Abstract
The retinaldehyde dehydrogenase (RALDH) enzymes, RALDH1, RALDH2, and RALDH3, catalyze the irreversible oxidation of retinaldehyde to all-trans-retinoic acid (ATRA). Despite the importance of the RALDH enzymes in embryonic development, postnatal growth and differentiation, and in several disease states, there are no commercially available inhibitors that specifically target these isozymes. We report here the development and characterization of a small molecule inhibitor dichloro-all-trans-retinone (DAR)1 that is an irreversible inhibitor of RALDH1, 2, and 3 that effectively inhibits RALDH1, 2, and 3 in the nanomolar range but has no inhibitory activity against mitochondrial ALDH2. These results provide support for the development of DAR as a specific ATRA synthesis inhibitor for a variety of experimental and clinical applications.
1. INTRODUCTION
The aldehyde dehydrogenase (ALDHs) superfamily catalyzes the NAD(P)+ dependent, irreversible oxidation of aldehydes into their respective carboxylic acids. Currently, there are 19 known ALDH protein coding genes and 3 pseudogenes present in the human genome.2-4 ALDH proteins are involved in a wide variety of physiological processes including aldehyde detoxification, alcohol metabolism, development, vision, and neurotransmission.5-8 Mutations and polymorphisms in ALDH genes have also been associated with a number diseases such as brain, breast, and lung cancer, late-onset Alzheimer’s disease, alcohol flushing syndrome, Sjögren-Larsson syndrome (SLS), and type II hyperprolinemia.4,9-19
The retinaldehyde dehydrogenase (RALDH, also ALDH1A) subfamily consists of three members, RALDH1 (ALDH1A1), RALDH2 (ALDH1A2), and RALDH3 (ALDH1A3), which synthesize all-trans-retinoic acid (ATRA) from retinaldehyde. All RALDH isozymes can utilize both all-trans-retinaldehyde and 9-cis-retinaldehyde as substrates; however, RALDH2 exhibits the highest specificity for all-trans-retinaldehyde.20, 21 RALDH proteins are essential for regulating ATRA signaling pre- and postnatally. In embryonic development, RALDH1, 2, and 3 are differentially expressed in a spatial-temporal manner to mediate organogenesis of the eye, brain, heart, kidney, lung, and reproductive organs.2, 22-24 Postnatally, RALDH isozymes have much more limited roles, but are involved in spermatogenesis,25-27 maintenance of lens transparency,28, 29 and stem cell differentiation.30-31 The RALDH subfamily has also been implicated in a variety of diseases through the modulation of all-trans-retinaldehyde and ATRA levels. RALDH1 has been linked to obesity, inflammation, and Crohn’s disease,32-36 while RALDH2 has been associated with abnormal ocular growth (myopia).37, 38 Changes in the expression of all three RALDH proteins in tumors have been associated with poor cancer Prognoses.39-44
Despite the involvement of various ALDH family members in numerous disease states, pharmacological inhibitors have been developed for only 3 of the ALDH isozymes: ALDH1A1, ALDH2, and ALDH3A1.3 Of these three, selective inhibitors have been developed for ALDH1A1 (CM026, CM037) and ALDH3A1 (CB29) for the treatment of oxazaphosphorine resistant cancers.45-47 Other inhibitors have been found to be effective against the above ALDHs; however, these compounds are non-specific and inhibit a range of ALDH family members.3 Non-specific ALDH inhibitors that have been effective at inhibiting the RALDH subfamily include 4-amino-4methyl-2pentyne-1-al (AMPAL), citral, 4-(diethylamino)benzaldehyde (DEAB), disulfiram (commercially known as Antabuse), and gossypol.3 DEAB is an irreversible inhibitor of ALDH1A2, ALDH2 and ALDH7A1 and has been utilized in studies of ALDH-mediated drug metabolism and in the treatment of cancer.48-51 Citral and gossypol are reversible noncompetitive inhibitors of ALDH1, ALDH2, and ALDH3 isozymes and have been used in experimental studies on embryonic development and cancer, respectively.52-56 AMPAL and disulfiram are irreversible inhibitors of ALDH1, ALDH2, and ALDH3 isoenzymes. AMPAL has been used to inhibit cell growth in tumors,57, 58 while disulfiram has been used commercially for the treatment of alcoholism, as an alcohol aversive agent.59, 60 Through inhibition of ALDH2, disulfiram administration before drinking alcoholic beverages results in unpleasant symptoms such as blurred vision, nausea, and flushing of the face and neck that are collectively known as the disulfiram ethanol reaction.61-63 Due to the lack of specificity of the aforementioned inhibitors, studies utilizing these non-specific inhibitors to gain insights into the physiologic roles of ALDH isoenzyme activity may be complicated by inhibition of multiple isoenzymes and lead to off-target effects.64-66
Recently, the bisdiamine, WIN 18446 (commercially known as Fertilysin), has been identified as a potent, irreversible RALDH2 inhibitor.67, 68 WIN 18446 has been used as a male contraceptive in a number of mammals67, 69-71 and was tested for its potential as a male contraceptive in human volunteers.63 In all of these studies, WIN 18446 was found to effectively and reversibly inhibit spermatogenesis, presumably through inhibition of RALDH2– mediated testicular ATRA synthesis, recently shown to be required for normal spermatogenesis.24, 63, 72 Although WIN 18446 was a promising male contraceptive, it was not marketed to the general public, as the disulfiram ethanol reaction was observed upon patient consumption of alcohol, indicating that WIN 18446 also inhibited ALDH2 and possibly other ALDH enzymes. Recently, the non-specificity of WIN 18446 has been confirmed in vitro.68 These results demonstrate the importance of developing small molecule inhibitors selective for the RALDH isozymes as such inhibitors would have application for the study and treatment of a variety of diseases and conditions. Using a structurally based drug design approach, we report here the development of a novel RALDH (ALDH1A) selective inhibitor, (3E,5E,7E,9E)-1,1-dichloro-4,8-dimethyl-10-(2,6,6-trimethylcyclohex-1-en-1-yl)deca-3,5,7,9-tetraen-2-one (“dichloro-all-trans-retinone” or “DAR”),1 that is an effective inhibitor of RALDH1 (IC50 = 434.70 ± 99.70 nM), RALDH2 (IC50 = 55.00 ± 10.71 nM), and RALDH3 (IC50 = 161.27 ± 16.57 nM), but has no activity on human mitochondrial ALDH2.
2. RESULTS AND DISCUSSION
2.1. STRUCTURE-BASED DESIGN OF DAR
WIN 18446 (Figure 1A) has been shown to inhibit RALDH1, 2, and 3, as well as ALDH2 and other members of the ALDH superfamily.67, 68, 74 Based on an x-ray crystal structure of WIN 18446 bound to human RALDH2, the ALDH active cysteine sulfhydryl group forms a covalent bond with the terminal carbon of WIN 18446, causing displacement of a chloride atom from WIN 18446.75 We postulated that the relatively large size of the substrate entrance tunnel for the RALDH enzymes, as compared with the other ALDH enzymes, would allow for the specificity of retinaldehyde (RAL) (Figure 1B) to the RALDH enzymes.20, 76 Using this knowledge, we suspected that a RALDH selective inhibitor could be generated by attaching a dichloro-methane moiety to the retinyl group (beta-ionone ring with an isoprenoid chain) of RAL, thereby forming dichloro-all-trans-retinone (DAR) (Figure 1C). Therefore, DAR (367.34 Da) was synthesized by reacting dichloromethyllithium with retinoic acid ethyl ester under strong basic conditions. DAR has no structural similarity to commercially available aldehyde dehydrogenase modulators or any small molecule in the CHemDiv, ZINC, or ChemSpider databases.
Figure 1. Design of Retinal Based Inhibitor.
The dichloro-methane moiety of WIN 18446 (A) was attached to the retinyl group of retinaldehyde (B) generating dichloro-all-trans-retinone (C). Numbering in (C) indicates the sequence of the carbon atoms.
α,α-Dichioro-all-trans-retinone (“DAR”) was produced in a one-step transformation of retinoic acid ethyl ester to DAR (Scheme 1). Dichloromethyllithium, generated in situ at low temperature,73 reacted with commercially available retinoic acid ethyl ester to produce the α,α-dichloro-retinoic acid in low yield (4 – 10%). A small amount of the starting material retinoic acid ethyl ester was recovered, with the remaining mass balance of this reaction being comprised of a complex mixture of unidentified isomeric byproducts. Extensive purification, including semi-preparative HPLC, produced analytically pure DAR, free from any contaminating byproducts. The low yield of this reaction is not surprising given the combination of the inherent reactivity of the extended conjugated system in DAR and the highly reactive dichloromethyllithium species.
Scheme 1. One-Step Synthesis of α, α-Dichloro-Retinal Analog.
Retinoic acid ethyl ester was reacted with dichloromethyllithium generated in situ from dichloromethane (DCM) and lithium diisopropylamide (LDA) in ethyl ether (Et2O) to generate α,α-dichloro-all-trans-retinone (DAR). 4 – 10% of DAR was produced in each reaction.
2.2. KINETIC CHARACTERIZATION OF DAR
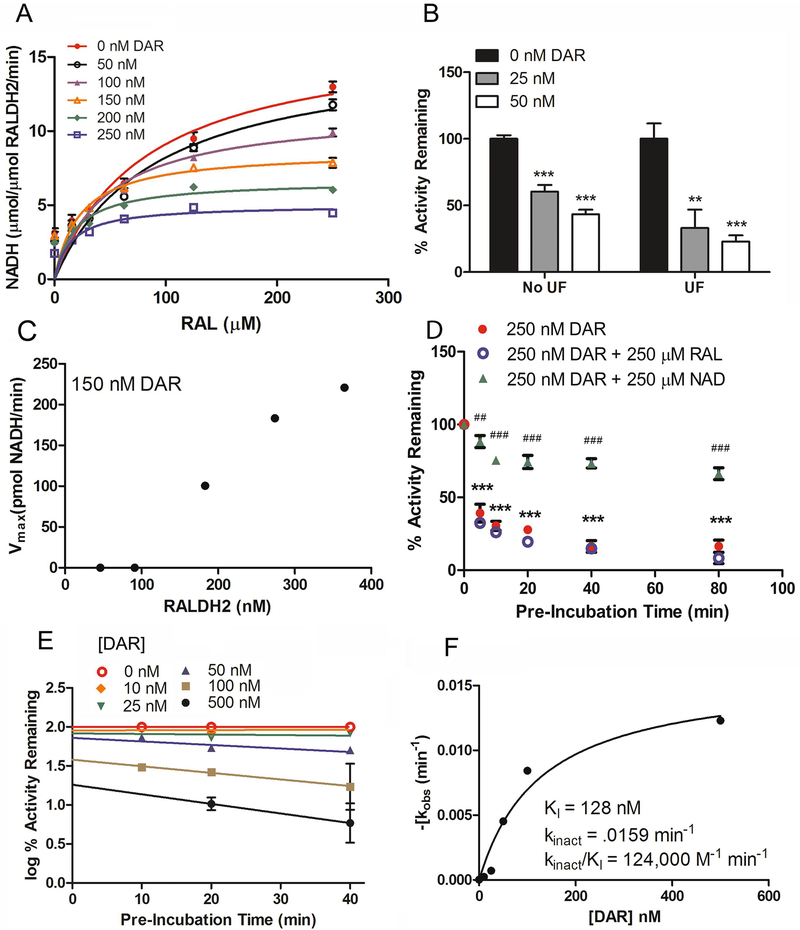
RALDH activity in the presence and absence of DAR was measured by quantifying the synthesis of NADH, as both NADH and ATRA are produced in equimolar amounts during the oxidation of all-trans-retinaldehyde.3 Due to overlap in the absorption spectrum of RAL and NADH (300 – 400 nm), a fluorescence-based NADH assay (AAT Bioquest, Inc.) was employed to measure RALDH enzyme activity (ATRA synthesis) utilizing RAL as the substrate (Figure 2). Additionally, ATRA was quantified by HPLC/spectrophotometric assay, and measurements of ATRA synthesis were compared between the fluorescence based and HPLC/spectrophotometric assays (Figure 2A). The fluorescence based assay exhibited high correlation with HPLC measurements (p < 0.001, Pearson’s correlation analysis) when recombinant chicken RALDH2 (2 μg) was incubated with RAL (0 – 100 μM) for 30 min. Therefore the effects of protein concentration, time, and substrate concentration on NADH synthesis, by chicken RALDH2 were examined (Figure 2B – D) as a proxy for ATRA synthesis. NADH synthesis was linear between 0 – 2 μg RALDH2 (Figure 2B) and 0 – 50 min of incubation time (Figure 2C); thus, all subsequent assays were conducted with 0.5 μg RALDH2 for 30 min, unless otherwise stated. The rate of NADH synthesis was saturated with increasing concentrations of RAL (0 – 250 μM) (Figure 2D). Under these experimental conditions, RALDH2 exhibited a Km of 80.70 ± 20.38 μM for retinaldehyde (determined from four independent experiments, each performed in triplicate). Previously reported Michaelis constants for RALDH2 (Km or K0.5 using retinaldehyde as the substrate) have ranged from 0.3 μM – 16 μM. 21, 67, 77, 78 Differences between these and our studies are most likely due to differences in the method of assessing RALDH2 activity (HPLC quantification of ATRA, NADH absorbance at 340 nM, and NADH quantification using the fluorescence-based recycling system in the present study).
Figure 2. Effects of protein, time, and substrate concentration on ATRA and NADH synthesis by recombinant chicken RALDH2.
(A) Measurements of NADH and ATRA synthesis exhibited high correlation between the fluorescence based assay of NADH synthesis and HPLC measurements of ATRA synthesis,***p < 0.001 (Pearson’s correlation analysis). (B–D) Production of NADH was determined with increasing RALDH2 concentration (0 – 3 μg) (B), increasing incubation time (0 – 60 min) (C), and increasing RAL concentration (0 – 250 μM). (D). All subsequent assays were conducted with 0.5 μg RALDH2 for 30 min, unless otherwise stated. Each data point represents the average ± SEM of triplicate samples. Errors bars are contained within data points in some instances.
To examine the effects of DAR on aldehyde dehydrogenase activity, DAR was tested on recombinant chicken RALDH1, RALDH2, RALDH3, human RALDH2 (hRALDH2) and recombinant human mitochondrial ALDH2 (hALDH2) (Figure 3, Table 1). RALDH1, 2, 3, and hRALDH2 were pre-incubated with DAR (0 – 10 μM) for 20 min at 37°C, after which RAL (250 μM) was added to the reaction mixture to initiate ATRA and NADH production. DAR was found to inhibit chicken RALDH1, 2, and 3 with IC50’s of 434.70 ± 99.70 nM, 55.00 ± 10.71 nM and 161.27 ± 16.57,nM, respectively, as well as human RALDH2 (hRALDH2; IC50 = 191.32 ± 36.77) (Figure 3A – D, Table 1). Comparison of the IC50 values for DAR on RALDH1, RALDH2, RALDH3 and hRALDH2 indicated significantly different inhibition for every pair of enzymes (p < 0.05) with the exception of RALDH3 as compared with hRALDH2 (one way ANOVA with Bonferroni correction for multiple comparisons). The effect of DAR on RALDH1 and RALDH3 activity demonstrated dose responses that were essentially step functions, and thus data were fit using the 4-parameter logistics equation and the Hill-slope. Using a spectrophotometric, absorbance assay measuring NADH production at 340 nm and propionaldehyde as the substrate (5 mM), DAR(0 – 100 μM, 1 hr pre-incubation) did not have an inhibitory effect on mitochondrial human aldehyde dehydrogenase 2 (hALDH2) (Figure 3E, Table 1). As expected, hALDH2 was effectively inhibited by WIN 18446 (0 – 10 μM) with an observed IC50 of 229.33 ± 40.95 nM, similar to that observed in other studies.67 These results indicate that DAR has selectivity for the RALDH enzymes with the most robust inhibition (lowest IC50) observed for RALDH2 in vitro in the presence of the natural substrate, RAL (inhibition of RALDH2 > RALDH3 > RALDH1).
Figure 3. Representative Data for Determining Effect of DAR on Chick and Human RALDH1a Isoforms and Human Mitochondrial ALDH2 (hALDH2).
(A - D) IC50 of DAR (0 – 10 μM) with chick RALDH1 (2 μg = 0.358 μM) (A), RALDH2 (0.5 μg = 0.091 μM) (B), RALDH3 (2 μg = 0.355 μM) and (C), human RALDH2 (hRALDH2; 1 μg = 0.176 μM). The RALDH isozymes were pre-incubated with DAR for 20 min, after which ATRA and NADH synthesis were initiated by addition of substrate and cofactor, as described in the experimental section. (E) No inhibition was observed when DAR (0 – 100 μM) was incubated with hALDH2 (25 μg). The non-specific inhibitor WIN 18446 served as the positive control. hALDH2 was pre-incubated with DAR or WIN 18446 for 1 hr prior to addition of substrate (propionaldehyde) and cofactor. Enzyme activity was using a spectrophotometric assay measuring the production of NADH at 340 nm. Data points (A- E) represent the average ± SEM for triplicate samples. The results of 3 – 5 independent determinations for IC50 and KI values for DAR are summarized in Table 1.
Table 1.
IC50and KI Values Determined for DAR
| ALDH Isoenzyme | IC50 * | KI (Morrison) |
|---|---|---|
| nM | nM | |
| RALDH1 (chick) | 434.70 ± 99.70 | 194.40 ± 99.32 |
| RALDH2 (chick) | 55.00 ± 10.71 | 5.10 ± 2.50 |
| RALDH3 (chick) | 161.27 ± 16.57 | 6.74 ± 1.73 |
| RALDH2 (human) | 191.32 ± 36.77 | 64.35 ± 10.76 |
| ALDH2 (human) | NI | NI |
KI’s were determined using the Morrison equation: Q=(Ki*(1+(S/Km))) and Y=Vo*(1-((((Et+X+Q)-(((Et+X+Q)^2)-4*Et*X)^0.5))/(2*Et))) (GraphPad Prism v5), where Et is the concentration of enzyme (RALDH1 = 0.358 μM; RALDH2 = 0.091 μM; RALDH3 = 0.355 μM; hRALDH2 = 0.176 μM; ALDH2 = 4.43 μM), S is the concentration of substrate (described in Experimental Section), and Km is the Michaelis-Menten constant (see supplemental Table S1).
Values are the mean ± SEM of triplicate determinations from 3 – 5 independent experiments. NI = no inhibition.
It is known that WIN 18446 inhibits human RALDH2 in a time-dependent and irreversible manner.68, 74, 79 To determine if DAR also acts as an irreversible inhibitor, the mechanism of inhibition for recombinant chicken RALDH2 was investigated using various methods (Figure 4). Increasing concentrations of DAR in the enzyme reaction resulted in a dose dependent decrease of Vmax (1665, 1563, 1148, 866.9, 666.5, 503.0 pmol/μM RALDH2/min) (Figure 4A, Table S1). A hallmark of irreversible inhibition is the persistence of inhibition following removal of unbound inhibitor.80, 81 When unbound inhibitor was removed by ultrafiltration (Figure 4B; “UF”) prior to addition of cofactor and substrate, RALDH2 activity was not restored. Significant inhibition was observed at 25 nM (33.02 ± 13.89% activity remaining) and 50 nM (22.97 ± 4.55% activity remaining) of DAR compared to vehicle control (100.00 ± 11.57% activity remaining) (p < 0.001, one-way ANOVA with Bonferroni’s test for multiple comparisons), similar to the results seen when unbound DAR remained present in the enzyme reaction (Figure 4B; “No UF”). In samples subjected to ultrafiltration, the average inhibition of RALDH2 by DAR was greater than predicted by stoichiometric irreversible inhibition. As can be observed in Figure 4B, the variability of ultracentrifuged (UF) samples was 2- 4x higher than that of samples not ultracentrifuged (No UF) prior to enzyme assays. This increased inhibition and variability is most likely due to variable loss of enzyme during recovery from the centrifugal filter and/or variable loss of activity as a consequence of ultracentrifugation. In addition, the Vmax for NADH synthesis was determined using increasing concentrations of RALDH2 (46 – 365 nM) in the presence of DAR(150 nM) (Figure 4C). A plot of Vmax vs. RALDH2 concentration indicated that RALDH2 activity was completely inhibited when enzyme concentrations (46 – 91 nM) were less than the concentration of DAR ([RALDH2] ≤ 150 nM). In the presence of 150 nM DAR, RALDH2 concentrations (183 – 365 nM) resulted in linear increases in Vmax for NADH synthesis (0.50 – 3.01 μM NADH/min). To further characterize the inhibitory effects of DAR, RALDH2 (0.5 μg = 0.091 μM) was pre-incubated with DAR (250 nM) for 0 – 80 min at 37 °C in the presence or absence of all-trans-retinaldehyde (250 μM) before the enzyme reaction was initiated by addition of all-trans-retinaldehyde (250 μM) and NAD+ (Figure 4D). RALDH2 activity was significantly decreased at 5 min (39.18 ± 6.10% activity remaining), 10 min (30.43 ± 3.09% activity remaining), and 20 min (27.80 ± 2.87% activity remaining) of pre-incubation, with maximal inhibition observed between 40 (16.31 ± 3.98% activity remaining) and 80 min (16.50 ± 4.21% activity remaining) (***p < 0.001, oneway ANOVA with Bonferroni’s multiple comparison test). Pre-incubation of RALDH2 with vehicle alone (5 – 80 min at 37°C) had no significant effect on RALDH2 activity. No protection of RALDH2 activity was observed when all-trans-retinaldehyde was included in the pre-incubation period (250 nM DAR + 250 μM RAL). However, pre-incubation of RALDH2 with DAR and NAD+ (250 μM) resulted in a recovery of 52 – 65% of RALDH2 activity as compared with pre-incubation with DAR and RAL or DAR alone (##p<0.01, ###p<0.001, Student’s t-test). Inhibitors that exhibit time dependence demonstrate a shift in IC50 values depending on the time of pre-incubation which allows for the determination of the rate of inactivation (kinact) and inhibition constant (KI).82-85 Therefore, IC50 shift assays were performed and the kobs determined from the slopes of lines calculated from the log of RALDH2 activity (log % Activity Remaining) following 10 – 40 min of pre-incubation with DAR at concentrations of 0 – 500 nM. From these slopes, the rate of inactivation (kobs) was determined to be .000028 ± 0.00086 – 0.012 ± 0.009 log % activity remaining/min for 0 – 500 nM DAR (Figure 4E). From the kobs values calculated in Figure 4E, the kinact and KI for RALDH2 by DAR were calculated to be 0.0159 min−1 and 128 nM, respectively with a time dependence ratio (kinact/KI) of 124,000 M−1 min−1 (Figure 4F). The time dependence ratio calculated for DAR inactivation of RALDH2 is within the range reported for the covalent inhibition of RALDH2 by DEAB (≈93,000 M−1 min−1 – 140,00,000 M−1 min−1).48 RALDH1 and RALDH3 also exhibited a decrease in Km and Vmax with increasing concentrations of DAR and time dependent inhibition, suggesting that DAR is an irreversible inhibitor for chick RALDH1 and RALDH3 as well (Figure S1, Table S1). Interestingly, pre-incubation of RALDH3 with retinaldehyde and DAR resulted in a significant increase in RALDH3 inactivation as compared with pre-incubation with DAR alone. These results suggest that the irreversible binding of DAR to RALDH3 is somehow promoted by the presence of retinaldehyde. At present, it is unclear as to the molecular basis for this interaction.
Figure 4. DAR is an Irreversible Inhibitor of RALDH2.
(A) All-trans-retinaldehyde concentration-dependent RALDH2 (91 nM) activity in the presence of increasing concentrations of DAR (0 – 250 nM; pre-incubated with RALDH2 for 20 minutes prior to addition of substrate and cofactor). (B) RALDH2 activity measured before and after ultrafiltration of DAR-inactivated RALDH2 (0 – 50 DAR). (C) Maximal rate (Vmax) of enzyme activity was determined at increasing concentrations of RALDH2 (46 – 365 nM) with DAR concentration remaining constant (150 nM). For (A – C) DAR was pre-incubated with RALDH2 for 20 min, after which NADH synthesis was initiated by addition of substrate and cofactor. (D) Time-dependent inhibition of RALDH2 (91 nM) by DAR (250 nM) was determined by pre-incubating the enzyme with DAR for predetermined times (0 – 80 min) in the presence and absence of all-trans-retinaldehyde (250 μM) or NAD+ (250 μM) before initiating the enzyme reaction by addition of substrate and/or cofactor. (E) The rate of inactivation (kobs) of RALDH2 activity by increasing concentrations of DAR (0 – 500 nM) at 10, 20, and 40 min of pre-incubation was determined by linear regression analysis of the logarithm of the percentage of activity remaining versus pre-incubation time. (F) The KI and kinact were calculated by nonlinear regression analysis of the kobs (E) versus concentration of DAR. *p < 0.05, **p < 0.01, ***p < 0.001 (one-way ANOVA with Bonferroni’s test for multiple comparisons); ##p<0.01 ###p<0.001 (Student’s t-test). Enzyme reactions were carried out with 250 μM all-trans-retinaldehyde, 4 mM NAD and 91 nM RALDH2 at 37°C for 30 min. Triplicate samples are shown (average ± SEM) for each data point.
2.3. EX VIVO EFFECTS OF COMPOUND 2
(Dox)RALDH2-eGFP, a HEK-293 cell line expressing doxycycline (DOX)-inducible chicken RALDH2 was employed to assess the effect of DAR on intracellular RALDH2 activity (Figure 5). Treatment of the (Dox)RALDH2-eGFP cells with DOX (5 μg/mL; 24 hrs) resulted in the induction of RALDH2-eGFP expression as compared to non-induced (Dox)RALDH2-eGFP cells (Figure 5A). In order to determine the toxicity of DAR on the (Dox)RALDH2-eGFP cells, cell viability assays measuring DNA content and ATP production were performed (Figure 5B). Following treatment with DAR (0 – 50 μM) for 24 hrs, toxicity curves were determined with IC50 values of 2.94 ± 1.58 μM and 3.09 ± 0.48 μM, for DNA and ATP production, respectively. Treatment of DOX-induced RALDH2-eGFP expressing cells with DAR (0 – 2 μM for 24 hrs) resulted in a significant inhibition of ATRA synthesis at 0.05 μM (↓42%, p<0.01), 0.50 μM (↓47%, p<0.001), 1 μM (↓92%, p<0.001), and 2 μM DAR (↓93%, p<0.001) compared to the vehicle treated control (one-way ANOVA with Bonferroni’s test for multiple comparisons) (Figure 5C). No ATRA synthesis was detected in cells not induced with DOX (-DOX). Normalization of results in Figure 5C to RALDH2 protein expression in each sample (relative density quantified from western blot, inset) indicated a dose-dependent decrease in ATRA synthesis with an IC50 value for the normalized inhibition curve calculated to be 187.20 ± 40.80 nM (Figure 5D).
Figure 5. DAR Inhibits ATRA Synthesis in a RALDH2 Expressing Cell Line.
(A) Induction of chicken RALDH2-eGFP expression in a stably transfected, doxycycline (DOX)-inducible HEK 293 cell line ((Dox)RALDH2-eGFP). RALDH2-eGFP expression is not detected in cell lysates not treated with DOX (-DOX). (Scale bar =100 pixels) (B) Toxicity curves measuring DNA content and ATP production in cells treated with DAR (0.01 – 50 μM) for 24 hrs. Data represent results of two experiments (n = 5 for each concentration/exp). (C) Inhibition of ATRA synthesis in (Dox)RALDH2-eGFP cells treated with DAR (0.05 – 2 μM) or vehicle (DMSO) for 24 hrs. Following isolation of cell lysates, the enzyme reaction was initiated by addition of NAD+ and all-trans-retinaldehyde (25 μM). ATRA was quantified by HPLC. No ATRA synthesis was detected in cells not induced with DOX (-DOX). (D) Results in (C) normalized to RALDH2 protein expression. Relative RALDH2 expression was quantified from western blots of (Dox)RALDH2-eGFP cell lysates (inset). **p < 0.01, ***p < 0.001 (one-way ANOVA with Bonferroni’s test for multiple comparisons). Data represent results of three independent experiments (n = 3 cultures/concentration/experiment).
We and others 37,38,86 have previously shown that RALDH2 protein expression and RALDH enzymatic activity are detectible in the choroids of chick eyes, with negligible amounts in the retina/RPE and sclera. Moreover, ATRA synthesis is increased in the chick choroid during recovery from form-deprivation induced myopia, which results from an increase in RALDH2 protein expression. Western blot analyses of choroidal lysates indicates that RALDH2 protein (~↑125% over contralateral control eyes). Increased RALDH2 protein expression in recovering choroids is responsible for an~ 136% increase in RALDH enzymatic activity in choroidal lysates in recovering eyes. We therefore employed this animal model to examine the effect of DAR on RALDH2 activity in choroids of recovering eyes and contralateral control eyes (Figure 6). Choroidal cytosol fractions from control and 4 day recovering chick eyes were isolated and pre-incubated with DAR (0 – 6 μM) for 20 min at 37°C prior to initiating the enzyme reaction by addition of NAD+ (4 mM) and all-trans-retinaldehyde (25 μM), and RALDH2 activity was measured by HPLC quantification of ATRA (Figure 6A). RALDH2 activity was significantly inhibited in cytosol fractions from control and recovering choroids with 0.10 – 6 μM DAR (92 – 99% inhibition in control lysates and 70 – 96% inhibition in recovering lysates compared to vehicle treated choroids, respectively (p < 0.001, one-way ANOVA with Bonferroni’s test for multiple comparisons). Non-linear regression analyses of ATRA synthesis by choroid cytosol fractions from recovering eyes following treatment with DAR indicated that DAR inhibited RALDH2 activity in recovering choroid cytosol fractions with an IC50 of 53.60 nM (Figure 6B). RALDH2 activity was also assessed, ex vivo, in choroidal lysates following overnight incubation of living, intact choroids with DAR (0 – 5 μM) (Figure 6C, D). Choroids were isolated from 4 day recovering eyes or contralateral control eyes and incubated with DAR (0 – 5 μM) in organ culture for 24 hrs, after which time choroid cytosol fractions were prepared and RALDH activity measured. In contrast to results in which DAR was directly applied to choroid cytosol fractions (in Figure 6A), DAR had no significant effect on ATRA synthesis in living, intact choroids from control eyes. ATRA synthesis was maximally inhibited in recovering choroids by 1 μM (42% inhibition), compared to vehicle treated choroids (p < 0.05, one-way ANOVA with Bonferroni’s test for multiple comparisons). The EC50 (relative IC50) for this 42% inhibition of ATRA by DAR was determined to be 119.70 nM when applied to living intact tissue, ex vivo (Figure 6D). For comparison, living, intact choroids from control and 4 day recovering eyes were incubated with WIN 18,446, a potent and irreversible ALDH1a inhibitor 67,68 (Figure 6E). Similar to results with DAR, WIN had no significant effect on ATRA synthesis in living, intact choroids from control eyes. However, ATRA synthesis was significantly inhibited in recovering choroids by 0.1 μM WIN (40% inhibition), 1 μM WIN (51% inhibition) and 10 μM WIN (76% inhibition) compared to vehicle treated choroids (p < 0.05, 0.01 and 0.001, respectively; one-way ANOVA with Bonferroni’s test for multiple comparisons). Non-linear regression analyses of ATRA synthesis by recovering choroids following treatment with WIN (Figure 6F) indicated that WIN inhibits RALDH2 activity in recovering choroid lysates with an EC50 of 44.47 nM, when applied to living intact tissue, ex vivo.
Figure 6. Effect of DAR and WIN on RALDH2 Activity in Control and Recovering Choroids.
(A) Inhibition of ATRA synthesis in control and recovering choroidal lysates following 20 min pre-incubation with DAR (0.01 – 6 μM) or vehicle (DMSO). After pre-incubation, the enzyme reaction was initiated by addition of NAD+ and RAL (25 μM) and allowed to proceed for 30 min at 37°C. Data are representative of results from two experiments (n = 3 – 5/experiment). (C) Inhibition of RALDH2 activity in living choroid tissue, “ex vivo” by DAR. Control and recovering choroids were placed in organ culture and treated with DAR (0.01 – 10 μM) or vehicle (DMSO) for 24 hrs at 37°C and 5% CO2. Following isolation of cytosol fractions, the enzyme reaction was initiated by addition of NAD+ and RAL (25 μM) (n = 9 −14 choroids/concentration). Data represent results of three independent experiments (n = 3 – 5 choroids/experiment) (E) Inhibition of RALDH2 activity in living choroid tissue, “ex vivo” by WIN 18,446 (WIN). Control and recovering choroids were placed in organ culture and treated with WIN (0.1 – 10 μM) or vehicle (DMSO) for 24 hrs at 37°C and 5% CO2. Following isolation of cytosol fractions, the enzyme reaction was initiated by addition of NAD+ and RAL (25 μM) (n = 6 choroids/concentration). ATRA was quantified using HPLC. Data represent results of two independent experiments (n = 3/choroids/experiment). (B, D, F) IC50 and EC50 values for DARand WIN were calculated by non-linear regression analyses of data from recovering choroids from figures A, C, and E, respectively. *p < 0.05, **p<0.01, ***p < 0.001 (one-way ANOVA with Bonferroni’s test for multiple comparisons).
2.4. MOLECULAR MODELING.
We performed a series of molecular docking simulations using a homology model of chicken RALDH2 constructed from the PDB crystal structure of sheep RALDH1 (PDB: 5ABM)45 in order to investigate possible binding interactions of DAR. Based on the proposed mechanism of inhibition by WIN 18446 of human RALDH2 75 and the known mechanism of aldehyde oxidation by the ALDH enzymes,3 we predicted that an interaction would occur between either the thiolate group of the catalytic cysteine of RALDH2 and either the carbonyl carbon or carbon 1 of DAR (see Figure 1) resulting in an irreversible inhibition of the enzyme. Binding poses were visualized with PyMOL to determine the most likely binding interaction by examining the Van der Waals radii and any overlap that occurred between DAR and residues of the RALDH2 active site (Figure 7). Three dimensional modeling of DAR in the binding pocket of RALDH2 (Figure 7A) predicts the cyclic end group of DAR to be located at the entrance of the substrate binding cavity, and carbon 1 of DAR with an attached chloride atom located deep in the binding cavity in association with Cys302 of RALDH2. Moreover, the most probable interaction occurs between the γ-sulfur of that catalytic cysteine and carbon 1 of DAR (cyan, stick representation), resulting in the carbonyl carbon and chloromethyl end-group of DAR being located deep in the substrate binding pocket, possibly participating in stabilizing hydrogen bonding interactions with the residues of NAD, Asn169, and Cys302 (Figure 7B,C). Interactions between the γ-sulfur of the catalytic cysteine of RALDH2 and the carbonyl carbon of DAR were ruled out as modeling predicted insufficient space in the substrate binding pocket to accommodate the additional methylene group of DAR. We predict that, similar to what has been observed with WIN 18446,75 upon nucleophilic attack by the catalytic cysteine, a chloride atom is displaced from DAR, resulting in an irreversible interaction (Figure 7C). Modeling of DAR in the binding pocket of RALDH2 using the hydride transfer configuration of NAD+ 87 indicated significant atomic collision between the nicotinamide ring of NAD+ with the dichloromethyl end group of compound 2, suggesting steric inhibition of DAR binding in the active site of RALDH2 when NAD+ is present (data not shown). This model is substantiated by results presented in Figure 4D, which demonstrated some protection of DAR inhibition by pre-incubation with NAD+.
Figure 7. Modeling of DAR into Active Site of RALDH2.
(A) Parallel view stereo pair images of DAR (cyan, stick representation) docked into the active site of RALDH2 (surface representation). The cyclic end group of DAR is located at the entrance of the substrate binding cavity (foreground) and carbon 1 of DAR with attached chloride atom (green) is located deep in the binding cavity in association with Cys302 of RALDH2 (as a yellow surface). (B) DAR in the active site of RALDH2 with the cyclic end group of DAR at the entrance of the substrate binding cavity (left) and the chloromethyl end group in close proximity to NAD+ (shown in stick) and enzyme residues Cys302 (yellow surface) and Asn169 (shown in stick). Surface colors apply to amino acids of RALDH2 as follows: green - polar, uncharged; red - acidic, negatively charged; orange - hydrophobic; yellow - cysteine; white - polypeptide backbone. (C) Parallel view stereo pair images of the proposed mechanism of RALDH2 inhibition by DAR. Carbon 1 of DAR (1), with attached chloride atom (green, “Cl”) forms a covalent bond with the γ-sulfur of Cys302 (yellow bond). The carbonyl oxygen of DAR may participate in hydrogen bond interactions (dotted lines with distances indicated in Angstroms) with NAD, Asn169, and Cys302. Key atoms are colored by element (chloride, green; nitrogen, indigo; oxygen, red; sulfur, yellow). Figure generated in PyMOL following energy minimization by AMBER v. 17.
3. CONCLUSIONS
Aldehyde dehydrogenases are essential for many biological processes and disease states. In particular, the RALDH members of the ALDH family have been linked to a variety of diseases such as cancer,39, 40, 42 obesity,32, 33 and myopia37, 38 due to their importance in modulating endogenous concentrations of ATRA. Of the three RALDH isoforms, RALDH2, and to a lesser extent, RALDH1 are responsible for the majority of ATRA synthesis in postnatal tissues.74, 79, 88 Therefore, the identification of inhibitors selective for the RALDH isozymes would enable investigations on the roles of these enzymes in normal and disease processes, as well as provide new therapeutic options for patients with the aforementioned diseases. By using an intelligent drug design approach based on enzyme structure and substrate affinity, we report here the development of a novel compound, DAR, that is effective at inhibiting the RALDH isozymes in the nanomolar range, with no inhibition observed for ALDH2.
DAR was most effective at inhibiting chick RALDH2, followed by chick RALDH3 and chick RALDH1, with IC50’s of 55 ± 10.71 nM, 161 ± 16.57 nM, and 435 ± 99.70 nM, respectively. Additionally, DAR inhibited human RALDH2 (IC50 = 191.32 ± 36.77 nM). This is not surprising, considering the full length amino acid sequences of chick and human RALDH2 are 95.8% identical (99.2% similar) and are 100% identical among residues lining the active site. Future studies comparing DAR on all human RALDH isoforms will provide necessary information on isoform specificity of DAR for application to specific clinical conditions. Based on the three dimensional structures of our chick RALDH models, RALDH1 contains a methionine (Met120) residue that partially projects into the active site tunnel, potentially limiting access of RALDH1 to DAR. Interestingly, this methionine is not found in RALDH2 or RALDH3 (valine and isoleucine occupy this position, respectively), indicating that the access tunnels of RALDH2 and RALDH3 are unobstructed and can readily accommodate DAR, resulting in the observed lower IC50 values for DAR on RALDH2 and RALDH3, as compared with RALDH1.
Several lines of evidence presented in this report indicate that DAR inhibits RALDH2 in an irreversible and time-dependent manner, similar to the mechanism described for WIN 18446. 67, 68 Based on the proposed mechanism of inhibition of WIN 18446 on human RALDH2,75 we predict that upon binding of DAR, nucleophilic attack on carbon 1 of DAR (see Figure 1) by the thiolate group of the catalytic cysteine may result in the displacement of one of the chloride atoms, resulting in the formation of a covalent, tetrahedral 2-chloro-2-(cysteinyl)acetaldehyde. However, the chemical electrophilic reactivity of the terminal carbon atom may be different between WIN 18446, which contains amide groups adjacent to the carbonyl carbons and DAR, which contains a conjugated double bond system. Mass spectrometry and protein crystallization experiments are underway to confirm the proposed mechanism of RALDH inhibition by DAR.
DAR was also evaluated in cell-based studies using a DOX-inducible chicken RALDH2 cell line ((Dox)RALDH2-eGFP), in choroid tissue homogenates, and in ex vivo studies with isolated chick choroids. In all of these systems, DAR was effective at inhibiting ATRA synthesis with IC50 values similar to that calculated in vitro for recombinant RALDH2 and well below concentrations determined to be toxic, both in vitro and ex vivo. Interestingly, when evaluating DAR on living intact choroids ex vivo, DAR was only effective on choroids isolated from chick eyes during recovery from induced myopia (“recovering choroids”). We have previously established that RALDH2 expression and activity are increased significantly in recovering choroids37 and that RALDH2 is responsible for elevated ATRA concentrations in recovering choroids.38, 86 Moreover, when placed in vitro, only recovering choroids are capable of de novo ATRA synthesis. 37 The EC50 value for inhibition of ATRA formation in living recovering choroids by compound 2 was estimated to be 119.7 nM, but DAR only inhibited a maximum of 51% of ATRA formation in recovering choroids (with maximum inhibition at 1 μM). We speculate that DAR may be partially metabolized in living choroids, similar to the metabolism of ATRA by the cytochrome p450 (CYP) 26 enzyme.89 This idea is strengthened by our observation that DAR is a more effective ATRA synthesis inhibitor in choroid cytosol fractions (in which microsome-associated enzymes were removed by centrifugation at 100,000 × g) of both control and recovering eyes (IC50 = 20.7 nM and 53.6 nM, respectively). Moreover, WIN 18,446 (WIN), a non-specific ALDH inhibitor of the bisdichloroacetyldiamine class of compounds, is significantly more effective at inhibiting ATRA synthesis by recovering choroids ex vivo (76% inhibition at 10 μM WIN; EC50 = 44.47 nM) as compared with DAR. DAR was more effective at inhibiting RALDH2 activity in the HEK-293 cell line, (Dox)RALDH2-eGFP, as compared with the level of RALDH2 inhibition in lysates of recovering choroids following incubation of living choroids with DAR, ex vivo. It is possible that one or more cell type in the chick choroid expresses higher levels of CYP 26 as compared with (Dox)RALDH2-eGFP, It is also possible that limitations in cellular uptake and chemical instability of DAR may contribute to its less effective action on living intact choroids. Therefore, these results suggest that for effective inhibition of ATRA synthesis in vivo, larger concentrations of DAR may be needed to maximize the inhibitory effect of DAR on RALDH2 and/or necessitate modifications to the structure of DAR to limit its metabolism by CYP degradation, improve compound stability and increase bioavailability.
4. EXPERIMENTAL SECTION
4.1. GENERAL METHODS FOR CHEMISTRY.
All reactions were performed in oven-dried glassware under a positive pressure of nitrogen unless noted otherwise. Flash column chromatography was performed as described by Still et al.90 employing E. Merck silica gel 60 (230 - 400 mesh ASTM). TLC analyses and preparative TLC (pTLC) purification was performed on 250μm Silica Gel 60 F254 plates purchased from EM Science (Hatfield, PA) and Fluka Analytical (St. Louis, MO). All solvents and chemicals were used as purchased without further purification. Methylene chloride (CH2Cl2) was prepared by distillation with CaH2. Infrared spectra were recorded on a Shimadzu IRAffinity-1 instrument (Shimadzu; Kyoto, Japan), and IR spectra peaks are reported in terms of frequency of absorption (cm−1). 1H and 13CNMR spectra were recorded on VNMRS 400 and VNMRS 500 MHz-NMR Spectrometers (Varian, Inc.; Paolo Alto, CA). Chemical shifts for proton and carbon resonances are reported in ppm (δ) relative to the residual proton or the specified carbon in chloroform (δ 7.27, proton; 77.23, carbon). High-resolution mass spectrometry (HRMS) analysis was performed using Agilent 6538 high-mass-resolution QTOF mass spectrometer (Santa Clara, CA). HPLC purification was performed on Shimadzu LCMS 2020 system [LC-20AP (pump), SPD-M20A (diode array detector), LCMS-2020 (mass spectrometer)]. Semi-preparative HPLC purification was performed using Phenomenex (Torrance, CA) Luna C-18(2) column, 5 μm particle size (250 mm × 10mm), supported by Phenomenex Security Guard cartridge kit C18 (4.0 mm × 3.0 mm); and HPLC-grade solvents. Purity was determined by reverse-phase HPLC (Phenomenex Luna 5μ C18(2) 100Ǻ, 250 mm × 10 mm). All compounds tested present a purity >95%. Method: acetonitrile/water with 0.1% formic acid gradient (flow rate: 2.5 mL/min). The HPLC run started with 96% acetonitrile for 3 minutes, followed by a gradient increase in acetonitrile from 96-98% over 9 minutes, followed 98% acetonitrile for 8 minutes.
4.2. (3E,5E,7E,9E)-1,1-DICHLORO-4,8-DIMETHYL-10-(2,6,6-TRIMETHYLCYCLOHEX-1-EN-1-YL)DECA-3,5,7,9-TETRAEN-2-ONE (DAR):
Note: DAR can photoisomerize under ambient light conditions. All steps were performed in a darkened fume hood, and DAR was wrapped in aluminum foil at all steps of purification to protect it from light. The stock solution of DAR dissolved in DMSO was also protected from light at all times.
Ethyl retinoate (69.5 mg, 0.211 mmol) was dissolved in dry ethyl ether (0.9 mL) under nitrogen, and dichloromethane (19.0 μL, 0.296 mmol) was added via gas-tight syringe. The resulting dark yellow solution was cooled to −78°C. Lithium diisopropylamide (LDA) solution [2.0 M in tetrahydrofuran (THF), 170 μL, 0.338 mmol] was then added dropwise slowly over 5 min. Upon addition of LDA, the color of the reaction mixture turned dark red/brown. The progress of the reaction was monitored by TLC (10% EtOAc/hexanes, CAM stain) every 2 min for 6 minutes. The reaction mixture was then stirred at −78°C for an additional 10 min, until no further reaction progress was observed with TLC. The reaction mixture was quenched by addition of 400 μL of 6N HCl. The mixture was then warmed to room temperature (RT), diluted with ethyl ether (30 mL), washed with 1N HCl (15 mL × 2), followed by washes with distilled water (15 mL × 2). The combined aqueous phase was back extracted with ethyl ether (5 mL × 2). The combined organic phase was then washed with brine (20 mL) and dried over Na2SO4. The solvent was removed under reduced pressure to afford the crude product as a dark brown oil (70 mg).
The reaction did not go to completion, and produced multiple products based on TLC analysis. The crude mixture was first separated using preparative silica gel TLC (3 plates) and eluted with 4.5% EtOAc/hexane. Band 1 (Rf = 0.54, 12.8 mg, desired product, DAR); band 2 (Rf = 0.48, 14.9 mg, recovered ethyl retinoate); band 3 (Rf = 0.37, 7 mg, unassigned product); band 4 (Rf = 0.22, 2.8 mg, mixture of compounds). The desired product, DAR, (Figure S2-S7) obtained from pTLC fraction 1 was purified with reverse-phase HPLC using a Phenomenex C18(2) Luna semiprep column with a 96 - 98% acetonitrile/0.1% aqueous formic acid solvent gradient. 2D-NMR methods (1H-1H COSY and 1H-13C HMBC) were used to assign NMR signals to the proposed structure. Pure desired product (DAR) was obtained as a yellow oil (7.7 mg, 10% yield, 12.6% borsm). DAR: Rf = 0.54 (4.5% EtOAc/hexanes); 1H NMR (500 MHz, CDCl3): δ = 7.21 (d, J = 15.0, 11.5 Hz, 1H, H6), 6.49 (s, 1H, H3), 6.42 (d, J= 15.1 Hz, 1H, H5), 6.37 (d, J= 18.0 Hz, 1H, H7), 6.20 (d, J= 11.5 Hz, 1H, H9), 6.17 (d, J= 15.9 Hz, 1H, H10), 5.84 (s, 1H, H1), 2.44 (s, 3H, H17), 2.05 (s, 3H, H18), 2.04 (m, 2H, H13), 1.73 (s, 3H, H19), 1.68 - 1.59 (m, 2H, H14), 1.52-1.46 (m, 2H, H15), 1.05 (s, 6H, H20 and H21). 13C NMR (126 MHz, CDC13): δ = 186.46 (C2), 159.02 (C4), 141.93 (C8), 137.63 (C11), 137.05 (C10), 134.81 (C5), 134.38 (C6), 130.63 (C12), 130.03 (C7), 129.40 (C9), 118.02 (C3), 71.02 (C1), 39.60 (C15), 34.28 (C16), 33.17 (C13), 28.97 (C20, C21), 21.77 (C19), 19.18 (C14), 14.96 (C17), 13.05 (C18). IR (NaCl, cm−1): 3440 (Csp3-H), 2920 (Csp2-H), 1680 (C=0), 1558 (C=C), 785 (C-Cl). HRMS (ESI): m/z calculated for C21H28Cl2O + H+ [M + H+]: 367.1595, found: 367.1596, Δ = 0.27 ppm. Purity was determined as >95% by HPLC. Rt: 16.8 min.
4.3. BIOLOGICAL TESTING OF DAR
DAR is prone to photoisomerization, and therefore, was protected from ambient light at all times. DAR was dissolved in dimethylsulfoxide (DMSO) to produce a 10 mM solution for biological testing. These stock solutions were protected from light and stored frozen at −20°C. Multiple small aliquots of DAR (10 mM) were prepared and used for biological testing to prevent refreezing of sample as DAR will degrade upon frequent freeze/thaw cycles and exposure to ambient light. DAR uniquely absorbs at 410 nm, and shifts in the spectral profile are indicative of degradation and loss of function. Therefore we have determined that this compound is stable, if stored in the dark and at −80° for at least three months. Fresh aliquots of DAR were used for each assay and DAR was stable under assay conditions (dark, 25°C and 37°C) for at least three days.
4.4. ALDH1A/ALDH2 EXPRESSION AND PURIFICATION
Chicken recombinant RALDH1 (ALDH1A1), RALDH2 (ALDH1A2), RALDH3 (ALDH1A3), were produced and purified, as previously described.38 Human recombinant ALDH2 (hALDH2) and human recombinant RALDH2 (hRALDH2) were prepared from plasmids generously provided by Dr. Jisun Paik (University of Washington, U.S.A.), and protein was produced as described for chicken recombinant proteins except that kanamycin (250 μg/ml) was used as the bacterial selection agent. Following purification on cobalt-agarose columns, RALDH proteins were dialyzed in 20 mM triethanolamine-HCl, pH 7.4 containing 0.1 mM EDTA (buffer degassed prior to use), while hALDH2 was dialyzed in 20 mM Hepes, pH 8.5 containing 150 mM KCl and 1 mM EDTA. After dialysis, 500 mM tris(2-carboxyethyl) phosphine (TCEP) (Thermo Fisher Scientific; Waltham, MA) was added to the protein solutions to a final concentration of 1 mM.67 Purified enzymes were stored at 4 °C for up to 6 months, and enzyme stability/activity was measured at least once a month. When stored in 1 mM TCEP, recombinant RALDH enzymes demonstrated a ≤ 40% loss of activity after 6-month storage at 4°C; therefore, enzyme assays were routinely carried out within 2 months following protein purification.
4.5. ENZYME ACTIVITY ASSAYS USING PROPIONALDEHYDE
Dehydrogenase activity of purified recombinant hALDH2 was assayed spectrophotometrically by monitoring the formation of NADH at 340 nm using a microplate reader (FLUOstar OPTIMA, BMG Labtech; Ortenberg, Germany). Recombinant hALDH2 (4.5 μM in dialysis buffer, see above) was added to 96 well plates with assay buffer [20 mM Hepes pH 8.3, 150 mM KCl, 1 mM EDTA, 2 mM dithiothreitol (DTT), 5 mM propionaldehyde in EtOH; prepared from a 2x stock] in a volume of 100 μL at 25°C. Reactions were initiated by adding NAD+ to a final concentration of 5 mM (from a 20 mM stock in dH2O), and the rate of NADH synthesis was monitored at 340 nm for 5 min at 25°C. When present, ALDH inhibitors (1 – 100 μM in DMSO, final concentration) or vehicle (DMSO) were added to 96 well microplates and pre-incubated (20 min – 1 hr) with hALDH2 and assay buffer prior to addition of NAD+ and monitoring of NADH synthesis at 340 nm.
4.6. ENZYME ACTIVITY ASSAYS USING RETINALDEHYDE
Due to overlap in the absorption spectrum of retinaldehyde and NADH, a fluorescence-based NADH assay (Amplite™ Fluorimetric NADH Assay Kit, AAT Bioquest, Inc; Sunnyvale, CA) was employed to measure dehydrogenase activity of purified recombinant RALDH isozymes with the natural substrate, retinaldehyde. Recombinant protein was diluted in dialysis buffer (see above) containing 2 mM DTT to a volume of 40 μL and added to amber microfuge tubes containing 50 μL of 2x assay buffer [5% DMSO, 8 mM NAD+, 64 mM tetrasodium pyrophosphate pH 8.2, 0.2 mM pyrazole, 10 mM reduced glutathione (GSH), 1.9 mM EDTA] at 25°C. Retinaldehyde was used directly from sealed ampules (Sigma, ≥98% purity, 25 mg/vial), stored at −20° and discarded after 1 week. Retinaldehyde was initially solubilized in ethanol and its concentration determined by absorption spectroscopy (λ max = 383 and ε = 42,880) of serial dilutions.91 The retinaldehyde stock solution was then diluted to 10 × concentrations (0.155 mM - 2.50 mM) in ethanol. Under dim red light, reactions were initiated by addition of retinaldehyde in EtOH (10 uL; final retinaldehyde concentration = 15.5 - 250 μM; final reaction volume = 100 μl), and reactions were incubated in a water bath for 30 min at 37°C. Under these conditions, retinaldehyde remained in solution in synthesis buffer at all concentrations. Reactions were stopped by immersion in ice water. For NADH quantification, a 50 μl aliquot of each reaction was added to wells of solid black 96 well microplates (Greiner Bio One; Kremsmunster, Austria) followed by 50 μl of the NADH reaction mixture (Amplite™ Fluorimetric NADH Assay Kit, according to manufacturer’s directions). Total NADH was quantified by fluorescence intensity at Ex/Em = 540/590 nm and compared with a standard curve (0.1 – 30 μM NADH). While an excess of NAD+ was added to the reactions, the amount of NADH generated in the 30 min was within the range of the standard curve. For inhibition assays, recombinant protein was pre-incubated with DAR at a final concentration of 0 – 10 μM in DMSO for 0 – 80 minutes at 37°C prior to addition of assay buffer and retinaldehyde (DMSO final concentration after addition of substrate and synthesis buffer ≤ 3.5%). The calculations for KI and kinact were performed as previously described.84, 85
4.7. CELL CULTURE AND DAR TREATMENT
A stably transfected HEK-293 cell line, (Dox)RALDH2-eGFP, that exhibits doxycycline-inducible expression of chicken RALDH2-eGFP was employed to assess the effect of DAR on intracellular RALDH2 activity. The (Dox)RALDH2-eGFP cell line was generated via lentiviral infection of HEK 293 cells as follows: The lentiviral transfer vector, #143, was generated by ligation of the chicken RALDH2 coding sequence (GeneBank Accession # NM_204995) together with the ribosomal entry site (IRES) and eGFP (obtained from pTet-IRES-EGFP; plasmid #64238 Addgene, Cambridge, MA) into the open reading frame of pInducer 20 92 (obtained as a generous gift from Dr. Thomas Westbrook, Baylor College of Medicine, Houston TX). This construct encoded for constitutive expression of the transactivator (rtTA3) and neomycin resistance (Neo) via the Ubc promoter as well as inducible expression of RALDH2-IRES-eGFP by a tetracycline responsive element (TRE2) in the presence of doxycycline. Third generation lentiviral particles were generated by the transient co-transfection of HEK 293T with the lentiviral transfer vector plasmid (#143; 8 μg/10 cm plate), packaging plasmids (pRSV-Rev and pMDL/pRRE; 2μg each/ 10 cm plate) and envelope plasmid DNA (pMD2.G; 2 μg/10 cm plate) together with an equal mass of PEI (14 μg/10 cm plate, from a 2 ug/ul stock) in Opti-MEM (Gibco® by Life Technologies, Carlsbad, CA) at a final volume of 1 ml DNA solution per 10 cm plate. DNA/PEI mixtures were incubated at room temperature for 15 min added to dropwise to plated 293T cells (1ml per 10 cm plate) and incubated at 37°C in a 5% CO2 humidified atmosphere. The culture supernatants containing lentiviral particles were collected at 48 – 72 hrs post-transfection, concentrated by ultracentrifugation at 20,000 rpm (Beckman rotor SW-28) for 2.5 hrs at 4 °C and stored at −80°C. A stable cell line expressing inducible chicken RALDH2 and eGFP was subsequently generated by lentiviral transduction of HEK 293 cells in the presence of polybrene (8 μg/mL). Transfected cells were monitored by eGFP fluorescence and sorted via flow cytometry using a BD Biosystems LSRII analyzer. RALDH2-eGFP positive cells were cultured in Dulbecco’s modified Eagle medium (DMEM)/high glucose (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) containing neomycin sulfate (1 μg/ml). The cells were maintained at 37°C and 5% CO2 in a humidified incubator (VWR International, LLC., Atlanta, GA, USA) and were harvested at 80–90% confluence.
For experiments testing DAR, (Dox)RALDH2-eGFP cells were seeded in 6- well plates at a density of 150,000 cells/well in culture medium [DMEM containing 10% fetal bovine serum (FBS) and doxycycline (5 μg/ml)] and incubated at 37°C and 5% CO2 for 25 hrs. Culture medium was replaced with low serum media (DMEM containing 0.1% FBS) containing 0.05 – 2 μM DAR in DMSO (< 0.004%) or vehicle, and cells were incubated for 8 hrs at 37°C and 5% CO2. Following incubation, media was removed, and cell layers were collected [1.0 ml + 0.5 ml homogenization buffer (20 mM triethanolamine-HCl pH 7.4, 1 mM DTT, 0.1 mM EDTA)] with the aid of a rubber policeman. Cells were lysed using a VirTis rotor-stator homogenizer (SP Industries; Gardiner, NY). Homogenates were then transferred to thick-walled microfuge tubes (polyallomer tubes; Beckman Coulter; Brea, CA) and ultracentrifuged (100,000 × g for 1 hr; Optimum MAX Ultracentrifuge, Beckman Coulter) at 4°C to isolate the cytosol fraction (supernatant) which was stored at −20°C until use. Relative RALDH2 protein expression was determined by western blot, as previously described.38 RALDH2 protein expression using the (Dox)RALDH2-eGFP 293 HEK cell line ranged from 25% – 37% following incubation with 0.05 – 2 μM DAR, as determined from three independent experiments, each performed in triplicate.
RALDH activity was determined by measuring the production of ATRA in vitro, as described previously.38 All procedures with retinaldehyde or ATRA were performed under dim red light. 200 μL of synthesis buffer [2.5% DMSO, 4 mM NAD, 32 mM tetrasodium pyrophosphate pH 8.2, 0.1 mM pyrazole, 5 mM reduced glutathione (GSH), 1 mM EDTA; prepared from a 2x stock] was added to 200 μL of cytosol from homogenized cells. The reaction was initiated by the addition of 50 μL of 250 μM all-trans-retinaldehyde (in synthesis buffer) to the reaction mixture (25 μM final concentration). The reactions were vigorously mixed and placed in a water bath at 37°C for 30 min, after which time, the reactions were stopped by immersion in ice water and addition of 3/2 volumes (675 μL) of methanol. Samples were stored at −80°C until time of use.
ATRA was extracted for quantification by reverse phase HPLC, as previously described91 with the following modifications: to 500 μL of sample, 60 μL of acetonitrile and 17 μL of 4M HCl were added to clear, microcentrifuge tubes and vortex mixed for ~ 15 sec. Hexane (600 μL) was added to each tube and vortex mixed for ~ 10 sec. Tubes were then centrifuged at RT for 3 min at 1000g to facilitate phase separation. The organic (top) phase was removed (400 μL), with great care taken to not disturb the bottom aqueous layer, and transferred to a new, clear microcentrifuge tube (SafeSeal; Sorenson BioScience Inc, Salt Lake City, UT). The organic phase was then evaporated under a gentle nitrogen stream at RT. Samples were then resuspended in 100 μL of acetonitrile and transferred to plastic HPLC vials (9 mm S/T vial with septa) (Supelco, Inc; Bellefonte, PA). All steps were performed in dim red light. ATRA was quantified as previously described90 with the following modifications: a Shimadzu LCMS 2020 (Shimadzu; Kyoto, Japan) with a Cortecs C18 column [2.7 μm particle size with VanGuard Cartridge (Waters; Milford, MA)] was used to achieve ATRA separation. The column compartment was maintained at 25°C, and the autosampler was maintained at 10°C. The following solvents were used: A, H2O with 0.1% formic acid; B, acetonitrile. The column was equilibrated in 70% acetonitrile prior to sample loading. 25 μL of sample was injected into the column, and separation was achieved at 400 μL/min with the following gradient: 0 – 3 min, hold at 70% B; 3 – 15 min, 70% B to 95% B; 15 – 20 min, hold at 95% B; 20 – 21 min, 95% B to 70% B; 21 – 37 min, re-equilibrate at 70% B. Elution of ATRA was monitored using a photodiode array detector at 350 nm, followed by MS detection. Retention time of ATRA was 20.6 min with an m/z of 300.25. ATRA was quantified from a standard curve (Figure S8) generated using known amounts of ATRA (0 – 1500 pmol) eluted with the above gradient.
For cell viability assays, aliquots of the cell homogenates (prior to ultracentrifugation) were digested with proteinase K (protease type XXVIII, Sigma; in 10 mM EDTA, 0.1 M sodium phosphate pH 6.5; 0.05% w/v) at 60°C overnight (O/N). DNA was measured directly from proteinase K digests using Pico-green (Invitrogen; Carlsbad, CA) and a mini-fluorometer (TBS-380; Turner Biosystems; Sunnyvale, CA), according to manufacturer’s protocols. Additionally, cell viability was determined by quantification of ATP in separate cell cultures. For ATP quantification, (Dox)RALDH2-eGFP 293 cells were plated at a density of 10,000 cells/well solid black, flat-bottomed 96 well microplates (Greiner Bio One; Kremsmünster, Austria) in culture medium (DMEM containing 10% FBS) and incubated at 37°C and 5% CO2 for 24 hrs. Culture medium was replaced with low serum media (DMEM containing 0.1% FBS) containing 0 – 50 μM DAR in DMSO (< 0.5% DMSO) and incubated for 24 hrs at 37°C and 5% CO2. ATP was quantified using the CelTiter-Glo Luminescent Cell Viability Assay (Promega; Madion, WI) according to manufacturer’s instructions. Luminescence was measured using a FLUOstar OPTIMA plate reader (BMG Labtech; Cary, NC). Viability was calculated as a percentage of vehicle-treated (0.5% DMSO v/v) control cells at each concentration of DAR.
4.8. ANIMALS
White Leghorn male chicks (Gallus gallus) were obtained as 2 day old hatchlings from Ideal Breeding Poultry Farms (Cameron, TX). Chicks were housed in temperature controlled brooders with a 12-hour light/dark cycle and were given food and water ad libitum. Form deprivation myopia (FDM) was induced in 3 day old chicks by applying translucent plastic goggles to one eye, as described previously.93 The contralateral eyes (left eyes) of all chicks were never goggled and used as controls. Chicks were checked daily for goggle condition. Goggles remained in place for 10 days, after which time the goggles were removed, and chicks were allowed to experience unrestricted vision (recover) for 4 days. Chicks were managed in accordance with the Animal Welfare Act and with the National Institutes of Health (NIH; Bethesda, MD, USA) Guidelines. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Oklahoma Health Sciences Center.
4.9. INHIBITION OF RALDH ACTIVITY IN TISSUE LYSATES
Choroids were isolated from eyes of normal chicks, as well as from control and treated eyes of chicks during recovery from induced myopia, as previously described.38 Briefly, an 8 mm punch was taken from the posterior pole of the chick eye using a dermal biopsy punch (Miltex, Inc.; York, PA). Punches were located nasal to the exit of the optic nerve, with care to exclude the optic nerve and pecten. With the aid of a dissecting microscope, the neural retina and RPE were removed from the underlying choroid and sclera using a drop of PBS (3 mM dibasic sodium phosphate, 1.5 mM monobasic sodium phosphate, 150 mM NaCl, pH 7.2) and gentle brushing. Choroids were dissected from the sclera with the aid of a rounded spatula and incubated in N2 medium [DMEM:Ham’s F-12 1:1 containing N2 supplement (Invitrogen; Carlsbad, CA) supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) and amphotericin B (1.5 μg/ml, Gibco® by Life Technologies, Carlsbad, CA)]. For tissue incubated with DAR, DAR was serially diluted (0.01 – 5 μM) in N2 media (initial dilution: 1:10,000 in N2 medium from a 10 mM stock in DMSO). For tissue incubated with WIN 18,446 (WIN; Cayman Chemical Company, Ann Arbor, MI), WIN was serially diluted (0.01 – 5 μM) in N2 media (initial dilution: 1:10,000 in N2 medium from a 10 mM stock in DMSO). Choroids were cultured for 24 hrs at 37°C and 5% CO2 in 48-well tissue culture plates (Costar; Corning, NY), with one choroid per well. Following incubation, choroids were placed in microfuge tubes and frozen at −20°C. Additional choroids were isolated from control and treated eyes as described above and immediately placed in microfuge tubes and frozen at −20°C. Choroids were homogenized using a VirTis rotor-stator homogenizer, and cytosol fractions were obtained by ultracentrifugation at 100,000g for 1 hr, as described above. In some cases, protein concentrations of cytosol fractions were determined by a Bradford assay (BioRad, Hercules, CA). RALDH activity was assessed in cytosol fractions of control and inhibitor treated choroids, as described for the (Dox)RALDH2-eGFP 293 cells (section 4.7 above).
4.10. MOLECULAR MODELING
All molecular modeling studies were performed on a 3.00 GHz Intel Duo CPU running Windows 7 Professional Operating System. A structural model of chicken RALDH2 (NP_990326.1) was generated via homology modeling using a crystal structure of sheep ALDH1A1 with NAD+ bound (PDB: 5abm)44 as the template and the Homology detection & structure prediction by HMM-HMM comparison (HHpred) server (Max-Planck Institute for Developmental Biology). The sheep ALDH1A1 structure was selected as the template for these studies since this structure includes all residues in the active site, has the highest resolution (1.7A), and excellent validation parameters. Ligand structures were built in InsightII/Discover (Accelrys Inc.; San Diego, CA). Modeling of DAR into the active site of RALDH2 was based on the crystal structure of human RALDH2 with bound WIN 18,446 retaining one chloride atom (residue # DAR 700-Cl1) as seen in the crystal structure.75 The retinyl group (beta-ionone ring with isoprenoid chain) was positioned and energy minimization was carried out using AMBER version 17. Binding poses were visualized and figures generated with PyMOL v. 1.8.6 (Schrodinger, Cambridge MA).
4.11. STATISTICS AND DATA ANALYSIS.
Analyses between groups were made using a 1-way ANOVA followed by a Bonferroni correction for multiple comparisons; analyses between pairs within a group were made using a paired or unpaired t-test (GraphPad Prism 5; La Jolla, CA). IC50 inhibition curves were generated using the activity of the ALDH isozymes following incubation with DAR, as described above. The inhibition curves were fit to the three-parameter inhibition dose response equation or four-parameter inhibition dose response equations using GraphPad Prism 5. Additionally, apparent Ki’s for DARwere generated by nonlinear regression using the Morrison equation for tight binding in GraphPad Prism 5.
Supplementary Material
Figure S1 – 8: Michaelis-Menten and Time Dependent Data for RALDH1 and RALDH3; Analysis of DAR Synthesis and Purification; Standard Curve for HPLC Quantification of RA
Table S1: Effect of DAR on Km and Vmax for RALDH 1, 2, 3
ACKNOWLEDGMENTS
This research was supported by National Eye Institute grant R01 EY09391 (JAS), National Eye Institute fellowship F31EY025168 (ARH), and by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences grant P20GM103640 (PI: Ann West). The authors are grateful to Dr. Jisun Paik (University of Washington, U.S.A.) for generously providing the hu-ALDH2 and hu-RALDH2 plasmids, to Dr. Thomas Westbrook (Baylor College of Medicine, Houston TX) for generously providing pInducer lentiviral constructs, and to John Moore (laboratory technician; University of Oklahoma Health Sciences Center, U.S.A.), for technical assistance.
ABBREVIATIONS
- RALDH
retinaldehyde dehydrogenase
- ATRA
all-trans-retinoic acid
- RAL
retinaldehyde
- DAR
dichloro-all-trans-retinone
- ALDH
aldehyde dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional Information
Declarations of Interest: none
REFERENCES
- 1.Summers JA, Harper AR, Mather T, Burgett A. Le ATQ. Inhibitors of Retinaldehyde Dehydrogenases And Methods Of Use. Patent No. WO 2017049110 A1, pending.
- 2.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 2008;4:697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol. Rev. 2012;64:520–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Priyadharshini CJ, George PD. Single amino acid polymorphism in aldehyde dehydrogenase gene superfamily. Front. Biosci. 2015;20:335–376. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida A, Hsu L, Dave V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 1991;46:239–244. [DOI] [PubMed] [Google Scholar]
- 6.Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–553. [DOI] [PubMed] [Google Scholar]
- 7.Kim KJ, Pearl PL, Jensen K, Snead OC, Malaspina P, Jakobs C, Gibson KM. Succinic semialdehyde dehydrogenase: biochemical–molecular–clinical disease mechanisms, redox regulation, and functional significance. Antioxid. Redox Signaling. 2011;15:691–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genomics. 2011;5:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Januchowski R, Wojtowicz K, Zabel M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharmacother. 2013;67:669–680. [DOI] [PubMed] [Google Scholar]
- 10.Jelski W, Szmitkowski M. Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer diseases. Clin. Chim. Acta. 2008;395:1–5. [DOI] [PubMed] [Google Scholar]
- 11.Balcz B, Kirchner L, Cairns N, Fountoulakis M, Lubec G. Increased brain protein levels of carbonyl reductase and alcohol dehydrogenase in Down syndrome and Alzheimer’s disease In Lubec G, ed. Protein Expression in Down Syndrome Brain. New York, NY: Springer; 2001: 193–201. [DOI] [PubMed] [Google Scholar]
- 12.Grünblatt E, Riederer P. Aldehyde dehydrogenase (ALDH) in Alzheimer’s and Parkinson’s disease. J. Neural Transm. 2016;123:83–90. [DOI] [PubMed] [Google Scholar]
- 13.Thomasson HR, Crabb DW, Edenberg HJ, Li TK. Alcohol and aldehyde dehydrogenase polymorphisms and alcoholism. Behav. Genet. 1993;23:131–136. [DOI] [PubMed] [Google Scholar]
- 14.De Laurenzi V, Rogers GR, Hamrock DJ, Marekov LN, Steinert PM, Compton JG, Markova N, Rizzo WB. Sjögren-Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat. Genet. 1996;12:52–57. [DOI] [PubMed] [Google Scholar]
- 15.Rizzo WB. Sjögren–Larsson syndrome: molecular genetics and biochemical pathogenesis of fatty aldehyde dehydrogenase deficiency. Mol. Genet. Metab. 2007;90:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valle DL, Phang JM, Goodman SI. Type 2 Hyperprolinemia: absence of delta1-pyrroline-5-carboxylic acid dehydrogenase activity. Science. 1974;185:1053–1054. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava D, Singh RK, Moxley MA, Henzl MT, Becker DF, Tanner JJ. The three-dimensional structural basis of type II hyperprolinemia. J. Mol. Biol. 2012;420:176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flahaut M, Jauquier N, Chevalier N, Nardou K, Bourloud KB, Joseph JM, Barras D, Widmann C, Gross N, Renella R, Mühlethaler-Mottet A. Aldehyde dehydrogenase activity plays a key role in the aggressive phenotype of neuroblastoma. BMC Cancer. 2016;16:781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, Liu H, Behrens C, Shay JW, Wistuba II, Minna JD. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin. Cancer Res. 2014;20:4154–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao D, McCaffery P, Ivins KJ, Neve RL, Hogan P, Chin WW, Drager UC. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur. J. Biochem. 1996;240:15–22. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Penzes P, Napoli JL. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J. Biol. Chem. 1996; 271:16288–16293. [DOI] [PubMed] [Google Scholar]
- 22.Haselbeck RJ, Hoffmann I, Duester G. Distinct functions for Aldh1 and Raldh2 in the control of ligand production for embryonic retinoid signaling pathways. Dev. Genet. 1999;25:353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol. Rev. 2000;80:1021–1054. [DOI] [PubMed] [Google Scholar]
- 24.Niederreither K, Fraulob V, Garnier JM, Chambon P, Dolle P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech. Dev. 2002; 110:165–171. [DOI] [PubMed] [Google Scholar]
- 25.Raverdeau M, Gely-Pernot A, Féret B, Dennefeld C, Benoit G, Davidson I, Chambon P, Mark M, Ghyselinck NB. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16582–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JW, Wang RY, Guo QS, Xu C. Expression of the retinoic acid-metabolizing enzymes RALDH2 and CYP26b1 during mouse postnatal testis development. Asian J. Androl. 2008;10:569–576. [DOI] [PubMed] [Google Scholar]
- 27.Hogarth CA, Griswold MD. The key role of vitamin A in spermatogenesis. J. Clin. Invest 2010;120:956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Thompson DC, Koppaka V, Jester JV, Vasiliou V. Ocular aldehyde dehydrogenases: protection against ultraviolet damage and maintenance of transparency for vision. Prog. Retinal Eye Res. 2013;33:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Koppaka V, Thompson DC, Vasiliou V. Focus on molecules: ALDH1A1: from lens and corneal crystallin to stem cell marker. Exp. Eye Res. 2012;102:105–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Curr. Stem Cell Res. Ther. 2008;3:237–246. [DOI] [PubMed] [Google Scholar]
- 31.Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155:215–227. [DOI] [PubMed] [Google Scholar]
- 32.Petrosino JM, Disilvestro D, Ziouzenkova O. Aldehyde dehydrogenase 1A1: friend or foe to female metabolism? Nutrients. 2014;6:950–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, Vogel S, Duester G, Plutzky J. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat. Med. 2007;13:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goswami S, Angkasekwinai P, Shan M, Greenlee KJ, Barranco WT, Polikepahad S, Seryshev A, Song LZ, Redding D, Singh B, Su S, Woodruff P, Dong C, Corry DB, Kheradmand F. Divergent functions for airway epithelial matrix metalloproteinase 7 and retinoic acid in experimental asthma. Nat. Immunol. 2009;10:496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito K, Zolfaghari R, Hao L, Ross AC. Inflammation rapidly modulates the expression of ALDH1A1 (RALDH1) and vimentin in the liver and hepatic macrophages of rats in vivo. Nutr. Metab. 2014;11:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders TJ, McCarthy NE, Giles EM, Davidson KL, Haltalli ML, Hazell S, Lindsay JO, Stagg AJ. Increased production of retinoic acid by intestinal macrophages contributes to their inflammatory phenotype in patients with Crohn's disease. Gastroenterology. 2014;146:1278–1288. [DOI] [PubMed] [Google Scholar]
- 37.Rada JA, Hollaway LY, Li N, Napoli J. Identification of RALDH2 as a visually regulated retinoic acid synthesizing enzyme in the chick choroid. Invest. Ophthalmol. Vis. Sci. 2012;53: 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harper AR, Wang X, Moiseyev G, Ma JX, Summers JA. Postnatal chick choroids exhibit increased retinaldehyde dehydrogenase activity during recovery from form deprivation induced myopia. Invest. Ophthalmol. Vis. Sci. 2016;57:4886–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreb JS. Muhoczy D, Ostmark B, Zucali JR. RNAi-mediated knockdown of aldehyde dehydrogenase class-1A1 and class-3A1 is specific and reveals that each contributes equally to the resistance against 4-hydroperoxycyclophosphamide. Cancer Chemother. Pharmacol. 2007; 59: 127–136. [DOI] [PubMed] [Google Scholar]
- 40.Sladek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Cancer Chemother. Pharmacol. 2002;49:309–321. [DOI] [PubMed] [Google Scholar]
- 41.De Luca LM. Retinoids and their receptors in differentiation, embryogenesis, and neoplasia. FASEB J. 1991; 5:2924–2933. [PubMed] [Google Scholar]
- 42.Kim H, Lapointe J, Kaygusuz G, Ong DE, Li C, van de Rijn M, Brooks JD, Pollack JR. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005; 65: 8118–8124. [DOI] [PubMed] [Google Scholar]
- 43.Rexer BN, Zheng WL, Ong DE. retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in MCF-7 cells. Cancer Res. 2001;61: 7065–7070. [PubMed] [Google Scholar]
- 44.Koch JG, Gu X, Han Y, El-Naggar AK, Olson MV, Medina D.; Jerry DJ, Blackburn AC, Peltz G, Amos CI, Lozano G. Mammary tumor modifiers in BALB/cJ mice heterozygous for p53. Mamm. Genome. 2007;18:300–309. [DOI] [PubMed] [Google Scholar]
- 45.Koch MF, Harteis S, Blank ID, Pestel G, Tietze LF, Ochsenfeld C, Schneider S, Sieber S. Structural, biochemical, and computational studies reveal the mechanism of selective aldehyde dehydrogenase 1A1 inhibition by cytotoxic duocarmycin analogues. Angew. Chem. Int. Ed. Engl. 2015;54:13550–13554. [DOI] [PubMed] [Google Scholar]
- 46.Parajuli B, Georgiadis TM, Fishel ML, Hurley TD. Development of selectiveinhibitors for human aldehyde dehydrogenase 3A1 (ALDH3A1) for the enhancement of cyclophosphamide cytotoxicity. Chembiochem. 2014;15:701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan CA, Hurley TD. Characterization of two distinct structural classes of selective aldehyde dehydrogenase 1A1 inhibitors. J. Med. Chem. 2015;58:1964–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan CA, Parajuli B, Buchman CD, Dria K, Hurley TD. N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem Biol Interact. 2015;234:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo M, Gates KS, Henzl MT, Tanner JJ. Diethylaminobenzaldehyde is a covalent, irreversible inactivator of ALDH7A1. ACS Chem Biol. 2015;10:693–697. [DOI] [PubMed] [Google Scholar]
- 50.Russo JE, Hauquitz D, Hilton J. Inhibition of mouse cytosolic aldehyde dehydrogenase by 4-(diethylamino) benzaldehyde. Biochemical pharmacology. 1988;37:1639–1642. [DOI] [PubMed] [Google Scholar]
- 51.Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res. Treat. 2012; 133: 75–87. [DOI] [PubMed] [Google Scholar]
- 52.Boyer CS, Petersen DR. The metabolism of 3, 7-dimethyl-2, 6-octadienal (citral) in rat hepatic mitochondrial and cytosolic fractions. Interactions with aldehyde and alcohol dehydrogenases. Drug Metab. Dispos. 1991;19:81–86. [PubMed] [Google Scholar]
- 53.Kikonyogo A, Abriola DP, Dryjanski M, Pietruszko R. Mechanism of inhibition of aldehyde dehydrogenase by citral, a retinoid antagonist. Eur. J. Biochem. 1999;262:704–712. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka M, Tamura K, Ide H. Citral, an inhibitor of retinoic acid synthesis, modifies chick limb development. Dev. Biol. 1996;175:239–247. [DOI] [PubMed] [Google Scholar]
- 55.Cengiz E, Karaca B, Kucukzeybek Y, Gorumlu G, Gul MK, Erten C, Atmaca H, Uzunoglu S, Karabulut B, Sanli UA, Uslu R. Overcoming drug resistance in hormone-and drug-refractory prostate cancer cell line, PC-3 by docetaxel and gossypol combination. Mol. Biol. Rep. 2010;37:1269–1277. [DOI] [PubMed] [Google Scholar]
- 56.Rekha GK, Sladek NE. Inhibition of human class 3 aldehyde dehydrogenase, and sensitization of tumor cells that express significant amounts of this enzyme to oxazaphosphorines, by the naturally occurring compound gossypol In: Weiner H, Duester G, Maser E, Plapp, eds. Enzymology and Molecular Biology of Carbonyl Metabolism. New York,m NY: Springer; 1996: 133–146. [DOI] [PubMed] [Google Scholar]
- 57.Quemener V, Quash G, Moulinoux J, Penlap V, Ripoll H, Havouis R, Doutheau A, Goré J. In vivo antitumor activiy of 4-amino 4-methyl 2-pentyne 1-al, an inhibitor of aldehyde dehydrogenase. In Vivo. 1988;3:325–330. [PubMed] [Google Scholar]
- 58.Canuto RA, Muzio G, Salvo RA, Maggiora M, Trombetta A, Chantepie J, Fournet G, Reichert U, Quash G. The effect of a novel irreversible inhibitor of aldehyde dehydrogenases 1 and 3 on tumour cell growth and death. Chem.-Biol. Interact. 2001;130:209–218. [DOI] [PubMed] [Google Scholar]
- 59.Fuller RK, Branchey L, Brightwell DR, Derman RM, Emrick CD, Iber FL, James KE, Lacoursiere RB, Lee KK, Lowenstam I. Disulfiram treatment of alcoholism: a Veterans Administration cooperative study. Jama. 1986;256:1449–1455. [PubMed] [Google Scholar]
- 60.Petrakis IL, Poling J, Levinson C, Nich C, Carroll K, Rounsaville B. Naltrexone and disulfiram in patients with alcohol dependence and comorbid psychiatric disorders. Biol. Psychiatry. 2005;57:1128–1137. [DOI] [PubMed] [Google Scholar]
- 61.Tottmar O, Hellström E. Blood pressure response to ethanol in relation to acetaldehyde levels and dopamine-β-hydroxylase activity in rats pretreated with disulfiram, cyanamide and coprine. Acta Pharmacol. Toxicol. 1979;45:272–281. [DOI] [PubMed] [Google Scholar]
- 62.Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011; 10:1378–1384. [DOI] [PubMed] [Google Scholar]
- 63.Heller CG, Moore DJ, Paulsen CA. Suppression of spermatogenesis and chronic toxicity in men by a new series of bis (dichloroacetyl) diamines. Toxicol. Appl. Pharmacol. 1961;3:1–11. [DOI] [PubMed] [Google Scholar]
- 64.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells, 2011;29:32–45. [DOI] [PubMed] [Google Scholar]
- 65.Moreb JS, Ucar D, Han S, Amory JK, Goldstein AS, Ostmark B, Chang LJ. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact. 2012;195:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bitzer M, Feldkaemper M, Schaeffel F. Visually induced changes in components of the retinoic acid system in fundal layers of the chick. Exp. Eye Res. 2000;70:97–106. [DOI] [PubMed] [Google Scholar]
- 67.Amory JK, Muller CH, Shimshoni JA, Isoherranen N, Paik J, Moreb JS, Amory DW, Evanoff R, Goldstein AS, Griswold MD. Suppression of spermatogenesis by bisdichloroacetyldiamines is mediated by inhibition of testicular retinoic acid biosynthesis. J. Androl. 2011;32:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paik J, Haenisch M, Muller CH, Goldstein AS, Arnold S, Isoherranen N, Brabb T, Treuting PM, Amory JK. Inhibition of retinoic acid biosynthesis by the bisdichloroacetyldiamine WIN 18,446 markedly suppresses spermatogenesis and alters retinoid metabolism in mice. J. Biol. Chem. 2014;289:15104–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asa CS, Zaneveld LJ, Munson L, Callahan M, Byers AP. Efficacy, safety, and reversibility of a bisdiamine male-directed oral contraceptive in gray wolves (Canis lupus). J Zoo Wildl Med. 1996;27:501–506. [Google Scholar]
- 70.Munson L, Chassy LM, Asa C. Efficacy, safety and reversibility of bisdiamine as a male contraceptive in cats. Theriogenology. 2004;62:81–92. [DOI] [PubMed] [Google Scholar]
- 71.Singh S, Dominic C. Effect of N, N'-bis (dichloroacetyl)-i, 8-octamethylenediamine (WIN 18446) on the testis+ epididymis of the musk shrew Suncus murinus L. Indian J. Exp. Biol. 1980;18:1217–1220. [PubMed] [Google Scholar]
- 72.Chung S, Wolgemuth D. Role of retinoid signaling in the regulation of spermatogenesis. Cytogenet. Genome Res. 2004;105:189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barluenga J, Llavona L, Yus M, Concellón JM. Reactivity of in situ generated dihalomethyllithium towards dicarboxylic acid diesters and lactones: Synthetic applications. Tetrahedron. 1991;47:7875–7886. [Google Scholar]
- 74.Arnold SLM, Kent T, Hogarth CA, Griswold MD, Amory JK, Isoherranen N. Pharmacological inhibition of ALDH1A in mice decreases all-trans retinoic acid concentrations in a tissue specific manner. Biochem. Pharmacol. 2015;95:177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y, Zhu JY, Hong KH, Mikles DC, Georg GI, Goldstein AS, Amory JK, Schonbrunn E. Structural Basis of ALDH1A2 Inhibition by Irreversible and Reversible Small Molecule Inhibitors. ACS Chem Biol. 2018;13:582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6:1541–1551. [DOI] [PubMed] [Google Scholar]
- 77.Gagnon I, Duester G, Bhat PV. Kinetic analysis of mouse retinal dehydrogenase type-2 (RALDH2) for retinal substrates. Biochim. Biophys. Acta. 2002;1596:156–162. [DOI] [PubMed] [Google Scholar]
- 78.Shabtai Y, Jubran H, Nassar T, Hirschberg J, Fainsod A. Kinetic characterization and regulation of the human retinaldehyde dehydrogenase 2 enzyme during production of retinoic acid. Biochem. J. 2016;473:1423–1431. [DOI] [PubMed] [Google Scholar]
- 79.Arnold SL, Kent T, Hogarth CA, Schlatt S, Prasad B, Haenisch M, Walsh T, Muller CH, Griswold MD, Amory JK, Isoherranen N. Importance of ALDH1A enzymes in determining human testicular retinoic acid concentrations. J. Lipid Res. 2015;56:342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silverman RB. Mechanism-based enzyme inactivators. Methods Enzymol. 1995;249: 240–283. [DOI] [PubMed] [Google Scholar]
- 81.Berry LM, Zhao Z. An examination of IC50 and IC50-shift experiments in assessing time-dependent inhibition of CYP3A4, CYP2D6 and CYP2C9 in human liver microsomes. Drug Metab. Lett. 2008;2:51–59. [DOI] [PubMed] [Google Scholar]
- 82.Perloff ES, Mason AK, Dehal SS, Blanchard AP, Morgan L, Ho T, Dandeneau A, Crocker RM, Chandler CM, Boily N, Crespi CL, Stresser DM. Validation of cytochrome P450 time-dependent inhibition assays: a two-time point IC50 shift approach facilitates kinact assay design. Xenobiotica. 2009;39:99–112. [DOI] [PubMed] [Google Scholar]
- 83.Kitz R, Wilson IB. Esters of methanesulfonic acid as irreversible inhibitors of acetylcholinesterase. J. Biol. Chem. 1962;237:3245–3249. [PubMed] [Google Scholar]
- 84.Filppula AM, Neuvonen PJ, Backman JT. In vitro assessment of time-dependent inhibitory effects on CYP2C8 and CYP3A activity by fourteen protein kinase inhibitors. Drug Metab. Dispos. 2014;42:1202–1209. [DOI] [PubMed] [Google Scholar]
- 85.Amory JK, Arnold S, Lardone MC, Piottante A, Ebensperger M, Isoherranen N, Muller CH, Walsh T, Castro A. Levels of the retinoic acid synthesizing enzyme aldehyde dehydrogenase-1A2 are lower in testicular tissue from men with infertility. Fertil. Steril. 2014; 101:960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mertz JR, Wallman J. Choroidal retinoic acid synthesis: a possible mediator between refractive error and compensatory eye growth. Exp. Eye Res. 2000;70:519–527. [DOI] [PubMed] [Google Scholar]
- 87.Perez-Miller SJ, Hurley TD. Coenyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry 2003;42:7100–7109. [DOI] [PubMed] [Google Scholar]
- 88.Asson-Batres MA, Smith WB. Localization of retinaldehyde dehydrogenases and retinoid binding proteins to sustentacular cells, glia, Bowman's gland cells, and stroma: potential sites of retinoic acid synthesis in the postnatal rat olfactory organ. J. Comp. Neurol. 2006;496:149–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ross AC, Zolfaghari R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu. Rev. Nutr. 2011;31:65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Still WC, Kahn M, Mitra A. Rapid chromatographic technique for preparative separations with moderate resolution. The Journal of Organic Chemistry. 1978;43:2923–2925. [Google Scholar]
- 91.Kane MA, Napoli JL. Quantification of endogenous retinoids. Methods Mol Biol. 2010; 652:1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meerbrey K, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108:3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 – 8: Michaelis-Menten and Time Dependent Data for RALDH1 and RALDH3; Analysis of DAR Synthesis and Purification; Standard Curve for HPLC Quantification of RA
Table S1: Effect of DAR on Km and Vmax for RALDH 1, 2, 3