Abstract
The functional significance of the chemokine receptor CCR5 in human breast cancer (BCa) epithelial cells is poorly understood. Here we report that CCR5 expression in human breast cancer correlates with poor outcome. CCR5+ BCa epithelial cells formed mammospheres and initiated tumors with >60-fold greater efficiency in mice. Reintroduction of CCR5 expression into CCR5-negative BCa cells promoted tumor metastases and induced DNA repair gene expression and activity. CCR5 antagonists Maraviroc and Vicriviroc dramatically enhanced cell killing mediated by DNA-damaging chemotherapeutic agents. Single cell analysis revealed CCR5 governs PI3K/Akt, ribosomal biogenesis, and cell survival signaling. As CCR5 augments DNA repair and is re-expressed selectively on cancerous but not normal breast epithelial cells, CCR5 inhibitors may enhance the tumor-specific activities of DDR-based treatments, allowing a dose reduction of standard chemotherapy and radiation.
Keywords: CCR5, cancer stem cell, breast, DNA damage/Repair, Maraviroc, Vicriviroc
INTRODUCTION
In 2012 more than 521,000 women died from breast cancer worldwide (1), and more than 40,000 women in the United States are predicted to die from breast cancer in 2017(2). Relapses occur in 20–30% of patients and patients die primarily from metastatic breast cancer (3). The basal breast cancer genetic subtype is associated with increased risk of metastasis and reduced survival rates compared with either luminal A or B tumors (4,5). Recent studies of more than 2,000 patients demonstrated that the G protein-coupled receptor family (GPCR) member CCR5 is over-expressed in >50% of human breast cancer and in most basal breast cancers (6). In normal physiology, CCR5 is restricted to a subset of immune cells. Oncogenic transformation of immortalized human breast cancer cells with a single oncogene (either Ha-Ras, RAS, c-Myc, v-Src, or ErbB2) is sufficient for the induction of CCR5 expression (6). Interrogation of microarray databases of 2,254 human breast cancers demonstrated that CCL5/CCR5 signaling is activated primarily in the basal and Her2 breast cancer subtypes (6). The CCR5 ligand CCL5 (RANTES) also correlates with disease progression in patients with breast cancer (7,8) and additional ligands for CCR5 have been described, many of which are secreted from breast tumor stroma (9).
Highly specific CCR5 inhibitors (Maraviroc and Vicriviroc) were developed for treatment of HIV, which deploys CCR5 as a co-receptor for cellular entry (10). Theses small molecular inhibitors have undergone extensive testing of their specificity in modeling, mutagenesis, crystallography, and subsequent testing in tissue culture, in animals and in humans (11–15). Extensive use of Maraviroc in the clinic has demonstrated the drug is well tolerated, and does not compromise immune responses. The discovery that CCR5 is selectively re-expressed on the surface of tumor cells during the dedifferentiation and transformation process (6) has led to interest in targeting CCR5 for cancer therapy. An analysis of CCR5 protein levels and the function of CCR5 in breast cancer epithelial cells remained to be determined.
The cancer stem cell (CSCs) concept proposes that a sub-population at the top of the tumor cell hierarchy contributes to tumor heterogeneity and are uniquely capable of seeding new tumors (16,17). CSCs grow in spheres in nutrient poor medium, are capable of initiating tumors in mice and contribute to metastasis and therapy resistance (18,19). The mechanisms by which CSC survive chemotherapy- and radiotherapy-induced death are multifactorial and correlate with mechanisms protecting genomic integrity (20). The ability of CSCs to survive stressful conditions correlates with prompt activation of the DNA damage sensor and repair machinery. High dose radiation and alkylating agents induce single strand breaks that are repaired by the base excision repair (BER) process and double strand breaks that are repaired by homologous recombination repair (HRR) and by non-homologous end joining (NHEJ). The basic excision repair (BER) system targets small chemical alterations (base modifications) and includes PCNA and LIG3 (ligase 3 DNA ATP-dependent polymerase) (DNA-directed).
CCR5 mRNA is overexpressed in ~50% of human breast cancers and contributes to the homing component of the breast cancer metastasis process (6). CCR5 inhibitors dramatically reduced breast cancer metastasis in vivo (6). Given the importance of cancer stem cells to the metastatic cancer phenotype we hypothesized that CCR5 may contribute stem cell-like characteristics and potentially enhance DNA repair.
MATERIALS AND METHODS
Reagents and antibodies
CCL5 (Cat. 278-RN) and anti-CCR5 APC antibody (Cat. FAB1802A) were purchased from R&D Systems (Minneapolis, MN). The anti-vinculin rabbit polyclonal antibody (H-300, SC-5573) was from Santa Cruz Biotechnology. Anti-γH2AX (S139) (20E3, #9718) and anti-pAkt1 (S473) (D7F10, #9018) rabbit monoclonal antibodies were from Cell Signaling. The plasmids used in DNA repair reporter assay, includes DR-GFP, SA-GFP, NZ-GFP (pCAGGS-NZEGFP), I-SceI (pCAGGS-I-SceI, called pCβASce), and empty vector (pCAGGS-BSKX) were obtained from Dr. Jeremy M. Stark (21). Doxorubicin was obtained from Sigma. Vicriviroc and Maraviroc were obtained from Selleck Chemicals (Houston, TX). Luciferin was obtained from Gold Biotechnology (St. Louis, MI). GDC-0068 (Ipatasertib) was obtained from Selleck Chemicals. For in vitro treatments, Maraviroc was dissolved in DMSO and diluted in culture medium. The final concentration of DMSO in treated and control cultures was 0.5%. Vicriviroc was dissolved in culture medium.
Cell lines
HCC70, HCC1395, HCC1569, HCC1937, MDA-MB-175VII, MDA-MB-231, and MDA-MB-436 cell lines were obtained from ATCC (Manassus, VA). SUM149, SUM1315MO2, and SUM159 cell lines were kindly provided by Dr. Stephen Ethier (Wayne State University). FC-IBC-02 Cells was generated in Dr. Massimo Cristofanilli’s lab. HCC70, HCC1395, HCC1569, HCC1937, MDA-MB-231, MDA-MB-436, SUM149, SUM1315MO2, and SUM159 cell lines were obtained in the early 2000’s and cultured as described previously (22). All of them were genotyped (Genetica DNA Laboratories, Burlington, NC) within the past year to confirm identity and tested to ensure absence of mycoplasma contamination using PCR based assays. FC-IBC-02 cell line was certified by ATCC STR profile testing in August 2017. MDA-MB-175VII cell line was purchased recently. The early passages of the cells were stored. The cells thawed from low passage stocks were used within one month of the initial thaw. During the experiments, the morphology of all cell lines was checked under phase contrast microscope routinely. All of the newly revived cells were treated with BM-cyclins (Roche) and the mycoplasma contamination was determined with Hoechst 33258 staining under high magnification fluorescent microscope routinely.
Doxorubicin resistant breast cancer cell lines were derived through growth survival selection in Doxorubicin. SUM-159 cells were grown in 10 nM for 1 month, then 20 nm for 1 month, and then 40 nM for 3 weeks, prior to analysis. FC-IBC-02 cells were grown in 40 nm Doxorubicin for 1 month prior to analysis. MDA-MB-231 cells were grown in 20 nM Doxorubicin for 1 month then 40 nM Doxorubicin for 3 weeks prior to analysis.
Viral Cell Transduction
A lentiviral vector encoding firefly luciferase 2 (Luc2)-eGFP fusion protein was a generous gift from Dr. Gambhir (School of Medicine, Stanford University) (23). Lentivirus propagation was performed following the protocol described by Zahler at al. (24). Breast cancer cell lines were transduced at a MOI of 20 in the presence of 8 mg/ml polybrene (Sigma, St. Louis MO) for 24 h (23,24).
Fluorescence Activated Cell Sorting (FACS) Analysis
Cell labeling and FACS analysis for CCR5 and breast stem cell markers were based on prior publications (6,25) with minor modifications. Before labeling, the cells were blocked with normal mouse IgG (1/100) and purified rat anti-mouse Fcγ III/II receptor antibody (1/100) (Pharmingen, San Diego, California) for 30 min and then incubated with either allophycocyanin (APC)-labeled CCR5 antibody (R&D Systems) alone or combining with antibodies of PE conjugated anti-human CD24 (ML5, BD-Pharmingen), FITC conjugated anti-human CD44 (G44-26, BD-Pharmingen) and PE/Cy7 conjugated anti-human EpCAM (G8.8, Biolegend). All experiments were conducted at 4°C. Sample analysis was performed on either FACSCalibur or FACSCanto flow cytometer (BD Biosciences, San Jose, CA). The data were analyzed with FlowJo software (Tree Star, Inc., Ashland, OR).
Tumor formation assay
12-week-old Female NCr nu/nu (NCI, Bethesda, MD) mice received 4000 FACS-sorted CCR5+ or CCR5− cells suspended in 50 µL of Dulbecco PBS lacking calcium and magnesium (DPBS) and 50 µL of BD Matrigel Basement Membrane Matrix (BD Biosciences) by subcutaneous injection at one dorsal flank. The injection was performed using 27.5-gauge needle. Tumor progression was followed by measurement of bioluminescence once a week until tumor excision, using the IVIS LUMINA XR system (Caliper Life Sciences). Briefly, for in vivo imaging, mice received the substrate of luciferase, d-Luciferin (Gold Biotechnology), at 15 mg/mL in PBS by intraperitoneal injection of 10 µL of luciferin stock solution per gram of body weight (manufacturer's recommendation) and were anesthetized by exposure to 3% isoflurane. At 10 to 15 minutes after d-luciferin injection, animals were placed inside the camera box of the IVIS Lumina XR and received continuous exposure to 2.5% isoflurane. Imaging time ranged from 5 minutes (for earlier time points) to 5 seconds (for later time points), depending on the bioluminescence of neoplastic lesion. Regions of interest (ROI) from displayed images were drawn around the tumor sites and quantified using the Living Image 3.0 software (Caliper Life Sciences). Tumor samples were harvested after 4 months. For the tumor formation by CCR5 and control vector stable transfected cells, 1 × 106 cells were subcutaneously injected into the mice and the tumor samples were harvested after 6 weeks. Before injection, the viability of the cells was checked by trypan-blue staining. All experiments involving mice were carried out under the approval of Institutional Animal Care and Use Committee (IACUC) of the Thomas Jefferson University (Philadelphia, PA).
Experimental Metastasis Assay and Bioluminescence Imaging
SUM-159 cells expressing Luc2-eGFP (called SUM-159.pFLUG for the rest of the paper) were detached with a non-enzymatic cell dissociation buffer (4 mM EDTA in Ca2+ and Mg2+-free PBS), resuspended in Dulbecco’s PBS without Ca2+ and Mg2+ and immediately injected intracardiac to 8-week old, female NOD/SCID mice (NCI, Bethesda, MD). Each mouse received 2 × 105 cells. Mice were treated by oral gavage with Maraviroc (8 mg/kg every 12 h) or vehicle (5% DMSO in acidified water) (26). Treatment was started immediately after injection. For in vivo bioluminescence imaging (BLI), mice were given an intraperitoneal (i.p.) injection with 200 µL of D-luciferin (30 mg/mL). Mice were anesthetized with isoflurane (2% in 1 L/min oxygen), and bioluminescence images were acquired 4–5 min after D-luciferin injection using the IVIS XR system (Caliper Life Sciences, Hopkinton MA). Acquisition times ranged from 10 s (for later time points) to 5 min (for early time points). Data are expressed as total photon flux and were analyzed using Living Image 3.0 software (Caliper Life Sciences). Animal experiments were approved by the Thomas Jefferson University’s Institutional Animal Care and Use Committee (IACUC).
Immunohistochemistry and Survival Analysis for CCR5
Quantitative immunofluorescence-based immunohistochemistry and survival analysis for CCR5 were performed as previously described (27,28) on tumors from a cohort of patients with node-negative breast cancer (27). Briefly, after deparaffinization and rehydration, antigen retrieval was performed by microwave treatment in citrate buffer (pH 9; DAKO). Sections were blocked with 10% goat serum and followed by incubation of primary anti-CCR5 (Abcam) at a dilution of 1:200 for 30 min. Sections were then washed thrice with TBS and subsequently incubated with anti-pan-cytokeratin antibody (Dako, Cat#AE1/AE3) for 1 h. Bound CCR5 antibody was detected using an anti-rabbit horseradish peroxidase–conjugated secondary antibody (DAKO EnVision-Plus), followed by incubation with Tyramide-Cy5 (Perkin-Elmer). Cytokeratin was visualized by further incubating the sections with an anti-mouse secondary antibody conjugated to Alexa 555 (Molecular Probes). Finally, all sections were stained with 4,6-diamidino-2-phenylindole (DAPI; Vector) for nuclear visualization. Slides were imaged on an Aperio Scanscope FL and quantitative expression levels were determined using Tissue Studio (Definiens) image analysis software. Analysis of overall survival was conducted using Xtile (28) to establish data-driven, optimal cutpoint for dichotomization (High vs. Low) of CCR5 levels in the cohort. SPSS software was used to evaluate the differences between patients with High vs. Low CCR5 levels using the Kaplan-Meier estimator of the survival curves and log-rank test, and Cox regression was used for multivariable analyses.
Drug Screens
We screened drug response to the CCR5 inhibitors alone and in combination using breast cancer cell lines as described previously (29,30). Briefly, cells were plated into 96-well plates and treated with CCR5 inhibitor (either Maraviroc or Vicriviroc), Doxorubicin, or at 1:1 molar ratio of the two drugs as described previously (29). Briefly, we prepared drug treatment plates that were randomized to minimize plate edge effects. Each drug was assessed at nine different concentrations that varied by two-fold, in triplicate. Cells were plated, allowed to adhere overnight, then treated with drug for 72 h. A measurement of cell number was made at both the time of treatment (time 0) and after drug treatment (time 72) using CTG reagent (Promega, Madison, WI) to allow for calculation of percent growth inhibition and the dose required to inhibit growth rate by 50% (GR50), as described recently (31). We used the online GR50 calculator tool for all GR50 calculations (see:http://www.grcalculator.org/grcalculator/).
DNA repair assays
The DNA repair reporter assays were previously described (21,32). The DR-GFP expression plasmid is repaired by the homology-directed repair (HDR) pathway. With DR-GFP, an I-SceI-induced DSB in the upstream SceGFP cassette, followed by HDR that uses the downstream homologous template (iGFP) to prime nascent DNA synthesis, restores the GFP+ cassette co-introduced with I-SceI. The number of GFP positive cells is determined. The repair of single strand breaks (single strand annealing) was assayed with the SA-GFP reporter. The SA-GFP reporter contains a GFP fragment separated 2.7 kilobases (kb) from a GFP fragment that contains an I-SceI recognition site (33). The two GFP fragments share 266 nt of homology that can bridge the I-SceI-induced DSB during SSA, thereby restoring a functional GFP+ cassette. NZ-GFP, a plasmid encoding stable expressed GFP, was used as transfection efficiency control. The DNA repair activity was showed as (RI-SceI-RpCAGGS)/RNZ-GFP. RI-SceI, RpCAGGS and RNZ-GFP represent the ratio of GFP positive cells in I-SceI, pCAGGS-BSKX and NZ-GFP transfected cells repectively.
Single-cell RNA-seq
CCR5+ and CCR5− cells were isolated by FACS sorting as described above. The single-cell RNA-seq libraries were constructed with the REPLI-g single-cell RNA library kit (Qiagen, USA). All single-cell libraries were sequenced on an IlluminaHiSeq 2000 platform (Illumina, USA). The raw reads generated were filtered according to sequencing quality and with regard to adaptor contamination and duplicated reads. Thus, only high-quality reads remained and were used in the genome assembly. The RNA-seq data were analyzed with Partek Flow version 4 (Partek Inc., USA). Bases with Phred score less than 20 were trimmed from both ends of the raw sequencing reads, and trimmed reads shorter than 25 nt were excluded from downstream analyses. Both pre- and post-alignment quality assessment and quality control was carried out with default settings as part of Flow workflow. Trimmed reads were mapped onto human genome hg38 using Tophat 2.0.8 as implemented in Flow with default settings, and using Gencode 20 annotation as guidance. The Gencode 20 annotation (www.gencodegenes.org) was used to quantify aligned reads to genes/transcripts using the method of Partek (34). Read counts per gene in all samples were normalized using Upper Quartile normalization (35) and analyzed for differential expression using Partek’s Gene Specific Analysis method (genes with less than 10 reads in any sample were excluded). To generate significantly differentially expressed genes among all samples, a cutoff of FDR was adjusted to p<0.05 (Poisson regression) with a >2-fold change applied. Principle component analysis of gene expression on all single-cells was performed with the Partek package. Pathway analysis was performed using the Ingenuity pathway analysis package (Qiagen Bioinformatics, USA).
RESULTS
CCR5+ breast cancer cells form mammospheres
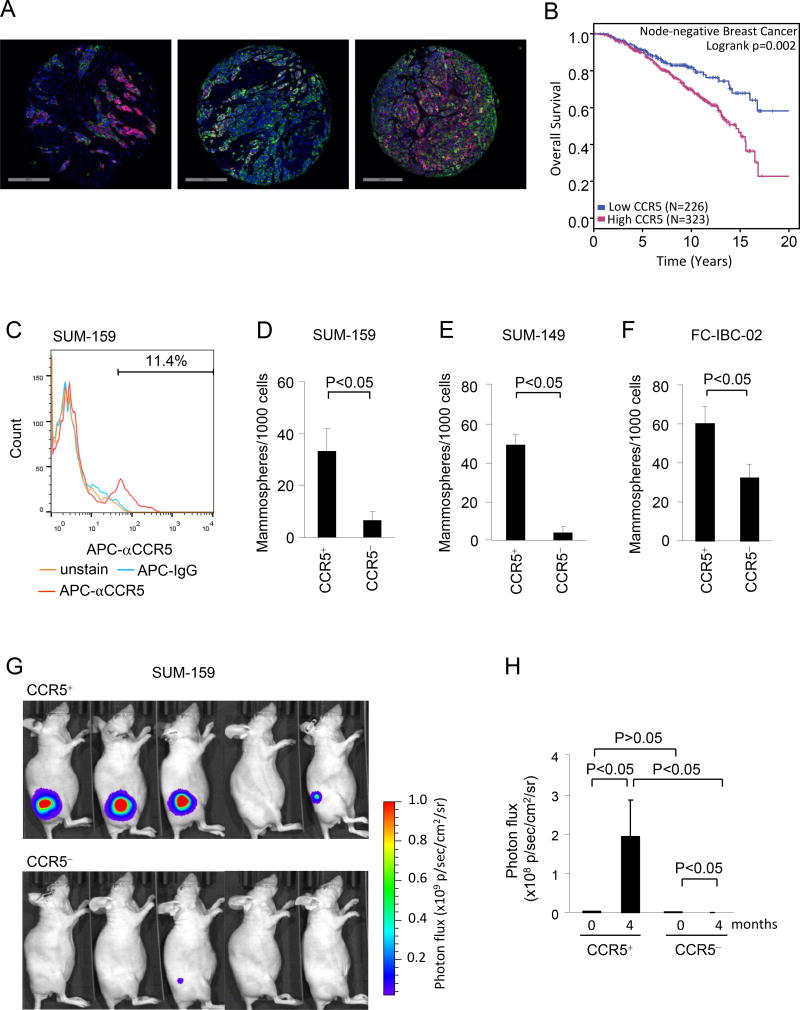
We had previously shown that CCR5 expression in human breast cancer is associated with increased metastatic progression and more aggressive disease (6). In order to extend these studies, we determined CCR5 protein IHC staining in 549 human breast cancers. CCR5 staining was heterogeneous within individual human breast cancer specimens as shown in three representative cases of breast adenocarcinomas immunostained for CCR5 (red), pan-Cytokeratin (green), and cell nuclei (DAPI; blue). Both cell-to-cell and region-to-region variability of CCR5 expression was observed within each tumor specimen (Figure 1A, Supplemental Figure 1). Consistent with a prometastatic role of CCR5 in breast cancer, node-negative patients whose tumors expressed the highest levels of CCR5 protein were at increased risk of death (Figure 1B). The patient population demographics are shown in Supplemental Table 1). High CCR5 remained an independent marker for unfavorable outcome in node-negative breast cancer patients after multivariable adjustment for patient demographic and pathological tumor features, including menopausal status and race, tumor grade and size, and pathological ERα, PR and Her2 status (Supplemental Table 2).
Figure 1. The CCR5+ population of SUM-159 cells is enriched with tumor initiating cells.
(A) Three different cases of breast adenocarcinomas immunostained for CCR5 (red), pan-Cytokeratin (green), and cell nuclei (DAPI; blue). Note cell-to-cell and region-to-region variability of CCR5 expression within carcinoma cells. (B) Kaplan-Meier plots of survival for high cytoplasmic CCR5 vs. low cytoplasmic CCR5. (C) Representative example of SUM-159 cell FACS analysis by CCR5 staining. (D-F) Mammosphere assays conducted with equal number of CCR5+ vs. CCR5− cells selected by FACS from SUM-159 (D), SUM-149 (E), or FC-IBC-02 cells (F). The mean number of mammospheres formed per 1000 cells are shown ±SEM for N=4. (G) Photos of photon flux from breast tumors in nude mice derived from injection of CCR5+ vs. CCR5− luc2 stable SUM-159 breast cancer cells. An equal number of cells were injected into each animal. (H) Quantitation of photon-flux of tumors from mice at time 0 months and 4 months shown as mean ± SEM for N=5 separate mice in linear scale.
Only a small subpopulation of cells within a breast tumor initiates tumor formation in mice. These tumor initiating cells correlate with increased propensity to metastasize (36). The ability of the cells to grow as a sphere under specific culture conditions has been shown to represent a propensity towards progenitor cell expansion and correlates with both tumor initiating ability and metastatic capacity (36). Mammospheres thus reflect the relative propensity for progenitor cell formation (19,37,38). In order to determine the role of CCR5+ cells within the heterogeneous tumors to form mammospheres, the basal breast cancer cell lines including SUM-159, SUM-149 and FC-IBC-02 were assessed. Consistent with the known heterogeneity of breast cancers, fluorescence activated cell sorting (FACS) identified a subset of CCR5 positive cells (about 1–10%) within the SUM-159 cell line (Figure 1C) as well as two other cell lines (SUM-149 and FC-IBC-02, Supplemental figure 2A). In order to examine the contribution of CCR5 to the formation of mammospheres, FACS sorting was conducted and equal number of the CCR5+ vs. CCR5− were assessed. The relative number of mammospheres was increased 5-fold in SUM-159, 12-fold in SUM-149 and 2-fold in FC-IBC-02 comparing the CCR5+ with CCR5− cells (Figure 1D, E and F. P<0.05 for all of cell lines) with representative morphology of CCR5− vs CCR5+ SUM-159 cell mammospheres shown in Supplemental Figure 2B. Both SUM149 and FC-IBC-02 cell lines were derived as the model of inflammatory breast (IBC). IBC has a high capacity to spread early with significant risk of disease recurrence and lower survival rates. The CCR5+ population of both IBC cell lines showed enhanced mammosphere formation.
Cell surface markers have been defined as an additional characteristic of cancer stem cells with enrichment of EpCAM+CD24−CD44+ correlating with stem cell characteristics (18,19). We therefore conducted CCR5-based FACS sorting of breast cancer cells, and subsequently examined the relative distribution of the EpCAM+CD24−CD44+ cell surface markers in the CCR5+ vs. CCR5− cells. In SUM-159 cells there was an approximately 20-fold increase in the relative proportion of EpCAM+CD24−CD44+ (Supplemental Figure 2C, 20.5% vs. 0.91%).
CCR5+ breast cancer cells show enhanced ability to initiate tumors in vivo
Breast cancers are thought to contain stem–like cells that contribute to tumor initiation and metastasis (36). In order to define the tumor initiating propensity of CCR5+ breast cancer cells, SUM-159 breast cancer cells stable expressing luciferase 2 (Luc2) were FACS sorted into CCR5+ vs. CCR5− populations based on APC labeled CCR5 staining. An equal number of CCR5+ or CCR5− cells were injected subcutaneously into the lower flank region of nude mice and the tumor formation was monitored with an in vivo bioluminescence imaging system (IVIS) (Figure 1G). The tumor volume is shown as photon flux of Luc2 labeled breast cancer cells. The CCR5+ subpopulation of SUM-159 cells developed substantial tumors, increasing 60-fold over 4 months (194 × 106 vs. 3.25 × 106, p<0.05, Figure 1H, Supplemental Figure 3A and B). In contrast, the CCR5− population declined in size in the same period resulting in a 770-fold difference in tumor volume assessed by photon flux at 4 months (Figure 1H, Supplemental Figure 3A and B). These studies are consistent with an important role for CCR5+ cells in the process of tumor initiation. In the animal in which CCR5− cells were injected subcutaneously into the lower flank region of nude mice and a tiny yet detectable tumor remained, immunohistochemical staining for CCR5 identified detectable heterogeneous staining CCR5, which may reflect either some contamination in the FACS sorting, or re-expression of CCR5 (Supplemental Figure 3C).
CCR5 antagonists block metastases of basal breast cancer in vivo
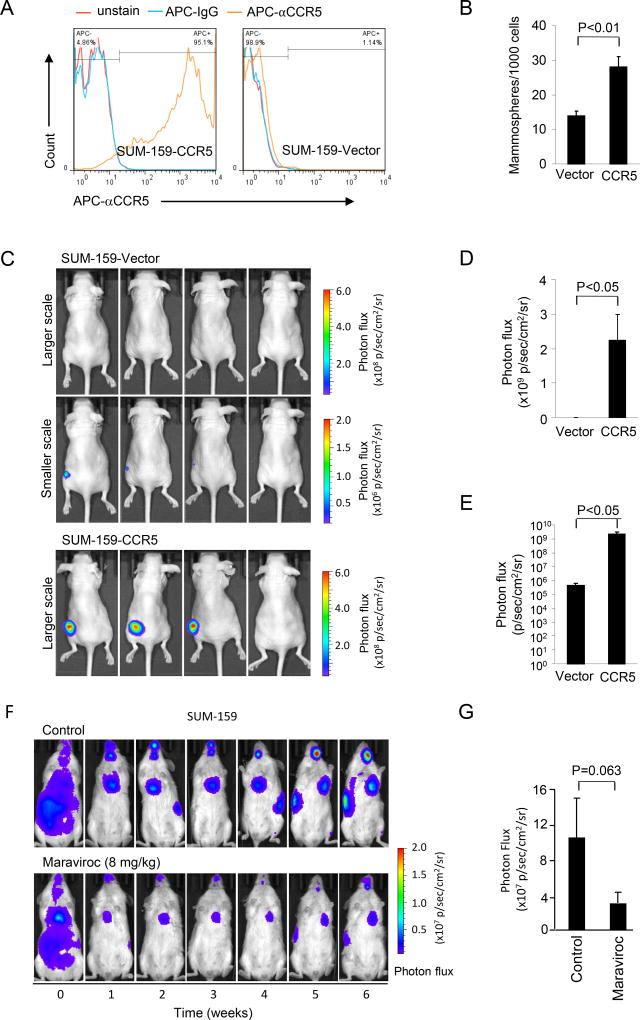
The SUM-159 cells were stably transfected with an expression vector encoding CCR5 or an empty control vector (Figure 2A). CCR5 expression increased mammosphere formation by 2-fold (Figure 2B). However, there was a more modest (23%) but significant increase in proliferation between CCR5 expressing and control vector transfected SUM-159 cells (Supplemental Figure 4A, P<0.001 at 96 hrs). An equal number of CCR5 expressing SUM-159 or its vector control cells were subcutaneously injected into the mice and tumor growth was examined over 6 weeks (Figure 2C, Supplemental Figure 4B). The mean size of tumor volume was determined using photon flux and expressed on a linear (Figure 2D) and a log scale (Figure 2E). The size of tumors was enhanced 10000-fold by CCR5 expression (Figure 2E). Together, these studies demonstrated both endogenous CCR5, and overexpression of CCR5 in breast cancer cells, is sufficient for the induction of basal breast cancer cellular tumor formation in vivo. Immunohistochemical staining of the tumors for CCR5 identified relatively homogeneous high-level expression of CCR5 in the SUM-159 cells stably transfected with the CCR5 expression vector, and minor heterogeneous staining for CCR5 in the empty control stable line tumors (Supplemental Figure 4C).
Figure 2. CCR5 overexpression in SUM-159 breast cancer cells generates breast tumors in mice and CCR5 antagonists block breast cancer metastasis in mice.
(A, B) Luc2-GFP SUM-159 cells stably transfected with either a CCR5 expression vector or control vector were analyzed by FACS in which red is unstained, blue is control IgG and orange is APC-CCR5 antibody (A) and then mammosphere formation assays were conducted in (B). CCR5 expressing cells showed 2-fold increased mammosphere formation. (C–E) Equal number of cells was injected subcutaneously into lower flank region of the nude mice and tumor size was determined by photon flux. Individual mouse tumors are shown as representative photon emission images at 6 weeks (C). Size of tumors for CCR5 re-expressing or vector control animals during five week shown as mean ± SEM of photon flux (×109 photon/sec/cm2/sr) for N=4. Note shown as either linear (D) or log scale (E) of mean tumor volume detected by photon image is significantly greater for CCR5+ vs. CCR5 vector control (2.24 × 109±0.75 × 109 vs. 4.63 × 105±1.49 × 105, p<0.05 with student t-test). (F) Representative timed photon emission of mice injected with SUM-159 cells treated with either vehicle control or CCR5 antagonist Maraviroc (8 mg/kg) for six weeks. Colorimetric scale of photon flux (×107 p/sec/cm2/sr) reflects tumor volume. (G) The size of tumors defined by photon flux for 6 animals in control group and 7 animals in the Maraviroc treated group shown at 5 weeks. Maraviroc treatment reduces mean lung tumor volume by 67% assessed by photon flux [(3.01 ± 1.16) × 107 vs. (10.05 ± 4.4) × 107, p = 0.063 with Mann-Whitney Test].
The CCR5 antagonist Maraviroc was previously approved by the US Food and Drug Administration (FDA) for the use in treatment-naïve adults with CCR5-trophic HIV. In order to determine the role of endogenous CCR5 in metastases, Luc2-expressing SUM-159 cells were introduced into NOD/SCID mice via intracardiac injection, and tumor volume was characterized by fluorescence of the cells using the IVIS system (Figure 2F). Animals were treated with the bioequivalent dose of Maraviroc that had been approved as safe and used in humans for treatment of HIV. Metastases quantified with photon flux, demonstrated a >65% decrease in breast cancer metastases in the Maraviroc-treated group compared to the control group (Figure 2F,G) (p=0.063).
CCR5 promotes DNA repair
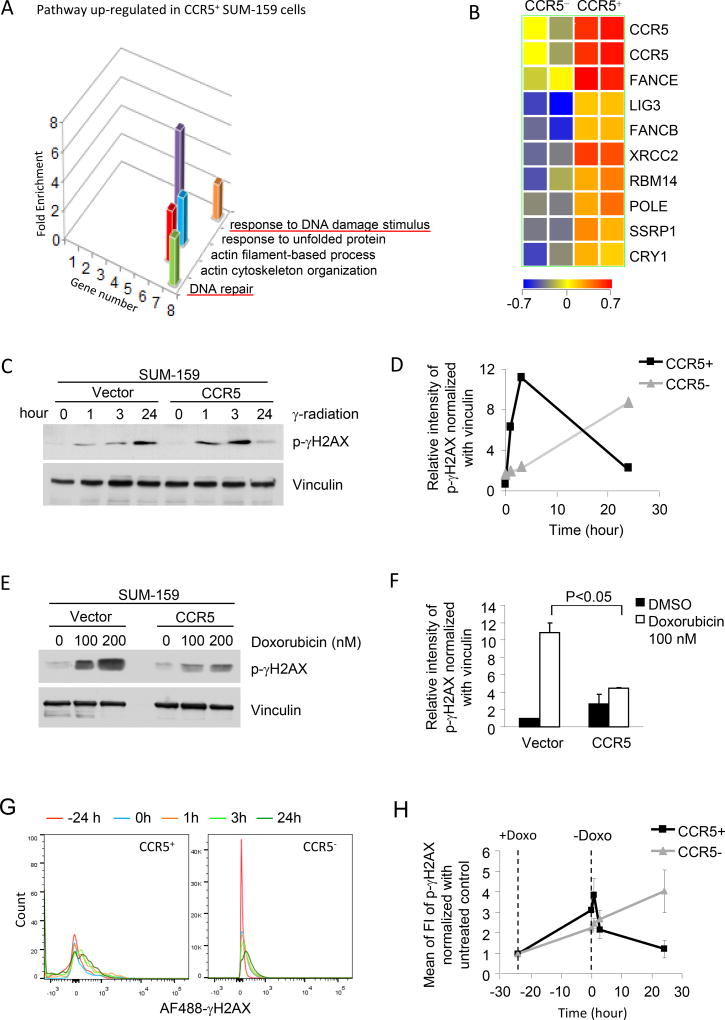
In order to characterize the functional pathways regulated by CCR5 within the SUM-159 basal breast cancer cells, both CCR5+ and CCR5− cells were separated by FACS sorting and subjected to microarray mRNA analysis (Supplemental Figure 5A). The “Gene Ontology” pathway analysis identified a subset of pathways enriched in CCR5+ breast cancer cells, including pathways involved in “DNA repair” and “response to DNA damage stimulus” (Figure 3A). The “DNA repair” related genes involved members of base excision repair (BER) and recombination repair (HR and NHEJ) (Figure 3B, Supplemental Figure 5B,C).
Figure 3. CCR5 increases repair of damaged DNA in SUM-159 breast cancer cells.
(A) Microarray gene expression was analyzed in CCR5+ vs. CCR5− cells separated from SUM-159 breast cancer cells by FACS sorting. Gene-Ontology pathway analysis demonstrates pathways regulated in CCR5+ vs. CCR5− cells. The “response to DNA damage stimulus” and “DNA repair” pathways are shown with number of genes and enrichment score (ES). Additional pathways include “response to unfolded proteins, “actin filament based process” and “actin cytoskeleton organization. (B) Heat map display of gene expression from the DNA damage repair signaling pathways. (C) SUM-159 cells stably expressing CCR5 or control vector were treated with γ-radiation (6.5 Gy). The samples were collected at 1, 3 and 24 hours after γ-radiation. (D) The kinetics of induction and subsequent reduction of phospho-γH2AX abundance was faster in CCR5-stable transfected cells than vector-control cells shown from quantitation of Western blotting. Data are representative of 3 separate experiments. (E) The CCR5 stably-transfected and vector-control SUM-159 cells were treated with the DNA damage inducing breast cancer therapeutic agent Doxorubicin for 7 days and analyzed by Western blot. DNA damage, shown by the abundance of phospho-γH2AX, was reduced in CCR5-transfected cells. (F) The relative intensity of γH2AX is shown as mean ± SEM of 3 separate experiments. (G, H) FACS analysis of phospho-γH2AX in CCR5+ and CCR5− SUM-159 cells. After Doxorubicin treatment, the kinetics of induction and subsequent reduction of phospho-γH2AX abundance was faster in CCR5+ cell than in CCR5− cells.
In view of the finding that CCR5+ cells were enriched for expression of genes involved in DNA repair, we examined the functional significance of CCR5 in response to DNA damage inducing agents that are used in treatment of breast cancer patients (γ-radiation and Doxorubicin). Histone H2AX phosphorylation at Serine 139 (γH2AX) recruits proteins that either sense or signal the presence of DNA damage and can be used as surrogate marker of DNA damage/repair. SUM-159 cells, either expressing CCR5 or a control vector, were compared for the DNA damage response. γ-irradiation of SUM-159 cells induced γH2AX, however CCR5 enriched cells showed reduced γH2AX at 24 hours, consistent with increased DNA repair (Figure 3C,D). Similar observations were made in MDA-MB-231 cells in which MDA-MB-231 cells stably expressing a CCR5 expression vector showed reduced γH2AX staining at 24 hrs after either γ irradiation or after Doxorubicin release (Supplemental Figure 6A,B).
The DNA intercalating anthracycline, Doxorubicin, is used for the treatment of human breast cancer. Treatment of SUM-159 cells with doxorubicin induced γH2AX phosphorylation at 100 and 200 nM after 24 hours, however CCR5 enriched cells showed reduced γH2AX when normalized to the protein loading control vinculin (Figure 3E,F).
In order to examine the DNA damage response of the CCR5+ cells to DNA damage within the heterogeneous tumor environment, Doxorubicin treatment was given at -24 hrs and removed at time 0 hrs (Figure 3G,H). As SUM-159 cells contain a heterogeneous population of CCR5+ and CCR5− cells, FACS sorting was conducted, and the two populations were examined for the relative abundance of γH2AX after treatment with Doxorubicin (Figure 3G,H). The relative abundance of γH2AX was enhanced in the CCR5+ cells after treatment with Doxorubicin (Figure 3H), which rapidly declined over the subsequent 24 hrs compared with CCR5− cells (Figure 3H).
CCR5 induces repair of double strand and single strand DNA damage
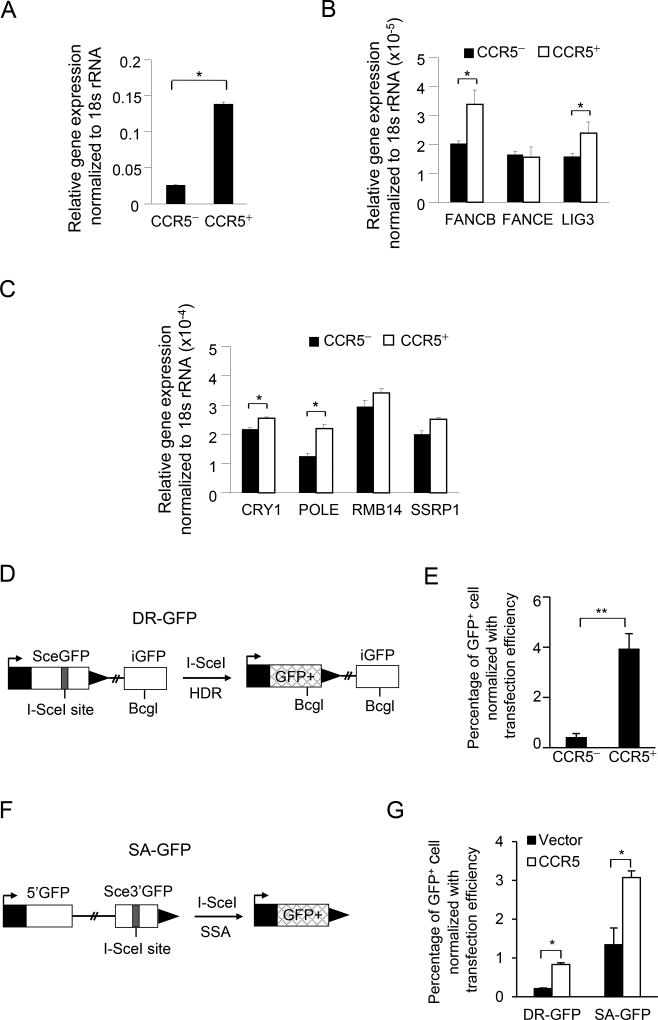
As microarray based gene expression had demonstrated CCR5+ cells were enriched for expression of pathways mediating DNA repair, we examined the levels of gene expression and assessed DNA repair activity mediated by CCR5 using surrogate reporter gene assays. We conducted further analyses of CCR5-mediated DNA repair by comparing CCR5+ vs. CCR5− cells after FACS separation. Quantitative RT-PCR analysis demonstrated the induction in the relative abundance of several genes that contribute to the repair of homologous DNA repair (HDR) (FANCB) base excision repair (BER) (LIG3, POLE) and nucleotide excision repair (NER) (CRY1) (Fig. 4A–C) (Supplemental Figure 7).
Figure 4. CCR5 enhances both homology-directed repair (HDR) and single strand annealing (SSA) DNA repair.
(A–C) Gene expression was determined from cells (CCR5+ vs. CCR5− cells derived by FACS sorting, Supplemental Figure 5) using Q-RT-PCR. The relative abundance of the transcripts participating in DNA damage/repair are shown as mean ± SEM for N=4. (D) Schematic representation of the DNA repair reporter (DR-GFP) for HDR. (E) HDR activity was increased in CCR5+ SUM-159 cells. The cells were co-transfected with the plasmid encoding I-SceI and the I-SceI based DNA repair reporter DR-GFP or (F), SA-GFP, SSA and stained with APC labeled anti-CCR5 antibody. GFP+ cells, generated by HDR of I-SceI induced double-strand DNA, were sorted by FACS into CCR5− and CCR5+ populations (Supplemental Fig. 8). The percentage of DR-GFP+ cells or SA-GFP positive cells were calculated and normalized with the transfection efficiency control (NZ-GFP). (G) The percentage of DR-GFP+ and SA-GFP+ cells was increased in CCR5 expressing cells compared with vector control cells.
In order to examine the effects of CCR5 on the DNA repair process, a DNA repair reporter assay (21) was deployed. The DR-GFP expression plasmid is repaired by the homology-directed repair (HDR) pathway. With DR-GFP, an I-SceI-induced double strand break (DSB) in the upstream SceGFP cassette, is followed by HDR that uses the downstream homologous template (iGFP) to prime nascent DNA synthesis and restores the GFP+ cassette (Figure 4D), when the plasmid is co-introduced with I-SceI into cultured cells. The number of GFP positive cells was determined. By FACS sorting for CCR5+ vs. CCR5−, we determined the role of CCR5 to HDR activity using the repair reporter assays (Supplemental Figure 8A). CCR5+ cells, reflecting endogenous CCR5, showed a 9-fold greater activity of DR-GFP (Figure 4E).
An additional mechanism for repairing double stranded DNA breaks (DSBs) induced by cytotoxic lesions, involves single strand annealing (SSA) which can be assayed with the SA-GFP reporter (Figure 4F). The SA-GFP reporter contains a GFP fragment separated 2.7 kilobases (kb) from a GFP fragment that contains an I-SceI recognition site (33). The two GFP fragments share 266 nt of homology that can bridge the I-SceI-induced DSB during SSA, thereby restoring a functional GFP+ cassette. We assayed the role of CCR5 in DNA repair using cells stably expressing CCR5 vs. vector control (Supplemental Figure 8B). CCR5 expression enhanced DR-GFP activity 4-fold (Figure 4E, N=3, P<0.048) and SA-GFP was enhanced 2-fold (Figure 4G, N=5, p<0.031). Thus the CCR5 enriched cells augment ability to repair double stranded DNA breaks (DSBs), which can be induced by cytotoxic lesions.
CCR5 antagonists enhance cell killing by DNA damage inducing chemotherapy agents used for breast cancer treatment
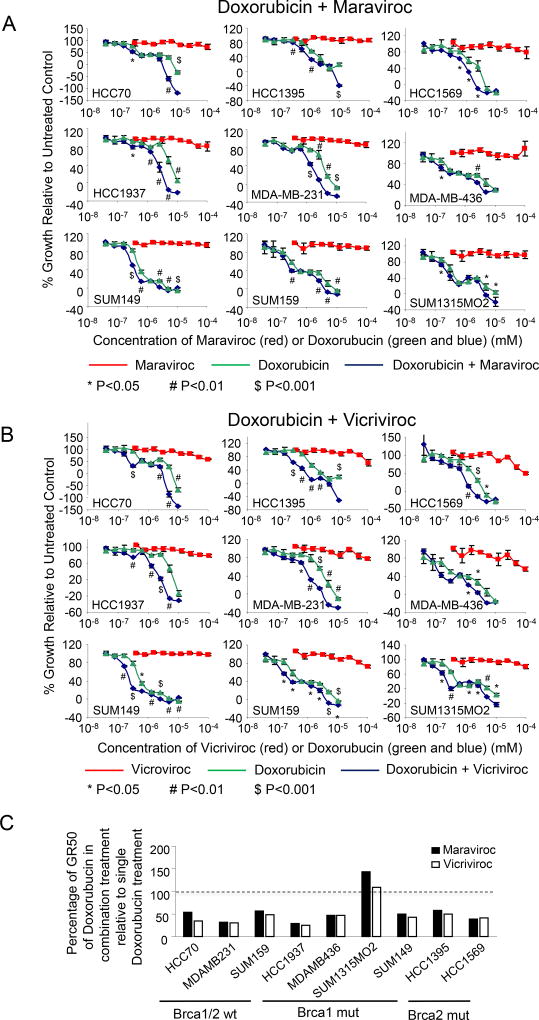
We reasoned that CCR5 inhibitors might sensitize cells to DNA damaging agents, allowing for chemotherapy dose reduction to reduce peripheral toxicity. To test this hypothesis, we treated nine different breast cancer cell lines with either Maraviroc (Figure 5A) or Vicroviroc (Figure 5B) in combination with Doxorubicin, an intercalating agent that disrupts topoisomerase II that causes DNA damage. Neither Vicroviroc nor Maraviroc caused significant cytotoxicity. Doxorubicin significantly reduced cell viability, producing GR50 values ranging from 0.4–6 µM for the five cell lines (Figure 5C). The addition of either Maraviroc and Vicroviroc to Doxorubicin resulted in substantially decreased cell viability as measured by GR50 value estimates, compared to the same dose of Doxorubicin alone in each cell line, except SUM1315MO2 (Figure 5C, Supplemental Table 3), shown by colorimetric scale for synergy of cell killing in Supplemental Figure 9 A,B. CCR5 inhibitor addition increased the GR50 of Doxorubicin-mediated cell killing by up to 4-fold.
Figure 5. CCR5 inhibitors enhance the cell killing of DNA damage inducing chemotherapy agents.
(A, B) Dose-response curves for the breast cancer cell lines treated with CCR5 inhibitors (Maraviroc (A) or Vicriviroc (B)), Doxorubicin, or a combination of CCR5 inhibitor plus doxorubicin. The combination treatment is plotted relative to the dose of doxorubicin used (CCR5 inhibitor concentration was 10 × higher than Doxorubicin). Data are shown as mean ± SEM for N= 3. (C) Percentage of GR50 with doxorubicin for CCR5 antagonist and Doxorubicin combined treatment relative to single doxorubicin treatment.
Single cell sequencing reveals volatility of gene expression in the CCR5+ breast cancer cellular population
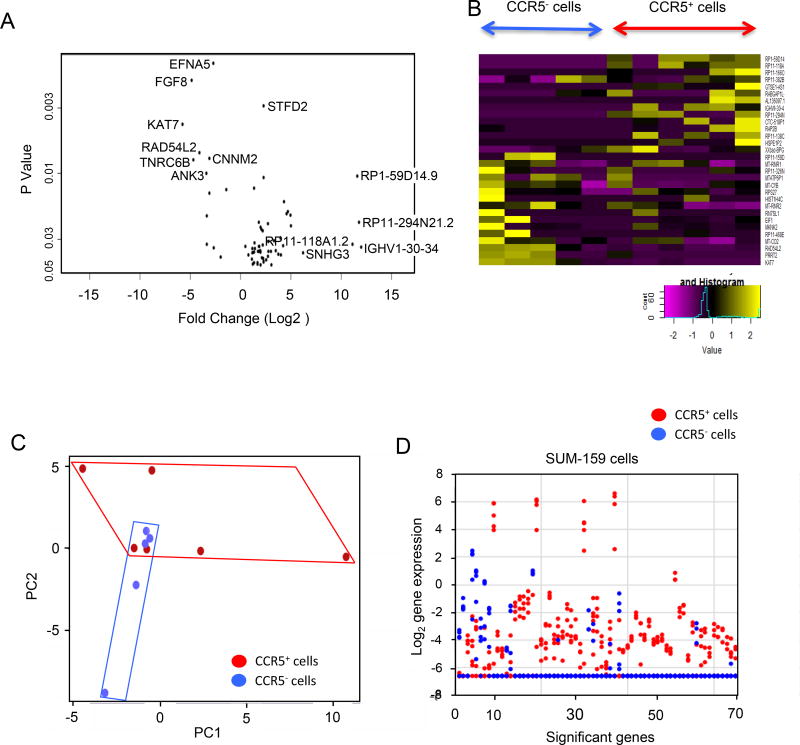
The stem cell hypothesis of cancer proposes that a single stem like cell is both capable of unlimited self-renewal and has the potential to differentiate into specialized types of cells. Our functional analysis conducted suggested CCR5+ cells have several features of “stem like cells” including the capacity to form mammospheres, the ability to initiate tumors and the ability to give rise to metastasis, when compared with the CCR5− cells. Sequencing of individual stem like cells has revealed tumors consist of heterogeneous populations. Single-cell RNA-seq has been used to dissect cellular heterogeneity within a tissue-specific stem cell population. In order to identify the regulatory relationships within the cell driven by CCR5 it is ideal to conduct single cell molecular analysis, for which we deployed the microfluidic approach (39). Single cell RNA sequencing studies were conducted of CCR5+ vs. CCR5− SUM-159 cell. The Volcano plot, which displays the mean differences in gene expression between CCR5+ vs. CCR5− cells, plotted as significance of differences (P<0.05, vs. Log2-fold change, showed a subset of genes that were induced between 25 (32)- to 210 (1000)-fold (Figure 6A). These genes are involved in ribosomal biogenesis. Heatmap display of individual cell RNA-seq showed difference in gene expression with the top genes involved in protein synthesis (Figure 6B). Principal component analysis (PCA) identified significant gene expression pattern differences between individual CCR5+ (red) and CCR5− (blue) cells (Figure 6C). The CCR5− cells were more homogeneous than the CCR5+ cells. These findings are consistent with greater heterogeneity in gene expression among the individual CCR5+ cells. Such heterogeneity is also seen when examining the display of altered gene expression changes for each of the 6 CCR5+ (red) cells sequenced. CCR5+ (red) cells exhibit great differences in levels of gene expression between cells within the CCR5+ group (Fig. 6D).
Figure 6. Single cell RNA sequencing of CCR5+ vs. CCR5− SUM-159 cells.
(A) Volcano plot displays the mean differences in gene expression between CCR5− (N=5) vs. CCR5+ (N=6) cells plotted as significance of differences vs. Log2-fold change. (B) Heatmap display of individual cell RNA-seq showing difference in gene expression levels. (C) Principal component analysis (PCA) illustrating significant differences between individual CCR5+ cells (red) and CCR5− cells (blue). CCR5+ cells are more diverse and spread out in a broader area of PC1 vs. PC2 than those of CCR5− cells. (D) Display of expression levels for 68 genes differentially expressed between CCR5+ and CCR5− cells. The expression levels of the 68 genes in each cell were plotted. For each gene on the x-axis, red dots represent the expression levels of expression in the 6 CCR5+ cells. Blue dots represent the expression levels of 5 CCR5− cells. The CCR5+ cells show a substantially larger number of genes with dramatically enhanced levels of gene expression when compared with the CCR5− cells.
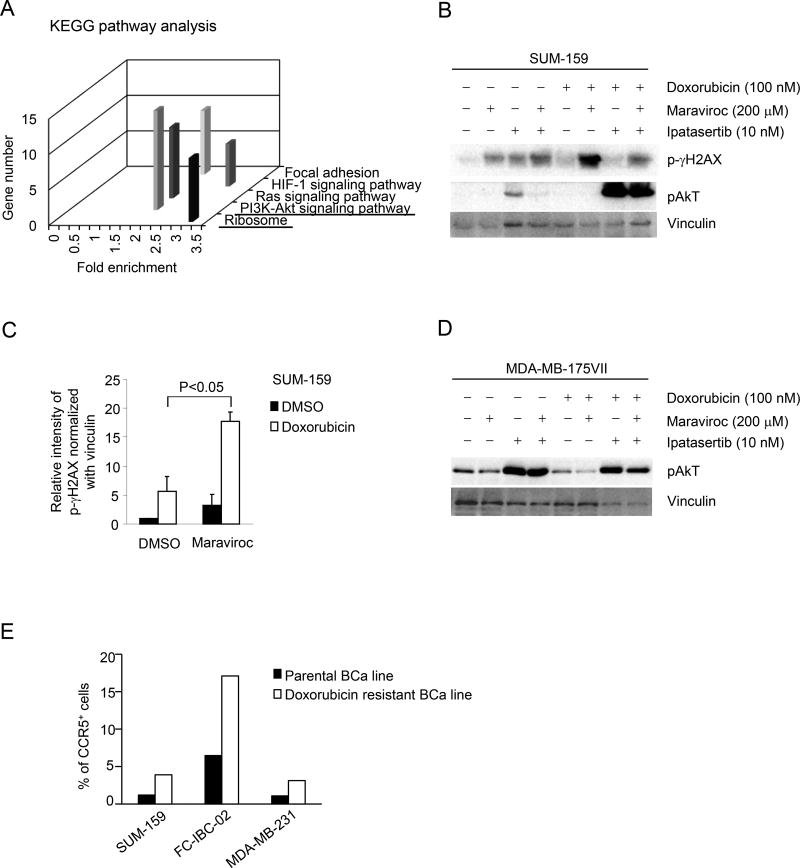
In order to examine the biological pathways governed by CCR5, unbiased interrogation was conducted using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) (Figure 7A and Supplemental Figure 10A,B). Substantial pathway enrichment was identified for ribosomal biogenesis, and the Akt-PI3 kinase-signaling pathway (Figure 7A). The induction of gene expression pathways involved in ribosomal biogenesis and Akt signaling in the CCR5+ population, are consistent with the known induction of Akt signaling by CCR5 (40) and the induction of ribosomal biogenesis by Akt signaling (41). In order to determine the potential role of the Akt signaling by CCR5 in the DNA damage response, we deployed the selective ATP-competitive pan-Akt inhibitor GDC-0068 (Ipatasertib). SUM-159 cells treated with Maraviroc showed an induction of γH2AX, which was augmented by the addition of Ipatasertib (10 nM) (Figure 7B). Ipatasertib induced pAkt, consistent with prior studies (42) and its mechanism of action as a selective ATP competitive inhibitor. Doxorubicin induced γH2AX compared with vehicle control, which was dramatically enhanced further by the addition of Maraviroc (Figure 7B and C). The addition of Ipatersertib to Doxorubicin provided no significant additional induction of γH2AX.
Figure 7. Single cell RNA sequencing of CCR5+ vs. CCR5− SUM-159 cells identified activation of PI3K/Akt signaling.
(A) The single cell sequencing analysis of CCR5+ vs. CCR5− SUM-159 cells was subjected to functional pathway analysis by KEGG (Kyoto Encyclopedia of Genes and Genomes) (Supplemental Figure 9). (B–E) The functional significance of the PI3K/Akt pathway in CCR5-mediated DNA damage signaling was assessed using the ATP-competitive, small-molecule pan-Akt inhibitor (Ipatasertib) and or the CCR5 inhibitor Maraviroc in Doxorubicin treated cells. Western blot was conducted as shown for SUM-159 cells (B) with data shown as mean ± SEM for the densitometry of N=2 separate experiments (C) or in (D) for MDA-MB-175VII (Doxorubicin resistant breast cancer cells). (E) The relative abundance of CCR5+ cells assessed by FACS analysis (Supplemental Figure 10) comparing parental and Doxorubicin resistant cells, derived as described in the Materials and Methods.
In order to determine whether CCR5-mediated Akt signaling in Doxorubicin resistant breast cancer cells MDA-MB-175VII (p53 wt) cell line was deployed. Doxorubicin was used at a dose well below the cell-killing threshold at 200 nM. Maraviroc reduced pAkt in the basal state, in the presence of Doxorubicin and upon the addition of Ipatasertib (Figure. 7D). These studies suggest that CCR5 mediates the induction of Akt activity in both SUM-159 and MDA-MB-175VII breast cancer cells.
In order to determine whether CCR5 remains a viable target for breast cancer treated with Doxorubicin, we selected Doxorubicin resistant breast cancer cell lines (Materials and Methods), and then conducted semi-quantitative analyses of CCR5 by FACS sorting (Fig. 7E, Supplemental Figure 11). The Doxorubicin resistant breast cancer cells showed a greater than 2-fold relative increase in the proportion of CCR5+ cells (Fig. 7E).
Together these studies suggest that CCR5 inhibition reduces pAkt and induces γH2AX. The finding that individual CCR5+ cells have such dramatic and variable induction of individual genes within this pathway indicates stochastic responsiveness within the CCR5+ population.
DISCUSSION
The current study identified novel functions of CCR5 in breast cancer that are relevant to patient therapy and suggest CCR5 may participate in certain characteristics of breast cancer stem cells in breast cancer. Firstly, human breast cancer and breast cancer cell lines were shown to express CCR5 with expression patterns that are heterogeneous within the tumor, and higher cytoplasmic CCR5 staining correlated with poor prognosis. Secondly, the CCR5+ expressing human breast cancer cells within the tumor initiate tumors in mice that grow ~60-fold larger than CCR5− cells. Thirdly, CCR5 antagonists reduced the ability of basal breast cancer cells to metastasize. Fourthly, CCR5 expression correlated with the ability of breast cancer cells to form mammospheres, a surrogate assay for tumor initiating cells. Fifth, functional analysis demonstrated endogenous CCR5 enhanced DNA repair (HDR and SSA) in response to DNA damaging agents used in chemotherapy for breast cancer and enhanced repair in response to high dose γ-irradiation and unbiased gene expression analysis of CCR5+ cells demonstrated enrichment of pathways governing DNA damage and DNA repair. Consequently, CCR5 antagonists substantially enhanced the cell killing of diverse human breast cancer cell lines in response to DNA damage inducing chemotherapy.
The role of p53 in CCR5-dependent proliferation is controversial (43) and the role of p53 in CCR5-dependent metastasis is not known. Human breast cancers harbor p53 mutations in ~40% of cases. Therefore understanding the function of CCR5 in p53− human breast cancer is of importance. The current studies demonstrated CCR5 promoted breast tumor metastasis and that CCR5 inhibitors block breast tumor metastasis in p53− SUM-159 cells, extending prior studies in MDA-MB-231, which are p53+ cells (22). In prior studies, inhibition of CCR5 expression by a CCR5Δ32 mutant enhanced BrdU uptake of breast tumor cells that were p53 wild type but not p53 mutant (43). The current studies were therefore conducted to determine the role of CCR5 in p53-independent growth and metastasis. In the current studies, CCR5 induced metastasis in p53− breast cancer cells in vivo. The difference in our findings compared with prior studies may relate to the different approaches used to inactivate CCR5 signaling in the two studies. In the previous publication expression of a CCR5Δ32 mutant was used to inactivate CCR5 (43). The current studies used complementary approaches of firstly CCR5 specific small molecular inhibitors, secondly FACS sorting of CCR5+ populations and thirdly engineering of CCR5 into CCR5− cells. Compared with MDA-MB-231 cells, SUM-159 cells exhibit a ~1 log order greater resistance to DNA damage agents (5-FdUR, 5-FU), DNA cross-linked compounds (Carboplatin), HSP90 inhibitors (17-AAG), and polyamine analogues (CGC11047) (30). Therefore, understanding whether CCR5 antagonists block growth of SUM-159 tumors, that are more resistant to current treatment, is of importance.
In the current studies, genomic pathway analysis of CCR5+ vs. CCR5− cells demonstrated the altered regulation of pathways involved in DNA repair. Using DNA damage functional reporter gene assays, CCR5 was shown to enhance the repair of DSBs by inducing HDR and SSA-based DNA repair. We deployed CCR5 antagonists in the presence of DNA damaging agents and the reporters DR-GFP, and SA-GFP (SSA) (33,44). Homology-directed repair (HDR) is essential to limit mutagenesis, chromosomal instability (CIN) and tumorigenesis. In mammalian cells, double-stranded breaks (DSB) may be repaired by either homology directed recombination (HDR), or non-homologous enjoining (NHEJ) and single-strand appealing (SSA). Defects in these repair mechanisms can result in chromosomal fusions, translocations and breaks (45). DNA damage and double strand breaks induce NHEJ and HR and oncogenes such as the c-Myc oncogene are known to disrupt the repair of double-strand DNA breaks, increasing chromosomal breaks (46). c-Myc inhibited the repair of DNA breaks and blocked the repair of single-strand breaks (46). The current studies demonstrate that CCR5 augments DNA repair.
In the current studies, CCR5+ cells demonstrated several features characteristic of breast cancer stem cells, including the increased formation of mammospheres, enhanced ability to initiate tumors, metastatic capacity, and enhanced DNA repair activity. CCR5+ SUM-159 cells gave rise to a greater proportion of mammospheres, which are considered a surrogate measure of breast cancer stem cells. Several lines of evidence have suggested an association between CSCs and enhanced DNA repair. The ability of CSCs to survive stressful conditions is correlated with protection of genomic integrity by activation of the DNA sensor and repair machinery (47). CD133+ glioblastoma stem cells activate Chk1 and ATM faster than CD133− cells (48). Significant increases in DNA repair gene expression has been observed in pancreatic CSCs (49). Colon and lung CSCs activate Chk1 more efficiently than parental (50) and enhanced DNA repair has been described in breast CSCs (19). Collectively these studies are consistent with a model in which CSCs are enriched for DNA repair activities, and that CCR5 induces both CSC and DNA repair activities independently of p53.
Our single-cell transcriptome analysis on CCR5+ and CCR5− cells revealed that levels of gene expression and volatility of gene expression, assessed through principal component analysis, are substantially increased in CCR5+ cells. Only a few studies addressed tumor transcriptome heterogeneity at the single-cell level of resolution (51–53). The molecular pathways activated in CCR5+ cells included ribosomal biogenesis and Akt-PI3K signaling. The induction of PI3K/Akt signaling in CCR5+ cells is consistent with prior studies in inflammatory cells (54), demonstrating CCR5 induces Akt. Furthermore, Akt is known to enhance ribosomal biogenesis and DNA repair (41). Together these studies are consistent with a model in which CCR5-mediated induction of Akt enhances ribosomal biogenesis and DNA repair.
The rational development of DNA repair inhibitors that function specifically in the tumorous but not normal cells is an important goal of cancer therapies. We showed that CCR5 is selectively overexpressed in BCa cells compared with normal tissues and >50% of human BCa overexpress CCR5,(6). The current studies demonstrate CCR5 inhibitors reduce DNA repair and enhance cell killing by DNA damage inducing agents in CCR5+ human breast cancer. In the current studies both Maraviroc and Vicriviroc increased the DNA damage induced cell killing by Doxorubicin in BRCA1 or BRCA2 defective cell lines.. Because CCR5 inhibitors selectively reduce DNA repair and enhance DNA damage in the tumor this study suggests CCR5 inhibitors may enhance the tumor specific activities of DDR-based treatments.
Supplementary Material
Acknowledgments
GRANT/FUNDING INFORMATION
This work was supported in part by grants from NIH R01CA70896, R01CA75503, R01CA86072 (R.G. Pestell), the Breast Cancer Research Foundation (R.G Pestell), the Dr. Ralph and Marian C. Falk Medical Research Trust (R.G. Pestell), and grants from the Pennsylvania Department of Health (R.G. Pestell). The Department specifically disclaims responsibility for analyses, interpretations or conclusions. Part of the work also supported by grants R01CA197903 and R01CA1645093 (H. Rui) from the National Institutes of Health, and CHE1213161 from the National Science Foundation, USA, and an internal grant from the University of Southern California (J.F. Zhong).
Footnotes
Conflicts of Interest: R.G. Pestell is founder of ProstaGene, LLC and owns issued patents.
References
- 1.Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and Mortality and Epidemiology of Breast Cancer in the World. Asian Pac J Cancer Prev. 2016;17:43–6. doi: 10.7314/apjcp.2016.17.s3.43. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 4.Meyers MO, Klauber-Demore N, Ollila DW, Amos KD, Moore DT, Drobish AA, et al. Impact of breast cancer molecular subtypes on locoregional recurrence in patients treated with neoadjuvant chemotherapy for locally advanced breast cancer. Ann Surg Oncol. 2011;18:2851–7. doi: 10.1245/s10434-011-1665-8. [DOI] [PubMed] [Google Scholar]
- 5.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–7. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 6.Velasco-Velazquez M, Jiao X, De La Fuente M, Pestell TG, Ertel A, Lisanti MP, et al. CCR5 antagonist blocks metastasis of basal breast cancer cells. Cancer Res. 2012;72:3839–50. doi: 10.1158/0008-5472.CAN-11-3917. [DOI] [PubMed] [Google Scholar]
- 7.Luboshits G, Shina S, Kaplan O, Engelberg S, Nass D, Lifshitz-Mercer B, et al. Elevated expression of the CC chemokine regulated on activation, normal T cell expressed and secreted (RANTES) in advanced breast carcinoma. Cancer Res. 1999;59:4681–7. [PubMed] [Google Scholar]
- 8.Zhang Y, Yao F, Yao X, Yi C, Tan C, Wei L, et al. Role of CCL5 in invasion, proliferation and proportion of CD44+/CD24- phenotype of MCF-7 cells and correlation of CCL5 and CCR5 expression with breast cancer progression. Oncol Rep. 2009;21:1113–21. [PubMed] [Google Scholar]
- 9.Pestell TG, Jiao X, Kumar M, Peck AR, Prisco M, Deng S, Li Z, Ertel A, Casimiro MC, Ju X, DiRocco A, DiSante G, Katiyar S, Shupp A, Lisanti MP, Jain P, Wu K, Rui H, Hooper DC, Yu Z, Goldman AR, Speicher DW, Laury-Kleintop L, Pestell RG. Stromal cyclin D1 promotes heterotypic immune signaling and breast cancer growth. Oncotarget. 2017;8:81754–75. doi: 10.18632/oncotarget.19953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westby M, van der Ryst E. CCR5 antagonists: host-targeted antivirals for the treatment of HIV infection. Antivir Chem Chemother. 2005;16:339–54. doi: 10.1177/095632020501600601. [DOI] [PubMed] [Google Scholar]
- 11.Kondru R, Zhang J, Ji C, Mirzadegan T, Rotstein D, Sankuratri S, et al. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol Pharmacol. 2008;73:789–800. doi: 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
- 12.Maeda K, Das D, Ogata-Aoki H, Nakata H, Miyakawa T, Tojo Y, et al. Structural and molecular interactions of CCR5 inhibitors with CCR5. J Biol Chem. 2006;281:12688–98. doi: 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- 13.Nishikawa M, Takashima K, Nishi T, Furuta RA, Kanzaki N, Yamamoto Y, et al. Analysis of binding sites for the new small-molecule CCR5 antagonist TAK-220 on human CCR5. Antimicrob Agents Chemother. 2005;49:4708–15. doi: 10.1128/AAC.49.11.4708-4715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–90. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kothandan G, Gadhe CG, Cho SJ. Structural insights from binding poses of CCR2 and CCR5 with clinically important antagonists: a combined in silico study. PLoS One. 2012;7:e32864. doi: 10.1371/journal.pone.0032864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–29. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18:55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velasco-Velazquez MA, Popov VM, Lisanti MP, Pestell RG. The role of breast cancer stem cells in metastasis and therapeutic implications. Am J Pathol. 2011;179:2–11. doi: 10.1016/j.ajpath.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maugeri-Sacca M, Bartucci M, De Maria R. DNA damage repair pathways in cancer stem cells. Mol Cancer Ther. 2012;11:1627–36. doi: 10.1158/1535-7163.MCT-11-1040. [DOI] [PubMed] [Google Scholar]
- 21.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol. 2012;920:379–91. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 22.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107:18115–20. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahler MH, Irani A, Malhi H, Reutens AT, Albanese C, Bouzahzah B, et al. The application of a lentiviral vector for gene transfer in fetal human hepatocytes. J Gene Med. 2000;2:186–93. doi: 10.1002/(SICI)1521-2254(200005/06)2:3<186::AID-JGM100>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Jiao X, Rizvanov AA, Cristofanilli M, Miftakhova RR, Pestell RG. Breast Cancer Stem Cell Isolation. Methods Mol Biol. 2016;1406:121–35. doi: 10.1007/978-1-4939-3444-7_10. [DOI] [PubMed] [Google Scholar]
- 26.Walker DK, Abel S, Comby P, Muirhead GJ, Nedderman AN, Smith DA. Species differences in the disposition of the CCR5 antagonist, UK-427,857, a new potential treatment for HIV. Drug Metab Dispos. 2005;33:587–95. doi: 10.1124/dmd.104.002626. [DOI] [PubMed] [Google Scholar]
- 27.Peck AR, Girondo MA, Liu C, Kovatich AJ, Hooke JA, Shriver CD, et al. Validation of tumor protein marker quantification by two independent automated immunofluorescence image analysis platforms. Mod Pathol. 2016 doi: 10.1038/modpathol.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peck AR, Witkiewicz AK, Liu C, Stringer GA, Klimowicz AC, Pequignot E, et al. Loss of nuclear localized and tyrosine phosphorylated Stat5 in breast cancer predicts poor clinical outcome and increased risk of antiestrogen therapy failure. J Clin Oncol. 2011;29:2448–58. doi: 10.1200/JCO.2010.30.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korkola JE, Collisson EA, Heiser L, Oates C, Bayani N, Itani S, et al. Decoupling of the PI3K Pathway via Mutation Necessitates Combinatorial Treatment in HER2+ Breast Cancer. PLoS One. 2015;10:e0133219. doi: 10.1371/journal.pone.0133219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724–9. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods. 2016;13:521–7. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casimiro MC, Di Sante G, Ju X, Li Z, Chen K, Crosariol M, et al. Cyclin D1 Promotes Androgen-Dependent DNA Damage Repair in Prostate Cancer Cells. Cancer Res. 2016;76:329–38. doi: 10.1158/0008-5472.CAN-15-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dontu G, Wicha MS. Survival of mammary stem cells in suspension culture: implications for stem cell biology and neoplasia. J Mammary Gland Biol Neoplasia. 2005;10:75–86. doi: 10.1007/s10911-005-2542-5. [DOI] [PubMed] [Google Scholar]
- 37.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–11. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 38.Yu Z, Pestell TG, Lisanti MP, Pestell RG. Cancer stem cells. The international journal of biochemistry & cell biology. 2012;44:2144–51. doi: 10.1016/j.biocel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Zhang C, Weiner LP, Zhang Y, Zhong JF. Molecular characterization of heterogeneous mesenchymal stem cells with single-cell transcriptomes. Biotechnol Adv. 2013;31:312–7. doi: 10.1016/j.biotechadv.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuster B, Pum D, Sleytr UB. S-layer stabilized lipid membranes (Review) Biointerphases. 2008;3:FA3. doi: 10.1116/1.2889067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SB, Kwon IS, Park J, Lee KH, Ahn Y, Lee C, et al. Ribosomal protein S3, a new substrate of Akt, serves as a signal mediator between neuronal apoptosis and DNA repair. J Biol Chem. 2010;285:29457–68. doi: 10.1074/jbc.M110.131367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y, Serra V, Prudkin L, Scaltriti M, Murli S, Rodriguez O, et al. Evaluation and clinical analyses of downstream targets of the Akt inhibitor GDC-0068. Clin Cancer Res. 2013;19:6976–86. doi: 10.1158/1078-0432.CCR-13-0978. [DOI] [PubMed] [Google Scholar]
- 43.Manes S, Mira E, Colomer R, Montero S, Real LM, Gomez-Mouton C, et al. CCR5 expression influences the progression of human breast cancer in a p53-dependent manner. J Exp Med. 2003;198:1381–9. doi: 10.1084/jem.20030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009;5:e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–75. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci U S A. 2003;100:9974–9. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maugeri-Sacca M, Bartucci M, De Maria R. Checkpoint kinase 1 inhibitors for potentiating systemic anticancer therapy. Cancer Treat Rev. 2013;39:525–33. doi: 10.1016/j.ctrv.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 49.Maugeri-Sacca M, Vigneri P, De Maria R. Cancer stem cells and chemosensitivity. Clin Cancer Res. 2011;17:4942–7. doi: 10.1158/1078-0432.CCR-10-2538. [DOI] [PubMed] [Google Scholar]
- 50.Mathews LA, Cabarcas SM, Farrar WL. DNA repair: the culprit for tumor-initiating cell survival? Cancer Metastasis Rev. 2011;30:185–97. doi: 10.1007/s10555-011-9277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–7. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–19. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyner JW, Uchida O, Kajiwara N, Kim EY, Patel AC, O'Sullivan MP, et al. CCL5-CCR5 interaction provides antiapoptotic signals for macrophage survival during viral infection. Nat Med. 2005;11:1180–7. doi: 10.1038/nm1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.