Abstract
Bacteria encode homooligomeric single-stranded (ss) DNA-binding proteins (SSBs) that coat and protect ssDNA intermediates formed during genome maintenance reactions. The prototypical Escherichia coli SSB tetramer can bind ssDNA using multiple modes that differ by the number of bases bound per tetramer and the magnitude of the binding cooperativity. Our understanding of the mechanisms underlying cooperative ssDNA binding by SSBs has been hampered by the limited amount of structural information available for interfaces that link adjacent SSB proteins on ssDNA. Here we present a crystal structure of Bacillus subtilis SsbA bound to ssDNA. The structure resolves SsbA tetramers joined together by a ssDNA “bridge” and identifies an interface, termed the “bridge interface”, that links adjacent SSB tetramers through an evolutionarily-conserved surface near the ssDNA binding site. E. coli SSB variants with altered bridge interface residues bind ssDNA with reduced cooperativity and with an altered distribution of DNA binding modes. These variants are also more readily displaced from ssDNA by RecA than wild-type SSB. In spite of these biochemical differences, each variant is able to complement deletion of the ssb gene in E. coli. Together our data suggest a model in which the bridge interface contributes to cooperative ssDNA binding and SSB function but that destabilization of the bridge interface is tolerated in cells.
Keywords: DNA replication, DNA repair, Protein-DNA interactions, SSB, RecA
Introduction:
Single-stranded (ss) DNA-binding proteins (SSBs) bind ssDNA intermediates formed during cellular processes such as DNA replication, recombination, and repair [1–5]. SSB binding protects ssDNA from damage and suppresses formation of secondary structures that can block genome maintenance reactions. In bacteria, SSBs most commonly exist as tetramers that coat ssDNA by binding cooperatively to shared ssDNA molecules [6–11]. However, the structural mechanisms responsible for cooperative interactions remain ill-defined. SSBs have a second function in which they interact directly with numerous genome maintenance proteins [1, 12–18]. These interactions provide access to SSB/ssDNA substrates for proteins that associate with SSB.
Bacterial SSB subunits, including those of the prototypical Escherichia coli SSB (EcSSB), are comprised of an N-terminal DNA binding/tetramerization domain and a C-terminal protein interaction motif (Fig. 1a). The N-terminal domain is made up of an oligosaccharide/oligonucleotide-binding (OB) fold whereas the C-terminal region is comprised of an intrinsically disordered linker (IDL) capped by a highly-conserved “SSBCt” protein-docking motif. Within the OB domain, the ssDNA-binding interface is defined by aromatic residues that stack with the ssDNA bases and basic and polar residues that interact electrostatically with the ssDNA backbone [19–24].
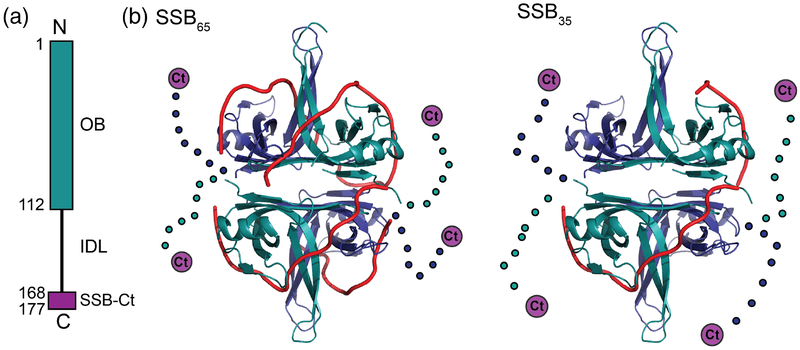
Figure 1. Bacterial SBB structure.
(a) Schematic diagram of a bacterial SSB. The N-terminal OB domain is shown along with the C-terminal IDL and the SSB-Ct (purple). (b) Ribbon diagrams of E. coli SSB65 (left) and SSB35 (right) ssDNA binding models[19]. The OB domains are shown in blue or teal, the IDL is shown schematically as dots, and the SSB-Ct is depicted as a purple circle, with ssDNA in red.
EcSSB binds DNA in multiple modes that vary by the number of ssDNA nucleotides that are occluded per SSB tetramer and intermolecular cooperativity [5, 9, 25–32]. One binding mode, called SSB65, wraps 65 nucleotides of ssDNA around each tetramer and residues from all four protomers contact the DNA [33–35] (Fig. 1b). The SSB65 mode is favored under high-salt conditions (>200 mM NaCl, >10 mM MgCl2) and low SSB/ssDNA ratios in vitro [9, 30]. A second mode, SSB35, wraps 35 nucleotides of ssDNA around each SSB tetramer, engaging an average of only two OB domains per tetramer for ssDNA binding (Fig. 1b). The SSB35 mode is favored under low-salt conditions (< 20 mM NaCl, <1 mM MgCl2) and high SSB/DNA ratios [11, 36]. In addition to these modes, a third mode (SSB56) is observed at intermediate salt concentrations [5, 26, 30]. The physiological concentration of SSB is estimated to range from ~500 tetramers/cell in minimal media to upwards of 3,600 tetramers in rich media [37, 38].
SSB modes also vary in their observed cooperative binding to ssDNA in vitro. The SSB65 mode has limited positive cooperativity, wrapping DNA in “beaded” octameric units [6, 28, 29, 39, 40]. In the SSB35 mode, DNA binding cooperativity is very highly positive [9] and SSB/ssDNA complexes can be visualized as large protein clusters by electron microscopy [40, 41]. Recently, it has been shown that highly cooperative binding of EcSSB to ssDNA is dependent on the IDL [10, 11]. This highly cooperative binding likely has contributions from interactions between non-nearest neighbor SSB tetramers and is known to be enhanced in the presence of glutamate, an abundant free metabolite in bacteria [10, 11, 72]. SSB binding modes have been shown to have differential effects on SSB/ssDNA substrate utilization by RecA, a recombinase that is dependent upon ssDNA binding for ATP hydrolysis. Specifically, the SSB65 mode stimulates RecA ATP hydrolysis in a condition-dependent manner in vitro [40, 42–45].
Although SSB binding modes and cooperativity have been investigated for decades, the inter-protein interfaces that mediate cooperative interactions in any binding mode remain poorly defined.
To better understand how bacterial SSBs bind ssDNA, we have determined X-ray crystal structures of free and ssDNA-bound forms of the essential SSB from Bacillus subtilis, BsSsbA. A ssDNA “bridge” was observed between BsSsbA tetramers in the DNA-bound structure and a novel intermolecular interface (termed the “bridge interface”) was formed near the ssDNA linking two BsSsbA tetramers. Two EcSSB bridge interface variants displayed reduced ssDNA binding cooperativity and altered distributions of DNA binding modes relative to the wild-type protein. These variants were also more readily displaced from ssDNA by RecA than EcSSB. Despite these differences, the EcSSB bridge interface variants complemented an ssb gene deletion in E. coli, with only a minor DNA repair defect observed for one of the variants. These results suggest that the bridge interface has a role in formation and stabilization of higher-order SSB/ssDNA complexes but that changes to these properties are tolerated in bacteria.
Results:
Crystal structures of B. subtilis SsbA in free and ssDNA-bound forms
To better understand the structural basis of bacterial SSB ssDNA binding, we crystallized BsSsbA (36% identical to EcSSB) in the presence of dT35 ssDNA. Two crystal forms were observed and both structures were determined by molecular replacement (Table 1). One crystal form diffracted to 2.2-Å resolution and contained two BsSsbA tetramers per asymmetric unit but lacked electron density for ssDNA. The second crystal form diffracted to 2.9-Å resolution – it contained a single BsSsbA tetramer per asymmetric unit and had clear electron density for ssDNA (Fig. 2 and 3). Differences between the free and ssDNA-bound BsSsbA structures were apparent in the L12 and L45 loops, which are involved in ssDNA binding and/or inter-tetramer interfaces (Fig. 4, and see below). These loops are named based on their positions between beta strands 1 and 2 (L12) or 4 and 5 (L45) within the OB fold [46]. Cα atoms in the L12 and L45 loops differ between the two structures with root mean square displacements of 1.4 and 3.6 Å, respectively.
Table 1:
X-ray data collection and structural refinement statistics
| Data collection | ||
| Structure | BsSsbA with DNA | BsSsbA |
| Wavelength (Å) | 0.97872 | 1.0782 |
| Resolution range (highest resolution bin) (Å) | 50 − 2.92 (2.97 − 2.92) | 50 − 2.2 (2.25 − 2.2) |
| Space group | P212121 | P41212 |
| Unit cell | ||
| a, b, c (Å) | 57.939, 93.681, 100.968 | 97.479, 97.479, 213.595 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Completeness (%) | 99.7 (98.4) | 100 (100) |
| Unique reflections/total collected | 12,270/93,003 | 52,594/846,636 |
| Redundancy | 7.6 (6.3) | 16.1 (15.5) |
| <I/σI> | 14.48 (2.29) | 36.4 (3.04) |
| Rmergea (%) | 12.8 (49.6) | 12.8 (132.0) |
| CC1/2 | Not determined | (0.814) |
| Refinement | ||
| Resolution range (highest resolution bin) (Å) | 44.28 − 2.94 | 40.22 − 2.21 |
| Rwork/Rfreeb (%) | 21.6/26.9 | 23.2/25.8 |
| r.m.s.c deviations | ||
| Bonds (Å) | 0.002 | 0.002 |
| Angles (Å) | 0.49 | 0.46 |
| Ramachandran statistics (%) | ||
| Favored | 95.2 | 96.42 |
| Allowed | 4.8 | 3.58 |
| Disallowed | 0.0 | 0.0 |
| Rotamer outliers (%) | 0.0 | 0.45 |
| No. atoms | ||
| Macromolecules | 3296 | 6322 |
| Ligands | - | 35 |
| Solvent | 31 | 256 |
| <B factor> (Å2) | ||
| Macromolecules | 43.45 | 46.25 |
| Ligands | - | 57.93 |
| Solvent | 32.8 | 42.91 |
Rmerge = ΣΣj|Ij-I|ΣIj, where Ij is the intensity measurement for reflection j and I is the mean intensity of multiple recorded reflections
Rwork/Rfree = Σ||Fobs|-|Fcalc||/|Fobs|, where the working the free R factors are calculated by using the working and free reflection sets, respectively. The free R reflections were held aside throughout refinement.
Root mean square
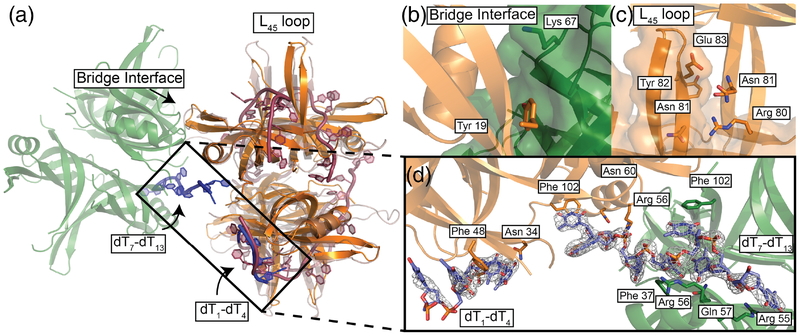
Figure 2. B. subtilis SsbA structure with ‘bridging’ ssDNA between tetramers.
(a) BsSsbA structure (orange) with ‘bridging’ ssDNA (blue) joining it to an adjacent symmetrically related tetramer (green). EcSSB (1EYG) structure (raspberry) alignment showing overlap in ssDNA between structures. The L45 loop and bridge interface are highlighted. (b) Bridge interface with Tyr19 and Lys67 highlighted. (c) L45 loop interface with Arg80, Asn81, Tyr82, and Glu83 highlighted. (d) Omit Fo-Fc electron density map (1.5 σ) of ssDNA ‘bridging’ SsbA tetramers with labeled interacting residues.
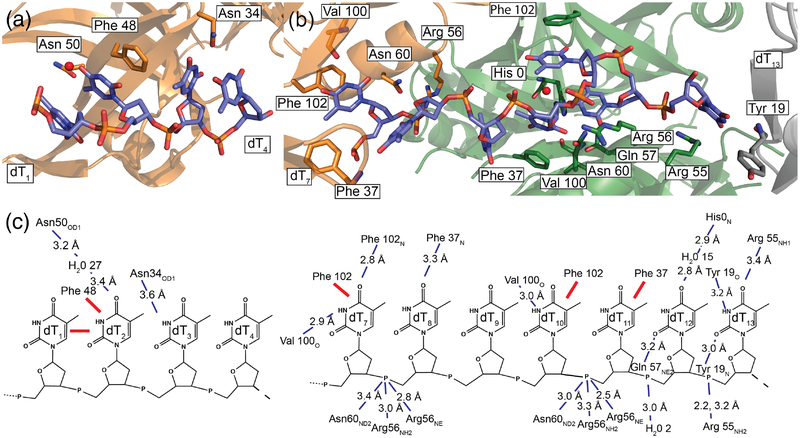
Figure 3. BsSsbA DNA interactions.
(a) dT1-dT4 with interacting residues highlighted. (b) dT7 -dT13 with interacting residues highlighted. (c) Diagram illustrating stacking interactions (red lines) and hydrogen/polar bonding with distances shown (blue dashed lines).
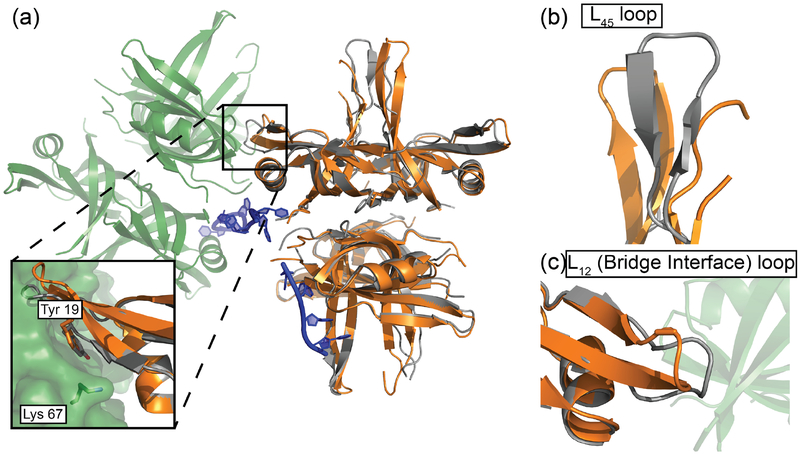
Figure 4. Superimposed DNA-bound and apo B. subtilis SsbA structures.
(a) BsSsbA (grey) asymmetric unit overlaid with DNA-bound BsSsbA (orange) and a symmetry mate (green). Inset shows bridge interfaces found in DNA-bound structure. Tyr19 and Lys67 are highlighted. (b) Overlay of L45 loops in BsSsbA/DNA and BsSsbA. (c) Overlay of L12 loops in BsSsbA/DNA and BsSsbA.
Electron density for two discrete segments of ssDNA was observed in the BsSsbA/ssDNA structure (Fig. 2d). The first segment (dT1-dT4) is bound by a single BsSsbA tetramer whereas the second (dT7-dT13) bridges two symmetrically-related BsSsbA tetramers. Each segment was shorter than the 35-base DNA ligand that was included in the crystallization conditions. Similar short segments have been resolved in other SSB/ssDNA crystal structures [19, 47, 48] and the segments likely represent stably bound portions of the longer ssDNA included in the crystallization conditions. Even though SSB is able to diffuse along ssDNA [44], the short DNA segments resolved in the structure likely represent base- or phosphodiester backbone-binding regions that are most significantly occupied within in the crystal system.
Apparent protein contacts to the phosphodiester backbone and bases of the ssDNA form the SSB/DNA interface (Fig. 2 and 3). The dT1-dT4 segment is contacted by the side chains of Asn50 (binding dT2 through a water molecule) and Asn34 (dT3). Additionally, Phe48 stacks with the base of dT2, which in turn stacks against dT1. The dT7-dT13 segment shows stacking interactions with Phe102 (dT7, dT10) and Phe37 (dT11). Interactions are also formed with the phosphodiester backbone via Asn60 and Arg56 (dT8, dT11), and Arg55 (dT13). Additional contacts are made by main-chain carbonyl or amide groups of Val100 (dT7, dT10), Phe102 (dT7), Phe37 (dT8), His0 (from His-tag, dT12), and Tyr19 (dT13). Finally, the side-chains of Gln57 (dT12) and Arg55 (dT13) contact bases.
Superimposition of a ssDNA-binding model derived from the EcSSB/ssDNA complex crystal structure [19] with the BsSsbA/ssDNA structure shows that a portion of the ssDNA binding groove in EcSSB corresponds to the dT1-dT4 segment (Fig. 2a). The EcSSB structure resolved more ssDNA however, with DNA wrapping observed around all four protomers. In EcSSB, the segment corresponding to dT1-dT4 is the entry/exit site in a model of its fully-wrapped SSB65 binding mode. This similarity suggests that the dT1-dT4 segment may also mark the exit/entry point for ssDNA wrapped around BsSsbA.
Bridge interface in ssDNA-bound BsSsbA
The ssDNA-bound BsSsbA structure revealed a protein interface that links adjacent BsSsbA molecules within the crystal lattice (Fig. 2b). The interface was formed near the ssDNA bridge where BsSsbA tetramers abut one another on ssDNA. We refer to this interface as the bridge interface. A second interface was formed between L45 loops of two BsSsbA tetramers docking against one another (Fig. 2c). However, since this interface is observed in both the free and ssDNA-bound BsSsbA structures and similar packing has been observed in many prior SSB structures either with or without bound ssDNA [19, 46, 49–57], our subsequent studies focused on the bridge interface.
The BsSsbA bridge interface buries 431 Å2 of surface area, comprised primarily of Tyr19 from one tetramer docking into a pocket formed by several residues (Arg10, Thr12, Ala32, and Phe48) from the adjacent tetramer (Fig. 2b, Fig. S1). Additionally, Lys67 contacts the main-chain carbonyl of Leu17 from the adjacent tetramer in the interface and Asn22 contacts Arg44 and Glu45 through side-chain and main-chain contacts, respectively. Most of the bridge interface residues are evolutionarily well conserved among bacterial SSBs (Fig. S1), suggesting that the interface is preserved broadly across bacteria. The average crystallographic B value for bridge interface residues in the BsSsbA/ssDNA structure was 45.1 Å2, compared to 47.5 Å2 for all residues from interfacing monomers. In contrast, the bridge interface residues had higher B values in the DNA-free structure (61.9 Å2), compared to the average value for all residues from the monomers containing these residues (50.5 Å2) (Fig. 4 and S1). This is consistent with stabilization of the bridge interface in the ssDNA-bound complex.
To examine possible roles of the bridge interface in ssDNA binding and cooperativity, we created variants that altered bridge interface residues in EcSSB (Fig. S2a). EcSSB was chosen to carry out these experiments due to its well-defined binding modes and cooperativity, the large number of biochemical assays that have been developed to analyze its ssDNA binding properties, and because the bridge interface was well conserved between BsSsbA and EcSSB. Bridge interface EcSSB variants included Tyr22Ala (equivalent to Tyr19 in BsSsbA), Lys73Glu (Lys67 in BsSsbA), and a Tyr22Ala Lys73Glu double variant.
The EcSSB variants were purified and their folding stabilities were assessed via differential scanning fluorimetry [58] (Fig. S2). EcSSB had a midpoint of denaturation (Tm) of 66°C, similar to earlier Tm measurements of 69 and 71°C [59, 60]. The EcSSB variants had modestly altered thermal stabilities, with Tm values of 56.3 to 61.7°C, and appeared to unfold cooperatively. Therefore, the EcSSB variants are folded at temperatures tested in this study.
Gel shift analysis of ssDNA binding by EcSSB interface variants
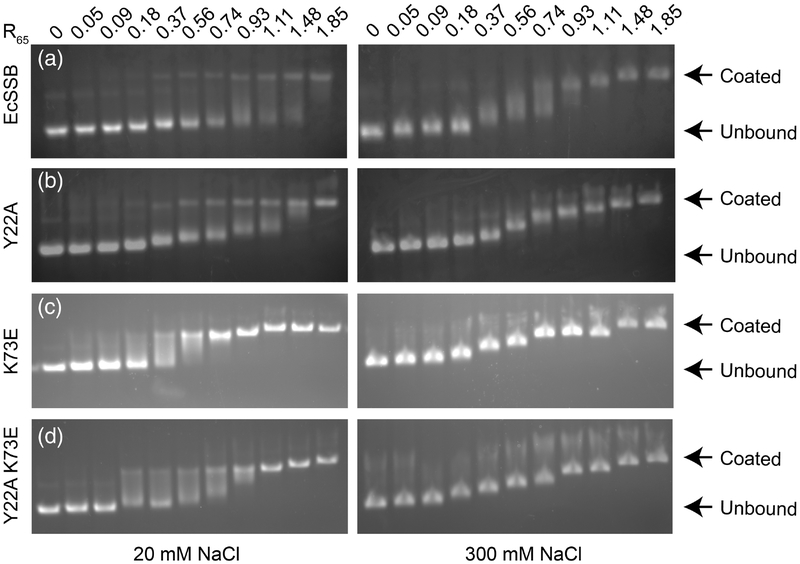
To assess the contributions of the bridge interface to ssDNA binding and intermolecular cooperativity, the DNA binding properties of the EcSSB variants were first investigated using an electrophoretic mobility shift assay (EMSA). Previous EMSA analyses of EcSSB have demonstrated a strong salt dependence in binding to long ssDNA such as the circular M13 bacteriophage genome (7.2 kb in length) [9, 48]. Consistent with published results, M13 ssDNA binding by SSB was found to be very highly cooperative in low salt conditions (20 mM NaCl), with the DNA migrating in a bimodal distribution reflecting DNA with little/no SSB bound in the same population with SSB-coated DNA (Fig. 5). In a higher salt concentration (300 mM NaCl), however, SSB binding to M13 was far less cooperative, with partially-coated SSB/ssDNA bands observed in an SSB concentration-dependent manner. Each of the EcSSB variants tested by EMSA displayed similar binding to that of EcSSB under high salt conditions. However, differences were observed in the low-salt binding assays. Each of the variants appeared to bind M13 ssDNA with mixed cooperativity, with the simultaneous appearance of both fully- and partially-coated ssDNA bands in subsaturating protein concentrations (Fig. 5). This difference is consistent with sequence changes in the bridge interface altering cooperative intermolecular interactions.
Figure 5. Electrophoretic mobility shift assays showing representative binding of SSB under low or high salt conditions.
EMSAs performed at 20 mM (left) and 300 mM NaCl (right) with EcSSB and bridge interface variants. Increasing protein:DNA ratios are shown from left to right with 0.54 being approximately half-saturation of 65-base sites by EcSSB tetramers. (a) EcSSB at 20 mM NaCl shows a bimodal binding distribution characteristic of cooperative binding. At 300 mM NaCl (right), EcSSB displays step-wise, less cooperative binding corresponding to the SSB65 mode. (b) Tyr22Ala, (c) Lys73Glu, and (d) Tyr22Ala Lys73Glu SSB variants at low (left) and high (right) salt concentrations.
Binding modes and cooperativity in the bridge interface EcSSB variants
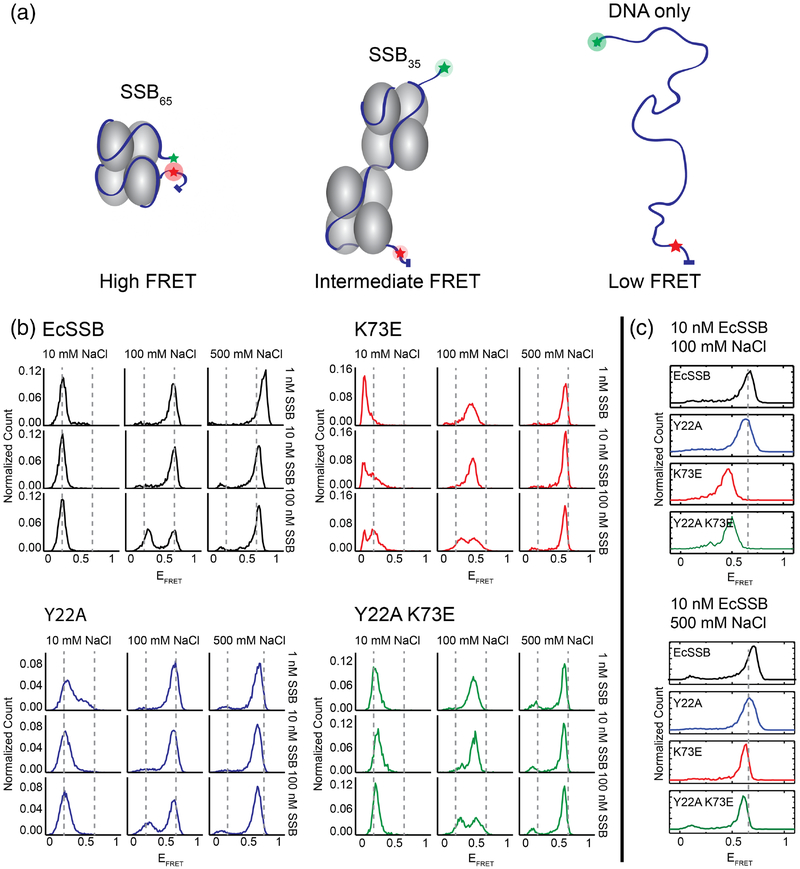
We next examined the impact of sequence changes in the EcSSB bridge interface on the cooperativity and site size of ssDNA binding. We first used a single-molecule (sm) FRET approach that monitors SSB binding modes on a ssDNA segment with Cy3 and Cy5 (donor and acceptor fluorophores) separated by 70 nt of ssDNA [27, 44] (Fig. 6a). In the absence of SSB, the average distance between the donor-acceptor pair is large, leading to a low FRET signal (EFRET ~0.1). Upon addition of EcSSB under conditions that favor the SSB65 mode, such as high salt (>200 mM NaCl) or low [SSB], a high FRET signal (EFRET ~ 0.65–0.75) is observed. This high FRET has been attributed to the FRET pair being brought close together near the entry/exit sites of the DNA in the SSB65 mode [27, 44, 61]. An intermediate FRET signal (EFRET ~ 0.2) is observed under low salt (10 mM NaCl) or high [SSB] conditions that favor the SSB35 mode. In this state, two SSB tetramers bind along the 70 nt ssDNA and the donor/acceptor pair are further apart than in the SSB65 mode but closer together than in the free ssDNA state.
Figure 6. Single-molecule FRET binding experiments of bridge interface variants.
(a) Schematic of SSB DNA binding leading to varied FRET efficiencies. (b) Normalized EFRET histograms of SSB binding to ssDNA with a range of NaCl and SSB concentrations. (c) Histograms for SSB variants showing shifted EFRET values. Lys73Glu and Tyr22Ala Lys73Glu variants show decreased EFRET values under conditions favoring the SSB65 mode (100 mM NaCl, 10 nM SSB condition is highlighted).
Two of the bridge interface variants differed from EcSSB in their binding behavior in the smFRET assay. When ssDNA was bound in intermediate NaCl concentrations (100 mM), binding by low concentrations of the Lys73Glu and Tyr22Ala Lys73Glu variants produced FRET signals (EFRET ~ 0.5) that were not consistent with either the SSB35 or the SSB65 state (Fig. 6b and 6c). Moreover, these EcSSB variants had a maximum FRET (EFRET ~ 0.6) under high salt conditions that differed from the SSB65 state for EcSSB. The Tyr22Ala variant behaved similarly to EcSSB under most of these conditions, except that with 100 nM Tyr22Ala EcSSB and 100 mM NaCl the variant appeared to show a somewhat stronger bias toward the SSB65 mode binding than EcSSB. Under high [SSB] and low salt conditions, the variants displayed SSB35-mode FRET efficiencies (EFRET ~ 0.2) normally observed with EcSSB, except that the Lys73Glu had an unusually low FRET signal that was below that of the free ssDNA (Fig 6b). These data are consistent with the Lys73Glu and Tyr22Ala Lys73Glu EcSSB variants binding ssDNA in an unusual, condition-specific manner. In particular, the lower FRET signals for the Lys73Glu and Tyr22Ala Lys73Glu variants (EFRET ~ 0.5–0.6) compared to EcSSB in the SSB65 state could be due to ssDNA fraying in an otherwise wrapped state, which would decrease the observed FRET signal.
Time-resolved smFRET traces of the Lys73Glu and Tyr22Ala Lys73Glu EcSSB further confirm that the variants access a lower maximum EFRET (~ 0.5–0.6) compared to EcSSB and Tyr22Ala EcSSB (~0.65 – 0.75), but that fluctuations between multiple FRET states are observed for all SSB variants examined (Fig. S3). The observation that Lys73Glu and Tyr22Ala Lys73Glu EcSSB reach a lower maximum EFRET may be due to the variants forming a frayed complex in which a portion of the ssDNA peels away from the SSB tetramer, rather than the fully wrapped mode in its maximal FRET state.
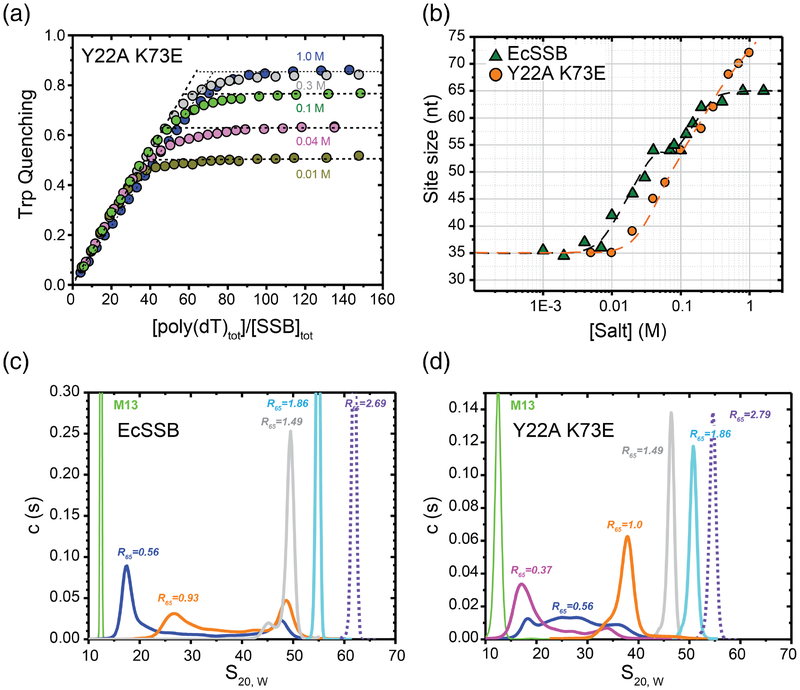
We next investigated the occluded site size and cooperativity of the Tyr22Ala Lys73Glu EcSSB variant. EcSSB tryptophan fluorescence quenching upon binding ssDNA is commonly used to estimate the occluded binding site size on ssDNA and to measure salt-dependent transitions between different binding modes [25, 26, 62]. Figures 7a and 7b show plots of the occluded site sizes (poly(dT) nucleotides occluded per SSB tetramer) as a function of [NaCl] for EcSSB and the Tyr22Ala Lys73Glu EcSSB variant. At low salt concentrations (<20 mM NaCl) EcSSB binds to poly(dT) in the SSB35 mode occluding ~35 nucleotides per tetramer accompanied by ~50% quenching of its Trp fluorescence. At high salt concentrations (>200 mM NaCl) the fluorescence is quenched by ~90% and with 65 nucleotides bound per tetramer (SSB65 mode) [25, 26, 62]. An intermediate SSB56 mode is populated for EcSSB at a narrow range of 50 to 100 mM NaCl. Titrations of the Tyr22Ala Lys73Glu variant with poly(dT) displayed similar [NaCl]-dependent Trp fluorescence quenching behavior (Fig. 7a) and similar occluded site sizes (Fig. 7b) with some notable differences. The [NaCl] required to transition from the SSB35 to the higher site size binding modes is notably higher, indicating a more stable SSB35 binding mode for the variant, and an explicit plateau reflecting the SSB56 mode is not observed for the variant (Fig. 7b). In addition, the binding site size appeared to increase beyond the 65 nt plateau observed for EcSSB at [NaCl] above 0.5M (Fig. 7a and b). These data indicate a deviation from the expected binding behavior at NaCl concentrations above 300 mM for the Tyr22Ala Lys73Glu EcSSB variant.
Figure 7. ssDNA binding properties of EcSSB versus the EcSSB Tyr22Ala Lys73Glu mutant.
(a) Occluded site sizes (nucleotides per tetramer) on poly(dT) plotted as a function of [NaCl] for the EcSSB Tyr22Ala Lys73Glu variant (orange circles) determined from analysis of stoichiometric titration curves, examples of which are shown in panel (b) (occluded site sizes for EcSSB from [24] are shown for comparison (green triangles). (b) Representative reverse equilibrium titrations of EcSSB Tyr22Ala Lys73Glu (0.2μM) with poly(dT) (monitoring intrinsic Trp fluorescence quenching) at different NaCl concentrations: 10mM – dark yellow, 40 mM – magenta, 100 mM – green, 300 mM – grey and 1 M – blue. The occluded site sizes were determined as described [57]. (c) and (d) Sedimentation coefficient distributions, c(s20,W), for EcSSB (c) and EcSSB Tyr22Ala Lys73Glu (d), in complex with M13 ssDNA in 10 mM NaCl at different protein to DNA ratios, R65, where R65=[SSBtetr, tot]×65/[M13ssDNAnts, tot]. Data were analyzed as described [10].
Sedimentation velocity experiments were next used to examine cooperative binding of the Tyr22Ala Lys73Glu EcSSB variant to M13 ssDNA. In these experiments, SSB binding to M13 ssDNA is detected as an increase in the average sedimentation coefficient of the DNA. The observation of a single peak in the distribution of sedimentation coefficients (s20, W) at less than saturating protein to DNA ratios is an indication of low cooperativity [10, 11]. At sub-saturating concentrations of EcSSB (R65 = 0.56, which is 56% saturating assuming a 65-nt binding site size) and low [NaCl] (0.01 M), EcSSB binds to M13 ssDNA with high cooperativity as indicated by the presence of a clear bimodal distribution of s20, W values of ~18S and ~45–50S (Fig. 7c). At a high SSB concentration (R65 = 1.49), only a single peak, corresponding to SSB-saturated ssDNA, is observed. In contrast, at sub-saturating concentrations (R65 = 0.56) of the Tyr22Ala Lys73Glu EcSSB variant a much broader distribution is observed (s20, W = ~20–40) indicating a decrease in cooperativity relative to EcSSB (Fig. 7d). Higher concentrations of the Tyr22Ala Lys73Glu EcSSB variant (R=1, 1.49 and 1.86) produced single peaks that shift to higher s20, w values, but these s20, w values were lower than observed for EcSSB at the equivalent ratios (Fig. 7c). This likely indicates that, in addition to a reduction in apparent cooperativity, the Tyr22Ala Lys73Glu EcSSB variant also has a lower overall affinity for M13 ssDNA.
The sedimentation velocity experiments were repeated in potassium glutamate, which enhances cooperativity of the SSB65 mode in EcSSB [10], to assess whether reduced cooperativity of the Tyr22Ala Lys73Glu EcSSB variant depends on the identity of the salt in the experimental condition. The data indicate that unlike EcSSB, which displayed cooperative binding, the variant did not appear to bind ssDNA cooperatively even in potassium glutamate (Fig. S4). These sedimentation velocity results are consistent with the EMSA data (Fig. 5) indicating a reduction in binding cooperativity in this bridge interface variant.
Impact of SSB bridge interface variants on use of SSB/ssDNA as a substrate by RecA
We next probed whether changes in the bridge interface that alter cooperativity and SSB binding modes have an impact on SSB function in an in vitro biochemical reaction. When bound first at saturating levels, SSB strongly inhibits RecA filament nucleation, leading to the observation of a long lag period in RecA-mediated ATP hydrolysis assays [63–66]. Conversely, if SSB is absent, RecA does not completely saturate ssDNA due to its poor binding to regions containing secondary structure (base pairing); subsequent addition of SSB stimulates RecA activity by melting out inhibitory DNA secondary structures that block RecA filament extension on ssDNA [42, 63]. Moreover, SSB is most effective in high MgCl2 concentrations that stabilize the SSB65 binding mode at equilibrium. SSB mode-dependent differences have been proposed to be due to SSB allowing RecA access to exposed ssDNA between bound SSBs along the DNA in the SSB65 binding mode but not in the more cooperative SSB35 mode [42, 67]. Based on the altered cooperativity of the Lys73Glu and Tyr22Ala Lys73Glu bridge interface SSB variants, we predicted that they would behave differently from EcSSB in RecA ATPase assays.
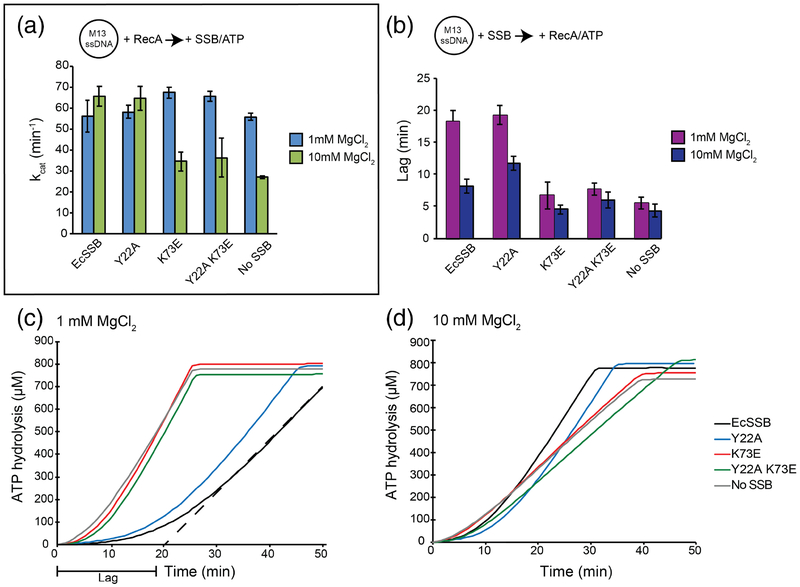
We first examined the impact of EcSSB or the bridge interface variants under assay conditions where RecA is preloaded onto M13 ssDNA, followed by the addition of ATP and SSB. Consistent with previously-observed SSB65-mode stimulation [42], the kcat for RecA ATPase activity in high (10 mM) MgCl2 was elevated ~2.4-fold by the addition of EcSSB (Fig. 8a and S6). With low (1 mM) MgCl2, which stabilizes the SSB35 mode, the kcat value was unaffected by the addition of EcSSB. At 10 mM MgCl2, the Tyr22Ala SSB variant stimulated RecA ATP hydrolysis (kcat = 64.7 ± 5.8 min−1), consistent with its unaltered binding cooperativity as observed in the smFRET assay. However, both the Lys73Glu and Tyr22Ala Lys73Glu variants were unable to stimulate RecA, closely matching results from conditions lacking EcSSB. These data support a model where the Lys73Glu and Tyr22Ala Lys73Glu bridge interface variants are destabilized on DNA leading to an inability to remove inhibitory secondary structures.
Figure 8. DNA-dependent ATPase activity and lag periods of RecA in the presence of SSB interface variants.
(a) M13 ssDNA was preincubated with RecA. After addition of ATP and SSB, ATP hydrolysis rates (kcat) were measured. Bar graph depicts the mean of three experiments with error bars representing the standard deviation. (b) M13 ssDNA preincubated with SSB was analyzed for a lag in ATP hydrolysis after the addition of ATP and RecA. Bar graph depicts the mean of three experiments with error bars representing the standard deviation. (c) and (d) Representative graphs of RecA lag periods with different SSB variants at 1 mM MgCl2 (c) 10 mM MgCl2 (d) concentrations. Representative dashed line in (c) depicts lag period calculation.
We next examined assays in which M13 ssDNA was first coated with SSB, followed by RecA and ATP addition. In this case, RecA must compete with SSB to find filament nucleation sites, leading to a prolonged lag time required for RecA to reach steady-state ATPase activity [42, 63–66, 68] (Fig. 8c and d). In the absence of SSB, RecA displayed lag times of 5.5 ± 1.0 or 4.3 ± 1.0 min in 1 or 10 mM MgCl2, respectively (Fig. 8b). Pre-binding of EcSSB to ssDNA increased RecA lag times to 18.4 ± 1.7 or 8.2 ± 1.1 min in 1 or 10 mM MgCl2. Consistent with its similar binding properties to EcSSB, the Tyr22Ala EcSSB variant showed lag times of 19.3 ± 1.5 min or 11.8 ± 1.1 min in 1 mM or 10 mM MgCl2. In contrast, both the Lys73Glu and Tyr22Ala Lys73Glu variants produced significantly shorter lags that were more similar to the SSB-free control reaction in both 1 and 10 mM MgCl2. These results are consistent with the altered ssDNA binding properties of the Lys73Glu and Tyr22Ala Lys73Glu bridge interface variants providing RecA greater access to the ssDNA and being displaced more readily by RecA.
Impact of SSB bridge interface variants in vivo
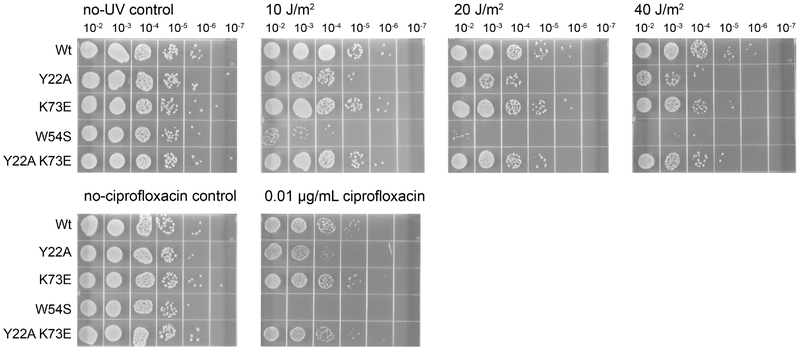
We next assessed whether the altered cooperativity and DNA-binding modes of the bridge interface variants affected SSB activity in vivo using a plasmid complementation assay. This assay takes advantage of an E. coli strain (RDP317) in which the genomic ssb gene has been deleted. Since ssb is essential, RPD317 requires a plasmid-borne copy of the ssb gene for viability [69]. This strain was transformed with compatible plasmids that included either wild-type ssb (positive control), vector alone (negative control), or an ssb gene encoding a bridge interface variant. Antibiotic selection for the variant plasmids and loss of the original wild-type ssb plasmid (not selected) was monitored by plating after several growth/dilution cycles. In addition, we analyzed a control SSB variant (Trp54Ser) that has an established ultraviolet (UV) light-sensitivity phenotype [70, 71]. With the exception of the vector-only negative control, all strains were able to survive without the original wild-type SSB-encoding plasmid, indicating that they were functional in vivo. Thus, the alterations to the bridge interface were tolerated in cells.
To determine whether DNA repair functions were compromised by the bridge interface changes, sensitivities to UV light were measured for the transformed RDP317 strains cured of the plasmid encoding EcSSB. As expected [70, 71], the Trp54Ser EcSSB variant-encoding E. coli strain was hypersensitive to UV light, even at the lowest level tested (10 J/m2) (Fig. 9). In addition, the strain carrying the Tyr22Ala SSB bridge interface variant was mildly sensitive to UV light exposure relative to the EcSSB control strain. UV light resistance of the Lys73Glu and Tyr22Ala Lys73Glu EcSSB strains was indistinguishable from the strain transformed with the wild-type ssb gene.
Figure 9. UV and ciprofloxacin sensitivity of EcSSB variants.
Strains contain EcSSB variants encoded by a pET21a-based plasmid under control of the ssb promoter. Trp54Ser EcSSB is used as a UV and ciprofloxacin-sensitive control.
To assess sensitivity to a different form of DNA damage, variant strains were also plated on media containing the DNA-damaging agent ciprofloxacin. The Trp54Ser EcSSB variant failed to grow in the presence of 0.01 μg/mL ciprofloxacin whereas the Tyr22Ala variant displayed a modest plating defect. In contrast, strains carrying the Lys73Glu or Tyr22Ala Lys73Glu bridge interface variants were indistinguishable from the wild-type strain (Fig. 9). Overall, the complementation results suggest that cells tolerate alterations to the SSB bridge interface, with only a modest sensitization to DNA-damaging agents for a single variant.
Discussion:
SSB proteins are essential for genome maintenance processes in all cells. Our understanding of how bacterial SSBs bind DNA in a cooperative manner and the impact of cooperativity on SSB function are not well understood. SSB binding modes display different cooperativities [5, 6, 9, 40] and there are likely multiple contributions to cooperative binding, including both nearest neighbor and non-nearest neighbor SSB interactions [10, 11, 36, 72]. This study examined the structural and biochemical basis of cooperative DNA binding in bacterial SSBs and tested the impact of altered ssDNA binding properties in cells. A crystal structure of ssDNA-bound BsSsbA identified an interface that formed near ssDNA to bridge symmetrically-related tetramers. Two EcSSB variants with altered residues in this bridge interface, Lys73Glu and Tyr22Ala Lys73Glu EcSSB, displayed a reduction in their cooperative binding behavior and an altered distribution of DNA binding modes. These variants were also more readily displaced from ssDNA by RecA than EcSSB. Interestingly, each of the variants complemented deletion of the ssb gene in E. coli. Our findings suggest that the bridge interface contributes to cooperative ssDNA binding and SSB function but that destabilization of the bridge interface is tolerated in bacterial cells.
The ssDNA-bound BsSsbA structure presented here identifies a ssDNA bridge linking two SSB tetramers. The SSB tetramers interact via a bridge interface that is evolutionarily well conserved among bacterial SSB proteins and is formed by residues from the L12 loop along with contributions from a small number of additional residues (Fig. 2 and S1). Previous SSB structures have shown L12 loops to be in proximity to one another within crystal lattices [50, 51, 53, 54, 73], but the present structure identified novel L12/L12 interactions that formed near ssDNA that links SSB tetramers. L45 loop interactions, which have previously been implicated in cooperative interactions [19, 46], were also observed between BsSsbA tetramers. The presence of these interfaces in the BsSsbA structure could indicate that both (and perhaps more interfaces) play roles in inter-SSB interactions. Indeed, sequence changes that affected the bridge interface only partially reduced cooperative binding as observed in each of our assays (Fig. 5 and 7). The observed intermediate cooperativity may mean that SSB variants require increased SSB concentrations on DNA to nucleate and allow for cooperativity. It is also likely that multiple interfaces are important for SSB/SSB interactions on ssDNA. In fact, it has recently been shown that the presence and composition of the C-terminal IDL that connects the DNA binding core to the SSB-Ct is essential for highly cooperative binding of EcSSB to ssDNA [11]. Hence, multiple regions of EcSSB contribute to cooperative ssDNA binding and interactions between both nearest neighbor (adjacent) SSBs and non-nearest neighbor SSBs likely contribute to the observed cooperativity [10, 11, 72].
Sequence changes in the bridge interface affect DNA wrapping and cooperativity of binding. Tryptophan quenching data showed that, compared to EcSSB, one of the bridge interface variants, Tyr22Ala Lys73Glu EcSSB, preserves the SSB35 mode in higher salt concentrations (Fig. 7). Further, single-molecule FRET data suggest that both the Lys73Glu and Tyr22Ala Lys73Glu EcSSB variants bind ssDNA in modes that differ from both the SSB35 and the SSB65 modes. Together these data show that both the Lys73Glu and Tyr22Ala Lys73Glu variants bind ssDNA using aberrant modes.
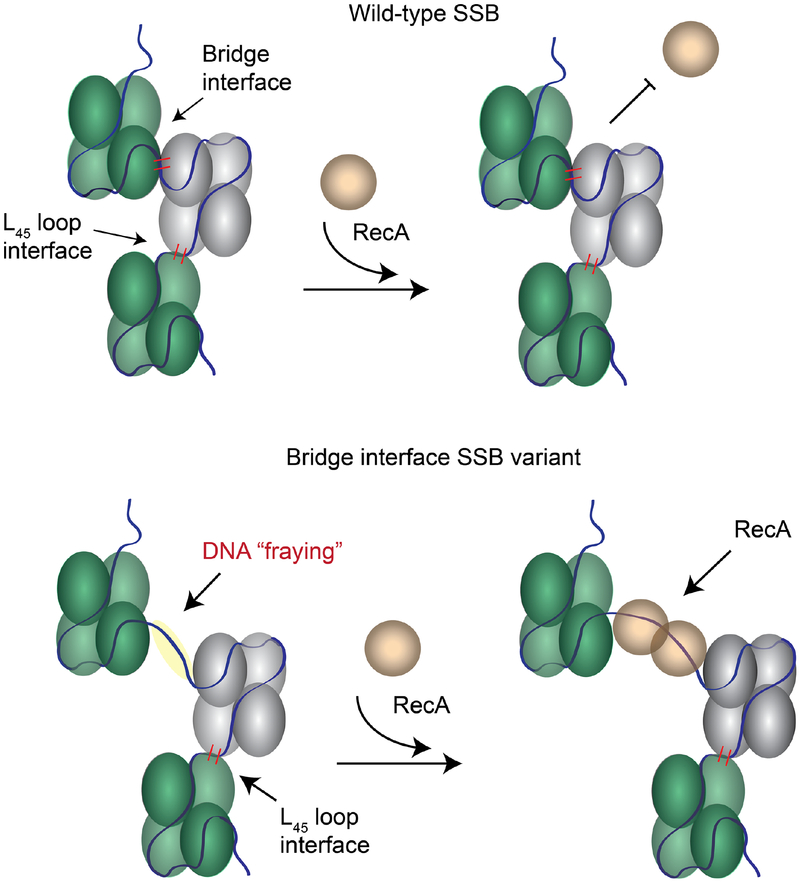
The altered cooperativity of the Lys73Glu and Tyr22Ala Lys73Glu bridge interface variants made them useful tools for examining the impact of cooperativity changes to SSB function. We tested the effect of these changes on the ssDNA-dependent ATPase activity of RecA. In RecA ATPase assays, both Lys73Glu and Tyr22Ala Lys73Glu failed to stimulate RecA ATP hydrolysis or to offer a barrier to RecA loading onto M13 ssDNA (Fig. 8). This may be due to their altered cooperativity and destabilized binding to M13 ssDNA. These altered properties would be expected to make displacement by RecA, which limits RecA nucleation on ssDNA [68], quite facile. The effects may also be related to the altered binding modes that were observed for each in smFRET experiments (Fig. 6 and S3). We noted that the FRET signal for both variants was unusually low for the SSB65 mode, which is consistent with the donor/acceptor pair being further apart than would be found in the EcSSB complex. This reduction could reflect fraying of ssDNA away from the surface of SSB at the entry/exit point between SSB tetramers. Such fraying may provide RecA greater access to ssDNA, allowing RecA to establish filaments more readily. Notably, the bridge interface includes Lys73, which has been shown to interact with DNA [24]. Sequence changes for Lys73 Lys73 could destabilize normal DNA wrapping around SSB leading to reduced cooperativity and affinity, and fraying ssDNA away from the SSB tetramer (Fig. 10).
Figure 10. Model of SSB cooperativity involving bridge and L45 interfaces.
DNA binding and direct protein interactions connect neighboring SSB tetramers (blue and green). (top) Both the L45 loop and bridge interface contribute to cooperative DNA binding, which can impede RecA access to ssDNA. (bottom) Disruption of the bridge interface disrupts cooperative binding, leading to DNA ‘fraying’ that presents readily-available ssDNA binding sites for RecA or other enzymes.
Genes encoding each of the EcSSB bridge interface variants were able to complement deletion of the native ssb gene. Only the Tyr22Ala variant showed a modest growth defect when stressed with DNA damaging agents (Fig. 9), although this variant appeared to bind DNA normally in vitro Thus, alterations in ssDNA binding properties of bridge interface variants observed in vitro are not sufficiently detrimental to render the protein non-functional in E. coli. This may be because cooperative ssDNA binding is not essential for SSB function in cells or because other self-interaction elements in SSB are sufficient to support the level of cooperativity needed for function in E. coli. Indeed, an EcSSB variant with a deleted IDL that binds ssDNA with low cooperativity can also complement ssb deletion in E. coli [11]. While additional factors beyond the bridge interface are clearly involved in modulating SSB ssDNA binding properties in vitro and in vivo, our studies have identified a new interface that plays a role in cooperative ssDNA binding by SSBs.
Materials and methods:
DNA substrates and plasmids
dT35 was purchased (IDT) and purified by polyacrylamide gel electrophoresis. For smFRET experiments, two DNA strands were annealed: 5’- /Cy5/GCC TCG CTG CCG TCG CCA -/biotin/ −3’ and 5’- TGG CGA CGG CAG CGA GGC (T)72-/Cy3/ −3’ [27]. Bacteriophage M13mp18 ssDNA was prepared as previously described [74–76]. Overexpression plasmids encoding EcSSB and N-terminally His-tagged BsSsbA were created by cloning the respective open reading frames into pET21a and pET11a, creating pET21a-EcSSB and pET11a-SsbA. Mutations of pET21a-EcSSB were made by single primer site-directed mutagenesis.
Proteins
SSBs:
E. coli BL21 (DE3) pLysS cells transformed with pET21a-SSB, a pET21a-SSB mutant, or pET11a-BsSsbA were grown to midlog phase (OD600 ~0.6) at 37°C in Lysogeny Broth (LB) medium [77] supplemented with 50 μg/mL ampicillin and 25 μg/mL chloramphenicol. Protein expression was induced by the addition of 1 mM isopropyl β-D-thiogalactopyranoside and cells were grown for an additional 3–4 hours. EcSSB and variants were purified as previously described [39]. Cells overexpressing BsSsbA were resuspended in Lysis buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 10% w/v glycerol, 20 mM imidazole, 2 mM phenylmethanesulfonyl fluoride, 2 mM benzamidine, and 1 protease cocktail inhibitor tablet (Pierce)) and disrupted using sonication. BsSsbA in the soluble fraction was incubated with Ni-NTA Agarose resin (GE) equilibrated with Lysis buffer. BsSsbA was washed with 0.1 L Lysis buffer and eluted after incubation with Elution buffer (10 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 0.5 M imidazole, 10% w/v glycerol). The concentrated BsSsbA eluent was further purified using a Sephacryl S-100 size exclusion column equilibrated with S100 buffer (10 mM Tris-HCl, pH 8.0, 1 M NaCl, 1 mM EDTA). Pure protein was pooled and concentrated. Protein concentrations were determined spectrophotometrically using ε280 = 2.8 × 104 M−1 cm−1 for monomeric SSB and Lys73Glu EcSSB, ε280 = 2.6 × 104 M−1 cm−1 for monomeric Tyr22Ala and Tyr22Ala Lys73Glu EcSSB.
RecA:
EcRecA was a gift from Michael M. Cox (UW Madison).
Crystallization and structure determination of BsSsbA
N-terminally His-tagged BsSsbA was prepared for crystallization studies as described above. BsSsbA was incubated with dT35 at a 1:2 BsSsbA tetramer:dT35 ratio, and α-chymotrypsin was added at a 100:1 (BsSsbA tetramer:α-chymotrypsin) ratio for 3 hours on ice. The reaction was stopped with 1 mM phenylmethanesulfonyl fluoride and diluted 4-fold with 10 mM Tris-HCl, pH 8.0. The sample was purified using a Sephacryl S-100 size exclusion column equilibrated with 10 mM Tris-HCl, pH 8.0, 0.1 M NaCl, 5 mM EDTA. The pure protein/DNA sample was pooled, concentrated to 10–30 mg/ml in 10 mM Tris-HCl, pH 8.0, 100 mM NaCl and crystallized by mixing proportionately with mother liquor #1 (50 mM MES-HCl pH 6.5, 7% v/v PEG 8000, 100 mM magnesium acetate, 200 mM KCl) using hanging-drop vapor-diffusion crystallization. BsSsbA/DNA crystals were transferred into 50 mM MES, pH 6.5, 100 mM magnesium acetate, 150 mM KCl, 8% v/v PEG 8000, 25% v/v ethylene glycol, and frozen in liquid nitrogen. Diffraction data were indexed and scaled using HKL2000 [78]. The BsSsbA/DNA structure was determined by molecular replacement with the previously determined BsSsbA structure (see below) using Phenix [79]. Iterative rounds of model building and refinement were carried out using Coot and Phenix [80].
The BsSsbA structure without visible DNA was determined as above, with the following deviations. B. subtilis SsbA was thrombin cleaved prior to DNA incubation and α-chymotrypsin digestion. The pure protein/DNA sample was pooled and concentrated (10–30) mg/ml in 10 mM Tris-HCl, pH 8.0, 100 mM NaCl). The protein sample was crystallized by mixing 1 μL protein with 1 μL mother liquor #2 (50 mM MES-HCl, pH 6.5, 5% v/v PEG 8000, 80 mM magnesium acetate, 200 mM KCl) using hanging-drop vapor-diffusion crystallization. BsSsbA crystals were transferred into above cryo protectant and frozen in liquid nitrogen. Diffraction data were indexed and scaled using HKL2000 [78]. The BsSsbA structure was determined by molecular replacement with the BsSsbB structure using Phenix [79]. Iterative rounds of model building and refinement were carries out using Coot and Phenix [79, 80].
Evolutionary conservation
Consurf was used to align 300 bacterial species with EcSSB chain A (E-value, 0.0001; maximum percent identity between sequences, 95; minimum percent identity between homologs, 35) (1EYG) [81–85]. The conservation scores were visualized with PyMol with an average pairwise distance of DNA sequences of 0.311 [86].
Electrophoretic mobility shift assay
EMSAs were performed as described [9], with the following changes. All reactions were completed in buffer T (10 mM Tris-HCl, pH 8.1, 0.1 mM EDTA), and specified [NaCl] with 4.7 nM (nucleotide) M13 mp18ssDNA in a 30 μL volume. Reactions were incubated at 22°C for 1 hour. Electrophoresis was carried out using 0.3% agarose gels at 22°C for 3 hours at 60 V. Gels were then soaked in buffer T with 1 M NaCl for 1–2 hours at 4°C, and stained with a 2 μg/ml solution of ethidium bromide in buffer T with 1 M NaCl for 30 min. Gels were destained for 1–2 hours at 4°C in buffer T prior to visualization.
Single-molecule FRET
We used a home-built two-color wide-field total internal reflection microscope [87]. DNA labeled with a donor and an acceptor was attached to a polymer-passivated surface and single molecule fluorescence images were acquired in the presence of SSB. Imaging was performed in 10 mM Tris-HCl (pH 8.0), 0.1 mg/mL BSA, 0.8 % (w/v) dextrose, 165 U/mL glucose oxidase, 2170 U/mL catalase, 2–3 mM Trolox, and indicated amounts of SSB and NaCl. Details of the experimental protocols and analysis methods are presented in [88].
Intrinsic tryptophan quenching
Titrations of Tyr22Ala Lys73Glu EcSSB with poly(dT) were performed by monitoring quenching of the intrinsic SSB tryptophan fluorescence and analyzed as described [11, 62]
Analytical sedimentation
Sedimentation velocity experiments were performed with an Optima XL-A analytical ultracentrifuge and An50Ti rotor (Beckman Instruments, Fullerton, CA) at 15000 rpm (25°C) as described [11].
RecA ATPase activity assay
RecA ATPase activity assays were performed as previously described [42], with the following changes. Five μM (nucleotides) M13mp18 ssDNA and 1.7 μM RecA were pre-incubated at 37°C for 10 min in buffer containing 20 mM Tris-HCl, pH 7.5, 440 μM KCl, 5% w/v glycerol, 1 mM dithiothreitol, 2.31 mM phosphoenolpyruvate, 1 mM NADH, 2.24 U/mL pyruvate kinase, 3.36 U/mL lactic dehydrogenase, and variable [MgCl2]. After incubation, 1 mM ATP and 0.97 μM SSB or SSB variant monomers were added to the reaction. The conversion of NADH to NAD+ was measured as a decrease in absorbance at 340 nM monitored every 45 seconds for 1.5 hours. ATP hydrolysis rate was assessed using the NADH extinction coefficient at 340 nm of 6.22 mM−1cm−1. kcat was calculated using equation 1:
| Eq. 1 |
The concentration of RecA bound to DNA and able to hydrolyze ATP is calculated by the DNA site size of RecA and total RecA concentration. In ATP saturating conditions, Vo is determined from the linear ATP hydrolysis rate. Data were collected on a BioTek Synergy 2 microplate reader using Gen5 version 1.11 software. Lag time RecA ATPase assays were performed as described above with SSB and M13 mp18 ssDNA pre-incubation, prior to addition of RecA and ATP. All experiments were completed in triplicate.
Plasmid complementation assay
In vivo plasmid complementation assays were performed as described [48, 69]. Briefly, ssb or ssb mutant genes were subcloned into pET21a (AmpR) with gene expression driven by the ssb promoter. The E. coli RDP317 strain, which carries a kan insertion in the genomic ssb locus but is complemented with a plasmid that contains the wild-type E. coli ssb gene and a tetR gene, were transformed with the test ssb or ssb mutant plasmids. Individual colonies were inoculated into 5 mL LB and grown at 37°C. Cells were passaged seven times, with 14–16 hours of growth for each passage, with Amp/Kan selection. Cells were diluted to 10−7, plated on LB plates supplemented with 50 μg/mL ampicillin and 50 μg/mL kanamycin, and grown overnight at 37°C. Plates were replica plated serially onto LB with 50 μg/mL ampicillin, LB with 34 μg/mL tetracycline, and then LB with 50 μg/mL kanamycin. Strains that are resistant to ampicillin and kanamycin but not tetracycline carry an active ssb gene. All experiments were completed in triplicate and plasmids were validated with sequencing after passaging.
UV radiation and ciprofloxacin sensitivity tests
Passaged E. coli RDP317 strains described above were tested for UV and ciprofloxacin sensitivity. Cells were grown overnight at 37°C in LB supplemented with 50 μg/mL ampicillin and 50 μg/mL kanamycin, diluted into fresh media, grown again, and diluted to an OD600 of ~1.0. Cells were serially diluted in 0.9% w/v sterile NaCl, and 10 μL of 10−2-10−7 were spotted onto LB agar or LB containing 0.1 μg/mL ciprofloxacin. For UV sensitivity, cells were exposed to shortwave light (254 nm) using a Spectrolinker XL-1000 UV crosslinker (Spectronics Corp) to the dose indicated after spot plating. Images were taken after growth at 37° overnight.
Differential scanning fluorimetry
Differential scanning fluorimetry experiments were performed as described previously [58]. Briefly, SSB or SSB variants were dialyzed overnight at 4°C into 10 mM Tris-HCl, pH 8.1, 0.1 mM EDTA, and 200 mM NaCl. SSB (10 μM) and 10× Sypro Orange dye (Thermo Fisher, 5000× stock) were combined and incubated at temperatures of 25°C to 90°C (1°C/minute ramp speed) on a Bio-Rad CFX Connect Real-Time PCR using CFX Manager Software, with measurements taken every 30 seconds. Tm values were determined as the minimum of the first derivative (CFX Manager Software).
Supplementary Material
Highlights:
An interface that links adjacent B. subtilis SsbA proteins on ssDNA is identified.
E. coli SSB cooperativity is affected by this inter-SSB contact.
Interface disruption facilitates SSB displacement from ssDNA by RecA.
Acknowledgements:
We thank Kenneth Satyshur and staff at the Advanced Photon Source (LS-CAT beamline) for assistance with data collection. We thank the Cox lab for RecA protein. We thank the Keck, Lohman, and Ha labs for critical evaluation of the manuscript.
Abbreviations:
- ssDNA
single-stranded DNA
- EcSSB
Escherichia coli ssDNA-binding protein
- BsSsbA
Bacillus subtilis ssDNA-binding protein A
- BsSsbB
Bacillus subtilis ssDNA-binding protein B
- OB
oligosaccharide/oligonucleotide-binding
- IDL
intrinsically disordered linker
- smFRET
single-molecule FRET
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers:
B. subtilis SsbA with DNA PDB ID: 6BHX; B. subtilis SsbA PDB ID: 6BHW; E. coli SSB with DNA PDB ID: 1EYG; B. subtilis SsbB PDB ID: 3VDY
References:
- [1].Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Critical reviews in biochemistry and molecular biology. 2008;43:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Molineux IJ, Pauli A, Gefter ML. Physical studies of the interaction between the Escherichia coli DNA binding protein and nucleic acids. Nucleic Acids Research. 1975;2:1821–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Molineux IJ, Gefter ML. Properties of the Escherichia coli DNA-binding (unwinding) protein interaction with nucleolytic enzymes and DNA. J Mol Biol. 1975;98:811–25. [DOI] [PubMed] [Google Scholar]
- [4].Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiological Reviews. 1990;54:342–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lohman TM, Ferrari ME. Escherichia Coli Single-Stranded DNA-Binding Protein: Multiple DNA-Binding Modes and Cooperativities. Annual Review of Biochemistry. 1994;63:527–70. [DOI] [PubMed] [Google Scholar]
- [6].Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:5803–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sigal N, Delius H, Kornberg T, Gefter ML, Alberts B. A DNA-Unwinding Protein Isolated from Escherichia coli: Its Interaction with DNA and with DNA Polymerases. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:3537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ruyechan WT, Wetmur JG. Studies on the cooperative binding of the Escherichia coli DNA unwinding protein to single-stranded DNA. Biochemistry. 1975;14:5529–34. [DOI] [PubMed] [Google Scholar]
- [9].Lohman TM, Overman LB, Datta S. Salt-dependent changes in the DNA binding cooperativity of Escherichia coli single strand binding protein. Journal of Molecular Biology.1986;187:603–15. [DOI] [PubMed] [Google Scholar]
- [10].Kozlov AG, Shinn MK, Weiland EA, Lohman TM. Glutamate promotes SSB protein–protein Interactions via intrinsically disordered regions. Journal of Molecular Biology. 2017;429:2790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kozlov AG, Weiland E, Mittal A, Waldman V, Antony E, Fazio N, et al. Intrinsically Disordered C-Terminal Tails of E. coli Single-Stranded DNA Binding Protein Regulate Cooperative Binding to Single-Stranded DNA. Journal of Molecular Biology. 2015;427:763–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proceedings of the National Academy of Sciences. 2008;105:9169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Petzold C, Marceau AH, Miller KH, Marqusee S, Keck JL. Interaction with Single-stranded DNA-binding Protein Stimulates Escherichia coli Ribonuclease HI Enzymatic Activity. Journal of Biological Chemistry. 2015;290:14626–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB Binding Site on E. coli RecQ Reveals a Conserved Surface for Binding SSB’s C Terminus. Journal of Molecular Biology. 2009;386:612–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shereda RD, Bernstein DA, Keck JL. A Central Role for SSB in Escherichia coli RecQ DNA Helicase Function. Journal of Biological Chemistry. 2007;282:19247–58. [DOI] [PubMed] [Google Scholar]
- [16].Chen SH, Byrne-Nash RT, Cox MM. Escherichia coli RadD Protein Functionally Interacts with the Single-stranded DNA-binding Protein. J Biol Chem. 2016;291:20779–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Page AN, George NP, Marceau AH, Cox MM, Keck JL. Structure and biochemical activities of Escherichia coli MgsA. J Biol Chem. 2011;286:12075–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kozlov AG, Eggington JM, Cox MM, Lohman TM. Binding of the Dimeric Deinococcus radiodurans Single-Stranded DNA Binding Protein to Single-Stranded DNA. Biochemistry. 2010;49:8266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Mol Biol. 2000;7:648–52. [DOI] [PubMed] [Google Scholar]
- [20].Bayer I, Fliess A, Greipel J, Urbanke C, Maass G. Modulation of the affinity of the single-stranded DNA-binding protein of Escherichia coli (E. coli SSB) to poly(dT) by site-directed mutagenesis. European Journal of Biochemistry. 1989;179:399–404. [DOI] [PubMed] [Google Scholar]
- [21].Merrill BM, Williams KR, Chase JW, Konigsberg WH. Photochemical cross-linking of the Escherichia coli single-stranded DNA-binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross-linking. Journal of Biological Chemistry. 1984;259:10850–6. [PubMed] [Google Scholar]
- [22].Casas-Finet JR, Khamis MI, Maki AH, Chase JW. Tryptophan 54 and phenylalanine 60 are involved synergistically in the binding of E. coli SSB protein to single-stranded polynucleotides. FEBS Letters. 1987;220:347–52. [DOI] [PubMed] [Google Scholar]
- [23].Khamis MI, Casas-Finet JR, Maki AH, Murphy JB, Chase JW. Role of tryptophan 54 in the binding of E. coli single-stranded DNA-binding protein to single-stranded polynucleotides. FEBS Letters. 1987;211:155–9. [DOI] [PubMed] [Google Scholar]
- [24].Chen J, Smith DL, Griep MA. The role of the 6 lysines and the terminal amine of Escherichia coli single-strand binding protein in its binding of single-stranded DNA. Protein Science: A Publication of the Protein Society. 1998;7:1781–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–603. [PubMed] [Google Scholar]
- [26].Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–802. [DOI] [PubMed] [Google Scholar]
- [27].Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. Journal of molecular biology. 2007;369:1244–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bujalowski W, Lohman TM. Limited co-operativity in protein-nucleic acid interactions. Journal of Molecular Biology. 1987;195:897–907. [DOI] [PubMed] [Google Scholar]
- [29].Overman LB, Bujalowski W, Lohman TM. Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry. 1988;27:456–71. [DOI] [PubMed] [Google Scholar]
- [30].Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. Journal of Biological Chemistry. 1988;263:4629–40. [PubMed] [Google Scholar]
- [31].Suksombat S, Khafizov R, Kozlov AG, Lohman TM, Chemla YR. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. eLife. 2015;4:e08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Waldman VM, Weiland E, Kozlov AG, Lohman TM. Is a fully wrapped SSB-DNA complex essential for Escherichia coli survival? Nucleic Acids Res. 2016;44:4317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lohman TM, Bujalowski W. Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry. 1988;27:2260–5. [DOI] [PubMed] [Google Scholar]
- [34].Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. Journal of Molecular Biology. 1989;207:249–68. [DOI] [PubMed] [Google Scholar]
- [35].Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. Journal of Molecular Biology. 1989;207:269–88. [DOI] [PubMed] [Google Scholar]
- [36].Ferrari ME, Bujalowski W, Lohman TM. Co-operative Binding of Escherichia coli SSB Tetramers to Single-stranded DNA in the (SSB)35 Binding Mode. Journal of Molecular Biology. 1994;236:106–23. [DOI] [PubMed] [Google Scholar]
- [37].Bobst EV, Bobst AM, Perrino FW, Meyer RR, Rein DC. Variability in the nucleic acid binding site size and the amount of single-stranded DNA-binding protein in Escherichia coli. FEBS Letters. 1985;181:133–7. [DOI] [PubMed] [Google Scholar]
- [38].Li G-W, Burkhardt D, Gross C, Weissman Jonathan S. Quantifying Absolute Protein Synthesis Rates Reveals Principles Underlying Allocation of Cellular Resources. Cell. 2014;157:624–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].George NP, Ngo KV, Chitteni-Pattu S, Norais CA, Battista JR, Cox MM, et al. Structure and Cellular Dynamics of Deinococcus radiodurans Single-stranded DNA (ssDNA)-binding Protein (SSB)-DNA Complexes. The Journal of Biological Chemistry. 2012;287:22123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Griffith JD, Harris LD, Register J. Visualization of SSB-ssDNA Complexes Active in the Assembly of Stable RecA-DNA Filaments. Cold Spring Harbor Symposia on Quantitative Biology. 1984;49:553–9. [DOI] [PubMed] [Google Scholar]
- [41].Hamon L, Pastre D, Dupaigne P, Le Breton C, Le Cam E, Pietrement O. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein--DNA complexes. Nucleic Acids Res. 2007;35:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Morrical SW, Cox MM. Stabilization of recA protein-ssDNA complexes by the single-stranded DNA binding protein of Escherichia coli. Biochemistry. 1990;29:837–43. [DOI] [PubMed] [Google Scholar]
- [43].Zhou R, Kozlov Alexander G, Roy R, Zhang J, Korolev S, Lohman Timothy M, et al. SSB Functions as a Sliding Platform that Migrates on DNA via Reptation. Cell. 2011;146:222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Roy R, Kozlov AG, Lohman TM, Ha T. SSB diffusion on single stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Muniyappa K, Williams K, Chase JW, Radding CM. Active nucleoprotein filaments of single-stranded binding protein and recA protein on single-stranded DNA have a regular repeating structure. Nucleic Acids Research. 1990;18:3967–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-Å resolution. Proceedings of the National Academy of Sciences. 1997;94:6652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yadav T, Carrasco B, Myers AR, George NP, Keck JL, Alonso JC. Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Research. 2012;40:5546–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Antony E, Weiland E, Yuan Q, Manhart CM, Nguyen B, Kozlov AG, et al. Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. Journal of molecular biology. 2013;425: 10.1016/j.jmb.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Matsumoto T, Morimoto Y, Shibata N, Kinebuchi T, Shimamoto N, Tsukihara T, et al. Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis. Journal of biochemistry. 2000;127:329–35. [DOI] [PubMed] [Google Scholar]
- [50].Saikrishnan K, Jeyakanthan J, Venkatesh J, Acharya N, Sekar K, Varshney U, et al. Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications. J Mol Biol. 2003;331:385–93. [DOI] [PubMed] [Google Scholar]
- [51].Jedrzejczak R, Dauter M, Dauter Z, Olszewski M, Dlugolecka A, Kur J. Structure of the single-stranded DNA-binding protein SSB from Thermus aquaticus. Acta crystallographica Section D, Biological crystallography. 2006;62:1407–12. [DOI] [PubMed] [Google Scholar]
- [52].Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J Mol Biol. 2009;388:508–19. [DOI] [PubMed] [Google Scholar]
- [53].Franklin MC, Cheung J, Rudolph MJ, Burshteyn F, Cassidy M, Gary E, et al. Structural genomics for drug design against the pathogen Coxiella burnetii. Proteins. 2015;83:2124–36. [DOI] [PubMed] [Google Scholar]
- [54].Kaushal PS, Singh P, Sharma A, Muniyappa K, Vijayan M. X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures. Acta crystallographica Section D, Biological crystallography. 2010;66:1048–58. [DOI] [PubMed] [Google Scholar]
- [55].Huang YH, Guan HH, Chen CJ, Huang CY. Staphylococcus aureus single-stranded DNA-binding protein SsbA can bind but cannot stimulate PriA helicase. PLoS One. 2017;12:e0182060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shishmarev D, Wang Y, Mason CE, Su XC, Oakley AJ, Graham B, et al. Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy. Nucleic Acids Res. 2014;42:2750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Savvides SN, Raghunathan S, Futterer K, Kozlov AG, Lohman TM, Waksman G. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols. 2007;2:2212–21. [DOI] [PubMed] [Google Scholar]
- [59].Olszewski M, Nowak M, Cyranka-Czaja A, Kur J. Identification and characterization of single-stranded DNA-binding protein from the facultative psychrophilic bacteria Pseudoalteromonas haloplanktis. Microbiological Research. 2014;169:139–47. [DOI] [PubMed] [Google Scholar]
- [60].Kozlov AG, Lohman TM. Adenine Base Unstacking Dominates the Observed Enthalpy and Heat Capacity Changes for the Escherichia coli SSB Tetramer Binding to Single-Stranded Oligoadenylates. Biochemistry. 1999;38:7388–97. [DOI] [PubMed] [Google Scholar]
- [61].Kozlov AG, Lohman TM. Kinetic Mechanism of Direct Transfer of Escherichia coli SSB Tetramers between Single-Stranded DNA Molecules†. Biochemistry. 2002;41:11611–27. [DOI] [PubMed] [Google Scholar]
- [62].Kozlov AG, Galletto R, Lohman TM. SSB–DNA Binding Monitored by Fluorescence Intensity and Anisotropy. Methods in molecular biology (Clifton, NJ). 2012;922:55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein: Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. Journal of Molecular Biology. 1987;193:97–113. [DOI] [PubMed] [Google Scholar]
- [64].Lavery PE, Kowalczykowski SC. Properties of recA441 protein-catalyzed DNA strand exchange can be attributed to an enhanced ability to compete with SSB protein. J Biol Chem. 1990;265:4004–10. [PubMed] [Google Scholar]
- [65].Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. Journal of Biological Chemistry. 1994;269:30005–13. [PubMed] [Google Scholar]
- [66].Bork JM, Cox MM, Inman RB. RecA protein filaments disassemble in the 5’ to 3’ direction on single-stranded DNA. J Biol Chem. 2001;276:45740–3. [DOI] [PubMed] [Google Scholar]
- [67].Lohman TM, Bujalowski W, Overman LB. E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends in Biochemical Sciences. 1988;13:5. [PubMed] [Google Scholar]
- [68].Kim T, Chitteni-Pattu S, Cox BL, Wood EA, Sandler SJ, Cox MM. Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination. PLoS Genet. 2015;11:e1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Porter RD, Black S. The single-stranded-DNA-binding protein encoded by the Escherichia coli F factor can complement a deletion of the chromosomal ssb gene. Journal of Bacteriology. 1991;173:2720–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Curth U, Greipel J, Urbanke C, Maass G. Multiple binding modes of the single-stranded DNA binding protein from Escherichia coli as detected by tryptophan fluorescence and site-directed mutagenesis. Biochemistry. 1993;32:2585–91. [DOI] [PubMed] [Google Scholar]
- [71].Ferrari ME, Fang J, Lohman TM. A mutation in E. coli SSB protein (W54S) alters intratetramer negative cooperativity and inter-tetramer positive cooperativity for single-stranded DNA binding. Biophysical Chemistry. 1997;64:235–51. [DOI] [PubMed] [Google Scholar]
- [72].Bell JC, Liu B, Kowalczykowski SC. Imaging and energetics of single SSB-ssDNA molecules reveal intramolecular condensation and insight into RecOR function. Elife. 2015;4:e08646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Chan K-W, Lee Y-J, Wang C-H, Huang H, Sun Y-J. Single-Stranded DNA-Binding Protein Complex from Helicobacter pylori Suggests an ssDNA-Binding Surface. Journal of Molecular Biology. 2009;388:508–19. [DOI] [PubMed] [Google Scholar]
- [74].Davis RW, Botstein D. Advanced bacterial genetics: a manual for genetic engineering. 1980.
- [75].Messing J [2] New M13 vectors for cloning Methods in enzymology: Elsevier; 1983. p. 20–78. [DOI] [PubMed] [Google Scholar]
- [76].Neuendorf SK, Cox MM. Exchange of recA protein between adjacent recA protein-single-stranded DNA complexes. Journal of Biological Chemistry. 1986;261:8276–82. [PubMed] [Google Scholar]
- [77].Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor: CSHL Press; 2001. [Google Scholar]
- [78].Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. [DOI] [PubMed] [Google Scholar]
- [79].Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D: Biological Crystallography. 2010;66:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
- [81].Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, et al. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Israel Journal of Chemistry. 2013;53:199–206. [Google Scholar]
- [84].Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–4. [DOI] [PubMed] [Google Scholar]
- [85].Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Delano WL. The PyMOL Molecular Graphics System. New York: Schrodinger, LLC; 2002. [Google Scholar]
- [87].Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature methods. 2008;5:507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Yang O, Ha T. Single-Molecule Studies of ssDNA-Binding Proteins Exchange. Methods Enzymol. 2018;600:463–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.