5, and SUV420H2 genes, and lower mRNA levels of PRDM6, SMYD1, and PRDM16 genes were significantly associated with reduced clinical outcome and PFS in patients with prostate cancer. Increased methylation levels of NSD1 was significantly associated with a reduced patient overall survival (OS).
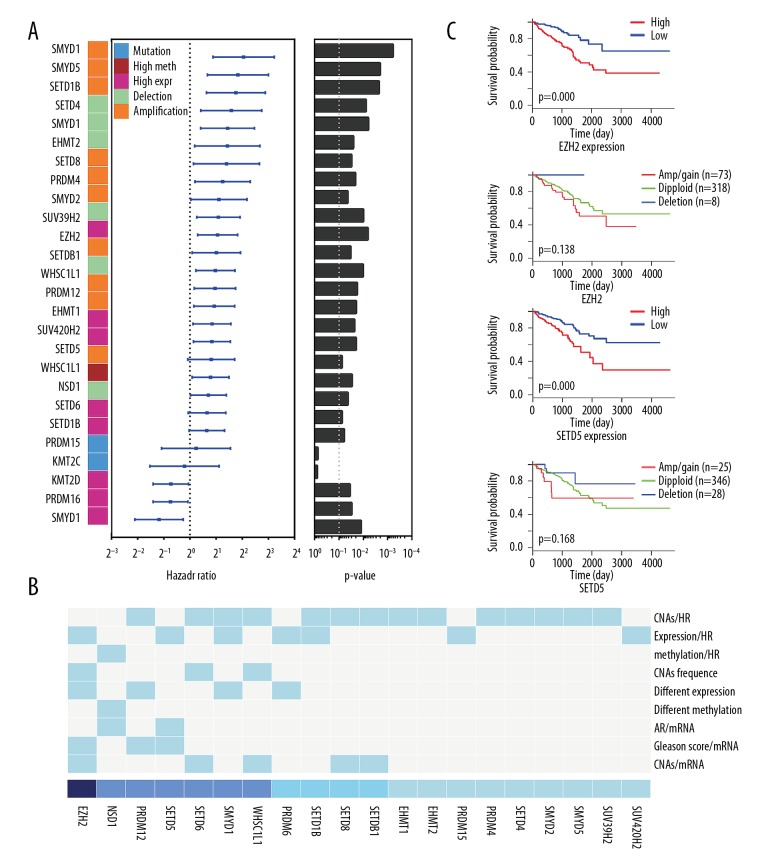
Of the 28 genetic alterations of HMT genes, mutations of KMT2C and KMT2D were investigated to show their clinical impact on prostate cancer samples using a forest plot (Figure 6A). The most relevant HMT genes were selected by integrating the score of mRNA, CNA and promoter methylation associated survival, CNA frequencies, differential expression and methylation, correlation of HMT gene expression and CNAs, androgen receptor expression and Gleason score (Figure 6B). Each of seven HMT genes, including EZH2, NSD1, RPDM12, SETD5, SETD6, SMYD1 and WHSC1L1, had a score of >3, indicating that these seven HMT genes might play important roles in oncogenesis in prostate cancer. For these seven HMT genes, Kaplan-Meier curves were plotted and the results showed that amplification of EZH2 and SETD5 were associated with clinical outcome (Figure 6C, Supplementary Figure 6).
Figure 6.
Genetic alterations of histone methyltransferases (HMTs) are associated with recurrence-free survival (RFS) of patients with prostate carcinoma. (A) The 28 genetic alterations (including mutation, amplification, deletion, increased gene expression, and methylation) of histone methyltransferases (HMTs) are shown by forest plot to demonstrate their impact on clinical survival, showing the hazard ratio (HR), 95% confidence interval (CI) and p-value (Wald test). (B) Critical HMT genes (EZH2, NSD1, RPDM12, SETD5, SETD6, SMYD1, and WHSC1L1) were analyzed using the mRNA score, copy number alterations (CNAs) and promoter methylation associated survival, CNA frequencies, differential expression and methylation, correlation of CNA, androgen receptor (AR) expression, and Gleason score with HMT expression. (C) Kaplan-Meier plot of the recurrence-free survival (RFS) show that two typical HMT genes (EZH2 and SETD5) have the same clinic outcome of amplification and increased expression or deletion and reduced expression.
The four-gene HMT prognostic signature included EZH2, SETD5, SMYD1, and SUV420H2
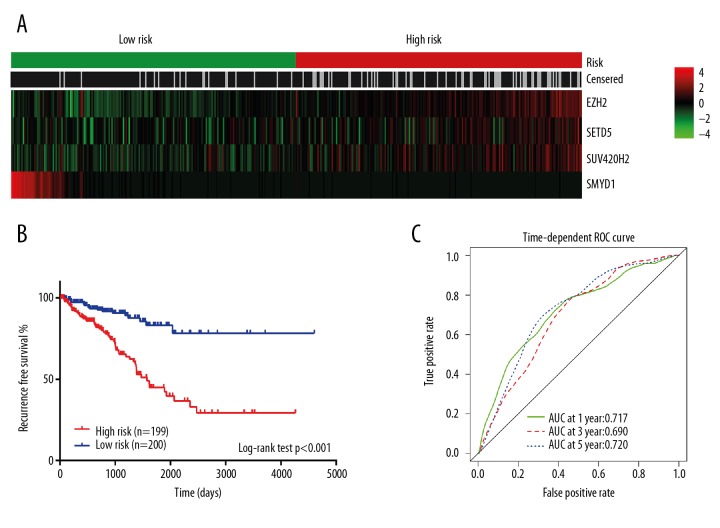
Expression of the four most significant HMT genes in terms of prognosis in prostate cancer, EZH2, SETD5, SMYD1, and SUV420H2, allowed a panel to be compiled to estimating their prognostic value for relapse of prostate cancer (Figure 7A). Kaplan-Meier curves were plotted to show the prognostic value of this gene panel in the Cancer Genome Atlas (TCGA) cohort (n=399), by dividing the samples a high-risk and low-risk group (Figure 7B). A time-dependent area under the receiver operating characteristics (AUROC) curve was used to assess the prognostic value of the four-gene HMT prognostic signature (EZH2, SETD5, SMYD1, and SUV420H2). The area under the curve (AUC) for the four-gene HMT prognostic signature model at one, three, and five years were 0.717, 0.690, and 0.720, respectively (Figure 7C).
Figure 7.
The histone methyltransferase (HMT) predictor-score analysis of 399 prostate cancer patients in the Cancer Genome Atlas (TCGA) cohort. (A) Expression profile of four histone methyltransferases (HMTs) in high-risk and low-risk patients. (B) Kaplan-Meier plot shows the recurrence-free survival (RFS) in high-risk and low-risk patients. (C) A time-dependent area under the receiver operating characteristics (AUROC) curve was used to assess the prognostic value of the four HMT gene prognostic signature (EZH2, SETD5, SMYD1, and SUV420H2). The AUC for the four HMT gene prognostic signature model at one, three, and five years were 0.717, 0.690, and 0.720, respectively via cross-validation (purple).
The SETD5 gene in the development and progression of prostate cancer
The SETD5 gene expression was studied in the 399 samples of prostate cancer from TCGA data. The study samples were divided into low-expression and high-expression groups for SETD5 in tissue samples of prostate cancer, with the median value as the cutoff point. Five characteristics were analyzed in the low-expression and high-expression groups. The results showed that high SETD5 expression was correlated with high Gleason scores, increase serum levels of prostate-specific antigen (PSA), advanced T-stage, and the presence of lymph node metastases (Table 4).
Table 4.
Association between SETD5 expression and clinical characteristics in prostate cancer from TCGA Data portal.
| Characteristics | Total (n=399) | SETD5 expression | p Value | |
|---|---|---|---|---|
| Higher | Lower | |||
| Age (years) | ||||
| ≤60 | 190 | 89 | 101 | 0.230 |
| >60 | 209 | 111 | 98 | |
| Gleason score | ||||
| G6–G7 | 256 | 109 | 147 | 0.000* |
| G8–G10 | 143 | 91 | 52 | |
| T-stage | ||||
| T1–T2 | 167 | 70 | 97 | 0.006* |
| T3–T4 | 232 | 130 | 102 | |
| Lymph node metastasis | ||||
| N0 | 288 | 142 | 146 | 0.039* |
| N1 | 54 | 35 | 19 | |
| Nx | 57 | 23 | 34 | |
| Grade | ||||
| I–II | 164 | 67 | 97 | 0.002* |
| III–IV | 235 | 133 | 102 | |
| PSA value | ||||
| 0–4 ng/ml | 375 | 182 | 193 | 0.019* |
| >4 ng/ml | 24 | 18 | 6 | |
Two-tailed fisher’s exact test was done with the SPSS software program to determine the statistical significance of the level of expression of SETD5 in different clinical characteristics,
p.value <0.05 was considered significant.
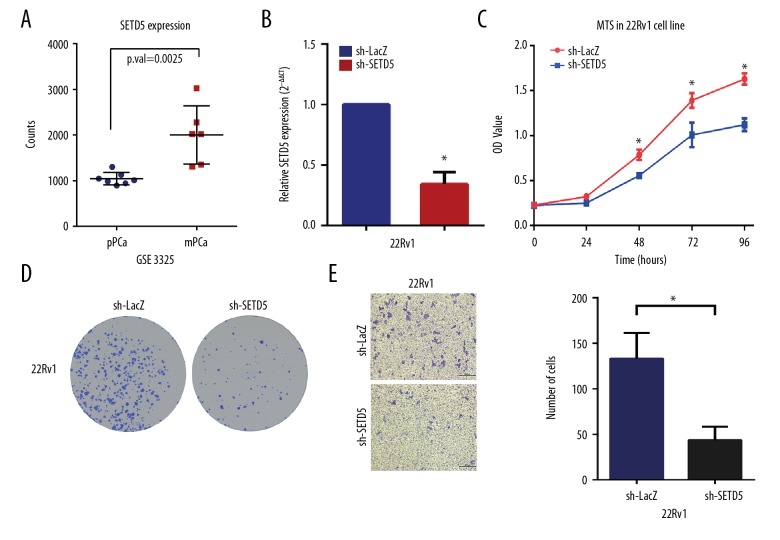
The validate these results in prostate cancer, SETD5 was selected. There was no difference in the expression of SETD5 between tumor and adjacent normal tissues, but there was a difference in the expression of SETD5 between tissue samples of primary prostate cancer and metastatic prostate cancer in the GSE3325 dataset [16]. Also, serial experiments were performed to validate the role of the SETD5 gene in the 22Rv1 prostate cancer cell line. Following the establishment of the SETD5 knockdown cell line, colony formation assay, the MTS proliferation assay, and the transwell assay, SETD5 gene knockdown reduced cancer cell growth and migration in the 22Rv1 cells (Figure 8).
Figure 8.
SETD5 gene knockdown reduced cell growth and migration in the 22Rv1 human prostate carcinoma cell line in vitro. (A) Differential expression of the SETD5 gene in primary prostate cancer (pPCa) and metastatic prostate cancer (mPCa) tissue samples. (B) The 22Rv1 human prostate carcinoma cells were stably transfected with lentivirus to knockdown the SETD5 gene. Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) detected the SETD5 gene expression level. (C) The stable transfection of the 22Rv1 cells underwent a colony formation assay. (D) The MTS cell proliferation assay was performed in stably transfected 22Rv1 cells. (E) A transwell assay was used to evaluate the migration capacity of stably transfected 22Rv1 cells.
Discussion
Integrated genomic analyses of 51 histone methyltransferase (HMT) genes were performed in prostate adenocarcinoma samples, which were collected from the Cancer Genome Atlas (TCGA) database. In this study, there were eight main findings. First, nine significant fold-change (FC) genes with false discovery rates (FDR) <0.05 were identified, including six 2.0-fold-change genes and three 1.5-fold-change genes. The expression of two HMT genes, PRDM6 and PRDM8, were significantly and negatively correlated with the Gleason score, while expression of the EZH2, PRDM12, SUV420H2, WHSC1, PRDM10 and SETD5 genes showed a significant positive correlation with the Gleason score. The mRNA levels of almost all HMT genes studied were significantly correlated with copy number, while expression of most HMT genes was negatively correlated with DNA methylation in prostate cancer in the cases in this study. An increased copy number alteration (CNA) in HMT genes in the prostate carcinoma tissue samples were positively correlated with a higher Gleason score and with a worse clinical outcome. Methylation of the PRDM8 gene was found to be significantly correlated with gene expression and clinical outcome. PRDM5, a methylation silencing gene, was identified as a potential gene for further investigation as a molecular target for prostate cancer. The SETD2 and ASH1L genes were found to have a potential role in the development of castration-resistant prostate cancer (CRPC). A panel of four HMT genes was identified as a gene prognostic signature, EZH2, SETD5, SMYD1, and SUV420H2, which could a potential prognostic biomarker panel. Also, knockdown of expression of the SETD5 gene reduced cell growth and migration of 22Rv1 human prostate carcinoma cells in vitro, indicating that the SETD5 gene may act as an oncogene in prostate cancer.
The HMT family includes histone lysine methyltransferases (HKMTs) and histone/protein arginine methyltransferases (PRMTs), which are enzymes that catalyze the transfer of methyl groups to specific substrates involving histones and specific proteins [7,17]. The expression of HMT gene alterations has been identified in several human cancers and are thought to have an oncogenic role, including in prostate cancer [8]. One of the most studied HMT genes is the enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), which is a protein-encoding gene. Genome and transcriptome alterations of EZH2 (amplification, overexpression, alternative splicing or mutation) have been validated to correlate with tumorigenesis and tumor progression in several types of cancers, including prostate cancer [18–21]. Also, increased EZH2 mRNA and protein levels have previously been shown to be associated with more rapid progression of prostate cancer, which may result from silencing of a specific cohort of genes by hypermethylation of specific histones [20,22]. Another example of a prognostic gene is WHSC1 (also known as NSD2), which is an H3K36 histone methyltransferase [23]. Current studies indicate that the WHSC1 gene is frequently overexpressed in several cancers, including prostate cancer, and has as an oncogenic role [24,25]. In the present study, overexpression of WHSC1 was more common in high Gleason score prostate cancers and negatively correlated with patient prognosis (Supplementary Figure 1, Supplementary Table 7). The findings of this study validated earlier studies that for individual HMT genes, several HMTs were identified as potential oncogene or tumor suppressor genes in prostate cancer.
Genetic alterations, including DNA methylation, to contribute to cellular transformation and carcinogenesis and are characteristic of most human malignancies [26]. Gene-specific DNA hypermethylation of promoter regions is associated with the inactivation of genes and downregulation of transcription [27,28]. The PRDM8 and PRDM2 genes have been previously shown to be downregulated in invasive pituitary adenomas by whole-exome sequencing (WES), and they are both associated with cell proliferation and tumor recurrence [29]. Aberrant methylation of PRDM8 has been shown to be correlated with congenital dyskeratosis [30], but has not been previously studied in malignant tumors.
PRDM5 is a recognized tumor suppressor gene. Abnormal methylation of PRDM5 and levels of expression has been associated with carcinogenesis was validated in several cancers including cervical, colorectal, lung, and gastric cancers [31–33]. In some studies, PRDM5 has been identified as a target of epigenetic silencing, which supports the findings of the present study [34]. Differentially methylated CpG sites of PRDM13, which may contribute to more aggressive behavior in prostate cancer, were identified in tumor tissues of African American men [35]. In the present study, promoter methylation levels of PRDM13 and PRDM14 were significantly associated with clinical survival (Supplementary Figure 7). Therefore, it is possible that when epigenetic modification caused by DNA methylation is reversible, carcinogenesis and cancer promotion caused by altered DNA methylation might be targeted by anti-methylation in cancer treatment.
The SETDB2 gene is one of HMT genes for histone H3 lysine 9 (H3K9) methylation and contains a commonly deleted gene at 13q in chronic lymphocytic leukemia (CLL) [36,37]. In the present study, SETDB2 was the most frequently deleted HMT gene in prostate cancer, and this deletion was significantly related to clinical outcome (Supplementary Figure 3). Studies in patients with CLL with deletion of SETDB2 have shown an associated with disease progression [37]. A recently reported study showed that overexpression of SETDB2 may contribute to disease progression in gastric cancer [38]. These previous studies support the findings of the present study (Supplementary Table 5), and raise the possibility that CNAs function in tumorigenesis by several methods, regardless of their effects on gene expression.
The PRDM1 gene, localized at 6q21, has been previously shown to be a tumor suppressor gene in several human malignancies, especially in lymphoma. In diffuse large B-cell lymphoma, the PRDM1 gene can be inactivated by multiple mechanisms, including homozygous deletions, or missense or truncating mutations, which contribute to lymphomagenesis by blocking plasma cell differentiation [39,40]. Additionally, deficiency of PRDM1 contributes to the maintenance of the phenotype and pathogenesis of gliomas [41]. PRDM14, a transcriptional regulator localized at 8q13, is overexpressed in several cancers and was also the most amplified HMT gene in the present study. However, as well as being associated with clinical survival, amplification of PRDM14 minimally contributed to its mRNA level (Figure 2B, Supplementary Figure 3). Overexpression of PRDM14 mRNA and protein as well as gene amplification have been reported in breast cancer, which correlated with disease progression and poor clinical outcome, indicating that alterations of PRDM14 could be a potential target for the treatment of breast cancer [42,43]. In pancreatic cancer, overexpression of PRDM14 accompanied by deregulation of miR-125a-3p has been shown to be involved in cancer stem-like phenotypes and liver metastasis [44]. Similar results were also found in cases of non-small cell lung cancer (NSCLC) and leukemia [45,46].
The KMT2C and KMT2D genes, the most commonly mutated HMT genes in human cancers, were also validated in the present study [47]. KMT2C and KMT2D are essential for regulation of enhancer activity by H3K4 mono-methylation, with mutation promoting tumorigenesis by deregulation of enhancer activity [48,49]. Reduced expression of KMT2C and KMT2D has previously been shown to significantly correlate with improved outcome in pancreatic ductal adenocarcinoma, and in vitro studies have previously shown that the depleted level of these two HMTs attenuated cell proliferation by blocking cell-cycle and diminishing progression from G0–G1 [50]. In endometrioid endometrial carcinoma, the mutational status of KMT2C and KMT2D has been shown to correlate with the degree of myometrial invasion, which could be used to predict prognostic clinical outcome [51]. Also, a high level of expression of the KMT2D gene was associated with poor prognosis in patients with breast cancer, and KMT2D knockdown impaired proliferation and invasion in human breast cancer xenografts in mice [52]. In the present study, the expression of KMT2D was significantly associated with clinical outcome (Supplementary Table 7), suggesting that KMT2D might function as an oncogene in prostate cancer.
Increasing published evidence has now shown that abnormal epigenetic regulation caused by genetic alterations of HMTs, including CNAs, mutation, ectopic expression, and methylation, contribute to tumorigenesis and progression in malignancy. Abnormal epigenetic dysregulation can be treated with targeted drugs or other treatments, and targeting these abnormalities provide potential personalized treatment approaches for cancer patients. Several inhibitors of selective HMTs, including DOT1L, EZH2, WHSC1, have been previously validated with targeted antitumor effects [53]. Also, because mixed-lineage leukemia (MLL) complex family members act as a crucial co-activator of the androgen receptor (AR) in advanced prostate cancer, a small-molecule inhibitor may improve the clinical outcome of patients with castration-resistant prostate cancer (CRPC) by targeting the MLL complex [15]. The results of the present study from the analysis 51 HMT genes systematically identified several HMT genes that have frequent genetic alterations or relate to clinical outcome in prostate cancer, and a four-gene HMT prognostic signature (EZH2, SETD5, SMYD1, and SUV420H2) was identified.
Conclusions
The findings of this study have shown that epigenetic regulation of histone methyltransferases (HMTs) play important roles in the genesis and progression of prostate cancer. Further studies are required to determine the roles of HMTs in the diagnosis, treatment, and prognosis in prostate cancer.
Supplementary Materials
mRNA levels of 51 HMTs in different Gleason score prostate cancer samples. (A) Pheatmap of 51 HMTs expression profile in totally 399 prostate cancer samples. Normalized gene expression values by rows were median-centered prior to rows clustering based on Euclidean distance. One way ANOVA analysis were performed to compare the differences between Gleason score 6, 7, 8, and 9 prostate tumors. Significantly different expressed genes in prostate tumors are shown at the top, potential cancer related genes(p<0.0001) are indicated by a red box. (B) Box plot show significantly four genes (PRDM6, PRDM8, PRDM12, EZH2) expression level(y-axis) positive or negative related to different Gleason score(x-axis).
mRNA levels of four HMTs in different Gleason score prostate cancers. Box plots show four genes (WHSC1, SUV420H2, PRDM10, SETD5) expression level (y-axis) significantly related to different Gleason score (x-axis).
CNAs frequencies of HMTs and their impact on clinical survival. (A, B) Bar graphs showed the most frequently altered HMTs. (C, D) Kaplan-Meier plotS of RFS proportion shows that deletion of SETDB2 and amplification of PRDM14 were significantly relevant to clinical outcome.
HMTs copy number alteration profiles in different Gleason score prostate samples. Pheatmap shows that CNAs frequencies of HMTs were tendentiously positive related to Gleason score, interestingly, both for amplification and deletion alterations.
HMTs copy number alteration profiles in different stage prostate cancers. (A) Pheatmap shows that increasing CNA events of 51 HMTs correlated with different stages of prostate cancers. (B) Histogram shown average CNAs per sample in different stage prostate cancers.
Kaplan-Meier plots of RFS proportion show that different impacts caused by CNAs and ectopic gene expression of significant HMTs (PRDM12, SETD6, SMYD1, WHSC1L1) on clinical survival.
Kaplan-Meier plots of RFS proportion show that different impactS caused by methylation level of significant HMTs (NSD1, PRDM13,PRDM14, KMT2B) on clinical survival.
Supplementary Table 1.
Summary of identified human HMTs and their histone substrates.
| Official symbol | Other aliases | Gene ID | Gene location | Histone substrates |
|---|---|---|---|---|
| ASH1L | KMT2H; ASH1L1 | 55870 | 1q22 | H3K4me1/3, H3K36me2 |
| DOT1L | DOT1; KMT4 | 84444 | 19p13.3 | H3K79me1/2/3 |
| EHMT1 | GLP1; KMT1D | 79813 | 9q34.3 | H3K9me1/2 |
| EHMT2 | G9A; KMT1C | 10919 | 6p21.31 | H3K9me1/2 |
| EZH1 | KMT6B | 2145 | 17q21.2 | H3K27me2/3 |
| EZH2 | KMT6A | 2146 | 7q36.1 | H3K27me2/3 |
| KMT2A | MLL | 4297 | 11q23.3 | H3K4me1/2/3 |
| KMT2B | MLL2 | 9757 | 19q13.12 | H3K4me1/2/3 |
| KMT2C | MLL3 | 58508 | 7q36.1 | H3K4me1/2/3 |
| KMT2D | MLL4 | 8085 | 12q13.12 | H3K4me1/2/3 |
| KMT2E | MLL5 | 55904 | 7q22.3 | H3K4 |
| MECOM | PRDM3; MDS1-EVI1 | 2122 | 3q26.2 | H3K9me1 |
| NSD1 | KMT3B | 64324 | 5q35.2 | H3K36me1/2 |
| PRDM1 | BLIMP16 | 639 | 6q21 | |
| PRDM10 | PFM7 | 56980 | 11q24.3 | |
| PRDM11 | PFM8 | 56981 | 11p11.2 | |
| PRDM12 | PFM9 | 59335 | 9q34.12 | |
| PRDM13 | PFM10 | 59336 | 6q16.2 | |
| PRDM14 | PFM11 | 63978 | 8q13.3 | |
| PRDM15 | PFM15 | 63977 | 21q22.3 | |
| PRDM16 | MEL1; PFM13 | 63976 | 1p36.32 | H3K9me1 |
| PRDM2 | RIZ; KMT8 | 7799 | 1p36.21 | H3K9me1/2/3 |
| PRDM4 | PFM1 | 11108 | 12q23.3 | |
| PRDM5 | PFM2 | 11107 | 4q27 | H3K9me2 |
| PRDM6 | 93166 | 5q23.2 | ||
| PRDM7 | PFM4; ZNF910 | 11105 | 16q24.3 | |
| PRDM8 | PFM5 | 56978 | 4q21.21 | H3k9me3 |
| PRDM9 | PFM6; MEISETZ | 56979 | 5p14.2 | H3K4me3 |
| SETD1A | KMT2F; SET1A | 9739 | 16p11.2 | H3K4me1/2/3 |
| SETD1B | KMT2G; SET1B | 23067 | 12q24.31 | H3K4me1/2/3 |
| SETD2 | HYPB; SET2 | 29072 | 3p21.31 | H3K36me1/2/3 |
| SETD3 | C14orf154 | 84193 | 14q32.2 | |
| SETD4 | C21orf18 | 54093 | 21q22.12 | |
| SETD5 | 55209 | 3p25.3 | ||
| SETD6 | 79918 | 16q21 | ||
| SETD7 | KMT7; SET7/9 | 80854 | 4q31.1 | H3K4me1 |
| SETD8 | KMT5A; PR-SETD7 | 387893 | 12q24.31 | H4K20me1 |
| SETDB1 | ESET; KMT1E | 9869 | 1q21.3 | H3K9me1/2/3 |
| SETDB2 | CLL8; KMT1F | 83852 | 13q14.2 | H3K9 |
| SETMAR | METNASE | 6419 | 3p26.1 | H3K36 |
| SMYD1 | KMT3D; ZMYND18 | 150572 | 2p11.2 | H3K4 |
| SMYD2 | KMT3C; ZMYND14 | 56950 | 1q32.3 | H3K4; H3K36me2 |
| SMYD3 | KMT3E; ZMYND1; | 64754 | 1q44 | H3K4me2/3 |
| SMYD4 | ZMYND21 | 114826 | 17p13.3 | |
| SMYD5 | RRG1; ZMYND23 | 10322 | 2p13.2 | |
| SUV39H1 | KMT1A | 6839 | Xp11.23 | H3K9me2/3 |
| SUV39H2 | KMT1B | 79723 | 10p13 | H3K9me2/3 |
| SUV420H1 | KMT5B; CGI-85 | 51111 | 11q13.2 | H4K20me2/3 |
| SUV420H2 | KMT5C | 84787 | 19q13.42 | H4K20me2/3 |
| WHSC1 | NSD2; MMSET | 7468 | 4p16.3 | H3K36me1/2 |
| WHSC1L1 | NSD3 | 54904 | 8p11.23 | H3K36me1/2; H3K4 |
Supplementary Table 2.
Number of samples from TCGA used in our analyses. For the RNA-Seq data and methylation data the number of tumor and normal samples are indicated. We also indicate the number of tumor samples with mutation data, and with copy number variation (CNV) data.
| Acronym | Tumor type | Data type | Paired samples | Total tumor samples | Metastatic PRAD | |
|---|---|---|---|---|---|---|
| Normal | Tumor | |||||
| PRAD | Prostate adenocarcinoma | RNA seq data | 52 | 52 | 497 | 0 |
| CpG data(46HMTs) | 50 | 50 | 499 | 0 | ||
| CNV data | 0 | 0 | 492 | 150 | ||
| Mutation data | 0 | 0 | 425 | 0 | ||
| Quadruple overlap | 0 | 0 | 399 | 0 | ||
Supplementary Table 3.
Summary of expression fold-changed genes with false discovery rates (FDR) <0.05.
| Gene | logFC | AveExpr | t | P.value | FDR | B | |
|---|---|---|---|---|---|---|---|
| 2.0FC | PRDM8 | −1.526 | 9.088 | −5.666 | 1.30E-07 | 8.08E-07 | 6.880 |
| MECOM | −1.216 | 8.501 | −6.737 | 9.03E-10 | 9.65E-09 | 11.736 | |
| PRDM16 | −1.129 | 4.769 | −4.080 | 8.80E-05 | 2.54E-04 | 0.592 | |
| SMYD1 | −1.033 | 1.099 | −2.254 | 2.63E-02 | 4.55E-02 | −4.668 | |
| PRDM12 | 1.016 | 1.348 | 7.227 | 8.37E-11 | 2.18E-09 | 14.071 | |
| EZH2 | 1.433 | 5.944 | 8.778 | 3.43E-14 | 1.78E-12 | 21.754 | |
| 1.5FC | PRDM5 | −0.932 | 5.892 | −5.874 | 5.10E-08 | 3.79E-07 | 7.790 |
| PRDM11 | −0.904 | 4.968 | −6.843 | 5.42E-10 | 9.40E-09 | 12.235 | |
| PRDM6 | −0.800 | 5.200 | −3.475 | 7.44E-04 | 1.93E-03 | −1.424 | |
| 1.2FC | EZH1 | −0.542 | 9.792 | −6.731 | 9.28E-10 | 9.65E-09 | 11.709 |
| SMYD4 | −0.371 | 8.315 | −4.951 | 2.84E-06 | 1.23E-05 | 3.885 | |
| PRDM15 | 0.320 | 7.400 | 5.011 | 2.21E-06 | 1.05E-05 | 4.127 | |
| PRDM10 | 0.337 | 7.990 | 4.292 | 3.96E-05 | 1.47E-04 | 1.353 | |
| WHSC1 | 0.367 | 9.820 | 5.064 | 1.77E-06 | 9.20E-06 | 4.345 | |
| SETD7 | 0.429 | 11.984 | 4.351 | 3.16E-05 | 1.26E-04 | 1.571 | |
| KMT2B | 0.432 | 11.025 | 3.438 | 8.42E-04 | 1.99E-03 | −1.540 | |
| SMYD2 | 0.433 | 8.849 | 4.118 | 7.65E-05 | 2.34E-04 | 0.725 | |
| SUV39H2 | 0.455 | 7.832 | 4.273 | 4.26E-05 | 1.48E-04 | 1.284 | |
| SMYD3 | 0.509 | 7.713 | 5.885 | 4.84E-08 | 3.79E-07 | 7.841 | |
| DOT1L | 0.567 | 8.862 | 5.649 | 1.40E-07 | 8.08E-07 | 6.807 |
Supplementary Table 4.
Summary for association of Gleason score and AR expression with HMTs expression profile.
| Gleason score/HMTs expression (one-way ANOVA, top 11 HMTs were filled with yellow color) | AR/HMTs expression (top 13 significant HMTs with the green padding were also in the same clustering) | |||
|---|---|---|---|---|
| Gene | p.Value | Gene | p.Value | Coefficients |
| ASH1L | 4.49E-01 | ASH1L | 4.94E-155 | 0.871 |
| DOT1L | 7.57E-04 | DOT1L | 1.43E-06 | 0.214 |
| EHMT1 | 2.78E-04 | EHMT1 | 9.08E-46 | 0.579 |
| EHMT2 | 8.27E-01 | EHMT2 | 2.19E-12 | 0.308 |
| EZH1 | 2.11E-02 | EZH1 | 9.07E-01 | 0.005 |
| EZH2 | 1.35E-12 | EZH2 | 1.23E-03 | 0.145 |
| KMT2A | 4.68E-01 | KMT2A | 5.77E-97 | 0.766 |
| KMT2B | 1.40E-01 | KMT2B | 1.24E-128 | 0.832 |
| KMT2C | 3.70E-01 | KMT2C | 5.65E-122 | 0.820 |
| KMT2D | 2.60E-05 | KMT2D | 5.63E-32 | 0.494 |
| KMT2E | 1.92E-01 | KMT2E | 4.56E-105 | 0.785 |
| MECOM | 2.21E-01 | MECOM | 1.31E-06 | 0.215 |
| NSD1 | 5.49E-02 | NSD1 | 8.13E-127 | 0.829 |
| PRDM1 | 3.20E-01 | PRDM1 | 7.06E-13 | 0.315 |
| PRDM10 | 4.92E-04 | PRDM10 | 1.21E-87 | 0.741 |
| PRDM11 | 2.71E-03 | PRDM11 | 6.96E-12 | 0.301 |
| PRDM12 | 5.82E-06 | PRDM12 | 1.80E-01 | 0.060 |
| PRDM13 | 4.62E-01 | PRDM13 | 2.76E-01 | 0.049 |
| PRDM14 | 4.33E-01 | PRDM14 | 7.13E-01 | 0.017 |
| PRDM15 | 5.90E-02 | PRDM15 | 6.48E-04 | 0.152 |
| PRDM16 | 6.83E-01 | PRDM16 | 7.48E-01 | 0.014 |
| PRDM2 | 3.85E-01 | PRDM2 | 2.77E-70 | 0.686 |
| PRDM4 | 4.69E-01 | PRDM4 | 3.72E-54 | 0.620 |
| PRDM5 | 9.29E-02 | PRDM5 | 3.45E-19 | 0.387 |
| PRDM6 | 6.89E-04 | PRDM6 | 4.26E-03 | 0.128 |
| PRDM7 | 3.84E-01 | PRDM7 | 5.38E-03 | 0.125 |
| PRDM8 | 6.63E-05 | PRDM8 | 8.01E-01 | 0.011 |
| PRDM9 | 5.35E-01 | PRDM9 | 7.02E-01 | 0.017 |
| SETD1A | 3.29E-01 | SETD1A | 3.63E-02 | 0.094 |
| SETD1B | 9.31E-03 | SETD1B | 1.11E-44 | 0.573 |
| SETD2 | 2.64E-02 | SETD2 | 3.47E-129 | 0.833 |
| SETD3 | 3.48E-01 | SETD3 | 8.77E-01 | 0.007 |
| SETD4 | 1.29E-03 | SETD4 | 2.56E-22 | 0.417 |
| SETD5 | 2.64E-05 | SETD5 | 6.88E-78 | 0.712 |
| SETD6 | 5.42E-02 | SETD6 | 7.32E-01 | 0.015 |
| SETD7 | 4.45E-01 | SETD7 | 9.24E-106 | 0.787 |
| SETD8 | 2.48E-03 | SETD8 | 5.01E-09 | 0.258 |
| SETDB1 | 1.24E-02 | SETDB1 | 1.60E-13 | 0.323 |
| SETDB2 | 3.50E-01 | SETDB2 | 1.61E-02 | 0.108 |
| SETMAR | 1.63E-01 | SETMAR | 2.40E-34 | 0.511 |
| SMYD1 | 1.97E-01 | SMYD1 | 9.40E-01 | 0.003 |
| SMYD2 | 4.78E-01 | SMYD2 | 2.22E-01 | 0.055 |
| SMYD3 | 5.14E-01 | SMYD3 | 1.14E-09 | 0.269 |
| SMYD4 | 3.06E-01 | SMYD4 | 9.99E-20 | 0.392 |
| SMYD5 | 9.71E-01 | SMYD5 | 8.87E-21 | 0.402 |
| SUV39H1 | 3.77E-01 | SUV39H1 | 4.30E-19 | 0.386 |
| SUV39H2 | 4.21E-02 | SUV39H2 | 1.03E-05 | 0.196 |
| SUV420H1 | 6.57E-03 | SUV420H1 | 3.81E-42 | 0.559 |
| SUV420H2 | 3.88E-04 | SUV420H2 | 7.67E-26 | 0.447 |
| WHSC1 | 2.89E-14 | WHSC1 | 4.06E-31 | 0.488 |
| WHSC1L1 | 5.19E-01 | WHSC1L1 | 6.19E-47 | 0.585 |
Supplementary Table 5.
Summary of correlation between CNAs frequencied and gene expression levels.
| Gene | Amplification | Deletion | Diploid | Total amplification | Total deletion | Amplification and expressed higher (%) | Amplification and expressed lower (%) | Deletion and expressed higher (%) | Deletion and expressed lower (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Higher | Lower | Higher | Lower | Higher | Lower | |||||||
| Expression | ||||||||||||
| ASH1L | 19 | 8 | 0 | 8 | 183 | 181 | 27 | 8 | 70.37 | 29.63 | 0.00 | 100.00 |
| DOT1L 4 | 1 | 11 | 19 | 179 | 185 | 5 | 30 | 80.00 | 20.00 | 36.67 | 63.33 | |
| EHMT1 | 35 | 11 | 3 | 5 | 183 | 162 | 46 | 8 | 76.09 | 23.91 | 37.50 | 62.50 |
| EHMT2 | 8 | 4 | 5 | 10 | 185 | 187 | 12 | 15 | 66.67 | 33.33 | 33.33 | 66.67 |
| EZH1 | 3 | 1 | 13 | 37 | 161 | 184 | 4 | 50 | 75.00 | 25.00 | 26.00 | 74.00 |
| EZH2 | 58 | 15 | 2 | 6 | 178 | 140 | 73 | 8 | 79.45 | 20.55 | 25.00 | 75.00 |
| KMT2A | 12 | 10 | 14 | 15 | 174 | 174 | 22 | 29 | 54.55 | 45.45 | 48.28 | 51.72 |
| KMT2B | 6 | 3 | 10 | 6 | 190 | 184 | 9 | 16 | 66.67 | 33.33 | 62.50 | 37.50 |
| KMT2C | 39 | 35 | 10 | 11 | 153 | 151 | 74 | 21 | 52.70 | 47.30 | 47.62 | 58.38 |
| KMT2D | 6 | 5 | 8 | 8 | 186 | 186 | 11 | 16 | 54.55 | 45.45 | 50.00 | 50.00 |
| KMT2E | 39 | 37 | 3 | 4 | 158 | 158 | 76 | 7 | 51.32 | 48.68 | 42.86 | 57.14 |
| MECOM | 36 | 26 | 3 | 4 | 169 | 161 | 62 | 7 | 58.06 | 41.94 | 42.86 | 57.14 |
| NSD1 | 16 | 6 | 5 | 5 | 188 | 179 | 22 | 10 | 72.73 | 27.27 | 50.00 | 50.00 |
| PRDM1 | 2 | 0 | 62 | 60 | 139 | 136 | 2 | 122 | 100.00 | 0.00 | 50.82 | 49.18 |
| PRDM10 | 18 | 6 | 6 | 13 | 180 | 176 | 24 | 19 | 75.00 | 25.00 | 31.58 | 68.42 |
| PRDM11 | 8 | 8 | 6 | 20 | 171 | 186 | 16 | 26 | 50.00 | 50.00 | 23.08 | 76.92 |
| PRDM12 | 33 | 11 | 1 | 9 | 179 | 166 | 44 | 10 | 75.00 | 25.00 | 10.00 | 90.00 |
| PRDM13 | 0 | 5 | 7 | 118 | 189 | 80 | 5 | 125 | 0.00 | 100.00 | 5.60 | 94.40 |
| PRDM14 | 16 | 96 | 0 | 2 | 270 | 15 | 112 | 2 | 14.29 | 85.71 | 0.00 | 100.00 |
| PRDM15 | 14 | 4 | 8 | 18 | 177 | 178 | 18 | 26 | 77.78 | 22.22 | 30.77 | 69.23 |
| PRDM16 | 3 | 4 | 13 | 16 | 179 | 184 | 7 | 29 | 42.86 | 57.14 | 44.83 | 55.17 |
| PRDM2 | 2 | 0 | 9 | 26 | 173 | 189 | 2 | 35 | 100.00 | 0.00 | 25.71 | 74.29 |
| PRDM4 | 18 | 2 | 6 | 13 | 184 | 176 | 20 | 19 | 90.00 | 10.00 | 31.58 | 68.42 |
| PRDM5 | 4 | 6 | 7 | 29 | 164 | 189 | 10 | 36 | 40.00 | 60.00 | 19.44 | 80.56 |
| PRDM6 | 7 | 6 | 6 | 26 | 167 | 187 | 13 | 32 | 53.85 | 46.15 | 18.75 | 81.25 |
| PRDM7 | 1 | 3 | 42 | 103 | 160 | 90 | 40 | 145 | 25.00 | 75.00 | 28.97 | 71.03 |
| PRDM8 | 4 | 7 | 5 | 10 | 182 | 191 | 11 | 15 | 36.36 | 63.64 | 33.33 | 66.67 |
| PRDM9 | 6 | 19 | 2 | 8 | 347 | 17 | 25 | 10 | 24.00 | 76.00 | 20.00 | 80.00 |
| SETD1A | 20 | 3 | 14 | 11 | 185 | 166 | 23 | 25 | 86.96 | 13.04 | 56.00 | 44.00 |
| SETD1B | 13 | 3 | 11 | 17 | 179 | 176 | 16 | 28 | 81.25 | 18.75 | 39.29 | 60.71 |
| SETD2 | 17 | 7 | 3 | 16 | 176 | 180 | 24 | 19 | 70.83 | 29.17 | 15.79 | 84.21 |
| SETD3 | 8 | 2 | 6 | 25 | 172 | 186 | 10 | 31 | 80.00 | 20.00 | 19.35 | 80.65 |
| SETD4 | 14 | 3 | 4 | 13 | 183 | 182 | 17 | 17 | 82.35 | 17.65 | 23.53 | 76.47 |
| SETD5 | 17 | 8 | 3 | 25 | 166 | 180 | 25 | 28 | 68.00 | 32.00 | 10.71 | 89.29 |
| SETD6 | 6 | 0 | 15 | 78 | 121 | 179 | 6 | 93 | 100.00 | 0.00 | 16.13 | 83.87 |
| SETD7 | 7 | 4 | 3 | 14 | 181 | 190 | 11 | 17 | 63.64 | 36.36 | 17.65 | 82.35 |
| SETD8 | 11 | 3 | 6 | 25 | 171 | 183 | 14 | 31 | 78.57 | 21.43 | 19.35 | 80.65 |
| SETDB1 | 26 | 2 | 1 | 1 | 196 | 173 | 28 | 2 | 92.86 | 7.14 | 50.00 | 50.00 |
| SETDB2 | 1 | 1 | 34 | 131 | 67 | 165 | 2 | 165 | 50.00 | 50.00 | 20.61 | 79.39 |
| SETMAR | 17 | 8 | 5 | 16 | 175 | 178 | 25 | 21 | 68.00 | 32.00 | 23.81 | 76.19 |
| SMYD1 | 2 | 10 | 4 | 16 | 279 | 88 | 12 | 20 | 16.67 | 83.33 | 20.00 | 80.00 |
| SMYD2 | 12 | 9 | 4 | 16 | 174 | 184 | 21 | 20 | 57.14 | 42.86 | 20.00 | 80.00 |
| SMYD3 | 18 | 3 | 3 | 11 | 185 | 179 | 21 | 14 | 85.71 | 14.29 | 21.43 | 78.57 |
| SMYD4 | 5 | 0 | 9 | 58 | 141 | 186 | 5 | 67 | 100.00 | 0.00 | 13.43 | 86.57 |
| SMYD5 | 11 | 1 | 5 | 19 | 179 | 184 | 12 | 24 | 91.67 | 8.33 | 20.83 | 79.17 |
| SUV39H1 | 4 | 5 | 13 | 7 | 187 | 183 | 9 | 20 | 44.44 | 55.56 | 65.00 | 35.00 |
| SUV39H2 | 12 | 2 | 4 | 29 | 168 | 184 | 14 | 33 | 85.71 | 14.29 | 12.12 | 87.88 |
| SUV420H1 | 26 | 7 | 0 | 6 | 186 | 174 | 33 | 6 | 174.79 | 21.21 | 0.00 | 100.0 |
| SUV420H2 | 13 | 1 | 10 | 4 | 194 | 177 | 14 | 14 | 92.86 | 7.114 | 71.43 | 28.57 |
| WHSC1 | 14 | 2 | 9 | 12 | 185 | 177 | 16 | 21 | 87.50 | 12.50 | 42.86 | 57.14 |
| WHSC1L1 | 44 | 7 | 18 | 111 | 81 | 138 | 51 | 129 | 86.27 | 13.73 | 13.95 | 86.05 |
Supplementary Table 6.
Summary for differential promoter methylation HMTs in paired normal and tumor tissues.
| Gene | Differential methylation analysis of 46 HMTs | Paired T-test | |||||
|---|---|---|---|---|---|---|---|
| logFC | AveExpr | t | P.value | adj.P.Val | B | ||
| PRDM14 | −0.201 | 0.320 | −11.345 | 1.30E-19 | 6.09E-18 | 33.276 | 1.75E-17 |
| NSD1 | −0.142 | 0.421 | −9.479 | 1.49E-15 | 3.50E-14 | 23.928 | 2.44E-13 |
| PRDM8 | −0.146 | 0.432 | −8.138 | 1.20E-12 | 1.89E-11 | 17.253 | 6.68E-12 |
| EZH1 | −0.024 | 0.103 | −7.529 | 2.41E-11 | 2.83E-10 | 14.277 | 1.61E-10 |
| PRDM13 | −0.121 | 0.306 | −7.276 | 8.19E-11 | 7.70E-10 | 13.062 | 6.16E-12 |
| PRDM6 | −0.065 | 0.158 | −6.626 | 1.81E-09 | 1.42E-08 | 9.998 | 6.49E-10 |
| PRDM12 | −0.047 | 0.119 | −6.586 | 2.18E-09 | 1.47E-08 | 9.813 | 3.76E-10 |
| SETD3 | 0.061 | 0.604 | 6.222 | 1.18E-08 | 6.93E-08 | 8.149 | 3.26E-08 |
| PRDM5 | −0.093 | 0.117 | −5.224 | 9.70E-07 | 5.07E-06 | 3.821 | 2.09E-06 |
| KMT2B | 0.057 | 0.551 | 4.548 | 1.54E-05 | 7.22E-05 | 1.136 | 5.32E-06 |
| PRDM15 | −0.012 | 0.157 | −4.393 | 2.81E-05 | 1.20E-04 | 0.554 | 2.38E-05 |
| SETD5 | 0.037 | 0.474 | 3.924 | 1.61E-04 | 6.31E-04 | −1.122 | 6.03E-05 |
| PRDM7 | 0.041 | 0.537 | 3.845 | 2.13E-04 | 7.71E-04 | −1.391 | 1.73E-04 |
| WHSC1 | 0.023 | 0.463 | 3.619 | 4.67E-04 | 1.57E-03 | −2.136 | 2.21E-04 |
| SETDB1 | 0.014 | 0.140 | 3.088 | 2.61E-03 | 8.19E-03 | −3.749 | 1.03E-04 |
| EHMT2 | 0.004 | 0.097 | 2.887 | 4.78E-03 | 1.40E-02 | −4.307 | 3.23E-04 |
| MECOM | −0.016 | 0.151 | −2.837 | 5.52E-03 | 1.53E-02 | −4.439 | 3.26E-03 |
| KMT2E | −0.031 | 0.417 | −2.721 | 7.69E-03 | 1.72E-02 | −4.741 | 5.31E-03 |
| SETD1A | 0.009 | 0.127 | 2.725 | 7.60E-03 | 1.72E-02 | −4.731 | 6.35E-03 |
| SMYD1 | 0.050 | 0.805 | 2.747 | 7.14E-03 | 1.72E-02 | −4.674 | 5.86E-03 |
| PRDM9 | 0.071 | 0.586 | 2.735 | 7.39E-03 | 1.72E-02 | −4.705 | 9.94E-04 |
| SUV420H1 | −0.008 | 0.225 | −2.700 | 8.16E-03 | 1.74E-02 | −4.795 | 2.43E-03 |
| PRDM2 | 0.007 | 0.431 | 2.583 | 1.13E-02 | 2.30E-02 | −5.087 | 3.74E-03 |
| PRDM11 | 0.019 | 0.845 | 2.497 | 1.42E-02 | 2.77E-02 | −5.293 | 9.22E-03 |
| PRDM4 | −0.004 | 0.062 | −2.420 | 1.73E-02 | 3.26E-02 | −5.473 | 1.23E-02 |
| PRDM10 | 0.025 | 0.564 | 2.390 | 1.87E-02 | 3.38E-02 | −5.542 | 2.59E-02 |
| DOT1L | 0.002 | 0.036 | 2.290 | 2.42E-02 | 4.20E-02 | −5.766 | 9.03E-03 |
| SMYD5 | 0.008 | 0.230 | 1.982 | 5.02E-02 | 8.43E-02 | −6.401 | 3.76E-02 |
| SETD7 | 0.003 | 0.057 | 1.902 | 6.00E-02 | 9.73E-02 | −6.551 | 2.41E-02 |
| SETMAR | −0.005 | 0.173 | −1.523 | 1.31E-01 | 2.05E-01 | −7.188 | 8.77E-02 |
| SETD4 | 0.001 | 0.032 | 1.425 | 1.57E-01 | 2.39E-01 | −7.331 | 1.30E-01 |
| KMT2D | 0.003 | 0.079 | 1.318 | 1.91E-01 | 2.71E-01 | −7.476 | 5.25E-02 |
| SETDB2 | 0.005 | 0.905 | 1.335 | 1.85E-01 | 2.71E-01 | −7.454 | 1.62E-01 |
| WHSC1L1 | −0.004 | 0.151 | −1.244 | 2.17E-01 | 2.99E-01 | −7.571 | 2.39E-01 |
| SUV39H2 | 0.001 | 0.041 | 1.191 | 2.36E-01 | 3.09E-01 | −7.634 | 2.32E-01 |
| AR | 0.005 | 0.316 | 1.191 | 2.36E-01 | 3.09E-01 | −7.634 | 1.74E-01 |
| PRDM1 | −0.014 | 0.148 | −1.147 | 2.54E-01 | 3.23E-01 | −7.686 | 2.81E-01 |
| SETD2 | 0.001 | 0.036 | 0.996 | 3.22E-01 | 3.98E-01 | −7.847 | 2.55E-01 |
| ASH1L | 0.003 | 0.473 | 0.978 | 3.31E-01 | 3.98E-01 | −7.865 | 2.85E-01 |
| SUV420H2 | −0.001 | 0.025 | −0.908 | 3.66E-01 | 4.30E-01 | −7.931 | 3.20E-01 |
| SMYD2 | −0.002 | 0.062 | −0.828 | 4.10E-01 | 4.70E-01 | −8.001 | 3.54E-01 |
| SMYD3 | −0.002 | 0.413 | −0.685 | 4.95E-01 | 5.29E-01 | −8.109 | 4.27E-01 |
| EZH2 | −0.001 | 0.295 | −0.703 | 4.84E-01 | 5.29E-01 | −8.097 | 3.90E-01 |
| SETD6 | −0.001 | 0.039 | −0.717 | 4.75E-01 | 5.29E-01 | −8.086 | 3.54E-01 |
| SETD8 | 0.000 | 0.030 | −0.373 | 7.10E-01 | 7.42E-01 | −8.275 | 6.77E-01 |
| KMT2A | 0.000 | 0.030 | 0.107 | 9.15E-01 | 9.35E-01 | −8.340 | 8.71E-01 |
| SUV39H1 | 0.000 | 0.115 | 0.057 | 9.55E-01 | 9.55E-01 | −8.344 | 9.52E-01 |
Supplementary Table 7A.
Summary of uniivariate analysis of RFS for amplification of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95% CI | Count amplification | Count diploid | Count percentage | |
|---|---|---|---|---|---|---|---|
| Lower | Higher | ||||||
| PRDM14 | 0.0081 | 1.860 | 1.185 | 2.919 | 112 | 285 | 28.2 |
| WHSC1L1 | 0.0051 | 2.401 | 1.330 | 4.332 | 51 | 219 | 18.9 |
| EHMT1 | 0.0001 | 3.133 | 1.876 | 5.232 | 46 | 345 | 11.8 |
| PRDM12 | 0.0002 | 3.022 | 1.795 | 5.090 | 44 | 345 | 11.3 |
| SUV420H1 | 0.0075 | 2.390 | 1.336 | 4.273 | 33 | 360 | 8.4 |
| SETDB1 | 0.0031 | 2.860 | 1.543 | 5.303 | 28 | 369 | 7.1 |
| SETD1A | 0.0157 | 2.520 | 1.286 | 4.938 | 23 | 351 | 6.1 |
| SMYD2 | 0.0188 | 2.723 | 1.304 | 5.686 | 21 | 358 | 5.5 |
| PRDM4 | 0.0015 | 3.808 | 1.882 | 7.703 | 20 | 360 | 5.3 |
| PRDM15 | 0.0445 | 2.214 | 1.095 | 4.478 | 18 | 355 | 4.8 |
| SETD1B | 0.0006 | 4.815 | 2.279 | 10.173 | 16 | 355 | 4.3 |
| WHSC1 | 0.0467 | 2.299 | 1.098 | 4.813 | 16 | 362 | 4.2 |
| SETD8 | 0.0100 | 3.722 | 1.596 | 8.682 | 14 | 354 | 3.8 |
| SUV420H2 | 0.0499 | 2.409 | 1.099 | 5.281 | 14 | 371 | 3.6 |
| SMYD5 | 0.0009 | 5.127 | 2.330 | 11.284 | 12 | 363 | 3.2 |
| SMYD1 | 0.0005 | 5.729 | 2.599 | 12.630 | 12 | 367 | 3.2 |
| EHMT2 | 0.0278 | 2.969 | 1.281 | 6.882 | 12 | 372 | 3.1 |
| KMT2B | 0.0311 | 3.242 | 1.300 | 8.084 | 9 | 374 | 2.3 |
| PRDM16 | 0.0029 | 7.508 | 2.674 | 21.079 | 7 | 363 | 1.9 |
| DOT1L | 0.0358 | 3.657 | 1.323 | 10.106 | 5 | 364 | 1.4 |
| PRDM1 | 0.0219 | 9.340 | 2.227 | 39.178 | 2 | 275 | 0.7 |
| SETD3 | 0.2315 | 0.362 | 0.050 | 2.622 | 10 | 358 | 2.7 |
| PRDM7 | 0.3568 | 0.000 | 0.000 | Inf | 4 | 250 | 1.6 |
| SETD4 | 0.0502 | 2.263 | 1.083 | 4.731 | 17 | 365 | 4.5 |
| PRDM8 | 0.0507 | 2.393 | 1.096 | 5.227 | 11 | 373 | 2.9 |
| PRDM13 | 0.0664 | 3.720 | 1.147 | 12.065 | 5 | 269 | 1.8 |
| ASH1L | 0.0749 | 1.998 | 0.993 | 4.017 | 27 | 364 | 6.9 |
| PRDM9 | 0.0840 | 1.953 | 0.971 | 3.926 | 25 | 364 | 6.4 |
| SETD6 | 0.0854 | 4.711 | 1.119 | 19.832 | 6 | 300 | 2.0 |
| PRDM2 | 0.1026 | 9.419 | 1.273 | 69.698 | 2 | 362 | 0.5 |
| NSD1 | 0.1187 | 1.887 | 0.905 | 3.937 | 22 | 367 | 5.7 |
| MECOM | 0.1318 | 1.543 | 0.898 | 2.653 | 62 | 330 | 15.8 |
| SMYD4 | 0.1402 | 3.589 | 0.868 | 14.839 | 5 | 327 | 1.5 |
| EZH2 | 0.1436 | 1.470 | 0.891 | 2.426 | 73 | 318 | 18.7 |
| KMT2E | 0.1859 | 1.418 | 0.857 | 2.347 | 76 | 316 | 19.4 |
| PRDM10 | 0.1868 | 1.649 | 0.821 | 3.311 | 24 | 356 | 6.3 |
| KMT2C | 0.1870 | 1.418 | 0.856 | 2.349 | 74 | 304 | 19.6 |
| SETMAR | 0.1931 | 1.746 | 0.801 | 3.809 | 25 | 353 | 6.6 |
| KMT2A | 0.1991 | 1.674 | 0.802 | 3.494 | 22 | 348 | 5.9 |
| SETD5 | 0.2196 | 1.688 | 0.774 | 3.679 | 25 | 346 | 6.7 |
| SMYD3 | 0.2284 | 1.742 | 0.754 | 4.024 | 21 | 364 | 5.5 |
| PRDM5 | 0.2589 | 1.894 | 0.688 | 5.217 | 10 | 353 | 2.8 |
| SUV39H1 | 0.2972 | 1.975 | 0.620 | 6.296 | 9 | 370 | 2.4 |
| SETD2 | 0.3418 | 1.538 | 0.666 | 3.551 | 24 | 356 | 6.3 |
| PRDM11 | 0.3802 | 1.543 | 0.620 | 3.839 | 16 | 357 | 4.3 |
| SETD7 | 0.6018 | 1.383 | 0.433 | 4.411 | 11 | 371 | 2.9 |
| SETDB2 | 0.6704 | 1.601 | 0.211 | 12.166 | 2 | 232 | 0.9 |
| PRDM6 | 0.6765 | 1.292 | 0.406 | 4.117 | 13 | 354 | 3.5 |
| EZH1 | 0.6774 | 1.570 | 0.216 | 11.403 | 4 | 345 | 1.1 |
| KMT2D | 0.7397 | 1.224 | 0.385 | 3.895 | 11 | 372 | 2.9 |
| SUV39H2 | 0.9204 | 0.943 | 0.295 | 3.016 | 14 | 352 | 3.8 |
Supplementary Table 7B.
Summary of uniivariate analysis of RFS for deletion of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95% CI | Count amplification | Count diploid | Count percentage | |
|---|---|---|---|---|---|---|---|
| Lower | Higher | ||||||
| DOT1L | 0.0363 | 2.053 | 1.103 | 3.821 | 30 | 364 | 7.6 |
| EHMT2 | 0.0356 | 2.801 | 1.211 | 6.478 | 15 | 372 | 3.9 |
| PRDM7 | 0.0055 | 1.881 | 1.205 | 2.934 | 145 | 250 | 36.7 |
| SETD4 | 0.0042 | 3.905 | 1.778 | 8.576 | 17 | 365 | 4.5 |
| SETD6 | 0.0016 | 2.161 | 1.364 | 3.423 | 93 | 300 | 23.7 |
| SETDB2 | 0.0082 | 1.833 | 1.164 | 2.886 | 165 | 232 | 41.6 |
| SMYD1 | 0.0102 | 2.847 | 1.409 | 5.756 | 20 | 367 | 5.2 |
| SUV39H2 | 0.0014 | 2.710 | 1.558 | 4.711 | 33 | 352 | 8.6 |
| SUV420H2 | 0.0121 | 2.947 | 1.411 | 6.159 | 14 | 371 | 3.6 |
| WHSC1L1 | 0.0154 | 1.886 | 1.129 | 3.151 | 129 | 219 | 37.1 |
| SETD5 | 0.1644 | 0.481 | 0.151 | 1.531 | 28 | 346 | 7.5 |
| PRDM8 | 0.3963 | 0.573 | 0.140 | 2.341 | 15 | 373 | 3.9 |
| EZH2 | 0.0977 | 0.000 | 0.000 | Inf | 8 | 318 | 2.5 |
| SUV420H1 | 0.1404 | 0.000 | 0.000 | Inf | 6 | 360 | 1.6 |
| SETDB1 | 0.1916 | 0.000 | 0.000 | Inf | 2 | 369 | 0.5 |
| EZH1 | 0.0528 | 1.783 | 1.026 | 3.096 | 50 | 345 | 12.7 |
| PRDM15 | 0.0550 | 2.129 | 1.053 | 4.307 | 26 | 355 | 6.8 |
| PRDM6 | 0.0598 | 2.008 | 1.031 | 3.914 | 32 | 354 | 8.3 |
| SETD1A | 0.0611 | 2.085 | 1.032 | 4.214 | 25 | 351 | 6.6 |
| PRDM1 | 0.0788 | 1.512 | 0.959 | 2.383 | 122 | 275 | 30.7 |
| SETD7 | 0.0805 | 2.162 | 0.989 | 4.727 | 17 | 371 | 4.4 |
| KMT2B | 0.0874 | 2.118 | 0.971 | 4.620 | 16 | 374 | 4.1 |
| PRDM4 | 0.0918 | 2.234 | 0.965 | 5.171 | 19 | 360 | 5.0 |
| PRDM9 | 0.0966 | 2.675 | 0.972 | 7.362 | 10 | 364 | 2.7 |
| PRDM13 | 0.1008 | 1.473 | 0.932 | 2.327 | 125 | 269 | 31.7 |
| SUV39H1 | 0.1021 | 2.042 | 0.937 | 4.449 | 20 | 370 | 5.1 |
| PRDM11 | 0.1118 | 1.846 | 0.916 | 3.719 | 26 | 357 | 6.8 |
| SMYD5 | 0.1304 | 1.855 | 0.884 | 3.892 | 24 | 363 | 6.2 |
| SETD1B | 0.1357 | 1.774 | 0.879 | 3.580 | 28 | 355 | 7.3 |
| SETD8 | 0.1572 | 1.672 | 0.856 | 3.269 | 31 | 354 | 8.1 |
| SMYD3 | 0.1598 | 2.058 | 0.828 | 5.119 | 14 | 364 | 3.7 |
| SETD3 | 0.1602 | 1.595 | 0.860 | 2.956 | 31 | 358 | 8.0 |
| EHMT1 | 0.2166 | 2.283 | 0.712 | 7.313 | 8 | 345 | 2.3 |
| PRDM12 | 0.2324 | 2.215 | 0.690 | 7.111 | 10 | 345 | 2.8 |
| SMYD4 | 0.2552 | 1.373 | 0.808 | 2.334 | 67 | 327 | 17.0 |
| NSD1 | 0.2914 | 1.994 | 0.625 | 6.357 | 10 | 367 | 2.7 |
| PRDM2 | 0.3186 | 1.405 | 0.740 | 2.668 | 35 | 362 | 8.8 |
| PRDM14 | 0.3336 | 3.197 | 0.436 | 23.424 | 2 | 285 | 0.7 |
| KMT2D | 0.3915 | 1.524 | 0.615 | 3.777 | 16 | 372 | 4.1 |
| ASH1L | 0.4391 | 1.635 | 0.513 | 5.211 | 8 | 364 | 2.2 |
| PRDM16 | 0.4450 | 1.327 | 0.658 | 2.676 | 29 | 363 | 7.4 |
| KMT2C | 0.5010 | 0.685 | 0.214 | 2.197 | 21 | 304 | 6.5 |
| SETD2 | 0.6543 | 1.218 | 0.526 | 2.817 | 19 | 356 | 5.1 |
| WHSC1 | 0.7368 | 0.844 | 0.307 | 2.321 | 21 | 362 | 5.5 |
| SETMAR | 0.7739 | 0.865 | 0.314 | 2.382 | 21 | 353 | 5.6 |
| PRDM5 | 0.7968 | 0.904 | 0.415 | 1.971 | 36 | 353 | 9.3 |
| SMYD2 | 0.8241 | 1.111 | 0.447 | 2.762 | 20 | 358 | 5.3 |
| KMT2A | 0.8987 | 1.056 | 0.457 | 2.441 | 29 | 348 | 7.7 |
| PRDM10 | 0.9515 | 0.965 | 0.303 | 3.077 | 19 | 356 | 5.1 |
| MECOM | 0.9671 | 1.025 | 0.318 | 3.305 | 7 | 330 | 2.1 |
| KMT2E | 0.9844 | 0.986 | 0.239 | 4.064 | 7 | 316 | 2.2 |
Supplementary Table 7C.
Summary of univariate analysis of RFS for methylation level of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95% CI | |
|---|---|---|---|---|
| Lower | Higher | |||
| PRDM14 | 0.0023 | 2.00E+01 | 2.93E+00 | 1.37E+02 |
| PRDM13 | 0.0181 | 1.17E+01 | 1.57E+00 | 8.66E+01 |
| PRDM6 | 0.0184 | 5.91E+01 | 2.28E+00 | 1.53E+03 |
| PRDM8 | 0.0211 | 1.58E+01 | 1.56E+00 | 1.60E+02 |
| KMT2B | 0.0383 | 1.56E−02 | 3.31E−04 | 7.31E−01 |
| NSD1 | 0.0441 | 3.10E+01 | 1.07E+00 | 8.99E+02 |
| SETDB2 | 0.0572 | 6.91E−02 | 4.83E−03 | 9.87E−01 |
| SETDB1 | 0.0580 | 3.52E−05 | 1.11E−09 | 1.11E+00 |
| SETD4 | 0.0873 | 1.31E+13 | 5.38E−02 | 3.18E+27 |
| EZH2 | 0.1045 | 7.80E−10 | 3.21E−21 | 1.90E+02 |
| KMT2E | 0.1152 | 1.45E+01 | 6.10E−01 | 3.45E+02 |
| PRDM4 | 0.1323 | 4.90E+03 | 1.71E−01 | 1.40E+08 |
| SUV39H1 | 0.1694 | 3.77E+01 | 2.58E−01 | 5.50E+03 |
| PRDM12 | 0.1911 | 1.19E+01 | 3.34E−01 | 4.23E+02 |
| SMYD3 | 0.2266 | 2.15E+04 | 1.62E−03 | 2.87E+11 |
| MECOM | 0.2358 | 4.09E+01 | 1.15E−01 | 1.45E+04 |
| DOT1L | 0.2610 | 3.25E+05 | 2.83E−04 | 3.73E+14 |
| SETD6 | 0.2710 | 4.95E−05 | 3.38E−13 | 7.26E+03 |
| SETD3 | 0.3030 | 1.56E−01 | 4.77E−03 | 5.09E+00 |
| PRDM9 | 0.3091 | 5.09E−01 | 1.39E−01 | 1.86E+00 |
| SETD5 | 0.3255 | 1.49E−01 | 3.32E−03 | 6.66E+00 |
| WHSC1L1 | 0.3297 | 1.86E+02 | 7.11E−03 | 4.86E+06 |
| PRDM1 | 0.3832 | 4.35E+00 | 1.70E−01 | 1.12E+02 |
| SUV420H2 | 0.3911 | 8.48E+07 | 1.16E−09 | 6.20E+24 |
| KMT2D | 0.4038 | 4.02E−04 | 3.62E−12 | 4.46E+04 |
| SUV39H2 | 0.4289 | 6.77E+06 | 1.22E−10 | 3.77E+23 |
| SETD1A | 0.4393 | 9.61E−03 | 6.83E−08 | 1.35E+03 |
| PRDM5 | 0.5427 | 1.69E+00 | 3.16E−01 | 9.06E+00 |
| SMYD2 | 0.5584 | 2.99E+01 | 1.02E−03 | 8.79E+05 |
| SETD7 | 0.5756 | 2.92E−04 | 1.06E−16 | 8.05E+08 |
| EZH1 | 0.6304 | 1.22E+01 | 4.84E−04 | 3.09E+05 |
| PRDM15 | 0.6484 | 6.84E−02 | 6.01E−07 | 7.78E+03 |
| PRDM11 | 0.6665 | 3.32E−01 | 2.30E−03 | 4.80E+01 |
| PRDM10 | 0.7125 | 4.99E−01 | 1.26E−02 | 1.98E+01 |
| SETD2 | 0.7127 | 5.25E−04 | 1.16E−21 | 2.37E+14 |
| SMYD5 | 0.7344 | 2.17E−01 | 3.15E−05 | 1.49E+03 |
| SETD8 | 0.7427 | 2.28E−03 | 7.04E−20 | 7.36E+13 |
| PRDM7 | 0.7496 | 5.91E−01 | 2.37E−02 | 1.48E+01 |
| SMYD1 | 0.7786 | 7.48E−01 | 1.00E−01 | 5.57E+00 |
| ASH1L | 0.8598 | 6.00E−01 | 1.78E−03 | 2.02E+02 |
| SETMAR | 0.8750 | 2.48E+00 | 3.02E−05 | 2.03E+05 |
| EHMT2 | 0.8931 | 1.75E−01 | 1.51E−12 | 2.02E+10 |
| SUV420H1 | 0.8952 | 2.14E+00 | 2.59E−05 | 1.77E+05 |
| WHSC1 | 0.9032 | 7.11E−01 | 2.96E−03 | 1.71E+02 |
| KMT2A | 0.9108 | 2.45E+01 | 1.39E−23 | 4.30E+25 |
| PRDM2 | 0.9111 | 2.29E+00 | 1.16E−06 | 4.51E+06 |
| EHMT1 | NA | NA | NA | NA |
| KMT2C | NA | NA | NA | NA |
| PRDM16 | NA | NA | NA | NA |
| SETD1B | NA | NA | NA | NA |
| SMYD4 | NA | NA | NA | NA |
Supplementary Table 7D.
Summary of uniivariate analysis of RFS for expression level of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95%CI | Coef. | Wald test | |
|---|---|---|---|---|---|---|
| Lower | Higher | |||||
| EZH2 | 0 | 2.897 | 1.755 | 4.78 | 1.064 | 17.31 |
| SETD5 | 3.00E-04 | 2.343 | 1.478 | 3.715 | 0.852 | 13.13 |
| WHSC1 | 4.00E-04 | 2.392 | 1.478 | 3.87 | 0.872 | 12.61 |
| SETD1B | 0.0054 | 1.931 | 1.215 | 3.07 | 0.658 | 7.74 |
| SUV420H2 | 0.0054 | 1.93 | 1.214 | 3.067 | 0.657 | 7.74 |
| PRDM6 | 0.0066 | 0.526 | 0.331 | 0.836 | −0.642 | 7.38 |
| PRDM15 | 0.0101 | 1.829 | 1.155 | 2.898 | 0.604 | 6.62 |
| KMT2D | 0.0119 | 1.793 | 1.137 | 2.827 | 0.584 | 6.32 |
| SMYD1 | 0.0207 | 0.471 | 0.249 | 0.891 | −0.753 | 5.35 |
| PRDM10 | 0.0235 | 1.682 | 1.073 | 2.639 | 0.52 | 5.13 |
| KMT2B | 0.0269 | 1.666 | 1.06 | 2.618 | 0.51 | 4.9 |
| PRDM16 | 0.0395 | 0.617 | 0.39 | 0.977 | −0.483 | 4.24 |
| DOT1L | 0.0397 | 1.617 | 1.023 | 2.556 | 0.481 | 4.23 |
| PRDM8 | 0.076 | 0.658 | 0.415 | 1.045 | −0.418 | 3.15 |
| PRDM12 | 0.0764 | 1.522 | 0.956 | 2.422 | 0.42 | 3.14 |
| SUV39H1 | 0.099 | 1.475 | 0.93 | 2.34 | 0.389 | 2.72 |
| KMT2C | 0.1048 | 1.448 | 0.926 | 2.264 | 0.37 | 2.63 |
| SETD8 | 0.1267 | 0.705 | 0.45 | 1.104 | −0.35 | 2.33 |
| KMT2E | 0.1411 | 1.402 | 0.894 | 2.2 | 0.338 | 2.17 |
| NSD1 | 0.1462 | 1.392 | 0.891 | 2.175 | 0.331 | 2.11 |
| EHMT1 | 0.1524 | 1.386 | 0.886 | 2.166 | 0.326 | 2.05 |
| SETD3 | 0.1529 | 0.722 | 0.461 | 1.129 | −0.326 | 2.04 |
| PRDM1 | 0.1609 | 1.378 | 0.88 | 2.156 | 0.32 | 1.97 |
| SETD4 | 0.1734 | 1.376 | 0.869 | 2.178 | 0.319 | 1.85 |
| PRDM11 | 0.2041 | 0.745 | 0.472 | 1.174 | −0.295 | 1.61 |
| SETDB2 | 0.2374 | 1.313 | 0.836 | 2.063 | 0.272 | 1.4 |
| MECOM | 0.2825 | 0.78 | 0.497 | 1.227 | −0.248 | 1.15 |
| SETDB1 | 0.2992 | 1.267 | 0.811 | 1.98 | 0.236 | 1.08 |
| SUV39H2 | 0.3051 | 0.788 | 0.5 | 1.242 | −0.238 | 1.05 |
| SETD2 | 0.3203 | 1.256 | 0.801 | 1.968 | 0.228 | 0.99 |
| PRDM14 | 0.3581 | 1.44 | 0.661 | 3.137 | 0.365 | 0.84 |
| SETD6 | 0.4063 | 1.207 | 0.774 | 1.883 | 0.188 | 0.69 |
| SMYD5 | 0.4071 | 1.209 | 0.772 | 1.892 | 0.19 | 0.69 |
| SETMAR | 0.4329 | 0.836 | 0.535 | 1.307 | −0.179 | 0.61 |
| SUV420H1 | 0.4691 | 1.179 | 0.755 | 1.841 | 0.165 | 0.52 |
| ASH1L | 0.5269 | 1.156 | 0.738 | 1.811 | 0.145 | 0.4 |
| KMT2A | 0.5758 | 1.136 | 0.727 | 1.775 | 0.127 | 0.31 |
| WHSC1L1 | 0.59 | 0.885 | 0.567 | 1.381 | −0.123 | 0.29 |
| PRDM5 | 0.6124 | 0.889 | 0.565 | 1.4 | −0.117 | 0.26 |
| EHMT2 | 0.631 | 1.115 | 0.715 | 1.741 | 0.109 | 0.23 |
| PRDM7 | 0.6638 | 0.899 | 0.555 | 1.455 | −0.107 | 0.19 |
| SMYD3 | 0.6794 | 1.098 | 0.704 | 1.714 | 0.094 | 0.17 |
| EZH1 | 0.7453 | 0.929 | 0.595 | 1.45 | −0.074 | 0.11 |
| PRDM9 | 0.8192 | 0.907 | 0.392 | 2.097 | −0.098 | 0.05 |
| SMYD4 | 0.8539 | 1.043 | 0.665 | 1.637 | 0.042 | 0.03 |
| SMYD2 | 0.8758 | 1.036 | 0.664 | 1.617 | 0.035 | 0.02 |
| SETD7 | 0.8837 | 1.034 | 0.657 | 1.629 | 0.034 | 0.02 |
| PRDM13 | 0.8883 | 1.038 | 0.619 | 1.741 | 0.037 | 0.02 |
| PRDM2 | 0.9321 | 0.981 | 0.626 | 1.536 | −0.019 | 0.01 |
| PRDM4 | 0.982 | 1.005 | 0.645 | 1.568 | 0.005 | 0 |
| SETD1A | 0.9988 | 1 | 0.641 | 1.562 | 0 | 0 |
Supplementary Table 8A.
Summary of multivariate analysis of RFS for amplification of 21 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | EHMT1 amplification | |
|---|---|---|---|---|---|---|
| Coefficient | −0.017 | 0.788 | 0.988 | 0.198 | 0.646 | |
| Hazard ratio | 0.983 | 2.198 | 2.687 | 1.219 | 1.908 | |
| 95%CI | Lower | 0.948 | 1.139 | 1.59 | 0.767 | 1.112 |
| Higher | 1.019 | 4.241 | 4.538 | 1.938 | 3.274 | |
| p-Value | 0.35 | 0.0189 | 0.0002 | 0.4019 | 0.0191 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | EHMT2 amplification | |
| Coefficient | −0.004 | 0.717 | 0.906 | 0.239 | 0.666 | |
| Hazard ratio | 0.996 | 2.049 | 2.476 | 1.27 | 1.947 | |
| 95%CI | Lower | 0.961 | 1.089 | 1.473 | 0.795 | 0.83 |
| Higher | 1.033 | 3.854 | 4.16 | 2.029 | 4.57 | |
| p-Value | 0.8407 | 0.0261 | 0.0006 | 0.3177 | 0.1257 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM12 amplification | |
| Coefficient | −0.007 | 0.676 | 0.921 | 0.325 | 0.665 | |
| Hazard ratio | 0.993 | 1.967 | 2.512 | 1.384 | 1.945 | |
| 95%CI | Lower | 0.959 | 1.037 | 1.492 | 0.871 | 1.125 |
| Higher | 1.029 | 3.73 | 4.23 | 2.2 | 3.362 | |
| p-Value | 0.7062 | 0.0383 | 0.0005 | 0.1691 | 0.0172 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM14 amplification | |
| Coefficient | −0.007 | 0.757 | 0.984 | 0.226 | 0.329 | |
| Hazard ratio | 0.993 | 2.131 | 2.675 | 1.254 | 1.39 | |
| 95%CI | Lower | 0.959 | 1.136 | 1.61 | 0.797 | 0.875 |
| Higher | 1.029 | 3.999 | 4.446 | 1.972 | 2.209 | |
| p-Value | 0.7034 | 0.0184 | 0.0001 | 0.3271 | 0.1637 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM15 amplification | |
| Coefficient | −0.005 | 0.654 | 0.952 | 0.132 | 0.436 | |
| Hazard ratio | 0.995 | 1.923 | 2.59 | 1.141 | 1.546 | |
| 95%CI | Lower | 0.958 | 1.019 | 1.534 | 0.704 | 0.743 |
| Higher | 1.033 | 3.629 | 4.372 | 1.849 | 3.22 | |
| p-Value | 0.7942 | 0.0436 | 0.0004 | 0.5931 | 0.2442 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM4 amplification | |
| Coefficient | −0.006 | 0.779 | 0.923 | 0.263 | 0.867 | |
| Hazard ratio | 0.994 | 2.179 | 2.517 | 1.301 | 2.379 | |
| 95%CI | Lower | 0.959 | 1.164 | 1.502 | 0.801 | 1.143 |
| Higher | 1.031 | 4.082 | 4.219 | 2.112 | 4.953 | |
| p-Value | 0.7471 | 0.0149 | 0.0005 | 0.2881 | 0.0205 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD1A amplification | |
| Coefficient | −0.015 | 0.872 | 0.893 | 0.073 | 0.391 | |
| Hazard ratio | 0.985 | 2.392 | 2.442 | 1.076 | 1.479 | |
| 95%CI | Lower | 0.947 | 1.217 | 1.43 | 0.662 | 0.722 |
| Higher | 1.024 | 4.702 | 4.171 | 1.749 | 3.029 | |
| p-Value | 0.4491 | 0.0114 | 0.0011 | 0.7672 | 0.2851 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD1B amplification | |
| Coefficient | −0.003 | 0.796 | 0.907 | 0.152 | 1.218 | |
| Hazard ratio | 0.997 | 2.216 | 2.477 | 1.164 | 3.381 | |
| 95%CI | Lower | 0.961 | 1.179 | 1.467 | 0.711 | 1.55 |
| Higher | 1.034 | 4.166 | 4.18 | 1.906 | 7.372 | |
| p-Value | 0.8521 | 0.0135 | 0.0007 | 0.5451 | 0.0022 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD8 Amplification | |
| Coefficient | 0 | 0.8 | 0.898 | 0.2 | 0.97 | |
| Hazard ratio | 1 | 2.226 | 2.456 | 1.221 | 2.638 | |
| 95%CI | Lower | 0.964 | 1.182 | 1.453 | 0.747 | 1.101 |
| Higher | 1.038 | 4.191 | 4.152 | 1.996 | 6.323 | |
| p-Value | 0.9897 | 0.0132 | 0.0008 | 0.4256 | 0.0296 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETDB1 amplification | |
| Coefficient | −0.009 | 0.812 | 0.941 | 0.158 | 0.701 | |
| Hazard ratio | 0.991 | 2.253 | 2.563 | 1.171 | 2.016 | |
| 95%CI | Lower | 0.957 | 1.209 | 1.559 | 0.744 | 1.062 |
| Higher | 1.027 | 4.2 | 4.214 | 1.842 | 3.826 | |
| p-Value | 0.6255 | 0.0106 | 0.0002 | 0.4956 | 0.0319 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD1 amplification | |
| Coefficient | −0.004 | 0.788 | 0.906 | 0.15 | 1.423 | |
| Hazard ratio | 0.996 | 2.198 | 2.475 | 1.162 | 4.149 | |
| 95%CI | Lower | 0.959 | 1.139 | 1.443 | 0.716 | 1.844 |
| Higher | 1.034 | 4.243 | 4.244 | 1.885 | 9.336 | |
| p-Value | 0.8369 | 0.0189 | 0.001 | 0.5434 | 0.0006 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD2 amplification | |
| Coefficient | 0 | 0.798 | 0.955 | 0.181 | 0.77 | |
| Hazard ratio | 1 | 2.221 | 2.6 | 1.198 | 2.159 | |
| 95%CI | Lower | 0.965 | 1.16 | 1.556 | 0.751 | 1.026 |
| Higher | 1.037 | 4.253 | 4.343 | 1.91 | 4.546 | |
| p-Value | 0.9835 | 0.0161 | 0.0003 | 0.4483 | 0.0426 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD5 Amplification | |
| Coefficient | 0.004 | 0.745 | 0.929 | 0.163 | 1.274 | |
| Hazard ratio | 1.004 | 2.106 | 2.532 | 1.177 | 3.574 | |
| 95%CI | Lower | 0.967 | 1.119 | 1.484 | 0.729 | 1.595 |
| Higher | 1.042 | 3.966 | 4.32 | 1.901 | 8.01 | |
| p-Value | 0.8462 | 0.0211 | 0.0007 | 0.5045 | 0.002 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H1 amplification | |
| Coefficient | −0.006 | 0.835 | 0.897 | 0.155 | 0.394 | |
| Hazard ratio | 0.994 | 2.304 | 2.452 | 1.168 | 1.483 | |
| 95%CI | Lower | 0.96 | 1.237 | 1.48 | 0.742 | 0.809 |
| Higher | 1.03 | 4.289 | 4.064 | 1.838 | 2.718 | |
| p-Value | 0.7365 | 0.0085 | 0.0005 | 0.5016 | 0.2028 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H2 amplification | |
| Coefficient | −0.014 | 0.885 | 0.927 | 0.072 | 0.422 | |
| Hazard ratio | 0.986 | 2.422 | 2.528 | 1.075 | 1.524 | |
| 95%CI | Lower | 0.949 | 1.263 | 1.51 | 0.664 | 0.673 |
| Higher | 1.025 | 4.642 | 4.231 | 1.741 | 3.455 | |
| p-Value | 0.4852 | 0.0077 | 0.0004 | 0.7691 | 0.3126 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1 amplification | |
| Coefficient | −0.014 | 0.786 | 1.051 | 0.152 | 0.439 | |
| Hazard ratio | 0.986 | 2.194 | 2.86 | 1.165 | 1.551 | |
| 95%CI | Lower | 0.951 | 1.142 | 1.702 | 0.733 | 0.732 |
| Higher | 1.022 | 4.218 | 4.806 | 1.851 | 3.286 | |
| p-Value | 0.4366 | 0.0184 | 0.0001 | 0.5191 | 0.2522 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1L1 amplification | |
| Coefficient | 0.001 | 0.486 | 1.027 | 0.21 | 0.565 | |
| Hazard ratio | 1.001 | 1.627 | 2.792 | 1.233 | 1.76 | |
| 95%CI | Lower | 0.956 | 0.709 | 1.378 | 0.686 | 0.946 |
| Higher | 1.049 | 3.732 | 5.657 | 2.216 | 3.272 | |
| p-Value | 0.95 | 0.251 | 0.0044 | 0.4836 | 0.0742 | |
Supplementary Table 8B.
Summary of multivariate analysis of RFS for deletion of 10 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | DOT1L deletion | |
|---|---|---|---|---|---|---|
| Coefficient | −0.003 | 0.742 | 0.971 | 0.304 | 0.441 | |
| Hazard ratio | 0.997 | 2.101 | 2.64 | 1.356 | 1.554 | |
| 95%CI | Lower | 0.962 | 1.118 | 1.585 | 0.847 | 0.815 |
| Higher | 1.033 | 3.945 | 4.397 | 2.17 | 2.963 | |
| p-Value | 0.8719 | 0.021 | 0.0002 | 0.2053 | 0.1802 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | EHMT2 deletion | |
| Coefficient | 0.004 | 0.775 | 0.951 | 0.176 | 0.992 | |
| Hazard ratio | 1.004 | 2.17 | 2.588 | 1.193 | 2.695 | |
| 95%CI | Lower | 0.968 | 1.156 | 1.548 | 0.743 | 1.135 |
| Higher | 1.042 | 4.074 | 4.327 | 1.916 | 6.398 | |
| p-Value | 0.81 | 0.016 | 0.0003 | 0.4662 | 0.0246 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM7 deletion | |
| Coefficient | 0.003 | 0.796 | 0.947 | 0.196 | 0.333 | |
| Hazard ratio | 1.003 | 2.217 | 2.577 | 1.216 | 1.395 | |
| 95%CI | Lower | 0.969 | 1.19 | 1.563 | 0.773 | 0.881 |
| Higher | 1.039 | 4.133 | 4.25 | 1.914 | 2.208 | |
| p-Value | 0.8706 | 0.0122 | 0.0002 | 0.397 | 0.1559 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD4 deletion | |
| Coefficient | −0.006 | 0.733 | 0.991 | 0.177 | 1.097 | |
| Hazard ratio | 0.994 | 2.081 | 2.694 | 1.194 | 2.996 | |
| 95%CI | Lower | 0.959 | 1.107 | 1.598 | 0.738 | 1.336 |
| Higher | 1.031 | 3.91 | 4.545 | 1.932 | 6.716 | |
| p-Value | 0.7515 | 0.0229 | 0.0002 | 0.4706 | 0.0077 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD6 deletion | |
| Coefficient | 0 | 0.762 | 0.911 | 0.24 | 0.491 | |
| Hazard ratio | 1 | 2.143 | 2.486 | 1.271 | 1.634 | |
| 95%CI | Lower | 0.966 | 1.146 | 1.486 | 0.805 | 1.018 |
| Higher | 1.035 | 4.006 | 4.159 | 2.008 | 2.622 | |
| p-Value | 0.9999 | 0.017 | 0.0005 | 0.3036 | 0.042 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETDB2 deletion | |
| Coefficient | −0.007 | 0.835 | 0.928 | 0.125 | 0.322 | |
| Hazard ratio | 0.993 | 2.305 | 2.531 | 1.133 | 1.38 | |
| 95%CI | Lower | 0.96 | 1.237 | 1.529 | 0.721 | 0.868 |
| Higher | 1.028 | 4.298 | 4.189 | 1.782 | 2.195 | |
| p-Value | 0.6953 | 0.0086 | 0.0003 | 0.5873 | 0.1739 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD1 deletion | |
| Coefficient | 0 | 0.755 | 0.96 | 0.309 | 0.998 | |
| Hazard ratio | 1 | 2.128 | 2.612 | 1.362 | 2.714 | |
| 95%CI | Lower | 0.964 | 1.135 | 1.558 | 0.848 | 1.329 |
| Higher | 1.038 | 3.99 | 4.378 | 2.189 | 5.543 | |
| p-Value | 0.9902 | 0.0185 | 0.0003 | 0.2012 | 0.0061 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV39H2 deletion | |
| Coefficient | −0.013 | 0.774 | 0.976 | 0.187 | 0.755 | |
| Hazard ratio | 0.987 | 2.169 | 2.655 | 1.205 | 2.129 | |
| 95%CI | Lower | 0.953 | 1.15 | 1.588 | 0.761 | 1.199 |
| Higher | 1.022 | 4.092 | 4.437 | 1.909 | 3.778 | |
| p-Value | 0.4618 | 0.0168 | 0.0002 | 0.4265 | 0.0099 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H2 deletion | |
| Coefficient | 0 | 0.77 | 0.89 | 0.289 | 0.553 | |
| Hazard ratio | 1 | 2.16 | 2.435 | 1.335 | 1.738 | |
| 95%CI | Lower | 0.964 | 1.147 | 1.443 | 0.831 | 0.808 |
| Higher | 1.037 | 4.071 | 4.108 | 2.144 | 3.736 | |
| p-Value | 0.9946 | 0.0172 | 0.0009 | 0.232 | 0.1572 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1L1 deletion | |
| Coefficient | −0.001 | 0.681 | 1.184 | 0.121 | 0.677 | |
| Hazard ratio | 0.999 | 1.976 | 3.267 | 1.129 | 1.968 | |
| 95%CI | Lower | 0.959 | 1.028 | 1.867 | 0.671 | 1.173 |
| Higher | 1.04 | 3.8 | 5.716 | 1.899 | 3.303 | |
| p-Value | 0.9534 | 0.0411 | 0 | 0.6489 | 0.0103 | |
Supplementary Table 8C.
Summary of multivariate analysis of RFS for higher promotor methylation of 6 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM14 high_meth | |
|---|---|---|---|---|---|---|
| Coefficient | −0.009 | 0.785 | 0.916 | 0.188 | 0.32 | |
| Hazard ratio | 0.991 | 2.193 | 2.498 | 1.207 | 1.377 | |
| 95%CI | Lower | 0.957 | 1.174 | 1.508 | 0.769 | 0.823 |
| Higher | 1.027 | 4.095 | 4.139 | 1.894 | 2.303 | |
| p-Value | 0.6314 | 0.0137 | 0.0004 | 0.4131 | 0.223 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM13 high_meth | |
| Coefficient | −0.006 | 0.791 | 0.971 | 0.171 | 0.317 | |
| Hazard ratio | 0.994 | 2.206 | 2.641 | 1.187 | 1.373 | |
| 95%CI | Lower | 0.96 | 1.182 | 1.608 | 0.755 | 0.86 |
| Higher | 1.029 | 4.119 | 4.338 | 1.864 | 2.19 | |
| p-Value | 0.7279 | 0.013 | 0.0001 | 0.4581 | 0.1839 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM6 high_meth | |
| Coefficient | −0.004 | 0.816 | 0.967 | 0.191 | 0.079 | |
| Hazard ratio | 0.996 | 2.262 | 2.631 | 1.211 | 1.082 | |
| 95%CI | Lower | 0.962 | 1.208 | 1.594 | 0.771 | 0.675 |
| Higher | 1.031 | 4.235 | 4.341 | 1.901 | 1.734 | |
| p-Value | 0.8298 | 0.0108 | 0.0002 | 0.4059 | 0.7441 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM8 high_meth | |
| Coefficient | −0.004 | 0.826 | 0.933 | 0.184 | 0.324 | |
| Hazard ratio | 0.996 | 2.285 | 2.542 | 1.201 | 1.383 | |
| 95%CI | Lower | 0.963 | 1.227 | 1.542 | 0.766 | 0.869 |
| Higher | 1.031 | 4.257 | 4.189 | 1.883 | 2.199 | |
| p-Value | 0.8314 | 0.0092 | 0.0003 | 0.4236 | 0.1712 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | KMT2B high_meth | |
| Coefficient | −0.003 | 0.86 | 0.897 | 0.229 | −0.417 | |
| Hazard ratio | 0.997 | 2.363 | 2.452 | 1.257 | 0.659 | |
| 95%CI | Lower | 0.963 | 1.274 | 1.486 | 0.8 | 0.409 |
| Higher | 1.033 | 4.382 | 4.045 | 1.975 | 1.061 | |
| p-Value | 0.8776 | 0.0064 | 0.0004 | 0.3215 | 0.086 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | NSD1 High_meth | |
| Coefficient | −0.006 | 0.852 | 0.889 | 0.161 | 0.547 | |
| Hazard ratio | 0.994 | 2.345 | 2.432 | 1.175 | 1.728 | |
| 95%CI | Lower | 0.961 | 1.26 | 1.476 | 0.749 | 1.06 |
| Higher | 1.028 | 4.364 | 4.009 | 1.843 | 2.818 | |
| p-Value | 0.7268 | 0.0071 | 0.0005 | 0.4839 | 0.0284 | |
Supplementary Table 8D.
Summary of multivariate analysis of RFS for higher mRNA level of 16 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | DOT1L high_exp | |
|---|---|---|---|---|---|---|
| Coefficient | −0.004 | 0.854 | 0.916 | 0.163 | 0.254 | |
| Hazard ratio | 0.996 | 2.35 | 2.5 | 1.177 | 1.289 | |
| 95%CI | Lower | 0.962 | 1.26 | 1.504 | 0.747 | 0.802 |
| Higher | 1.032 | 4.382 | 4.157 | 1.855 | 2.071 | |
| p-Value | 0.8373 | 0.0072 | 0.0004 | 0.4813 | 0.2941 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | EZH2 high_exp | |
| Coefficient | −0.006 | 0.858 | 0.731 | 0.185 | 0.736 | |
| Hazard ratio | 0.994 | 2.357 | 2.076 | 1.203 | 2.088 | |
| 95%CI | Lower | 0.961 | 1.263 | 1.23 | 0.768 | 1.23 |
| Higher | 1.028 | 4.4 | 3.506 | 1.887 | 3.544 | |
| p-Value | 0.7334 | 0.0071 | 0.0063 | 0.4195 | 0.0064 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | KMT2B high_exp | |
| Coefficient | −0.004 | 0.877 | 0.923 | −0.026 | 0.387 | |
| Hazard ratio | 0.996 | 2.403 | 2.516 | 0.975 | 1.472 | |
| 95%CI | Lower | 0.962 | 1.288 | 1.526 | 0.563 | 0.839 |
| Higher | 1.031 | 4.484 | 4.149 | 1.688 | 2.584 | |
| p-Value | 0.8266 | 0.0059 | 0.0003 | 0.9268 | 0.1781 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | KMT2D high_exp | |
| Coefficient | −0.004 | 0.857 | 0.906 | 0.087 | 0.333 | |
| Hazard ratio | 0.996 | 2.357 | 2.474 | 1.091 | 1.396 | |
| 95%CI | Lower | 0.962 | 1.264 | 1.492 | 0.677 | 0.851 |
| Higher | 1.031 | 4.393 | 4.102 | 1.758 | 2.289 | |
| p-Value | 0.8135 | 0.007 | 0.0004 | 0.721 | 0.1865 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM10 high_exp | |
| Coefficient | −0.003 | 0.829 | 0.94 | 0.054 | 0.266 | |
| Hazard ratio | 0.997 | 2.291 | 2.56 | 1.055 | 1.304 | |
| 95%CI | Lower | 0.963 | 1.23 | 1.551 | 0.617 | 0.755 |
| Higher | 1.033 | 4.267 | 4.227 | 1.804 | 2.255 | |
| p-Value | 0.878 | 0.009 | 0.0002 | 0.8443 | 0.3412 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM12 high_exp | |
| Coefficient | −0.005 | 0.841 | 0.923 | 0.231 | 0.224 | |
| Hazard ratio | 0.995 | 2.319 | 2.517 | 1.26 | 1.251 | |
| 95%CI | Lower | 0.962 | 1.245 | 1.512 | 0.799 | 0.773 |
| Higher | 1.03 | 4.322 | 4.188 | 1.988 | 2.024 | |
| p-Value | 0.7926 | 0.0081 | 0.0004 | 0.3195 | 0.362 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM15 high_exp | |
| Coefficient | −0.003 | 0.856 | 0.909 | 0.153 | 0.448 | |
| Hazard ratio | 0.997 | 2.354 | 2.483 | 1.165 | 1.566 | |
| 95%CI | Lower | 0.964 | 1.264 | 1.507 | 0.742 | 0.981 |
| Higher | 1.033 | 4.383 | 4.092 | 1.83 | 2.5 | |
| p-Value | 0.8865 | 0.007 | 0.0004 | 0.5069 | 0.0604 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM16 high_exp | |
| Coefficient | −0.006 | 0.898 | 0.948 | 0.175 | −0.514 | |
| Hazard ratio | 0.994 | 2.454 | 2.581 | 1.191 | 0.598 | |
| 95%CI | Lower | 0.96 | 1.315 | 1.571 | 0.759 | 0.377 |
| Higher | 1.029 | 4.579 | 4.24 | 1.87 | 0.949 | |
| p-Value | 0.747 | 0.0048 | 0.0002 | 0.4462 | 0.0293 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM6 high_exp | |
| Coefficient | −0.003 | 0.851 | 0.897 | 0.208 | −0.506 | |
| Hazard ratio | 0.997 | 2.343 | 2.452 | 1.231 | 0.603 | |
| 95%CI | Lower | 0.963 | 1.259 | 1.486 | 0.785 | 0.377 |
| Higher | 1.032 | 4.36 | 4.046 | 1.931 | 0.964 | |
| p-Value | 0.8603 | 0.0072 | 0.0004 | 0.366 | 0.0345 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM8 high_exp | |
| Coefficient | −0.005 | 0.857 | 0.916 | 0.208 | −0.284 | |
| Hazard ratio | 0.995 | 2.356 | 2.499 | 1.231 | 0.753 | |
| 95%CI | Lower | 0.962 | 1.261 | 1.504 | 0.785 | 0.47 |
| Higher | 1.03 | 4.401 | 4.155 | 1.93 | 1.206 | |
| p-Value | 0.7952 | 0.0072 | 0.0004 | 0.3658 | 0.2375 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD1B high_exp | |
| Coefficient | −0.008 | 0.861 | 0.903 | 0.059 | 0.453 | |
| Hazard ratio | 0.992 | 2.365 | 2.468 | 1.06 | 1.573 | |
| 95%CI | Lower | 0.958 | 1.271 | 1.496 | 0.661 | 0.957 |
| Higher | 1.028 | 4.401 | 4.071 | 1.701 | 2.584 | |
| p-Value | 0.6715 | 0.0066 | 0.0004 | 0.8076 | 0.0738 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD5 high_exp | |
| Coefficient | −0.005 | 0.784 | 0.903 | 0.033 | 0.584 | |
| Hazard ratio | 0.995 | 2.191 | 2.466 | 1.033 | 1.794 | |
| 95%CI | Lower | 0.961 | 1.176 | 1.496 | 0.646 | 1.1 |
| Higher | 1.031 | 4.082 | 4.067 | 1.651 | 2.926 | |
| p-Value | 0.7935 | 0.0135 | 0.0004 | 0.8915 | 0.0193 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD1 high_exp | |
| Coefficient | −0.009 | 0.862 | 1.002 | 0.191 | −0.819 | |
| Hazard ratio | 0.991 | 2.369 | 2.723 | 1.21 | 0.441 | |
| 95%CI | Lower | 0.957 | 1.269 | 1.662 | 0.772 | 0.232 |
| Higher | 1.027 | 4.423 | 4.461 | 1.896 | 0.838 | |
| p-Value | 0.623 | 0.0068 | 0.0001 | 0.405 | 0.0124 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV39H1 high_exp | |
| Coefficient | −0.007 | 0.86 | 0.925 | 0.22 | 0.329 | |
| Hazard ratio | 0.993 | 2.363 | 2.521 | 1.246 | 1.39 | |
| 95%CI | Lower | 0.958 | 1.267 | 1.525 | 0.792 | 0.866 |
| Higher | 1.028 | 4.407 | 4.167 | 1.958 | 2.23 | |
| p-Value | 0.6758 | 0.0068 | 0.0003 | 0.3412 | 0.1728 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H2 high_exp | |
| Coefficient | −0.009 | 0.852 | 0.847 | 0.382 | 0.586 | |
| Hazard ratio | 0.991 | 2.344 | 2.334 | 1.465 | 1.796 | |
| 95%CI | Lower | 0.957 | 1.257 | 1.402 | 0.909 | 1.087 |
| Higher | 1.027 | 4.37 | 3.884 | 2.361 | 2.966 | |
| p-Value | 0.6247 | 0.0073 | 0.0011 | 0.1165 | 0.0222 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1 high_exp | |
| Coefficient | −0.005 | 0.815 | 0.831 | 0.146 | 0.429 | |
| Hazard ratio | 0.995 | 2.259 | 2.297 | 1.157 | 1.535 | |
| 95%CI | Lower | 0.961 | 1.21 | 1.357 | 0.735 | 0.913 |
| Higher | 1.03 | 4.217 | 3.888 | 1.821 | 2.581 | |
| p-Value | 0.7789 | 0.0105 | 0.002 | 0.5276 | 0.1057 | |
Footnotes
Conflict of interest
None.
Source of support: This work was supported by the National Natural Science Foundation of China (81602236) and the National Major Scientific and Technological Special Project for Significant New Drugs Development (2017ZX09304022)
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spratt DE, Zumsteg ZS, Feng FY, Tomlins SA. Translational and clinical implications of the genetic landscape of prostate cancer. Nat Rev Clin Oncol. 2016;13:597–610. doi: 10.1038/nrclinonc.2016.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 7.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–20. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Tian X, Zhang S, Liu HM, et al. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: New targets for cancer therapy and prevention. Curr Cancer Drug Targets. 2013;13:558–79. doi: 10.2174/1568009611313050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, Memarzadeh S, Stoyanova T, et al. Collaboration of Kras and androgen receptor signaling stimulates EZH2 expression and tumor-propagating cells in prostate cancer. Cancer Res. 2012;72:4672–81. doi: 10.1158/0008-5472.CAN-12-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu K, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–69. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–76. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 14.Zhu X, He F, Zeng H, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet. 2014;46:287–93. doi: 10.1038/ng.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik R, Khan AP, Asangani IA, et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med. 2015;21:344–52. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Herz HM, Garruss A, Shilatifard A. SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013;38:621–39. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–18. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 19.Wagener N, Holland D, Bulkescher J, et al. The enhancer of zeste homolog 2 gene contributes to cell proliferation and apoptosis resistance in renal cell carcinoma cells. Int J Cancer. 2008;123:1545–50. doi: 10.1002/ijc.23683. [DOI] [PubMed] [Google Scholar]
- 20.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–29. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 21.Chen K, Xiao H, Zeng J, et al. Alternative splicing of EZH2 pre-mRNA by SF3B3 contributes to the tumorigenic potential of renal cancer. Clin Cancer Res. 2017;23:3428–41. doi: 10.1158/1078-0432.CCR-16-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asangani IA, Ateeq B, Cao Q, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol Cell. 2013;49:80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo AJ, Cheung P, Chen K, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–20. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kassambara A, Klein B, Moreaux J. MMSET is overexpressed in cancers: Link with tumor aggressiveness. Biochem Biophys Res Commun. 2008;379:840–45. doi: 10.1016/j.bbrc.2008.12.093. [DOI] [PubMed] [Google Scholar]
- 25.Kang HB, Choi Y, Lee JM, et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009;583:1880–86. doi: 10.1016/j.febslet.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 26.Dobosy JR, Roberts JL, Fu VX, Jarrard DF. The expanding role of epigenetics in the development, diagnosis and treatment of prostate cancer and benign prostatic hyperplasia. J Urol. 2007;177:822–31. doi: 10.1016/j.juro.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 27.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 28.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Ann Rev Pharmacol Toxicol. 2005;45:629–56. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 29.Lan X, Gao H, Wang F, et al. Whole-exome sequencing identifies variants in invasive pituitary adenomas. Oncol Lett. 2016;12:2319–28. doi: 10.3892/ol.2016.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidner CI, Lin Q, Birkhofer C, et al. DNA methylation in PRDM8 is indicative for dyskeratosis congenita. Oncotarget. 2016;7:10765–72. doi: 10.18632/oncotarget.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan SX, Hu RC, Tan YL, et al. Promoter methylation-mediated downregulation of PRDM5 contributes to the development of lung squamous cell carcinoma. Tumour Biol. 2014;35:4509–16. doi: 10.1007/s13277-013-1593-2. [DOI] [PubMed] [Google Scholar]
- 32.Deng Q, Huang S. PRDM5 is silenced in human cancers and has growth suppressive activities. Oncogene. 2004;23:4903–10. doi: 10.1038/sj.onc.1207615. [DOI] [PubMed] [Google Scholar]
- 33.Shu XS, Geng H, Li L, et al. The epigenetic modifier PRDM5 functions as a tumor suppressor through modulating WNT/beta-catenin signaling and is frequently silenced in multiple tumors. PLoS One. 2011;6:e27346. doi: 10.1371/journal.pone.0027346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe Y, Toyota M, Kondo Y, et al. PRDM5 identified as a target of epigenetic silencing in colorectal and gastric cancer. Clin Cancer Res. 2007;13:4786–94. doi: 10.1158/1078-0432.CCR-07-0305. [DOI] [PubMed] [Google Scholar]
- 35.Rubicz R, Zhao S, Geybels M, et al. DNA methylation profiles in African American prostate cancer patients in relation to disease progression. Genomics. 2016 doi: 10.1016/j.ygeno.2016.02.004. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falandry C, Fourel G, Galy V, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem. 2010;285:20234–41. doi: 10.1074/jbc.M109.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker H, Rose-Zerilli MJ, Parker A, et al. 13q deletion anatomy and disease progression in patients with chronic lymphocytic leukemia. Leukemia. 2011;25:489–97. doi: 10.1038/leu.2010.288. [DOI] [PubMed] [Google Scholar]
- 38.Nishikawaji T, Akiyama Y, Shimada S, et al. Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer. Oncotarget. 2016;7:67251–65. doi: 10.18632/oncotarget.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam W, Gomez M, Chadburn A, et al. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 40.Mandelbaum J, Bhagat G, Tang H, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–79. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Wang K, Han L, et al. PRDM1 is directly targeted by miR-30a-5p and modulates the Wnt/beta-catenin pathway in a Dkk1-dependent manner during glioma growth. Cancer Lett. 2013;331:211–19. doi: 10.1016/j.canlet.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Nishikawa N, Toyota M, Suzuki H, et al. Gene amplification and overexpression of PRDM14 in breast cancers. Cancer Res. 2007;67:9649–57. doi: 10.1158/0008-5472.CAN-06-4111. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi H, Hoshino D, Moriya C, et al. Silencing PRDM14 expression by an innovative RNAi therapy inhibits stemness, tumorigenicity, and metastasis of breast cancer. Oncotarget. 2017;8(29):46856–74. doi: 10.18632/oncotarget.16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriya C, Taniguchi H, Miyata K, et al. Inhibition of PRDM14 expression in pancreatic cancer suppresses cancer stem-like properties and liver metastasis in mice. Carcinogenesis. 2017;38(6):638–48. doi: 10.1093/carcin/bgx040. [DOI] [PubMed] [Google Scholar]
- 45.Zhang T, Meng L, Dong W, et al. High expression of PRDM14 correlates with cell differentiation and is a novel prognostic marker in resected non-small cell lung cancer. Med Oncol. 2013;30:605. doi: 10.1007/s12032-013-0605-9. [DOI] [PubMed] [Google Scholar]
- 46.Dettman EJ, Simko SJ, Ayanga B, et al. Prdm14 initiates lymphoblastic leukemia after expanding a population of cells resembling common lymphoid progenitors. Oncogene. 2011;30:2859–73. doi: 10.1038/onc.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–39. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu D, Gao X, Morgan MA, et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases and enhancers. Mol Cell Biol. 2013;33:4745–54. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorighi KM, Swigut T, Henriques T, et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell. 2017;66:568–76.e.4. doi: 10.1016/j.molcel.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dawkins JB, Wang J, Maniati E, et al. Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Res. 2016;76:4861–71. doi: 10.1158/0008-5472.CAN-16-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Sanz P, Trivino JC, Mota A, et al. Chromatin remodelling and DNA repair genes are frequently mutated in endometrioid endometrial carcinoma. Int J Cancer. 2017;140:1551–63. doi: 10.1002/ijc.30573. [DOI] [PubMed] [Google Scholar]
- 52.Kim JH, Sharma A, Dhar SS, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–17. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Copeland RA, Moyer MP, Richon VM. Targeting genetic alterations in protein methyltransferases for personalized cancer therapeutics. Oncogene. 2013;32:939–46. doi: 10.1038/onc.2012.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA levels of 51 HMTs in different Gleason score prostate cancer samples. (A) Pheatmap of 51 HMTs expression profile in totally 399 prostate cancer samples. Normalized gene expression values by rows were median-centered prior to rows clustering based on Euclidean distance. One way ANOVA analysis were performed to compare the differences between Gleason score 6, 7, 8, and 9 prostate tumors. Significantly different expressed genes in prostate tumors are shown at the top, potential cancer related genes(p<0.0001) are indicated by a red box. (B) Box plot show significantly four genes (PRDM6, PRDM8, PRDM12, EZH2) expression level(y-axis) positive or negative related to different Gleason score(x-axis).
mRNA levels of four HMTs in different Gleason score prostate cancers. Box plots show four genes (WHSC1, SUV420H2, PRDM10, SETD5) expression level (y-axis) significantly related to different Gleason score (x-axis).
CNAs frequencies of HMTs and their impact on clinical survival. (A, B) Bar graphs showed the most frequently altered HMTs. (C, D) Kaplan-Meier plotS of RFS proportion shows that deletion of SETDB2 and amplification of PRDM14 were significantly relevant to clinical outcome.
HMTs copy number alteration profiles in different Gleason score prostate samples. Pheatmap shows that CNAs frequencies of HMTs were tendentiously positive related to Gleason score, interestingly, both for amplification and deletion alterations.
HMTs copy number alteration profiles in different stage prostate cancers. (A) Pheatmap shows that increasing CNA events of 51 HMTs correlated with different stages of prostate cancers. (B) Histogram shown average CNAs per sample in different stage prostate cancers.
Kaplan-Meier plots of RFS proportion show that different impacts caused by CNAs and ectopic gene expression of significant HMTs (PRDM12, SETD6, SMYD1, WHSC1L1) on clinical survival.
Kaplan-Meier plots of RFS proportion show that different impactS caused by methylation level of significant HMTs (NSD1, PRDM13,PRDM14, KMT2B) on clinical survival.
Supplementary Table 1.
Summary of identified human HMTs and their histone substrates.
| Official symbol | Other aliases | Gene ID | Gene location | Histone substrates |
|---|---|---|---|---|
| ASH1L | KMT2H; ASH1L1 | 55870 | 1q22 | H3K4me1/3, H3K36me2 |
| DOT1L | DOT1; KMT4 | 84444 | 19p13.3 | H3K79me1/2/3 |
| EHMT1 | GLP1; KMT1D | 79813 | 9q34.3 | H3K9me1/2 |
| EHMT2 | G9A; KMT1C | 10919 | 6p21.31 | H3K9me1/2 |
| EZH1 | KMT6B | 2145 | 17q21.2 | H3K27me2/3 |
| EZH2 | KMT6A | 2146 | 7q36.1 | H3K27me2/3 |
| KMT2A | MLL | 4297 | 11q23.3 | H3K4me1/2/3 |
| KMT2B | MLL2 | 9757 | 19q13.12 | H3K4me1/2/3 |
| KMT2C | MLL3 | 58508 | 7q36.1 | H3K4me1/2/3 |
| KMT2D | MLL4 | 8085 | 12q13.12 | H3K4me1/2/3 |
| KMT2E | MLL5 | 55904 | 7q22.3 | H3K4 |
| MECOM | PRDM3; MDS1-EVI1 | 2122 | 3q26.2 | H3K9me1 |
| NSD1 | KMT3B | 64324 | 5q35.2 | H3K36me1/2 |
| PRDM1 | BLIMP16 | 639 | 6q21 | |
| PRDM10 | PFM7 | 56980 | 11q24.3 | |
| PRDM11 | PFM8 | 56981 | 11p11.2 | |
| PRDM12 | PFM9 | 59335 | 9q34.12 | |
| PRDM13 | PFM10 | 59336 | 6q16.2 | |
| PRDM14 | PFM11 | 63978 | 8q13.3 | |
| PRDM15 | PFM15 | 63977 | 21q22.3 | |
| PRDM16 | MEL1; PFM13 | 63976 | 1p36.32 | H3K9me1 |
| PRDM2 | RIZ; KMT8 | 7799 | 1p36.21 | H3K9me1/2/3 |
| PRDM4 | PFM1 | 11108 | 12q23.3 | |
| PRDM5 | PFM2 | 11107 | 4q27 | H3K9me2 |
| PRDM6 | 93166 | 5q23.2 | ||
| PRDM7 | PFM4; ZNF910 | 11105 | 16q24.3 | |
| PRDM8 | PFM5 | 56978 | 4q21.21 | H3k9me3 |
| PRDM9 | PFM6; MEISETZ | 56979 | 5p14.2 | H3K4me3 |
| SETD1A | KMT2F; SET1A | 9739 | 16p11.2 | H3K4me1/2/3 |
| SETD1B | KMT2G; SET1B | 23067 | 12q24.31 | H3K4me1/2/3 |
| SETD2 | HYPB; SET2 | 29072 | 3p21.31 | H3K36me1/2/3 |
| SETD3 | C14orf154 | 84193 | 14q32.2 | |
| SETD4 | C21orf18 | 54093 | 21q22.12 | |
| SETD5 | 55209 | 3p25.3 | ||
| SETD6 | 79918 | 16q21 | ||
| SETD7 | KMT7; SET7/9 | 80854 | 4q31.1 | H3K4me1 |
| SETD8 | KMT5A; PR-SETD7 | 387893 | 12q24.31 | H4K20me1 |
| SETDB1 | ESET; KMT1E | 9869 | 1q21.3 | H3K9me1/2/3 |
| SETDB2 | CLL8; KMT1F | 83852 | 13q14.2 | H3K9 |
| SETMAR | METNASE | 6419 | 3p26.1 | H3K36 |
| SMYD1 | KMT3D; ZMYND18 | 150572 | 2p11.2 | H3K4 |
| SMYD2 | KMT3C; ZMYND14 | 56950 | 1q32.3 | H3K4; H3K36me2 |
| SMYD3 | KMT3E; ZMYND1; | 64754 | 1q44 | H3K4me2/3 |
| SMYD4 | ZMYND21 | 114826 | 17p13.3 | |
| SMYD5 | RRG1; ZMYND23 | 10322 | 2p13.2 | |
| SUV39H1 | KMT1A | 6839 | Xp11.23 | H3K9me2/3 |
| SUV39H2 | KMT1B | 79723 | 10p13 | H3K9me2/3 |
| SUV420H1 | KMT5B; CGI-85 | 51111 | 11q13.2 | H4K20me2/3 |
| SUV420H2 | KMT5C | 84787 | 19q13.42 | H4K20me2/3 |
| WHSC1 | NSD2; MMSET | 7468 | 4p16.3 | H3K36me1/2 |
| WHSC1L1 | NSD3 | 54904 | 8p11.23 | H3K36me1/2; H3K4 |
Supplementary Table 2.
Number of samples from TCGA used in our analyses. For the RNA-Seq data and methylation data the number of tumor and normal samples are indicated. We also indicate the number of tumor samples with mutation data, and with copy number variation (CNV) data.
| Acronym | Tumor type | Data type | Paired samples | Total tumor samples | Metastatic PRAD | |
|---|---|---|---|---|---|---|
| Normal | Tumor | |||||
| PRAD | Prostate adenocarcinoma | RNA seq data | 52 | 52 | 497 | 0 |
| CpG data(46HMTs) | 50 | 50 | 499 | 0 | ||
| CNV data | 0 | 0 | 492 | 150 | ||
| Mutation data | 0 | 0 | 425 | 0 | ||
| Quadruple overlap | 0 | 0 | 399 | 0 | ||
Supplementary Table 3.
Summary of expression fold-changed genes with false discovery rates (FDR) <0.05.
| Gene | logFC | AveExpr | t | P.value | FDR | B | |
|---|---|---|---|---|---|---|---|
| 2.0FC | PRDM8 | −1.526 | 9.088 | −5.666 | 1.30E-07 | 8.08E-07 | 6.880 |
| MECOM | −1.216 | 8.501 | −6.737 | 9.03E-10 | 9.65E-09 | 11.736 | |
| PRDM16 | −1.129 | 4.769 | −4.080 | 8.80E-05 | 2.54E-04 | 0.592 | |
| SMYD1 | −1.033 | 1.099 | −2.254 | 2.63E-02 | 4.55E-02 | −4.668 | |
| PRDM12 | 1.016 | 1.348 | 7.227 | 8.37E-11 | 2.18E-09 | 14.071 | |
| EZH2 | 1.433 | 5.944 | 8.778 | 3.43E-14 | 1.78E-12 | 21.754 | |
| 1.5FC | PRDM5 | −0.932 | 5.892 | −5.874 | 5.10E-08 | 3.79E-07 | 7.790 |
| PRDM11 | −0.904 | 4.968 | −6.843 | 5.42E-10 | 9.40E-09 | 12.235 | |
| PRDM6 | −0.800 | 5.200 | −3.475 | 7.44E-04 | 1.93E-03 | −1.424 | |
| 1.2FC | EZH1 | −0.542 | 9.792 | −6.731 | 9.28E-10 | 9.65E-09 | 11.709 |
| SMYD4 | −0.371 | 8.315 | −4.951 | 2.84E-06 | 1.23E-05 | 3.885 | |
| PRDM15 | 0.320 | 7.400 | 5.011 | 2.21E-06 | 1.05E-05 | 4.127 | |
| PRDM10 | 0.337 | 7.990 | 4.292 | 3.96E-05 | 1.47E-04 | 1.353 | |
| WHSC1 | 0.367 | 9.820 | 5.064 | 1.77E-06 | 9.20E-06 | 4.345 | |
| SETD7 | 0.429 | 11.984 | 4.351 | 3.16E-05 | 1.26E-04 | 1.571 | |
| KMT2B | 0.432 | 11.025 | 3.438 | 8.42E-04 | 1.99E-03 | −1.540 | |
| SMYD2 | 0.433 | 8.849 | 4.118 | 7.65E-05 | 2.34E-04 | 0.725 | |
| SUV39H2 | 0.455 | 7.832 | 4.273 | 4.26E-05 | 1.48E-04 | 1.284 | |
| SMYD3 | 0.509 | 7.713 | 5.885 | 4.84E-08 | 3.79E-07 | 7.841 | |
| DOT1L | 0.567 | 8.862 | 5.649 | 1.40E-07 | 8.08E-07 | 6.807 |
Supplementary Table 4.
Summary for association of Gleason score and AR expression with HMTs expression profile.
| Gleason score/HMTs expression (one-way ANOVA, top 11 HMTs were filled with yellow color) | AR/HMTs expression (top 13 significant HMTs with the green padding were also in the same clustering) | |||
|---|---|---|---|---|
| Gene | p.Value | Gene | p.Value | Coefficients |
| ASH1L | 4.49E-01 | ASH1L | 4.94E-155 | 0.871 |
| DOT1L | 7.57E-04 | DOT1L | 1.43E-06 | 0.214 |
| EHMT1 | 2.78E-04 | EHMT1 | 9.08E-46 | 0.579 |
| EHMT2 | 8.27E-01 | EHMT2 | 2.19E-12 | 0.308 |
| EZH1 | 2.11E-02 | EZH1 | 9.07E-01 | 0.005 |
| EZH2 | 1.35E-12 | EZH2 | 1.23E-03 | 0.145 |
| KMT2A | 4.68E-01 | KMT2A | 5.77E-97 | 0.766 |
| KMT2B | 1.40E-01 | KMT2B | 1.24E-128 | 0.832 |
| KMT2C | 3.70E-01 | KMT2C | 5.65E-122 | 0.820 |
| KMT2D | 2.60E-05 | KMT2D | 5.63E-32 | 0.494 |
| KMT2E | 1.92E-01 | KMT2E | 4.56E-105 | 0.785 |
| MECOM | 2.21E-01 | MECOM | 1.31E-06 | 0.215 |
| NSD1 | 5.49E-02 | NSD1 | 8.13E-127 | 0.829 |
| PRDM1 | 3.20E-01 | PRDM1 | 7.06E-13 | 0.315 |
| PRDM10 | 4.92E-04 | PRDM10 | 1.21E-87 | 0.741 |
| PRDM11 | 2.71E-03 | PRDM11 | 6.96E-12 | 0.301 |
| PRDM12 | 5.82E-06 | PRDM12 | 1.80E-01 | 0.060 |
| PRDM13 | 4.62E-01 | PRDM13 | 2.76E-01 | 0.049 |
| PRDM14 | 4.33E-01 | PRDM14 | 7.13E-01 | 0.017 |
| PRDM15 | 5.90E-02 | PRDM15 | 6.48E-04 | 0.152 |
| PRDM16 | 6.83E-01 | PRDM16 | 7.48E-01 | 0.014 |
| PRDM2 | 3.85E-01 | PRDM2 | 2.77E-70 | 0.686 |
| PRDM4 | 4.69E-01 | PRDM4 | 3.72E-54 | 0.620 |
| PRDM5 | 9.29E-02 | PRDM5 | 3.45E-19 | 0.387 |
| PRDM6 | 6.89E-04 | PRDM6 | 4.26E-03 | 0.128 |
| PRDM7 | 3.84E-01 | PRDM7 | 5.38E-03 | 0.125 |
| PRDM8 | 6.63E-05 | PRDM8 | 8.01E-01 | 0.011 |
| PRDM9 | 5.35E-01 | PRDM9 | 7.02E-01 | 0.017 |
| SETD1A | 3.29E-01 | SETD1A | 3.63E-02 | 0.094 |
| SETD1B | 9.31E-03 | SETD1B | 1.11E-44 | 0.573 |
| SETD2 | 2.64E-02 | SETD2 | 3.47E-129 | 0.833 |
| SETD3 | 3.48E-01 | SETD3 | 8.77E-01 | 0.007 |
| SETD4 | 1.29E-03 | SETD4 | 2.56E-22 | 0.417 |
| SETD5 | 2.64E-05 | SETD5 | 6.88E-78 | 0.712 |
| SETD6 | 5.42E-02 | SETD6 | 7.32E-01 | 0.015 |
| SETD7 | 4.45E-01 | SETD7 | 9.24E-106 | 0.787 |
| SETD8 | 2.48E-03 | SETD8 | 5.01E-09 | 0.258 |
| SETDB1 | 1.24E-02 | SETDB1 | 1.60E-13 | 0.323 |
| SETDB2 | 3.50E-01 | SETDB2 | 1.61E-02 | 0.108 |
| SETMAR | 1.63E-01 | SETMAR | 2.40E-34 | 0.511 |
| SMYD1 | 1.97E-01 | SMYD1 | 9.40E-01 | 0.003 |
| SMYD2 | 4.78E-01 | SMYD2 | 2.22E-01 | 0.055 |
| SMYD3 | 5.14E-01 | SMYD3 | 1.14E-09 | 0.269 |
| SMYD4 | 3.06E-01 | SMYD4 | 9.99E-20 | 0.392 |
| SMYD5 | 9.71E-01 | SMYD5 | 8.87E-21 | 0.402 |
| SUV39H1 | 3.77E-01 | SUV39H1 | 4.30E-19 | 0.386 |
| SUV39H2 | 4.21E-02 | SUV39H2 | 1.03E-05 | 0.196 |
| SUV420H1 | 6.57E-03 | SUV420H1 | 3.81E-42 | 0.559 |
| SUV420H2 | 3.88E-04 | SUV420H2 | 7.67E-26 | 0.447 |
| WHSC1 | 2.89E-14 | WHSC1 | 4.06E-31 | 0.488 |
| WHSC1L1 | 5.19E-01 | WHSC1L1 | 6.19E-47 | 0.585 |
Supplementary Table 5.
Summary of correlation between CNAs frequencied and gene expression levels.
| Gene | Amplification | Deletion | Diploid | Total amplification | Total deletion | Amplification and expressed higher (%) | Amplification and expressed lower (%) | Deletion and expressed higher (%) | Deletion and expressed lower (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Higher | Lower | Higher | Lower | Higher | Lower | |||||||
| Expression | ||||||||||||
| ASH1L | 19 | 8 | 0 | 8 | 183 | 181 | 27 | 8 | 70.37 | 29.63 | 0.00 | 100.00 |
| DOT1L 4 | 1 | 11 | 19 | 179 | 185 | 5 | 30 | 80.00 | 20.00 | 36.67 | 63.33 | |
| EHMT1 | 35 | 11 | 3 | 5 | 183 | 162 | 46 | 8 | 76.09 | 23.91 | 37.50 | 62.50 |
| EHMT2 | 8 | 4 | 5 | 10 | 185 | 187 | 12 | 15 | 66.67 | 33.33 | 33.33 | 66.67 |
| EZH1 | 3 | 1 | 13 | 37 | 161 | 184 | 4 | 50 | 75.00 | 25.00 | 26.00 | 74.00 |
| EZH2 | 58 | 15 | 2 | 6 | 178 | 140 | 73 | 8 | 79.45 | 20.55 | 25.00 | 75.00 |
| KMT2A | 12 | 10 | 14 | 15 | 174 | 174 | 22 | 29 | 54.55 | 45.45 | 48.28 | 51.72 |
| KMT2B | 6 | 3 | 10 | 6 | 190 | 184 | 9 | 16 | 66.67 | 33.33 | 62.50 | 37.50 |
| KMT2C | 39 | 35 | 10 | 11 | 153 | 151 | 74 | 21 | 52.70 | 47.30 | 47.62 | 58.38 |
| KMT2D | 6 | 5 | 8 | 8 | 186 | 186 | 11 | 16 | 54.55 | 45.45 | 50.00 | 50.00 |
| KMT2E | 39 | 37 | 3 | 4 | 158 | 158 | 76 | 7 | 51.32 | 48.68 | 42.86 | 57.14 |
| MECOM | 36 | 26 | 3 | 4 | 169 | 161 | 62 | 7 | 58.06 | 41.94 | 42.86 | 57.14 |
| NSD1 | 16 | 6 | 5 | 5 | 188 | 179 | 22 | 10 | 72.73 | 27.27 | 50.00 | 50.00 |
| PRDM1 | 2 | 0 | 62 | 60 | 139 | 136 | 2 | 122 | 100.00 | 0.00 | 50.82 | 49.18 |
| PRDM10 | 18 | 6 | 6 | 13 | 180 | 176 | 24 | 19 | 75.00 | 25.00 | 31.58 | 68.42 |
| PRDM11 | 8 | 8 | 6 | 20 | 171 | 186 | 16 | 26 | 50.00 | 50.00 | 23.08 | 76.92 |
| PRDM12 | 33 | 11 | 1 | 9 | 179 | 166 | 44 | 10 | 75.00 | 25.00 | 10.00 | 90.00 |
| PRDM13 | 0 | 5 | 7 | 118 | 189 | 80 | 5 | 125 | 0.00 | 100.00 | 5.60 | 94.40 |
| PRDM14 | 16 | 96 | 0 | 2 | 270 | 15 | 112 | 2 | 14.29 | 85.71 | 0.00 | 100.00 |
| PRDM15 | 14 | 4 | 8 | 18 | 177 | 178 | 18 | 26 | 77.78 | 22.22 | 30.77 | 69.23 |
| PRDM16 | 3 | 4 | 13 | 16 | 179 | 184 | 7 | 29 | 42.86 | 57.14 | 44.83 | 55.17 |
| PRDM2 | 2 | 0 | 9 | 26 | 173 | 189 | 2 | 35 | 100.00 | 0.00 | 25.71 | 74.29 |
| PRDM4 | 18 | 2 | 6 | 13 | 184 | 176 | 20 | 19 | 90.00 | 10.00 | 31.58 | 68.42 |
| PRDM5 | 4 | 6 | 7 | 29 | 164 | 189 | 10 | 36 | 40.00 | 60.00 | 19.44 | 80.56 |
| PRDM6 | 7 | 6 | 6 | 26 | 167 | 187 | 13 | 32 | 53.85 | 46.15 | 18.75 | 81.25 |
| PRDM7 | 1 | 3 | 42 | 103 | 160 | 90 | 40 | 145 | 25.00 | 75.00 | 28.97 | 71.03 |
| PRDM8 | 4 | 7 | 5 | 10 | 182 | 191 | 11 | 15 | 36.36 | 63.64 | 33.33 | 66.67 |
| PRDM9 | 6 | 19 | 2 | 8 | 347 | 17 | 25 | 10 | 24.00 | 76.00 | 20.00 | 80.00 |
| SETD1A | 20 | 3 | 14 | 11 | 185 | 166 | 23 | 25 | 86.96 | 13.04 | 56.00 | 44.00 |
| SETD1B | 13 | 3 | 11 | 17 | 179 | 176 | 16 | 28 | 81.25 | 18.75 | 39.29 | 60.71 |
| SETD2 | 17 | 7 | 3 | 16 | 176 | 180 | 24 | 19 | 70.83 | 29.17 | 15.79 | 84.21 |
| SETD3 | 8 | 2 | 6 | 25 | 172 | 186 | 10 | 31 | 80.00 | 20.00 | 19.35 | 80.65 |
| SETD4 | 14 | 3 | 4 | 13 | 183 | 182 | 17 | 17 | 82.35 | 17.65 | 23.53 | 76.47 |
| SETD5 | 17 | 8 | 3 | 25 | 166 | 180 | 25 | 28 | 68.00 | 32.00 | 10.71 | 89.29 |
| SETD6 | 6 | 0 | 15 | 78 | 121 | 179 | 6 | 93 | 100.00 | 0.00 | 16.13 | 83.87 |
| SETD7 | 7 | 4 | 3 | 14 | 181 | 190 | 11 | 17 | 63.64 | 36.36 | 17.65 | 82.35 |
| SETD8 | 11 | 3 | 6 | 25 | 171 | 183 | 14 | 31 | 78.57 | 21.43 | 19.35 | 80.65 |
| SETDB1 | 26 | 2 | 1 | 1 | 196 | 173 | 28 | 2 | 92.86 | 7.14 | 50.00 | 50.00 |
| SETDB2 | 1 | 1 | 34 | 131 | 67 | 165 | 2 | 165 | 50.00 | 50.00 | 20.61 | 79.39 |
| SETMAR | 17 | 8 | 5 | 16 | 175 | 178 | 25 | 21 | 68.00 | 32.00 | 23.81 | 76.19 |
| SMYD1 | 2 | 10 | 4 | 16 | 279 | 88 | 12 | 20 | 16.67 | 83.33 | 20.00 | 80.00 |
| SMYD2 | 12 | 9 | 4 | 16 | 174 | 184 | 21 | 20 | 57.14 | 42.86 | 20.00 | 80.00 |
| SMYD3 | 18 | 3 | 3 | 11 | 185 | 179 | 21 | 14 | 85.71 | 14.29 | 21.43 | 78.57 |
| SMYD4 | 5 | 0 | 9 | 58 | 141 | 186 | 5 | 67 | 100.00 | 0.00 | 13.43 | 86.57 |
| SMYD5 | 11 | 1 | 5 | 19 | 179 | 184 | 12 | 24 | 91.67 | 8.33 | 20.83 | 79.17 |
| SUV39H1 | 4 | 5 | 13 | 7 | 187 | 183 | 9 | 20 | 44.44 | 55.56 | 65.00 | 35.00 |
| SUV39H2 | 12 | 2 | 4 | 29 | 168 | 184 | 14 | 33 | 85.71 | 14.29 | 12.12 | 87.88 |
| SUV420H1 | 26 | 7 | 0 | 6 | 186 | 174 | 33 | 6 | 174.79 | 21.21 | 0.00 | 100.0 |
| SUV420H2 | 13 | 1 | 10 | 4 | 194 | 177 | 14 | 14 | 92.86 | 7.114 | 71.43 | 28.57 |
| WHSC1 | 14 | 2 | 9 | 12 | 185 | 177 | 16 | 21 | 87.50 | 12.50 | 42.86 | 57.14 |
| WHSC1L1 | 44 | 7 | 18 | 111 | 81 | 138 | 51 | 129 | 86.27 | 13.73 | 13.95 | 86.05 |
Supplementary Table 6.
Summary for differential promoter methylation HMTs in paired normal and tumor tissues.
| Gene | Differential methylation analysis of 46 HMTs | Paired T-test | |||||
|---|---|---|---|---|---|---|---|
| logFC | AveExpr | t | P.value | adj.P.Val | B | ||
| PRDM14 | −0.201 | 0.320 | −11.345 | 1.30E-19 | 6.09E-18 | 33.276 | 1.75E-17 |
| NSD1 | −0.142 | 0.421 | −9.479 | 1.49E-15 | 3.50E-14 | 23.928 | 2.44E-13 |
| PRDM8 | −0.146 | 0.432 | −8.138 | 1.20E-12 | 1.89E-11 | 17.253 | 6.68E-12 |
| EZH1 | −0.024 | 0.103 | −7.529 | 2.41E-11 | 2.83E-10 | 14.277 | 1.61E-10 |
| PRDM13 | −0.121 | 0.306 | −7.276 | 8.19E-11 | 7.70E-10 | 13.062 | 6.16E-12 |
| PRDM6 | −0.065 | 0.158 | −6.626 | 1.81E-09 | 1.42E-08 | 9.998 | 6.49E-10 |
| PRDM12 | −0.047 | 0.119 | −6.586 | 2.18E-09 | 1.47E-08 | 9.813 | 3.76E-10 |
| SETD3 | 0.061 | 0.604 | 6.222 | 1.18E-08 | 6.93E-08 | 8.149 | 3.26E-08 |
| PRDM5 | −0.093 | 0.117 | −5.224 | 9.70E-07 | 5.07E-06 | 3.821 | 2.09E-06 |
| KMT2B | 0.057 | 0.551 | 4.548 | 1.54E-05 | 7.22E-05 | 1.136 | 5.32E-06 |
| PRDM15 | −0.012 | 0.157 | −4.393 | 2.81E-05 | 1.20E-04 | 0.554 | 2.38E-05 |
| SETD5 | 0.037 | 0.474 | 3.924 | 1.61E-04 | 6.31E-04 | −1.122 | 6.03E-05 |
| PRDM7 | 0.041 | 0.537 | 3.845 | 2.13E-04 | 7.71E-04 | −1.391 | 1.73E-04 |
| WHSC1 | 0.023 | 0.463 | 3.619 | 4.67E-04 | 1.57E-03 | −2.136 | 2.21E-04 |
| SETDB1 | 0.014 | 0.140 | 3.088 | 2.61E-03 | 8.19E-03 | −3.749 | 1.03E-04 |
| EHMT2 | 0.004 | 0.097 | 2.887 | 4.78E-03 | 1.40E-02 | −4.307 | 3.23E-04 |
| MECOM | −0.016 | 0.151 | −2.837 | 5.52E-03 | 1.53E-02 | −4.439 | 3.26E-03 |
| KMT2E | −0.031 | 0.417 | −2.721 | 7.69E-03 | 1.72E-02 | −4.741 | 5.31E-03 |
| SETD1A | 0.009 | 0.127 | 2.725 | 7.60E-03 | 1.72E-02 | −4.731 | 6.35E-03 |
| SMYD1 | 0.050 | 0.805 | 2.747 | 7.14E-03 | 1.72E-02 | −4.674 | 5.86E-03 |
| PRDM9 | 0.071 | 0.586 | 2.735 | 7.39E-03 | 1.72E-02 | −4.705 | 9.94E-04 |
| SUV420H1 | −0.008 | 0.225 | −2.700 | 8.16E-03 | 1.74E-02 | −4.795 | 2.43E-03 |
| PRDM2 | 0.007 | 0.431 | 2.583 | 1.13E-02 | 2.30E-02 | −5.087 | 3.74E-03 |
| PRDM11 | 0.019 | 0.845 | 2.497 | 1.42E-02 | 2.77E-02 | −5.293 | 9.22E-03 |
| PRDM4 | −0.004 | 0.062 | −2.420 | 1.73E-02 | 3.26E-02 | −5.473 | 1.23E-02 |
| PRDM10 | 0.025 | 0.564 | 2.390 | 1.87E-02 | 3.38E-02 | −5.542 | 2.59E-02 |
| DOT1L | 0.002 | 0.036 | 2.290 | 2.42E-02 | 4.20E-02 | −5.766 | 9.03E-03 |
| SMYD5 | 0.008 | 0.230 | 1.982 | 5.02E-02 | 8.43E-02 | −6.401 | 3.76E-02 |
| SETD7 | 0.003 | 0.057 | 1.902 | 6.00E-02 | 9.73E-02 | −6.551 | 2.41E-02 |
| SETMAR | −0.005 | 0.173 | −1.523 | 1.31E-01 | 2.05E-01 | −7.188 | 8.77E-02 |
| SETD4 | 0.001 | 0.032 | 1.425 | 1.57E-01 | 2.39E-01 | −7.331 | 1.30E-01 |
| KMT2D | 0.003 | 0.079 | 1.318 | 1.91E-01 | 2.71E-01 | −7.476 | 5.25E-02 |
| SETDB2 | 0.005 | 0.905 | 1.335 | 1.85E-01 | 2.71E-01 | −7.454 | 1.62E-01 |
| WHSC1L1 | −0.004 | 0.151 | −1.244 | 2.17E-01 | 2.99E-01 | −7.571 | 2.39E-01 |
| SUV39H2 | 0.001 | 0.041 | 1.191 | 2.36E-01 | 3.09E-01 | −7.634 | 2.32E-01 |
| AR | 0.005 | 0.316 | 1.191 | 2.36E-01 | 3.09E-01 | −7.634 | 1.74E-01 |
| PRDM1 | −0.014 | 0.148 | −1.147 | 2.54E-01 | 3.23E-01 | −7.686 | 2.81E-01 |
| SETD2 | 0.001 | 0.036 | 0.996 | 3.22E-01 | 3.98E-01 | −7.847 | 2.55E-01 |
| ASH1L | 0.003 | 0.473 | 0.978 | 3.31E-01 | 3.98E-01 | −7.865 | 2.85E-01 |
| SUV420H2 | −0.001 | 0.025 | −0.908 | 3.66E-01 | 4.30E-01 | −7.931 | 3.20E-01 |
| SMYD2 | −0.002 | 0.062 | −0.828 | 4.10E-01 | 4.70E-01 | −8.001 | 3.54E-01 |
| SMYD3 | −0.002 | 0.413 | −0.685 | 4.95E-01 | 5.29E-01 | −8.109 | 4.27E-01 |
| EZH2 | −0.001 | 0.295 | −0.703 | 4.84E-01 | 5.29E-01 | −8.097 | 3.90E-01 |
| SETD6 | −0.001 | 0.039 | −0.717 | 4.75E-01 | 5.29E-01 | −8.086 | 3.54E-01 |
| SETD8 | 0.000 | 0.030 | −0.373 | 7.10E-01 | 7.42E-01 | −8.275 | 6.77E-01 |
| KMT2A | 0.000 | 0.030 | 0.107 | 9.15E-01 | 9.35E-01 | −8.340 | 8.71E-01 |
| SUV39H1 | 0.000 | 0.115 | 0.057 | 9.55E-01 | 9.55E-01 | −8.344 | 9.52E-01 |
Supplementary Table 7A.
Summary of uniivariate analysis of RFS for amplification of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95% CI | Count amplification | Count diploid | Count percentage | |
|---|---|---|---|---|---|---|---|
| Lower | Higher | ||||||
| PRDM14 | 0.0081 | 1.860 | 1.185 | 2.919 | 112 | 285 | 28.2 |
| WHSC1L1 | 0.0051 | 2.401 | 1.330 | 4.332 | 51 | 219 | 18.9 |
| EHMT1 | 0.0001 | 3.133 | 1.876 | 5.232 | 46 | 345 | 11.8 |
| PRDM12 | 0.0002 | 3.022 | 1.795 | 5.090 | 44 | 345 | 11.3 |
| SUV420H1 | 0.0075 | 2.390 | 1.336 | 4.273 | 33 | 360 | 8.4 |
| SETDB1 | 0.0031 | 2.860 | 1.543 | 5.303 | 28 | 369 | 7.1 |
| SETD1A | 0.0157 | 2.520 | 1.286 | 4.938 | 23 | 351 | 6.1 |
| SMYD2 | 0.0188 | 2.723 | 1.304 | 5.686 | 21 | 358 | 5.5 |
| PRDM4 | 0.0015 | 3.808 | 1.882 | 7.703 | 20 | 360 | 5.3 |
| PRDM15 | 0.0445 | 2.214 | 1.095 | 4.478 | 18 | 355 | 4.8 |
| SETD1B | 0.0006 | 4.815 | 2.279 | 10.173 | 16 | 355 | 4.3 |
| WHSC1 | 0.0467 | 2.299 | 1.098 | 4.813 | 16 | 362 | 4.2 |
| SETD8 | 0.0100 | 3.722 | 1.596 | 8.682 | 14 | 354 | 3.8 |
| SUV420H2 | 0.0499 | 2.409 | 1.099 | 5.281 | 14 | 371 | 3.6 |
| SMYD5 | 0.0009 | 5.127 | 2.330 | 11.284 | 12 | 363 | 3.2 |
| SMYD1 | 0.0005 | 5.729 | 2.599 | 12.630 | 12 | 367 | 3.2 |
| EHMT2 | 0.0278 | 2.969 | 1.281 | 6.882 | 12 | 372 | 3.1 |
| KMT2B | 0.0311 | 3.242 | 1.300 | 8.084 | 9 | 374 | 2.3 |
| PRDM16 | 0.0029 | 7.508 | 2.674 | 21.079 | 7 | 363 | 1.9 |
| DOT1L | 0.0358 | 3.657 | 1.323 | 10.106 | 5 | 364 | 1.4 |
| PRDM1 | 0.0219 | 9.340 | 2.227 | 39.178 | 2 | 275 | 0.7 |
| SETD3 | 0.2315 | 0.362 | 0.050 | 2.622 | 10 | 358 | 2.7 |
| PRDM7 | 0.3568 | 0.000 | 0.000 | Inf | 4 | 250 | 1.6 |
| SETD4 | 0.0502 | 2.263 | 1.083 | 4.731 | 17 | 365 | 4.5 |
| PRDM8 | 0.0507 | 2.393 | 1.096 | 5.227 | 11 | 373 | 2.9 |
| PRDM13 | 0.0664 | 3.720 | 1.147 | 12.065 | 5 | 269 | 1.8 |
| ASH1L | 0.0749 | 1.998 | 0.993 | 4.017 | 27 | 364 | 6.9 |
| PRDM9 | 0.0840 | 1.953 | 0.971 | 3.926 | 25 | 364 | 6.4 |
| SETD6 | 0.0854 | 4.711 | 1.119 | 19.832 | 6 | 300 | 2.0 |
| PRDM2 | 0.1026 | 9.419 | 1.273 | 69.698 | 2 | 362 | 0.5 |
| NSD1 | 0.1187 | 1.887 | 0.905 | 3.937 | 22 | 367 | 5.7 |
| MECOM | 0.1318 | 1.543 | 0.898 | 2.653 | 62 | 330 | 15.8 |
| SMYD4 | 0.1402 | 3.589 | 0.868 | 14.839 | 5 | 327 | 1.5 |
| EZH2 | 0.1436 | 1.470 | 0.891 | 2.426 | 73 | 318 | 18.7 |
| KMT2E | 0.1859 | 1.418 | 0.857 | 2.347 | 76 | 316 | 19.4 |
| PRDM10 | 0.1868 | 1.649 | 0.821 | 3.311 | 24 | 356 | 6.3 |
| KMT2C | 0.1870 | 1.418 | 0.856 | 2.349 | 74 | 304 | 19.6 |
| SETMAR | 0.1931 | 1.746 | 0.801 | 3.809 | 25 | 353 | 6.6 |
| KMT2A | 0.1991 | 1.674 | 0.802 | 3.494 | 22 | 348 | 5.9 |
| SETD5 | 0.2196 | 1.688 | 0.774 | 3.679 | 25 | 346 | 6.7 |
| SMYD3 | 0.2284 | 1.742 | 0.754 | 4.024 | 21 | 364 | 5.5 |
| PRDM5 | 0.2589 | 1.894 | 0.688 | 5.217 | 10 | 353 | 2.8 |
| SUV39H1 | 0.2972 | 1.975 | 0.620 | 6.296 | 9 | 370 | 2.4 |
| SETD2 | 0.3418 | 1.538 | 0.666 | 3.551 | 24 | 356 | 6.3 |
| PRDM11 | 0.3802 | 1.543 | 0.620 | 3.839 | 16 | 357 | 4.3 |
| SETD7 | 0.6018 | 1.383 | 0.433 | 4.411 | 11 | 371 | 2.9 |
| SETDB2 | 0.6704 | 1.601 | 0.211 | 12.166 | 2 | 232 | 0.9 |
| PRDM6 | 0.6765 | 1.292 | 0.406 | 4.117 | 13 | 354 | 3.5 |
| EZH1 | 0.6774 | 1.570 | 0.216 | 11.403 | 4 | 345 | 1.1 |
| KMT2D | 0.7397 | 1.224 | 0.385 | 3.895 | 11 | 372 | 2.9 |
| SUV39H2 | 0.9204 | 0.943 | 0.295 | 3.016 | 14 | 352 | 3.8 |
Supplementary Table 7B.
Summary of uniivariate analysis of RFS for deletion of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95% CI | Count amplification | Count diploid | Count percentage | |
|---|---|---|---|---|---|---|---|
| Lower | Higher | ||||||
| DOT1L | 0.0363 | 2.053 | 1.103 | 3.821 | 30 | 364 | 7.6 |
| EHMT2 | 0.0356 | 2.801 | 1.211 | 6.478 | 15 | 372 | 3.9 |
| PRDM7 | 0.0055 | 1.881 | 1.205 | 2.934 | 145 | 250 | 36.7 |
| SETD4 | 0.0042 | 3.905 | 1.778 | 8.576 | 17 | 365 | 4.5 |
| SETD6 | 0.0016 | 2.161 | 1.364 | 3.423 | 93 | 300 | 23.7 |
| SETDB2 | 0.0082 | 1.833 | 1.164 | 2.886 | 165 | 232 | 41.6 |
| SMYD1 | 0.0102 | 2.847 | 1.409 | 5.756 | 20 | 367 | 5.2 |
| SUV39H2 | 0.0014 | 2.710 | 1.558 | 4.711 | 33 | 352 | 8.6 |
| SUV420H2 | 0.0121 | 2.947 | 1.411 | 6.159 | 14 | 371 | 3.6 |
| WHSC1L1 | 0.0154 | 1.886 | 1.129 | 3.151 | 129 | 219 | 37.1 |
| SETD5 | 0.1644 | 0.481 | 0.151 | 1.531 | 28 | 346 | 7.5 |
| PRDM8 | 0.3963 | 0.573 | 0.140 | 2.341 | 15 | 373 | 3.9 |
| EZH2 | 0.0977 | 0.000 | 0.000 | Inf | 8 | 318 | 2.5 |
| SUV420H1 | 0.1404 | 0.000 | 0.000 | Inf | 6 | 360 | 1.6 |
| SETDB1 | 0.1916 | 0.000 | 0.000 | Inf | 2 | 369 | 0.5 |
| EZH1 | 0.0528 | 1.783 | 1.026 | 3.096 | 50 | 345 | 12.7 |
| PRDM15 | 0.0550 | 2.129 | 1.053 | 4.307 | 26 | 355 | 6.8 |
| PRDM6 | 0.0598 | 2.008 | 1.031 | 3.914 | 32 | 354 | 8.3 |
| SETD1A | 0.0611 | 2.085 | 1.032 | 4.214 | 25 | 351 | 6.6 |
| PRDM1 | 0.0788 | 1.512 | 0.959 | 2.383 | 122 | 275 | 30.7 |
| SETD7 | 0.0805 | 2.162 | 0.989 | 4.727 | 17 | 371 | 4.4 |
| KMT2B | 0.0874 | 2.118 | 0.971 | 4.620 | 16 | 374 | 4.1 |
| PRDM4 | 0.0918 | 2.234 | 0.965 | 5.171 | 19 | 360 | 5.0 |
| PRDM9 | 0.0966 | 2.675 | 0.972 | 7.362 | 10 | 364 | 2.7 |
| PRDM13 | 0.1008 | 1.473 | 0.932 | 2.327 | 125 | 269 | 31.7 |
| SUV39H1 | 0.1021 | 2.042 | 0.937 | 4.449 | 20 | 370 | 5.1 |
| PRDM11 | 0.1118 | 1.846 | 0.916 | 3.719 | 26 | 357 | 6.8 |
| SMYD5 | 0.1304 | 1.855 | 0.884 | 3.892 | 24 | 363 | 6.2 |
| SETD1B | 0.1357 | 1.774 | 0.879 | 3.580 | 28 | 355 | 7.3 |
| SETD8 | 0.1572 | 1.672 | 0.856 | 3.269 | 31 | 354 | 8.1 |
| SMYD3 | 0.1598 | 2.058 | 0.828 | 5.119 | 14 | 364 | 3.7 |
| SETD3 | 0.1602 | 1.595 | 0.860 | 2.956 | 31 | 358 | 8.0 |
| EHMT1 | 0.2166 | 2.283 | 0.712 | 7.313 | 8 | 345 | 2.3 |
| PRDM12 | 0.2324 | 2.215 | 0.690 | 7.111 | 10 | 345 | 2.8 |
| SMYD4 | 0.2552 | 1.373 | 0.808 | 2.334 | 67 | 327 | 17.0 |
| NSD1 | 0.2914 | 1.994 | 0.625 | 6.357 | 10 | 367 | 2.7 |
| PRDM2 | 0.3186 | 1.405 | 0.740 | 2.668 | 35 | 362 | 8.8 |
| PRDM14 | 0.3336 | 3.197 | 0.436 | 23.424 | 2 | 285 | 0.7 |
| KMT2D | 0.3915 | 1.524 | 0.615 | 3.777 | 16 | 372 | 4.1 |
| ASH1L | 0.4391 | 1.635 | 0.513 | 5.211 | 8 | 364 | 2.2 |
| PRDM16 | 0.4450 | 1.327 | 0.658 | 2.676 | 29 | 363 | 7.4 |
| KMT2C | 0.5010 | 0.685 | 0.214 | 2.197 | 21 | 304 | 6.5 |
| SETD2 | 0.6543 | 1.218 | 0.526 | 2.817 | 19 | 356 | 5.1 |
| WHSC1 | 0.7368 | 0.844 | 0.307 | 2.321 | 21 | 362 | 5.5 |
| SETMAR | 0.7739 | 0.865 | 0.314 | 2.382 | 21 | 353 | 5.6 |
| PRDM5 | 0.7968 | 0.904 | 0.415 | 1.971 | 36 | 353 | 9.3 |
| SMYD2 | 0.8241 | 1.111 | 0.447 | 2.762 | 20 | 358 | 5.3 |
| KMT2A | 0.8987 | 1.056 | 0.457 | 2.441 | 29 | 348 | 7.7 |
| PRDM10 | 0.9515 | 0.965 | 0.303 | 3.077 | 19 | 356 | 5.1 |
| MECOM | 0.9671 | 1.025 | 0.318 | 3.305 | 7 | 330 | 2.1 |
| KMT2E | 0.9844 | 0.986 | 0.239 | 4.064 | 7 | 316 | 2.2 |
Supplementary Table 7C.
Summary of univariate analysis of RFS for methylation level of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95% CI | |
|---|---|---|---|---|
| Lower | Higher | |||
| PRDM14 | 0.0023 | 2.00E+01 | 2.93E+00 | 1.37E+02 |
| PRDM13 | 0.0181 | 1.17E+01 | 1.57E+00 | 8.66E+01 |
| PRDM6 | 0.0184 | 5.91E+01 | 2.28E+00 | 1.53E+03 |
| PRDM8 | 0.0211 | 1.58E+01 | 1.56E+00 | 1.60E+02 |
| KMT2B | 0.0383 | 1.56E−02 | 3.31E−04 | 7.31E−01 |
| NSD1 | 0.0441 | 3.10E+01 | 1.07E+00 | 8.99E+02 |
| SETDB2 | 0.0572 | 6.91E−02 | 4.83E−03 | 9.87E−01 |
| SETDB1 | 0.0580 | 3.52E−05 | 1.11E−09 | 1.11E+00 |
| SETD4 | 0.0873 | 1.31E+13 | 5.38E−02 | 3.18E+27 |
| EZH2 | 0.1045 | 7.80E−10 | 3.21E−21 | 1.90E+02 |
| KMT2E | 0.1152 | 1.45E+01 | 6.10E−01 | 3.45E+02 |
| PRDM4 | 0.1323 | 4.90E+03 | 1.71E−01 | 1.40E+08 |
| SUV39H1 | 0.1694 | 3.77E+01 | 2.58E−01 | 5.50E+03 |
| PRDM12 | 0.1911 | 1.19E+01 | 3.34E−01 | 4.23E+02 |
| SMYD3 | 0.2266 | 2.15E+04 | 1.62E−03 | 2.87E+11 |
| MECOM | 0.2358 | 4.09E+01 | 1.15E−01 | 1.45E+04 |
| DOT1L | 0.2610 | 3.25E+05 | 2.83E−04 | 3.73E+14 |
| SETD6 | 0.2710 | 4.95E−05 | 3.38E−13 | 7.26E+03 |
| SETD3 | 0.3030 | 1.56E−01 | 4.77E−03 | 5.09E+00 |
| PRDM9 | 0.3091 | 5.09E−01 | 1.39E−01 | 1.86E+00 |
| SETD5 | 0.3255 | 1.49E−01 | 3.32E−03 | 6.66E+00 |
| WHSC1L1 | 0.3297 | 1.86E+02 | 7.11E−03 | 4.86E+06 |
| PRDM1 | 0.3832 | 4.35E+00 | 1.70E−01 | 1.12E+02 |
| SUV420H2 | 0.3911 | 8.48E+07 | 1.16E−09 | 6.20E+24 |
| KMT2D | 0.4038 | 4.02E−04 | 3.62E−12 | 4.46E+04 |
| SUV39H2 | 0.4289 | 6.77E+06 | 1.22E−10 | 3.77E+23 |
| SETD1A | 0.4393 | 9.61E−03 | 6.83E−08 | 1.35E+03 |
| PRDM5 | 0.5427 | 1.69E+00 | 3.16E−01 | 9.06E+00 |
| SMYD2 | 0.5584 | 2.99E+01 | 1.02E−03 | 8.79E+05 |
| SETD7 | 0.5756 | 2.92E−04 | 1.06E−16 | 8.05E+08 |
| EZH1 | 0.6304 | 1.22E+01 | 4.84E−04 | 3.09E+05 |
| PRDM15 | 0.6484 | 6.84E−02 | 6.01E−07 | 7.78E+03 |
| PRDM11 | 0.6665 | 3.32E−01 | 2.30E−03 | 4.80E+01 |
| PRDM10 | 0.7125 | 4.99E−01 | 1.26E−02 | 1.98E+01 |
| SETD2 | 0.7127 | 5.25E−04 | 1.16E−21 | 2.37E+14 |
| SMYD5 | 0.7344 | 2.17E−01 | 3.15E−05 | 1.49E+03 |
| SETD8 | 0.7427 | 2.28E−03 | 7.04E−20 | 7.36E+13 |
| PRDM7 | 0.7496 | 5.91E−01 | 2.37E−02 | 1.48E+01 |
| SMYD1 | 0.7786 | 7.48E−01 | 1.00E−01 | 5.57E+00 |
| ASH1L | 0.8598 | 6.00E−01 | 1.78E−03 | 2.02E+02 |
| SETMAR | 0.8750 | 2.48E+00 | 3.02E−05 | 2.03E+05 |
| EHMT2 | 0.8931 | 1.75E−01 | 1.51E−12 | 2.02E+10 |
| SUV420H1 | 0.8952 | 2.14E+00 | 2.59E−05 | 1.77E+05 |
| WHSC1 | 0.9032 | 7.11E−01 | 2.96E−03 | 1.71E+02 |
| KMT2A | 0.9108 | 2.45E+01 | 1.39E−23 | 4.30E+25 |
| PRDM2 | 0.9111 | 2.29E+00 | 1.16E−06 | 4.51E+06 |
| EHMT1 | NA | NA | NA | NA |
| KMT2C | NA | NA | NA | NA |
| PRDM16 | NA | NA | NA | NA |
| SETD1B | NA | NA | NA | NA |
| SMYD4 | NA | NA | NA | NA |
Supplementary Table 7D.
Summary of uniivariate analysis of RFS for expression level of 51 HMTs in prostate cancer.
| Gene | p-Value | HR | 95%CI | Coef. | Wald test | |
|---|---|---|---|---|---|---|
| Lower | Higher | |||||
| EZH2 | 0 | 2.897 | 1.755 | 4.78 | 1.064 | 17.31 |
| SETD5 | 3.00E-04 | 2.343 | 1.478 | 3.715 | 0.852 | 13.13 |
| WHSC1 | 4.00E-04 | 2.392 | 1.478 | 3.87 | 0.872 | 12.61 |
| SETD1B | 0.0054 | 1.931 | 1.215 | 3.07 | 0.658 | 7.74 |
| SUV420H2 | 0.0054 | 1.93 | 1.214 | 3.067 | 0.657 | 7.74 |
| PRDM6 | 0.0066 | 0.526 | 0.331 | 0.836 | −0.642 | 7.38 |
| PRDM15 | 0.0101 | 1.829 | 1.155 | 2.898 | 0.604 | 6.62 |
| KMT2D | 0.0119 | 1.793 | 1.137 | 2.827 | 0.584 | 6.32 |
| SMYD1 | 0.0207 | 0.471 | 0.249 | 0.891 | −0.753 | 5.35 |
| PRDM10 | 0.0235 | 1.682 | 1.073 | 2.639 | 0.52 | 5.13 |
| KMT2B | 0.0269 | 1.666 | 1.06 | 2.618 | 0.51 | 4.9 |
| PRDM16 | 0.0395 | 0.617 | 0.39 | 0.977 | −0.483 | 4.24 |
| DOT1L | 0.0397 | 1.617 | 1.023 | 2.556 | 0.481 | 4.23 |
| PRDM8 | 0.076 | 0.658 | 0.415 | 1.045 | −0.418 | 3.15 |
| PRDM12 | 0.0764 | 1.522 | 0.956 | 2.422 | 0.42 | 3.14 |
| SUV39H1 | 0.099 | 1.475 | 0.93 | 2.34 | 0.389 | 2.72 |
| KMT2C | 0.1048 | 1.448 | 0.926 | 2.264 | 0.37 | 2.63 |
| SETD8 | 0.1267 | 0.705 | 0.45 | 1.104 | −0.35 | 2.33 |
| KMT2E | 0.1411 | 1.402 | 0.894 | 2.2 | 0.338 | 2.17 |
| NSD1 | 0.1462 | 1.392 | 0.891 | 2.175 | 0.331 | 2.11 |
| EHMT1 | 0.1524 | 1.386 | 0.886 | 2.166 | 0.326 | 2.05 |
| SETD3 | 0.1529 | 0.722 | 0.461 | 1.129 | −0.326 | 2.04 |
| PRDM1 | 0.1609 | 1.378 | 0.88 | 2.156 | 0.32 | 1.97 |
| SETD4 | 0.1734 | 1.376 | 0.869 | 2.178 | 0.319 | 1.85 |
| PRDM11 | 0.2041 | 0.745 | 0.472 | 1.174 | −0.295 | 1.61 |
| SETDB2 | 0.2374 | 1.313 | 0.836 | 2.063 | 0.272 | 1.4 |
| MECOM | 0.2825 | 0.78 | 0.497 | 1.227 | −0.248 | 1.15 |
| SETDB1 | 0.2992 | 1.267 | 0.811 | 1.98 | 0.236 | 1.08 |
| SUV39H2 | 0.3051 | 0.788 | 0.5 | 1.242 | −0.238 | 1.05 |
| SETD2 | 0.3203 | 1.256 | 0.801 | 1.968 | 0.228 | 0.99 |
| PRDM14 | 0.3581 | 1.44 | 0.661 | 3.137 | 0.365 | 0.84 |
| SETD6 | 0.4063 | 1.207 | 0.774 | 1.883 | 0.188 | 0.69 |
| SMYD5 | 0.4071 | 1.209 | 0.772 | 1.892 | 0.19 | 0.69 |
| SETMAR | 0.4329 | 0.836 | 0.535 | 1.307 | −0.179 | 0.61 |
| SUV420H1 | 0.4691 | 1.179 | 0.755 | 1.841 | 0.165 | 0.52 |
| ASH1L | 0.5269 | 1.156 | 0.738 | 1.811 | 0.145 | 0.4 |
| KMT2A | 0.5758 | 1.136 | 0.727 | 1.775 | 0.127 | 0.31 |
| WHSC1L1 | 0.59 | 0.885 | 0.567 | 1.381 | −0.123 | 0.29 |
| PRDM5 | 0.6124 | 0.889 | 0.565 | 1.4 | −0.117 | 0.26 |
| EHMT2 | 0.631 | 1.115 | 0.715 | 1.741 | 0.109 | 0.23 |
| PRDM7 | 0.6638 | 0.899 | 0.555 | 1.455 | −0.107 | 0.19 |
| SMYD3 | 0.6794 | 1.098 | 0.704 | 1.714 | 0.094 | 0.17 |
| EZH1 | 0.7453 | 0.929 | 0.595 | 1.45 | −0.074 | 0.11 |
| PRDM9 | 0.8192 | 0.907 | 0.392 | 2.097 | −0.098 | 0.05 |
| SMYD4 | 0.8539 | 1.043 | 0.665 | 1.637 | 0.042 | 0.03 |
| SMYD2 | 0.8758 | 1.036 | 0.664 | 1.617 | 0.035 | 0.02 |
| SETD7 | 0.8837 | 1.034 | 0.657 | 1.629 | 0.034 | 0.02 |
| PRDM13 | 0.8883 | 1.038 | 0.619 | 1.741 | 0.037 | 0.02 |
| PRDM2 | 0.9321 | 0.981 | 0.626 | 1.536 | −0.019 | 0.01 |
| PRDM4 | 0.982 | 1.005 | 0.645 | 1.568 | 0.005 | 0 |
| SETD1A | 0.9988 | 1 | 0.641 | 1.562 | 0 | 0 |
Supplementary Table 8A.
Summary of multivariate analysis of RFS for amplification of 21 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | EHMT1 amplification | |
|---|---|---|---|---|---|---|
| Coefficient | −0.017 | 0.788 | 0.988 | 0.198 | 0.646 | |
| Hazard ratio | 0.983 | 2.198 | 2.687 | 1.219 | 1.908 | |
| 95%CI | Lower | 0.948 | 1.139 | 1.59 | 0.767 | 1.112 |
| Higher | 1.019 | 4.241 | 4.538 | 1.938 | 3.274 | |
| p-Value | 0.35 | 0.0189 | 0.0002 | 0.4019 | 0.0191 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | EHMT2 amplification | |
| Coefficient | −0.004 | 0.717 | 0.906 | 0.239 | 0.666 | |
| Hazard ratio | 0.996 | 2.049 | 2.476 | 1.27 | 1.947 | |
| 95%CI | Lower | 0.961 | 1.089 | 1.473 | 0.795 | 0.83 |
| Higher | 1.033 | 3.854 | 4.16 | 2.029 | 4.57 | |
| p-Value | 0.8407 | 0.0261 | 0.0006 | 0.3177 | 0.1257 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM12 amplification | |
| Coefficient | −0.007 | 0.676 | 0.921 | 0.325 | 0.665 | |
| Hazard ratio | 0.993 | 1.967 | 2.512 | 1.384 | 1.945 | |
| 95%CI | Lower | 0.959 | 1.037 | 1.492 | 0.871 | 1.125 |
| Higher | 1.029 | 3.73 | 4.23 | 2.2 | 3.362 | |
| p-Value | 0.7062 | 0.0383 | 0.0005 | 0.1691 | 0.0172 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM14 amplification | |
| Coefficient | −0.007 | 0.757 | 0.984 | 0.226 | 0.329 | |
| Hazard ratio | 0.993 | 2.131 | 2.675 | 1.254 | 1.39 | |
| 95%CI | Lower | 0.959 | 1.136 | 1.61 | 0.797 | 0.875 |
| Higher | 1.029 | 3.999 | 4.446 | 1.972 | 2.209 | |
| p-Value | 0.7034 | 0.0184 | 0.0001 | 0.3271 | 0.1637 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM15 amplification | |
| Coefficient | −0.005 | 0.654 | 0.952 | 0.132 | 0.436 | |
| Hazard ratio | 0.995 | 1.923 | 2.59 | 1.141 | 1.546 | |
| 95%CI | Lower | 0.958 | 1.019 | 1.534 | 0.704 | 0.743 |
| Higher | 1.033 | 3.629 | 4.372 | 1.849 | 3.22 | |
| p-Value | 0.7942 | 0.0436 | 0.0004 | 0.5931 | 0.2442 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM4 amplification | |
| Coefficient | −0.006 | 0.779 | 0.923 | 0.263 | 0.867 | |
| Hazard ratio | 0.994 | 2.179 | 2.517 | 1.301 | 2.379 | |
| 95%CI | Lower | 0.959 | 1.164 | 1.502 | 0.801 | 1.143 |
| Higher | 1.031 | 4.082 | 4.219 | 2.112 | 4.953 | |
| p-Value | 0.7471 | 0.0149 | 0.0005 | 0.2881 | 0.0205 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD1A amplification | |
| Coefficient | −0.015 | 0.872 | 0.893 | 0.073 | 0.391 | |
| Hazard ratio | 0.985 | 2.392 | 2.442 | 1.076 | 1.479 | |
| 95%CI | Lower | 0.947 | 1.217 | 1.43 | 0.662 | 0.722 |
| Higher | 1.024 | 4.702 | 4.171 | 1.749 | 3.029 | |
| p-Value | 0.4491 | 0.0114 | 0.0011 | 0.7672 | 0.2851 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD1B amplification | |
| Coefficient | −0.003 | 0.796 | 0.907 | 0.152 | 1.218 | |
| Hazard ratio | 0.997 | 2.216 | 2.477 | 1.164 | 3.381 | |
| 95%CI | Lower | 0.961 | 1.179 | 1.467 | 0.711 | 1.55 |
| Higher | 1.034 | 4.166 | 4.18 | 1.906 | 7.372 | |
| p-Value | 0.8521 | 0.0135 | 0.0007 | 0.5451 | 0.0022 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD8 Amplification | |
| Coefficient | 0 | 0.8 | 0.898 | 0.2 | 0.97 | |
| Hazard ratio | 1 | 2.226 | 2.456 | 1.221 | 2.638 | |
| 95%CI | Lower | 0.964 | 1.182 | 1.453 | 0.747 | 1.101 |
| Higher | 1.038 | 4.191 | 4.152 | 1.996 | 6.323 | |
| p-Value | 0.9897 | 0.0132 | 0.0008 | 0.4256 | 0.0296 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETDB1 amplification | |
| Coefficient | −0.009 | 0.812 | 0.941 | 0.158 | 0.701 | |
| Hazard ratio | 0.991 | 2.253 | 2.563 | 1.171 | 2.016 | |
| 95%CI | Lower | 0.957 | 1.209 | 1.559 | 0.744 | 1.062 |
| Higher | 1.027 | 4.2 | 4.214 | 1.842 | 3.826 | |
| p-Value | 0.6255 | 0.0106 | 0.0002 | 0.4956 | 0.0319 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD1 amplification | |
| Coefficient | −0.004 | 0.788 | 0.906 | 0.15 | 1.423 | |
| Hazard ratio | 0.996 | 2.198 | 2.475 | 1.162 | 4.149 | |
| 95%CI | Lower | 0.959 | 1.139 | 1.443 | 0.716 | 1.844 |
| Higher | 1.034 | 4.243 | 4.244 | 1.885 | 9.336 | |
| p-Value | 0.8369 | 0.0189 | 0.001 | 0.5434 | 0.0006 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD2 amplification | |
| Coefficient | 0 | 0.798 | 0.955 | 0.181 | 0.77 | |
| Hazard ratio | 1 | 2.221 | 2.6 | 1.198 | 2.159 | |
| 95%CI | Lower | 0.965 | 1.16 | 1.556 | 0.751 | 1.026 |
| Higher | 1.037 | 4.253 | 4.343 | 1.91 | 4.546 | |
| p-Value | 0.9835 | 0.0161 | 0.0003 | 0.4483 | 0.0426 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD5 Amplification | |
| Coefficient | 0.004 | 0.745 | 0.929 | 0.163 | 1.274 | |
| Hazard ratio | 1.004 | 2.106 | 2.532 | 1.177 | 3.574 | |
| 95%CI | Lower | 0.967 | 1.119 | 1.484 | 0.729 | 1.595 |
| Higher | 1.042 | 3.966 | 4.32 | 1.901 | 8.01 | |
| p-Value | 0.8462 | 0.0211 | 0.0007 | 0.5045 | 0.002 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H1 amplification | |
| Coefficient | −0.006 | 0.835 | 0.897 | 0.155 | 0.394 | |
| Hazard ratio | 0.994 | 2.304 | 2.452 | 1.168 | 1.483 | |
| 95%CI | Lower | 0.96 | 1.237 | 1.48 | 0.742 | 0.809 |
| Higher | 1.03 | 4.289 | 4.064 | 1.838 | 2.718 | |
| p-Value | 0.7365 | 0.0085 | 0.0005 | 0.5016 | 0.2028 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H2 amplification | |
| Coefficient | −0.014 | 0.885 | 0.927 | 0.072 | 0.422 | |
| Hazard ratio | 0.986 | 2.422 | 2.528 | 1.075 | 1.524 | |
| 95%CI | Lower | 0.949 | 1.263 | 1.51 | 0.664 | 0.673 |
| Higher | 1.025 | 4.642 | 4.231 | 1.741 | 3.455 | |
| p-Value | 0.4852 | 0.0077 | 0.0004 | 0.7691 | 0.3126 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1 amplification | |
| Coefficient | −0.014 | 0.786 | 1.051 | 0.152 | 0.439 | |
| Hazard ratio | 0.986 | 2.194 | 2.86 | 1.165 | 1.551 | |
| 95%CI | Lower | 0.951 | 1.142 | 1.702 | 0.733 | 0.732 |
| Higher | 1.022 | 4.218 | 4.806 | 1.851 | 3.286 | |
| p-Value | 0.4366 | 0.0184 | 0.0001 | 0.5191 | 0.2522 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1L1 amplification | |
| Coefficient | 0.001 | 0.486 | 1.027 | 0.21 | 0.565 | |
| Hazard ratio | 1.001 | 1.627 | 2.792 | 1.233 | 1.76 | |
| 95%CI | Lower | 0.956 | 0.709 | 1.378 | 0.686 | 0.946 |
| Higher | 1.049 | 3.732 | 5.657 | 2.216 | 3.272 | |
| p-Value | 0.95 | 0.251 | 0.0044 | 0.4836 | 0.0742 | |
Supplementary Table 8B.
Summary of multivariate analysis of RFS for deletion of 10 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | DOT1L deletion | |
|---|---|---|---|---|---|---|
| Coefficient | −0.003 | 0.742 | 0.971 | 0.304 | 0.441 | |
| Hazard ratio | 0.997 | 2.101 | 2.64 | 1.356 | 1.554 | |
| 95%CI | Lower | 0.962 | 1.118 | 1.585 | 0.847 | 0.815 |
| Higher | 1.033 | 3.945 | 4.397 | 2.17 | 2.963 | |
| p-Value | 0.8719 | 0.021 | 0.0002 | 0.2053 | 0.1802 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | EHMT2 deletion | |
| Coefficient | 0.004 | 0.775 | 0.951 | 0.176 | 0.992 | |
| Hazard ratio | 1.004 | 2.17 | 2.588 | 1.193 | 2.695 | |
| 95%CI | Lower | 0.968 | 1.156 | 1.548 | 0.743 | 1.135 |
| Higher | 1.042 | 4.074 | 4.327 | 1.916 | 6.398 | |
| p-Value | 0.81 | 0.016 | 0.0003 | 0.4662 | 0.0246 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM7 deletion | |
| Coefficient | 0.003 | 0.796 | 0.947 | 0.196 | 0.333 | |
| Hazard ratio | 1.003 | 2.217 | 2.577 | 1.216 | 1.395 | |
| 95%CI | Lower | 0.969 | 1.19 | 1.563 | 0.773 | 0.881 |
| Higher | 1.039 | 4.133 | 4.25 | 1.914 | 2.208 | |
| p-Value | 0.8706 | 0.0122 | 0.0002 | 0.397 | 0.1559 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD4 deletion | |
| Coefficient | −0.006 | 0.733 | 0.991 | 0.177 | 1.097 | |
| Hazard ratio | 0.994 | 2.081 | 2.694 | 1.194 | 2.996 | |
| 95%CI | Lower | 0.959 | 1.107 | 1.598 | 0.738 | 1.336 |
| Higher | 1.031 | 3.91 | 4.545 | 1.932 | 6.716 | |
| p-Value | 0.7515 | 0.0229 | 0.0002 | 0.4706 | 0.0077 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD6 deletion | |
| Coefficient | 0 | 0.762 | 0.911 | 0.24 | 0.491 | |
| Hazard ratio | 1 | 2.143 | 2.486 | 1.271 | 1.634 | |
| 95%CI | Lower | 0.966 | 1.146 | 1.486 | 0.805 | 1.018 |
| Higher | 1.035 | 4.006 | 4.159 | 2.008 | 2.622 | |
| p-Value | 0.9999 | 0.017 | 0.0005 | 0.3036 | 0.042 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETDB2 deletion | |
| Coefficient | −0.007 | 0.835 | 0.928 | 0.125 | 0.322 | |
| Hazard ratio | 0.993 | 2.305 | 2.531 | 1.133 | 1.38 | |
| 95%CI | Lower | 0.96 | 1.237 | 1.529 | 0.721 | 0.868 |
| Higher | 1.028 | 4.298 | 4.189 | 1.782 | 2.195 | |
| p-Value | 0.6953 | 0.0086 | 0.0003 | 0.5873 | 0.1739 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD1 deletion | |
| Coefficient | 0 | 0.755 | 0.96 | 0.309 | 0.998 | |
| Hazard ratio | 1 | 2.128 | 2.612 | 1.362 | 2.714 | |
| 95%CI | Lower | 0.964 | 1.135 | 1.558 | 0.848 | 1.329 |
| Higher | 1.038 | 3.99 | 4.378 | 2.189 | 5.543 | |
| p-Value | 0.9902 | 0.0185 | 0.0003 | 0.2012 | 0.0061 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV39H2 deletion | |
| Coefficient | −0.013 | 0.774 | 0.976 | 0.187 | 0.755 | |
| Hazard ratio | 0.987 | 2.169 | 2.655 | 1.205 | 2.129 | |
| 95%CI | Lower | 0.953 | 1.15 | 1.588 | 0.761 | 1.199 |
| Higher | 1.022 | 4.092 | 4.437 | 1.909 | 3.778 | |
| p-Value | 0.4618 | 0.0168 | 0.0002 | 0.4265 | 0.0099 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H2 deletion | |
| Coefficient | 0 | 0.77 | 0.89 | 0.289 | 0.553 | |
| Hazard ratio | 1 | 2.16 | 2.435 | 1.335 | 1.738 | |
| 95%CI | Lower | 0.964 | 1.147 | 1.443 | 0.831 | 0.808 |
| Higher | 1.037 | 4.071 | 4.108 | 2.144 | 3.736 | |
| p-Value | 0.9946 | 0.0172 | 0.0009 | 0.232 | 0.1572 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1L1 deletion | |
| Coefficient | −0.001 | 0.681 | 1.184 | 0.121 | 0.677 | |
| Hazard ratio | 0.999 | 1.976 | 3.267 | 1.129 | 1.968 | |
| 95%CI | Lower | 0.959 | 1.028 | 1.867 | 0.671 | 1.173 |
| Higher | 1.04 | 3.8 | 5.716 | 1.899 | 3.303 | |
| p-Value | 0.9534 | 0.0411 | 0 | 0.6489 | 0.0103 | |
Supplementary Table 8C.
Summary of multivariate analysis of RFS for higher promotor methylation of 6 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM14 high_meth | |
|---|---|---|---|---|---|---|
| Coefficient | −0.009 | 0.785 | 0.916 | 0.188 | 0.32 | |
| Hazard ratio | 0.991 | 2.193 | 2.498 | 1.207 | 1.377 | |
| 95%CI | Lower | 0.957 | 1.174 | 1.508 | 0.769 | 0.823 |
| Higher | 1.027 | 4.095 | 4.139 | 1.894 | 2.303 | |
| p-Value | 0.6314 | 0.0137 | 0.0004 | 0.4131 | 0.223 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM13 high_meth | |
| Coefficient | −0.006 | 0.791 | 0.971 | 0.171 | 0.317 | |
| Hazard ratio | 0.994 | 2.206 | 2.641 | 1.187 | 1.373 | |
| 95%CI | Lower | 0.96 | 1.182 | 1.608 | 0.755 | 0.86 |
| Higher | 1.029 | 4.119 | 4.338 | 1.864 | 2.19 | |
| p-Value | 0.7279 | 0.013 | 0.0001 | 0.4581 | 0.1839 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM6 high_meth | |
| Coefficient | −0.004 | 0.816 | 0.967 | 0.191 | 0.079 | |
| Hazard ratio | 0.996 | 2.262 | 2.631 | 1.211 | 1.082 | |
| 95%CI | Lower | 0.962 | 1.208 | 1.594 | 0.771 | 0.675 |
| Higher | 1.031 | 4.235 | 4.341 | 1.901 | 1.734 | |
| p-Value | 0.8298 | 0.0108 | 0.0002 | 0.4059 | 0.7441 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM8 high_meth | |
| Coefficient | −0.004 | 0.826 | 0.933 | 0.184 | 0.324 | |
| Hazard ratio | 0.996 | 2.285 | 2.542 | 1.201 | 1.383 | |
| 95%CI | Lower | 0.963 | 1.227 | 1.542 | 0.766 | 0.869 |
| Higher | 1.031 | 4.257 | 4.189 | 1.883 | 2.199 | |
| p-Value | 0.8314 | 0.0092 | 0.0003 | 0.4236 | 0.1712 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | KMT2B high_meth | |
| Coefficient | −0.003 | 0.86 | 0.897 | 0.229 | −0.417 | |
| Hazard ratio | 0.997 | 2.363 | 2.452 | 1.257 | 0.659 | |
| 95%CI | Lower | 0.963 | 1.274 | 1.486 | 0.8 | 0.409 |
| Higher | 1.033 | 4.382 | 4.045 | 1.975 | 1.061 | |
| p-Value | 0.8776 | 0.0064 | 0.0004 | 0.3215 | 0.086 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | NSD1 High_meth | |
| Coefficient | −0.006 | 0.852 | 0.889 | 0.161 | 0.547 | |
| Hazard ratio | 0.994 | 2.345 | 2.432 | 1.175 | 1.728 | |
| 95%CI | Lower | 0.961 | 1.26 | 1.476 | 0.749 | 1.06 |
| Higher | 1.028 | 4.364 | 4.009 | 1.843 | 2.818 | |
| p-Value | 0.7268 | 0.0071 | 0.0005 | 0.4839 | 0.0284 | |
Supplementary Table 8D.
Summary of multivariate analysis of RFS for higher mRNA level of 16 HMTs in prostate cancer.
| Multivariate | Age | Pathology T | Gleason score | AR expression | DOT1L high_exp | |
|---|---|---|---|---|---|---|
| Coefficient | −0.004 | 0.854 | 0.916 | 0.163 | 0.254 | |
| Hazard ratio | 0.996 | 2.35 | 2.5 | 1.177 | 1.289 | |
| 95%CI | Lower | 0.962 | 1.26 | 1.504 | 0.747 | 0.802 |
| Higher | 1.032 | 4.382 | 4.157 | 1.855 | 2.071 | |
| p-Value | 0.8373 | 0.0072 | 0.0004 | 0.4813 | 0.2941 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | EZH2 high_exp | |
| Coefficient | −0.006 | 0.858 | 0.731 | 0.185 | 0.736 | |
| Hazard ratio | 0.994 | 2.357 | 2.076 | 1.203 | 2.088 | |
| 95%CI | Lower | 0.961 | 1.263 | 1.23 | 0.768 | 1.23 |
| Higher | 1.028 | 4.4 | 3.506 | 1.887 | 3.544 | |
| p-Value | 0.7334 | 0.0071 | 0.0063 | 0.4195 | 0.0064 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | KMT2B high_exp | |
| Coefficient | −0.004 | 0.877 | 0.923 | −0.026 | 0.387 | |
| Hazard ratio | 0.996 | 2.403 | 2.516 | 0.975 | 1.472 | |
| 95%CI | Lower | 0.962 | 1.288 | 1.526 | 0.563 | 0.839 |
| Higher | 1.031 | 4.484 | 4.149 | 1.688 | 2.584 | |
| p-Value | 0.8266 | 0.0059 | 0.0003 | 0.9268 | 0.1781 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | KMT2D high_exp | |
| Coefficient | −0.004 | 0.857 | 0.906 | 0.087 | 0.333 | |
| Hazard ratio | 0.996 | 2.357 | 2.474 | 1.091 | 1.396 | |
| 95%CI | Lower | 0.962 | 1.264 | 1.492 | 0.677 | 0.851 |
| Higher | 1.031 | 4.393 | 4.102 | 1.758 | 2.289 | |
| p-Value | 0.8135 | 0.007 | 0.0004 | 0.721 | 0.1865 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM10 high_exp | |
| Coefficient | −0.003 | 0.829 | 0.94 | 0.054 | 0.266 | |
| Hazard ratio | 0.997 | 2.291 | 2.56 | 1.055 | 1.304 | |
| 95%CI | Lower | 0.963 | 1.23 | 1.551 | 0.617 | 0.755 |
| Higher | 1.033 | 4.267 | 4.227 | 1.804 | 2.255 | |
| p-Value | 0.878 | 0.009 | 0.0002 | 0.8443 | 0.3412 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM12 high_exp | |
| Coefficient | −0.005 | 0.841 | 0.923 | 0.231 | 0.224 | |
| Hazard ratio | 0.995 | 2.319 | 2.517 | 1.26 | 1.251 | |
| 95%CI | Lower | 0.962 | 1.245 | 1.512 | 0.799 | 0.773 |
| Higher | 1.03 | 4.322 | 4.188 | 1.988 | 2.024 | |
| p-Value | 0.7926 | 0.0081 | 0.0004 | 0.3195 | 0.362 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM15 high_exp | |
| Coefficient | −0.003 | 0.856 | 0.909 | 0.153 | 0.448 | |
| Hazard ratio | 0.997 | 2.354 | 2.483 | 1.165 | 1.566 | |
| 95%CI | Lower | 0.964 | 1.264 | 1.507 | 0.742 | 0.981 |
| Higher | 1.033 | 4.383 | 4.092 | 1.83 | 2.5 | |
| p-Value | 0.8865 | 0.007 | 0.0004 | 0.5069 | 0.0604 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM16 high_exp | |
| Coefficient | −0.006 | 0.898 | 0.948 | 0.175 | −0.514 | |
| Hazard ratio | 0.994 | 2.454 | 2.581 | 1.191 | 0.598 | |
| 95%CI | Lower | 0.96 | 1.315 | 1.571 | 0.759 | 0.377 |
| Higher | 1.029 | 4.579 | 4.24 | 1.87 | 0.949 | |
| p-Value | 0.747 | 0.0048 | 0.0002 | 0.4462 | 0.0293 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM6 high_exp | |
| Coefficient | −0.003 | 0.851 | 0.897 | 0.208 | −0.506 | |
| Hazard ratio | 0.997 | 2.343 | 2.452 | 1.231 | 0.603 | |
| 95%CI | Lower | 0.963 | 1.259 | 1.486 | 0.785 | 0.377 |
| Higher | 1.032 | 4.36 | 4.046 | 1.931 | 0.964 | |
| p-Value | 0.8603 | 0.0072 | 0.0004 | 0.366 | 0.0345 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | PRDM8 high_exp | |
| Coefficient | −0.005 | 0.857 | 0.916 | 0.208 | −0.284 | |
| Hazard ratio | 0.995 | 2.356 | 2.499 | 1.231 | 0.753 | |
| 95%CI | Lower | 0.962 | 1.261 | 1.504 | 0.785 | 0.47 |
| Higher | 1.03 | 4.401 | 4.155 | 1.93 | 1.206 | |
| p-Value | 0.7952 | 0.0072 | 0.0004 | 0.3658 | 0.2375 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD1B high_exp | |
| Coefficient | −0.008 | 0.861 | 0.903 | 0.059 | 0.453 | |
| Hazard ratio | 0.992 | 2.365 | 2.468 | 1.06 | 1.573 | |
| 95%CI | Lower | 0.958 | 1.271 | 1.496 | 0.661 | 0.957 |
| Higher | 1.028 | 4.401 | 4.071 | 1.701 | 2.584 | |
| p-Value | 0.6715 | 0.0066 | 0.0004 | 0.8076 | 0.0738 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SETD5 high_exp | |
| Coefficient | −0.005 | 0.784 | 0.903 | 0.033 | 0.584 | |
| Hazard ratio | 0.995 | 2.191 | 2.466 | 1.033 | 1.794 | |
| 95%CI | Lower | 0.961 | 1.176 | 1.496 | 0.646 | 1.1 |
| Higher | 1.031 | 4.082 | 4.067 | 1.651 | 2.926 | |
| p-Value | 0.7935 | 0.0135 | 0.0004 | 0.8915 | 0.0193 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SMYD1 high_exp | |
| Coefficient | −0.009 | 0.862 | 1.002 | 0.191 | −0.819 | |
| Hazard ratio | 0.991 | 2.369 | 2.723 | 1.21 | 0.441 | |
| 95%CI | Lower | 0.957 | 1.269 | 1.662 | 0.772 | 0.232 |
| Higher | 1.027 | 4.423 | 4.461 | 1.896 | 0.838 | |
| p-Value | 0.623 | 0.0068 | 0.0001 | 0.405 | 0.0124 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV39H1 high_exp | |
| Coefficient | −0.007 | 0.86 | 0.925 | 0.22 | 0.329 | |
| Hazard ratio | 0.993 | 2.363 | 2.521 | 1.246 | 1.39 | |
| 95%CI | Lower | 0.958 | 1.267 | 1.525 | 0.792 | 0.866 |
| Higher | 1.028 | 4.407 | 4.167 | 1.958 | 2.23 | |
| p-Value | 0.6758 | 0.0068 | 0.0003 | 0.3412 | 0.1728 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | SUV420H2 high_exp | |
| Coefficient | −0.009 | 0.852 | 0.847 | 0.382 | 0.586 | |
| Hazard ratio | 0.991 | 2.344 | 2.334 | 1.465 | 1.796 | |
| 95%CI | Lower | 0.957 | 1.257 | 1.402 | 0.909 | 1.087 |
| Higher | 1.027 | 4.37 | 3.884 | 2.361 | 2.966 | |
| p-Value | 0.6247 | 0.0073 | 0.0011 | 0.1165 | 0.0222 | |
| Multivariate | Age | Pathology T | Gleason score | AR expression | WHSC1 high_exp | |
| Coefficient | −0.005 | 0.815 | 0.831 | 0.146 | 0.429 | |
| Hazard ratio | 0.995 | 2.259 | 2.297 | 1.157 | 1.535 | |
| 95%CI | Lower | 0.961 | 1.21 | 1.357 | 0.735 | 0.913 |
| Higher | 1.03 | 4.217 | 3.888 | 1.821 | 2.581 | |
| p-Value | 0.7789 | 0.0105 | 0.002 | 0.5276 | 0.1057 | |