Abstract
Drug resistance is a major cause of cancer-related mortality. Glucuronidation of drugs via elevation of UDP-glucuronosyltransferases (UGT1As) correlates with clinical resistance. The nine UGT1A family members have broad substrate specificities attributed to their variable N-terminal domains and share a common C-terminal domain. Development of UGT1As as pharmacological targets has been hampered by toxicity of pan-UGT inhibitors and by difficulty in isolating pure N-terminal domains or full-length proteins. Here, we developed a strategy to target selected UGT1As which exploited the biochemical tractability of the C-domain and its ability to allosterically communicate with the catalytic site. By combining NMR fragment screening with in vitro glucuronidation assays, we identified inhibitors selective for UGT1A4. Significantly, these compounds selectively restored sensitivity in resistant cancer cells only for substrates of the targeted UGT1A. This strategy represents a crucial first step towards developing compounds to overcome unwanted glucuronidation thereby reversing resistance in patients.
Introduction.
Drug resistance remains a major challenge in cancer therapies [1]. Many patients who initially respond to treatments eventually become resistant and relapse. Cancer cells adopt a variety of strategies to evade the effects of chemotherapies including: loss of drug receptors, elevated efflux of drugs through multidrug resistance proteins and/or genetic re-wiring to eliminate the dependence of a cancer cell on the targeted pathway [1–4]. While studying the efficacy of ribavirin targeting of dysregulated eIF4E activity in acute myeloid leukemia (AML) patients, we found that leukemia cells developed the means to deactivate not only ribavirin but also cytarabine (Ara-C), the cornerstone of AML therapy [5]. Specifically, resistant cells had elevated glioma-associated protein 1 (Gli1) which led to elevation of the UDP-glucuronosyltransferases (UGT; EC 2.4.1.17) [5]. UGTs catalyze the transfer of glucuronic acid from UDP-glucuronic acid (UDP-GlcA) to substrates with nitrogens, oxygens and sulphurs available for nucleophilic attack [6–9]. Glucuronidation plays important roles in clearing of metabolites as well as in drug detoxification [6–9].
For the case of Gli1-inducible glucuronidation, UGT elevation led to the formation of ribavirin- and cytarabine-glucuronides and ultimately, to drug deactivation [5, 10]. These observations suggest that “turning on” UGT1A1 activity could be the basis for a multi-drug resistance mechanism [11]. Indeed, at least 40 other drugs are similarly targeted by Gli1-inducible drug glucuronidation including commonly used chemotherapies such as methotrexateas well as newer generation drugs such as Sunitinib and Venetoclax [12]. Drug sensitivity was restored by RNAi knockdown of Gli1 or through its pharmacological inhibition with GDC-0449 (also known as Vismodegib) [5], an inhibitor of the extracellular receptor Smoothened (SMO) which is an upstream activator of Gli1 [13, 14]. The efficacy of GDC-0449 reversing ribavirin resistance is being tested in an ongoing clinical trial in AML patients (ClinicalTrials.gov NCT02073838). However, GDC-0449 is associated with significant toxicity and resistance tothis agent arises due to mutations in SMO [13, 14]. In addition to AML and head and neckcancer cells, elevation of UGTs correlates with drug resistance in chronic lymphoid leukemia patients and colon cancer cells, although whether this is mediated via Gli1 or another pathway is unknown[15, 16]. Furthermore, polymorphisms in UGT1A1 which result in reduced enzymatic activitycorrelate with better response to Ara-C in patients [17]. Approximately 35% of all medicationsare glucuronidated highlighting the impact of this pathway on drug metabolism [18], thus developing means to directly target selected UGTs would be beneficial for improving therapeutic responses. However, it is also important to strike a balance by targeting specific glucuronidationto avoid the severe toxicities associated with a global loss of this pathway.
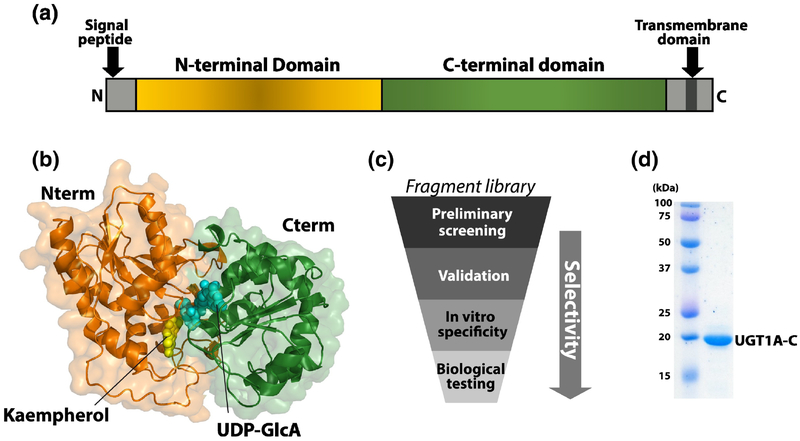
The UGT family are the major enzymes responsible for glucuronidation and is comprised of 22 transmembrane proteins located primarily in the endoplasmic reticulum [7–9, 19, 20]. There are two main families: UGT1A and UGT2B [7, 21]. The UGT1A family, but not UGT2B, is elevated by Gli1 overexpression and thus is the focus of our studies [5]. Different UGT1A family members have broadly overlapping substrate specificities with some preferences for glucuronidation of nitrogens by UGT1A4 whereas other enzymes show preferences for other chemical groups [7, 8, 21, 22]. UGTs can be divided into N- and C-terminal domains [7, 8, 21] (Fig. 1A). For UGT1As, the C-domains are derived from an exon-sharing mechanism and thus are identical for all family members [7, 21]. The N-domains are conserved, but not identical, at the sequence level leading to the hypothesis that this domain acts in substrate selection. However, the dearth of structural data for UGTs has made it difficult to dissect the molecular underpinnings of substrate specificity or the relative contribution of each domain to this process. There is no full-length structure of any UGT: the only structural information is an X-ray structure for the C-domain of UGT2B7 (PDB code 2O6L), which is highly homologous to the C-domain of UGT1As (~60% sequence identity and ~70% similarity) [23, 24] (Supplementary Fig. S1). However, UGTs are members of the glucosyltransferase GT1 superfamily for which X-ray structures have been determined [25, 26]. Homology modelling of UGT1A’s suggest UDP-GlcA and substrates bind at the interface between the N- and C-domains (Fig. 1B), supporting a mechanism whereby both domains contribute to catalysis and specificity [25–29]. To date, the design of UGT inhibitors have focussed on uracil or transition-state analogues both of which act as pan-UGT inhibitors [30].
Figure 1.
Characterization of UGT1As. (A) Schematic of domain structure of UGT1A enzymes. (B) Model of full-length UGT1A4 protein. UDP-GlcA (blue) and the substrate kaempherol (yellow) from the original VvGT1 x-ray structure (pdb 2C1Z [26]) are shown. Each subdomain is represented in the same color as in panel A. (C) Schematic of workflow for development of selective glucuronidation inhibitors. (D) UGT1A-C protein after purification by Coomassie blue stained SDS-PAGE gel. Molecular weight markers are shown.
Given the role of Gli1-inducible drug glucuronidation in resistance in patients and thegeneral importance of UGTs to clearance of metabolites in humans, we set out to identify compounds that targeted selected UGT1As. Specifically, we focussed on UGT1A4 given its rolein Gli1-inducible ribavirin glucuronidation [5, 10]. In these earlier studies, ribavirin glucuronidation was observed using mass spectrometry and supersomes indicating that ribavirin triphosphate (RTP) was glucuronidated by supersomes expressing: UGT1A1, UGT1A4, UGT1A6 and UGTIA9, but not UGT1A1 alone. Given the observations that RTP was glucuronidated on its carboxyamide and the role of UGT1A4 in glucuronidation of such groups, we prioritized this enzymefor these studies. We opted for a multi-faceted approach using NMR fragment screening techniques to detect fragments binding the C-terminus of UGT1A and subsequently submit these compoundsto biochemical assays to counter screen for selectivity (Fig. 1C). This approach yieldedfragments selective for inhibition of UGT1A4 glucuronidation. Notably, our NMR experimentson the UGT1A C-terminus show the fragments likely act via allostery, rather than direct bindingat the predicted active site. Significantly, these fragments selectively restored drug sensitivity in high-Gli1 resistant human cancer cells highlighting the strength of this approach to identify selective UGT1A inhibitors. In all, our studies provide a proof-of-concept that exploits the malleability of the C-terminus of UGT1A coupled with biochemical screens to identify selective inhibitors. This pipeline can be exploited to identify a broad range of selective inhibitors enabling restoration of drug sensitivity to many agents.
Results
NMR characterization of UGT1A4 and its binding to UDP-GlcA
We initiated studies by expressing and purifying the UGT1A4 protein from bacterial cells which had never been reported. As a first step, we generated several UGT1A4 constructs including those containing the N-domain, the C-domain and the full-length protein. Bacterial expression constructs were validated by DNA sequencing and molecular weight of the purified protein was verified by mass spectrometry. While we produced the N-domain and full-length proteins from E. coli, these proteins had poor solution behaviours including high propensities for aggregation and precipitation making them difficult to use in the NMR-based fragment screens. By contrast, the C-domain construct (UGT1A-C; residues 284–450) was relatively well-behaved at concentrations below 200 μM (Fig. 1D). While there is no structure of any full-length UGT protein, X-ray structures for members of the GT1 family (~ 20% homology) have been solved, including the plant glucosyltransferase VvGT1 in the presence of its substrate (kaempherol) and a non-hydrolysable analogue of UDP-GlcA [25, 26]. This structure shows an extensive interface between the N and C domains which form the active site. These interactions are confirmed in UGT1A homology models [27–29] including our model for UGT1A4 based on this structure and our NMR data (see below) (Fig. 1B). Whilst the active site is based on homology models, we do note that such models have accurately predicted substrate selectivity for UGT1A9 [28, 29]. Given these models, we postulated that studies into the C-domain would provide an opportunity to identify compounds that affect this interface and thus a means to selectively inhibit UGT1As.
We generated isotopically labelled UGT1A-C protein by growth in minimal media for NMR studies. The 1H-15N-HSQC spectrum was well-dispersed indicating that the protein was folded (Supplementary Fig. S2). We assigned signals for the backbone 1H, 13C and 15N atoms for ~90% of the residues via standard triple resonance techniques [31]. Analysis of the 13C chemical shift index from UGT1A-C revealed that the position of the secondary structure elements agreed remarkably well with those observed for the X-ray structure of the UGT2B7 C-domain [31], validating our structural model for UGT1A-C based on UGT2B7-C (Fig. 2A).
Figure 2.
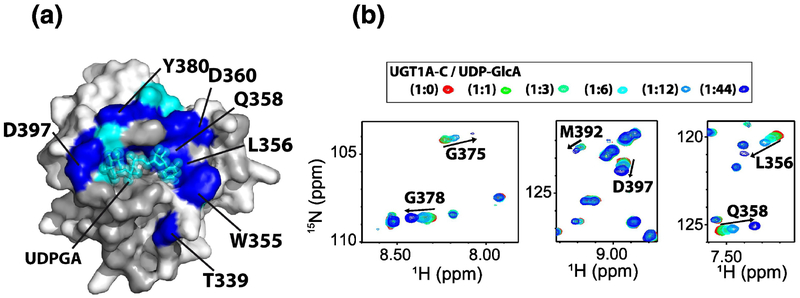
NMR characterization and UDP-GlcA binding to UGT1A-C. (A) Chemical shift perturbation (CSP) upon UDP-GlcA addition mapped onto the UGT1A-C model. Light blue: CSP > mean CSP + 0.2*standard deviation; dark blue, CSP > mean CSP + 0.5* standard deviation. UDP-GlcA is in cyan and is positioned based on homology to the VvGT1 binding site [26]. Unassigned residues are dark gray (including the 307–318 loop discussed in the text). (B) Regions of 1H-15N HSQC spectra of UGT1A-C at different UDP-GlcA ratios. Full size HSQC is in Supplementary Fig. S2.
To fully appreciate the effects of any fragments that bind UGT1A-C, it is important to have a strong grasp of the location of the UDP-GlcA-binding site. While the location of the UDP-GlcA binding site was predicted based on mutational data for UGT1As, the UGT2B7 C-terminal structure and homology modelling to VvGT1 [22–29, 32], this site has never been directly observed experimentally in either UGT1As or UGT2Bs. We used 1H/15N labelled UGT1A-C and monitored chemical shift perturbation (CSP) upon addition of UDP-GlcA (Fig. 2; Supplementary Fig. S2A). Major shifts were observed for regions predicted to interact with UDP-GlcA (Fig. 2A; Supplementary Fig. S2) including residues 396–398 at the glucuronic acid site, residues 374–375 at the diphosphate site and residues 378–380, 355–359 and 339 close to the uracil moiety. We note that the amide resonances for residues 307–318 which bordered the UDPGlcA binding site were not assigned in UGT1A-C likely due to conformational exchange (Fig. 2A). This region has large B-factors in the UGT2B7 X-ray structure and residues 314–317 (UGT1A4 numbering) in one of the two models are missing again suggesting flexibility.
Thus our NMR data delineates the UDP-GlcA binding site for the first time in any UGT1A protein and demonstrates that this overlaps well with the binding sites identified by crystallography for other glycosyltransferases [25, 26], and predicted from the UGT2B7 structure [23, 24] and homology models [27–29].
Fragment screening to identify binders to UGT1A-C
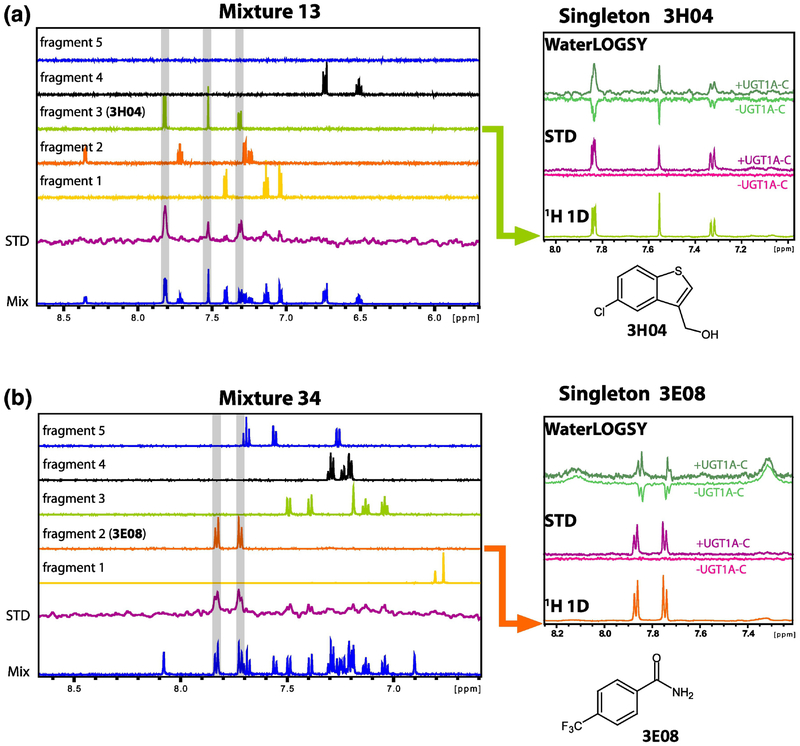
Fragment screening is an attractive alternative to high throughput screening to identify small molecules binding to target proteins [33, 34]. Typically small molecular weight compounds (<300 Da), with favourable physical properties are screened. These small molecules (“fragments”) cover chemical space more efficiently than drug-sized molecules: thus fragment libraries tend to be relatively small (300–3000 fragments) but have high hit rates [33, 34]. Although fragments tend to be weak binders they serve as a starting point for the design of more potent molecules as in the case of the FDA approved drugs vemurafenib and venetoclax [33, 34]. As a first step to identify selective binders to UGT1A4, we carried out an NMR-based fragment screen to identify compounds that could bind UGT1A-C. We screened curated compounds derived from an in-house inventory supplemented with compounds from Maybridge. Pan-interference (PAINS) compounds were eliminated from the libraries [35]. Fragment identity, quality, aggregation state and aqueous solubility were assessed by NMR for 570 compounds: only 385 were used in the screen based on satisfying these criteria (see materials and methods). Screens were carried out as pools of 5 fragments per sample and mixtures designed to minimise signal overlap. We monitored ligand binding using saturation transfer difference (STD) NMR [36] (Fig. 3). A total of 80 fragments were identified (hit rate ~ 20%) which were then confirmed as singletons using STD and WaterLOGSY [37] techniques (Fig. 3, right panels). Two compounds were removed due to aggregation as singletons. The remaining 78 compounds induced CSP’s in BEST 1H-15N HSQC NMR experiments [38] (see below) confirming binding for these fragments.
Figure 3.
Examples of fragment screening pools by NMR and identification of compounds 3H04 (A) and 3E08(B). Left panels summarise analysis for two mixtures (13 and 34) and show the aromatic region of the 1H NMR spectrum. The 1D 1H NMR spectra for the mixture of 5 compounds (0.5 mM each) in the presence of 10 μM UGT1A-C is shown in blue. The saturation transfer difference (STD) spectrum is shown in purple. The 1H 1D spectra of isolated compounds in the mixture are shown above (fragment 1 to fragment 5). Fragments binding to UGT1A-C will give a signal in the STD spectrum (fragment 3, 3H04, in A, and fragment 2, 3E08, in B). Right panels show binding of 3H04 and 3E08 (1 mM) as singletons with UGT1A-C protein (20 μM). For the STD, there was no signal in the absence of protein (dark pink) indicating that their significant magnetization is only transferred to the compound in the presence of the protein (purple). Water-LOGSY experiments also revealed binding of 3E08 and 3H04 in which light green is the negative signal associated with unbound fragment acquired in the absence of UGT1A-C, and the dark green spectrum is acquired in the presence of the protein and shows the characteristic positive signal associated with bound fragment. 1D 1H NMR spectra of the fragments alone as well as their chemical structure are shown. Fragment binding was further confirmed by 1H-15N HSQC titration using 15N-labelled UGT1A-C and unlabeled fragments (Fig. 5,6, Supplementry Fig. S4,5). STD and WaterLOGSY experiments were done on the same sample with or without protein.
In vitro glucuronidation assays identify fragments selective for UGT1A4 versus UGT1A1
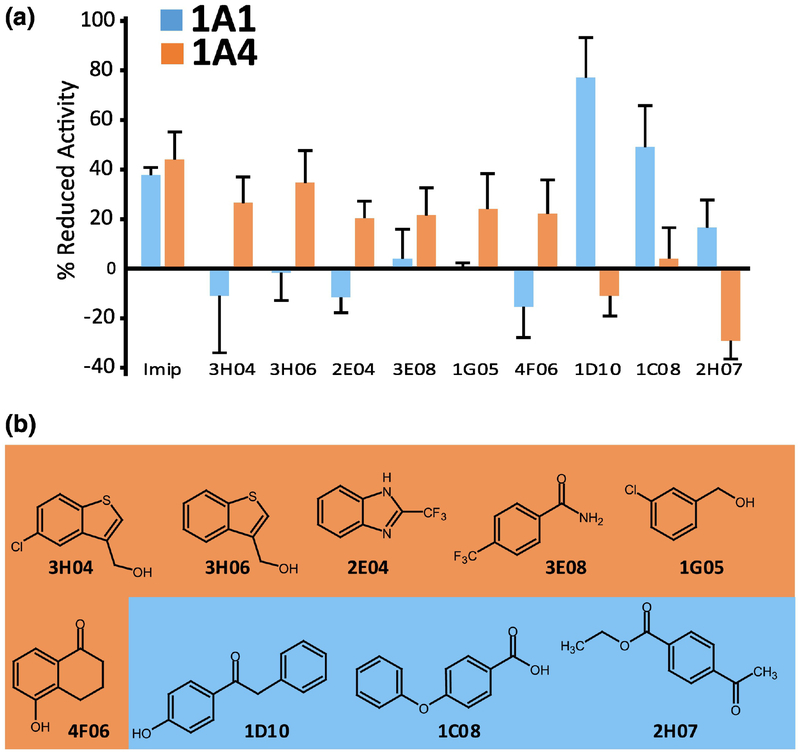
Having identified fragments that bound to UGT1A-C, the question arose whether these compounds could inhibit glucuronidation for full-length UGT1A family members. Therefore, we carried out glucuronidation assays as a function of fragment addition in supersomes expressing either UGT1A4 or UGT1A1 (Fig. 4) as a proof of concept. We monitored glucuronidation of proluciferin substrates specific for UGT1A4 or UGT1A1 using the commercially available assay UGT-Glo (Promega, V2082). Enzyme inhibition by the compounds was quantified by the consumption of proluciferin substrates and measured relative to untreated controls using chemiluminescence. Imipramine is a multi-UGT inhibitor [30] and efficiently inhibited both UGT1A1 and UGT1A4 activity by 40–45%. The effects of 44 fragments were examined, because these were readily available. We defined selectivity between UGT1A1 and UGT1A4 as follows: fragments with >20% inhibition activity for UGT1A4 and with < 5% inhibition of UGT1A1 were considered selective for UGT1A4. The converse specifications were used to classify fragments as selective for UGT1A1. There were 8 selective UGT1A4 fragments (3H04, 3H06, 3E08, 4F06, 2E04, 1A10, 1G05, 4E09), 6 for UGT1A1 (1D10, 3C08, 1C08, 1D05, 4F09, 2H07), 6 fragments had no activity (3F08, 3H10, 4A11, 2C03, 2H04, 3C05) and the remainder significantly inhibited activity for both enzymes (Fig. 4). Interestingly, some fragments that inhibited UGT1A4 actually activated UGT1A1 in this assay e.g. 3H04, and conversely, some that inhibited UGT1A1 activated UGT1A4 e.g. 1D10. Analysis of the fragments suggested some common chemical scaffolds. For instance, many fragments that selectively targeted UGT1A4 had bicyclic aromatic ring systems. Indeed, 3H04 and 3H06 are homologues with their only difference being the chlorine group (Fig. 4B). However, in general there was substantial diversity in both the UGT1A4 and UGT1A1 selective fragment groups. Interestingly, none of the scaffolds resembled UDP-GlcA (see the inset in Supplementary Fig. S2B), glucuronic acid or previously designed uracil-based or UGT transition-state inhibitors. Further, fragment selectivity did not appear to be based on the presence of particular active groups. For instance, substrate selectivity is expected to be roughly partitioned on the preference of amino groups for UGT1A4 and hydroxyl groups for UGT1A1. However, inspection of the structures revealed that many fragments contain hydroxyl groups for both the UGT1A4 and UGT1A1 selective fragments (e.g. 3H04, 1G05, 4F06, 1D10 etc; Fig. 4B).
Figure 4.
Fragment activity in in vitro glucuronidation assays. (A) The percent inhibition of glucuronidation for UGT1A1 (blue) and UGT1A4 (orange) proluciferin substrates using full-length UGT1A1 and UGT1A4 proteins expressed in the supersomes as indicated. Values are averages of three biological replicates each conducted in triplicate and error bars represent standard deviations. Imipramine is a multi-UGT inhibitor and served as a positive control for the inhibition of glucuronidation for both UGT1A1 and UGT1A4 substrates. Some fragments stimulated activity as observed by their negative inhibition. Results are relative to untreated controls which is set to 0% inhibition. (B) Structures of the fragments used in panel A: UGT1A4 specific and UGT1A1 specific fragments are highlighted with orange and blue, respectively.
We next examined whether these selective inhibitors were competitive inhibitors, i.e. if they could be glucuronidated. We studied a subset of these inhibitors as a proof-of-principle: 2E04, 3E08, 3H04 and 3H06 as examples of the UGT1A4 selective group, and 1C08 as an example for the UGT1A1. We monitored glucuronidation of these fragments using mass spectrometry under the same conditions used for the UGT-Glo assay with the exception that we did not add the reagent for luminescence. For positive controls, we used the specific UGT1A4 and UGT1A1 substrates provided in the UGT-Glo kit and imipramine. As expected our positive controls (imipramine in both UGT1A1 and UGT1A4 supersomes, and the UGT1A4 and UGT1A1 specific substrates) formed glucuronides by detection of peaks 176.0321 Da greater than the mass of the parent compound (Supplementary Fig. S3A–D, and data not shown). In contrast, we did not observe glucuronides for the fragments that were readily detectable by mass spectrometry i.e. 1C08 and 3E08 (tested for in both UGT1A1 and UGT1A4 supersomes, Supplementary Fig. S3E–H) and 2E04 (tested for in UGT1A4 supersomes). We note that 3H04 and 3H06, were not visible by mass spectrometry due to their non-polar characteristics and thus, poor ionization. So while we did not observe their glucuronides, we could also not detect the parent fragments. The status of all the fragments was verified by NMR and showed no degradation. In all, our studies demonstrate that 1C08, 3E08 and 2E04 are not glucuronidated suggesting under these conditions these fragments are not acting as competitive substrates. By analogy, these studies suggest that 3H06 and 3H04 are similarly not competitive inhibitors; however we note that problems with ionization of these fragments mean that these latter findings must be interpreted with caution. These findings also validate the UGT-Glo assay we used as our counter-screen.
Identification of Fragment Binding Sites on UGT1A-C
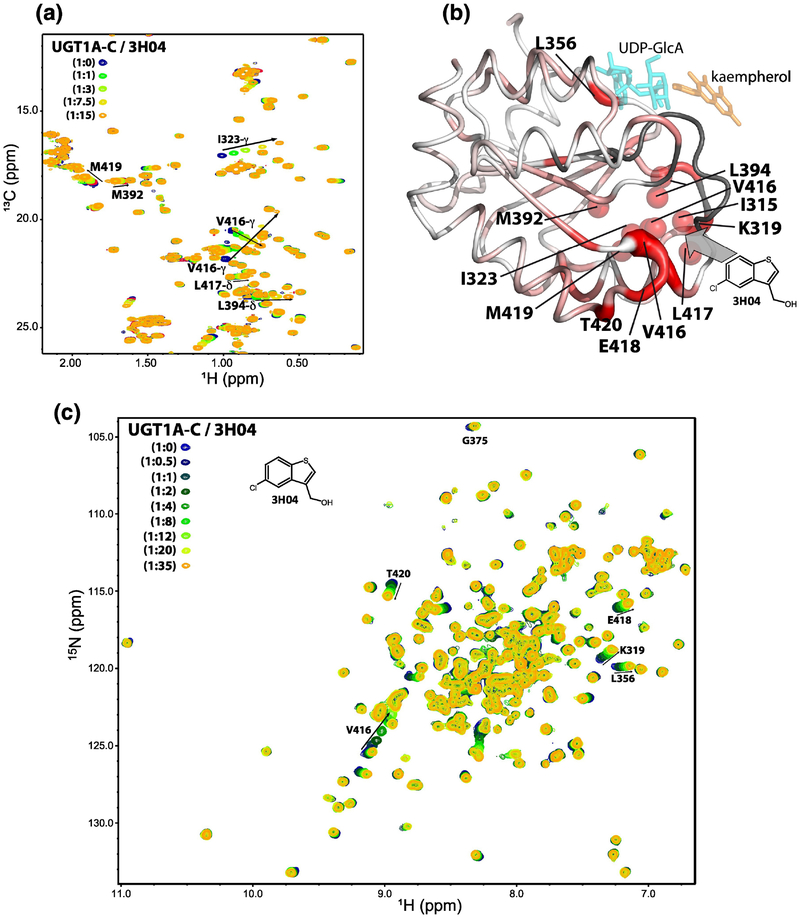
We set out to determine the binding site of these fragments on UGT1A-C. We prioritized three of these fragments for this analysis: the homologues 3H04 and 3H06 and the chemically distinct 3E08. Delineation of binding sites by NMR methods is best defined by NOESY experiments. These experiments detect spatial interactions, typically 5 – 6 Å or less, and thus are excellent for determining direct interactions in contrast to more typically used 1H-15N HSQC CSP’s which can arise due to either direct binding or from allosteric processes. We employed the 13C-edited, 13C/15N filtered NOESY experiments (Fig. 5A) to identify intermolecular NOEs between the fragments and 13C/15N labelled UGT1A-C (i.e. detect short distances between the fragments and UGT1A-C). Given the weak nature of the complexes which is expected for fragments, we tailored experiments for the methyl resonances of UGT1A-C and ran under conditions of high molar excess of each fragment (~1:14, UGT1A-C:fragment) to improve the sensitivity of the experiment. We observed intermolecular NOEs between the methyl side-chains of I323, I315, V416, L417, V394, M392 and M419 of UGT1A-C and the aromatic protons of 3E08 (Fig. 5A). As expected these residues on UGT1A-C are in close proximity to each other; the location of the methyl groups are shown as green balls mapped onto the UGT1A-C structure model (Fig. 5B). Consistently, these methyl groups were among the most perturbed in the 1H-13C-HSQC upon addition of 3E08 (Supplementary Fig. S4A). The same region of UGT1A-C exhibited intermolecular NOE’s to 3H06 (Supplementary Fig. S5A and B) indicating that 3E08 and 3H06 bound the same region of UGT1A-C. Attempts to detect intermolecular NOEs to 3H04 were unsuccessful due to aggregation of 3H04 at higher concentrations and the higher affinity of 3H04 to UGT1A-C leading to broadening of signals. However, fragments 3H04 and 3H06 share a common scaffold (Fig. 4B) and induced similar 13C CSP upon addition to UGT1A-C, most obviously V416 and I323 in the same fragment-binding pocket supporting a similar binding profile (Fig. 6A, Supplementary Fig. S5C). Thus 3E08, 3H04 and 3H06 bind the same region of UGT1A-C. Significantly, these residues do not overlap with the UDP-GlcA site defined above (Fig. 2 and S2). Indeed our fragments did not exhibit intermolecular NOEs to the methyl groups of L356 or V353, which are at the UDP-GlcA binding site we have defined, neither were these residues significantly perturbed in the 1H-13C HSQC. Conversely, addition of UDP-GlcA perturbed these peaks in the 1H-13C HSQC but not any of the resonances significantly perturbed by addition of our fragments (data not shown). Our data indicates that there were no spatial interactions between 3H06 or 3E08 and the NMR-defined UDP-GlcA binding site. The question remains if our fragments bind at the predicted substrate binding site? The precise site for substrate binding has not been determined directly for human UGT’s, but can be deduced from homology modelling [27–29] (Fig. 1B). Homology models to VvGT1 and other glycosyltransferases in complex with substrate and donor predicts contacts with the C-terminus including residues F395 and D397 [27–29]. These sites are close to glucuronic acid binding site we have defined with our CSP data described above. Our NOE-defined fragment binding-site would be too far from the glucuronic acid moiety of the UDP-GlcA donor to be directly involved in catalysis. This is consistent with the fact we did not observe glucuronidation of these fragments (Supplementary Fig. S3). A summary of the UDP-GlcA sites and fragment NOE’s to UGT1A-C are shown on the sequence alignment of UGT1A-C and UGT2B7 (Supplementary Fig. S6B) showing little overlap between these sites. Interestingly, the fragment binding site around V416 and L417 constitutes the most poorly conserved region between the sequences. This observation coupled with our NMR data (data not shown) suggests our fragments do not bind to UGT2B7-C.
Figure 5.
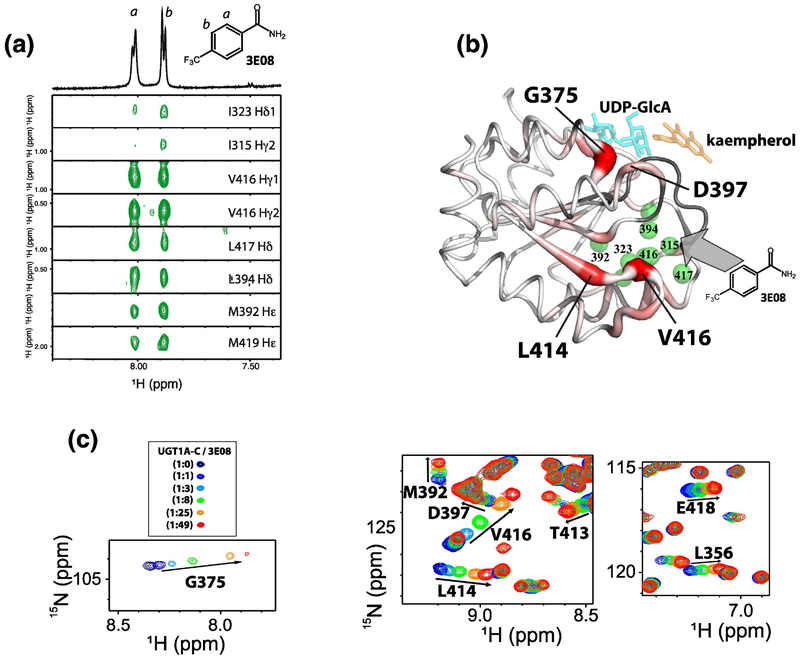
Definition of the 3E08 binding site on 13C/15N-UGT1A-C. (A) 1H-1H strips from the 13C-edited, 13C/15N filtered NOESY showing intermolecular NOEs between methyl resonances of 13C/15N UGT1A-C and 3E08. On top, the 1D 1H NMR spectrum of 3E08 and its chemical structure are shown. Protons are labelled a or b. (B) UGT1A-C model structure showing the fragment binding site via NOEs (green balls) and binding and allosteric sites with 15N chemical shift perturbation (red thick tubes). The darker red and thicker tube indicates larger chemical shifts. Unassigned residues are shown in gray. (C) CSP for selected residues of 15N-UGT1A-C at different 3E08 ratios by 1H-15N HSQC. The full HSQC is in Fig. S4B.
Figure 6.
NMR analysis of 3H04 binding to UGT1A-C. (A) 1H-13C HSQC showing CSP upon 3H04 addition. 13C atoms are less sensitive to allosteric shifts generally and thus this serves as a surrogate for the NOESY experiment. The peaks that shift overlay well with the fragment binding sites for 3H06 and 3E08 determined by NOEs. See text for details. (B) 1H-15N HSQC CSP was mapped onto the UGT1A-C model structure. Magnitude of chemical shift is shown by darker red and thickness of backbone. Unassigned residues are shown in gray. Methyls showing CSP are shown as red balls. The active site is marked by UDP-GlcA (cyan) and kaempherol (orange) to provide orientation, but these compounds were not present in the experiments. (C) 1H-15N HSQC showing chemical shift perturbation upon addition of increasing concentration of 3H04.
Given there was no direct overlap with the NMR-defined UDP-GlcA binding site or the predicted substrate site, we postulated that these fragments induced allosteric changes in UGT1A-C which in turn affected activity. Amide chemical shifts are exquisitely sensitive to allosteric processes; thus we monitored the effects of fragments on the 1H-15N HSQC spectra of 15N-UGT1A-C. Addition of 3E08 led to CSP’s at the fragment-binding site, as highlighted by shifts for V416, L414 and M392 which are the most perturbed. Perturbations are also observed at the UDP-GlcA binding site (Fig. 2 and S2) e.g. L356, G375, D397 and at the substrate site predicted from our homology model e.g. D397 (Fig. 5C; Supplementary Fig. S4B), indicating an allosteric mechanism affecting these sites that was transmitted from the fragment binding site. These CSP’s were mapped onto the UGT1A-C model structure in red (Fig. 5B). In this figure, the substrate kaempherol (orange) and UDP-GlcA (cyan) provide markers of the NMR-defined UDP-GlcA binding site and the predicted substrate site, although these compounds were absent from the experiments (Fig. 5B). The effected residues were very similar for 3E08 and 3H04 (Fig. 6B and C). One major difference between 3H04 and 3E08 is that only 3E08 induced CSP for G375 in the UDP-GlcA binding site suggesting it could have a greater allosteric effect than the other two fragments (Fig. 5C; Fig. 6C). Overall our NMR data suggest an allosteric model for fragment inhibition of UGT1A activity in which the fragments bind a region below the UDPGlcA and proposed substrate binding sites (centred around V416, L417, Fig. 5 and 6), which induces NMR-detectable allosteric routes to the predicted active site and N-C interface or the UDP-GlcA site (e.g. D397, G375, L356). These allosteric effects likely underpinned the selectivity observed in our biochemical assays. Importantly, 3E08, 3H04 and 3H06 binding sites, while distinct from the predicted active site, would also be positioned to interface with the N-domain (Supplementary Fig. S6A). In all, our NMR data suggest a model in which the fragments influence selectivity through allosteric effects transmitted to the N-C interface in the full-length protein. This model is validated by our NMR data defining the UGP-GlcA site. However in the absence of a full-length structure of a UGT1A we cannot definitively rule out that the substrate binding site is far removed from that predicted in these models. Irrespective of a mechanism for inhibition, our biochemical screens data show we can identify compounds that can selectively inhibit UGT1A1 and UGT1A4 in microsomes. We next examined if these fragments had any biological effects on ribavirin sensitivity in cells.
Fragments restored drug sensitivity in high-Gli1 cells
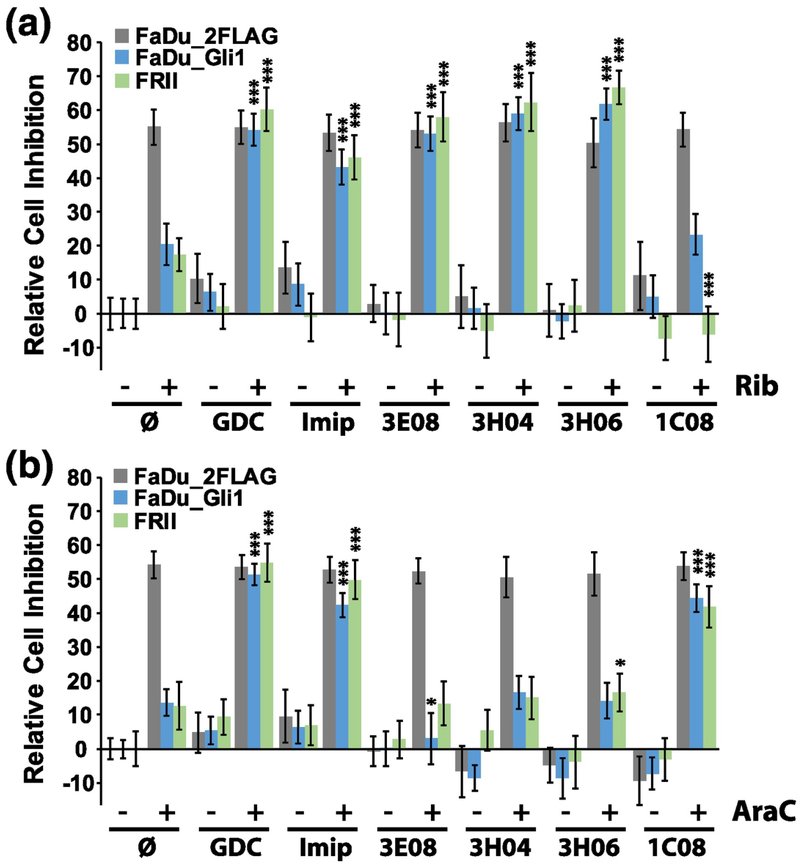
We determined whether the UGT1A4 specific fragments reversed Gli1-dependent ribavirin glucuronidation in human cancer cells (Fig. 7A). We used head and neck cancer cells (FaDu) stably overexpressing Gli1 (FaDu-Gli1), FaDu vector control cells (FaDu-2FLAG) or resistant FaDu cells (FRII). FRII cells were derived from FaDu cells after long-term exposure to ribavirin [5]. FRII cells are characterized by elevated Gli1 and UGT1A protein levels [5]. Thus, FRII and FaDu-Gli1 cells both utilize Gli1-inducible drug glucuronidation to mediate ribavirin resistance [5]. By contrast, FaDu-2FLAG cells are sensitive to ribavirin, as observed previously [5]. Cells were treated with ribavirin to achieve 50% inhibition of cell growth in vector controls. As expected, the proliferation of FaDu-Gli1 and FRII cells was only modestly affected by ribavirin at these concentrations (10–20% inhibition) consistent with these cells being resistant to the drug. We then examined the ability of our fragments to reverse ribavirin resistance. As a positive control for re-sensitization, we used the Gli1 pathway inhibitor GDC-0449. We observed that GDC-0449 fully reversed ribavirin resistance, with the result that FaDu-Gli1 and FRII cells were just as sensitive to ribavirin as the vector controls (Fig. 7A). Similarly, the multi-UGT inhibitor imipramine also completely reversed drug sensitivity to ribavirin. Strikingly, 3E08, 3H04 and 3H06 all re-sensitized cells to ribavirin, restoring sensitivity to levels observed in vector control cells. We also examined the effects of 1C08, a fragment that did not inhibit UGT1A4 but did inhibit UGT1A1 in our biochemical assays. Strikingly, this fragment did not restore drug sensitivity of FRII or FaDu-Gli1 cells to ribavirin. Importantly, none of these compounds alone inhibited growth of any of these cells. Thus, the increased inhibition of proliferation was not a result of overt toxicity of fragments alone.
Figure 7.
Selected fragments restored drug sensitivity to ribavirin (Rib, A) or Ara-C (B) in the human head and neck cancer FaDu cell lines including FaDu-2FLAG (gray); FaDu-Gli1 (blue) and FRII (green). The percent cell inhibition versus untreated FaDu-2FLAG is shown with untreated controls definned as 0% inhibition (null sign). Drug concentrations were as follows: ribavirin 20 μM, Ara-C 200 nM, GDC-0449 (GDC) 200 nM, imipramine 10 μM, and fragments 100 μM each. Ribavirin and Ara-C were used at their respective IC50s for FaDu-2FLAG cells and fragment concentrations were used at concentrations that were not toxic to the cell as shown. The average of three biological replicates (each conducted in triplicate) are shown with error bars indicating standard deviations. *** indicates P-values <0.001 and * P<0.05. Averages are compared between the given cell type and its ribavirin or Ara-C treated control to obtain P-values.
We extended our studies to determine whether any of these fragments would also reverse drug resistance to Ara-C (Fig. 7B). Our previous studies showed that while Ara-C glucuronidation was Gli1-dependent, supersomes expressing the following four UGT1As (UGT1A1, UGT1A4, UGT1A6, UGT1A9) were not sufficient to glucuronidate Ara-C or its metabolite CTP [5]. It is important to note that many glucuronidation enzymes heterodimerize [19, 39]. Thus, these enzymes might be required, but were not sufficient alone, to recapitulate the effect observed in cells where glucuronidation was readily detected by mass spectrometry [5]. Here, we used Ara-C, which is not a target of UGT1A4, as a selectivity control for the activity of our fragments that were selective for UGT1A4. Cells were treated with Ara-C so that proliferation was inhibited ~50% relative to untreated vector controls. As expected the FaDu-Gli1 and FRII cells were resistant to Ara-C with modest inhibition of 5–10% relative to untreated vector controls. The addition of either GDC-0449 or imipramine completely restored drug sensitivity to Ara-C. Importantly, 3E08, 3H04 and 3H06 had no affect i.e. Ara-C sensitivity was not restored by addition of these fragments. By contrast, 1C08 completely reversed drug resistance to Ara-C consistent with recent data in AML patients that strongly suggest that UGT1A1 polymorphisms play an important role in Ara-C metabolism [17], likely in combination with other UGT1As since UGT1A1 supersomes were not sufficient to glucuronidate Ara-C in vitro (12). Thus, the fragments restored drug sensitivity in a manner consistent with the in vitro glucuronidation assays with 3E08, 3H04 and 3H06 only effective against ribavirin which was UGT1A4-dependent while these did not restore sensitivity to Ara-C, which is not an UGT1A4 target. In all, these observations highlighted both efficacy and selectivity of our strategy.
Discussion
Here, we developed a path forward to identify selective inhibitors of the UGT1As. To do this, we exploited two key facets we discovered for these enzymes: 1. the biochemical tractability of the C-domain and 2. the allosteric connections between the fragment-binding site and the catalytic site at the N-C interface. Our strategy rested on the use of a combination of fragment and functional screens to find compounds that selectively inhibited UGT1A4 or UGT1A1, and restored drug sensitivity in human cancer cells. The premise of this approach is a substantial departure from the traditional view where it is usually considered that the N-domain governs substrate selectivity [7, 21]. This perspective is based on the fact that the C-domain of UGT1As is identical for all family members [7, 21] and that mutations in the N-domain is sufficient to swap selectivity for given enzymes [40, 41]. While these reasons are compelling, our studies showed that the C-domain alone can be used to identify selective inhibitors.
Mutational studies in the UDP-GlcA binding site support the notion that the C-domain influences selectivity. For example in UGT1A10, the Q394A mutation (using 1A10 numbering) in the UDP-GlcA pocket increased the glucuronidation of tetrachlorocatechol and genestein but at the same time reduced glucuronidation of chrysin with no effect on the estrogen metabolite 2-OH-E1 [24]. The same mutation Q396A in UGT1A6 (using 1A6 numbering) increased glucuronidation of 4-methylumbelliferone but did not alter glucuronidation of 1-Napthol [24]. In UGT1A4, this residue (Q398) is one of the endpoints of the allosteric route we identified emanating from the 3E08, 3H04 and 3H06 fragment-binding site, as seen by the CSP for D397 (Fig. 5). Importantly, our NMR data includes assignments for all of these residues and thus, allows us to directly monitor these residues upon UDP-GlcA or fragment binding. We do note, however, that in cells and in the biochemical assays, full-length UGT1As are used, and thus, it is possible that there may be additional binding sites for these fragments other than the single site observed with UGT1A-C. Investigations with the full-length protein are challenging but an important future direction for investigation. Further, some fragments may indeed interact with more than one UGT1A. In other words, fragments may bind a subgroup of UGT1As still providing selectivity and thus, potentially, less toxicity. Even this would be an important first step forward in the development of strategies to selectively target these enzymes.
Although glucuronidation deactivates many drugs, valid concerns over toxicity have impeded development of UGTs as drug targets. The development of selective inhibitors is an important first step towards demonstrating that drugging this pathway is feasible. Prior studies focused on active-site inhibitors. Indeed, UDP, UTP, UDP-N-acetyl glucosamine, UDP-glucose, UDP-based transition analogues and aglycone substrates all act as UGT1A inhibitors [22, 30]. The therapeutic application of these inhibitors is limited because they lack selectivity and also many impact on other aspects of UDP-nucleoside metabolism potentially leading to additional toxicities [30]. Importantly, our selective fragments did not resemble these active site inhibitors nor did they bind the predicted active site but rather appear to allosterically modulate enzyme selectivity. In this way, the chemical scaffolds and their mechanism of action represent a significant departure from previously reported UGT inhibitors. Our studies suggest that selective targeting of UGT1As activity could eventually be implemented clinically for patients where unwanted glucuronidation leads to drug inactivation and therapeutic resistance. Indeed, we plan to build on this work using traditional fragment based drug design approaches [42] in which precise structural information of the complexes will be used to grow fragments to increase affinity, whilst maintaining selectivity.
Materials and Methods
Protein expression and purification.
The full-length UGT1A4 construct was purchased from Origin (cat sc115733). The C-terminal domain from human UGT1A4 (residues 284–450) was inserted into the pET-26b vector between the EcoRI and XhoI restriction sites. In addition, a TEV protease cleavage site was introduced upstream to the C-terminal His-tag leaving the sequence Glu-Asn-Leu-Tyr-Phe-Gln attached to the C-terminus of the protein. The plasmid was overexpressed in Escherichia coli BL21(DE3)-RIPL competent cells in minimum media (see below) at 37°C and induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 20°C for 24 h. Cells were harvested by centrifugation and stored at −80°C until use. The frozen cells were resuspended in PBS buffer supplemented with 360 mM NaCl, 0.5 mM DTT, 0.3% Sarkosyl, 1mg/ml lysozyme, cocktail of protease inhibitor (GE), 15 mM imidazole, and lysed by sonication. After adding DNase and RNase, the lysate was mixed overnight end-over-end at 4°C. The lysate was cleared by centrifugation (30 min, 20,000 rpm, 4°C) and purified over Ni-NTA beads (Qiagen) onto a gravity flow column. After the resin was extensively washed in PBS, 360 mM NaCl, 0.5 mM DTT, 30 mM imidazole, the protein was unfolded on the beads with 8M urea in 50 mM Tris pH 7.5, 10 mM DTT, 30 mM imidazole and eluted with the same buffer containing 500 mM imidazole. The protein was refolded by dialysis with a gradient of decreasing concentration of urea (4M, 2M, 1M, 0.5M) in 20 mM Tris, pH 7.5 and 5 mM DTT. The UGT1A-C concentration was kept lower than 1mg/ml. The protein was finally dialyzed against 50 mM sodium phosphate (pH 7.2), 300 mM NaCl, 1 mM DTT. The tag was further removed by the TEV protease. High level of purity (>95%) was achieved by gel filtration on Superdex 75pg column (GE-Biosciences) in 50 mM sodium phosphate (pH 7.2), 300 mM NaCl, 1 mM DTT. The mass of UGT1A-C was confirmed by mass spectrometry after solubilising the purified protein in 50% methanol, 0.2% formic acid to a final concentration of 1mg/mL. The sample was electrosprayed directly onto an Agilent Q-TOF 6520. Intact mass profile was reconstructed with the Masshunter software.
Isotope labelling schemes.
Doubly or singly labelled sample was obtained by growing the cells in M9 minimal medium containing 2g/L of 15NH4Cl and/or 2g of 13 C-labelled glucose as the sole source of nitrogen and/or carbon (Sigma-Aldrich). For the methyl-labelled UGT1A-C, E. coli strains were adapted to deuterated minimal medium by gradually increasing the deuterium content [43]. Briefly, freshly transformed colonies were used to start a 25ml culture in LB medium/H2O, followed by successive pre-cultures in M9 media containing 0%, 50% and 100% D2O. Each pre-culture was initiated to an OD600 of 0.25. In the final culture, the 13C-labelled precursors α-ketoisovalerate and α-ketobutyrate (Sigma-Alrich) were added 1h before induction at 100 mg/L and 60 mg/L, respectively.
NMR Spectroscopy.
All NMR experiments were acquired on a 600MHz Bruker Avance III spectrometer equipped with a 5 mm QCIP cryoprobe with Z gradients and a samplejet autosampler unless otherwise specified. NMR experiments were acquired at 20°C. When indicated, data was acquired on a Bruker 800 MHz spectrometer equipped with TCI cryogenic probes. NMR assignments for UGT1A-C were deposited in the BMRB (Accession number 27446) and reported previously [31]. All uniform sampled spectra were processed with NMRPipe [44]. Non-uniform sampled spectra [45] were processed using SMILE [46] and NMRPipe [44]. Data were analyzed with NMRView [47].
Fragment library and screen.
570 compounds were selected for library curation based on the Rule of 3 selection criteria that were part of the Maybridge fragment library: of these 290 compounds were available from our in-house inventory, the remaining 280 were purchased from Maybridge. PAINS compounds [35] were not included. Each compound was tested for purity (> 95%), solubility (at 0.5 mM of compound) and aggregation state by running 1D 1H NMR spectra and a series of T2 (10, 50, 200, 400 and 800 ms) spectra (using excitation sculpting [48] to suppress water) in 50 mM sodium phosphate, 50 mM NaCl, 0.5 mM DTT, 95%H2O/5%D2O, pH 7.2 buffer. Compounds were added as 25 mM DMSO stocks to a final concentration of 0.5 mM compound (2% final concentration DMSO). Samples were submitted using the CMCq module in TOPSIN3.2 (Bruker Biospin) and analysed with the CMCa software (Bruker Biospin) using 0.5 mM maleic acid in the same buffer as an external reference control for concentration estimation. The CMCa software produces estimations of purity, concentration and compound identification, but was designed for organic solvents. Thus manual inspection of the results was necessary for our compounds that were acquired in aqueous buffer. 385 compounds passed quality control, the main reason for failure was low solubility.
The 385-fragment library was screened as 78 pools of up to 5 fragments per pool. Each pool was chosen to minimise overlap by manual visualisation in 1D 1H NMR spectra. Screens were ran in 3 mm samplejet tubes, in which 3.2 μL of 25 mM pooled compounds in DMSO was added to 356.8 μL of 10 μM UGT1A-C in NMR buffer (50 mM sodium phosphate, 50 mM NaCl, 0.5 mM DTT, pH 7.2, 95% H2O/5% D2O). Final concentrations of each fragment were ~ 0.5 mM, and DMSO was 2% (vol/vol). STD–NMR experiments [36] were ran at 293K using excitation sculpting to suppress H2O. On resonance and off resonance spectra were recorded with a 3 sec train of Gaussian pulses at −0.18 ppm and 30 ppm, respectively, and a recycle delay of 1 sec leading to a total acquisition time of ~ 15 min 30 sec. Spectra were processed and analysed with TOPSPIN 3.2 (Bruker Biopsin) and in house macros facilitating visualisation of STD spectra, pool mixtures and 1D spectra of compounds to identify hits.
Singleton Validation.
Compounds identified as hits from the fragment screen were tested as single compounds (singletons) in 3 mm samplejet tubes. Buffer conditions were identical to the screening conditions but using higher NaCl (300 mM) concentration to help stabilise protein aggregation. 20 μM protein and 1 mM fragment were used. WaterLOGSY experiments were performed as described [37] using excitation sculpting for water suppression and a 7.5 msec selective Gaussian 180° pulse for solvent excitation, total acquisition time was ~ 20 min. BEST 1H-15N HSQC NMR spectra [38] were acquired on samples containing 41 μM UGT1A-C and 2 mM fragment using 80 t1 increments.
NMR titrations.
Several titrations were run using different fragments and labelled versions of UGT1A-C. All experiments were run in NMR buffer (300 mM NaCl) at 600 MHz unless specified. 1H-15N NMR titrations were ran for fragments 3E08, 3H04 and 3H06 with a fixed concentration of 41 μM UGT1A-C and varying concentration of fragments from 0 up to 2 mM for fragments 3E08, 3H06 and 1 mM for 3H04. Ratios are provided in the figure legends. For UDP-GlcA, the salt concentration was reduced to 50 mM, UGT1A-C was 45μM and UDP-GlcA ranged from 0 to 2 mM. Chemical shift perturbation (CSP) mapping was based on the following formula: .
Constant time 1H-13C HSQC titration experiments with 3H04 and 3E08 samples were run at 800 MHz on a TCI cryoprobe. Protein concentrations were 50 μM and fragment ratios varied from 0–1 mM for 3E08 and 0–500 μM for 3H04 because of its reduced solubility. For 3H06, 1H-13C HSQC titration experiments were acquired at 600 MHz with 145 μM 13C labelled protein and the fragment concentration ranged from 0 to 2.1 mM. All 2D HSQC experiments were processed with NMRPipe and visualised with NMRView.
Intermolecular NOEs.
3D 13C-edited, 13C/15N filtered NOESY experiments were ran in NMR buffer to detect direct interactions between UGT1A-C and the 3E08 and 3H06 fragments using the Bruker pulse program hsqcgpnowgx33d [49, 50]. A 13C/15N double labelled UGT1A-C sample was used at 145 μM, the fragment concentration was 2.1 mM. Experiments were acquired with a NOESY mixing time of 120 msec with 30% non-uniform sampling corresponding to a total of 120 and 84 points in t2 and t1, respectively. Spectra were processed using SMILE. Additional quick 2D 1H-1H planes were also ran at 10 msec to check for artefactual peaks that might arise from the large excess of fragments. No artefacts were detected.
Methyl NMR Assignments.
Assignments for the perturbed methyl resonances were facilitated by a variety of NMR experiments including the 3D CCONH_TOCSY [51], 3D HCCONHTOCSY and the 4D methyl–methyl 13C-resolved HMQC–NOESY–HMQC experiment. Experiments were acquired on both uniformly labelled 13C/15N samples and ILV-labelled UGT1A-C. All experiments were acquired using non-inform sampling (typically 15–20% for 3D experiments). The 4D NOESY’s were acquired using a 4% non-uniform sampling schedule and a NOESY mixing time of 160 msec and 190 msec, respectively, for the doubly labelled and ILV-labelled samples.
In vitro glucuronidation assays.
UGT activity was determined using the UGT-Glo assay from Promega according to the manufacturer’s instructions using proluciferin substrates specific to UGT1A4 or a substrate targeting UGT1A1 but not UGT1A4 (Promega, V2082). UGT1A1 supersomes (Corning, 456411) and UGT1A4 supersomes (Corning, 456414) were used as indicated (at a final concentration of 0.2 mg/mL). Specifically for UGT1A1 experiments, 20 μM substrate was used and the reaction proceeded for 70 min. For UGT1A4, 50 μM of the UGT1A4 substrate was used in a reaction which proceeded for 3 hours. Alamethicin (SIGMA, A4665) was added (25 μg/mL) to enable membrane permeabilization. Reactions were performed at 37°C. Experiments were done in the presence or absence of UDP-GlcA. Concentrations were as follows: 1 mM fragment, 4 mM UDP-GlcA and 1% DMSO. Controls with 1% DMSO were carried out in parallel to ensure activities were not dependent on DMSO. Luciferase activity is reduced when the substrate is glucuronidated (indicating high UGT1A1 or UGT1A4 activity), and thus luciferase activity was measured using the M Star (BMG labtech) plate reader. The activity of UGT1As with no compound was normalized to 0% reduced activity. The percent of activity reduced relative to the measured activity of UGT1A in absence of compound is plotted on the graph and is relative to UGT observed inhibition. Results are the average of three biological replicates each conducted in triplicate.
Identification of glucuronides via mass spectrometry.
Selected fragments were tested for glucuronidation in UGT1A4 supersomes (1C08, 2E04, 3E08, 3H04 and 3H06) and UGT1A1 supersomes (1C08 and 3E08). Conditions were identical to conditions for the UGT-Glo assay described above, with the exception that no UGT specific substrate was added with the compounds. As controls, imipramine and the specific substrates for UGT1A4 and UGT1A1 supplied in UGT-Glo kit were also tested. Briefly, a reaction mix containing UGT1A4 (or UGT1A1) supersomes (0.2 mg/mL final concentration), UGT-glo buffer (50 mM TES, 10 mM MgCl2, pH 7.5) and alamethicin (25 μg/mL) was added to eppendorfs containing the tested compounds (1 mM and 100 μM) in the absence and presence of 4 mM UDP-GlcA. All concentrations are reported as the final concentration. The reaction was left at 37°C for 3 hours for UGT1A4 (70 minutes for UGT1A1) and quenched with a 2:1 (v/v) ratio of cold acetonitrile and spun down at 15K rpm for 15 minutes. The supernatant was used for further analysis. To identify possible formation of glucuronides UHPLC was used in conjunction with mass spectrometry. 2 μL (for 1 mM fragment incubations) or 6 μL (for 100 μM fragment incusbations) injections were run on a Waters Acquity I UHPLC coupled to a Waters Xevo G2-XS QTof (Waters Co., Milford, MA. USA) with an electrospray ionization interface (ESI) source. The UHPLC columns used were a Waters Acquity HSS T3 C18, 100 × 2.1 mm, 1.8 μm. The mobile phases were solvent A (100% water + 0.1% formic acid) and solvent B (100% acetonitrile + 0.1% formic acid) with the following gradient: of 3% B (0.8 min), 3–90% B (0.8–9 min), 95% B (9–9.5 min) and 3% B (9.5–13 min). A flow rate of 0.5 mL/min was used. The MS data were collected from m/z 100 to 1200 Da in positive or negative ion MSE continuum mode. Glucuronide formation was tested for by searching for the mass of the m/z of the parent ion +176.0321 Da (± 5ppm).
Mammalian Cell Culture.
FaDu cells (ATCC® HTB-43™) were maintained in Minimal Essential Medium (MEM) supplemented with 1% MEM Non-Essential Amino Acids, 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (Invitrogen). 2Flag-Gli1 or 2Flag vector control FaDu cell lines were generated using TransIT®-LT1 Transfection Reagent (Mirus), according to manufacturer guidelines and were selected using Geneticin® Selective Antibiotic (G418 Sulfate) (Invitrogen) [5]. FRII cell lines were generated by extended growth in ribavirin as described in [5]. All cell lines were routinely checked to ensure that there was no mycoplasma contamination using MycoAlert Mycoplasma Detection kit (Lonza,NY, U.S.A, Cat# LT07–418).
Drug treatments and cell viability assay.
Cell viability assays were carried out as previously described [5]. Briefly, 4 ×104 cells/100 μL were seeded in triplicates/condition in 96-well cell culture plates (BD Biosciences catalog number 353916). 24 hours post-plating, cells were treated as indicated. Working drug concentrations are as follows: ribavirin 20 μM, Ara-C 200 nM, GDC-0449 200 nM, imipramine 10 μM, and fragments 100 μM (note that we determined that this concentration of fragments was not toxic prior to these experiments). Treatments were for 96h at 37°C, 5% CO2. Cell viability was assessed using alamarBlue reagent (ThermoFisher Scientific, Catalog # DAL1100) according to manufacturer’s instructions. 2 hours following reagent addition, absorption was measured at 570 nm and was normalized by the reference absorption at 600 nm. Absorption of media alone was determined and compensated for. All values were normalized to DMSO treated control. Values are from three independent biological replicates each carried out in triplicate. P-values were determined using Student’s t-test.
Supplementary Material
Highlights.
The UGT1A (UDP-glucuronosyltransferase) family is involved in drug resistance
NMR fragment screens found UGT1A binders using the common C-terminus
Biochemical assays showed fragments selectively inhibited different UGT1A members
These fragments reduced glucuronidation in a target specific manner in cancer cells
We present a method to identify selective inhibition of the UGT1A family members
Acknowledgements.
We thank Drs. Steven LaPlante and Edward Ruediger for helpful discussions. 800 MHz NMR data were acquired at QANUC and 600 MHz NMR data at IRIC which were both supported in part by the Canadian Foundation for Innovation. This research was supported by the following grants to KLBB: IRICoR, CDRD, Merck, LLS USA, NIH 98571 and NIH 80728. KLBB holds a Canada Research Chair in Molecular Biology of the Cell Nucleus.
Abbreviations
- UGT
UDP-glucuronosyltransferase
- UGT1A
UDP-glucuronosyltransferase isoform 1A family
- UDP-GlcA
UDP-glucuronic acid
- UDP
uridine diphosphate
- AML
acute myeloid leukemia
- Gli1
glioma-associated protein 1
- CSP
chemical shift perturbation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests.
The authors have no competing interests.
REFERENCES
- 1.Zahreddine H, Borden KL (2013). Mechanisms and insights into drug resistance in cancer. Front. Pharmacol 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borst P (2012). Cancer drug pan-resistance: pumps, cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open. Biol 2:120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrethsen J, Angeletti RH, Horwitz SB, Yang CP (2014). Proteomics of cancer cell lines resistant to microtubule-stabilizing agents. Mol. Cancer Ther 13:260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orr GA, Verdier-Pinard P, McDaid H, Horwitz SB (2003). Mechanisms of Taxol resistance related to microtubules. Oncogene. 22:7280–7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, et al. (2014). The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 511:90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tukey RH, Strassburg CP (2000). Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol 40:581–616. [DOI] [PubMed] [Google Scholar]
- 7.Rowland A, Miners JO, Mackenzie PI (2013). The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol 45:1121–1132. [DOI] [PubMed] [Google Scholar]
- 8.Dutton G (1980). Glucuronidation of Drugs and Other Compounds. Boca Rotan, Florida: CRC Press. [Google Scholar]
- 9.Oda S, Fujiwara R, Kutsuno Y, Fukami T, Itoh T, Yokoi T, et al. (2015). Targeted screen for human UDP-glucuronosyltransferases inhibitors and the evaluation of potential drug-drug interactions with zafirlukast. Drug Metab. Dispos 43:812–818. [DOI] [PubMed] [Google Scholar]
- 10.Assouline S, Culjkovic-Kraljacic B, Bergeron J, Caplan S, Cocolakis E, Lambert C, et al. (2015). A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haemat. 100:e7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borden KL (2014). When Will Resistance Be Futile? Cancer Res. 74:7175–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahreddine H, Culjkovic-Kraljacic B, Gasiorek J, Duchaine J, Borden KLB. (submitted). Glioma associated protein 1 (Gli1) induces resistance to a broad spectrum of drugs.
- 13.Dijkgraaf GJ, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, et al. (2011). Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 71:435–444. [DOI] [PubMed] [Google Scholar]
- 14.LoRusso PM, Rudin CM, Reddy JC, Tibes R, Weiss GJ, Borad MJ, et al. (2011). Phase I trial of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin. Cancer Res 17:2502–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber M, Bellemare J, Hoermann G, Gleiss A, Porpaczy E, Bilban M, et al. (2013). Overexpression of uridine diphospho glucuronosyltransferase 2B17 in high-risk chronic lymphocytic leukemia. Blood. 121:1175–1183. [DOI] [PubMed] [Google Scholar]
- 16.Landmann H, Proia DA, He S, Ogawa LS, Kramer F, Beissbarth T, et al. (2014). UDP glucuronosyltransferase 1A expression levels determine the response of colorectal cancer cells to the heat shock protein 90 inhibitor ganetespib. Cell Death Dis. 5:e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P, Zhu KW, Zhang DY, Yan H, Liu H, Liu YL, et al. (2018). Influence of UGT1A1 polymorphisms on the outcome of acute myeloid leukemia patients treated with cytarabine-base regimens. J Transl Med. 16:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutsuno Y, Sumida K, Itoh T, Tukey RH, Fujiwara R (2013). Glucuronidation of drugs in humanized UDP-glucuronosyltransferase 1 mice: Similarity with glucuronidation in human liver microsomes. Pharmacol. Res. & Perspect 1:e00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara R, Yokoi T, Nakajima M (2016). Structure and Protein-Protein Interactions of Human UDP-Glucuronosyltransferases. Front. Pharmacol 7:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillemette C, Levesque E, Rouleau M (2014). Pharmacogenomics of human uridine diphospho-glucuronosyltransferases and clinical implications. Clin Pharmacol Ther. 96:324–339. [DOI] [PubMed] [Google Scholar]
- 21.Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI (1999). Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab. Rev 31:817–899. [DOI] [PubMed] [Google Scholar]
- 22.Tripathi SP, Bhadauriya A, Patil A, Sangamwar AT (2013). Substrate selectivity of human intestinal UDP-glucuronosyltransferases (UGTs): in silico and in vitro insights. Drug Metab. Rev 45:231–252. [DOI] [PubMed] [Google Scholar]
- 23.Miley MJ, Zielinska AK, Keenan JE, Bratton SM, Radominska-Pandya A, Redinbo MR (2007). Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J. Mol. Biol 369:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radominska-Pandya A, Bratton SM, Redinbo MR, Miley MJ (2010). The crystal structure of human UDP-glucuronosyltransferase 2B7 C-terminal end is the first mammalian UGT target to be revealed: the significance for human UGTs from both the 1A and 2B families. Drug Metab. Rev 42:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X (2005). Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant. Cell 17:3141–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, et al. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25:1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laakkonen L, Finel M (2010). A molecular model of the human UDP-glucuronosyltransferase 1A1, its membrane orientation, and the interactions between different parts of the enzyme. Mol. Pharmacol 77:931–939. [DOI] [PubMed] [Google Scholar]
- 28.Wu B, Morrow JK, Singh R, Zhang S, Hu M (2011). Three-dimensional quantitative structure-activity relationship studies on UGT1A9-mediated 3-O-glucuronidation of natural flavonols using a pharmacophore-based comparative molecular field analysis model. J. Pharmacol. Exp. Ther 336:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu B, Wang X, Zhang S, Hu M (2012). Accurate prediction of glucuronidation of structurally diverse phenolics by human UGT1A9 using combined experimental and in silico approaches. Pharm Res. 29:1544–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grancharov K, Naydenova Z, Lozeva S, Golovinsky E (2001). Natural and synthetic inhibitors of UDP-glucuronosyltransferase. Pharmacol. Ther 89:171–186. [DOI] [PubMed] [Google Scholar]
- 31.Osborne M, Coutinho de Oliveira L, Volpon L, Borden KLB. (accepted). Assignment of the apo-form of the human C-terminal domain of UDP-Glucuronsyltransferase A1 (UGT1A) Biomol. NMR Assign [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong D, Ako R, Hu M, Wu B (2012). Understanding substrate selectivity of human UDP-glucuronosyltransferases through QSAR modeling and analysis of homologous enzymes. Xenobiotica. 42:808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harner MJ, Frank AO, Fesik SW (2013). Fragment-based drug discovery using NMR spectroscopy. J. Biomol. NMR 56:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petros AM, Dinges J, Augeri DJ, Baumeister SA, Betebenner DA, Bures MG, et al. (2006). Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J. Med. Chem 49:656–663. [DOI] [PubMed] [Google Scholar]
- 35.Baell JB, Holloway GA (2010). New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem 53:2719–2740. [DOI] [PubMed] [Google Scholar]
- 36.Mayer M, Meyer B (1999). Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. Engl 38:1784–1788. [DOI] [PubMed] [Google Scholar]
- 37.Dalvit C, Fogliatto G, Stewart A, Veronesi M, Stockman B (2001). WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J. Biomol. NMR 21:349–359. [DOI] [PubMed] [Google Scholar]
- 38.Lescop E, Schanda P, Brutscher B (2007). A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson 187:163–169. [DOI] [PubMed] [Google Scholar]
- 39.Operana TN, Tukey RH (2007). Oligomerization of the UDP-glucuronosyltransferase 1A proteins: homo- and heterodimerization analysis by fluorescence resonance energy transfer and co-immunoprecipitation. J Biol Chem. 282:4821–4829. [DOI] [PubMed] [Google Scholar]
- 40.Kubota T, Lewis BC, Elliot DJ, Mackenzie PI, Miners JO (2007). Critical roles of residues 36 and 40 in the phenol and tertiary amine aglycone substrate selectivities of UDP-glucuronosyltransferases 1A3 and 1A4. Mol. Pharmacol 72:1054–1062. [DOI] [PubMed] [Google Scholar]
- 41.Kerdpin O, Mackenzie PI, Bowalgaha K, Finel M, Miners JO (2009). Influence of N-terminal domain histidine and proline residues on the substrate selectivities of human UDP-glucuronosyltransferase 1A1, 1A6, 1A9, 2B7, and 2B10. Drug Metab. Dispos 37:1948–1955. [DOI] [PubMed] [Google Scholar]
- 42.Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H (2016). Twenty years on: the impact of fragments on drug discovery. Nat. Rev. Drug Discov 15:605–619. [DOI] [PubMed] [Google Scholar]
- 43.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE (1999). A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR. 13:369–374. [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 45.Hyberts SG, Robson SA, Wagner G (2013). Exploring signal-to-noise ratio and sensitivity in non-uniformly sampled multi-dimensional NMR spectra. J Biomol NMR. 55:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ying J, Delaglio F, Torchia DA, Bax A (2017). Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J. Biomol. NMR 68:101–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson BA, Blevins RA (1994). NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603–614. [DOI] [PubMed] [Google Scholar]
- 48.Hwang TL, Shaka AJ (1995). Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson. A 112:275–279. [Google Scholar]
- 49.Breeze A (2000). Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Magn. Reson. Spectrosc 36:323–372. [Google Scholar]
- 50.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE (1997). Methods for Measurement of Intermolecular NOEs by Multinuclear NMR Spectroscopy: Application to a Bacteriophage λ N-Peptide/boxB RNA Complex. JACS. 119:6711–6721. [Google Scholar]
- 51.Kovacs H, Gossert A (2014). Improved NMR experiments with (1)(3)C-isotropic mixing for assignment of aromatic and aliphatic side chains in labeled proteins. J Biomol NMR. 58:101–112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.