Abstract
The Cre/loxP system is a widely applied technology for site-specific genetic manipulation in mice. This system allows for deletion of the genes of interest in specific cells, tissues and the whole organism to generate a diversity of conditional knockout mouse strains. Additionally, the Cre/loxP system is useful for development of cell- and tissue-specific reporter mice for lineage tracing, and cell-specific conditional depletion models in mice. Recently, the Cre/loxP technique was extensively adopted to characterize the monocyte/macrophage biology in mouse models. Compared to other relatively homogenous immune cell types such as neutrophils, mast cells and basophils, monocytes/macrophages represent a highly heterogeneous population which lack specific markers or transcriptional factors. Though great efforts have been made towards establishing macrophage-specific Cre driver mice in the past decade, all of the current available strains are not perfect with regards to their depletion efficiency and targeting specificity for endogenous macrophages. Here we overview the commonly used Cre driver mouse strains targeting macrophages and discuss their major applications and limitations.
Keywords: Macrophages, Monocytes, Cre/loxP, Macrophage reporter mice, Macrophage-specific conditional knockout
1. Introduction
Cre/loxP system is a site-specific genetic modulation technique which has been extensively applied to create gene deletions, insertions, inversions and translocations at the specified DNA sites in mice (1). This system contains two major components: the enzyme Cre recombinase and the loxP sites. The Cre recombinase, which is originally derived from P1 bacteriophage, specifically recognizes the loxP sites, the 34-base pair asymmetric DNA sequences with directionality. Depending on the orientation of the two loxP sites, Cre recombinase either excises or inverts the transgene sequences inserted between the two loxP sites. Therefore, the Cre/loxP system allows for making a series of DNA sequence rearrangements, which helps to build one of the most versatile genetic tools in mice (2,3).
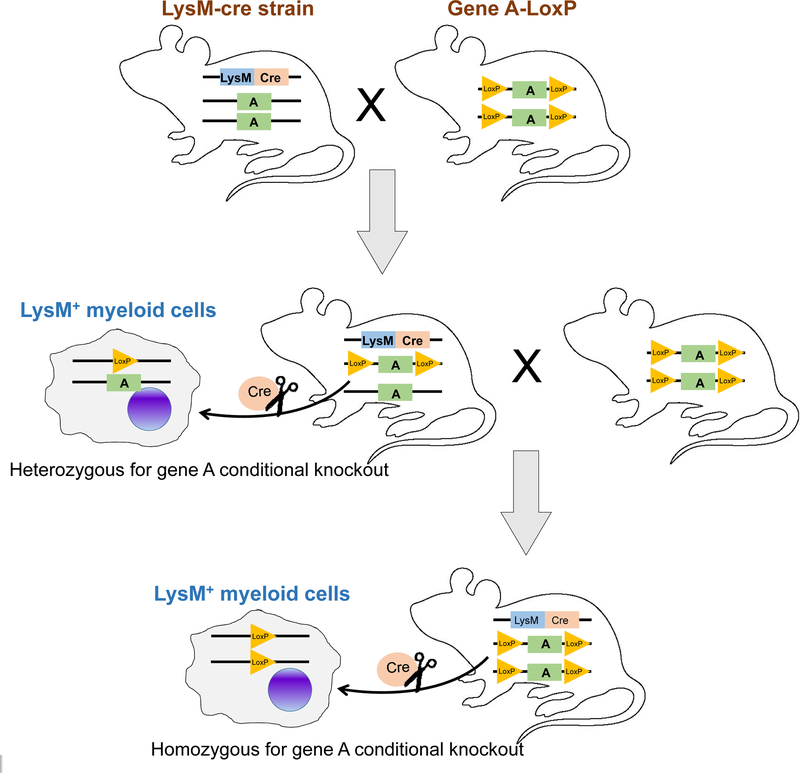
The major application of Cre/loxP system is generation of cell- or tissue-specific conditional knockout alleles in mice (4). This is achieved by knocking in two loxP sites at distant introns within the gene of interest, and such a knock-in strain is then crossed with another mouse strain that contains the Cre recombinase transgene under the direction of a cell or tissue-specific gene promoter. In the specific cells or tissues expressing Cre, the gene of interest will then be excised leading to a conditional gene knockout. As an example shown in Figure 1, when the myelomonoytic cell-specific Lysozyme M (LysM)-Cre mice are crossed with homozygous loxP-flanked transgenic strain of interest (gene A), mice heterozygous for the loxP allele will be generated in the first generation. The experimental mice, homozygous for the loxP-flanked gene A with Cre activity, will be obtained by further crossing the heterozygous mice back to the homozygous loxP-flanked mice. In the LysM+ myelomonoytic cells of the experimental mice, the transgene A will be knocked out while LysM− cells remain unaffected. This type of cell- or tissue-specific gene knockout models is very helpful for a precise understanding of any genes of interest in a given cell type or tissue. When fusing the Cre recombinase gene with the gene encoding estrogen receptor (ER), such Cre driver mice can be used for a temporal control of loxP-flanked transgene expression upon administration of tamoxifen, the estrogen antagonist (5). This is particularly critical when the gene knockout in specific cells or tissues causes embryonic lethality.
Figure 1. Generation of LysM+ cell-specific conditional knockout mice using Cre/loxP technology.
LysM-Cre transgenic mouse is first crossed with the mouse homozygous for loxP-flanked gene A. Approximately 50% of the offspring will be heterozygous for the loxP-flanked gene A and hemizygous/heterozygous for the Cre transgene. In this F1 generation, the gene A is heterozygous knockout specific for LysM+ cells. A further cross of this F1 generation with the parental mice homozygous for loxP-flanked gene A will lead to generation of about 25% of the progeny (experimental mice) homozygous for the loxP-flanked gene A and hemizygous/heterozygous for the Cre transgene. In this F2 generation of mice, Cre-mediated excision causes the homozygous knockout for gene A specifically in LysM+ cells. Other F2 littermates, such as mice homozygous for loxP with no Cre transgene, and mice heterozygous for loxP and hemizygous/heterozygous for the Cre transgene, can serve as controls.
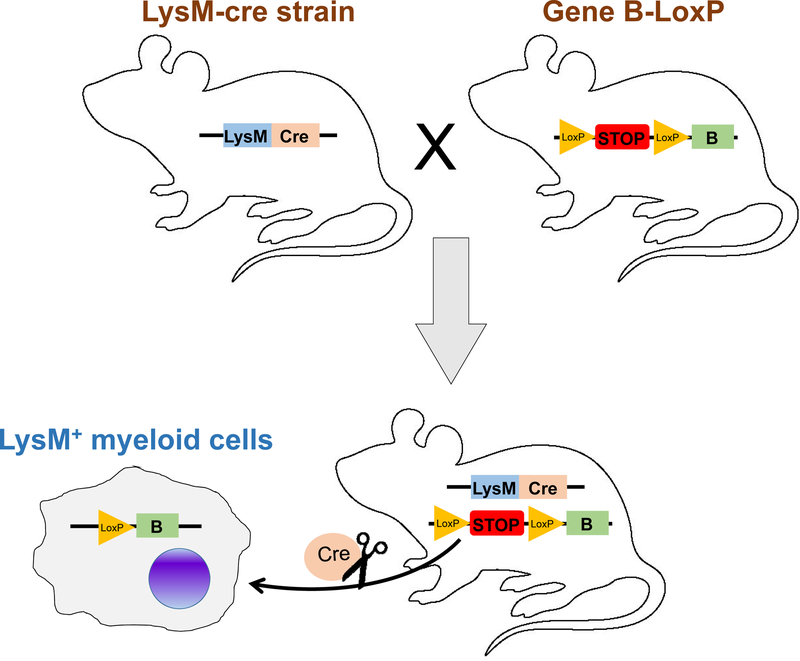
In addition, the Cre/loxP system is widely utilized for development of mouse strains with conditional gene activation/overexpression, reporter mice for cell tracking or lineage tracing, and cell-specific depletion models (6). For example, to generate a conditional gene activation strain as shown in Figure 2, the transcriptional stop sequence, such as a polyadenylation signal, is inserted upstream of the gene of interest (gene B) to prevent the transgene expression. As the “stop” sequence is flanked by the two loxP sites (“lox-stop-lox” cassette), when crossing this strain with LysM-Cre transgenic mice, Cre-mediated deletion of the “stop” sequence drives the expression of gene B only in LysM+ myelomonoytic cells. This gene activation or overexpression model offers a gain-of-function approach to precisely dissect the function of any genes of interest in a specific cell type or tissue.
Figure 2. Application of Cre/loxP system for generation of mouse lines with gene conditional activation.
The gene B-loxP transgenic mouse is created by inserting a loxP-flanked transcriptional “STOP” sequence between the promoter and the gene B coding sequence, which prevents the expression of gene B. To cross this gene B-loxP transgenic mouse with LysM-Cre mouse will generate the progeny with gene B specifically expressed in LysM+ cells. Such a system helps to develop cell-specific reporter mice, gene activation/overexpression and conditional depletion mouse lines.
Following a similar strategy, the myelomonoytic cell-specific reporter strains can be developed if the transgene B (Figure 2) is a visible marker, such as green fluorescent protein (GFP), red fluorescent protein (RFP), tdTomato fluorescent protein and LacZ (6). The reporter mice are good tools for spatial and temporal tracking of the specific cells in vivo, and also benefit cell lineage tracing to determine the developmental origins of specific cell types since the Cre-mediated reporter gene expression is irreversible from ancestor to descendant cells. Moreover, the strategy shown in Figure 2 also helps to generate cell-specific conditional depletion models if the transgene B is diphtheria toxin receptor (DTR) gene or other similar constructs (7). In this model, Cre-induced deletion of the “lox-stop-lox” cassette leads to expression of the DTR gene which in turn forms an inducible ablation system specific for LysM+ cells upon receiving diphtheria toxin (DT). The cell depletion models facilitate us to understand the systemic functions of a given cell type in physiological and pathological conditions.
One of the major limitations of Cre/loxP system is its off-target effects. It has been frequently reported that many Cre transgenic mouse strains express Cre recombinase in non-target cells or tissues. Such off-target effects may confound the results from gene conditional knockout, reporter and cell depletion models, as well as other Cre/loxP-based genetic tools. Thus, a thorough analysis of each Cre transgenic strain with regards to their Cre sensitivity (efficiency) and expression pattern (specificity) should be carefully performed and recorded. To this end, the Jackson Laboratory (USA) has developed a Cre portal which aims to provide the scientific community with a comprehensive well-characterized Cre transgenic mouse strains and high throughput data related to these strains (2).
The currently available Cre driver mice targeting macrophages are primarily designed based on the monocyte/macrophage markers such as LysM, colony-stimulating factor 1 receptor (CSF1R or CD115), CD11b, F4/80 and CX3C chemokine receptor 1 (CX3CR1) (8), although none of them are perfectly specific. On the basis of these Cre driver mice, a diversity of conditional gene knockout strains, reporter strains and cell depletion strains were accordingly developed towards elucidating the macrophage lineage development in prenatal and adult life, as well as their functional contribution to tissue homeostasis and pathological processes in mice. The main applications and specificity of these Cre driver mouse strains are described below. For each strain, its number (s) recorded in the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/) is also included and more information about these strains can be obtained from this database.
2. Overview of The Commonly Used Cre Driver Mice for Macrophages
2.1. LysM-Cre (MGI: 1934631)
Lysozymes are antimicrobial enzymes widely produced by the innate immune cells in eukaryota. As a type of glycoside hydrolase, lysozymes function to cleave the peptidoglycan of bacterial cell wall as their antimicrobial mechanism (9). In mice, there are two forms of lysozyme genes, Lyz1 and Lyz2, which encode lysozyme P (LysP) and lysozyme M (LysM), respectively. While LysP is expressed in the Paneth cells of the small intestine, LysM is exclusively expressed in myelomonoytic cells including monocytes, macrophages and granulocytes in mice (10). LysM is therefore served as a marker for myelomonoytic cells.
In 1999, Clausen et al. generated the LysM-Cre mouse by targeted inserting the Cre cDNA into the endogenous LysM gene locus (11). When crossing the LysM-Cre mice with two mouse strains with different loxP-flanked targeted genes (β-polymerase and transcription factor RX5), there was a highly efficient gene depletion in mature macrophages and granulocytes isolated from peritoneal cavity or derived from bone marrow (BM). No depletion was detected in lymphoid cells such as B and T lymphocytes (11). Later on, Faust et al. Created a LysM-reporter strain by knocking the enhanced GFP (EGFP) gene into the LysM gene locus together with a targeting vector, which contains a neomycin resistant (neo) gene flanked by loxP sites (12). Using this reporter strain, the EGFP+ cells were confirmed to be mostly myelomonocytic cells in peripheral blood, peritoneal cavity and BM (12). A further cross of this LsyM-reporter strain with Cre-expressing mice, Cre-mediated removal of the loxP-flanked neo gene led to generation of LysM-deficient mice in the offspring (13). In these LysM-deficient mice, a prolonged and more robust inflammatory response was observed compared to the wild type (WT) controls when challenged by normally non-pathogenic bacteria. This suggested a critical role of myelomonocytic cells in a timely elimination of bacteria and their products which elicit immune responses (13).
Since the creation of this LysM-Cre strain, it has been extensively applied to develop reporter mice for in vivo tracking or lineage tracing of macrophages and other myelomonocytic cells in normal and diseased conditions. For example, in the myelomonocytic cell reporter mice generated through crossing LysM-Cre mice with ROSA26-flox-stop-flox-EYFP (Rosa26-LSL-EYFP) mice, yellow fluorescent protein (YFP) was found to be expressed not only in myelomonocytic cells, but also in a fraction of long-term hematopoietic stem cells (LT-HSCs) as well as megakaryocyte/erythrocyte progenitor (MEP) cells suggesting a universal expression of LysM at the hematopoietic stem/progenitor cell (HSPC) stages in mice. Differentiation of HSPCs towards myelomonocytic cells is presumably as a default while their differentiation to other lineages might be dominated by additional transcriptional factors (14).
A series of macrophage-specific conditional gene knockout mice were generated by crossing the LysM-Cre line with different “floxed” strains, which highly accelerate identification of numerous myeloid genes which play key roles in a variety of inflammatory disorders such as infections, autoimmune diseases, fibrosis and cancer progression (15–19). As well, the LysM-Cre strain was also crossed with loxP-flanked DTR mice to develop the inducible macrophage depletion model-LysM-Cre/iDTR line to characterize the systemic functions of endogenous macrophages in wound healing, fibrotic diseases, arterial hypertension and vascular dysfunction in mouse models (20–23).
In spite of a high efficiency of LysM-Cre strain in manipulating endogenous macrophages, LysM is not a specific marker for macrophages. In addition to monocytes and mature macrophages, LysM is also expressed in most granulocytes and few CD11c+ dendritic cells (DCs), as well as a small percentage of non-hematopoietic cells such as type II lung alveolar cells in mice (12,23). In a comparative analysis of multiple myeloid cell-specific Cre reporter strains, about 60–80% neutrophils in spleen, peripheral blood and BM were shown as LysM+ (8). For mature macrophage populations, about 90–100% alveolar macrophages and peritoneal macrophages were LysM+, whereas only 40% of spleen red pulp or spleen marginal zone macrophages had LysM-Cre activity implicating the heterogeneous origins of tissue resident macrophages (8). Overall, the LysM-Cre mouse strain is a broadly used tool to investigate the endogenous myelomonocytic cells.
2.2. Csf1r-Cre (MGI: 4429470)
CSF1R, also known as macrophage colony-stimulating factor receptor (M-CSFR), and CD115, is the receptor for the major monocyte/macrophage lineage differentiation factor CSF1. It is a tyrosine kinase encoded by the proto-oncogene c-fms and expressed in all monocytic cells including monocytes and macrophages in mice.
Prior to generation of the Csf1r-Cre strain, the Csf1r-EGFP reporter mice (MacGreen mice) had been created and were successfully utilized for tracing the endogenous macrophages in various tissues (24). The Csf1r-Cre line was developed in 2010 which was used to identify the role of signal transducer and activator of transcription 3 (STAT3) in macrophages (25). In the conditional STAT3-knockout mice generated via mating the Csf1-Cre strain with the STAT3flox/flox strain, spontaneous colitis and tumor formation in the inflamed colon and cecum were detected indicating a key contribution of myeloid signaling to malignancy in mouse models (25).
With this Csf1r-Cre strain, several groups generated macrophage reporter mice by crossing the Csf1r-Cre mice with Rosa26-LSL-YFP mice to determine the macrophage lineage development during organogenesis in the mouse prenatal stage (26–28). From these lineage tracing studies, the yolk-sac macrophages, originated from yolk-sac-derived erythro-myeloid progenitors, were shown to give rise to some F4/80bright tissue resident macrophages such as liver Kupffer cells, epidermal Langerhans cells and microglial cells (27,28). In mouse models of wound healing, autoimmune encephalitis and breast carcinoma, the roles of macrophage-associated factors such as neuropilin-1, porcupine, Wnt family member 7B (Wnt 7B) and vascular endothelial growth factor receptor 1 (VEGFR1), were characterized with their respective Csf1r-specific gene knockout strains (29–33).
Therefore, the Csf1r-Cre strain is a good tool for fate mapping of macrophages in development and identification of macrophage regulatory genes essential for both steady status and inflammatory conditions. The Csf1r-Cre-based gene deletion was detected in macrophages of various tissues including liver, spleen, intestine, heart, kidney, and muscle with a high efficiency in mice. However, the specificity of Csf1r-Cre is not high and Cre-mediated deletion could be detected in DCs, granulocytes and T lymphocytes (25).
2.3. CD11b-Cre (MGI: 3577104; 3629092)
CD11b, also known as integrin alpha M (ITGAM), is an integrin family member which pairs with CD18 to form the heterodimeric integrin alpha M beta-2 (αMβ2) molecule (macrophage-1 antigen, Mac-1). It is widely expressed in monocytes, macrophages, granulocytes and natural killer (NK) cells. Functionally, CD11b regulates leukocyte adhesion and migration which is thus a key molecule in inflammatory responses.
In 2005, Ferron et al. created the CD11b-Cre mouse strain aiming to establish the conditional gene ablation in the hematopoietic myeloid-osteoclast lineage (34). In this study, the CD11b-reporter line was generated by crossing the CD11b-Cre strain with the Z/EG (lacZ/EGFP) double reporter strain to determine the Cre specificity. As expected, the Cre activity was detected in most peritoneal macrophages, and a large portion of macrophages and granulocytes in BM and spleen. In addition, mature osteoclast cells derived from BM and spleen progenitors of the reporter mice were mostly EGFP+ (34).
Afterwards, the CD11b-Cre strain was applied for identifying the key myeloid genes through its cross with the respective loxP-flanked transgenic mice. In particular, a plethora of conditional knockout models were constructed to study the microglia-associated factors such as prostaglandin E2 (PGE2) receptors EP2 and EP4, inhibitor of nuclear factor kappa-B kinase subunit beta (IKK-β), brain-derived neurotrophic factor (BDNF), progranulin and superoxide dismutase (SOD1) in induction of Alzheimer’s disease, autoimmune encephalitis, hyperalgesia, neuroinflammation, amyotrophic lateral sclerosis (ALS) and hypertension in mice (35–40). Using the CD11b-Cre mice, the roles of serine/threonine-protein kinase/endoribonuclease IRE1a, myeloid differentiation primary response 88 (Myd88), huntingtin, cathepsin K were also characterized in inflammatory responses, Huntington’s disease, bone formation and other pathological processes in tissue macrophages (41–44).
In comparison to other Cre driver mice targeting macrophages, the CD11b-Cre strain appeared to have a relatively low efficiency (less than 50%) to mark macrophages in bronchoalveolar lavage (BAL), peritoneal cavity, spleen and BM (8). As well, the CD11b-Cre activity was detected not only in monocytes and macrophages, but also in neutrophils and DCs suggesting a low level of specificity for macrophages (8). Lastly, this Cre transgenic strain was reported to be an unreliable line as inconsistent deletion among littermates had been observed (8).
2.4. F4/80-Cre (MGI: 2429642)
F4/80, also known as EGF-like module-containing mucin-like hormone receptor-like 1 (EMR1), is encoded by adhesion G protein-coupled receptor E1 (ADGRE1) gene in mice. As a cell surface glycoprotein, F4/80 is expressed in tissue macrophages such as liver Kupffer cells and the spleen red pulp macrophages in mice (45).
The F4/80-Cre strain was generated in 2002 in which the Cre recombinase cDNA is introduced into the first coding exon of F4/80 (46). This line has been crossed with loxP-flanked sphingosine-1-phosphate receptor 1 (S1PR1) and ferroportin (Fpn1) transgenic mice to determine their respective roles in peritonitis and iron homeostasis (47,48). As well, the F4/80-Cre line was used to create reporter mice to monitor the interstitial lung macrophage development after crossing with Rosa26-LSL-Tdtomato strain (49).
As the F4/80-Cre activity was revealed to be associated with limited types of tissue macrophages such as peritoneal macrophages (8), this strain was not often applied for developing genetic manipulation models compared to other Cre driver mice for macrophages.
2.5. CX3CR1-Cre (MGI: 5311737; 5467983; 5450813; 5467985)
CX3C chemokine receptor 1 (CX3CR1), also called fractalkine receptor or G-protein coupled receptor 13 (GPR13), is the receptor for chemokine CX3CL1 and exclusively expressed in the mononuclear phagocyte system. In 2013, two groups reported their generation of Cx3cr1-Cre and Cx3cr1-CreER mice with constitutive or tamoxifen-inducible Cre recombinase inserted in the Cx3cr1 loci (50,51). The Cx3cr1-CreER line is particularly useful as it allows for a temporal control of the transgene knockout/activation in macrophages.
There are three main applications for the Cx3cr1-Cre and Cx3cr1-CreER strains. First, these strains can be crossed with fluorescent reporters such as Rosa26-LSL-YFP, Rosa26-LSL-RFP, Rosa26-LSL-GFP and Rosa26-LSL-Tdtomato lines to generate reporter mice for fate mapping of fetal monocytes/macrophages during prenatal development and the adult macrophages in heart, ileum, colon, peritoneal cavity and liver during tissue homeostasis (50,52–57). Second, by mating the Cx3cr1-Cre or Cx3cr1-CreER strain with loxP-flanked transgenic mice, a diversity of lines of macrophage-specific conditional knockout mice were developed. These mice advanced our knowledge about the macrophage-associated genes such as TGF-beta activated kinase 1 (TAK1), ubiquitin specific peptidase 18 (Usp18), transmembrane protein 16F (TMEM16F), tumor necrosis factor alpha (TNFα) and channelrhodopsin-2 (ChR2) in maintenance of brain and heart functions and their involvement in inflammation and autoimmune disorders (58–65). Lastly, through crossing with the Rosa26iDTR strain, the Cx3cr1-Cre and Cx3cr1-CreER mice were also utilized to build macrophage-specific conditional depletion models in which the macrophages can be ablated upon injection of DT toxin. These models showed a high efficiency in depleting microglia and intestinal macrophages in mice (52,66).
With regards to the specificity, although Cx3cr1-Cre does not significantly mark neutrophils, this strain provides deletion in mast cells and DCs in addition to monocytes/macrophages (8,53). The deletion efficiency was high for peritoneal macrophages (70–80%), but relatively low (40–60%) for BAL and splenic macrophages, as well as peripheral blood monocytes (8).
3. Concluding Remarks
In this chapter, we mainly discussed the commonly used Cre driver mice targeting macrophages. Besides these strains, there are some other myeloid transgenes specific for certain subpopulations of tissue macrophages, which had also been applied to generate Cre transgenic mice. For example, the c-type lectin CD207a (langerin) is exclusively expressed in epidermal macrophages (Langerhans cells) and the CD207a-Cre strain was accordingly created for studying the endogenous Langerhans cells in mice (67,68). Furthermore, in examining macrophage development at the early prenatal stages, several Cre transgenic mice under the control of promoters of hematopoietic cell differentiation genes such as runt-related transcription factor 1 (Runx1) and FMS-like tyrosine kinase 3 (Flt3), were also applied (28,69,70).
Macrophages are a highly heterogeneous and plastic myeloid cell population in both steady status and pathological conditions. Their gene expression patterns dynamically change during the prenatal development, adult tissue homeostasis and in various inflammatory diseases. Moreover, such gene expression profiles are largely overlapped among monocytes/macrophages, granulocytes and DCs due to their close lineage relationship. Therefore, a Cre-transgenic line perfectly specific for macrophages is intrinsically impossible. The usage of these Cre driver mouse lines must be accompanied by other technical analyses such as flow cytometry, immunostaining and functional genomics before drawing any affirmative conclusions about the macrophage biology.
Acknowledgements
This work was supported by grants from the National Institutes of Health (R00 CA188093 to G.R. and P30 CA034196 to E.L.) of the United States. We apologize to our colleagues whose works are not cited in this manuscript due to space limitations.
4. References
- 1.Nagy A (2000) Cre recombinase: the universal reagent for genome tailoring. Genesis 26 (2):99–109 [PubMed] [Google Scholar]
- 2.Heffner CS, Herbert Pratt C, Babiuk RP et al. (2012) Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nature communications 3:1218. doi: 10.1038/ncomms2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn R, Torres RM (2002) Cre/loxP recombination system and gene targeting. Methods in molecular biology 180:175–204. doi: 10.1385/1-59259-178-7:175 [DOI] [PubMed] [Google Scholar]
- 4.Kwan KM (2002) Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis 32 (2):49–62 [DOI] [PubMed] [Google Scholar]
- 5.Danielian PS, Muccino D, Rowitch DH et al. (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current biology : CB 8 (24):1323–1326 [DOI] [PubMed] [Google Scholar]
- 6.Srinivas S, Watanabe T, Lin CS et al. (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buch T, Heppner FL, Tertilt C et al. (2005) A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nature methods 2 (6):419–426. doi: 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- 8.Abram CL, Roberge GL, Hu Y et al. (2014) Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. Journal of immunological methods 408:89–100. doi: 10.1016/j.jim.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Short ML, Nickel J, Schmitz A et al. (1996) Lysozyme gene expression and regulation. Exs 75:243–257 [DOI] [PubMed] [Google Scholar]
- 10.Cross M, Renkawitz R (1990) Repetitive sequence involvement in the duplication and divergence of mouse lysozyme genes. The EMBO journal 9 (4):1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clausen BE, Burkhardt C, Reith W et al. (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic research 8 (4):265–277 [DOI] [PubMed] [Google Scholar]
- 12.Faust N, Varas F, Kelly LM et al. (2000) Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood 96 (2):719–726 [PubMed] [Google Scholar]
- 13.Ganz T, Gabayan V, Liao HI et al. (2003) Increased inflammation in lysozyme M-deficient mice in response to Micrococcus luteus and its peptidoglycan. Blood 101 (6):2388–2392. doi: 10.1182/blood-2002-07-2319 [DOI] [PubMed] [Google Scholar]
- 14.Ye M, Iwasaki H, Laiosa CV et al. (2003) Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 19 (5):689–699 [DOI] [PubMed] [Google Scholar]
- 15.Yasukawa H, Ohishi M, Mori H et al. (2003) IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nature immunology 4 (6):551–556. doi: 10.1038/ni938 [DOI] [PubMed] [Google Scholar]
- 16.Herbert DR, Holscher C, Mohrs M et al. (2004) Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 20 (5):623–635 [DOI] [PubMed] [Google Scholar]
- 17.Vannella KM, Barron L, Borthwick LA et al. (2014) Incomplete deletion of IL-4Ralpha by LysM(Cre) reveals distinct subsets of M2 macrophages controlling inflammation and fibrosis in chronic schistosomiasis. PLoS pathogens 10 (9):e1004372. doi: 10.1371/journal.ppat.1004372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greten FR, Eckmann L, Greten TF et al. (2004) IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118 (3):285–296. doi: 10.1016/j.cell.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 19.Kanter JE, Kramer F, Barnhart S et al. (2012) Diabetes promotes an inflammatory macrophage phenotype and atherosclerosis through acyl-CoA synthetase 1. Proceedings of the National Academy of Sciences of the United States of America 109 (12):E715–724. doi: 10.1073/pnas.1111600109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goren I, Allmann N, Yogev N et al. (2009) A transgenic mouse model of inducible macrophage depletion: effects of diphtheria toxin-driven lysozyme M-specific cell lineage ablation on wound inflammatory, angiogenic, and contractive processes. The American journal of pathology 175 (1):132–147. doi: 10.2353/ajpath.2009.081002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas T, Waisman A, Ranjan R et al. (2010) Differential roles of macrophages in diverse phases of skin repair. Journal of immunology 184 (7):3964–3977. doi: 10.4049/jimmunol.0903356 [DOI] [PubMed] [Google Scholar]
- 22.Meng XM, Wang S, Huang XR et al. (2016) Inflammatory macrophages can transdifferentiate into myofibroblasts during renal fibrosis. Cell death & disease 7 (12):e2495. doi: 10.1038/cddis.2016.402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyake Y, Kaise H, Isono K et al. (2007) Protective role of macrophages in noninflammatory lung injury caused by selective ablation of alveolar epithelial type II Cells. Journal of immunology 178 (8):5001–5009 [DOI] [PubMed] [Google Scholar]
- 24.Sasmono RT, Oceandy D, Pollard JW et al. (2003) A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101 (3):1155–1163. doi: 10.1182/blood-2002-02-0569 [DOI] [PubMed] [Google Scholar]
- 25.Deng L, Zhou JF, Sellers RS et al. (2010) A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. The American journal of pathology 176 (2):952–967. doi: 10.2353/ajpath.2010.090622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mass E, Ballesteros I, Farlik M et al. (2016) Specification of tissue-resident macrophages during organogenesis. Science 353 (6304). doi: 10.1126/science.aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz C, Gomez Perdiguero E, Chorro L et al. (2012) A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336 (6077):86–90. doi: 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- 28.Gomez Perdiguero E, Klapproth K, Schulz C et al. (2015) Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518 (7540):547–551. doi: 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SL, Li B, Rao S et al. (2010) Macrophage Wnt7b is critical for kidney repair and regeneration. Proceedings of the National Academy of Sciences of the United States of America 107 (9):4194–4199. doi: 10.1073/pnas.0912228107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nissen JC, Tsirka SE (2016) Tuftsin-driven experimental autoimmune encephalomyelitis recovery requires neuropilin-1. Glia 64 (6):923–936. doi: 10.1002/glia.22972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha S, Aranda E, Hayakawa Y et al. (2016) Macrophage-derived extracellular vesicle-packaged WNTs rescue intestinal stem cells and enhance survival after radiation injury. Nature communications 7:13096. doi: 10.1038/ncomms13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefater JA 3rd, Rao S, Bezold K et al. (2013) Macrophage Wnt-Calcineurin-Flt1 signaling regulates mouse wound angiogenesis and repair. Blood 121 (13):2574–2578. doi: 10.1182/blood-2012-06-434621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo EJ, Cassetta L, Qian BZ et al. (2014) Myeloid WNT7b mediates the angiogenic switch and metastasis in breast cancer. Cancer research 74 (11):2962–2973. doi: 10.1158/0008-5472.CAN-13-2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferron M, Vacher J (2005) Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis 41 (3):138–145. doi: 10.1002/gene.20108 [DOI] [PubMed] [Google Scholar]
- 35.Boillee S, Yamanaka K, Lobsiger CS et al. (2006) Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312 (5778):1389–1392. doi: 10.1126/science.1123511 [DOI] [PubMed] [Google Scholar]
- 36.Evangelidou M, Karamita M, Vamvakas SS et al. (2014) Altered expression of oligodendrocyte and neuronal marker genes predicts the clinical onset of autoimmune encephalomyelitis and indicates the effectiveness of multiple sclerosis-directed therapeutics. Journal of immunology 192 (9):4122–4133. doi: 10.4049/jimmunol.1300633 [DOI] [PubMed] [Google Scholar]
- 37.Ferrini F, Trang T, Mattioli TA et al. (2013) Morphine hyperalgesia gated through microglia-mediated disruption of neuronal Cl(−) homeostasis. Nature neuroscience 16 (2):183–192. doi: 10.1038/nn.3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson JU, Woodling NS, Wang Q et al. (2015) Prostaglandin signaling suppresses beneficial microglial function in Alzheimer’s disease models. The Journal of clinical investigation 125 (1):350–364. doi: 10.1172/JCI77487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang MZ, Yao B, Wang Y et al. (2015) Inhibition of cyclooxygenase-2 in hematopoietic cells results in salt-sensitive hypertension. The Journal of clinical investigation 125 (11):4281–4294. doi: 10.1172/JCI81550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martens LH, Zhang J, Barmada SJ et al. (2012) Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. The Journal of clinical investigation 122 (11):3955–3959. doi: 10.1172/JCI63113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwan W, Trager U, Davalos D et al. (2012) Mutant huntingtin impairs immune cell migration in Huntington disease. The Journal of clinical investigation 122 (12):4737–4747. doi: 10.1172/JCI64484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robblee MM, Kim CC, Porter Abate J et al. (2016) Saturated Fatty Acids Engage an IRE1alpha-Dependent Pathway to Activate the NLRP3 Inflammasome in Myeloid Cells. Cell reports 14 (11):2611–2623. doi: 10.1016/j.celrep.2016.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugatani T, Hruska KA (2009) Impaired micro-RNA pathways diminish osteoclast differentiation and function. The Journal of biological chemistry 284 (7):4667–4678. doi: 10.1074/jbc.M805777200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu M, Zhou H, Zhao J et al. (2014) MyD88-dependent interplay between myeloid and endothelial cells in the initiation and progression of obesity-associated inflammatory diseases. The Journal of experimental medicine 211 (5):887–907. doi: 10.1084/jem.20131314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davies LC, Jenkins SJ, Allen JE et al. (2013) Tissue-resident macrophages. Nature immunology 14 (10):986–995. doi: 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaller E, Macfarlane AJ, Rupec RA et al. (2002) Inactivation of the F4/80 glycoprotein in the mouse germ line. Molecular and cellular biology 22 (22):8035–8043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weichand B, Weis N, Weigert A et al. (2013) Apoptotic cells enhance sphingosine-1-phosphate receptor 1 dependent macrophage migration. European journal of immunology 43 (12):3306–3313. doi: 10.1002/eji.201343441 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, Zhang F, An P et al. (2011) Ferroportin1 deficiency in mouse macrophages impairs iron homeostasis and inflammatory responses. Blood 118 (7):1912–1922. doi: 10.1182/blood-2011-01-330324 [DOI] [PubMed] [Google Scholar]
- 49.Tan SY, Krasnow MA (2016) Developmental origin of lung macrophage diversity. Development 143 (8):1318–1327. doi: 10.1242/dev.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yona S, Kim KW, Wolf Y et al. (2013) Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38 (1):79–91. doi: 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parkhurst CN, Yang G, Ninan I et al. (2013) Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155 (7):1596–1609. doi: 10.1016/j.cell.2013.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aychek T, Mildner A, Yona S et al. (2015) IL-23-mediated mononuclear phagocyte crosstalk protects mice from Citrobacter rodentium-induced colon immunopathology. Nature communications 6:6525. doi: 10.1038/ncomms7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cain DW, O’Koren EG, Kan MJ et al. (2013) Identification of a tissue-specific, C/EBPbeta-dependent pathway of differentiation for murine peritoneal macrophages. Journal of immunology 191 (9):4665–4675. doi: 10.4049/jimmunol.1300581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim KW, Williams JW, Wang YT et al. (2016) MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes. The Journal of experimental medicine 213 (10):1951–1959. doi: 10.1084/jem.20160486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molawi K, Wolf Y, Kandalla PK et al. (2014) Progressive replacement of embryo-derived cardiac macrophages with age. The Journal of experimental medicine 211 (11):2151–2158. doi: 10.1084/jem.20140639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theurl I, Hilgendorf I, Nairz M et al. (2016) On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nature medicine 22 (8):945–951. doi: 10.1038/nm.4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeFalco T, Bhattacharya I, Williams AV et al. (2014) Yolk-sac-derived macrophages regulate fetal testis vascularization and morphogenesis. Proceedings of the National Academy of Sciences of the United States of America 111 (23):E2384–2393. doi: 10.1073/pnas.1400057111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Batti L, Sundukova M, Murana E et al. (2016) TMEM16F Regulates Spinal Microglial Function in Neuropathic Pain States. Cell reports 15 (12):2608–2615. doi: 10.1016/j.celrep.2016.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldmann T, Wieghofer P, Muller PF et al. (2013) A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nature neuroscience 16 (11):1618–1626. doi: 10.1038/nn.3531 [DOI] [PubMed] [Google Scholar]
- 60.Goldmann T, Zeller N, Raasch J et al. (2015) USP18 lack in microglia causes destructive interferonopathy of the mouse brain. The EMBO journal 34 (12):1612–1629. doi: 10.15252/embj.201490791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewitus GM, Konefal SC, Greenhalgh AD et al. (2016) Microglial TNF-alpha Suppresses Cocaine-Induced Plasticity and Behavioral Sensitization. Neuron 90 (3):483–491. doi: 10.1016/j.neuron.2016.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li D, Wang C, Yao Y et al. (2016) mTORC1 pathway disruption ameliorates brain inflammation following stroke via a shift in microglia phenotype from M1 type to M2 type. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 30 (10):3388–3399. doi: 10.1096/fj.201600495R [DOI] [PubMed] [Google Scholar]
- 63.Wolf Y, Shemer A, Polonsky M et al. (2017) Autonomous TNF is critical for in vivo monocyte survival in steady state and inflammation. The Journal of experimental medicine 214 (4):905–917. doi: 10.1084/jem.20160499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hulsmans M, Clauss S, Xiao L et al. (2017) Macrophages Facilitate Electrical Conduction in the Heart. Cell 169 (3):510–522 e520. doi: 10.1016/j.cell.2017.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao H, Danzi MC, Choi CS et al. (2017) Opposing Functions of Microglial and Macrophagic TNFR2 in the Pathogenesis of Experimental Autoimmune Encephalomyelitis. Cell reports 18 (1):198–212. doi: 10.1016/j.celrep.2016.11.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng J, Gu N, Zhou L et al. (2016) Microglia and monocytes synergistically promote the transition from acute to chronic pain after nerve injury. Nature communications 7:12029. doi: 10.1038/ncomms12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan DH, Li MO, Jenison MC et al. (2007) Autocrine/paracrine TGFbeta1 is required for the development of epidermal Langerhans cells. The Journal of experimental medicine 204 (11):2545–2552. doi: 10.1084/jem.20071401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zahner SP, Kel JM, Martina CA et al. (2011) Conditional deletion of TGF-betaR1 using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity. Journal of immunology 187 (10):5069–5076. doi: 10.4049/jimmunol.1101880 [DOI] [PubMed] [Google Scholar]
- 69.Samokhvalov IM, Samokhvalova NI, Nishikawa S (2007) Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 446 (7139):1056–1061. doi: 10.1038/nature05725 [DOI] [PubMed] [Google Scholar]
- 70.Boiers C, Carrelha J, Lutteropp M et al. (2013) Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell stem cell 13 (5):535–548. doi: 10.1016/j.stem.2013.08.012 [DOI] [PubMed] [Google Scholar]