Abstract
Drugging large protein pockets is a challenge due to the need for higher molecular weight ligands, which generally possess undesirable physicochemical properties. In this communication, we highlight a strategy leveraging small molecule active site dimers to inhibit the large symmetric binding pocket in the STING protein. By taking advantage of the 2:1 binding stoichiometry, maximal buried interaction with STING protein can be achieved while maintaining the ligand physicochemical properties necessary for oral exposure. This mode of binding requires unique considerations for potency optimization including simultaneous optimization of protein–ligand as well as ligand–ligand interactions. Successful implementation of this strategy led to the identification of 18, which exhibits good oral exposure, slow binding kinetics, and functional inhibition of STING-mediated cytokine release.
Keywords: STING, active site dimers, ligand−ligand interactions, permeability, inflammatory disease, druggability
The activation of the innate immune system through nucleic acid sensing is a key mechanism of host defense from viruses and bacteria.1,2 Recent discoveries in the cyclic GMP-AMP synthase–stimulator of interferon genes (cGAS–STING) pathway have captured the attention of the pharmaceutical industry due to the pathway’s role in host pathogen defense, immuno-oncology, and autoimmune diseases.3 Upon activation by double-stranded DNA (dsDNA), cGAS synthesizes the cyclic dinucleotide secondary messenger 2′,3′-cyclic GMP-AMP (cGAMP), which then activates the endoplasmic reticulum (ER)–membrane adaptor protein STING.4,5 Activated STING initiates a cascade that ultimately primes the immune system to restrict viral spread through the activation and production of type 1 interferons (IFN) and pro-inflammatory cytokines such as tumor necrosis factor-α (TNF-α).
While stimulation of STING and the production of type 1 interferons is an important mechanism for pathogen defense and tumor control, failure to regulate chronic inflammatory signaling can lead to autoimmunity.6 The mislocalization of nuclear and mitochondrial dsDNA in the cytoplasm combined with the inability of cGAS to differentiate foreign from self-dsDNA is a potential trigger for type 1 interferon production and autoinflammation.7−9 Type 1 interferon and mislocalized dsDNA are hallmarks and key drivers for the pathogenesis of autoimmune diseases such as systemic lupus erythematosus (SLE).10 Additionally, loss of function mutations in the DNA exonuclease TREX1 lead to the excessive type 1 interferon signature found in Aicardi–Goutières syndrome (AGS) and SLE, implicating the role of self-dsDNA in autoinflammation.11 Furthermore, monogenic Mendelian diseases with STING gain of function mutations such as familial chilblain lupus (FCL) support the role of STING in autoimmune disease.12 These studies collectively suggest that inhibition of STING might regulate DNA-driven inflammatory diseases.
Although the biological rationale linking STING to inflammatory diseases supports the development of a STING antagonist, the general lack of biochemical mechanistic understanding of STING activation combined with the absence of known small molecule tools suggested that identification of binders to the STING protein might pose a significant challenge.13,14 Based on biophysical and X-ray crystallographic data, the STING C-terminal domain (referred to as STING henceforth) exists as a symmetrical dimer with the ligand binding site located at the interface between the two monomers, which has been shown to be the binding site of the natural agonist cGAMP and the mouse-specific agonist DMXAA.15 Comparison of the structures of STING apoprotein with cGAMP-bound protein highlights two distinct conformations (Figure 1). The STING apoprotein adopts an “open” conformation, with residues 226 to 241 in each monomer. In contrast, STING complexed with an agonist adopts a “closed” conformation wherein the central α-2 helix from each monomer (approximately residues 171 to 185) tilt toward each other by roughly 15°, and residues 226 to 241 in each monomer, disordered in the “open” conformation, adopt a four-stranded β-sheet. These distinct conformational states suggest that the stabilization of the “open” conformation might lead to inactivation, whereas stabilization of the “closed” conformation might lead to activation of the protein.15−17
Figure 1.

Conformational comparison of STING apoprotein (“open” conformation, green, PDB 6MX0) with a STING:cGAMP complex (“closed”, white, from PDB 4KSY). Residues that become ordered upon cGAMP binding are shown as dark gray β-strands in the closed conformation and green dashes in the open conformation. cGAMP is shown as sticks in the center of the STING homodimer. Arrows indicate the direction of movement of the central α-2 helix upon cGAMP binding and the ordering of the central β-sheet.
With the knowledge of the ligand binding site, a druggability assessment was performed on the apoprotein to probe the feasibility of discovering a suitable ligand, which might preferentially bind to the “open” conformation.18 The total protein solvent accessible surface area (SASA) is calculated to be 390 Å2, which would require a ligand of ∼700 Da to occupy.19 Of the total SASA, 60% is polar SASA, which limits maximal binding affinity driven by hydrophobic interactions. Finally, the calculated volume of the ligand binding site is 952 Å3, suggesting the possibility for binding a large complex ligand. Unfortunately, physicochemical properties of ligands of this nature are not generally consistent with orally bioavailable drug-like small molecules.20 Another complexity in targeting the STING protein is the requirement for a small molecule antagonist to be competitive with the high molecular weight (674 Da), high affinity endogenous ligand (Kd = 4.6 nM), cGAMP.16 Together, these considerations highlight the challenges for identifying small molecule STING antagonists with the appropriate properties necessary for oral exposure.
The large ligand binding surfaces and volumes calculated for STING necessitated a unique approach. Our investigation was inspired by the recognition that two molecules of the mouse-specific STING agonist DMXAA are bound to a single mouse STING homodimer (Figure 2).15 This 2:1 binding ratio is possible due to the C2 symmetry of the STING protein, which allows each DMXAA molecule to interact with a single STING monomer. Our strategy to maximize binding efficiency was to exploit the intrinsic symmetry of the STING protein by identifying small molecules that can bind to the “open” conformation in a 2:1 ratio to the STING homodimer. Such a binding stoichiometry could afford fuller occupation of the large binding site, and thus effectively compete with cGAMP, while still maintaining physiochemical properties compatible with oral drugs.
Figure 2.

X-ray structure of DMXAA molecules bound to mouse STING in the “closed” conformation (PDB 4LOL).15 STING protein is colored green, and DMXAA is shown in blue sticks. Intermolecular interactions between DMXAA and Thr-262 and Thr-266 are indicated by black dashes. There is also a salt bridge between the DMXAA carboxylate group and Arg-237 (not shown; corresponds to Arg-238 in human STING).
Encouraged by the possibility of utilizing 2:1 binding stoichiometry to offset the challenge of a large binding pocket, we turned to our Automated Ligand Identification System (ALIS), which has emerged as a robust platform for hit identification.21 Compound 1 was initially identified as a low activity hit (IC50 = 7300 nM; Table 1).
Table 1. Profile of Screening Hit Compound 1.

Values in this table are determined by the HAQ STING cGAMP displacement assay and are the means from at least n = 2 experiments.
Values are calculated from pIC50 – ALogP98.
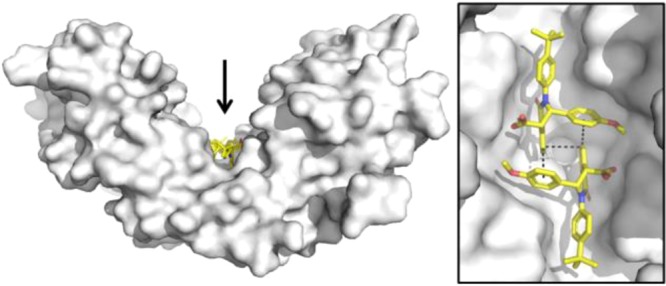
Crystallographic studies confirmed 2:1 binding. Not only do two molecules of 1 complement each other’s shape and bind to one dimer of STING, but they also bind to the “open” conformation (Figure 3) suggesting these molecules might function as an antagonist. Two interlocking copies of the compound are bound in the central cleft of the STING homodimer, and the absolute configuration is unambiguously assigned as (S, S).
Figure 3.
X-ray structure of 1 in STING protein showing 2:1 binding and ligand interactions (PDB 6MX3). Top: STING is shown as a white surface, with 1 colored yellow. The inset shows a close-up view of the two copies of 1 from the direction indicated by the arrow. Bottom: close-up of polar interactions between 1 and side chains of Thr-263 and Thr-267 of both STING chains. Side chain atoms of Tyr-167 are also shown as sticks. Dashed lines indicate selected van der Waals interactions and hydrogen bonds.
Compound 1 makes predominantly hydrophobic interactions, punctuated by several polar contacts. The carboxylate group forms a hydrogen bond with the side chain of Thr-263, and the exocyclic oxygen forms a hydrogen bond with the side chain of Thr-267. The methoxyphenyl group maintains van der Waals contact with the protein, and the para-tert-butyl group projects toward open solvent. There are a number of disordered residues that have been omitted from the crystallographic model, including many of which become ordered upon agonist binding. In addition, there are various interactions between the two molecules of 1 found in the ligand binding site. The aromatic ring of the isoquinolone not only engages in face-to-face π-stacking with the aromatic ring of the other copy of 1 but also engages in edge-to-face π-stacking with the methoxyphenyl group.22 This network of interlocking π-stacking interactions between the two copies of the molecules reinforces binding to the STING protein and is likely the source of observed cooperative binding (Supporting Information, Figure S1). The two monomers form a substantial dimer interface within the active site, burying approximately 274 Å2 of water accessible surface area and emphasizing the importance of ligand–ligand interactions in the observed binding mode.23
An advantage of the tetrahydroisoquinolone series is the flexibility of the synthesis, which allows for rapid diversity generation in a one-pot synthesis. From the initial hit, SAR was generated using the procedures highlighted in Scheme 1. In this initial approach, aniline (2), homopthalic anhydride (4), and benzaldehyde (3) were heated in a microwave reactor with cesium carbonate (Cs2CO3) and indium chloride (InCl3) as a catalyst. However, the yields were poor and the purification was complicated by mixtures of side products. By switching to the diacid (5) and removing InCl3, reactions proceeded more cleanly to desired products.
Scheme 1. Synthesis of Tetrahydroisoquinolone Analogs.
InCl3, Cs2CO3, MeCN, microwave 100 °C, 2–38%;
toluene, reflux, 31–81%.
Despite a clear view of the compound–protein interactions, early SAR highlighted the challenge of designing molecules with improved affinity. Table 2 lists a few examples to illustrate initial struggles to improve potency. While computational studies supported the replacement of the methoxyphenyl with various substitutions, these modifications did not lead to improvements in activity with the exception of the benzodioxane compound 7. Because these compounds are bound to STING as dimers, special consideration is required to rationalize the observed SAR. In addition to stabilizing the protein–ligand complex relative to the unbound state, the interaction between the bound monomers must also be considered. To this end, we redesigned our computational models to include an evaluation of dimer association energy using density functional theory (DFT) to estimate active site ligand–ligand complementarity in addition to docking studies to model protein–ligand interactions.24
Table 2. Initial Representative SAR on Tetrahydroisoquinoline Screening Hit.
Values in this table are determined by the HAQ STING cGAMP displacement assay and are the means from at least n = 2 experiments.
Consistent with previous observations regarding substituent effects on edge-to-face aromatic interactions, the predicted dimer association energies track with the electron richness of the accepting ring as well as steric interaction/repulsion across the dimer interface (see SI).25 For example, the electron-rich benzodioxyl 7 has a more favorable predicted dimer association energy, which may explain its improved binding activity, while naphthyl 8 and p-cyanophenyl 10, being more electron poor, are expected to exhibit less self-interaction, which may contribute to their weaker STING binding. Difluorobenzodioxole 12 is predicted to have a steric clash across the dimer interface where fluorine on one monomer is in close proximity to the carboxylate group of the other monomer.
Encouraged by these calculations, efforts were made to simultaneously design subtle variations of the aryl ring to enhance both ligand–ligand and protein–ligand interactions. Toward that end, various heteroatoms were incorporated to increase the ligand−π interactions. In order to compare the analogs as matched pairs, the compounds were synthesized as single enantiomers with the more potent chloro t-butyl phenyl solvent piece (Table 3). Although none of these analogs were dramatically differentiated from compound 13, the experimental results were consistent with our hypothesis that activity was dependent on both ligand and protein interactions when compared to Table 2.
Table 3. Benzodioxane Variants.
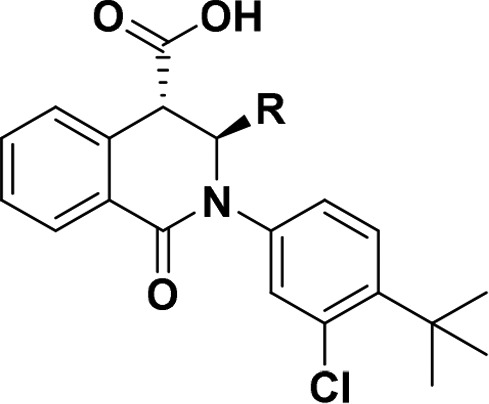
Values in this table are determined by the HAQ STING cGAMP displacement assay and are the means from at least n = 2 experiments.
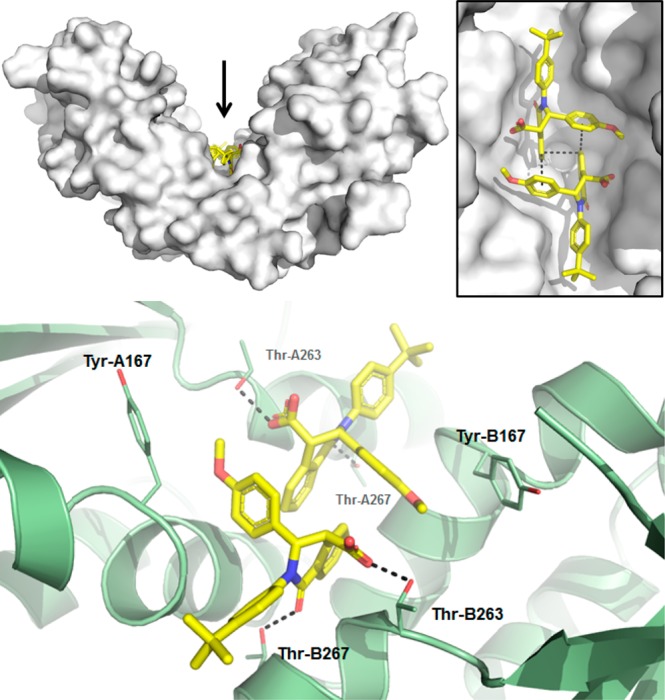
Although 13 exhibited potent activity in the cell-free ligand displacement assay, it displayed low oral bioavailability (%F = 5), which may be a consequence of the modest permeability (MDCK Papp = 9 × 10–6 cm/s).26 We postulated that an increased ionization of 13 negatively impacted the passive permeability. The pKa is calculated to be 3.4, lower than the typical value (pKa = 4–5) for an aliphatic carboxylic acid, likely due to an inductive effect from the aromatic isoquinolone core. Accordingly, extension of the carboxylic acid to the homologated acid (compound 18) preserved activity and increased the calculated pKa to 4.3. To rule out any unexpected conformational change induced by 18, crystallographic studies were conducted (Figure 4). As with 1, two molecules of extended acid 18 bind a single STING homodimer in the “open” conformation. The extended acid group and the lone pair of the isoquinonlone amide maintain the same interactions with Thr-263 and Thr-267, which are found in the identical conformation as in the complex with 1. Unique to this structure is the benzodioxane stacking with Tyr-167. The aromatic fluorine fills a pocket created by Gly-158 and Leu-159 at the bottom of the central binding groove, while the aromatic chlorine further fills the pocket created by Ile-165 and Ala-270.
Figure 4.

Cocrystal structure of 18 bound to STING protein (PDB 6MXE) showing interactions with Thr-263 and Thr-267, with side chain atoms depicted as sticks. As observed with 1, the compound binds in a 2:1 ratio to the STING homodimer and makes the same interactions, with additional van der Waals contact with the side chain phenol of Tyr-167. The homologated carboxylate group maintains a hydrogen bond interaction with Thr-263.
Consistent with our hypothesis, the increase in calculated pKa translated to a combination of improved permeability (Papp = 18 × 10–6 cm/s) as well as improved oral bioavailability (%F = 60; solubility and Clint remain constant, allowing for the interpretation that permeability is the main driver for improvement, Table 4). Overall, the pharmacokinetics properties for compound 18 are modest with high intrinsic clearance. Kinetic binding assessment of 13 and 18 by surface plasmon resonance (Biacore) revealed a slow koff (13, t1/2 = 35 min; 18, t1/2 = 62 min). Moreover, consistent with cooperative binding, data fitting required a two-step model (Figure S1A–C and Supporting Information). These observations may reflect an overall increase in stability when a second molecule binds to STING.
Table 4. In Vitro Profile of Lead STING Ligands.
Values in this table are determined by the HAQ STING cGAMP displacement assay and are the means from at least n = 2 experiments.
Values are calculated from pIC50 – ALogP98.
Values calculated from ACD LABORATORIES 11.0.
Dose; rat iv 0.5 mpk as a solution in PEG400/H2O (60:40 (v/v)), po: 1 mpk as a solution in PEG400/H2O (60:40 (v/v)).
Values in this table are determined by inhibiting IFNβ production in cGAMP-stimulated THP1 cells and are the means from at least n = 2 experiments.
Although we have identified molecules that are able to displace cGAMP and stabilize the “open” conformation of STING, a key assumption is that stabilizing the “open” conformation will prevent all STING signaling. To test this hypothesis, THP1 cells were incubated with 13 and 18, with and without cGAMP stimulation. The compounds did not stimulate IFNβ production (13 EC50 = >30000 nM; 18 EC50 = >30000 nM; Table 4), but modestly inhibited cGAMP-induced IFNβ production (13 IC50 = 11500 nM; 18 IC50 = 11000 nM; Table 4), with a >100-fold shift in potency from binding to the functional cell assay. Our observations are consistent with these compounds functioning as STING antagonists. Considering that the assay is stimulated with nonphysiological levels of cGAMP, the cellular IC50 may not be an accurate representation of antagonist potency.27
In conclusion, we have identified weak antagonists of STING-mediated signaling that binds to the cGAMP binding site in the inactive “open” conformation. By exploiting the natural symmetry of the STING protein and utilizing 2:1 binding stoichiometry, these compounds are able to fully occupy the binding pocket while mitigating the undesirable physicochemical properties associated with larger ligands. As a consequence of the 2:1 binding ratio, two-dimensional optimization of the protein–ligand and ligand–ligand interactions was necessary to improve potency. This approach led to the discovery of compound 18 with slow dissociation kinetics, good oral bioavailability, and ability to inhibit cGAMP-dependent signaling in vitro.
Acknowledgments
We thank J. Presland for helpful discussions and P. Schwartz for assistance with developing the kinetic model for Biacore fitting.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00466.
Synthetic procedures and analytical data of selected compounds, conditions for all biological assays, X-ray crystallographic methods and statistics, DFT methods, surface plasmon resonance and protein preparation, off-target profile (PDF)
Accession Codes
STING apoprotein, 6MX0; STING complex with 1, 6MX3; STING complex with 18, 6MXE.
The authors declare no competing financial interest.
Supplementary Material
References
- Ahlers L. R.; Goodman A. G. Nucleic acid sensing and innate immunity: signaling pathways controlling viral pathogenesis and autoimmunity. Curr. Clin Microbiol Rep 2016, 3 (3), 132–141. 10.1007/s40588-016-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Chen Z. J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014, 32, 461–88. 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Junt T.; Barchet W. Translating nucleic acid-sensing pathways into therapies. Nat. Rev. Immunol. 2015, 15 (9), 529–44. 10.1038/nri3875. [DOI] [PubMed] [Google Scholar]
- Sun L.; Wu J.; Du F.; Chen X.; Chen Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339 (6121), 786–91. 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.; Sun L.; Chen Z. J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17 (10), 1142–9. 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Ahn J.; Barber G. N. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr. Opin. Immunol. 2014, 31, 121–6. 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Ahn J.; Gutman D.; Saijo S.; Barber G. N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (47), 19386–91. 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J.; Ruiz P.; Barber G. N. Intrinsic self-DNA triggers inflammatory disease dependent on STING. J. Immunol. 2014, 193 (9), 4634–42. 10.4049/jimmunol.1401337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D.; Li T.; Li X. D.; Chen X.; Li Q. Z.; Wight-Carter M.; Chen Z. J. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (42), E5699–705. 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumore P. M.; Steinman C. R. Endogenous circulating DNA in systemic lupus erythematosus. Occurrence as multimeric complexes bound to histone. J. Clin. Invest. 1990, 86 (1), 69–74. 10.1172/JCI114716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G. I.; Rodero M. P.; Crow Y. J. Human disease phenotypes associated with mutations in TREX1. J. Clin. Immunol. 2015, 35 (3), 235–43. 10.1007/s10875-015-0147-3. [DOI] [PubMed] [Google Scholar]
- Konig N.; Fiehn C.; Wolf C.; Schuster M.; Cura Costa E.; Tungler V.; Alvarez H. A.; Chara O.; Engel K.; Goldbach-Mansky R.; Gunther C.; Lee-Kirsch M. A. Familial chilblain lupus due to a gain-of-function mutation in STING. Ann. Rheum. Dis. 2017, 76 (2), 468–472. 10.1136/annrheumdis-2016-209841. [DOI] [PubMed] [Google Scholar]
- Koch P. D.; Miller H. R.; Yu G.; Tallarico J. A.; Sorger P. K.; Wang Y.; Feng Y.; Thomas J. R.; Ross N. T.; Mitchison T. A High Content Screen in Macrophages Identifies Small Molecule Modulators of STING-IRF3 and NFkB Signaling. ACS Chem. Biol. 2018, 13 (4), 1066–1081. 10.1021/acschembio.7b01060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag S. M.; Gulen M. F.; Reymond L.; Gibelin A.; Abrami L.; Decout A.; Heymann M.; van der Goot F. G.; Turcatti G.; Behrendt R.; Ablasser A. Targeting STING with covalent small-molecule inhibitors. Nature 2018, 559 (7713), 269–273. 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- Gao P.; Ascano M.; Zillinger T.; Wang W.; Dai P.; Serganov A. A.; Gaffney B. L.; Shuman S.; Jones R. A.; Deng L.; Hartmann G.; Barchet W.; Tuschl T.; Patel D. J. Structure-function analysis of STING activation by c[G(2’,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 2013, 154 (4), 748–62. 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Shi H.; Wu J.; Zhang X.; Sun L.; Chen C.; Chen Z. J. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 2013, 51 (2), 226–35. 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanjulu J. M.; Pesiridis G. S.; Yang J.; Concha N.; Singhaus R.; Zhang S. Y.; Tran J. L.; Moore P.; Lehmann S.; Eberl H. C.; Muelbaier M.; Schneck J. L.; Clemens J.; Adam M.; Mehlmann J.; Romano J.; Morales A.; Kang J.; Leister L.; Graybill T. L.; Charnley A. K.; Ye G.; Nevins N.; Behnia K.; Wolf A. I.; Kasparcova V.; Nurse K.; Wang L.; Li Y.; Klein M.; Hopson C. B.; Guss J.; Bantscheff M.; Bergamini G.; Reilly M. A.; Lian Y.; Duffy K. J.; Adams J.; Foley K. P.; Gough P. J.; Marquis R. W.; Smothers J.; Hoos A.; Bertin J., Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 2018. 10.1038/s41586-018-0705-y [DOI] [PubMed] [Google Scholar]
- Le Guilloux V.; Schmidtke P.; Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinf. 2009, 10, 168. 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. C.; Coleman R. G.; Smyth K. T.; Cao Q.; Soulard P.; Caffrey D. R.; Salzberg A. C.; Huang E. S. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007, 25 (1), 71–5. 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46 (1–3), 3–26. 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Annis D. A.; Nickbarg E.; Yang X.; Ziebell M. R.; Whitehurst C. E. Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Curr. Opin. Chem. Biol. 2007, 11 (5), 518–26. 10.1016/j.cbpa.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Meyer E. A.; Castellano R. K.; Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem., Int. Ed. 2003, 42 (11), 1210–50. 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- Buried accessible surface area was calculated using MOE by taking the difference in accessible surface area between the dimer and two monomers. Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group ULC: Montreal, QC, Canada, 2018. [Google Scholar]
- See Supporting Information for methods and free energy calculations of analogs tested.
- Sinnokrot M. O.; Sherrill C. D. Substituent effects in pi-pi interactions: sandwich and T-shaped configurations. J. Am. Chem. Soc. 2004, 126 (24), 7690–7. 10.1021/ja049434a. [DOI] [PubMed] [Google Scholar]
- Irvine J. D.; Takahashi L.; Lockhart K.; Cheong J.; Tolan J. W.; Selick H. E.; Grove J. R. MDCK (Madin-Darby canine kidney) cells: A tool for membrane permeability screening. J. Pharm. Sci. 1999, 88 (1), 28–33. 10.1021/js9803205. [DOI] [PubMed] [Google Scholar]
- Cell viability has been accounted for in the THP1 assay; for list of off-target hits for 13, see Supporting Information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.