Abstract
Purpose:
Neurite orientation dispersion and density imaging (NODDI) is a clinically feasible approach to measure intra-neurite volume fraction (fin). However, the sophisticated fitting procedure takes several hours. And the NODDI model relied on several questionable assumptions. Recent analytical work demonstrated that fin could be simply calculated from the spherical mean signal (MEANS) averaged over all gradient directions with a more solid theoretical foundation. The current study aims to compare NODDI and MEANS for measuring fin in human brain and investigate the potential of MEANS as a fast approach in clinics.
Methods:
NODDI fin and MEANS fin were measured and compared on the same dataset. NODDI fin was obtained using the NODDI MATLAB Toolbox. MEANS fin is the product of the spherical mean signal and where D is the intra-neurite intrinsic diffusivity.
Results:
NODDI fin and MEANS fin maps are similar. The voxel-by-voxel correlation suggests that NODDI fin and MEANS fin are approximately equivalent to each other.
Conclusion:
MEANS may have potential to serve a fast and simple approach to estimate fin in clinics.
Keywords: Neurite orientation dispersion and density imaging (NODDI), Intra-neurite volume fraction, Spherical mean signal
Introduction
Diffusion MRI has been widely used to measure tissue microstructure non-invasively. The conventional diffusion tensor imaging (DTI) technique models restricted water diffusion as a simple ellipsoid with three eigenvectors and corresponding eigenvalues [1]. The principal eigenvector is assumed to be the fiber orientation and the calculated fractional anisotropy (FA) is assumed to be an indicator of fiber density. However, it is well known that DTI is not an appropriate model in situations with crossing fibers, which commonly exist in brain white matter [2–4]. Based on the concept of high angular resolution diffusion imaging (HARDI), a large number of methods have been developed to estimate fiber orientation distribution [5–8] and fiber density [9–13]. Among them, neurite orientation dispersion and density imaging (NODDI) has been widely used to detect axonal injury in various neurological diseases [14–19] and assess neurite density changes in brain development [20–22].
NODDI is based on a three-compartment tissue model with distinct diffusion properties for each compartment [11]. The intra-neurite compartment is highly restricted for water diffusion perpendicular to neurites, and thus the intra-neurite perpendicular diffusivity is reasonably assumed to be 0. The extra-neurite compartment is also anisotropic but less restricted than the intra-neurite compartment. The third compartment is isotropic to mimic the diffusion behavior of free water. The diffusion weighted signals are measured along different gradient directions at multiple b-values and then fit to the analytical model. From the fitting, several parameters can be derived including the intra-neurite volume fraction. Note that the number of fitted parameters depends on the complexity of the model [23].
Instead of fitting the measured signals individually, recent works have proposed to focus on the spherical mean signal averaged over all gradient directions at the same b-value [24,25]. The reason is that the spherical mean signal is independent of the fiber orientation distribution [8]. In other word, the spherical mean signal based analysis can factor out the confounding fiber orientation distribution information and reduce the number of unknown parameters in the fitting, which in turn makes the fitting procedure more robust. Because of its substantial advantage in estimating tissue microstructure, the spherical mean signal based analysis has been applied in several recent mathematical modeling studies [26–32]. The spherical mean signal based NODDI fitting was implemented efficiently and resulted in similar results with the standard NODDI fitting [27,31]. Moreover, if the diffusion weighting b-value is sufficiently large, the intra-neurite volume fraction is simply the product of the spherical mean signal and, where D is the intra-neurite intrinsic diffusivity [33].
If the intrinsic diffusivity D is constant, as assumed in the NODDI model, the intra-neurite volume fraction can be calculated directly from the spherical mean signal without any fitting procedures. Besides, NODDI is based on several other assumptions. The fiber orientation distribution is assumed to be a single Watson distribution which could not account for fiber crossings. The extra-neurite perpendicular diffusivity is simplified with a simple tortuosity model and the extra-neurite water is modeled in fast exchange over all fiber orientations. The spherical mean signal is not affected by these assumptions. The current study aims to compare NODDI and spherical mean signal for measuring intra-neurite volume fraction and investigate the potential of spherical mean signal as a fast approach in clinics.
Materials and methods
Human Connectome Project (HCP) data
High-quality HCP data from 6 healthy adults, as part of the MGH-USC Adult Diffusion Dataset, were downloaded from ConnectomeDB (http://db.humanconnectome.org). Diffusion data were acquired with 4 different b-values ranging from 1 ms/μm2 to 10 ms/μm2, but only b = 1 ms/μm2 and b = 3 ms/μm2 were used in the current study. The number of gradient directions was 64 at each shell and the number of b = 0 images was 10. Other imaging parameters were: repetition time (TR) = 8800 ms, echo time (TE) = 57 ms, gradient duration (δ) = 12.9 ms, gradient separation (Δ) = 21.8 ms, image resolution = 1.5 × 1.5 × 1.5 mm3, parallel imaging acceleration factor = 3, and multiband factor = 1. The data were preprocessed with corrections for gradient nonlinearity distortions, head motion, and eddy current artifacts [34]. All the 10 b = 0 images were averaged and then segmented to gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) using FSL fast command [35,36].
Data analysis
As modeled in NODDI, the diffusion weighted signal (S) measured along gradient direction g can be written as
| (1) |
where S0 is the signal for b = 0; Sin and fin are the normalized signal and volume fraction of the intra-neurite compartment; Sex is the normalized signal of the extra-neurite compartment; and Siso and fiso are the normalized signal and volume fraction of the CSF compartment. The intra-neurite intrinsic diffusivity D is fixed as 1.7 μm2/ms. Details of other assumptions can be found in the original work [11], which have also been discussed in several recent studies [26,27,37,38]. NODDI fin was obtained using the NODDI MATLAB Toolbox.
When the diffusion weighting b-value is sufficiently large (b ≥ 3 ms/μm2) [33], the extra-neurite water contribution is negligible and the spherical mean diffusion weighted signal can be expressed as
| (2) |
Here the -based fin is termed MEANS fin. Only b = 3 ms/μm2 was used in Eq. (2) to calculate MEANS fin. The Rician bias was reduced by using the adjusted signal [25,39], where σ is the median of the standard deviation calculated voxel-by-voxel from all the b = 0 images.
Clinical data
As a supplementary analysis, NODDI fin and MEANS fin were further compared on clinical data. Three children (12–36 months, 2 boys) with focal cortical dysplasia at Nemours/Alfred I. duPont Hospital for Children were retrospectively selected for the analysis. Two children were scanned on a 3T GE MR750 scanner, and the other one was scanned on a GE SIGNA PET/MR scanner using the same diffusion protocol. The b-values were 1, 2 and 3 ms/μm2 with 15, 20 and 25 gradient directions, respectively. The number of b = 0 images was 6. Other diffusion imaging parameters were: TR = 9700 ms, TE = 99 ms, number of slices = 56, slice thickness = 2.5 mm, field of view = 240 × 240 mm2, in-plane image resolution = 2.5 × 2.5 mm2, and parallel imaging acceleration factor = 2. Diffusion images were preprocessed with corrections for head motion, and eddy current artifacts using FSL eddy command [40]. NODDI fin and MEANS fin were obtained subsequently following the above data analysis. T1 weighted, T2 weighted and T2 FLAIR structural images were co-registered to the mean b = 0 image using FSL flirt command [41]. The study was approved by local Institutional Review Board.
Results
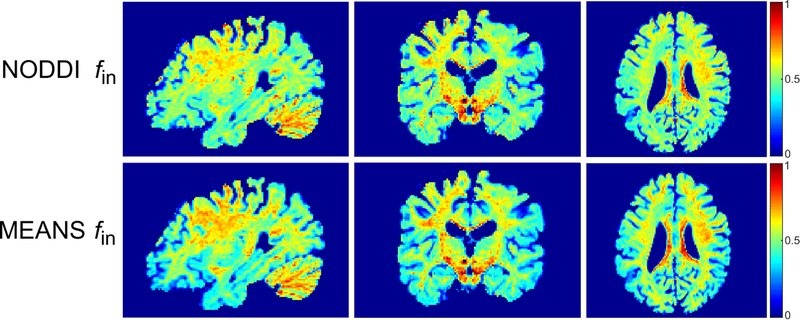
Fig. 1 shows the NODDI fin and MEANS fin maps acquired from a representative HCP subject at three anatomical planes. The two methods result in similar contrast. Consistent with previous NODDI studies [11,27,31], the value of fin is higher in white matter than in gray matter.
Figure 1.
The top row shows the fitted NODDI fin maps from a representative HCP subject at sagittal, coronal and axial planes. The bottom row shows the corresponding MEANS fin maps.
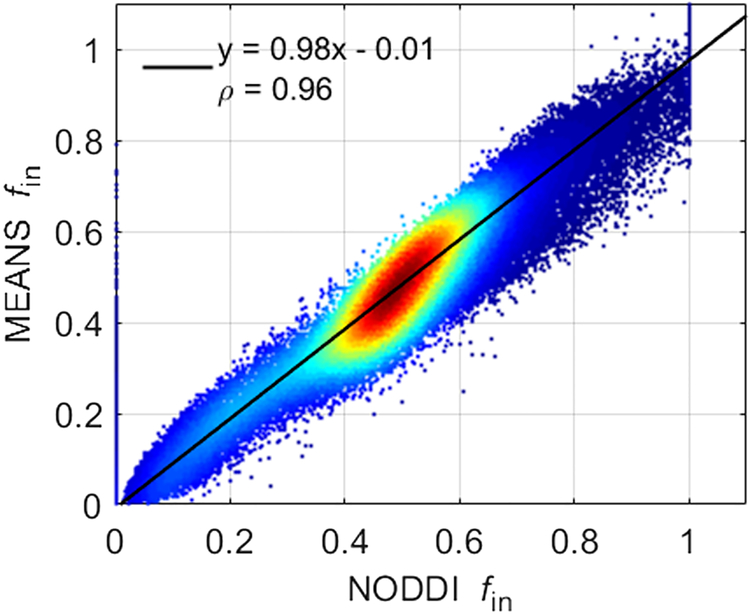
Fig. 2 shows the correlation between NODDI fin and MEANS fin. The density plot is from all gray matter and white matter voxels of the same subject shown in Fig. 1. Red denotes higher density of points, and blue denotes lower density. The solid line indicates the result of linear least squares fitting. The fitting slope is close to 1 and the intercept is close to 0. Pearson’s linear correlation coefficient (ρ) is also provided. The correlation coefficient ρ = 0.96 ± 0.01 over six HCP subjects. Fig. 2 suggests that NODDI fin and MEANS fin are approximately equivalent to each other.
Figure 2.
Density scatter plot and Pearson correlation between NODDI fin and MEANS fin using all gray matter and white matter voxels of the same subject shown in Figure 1. Red denotes higher density of points, and blue denotes lower density. The solid line indicates the result of linear least squares fitting.
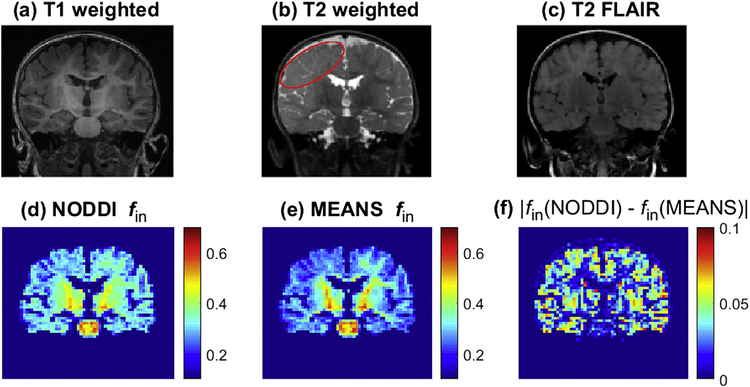
Fig. 3 shows representative structural images, NODDI fin, MEANS fin, and the absolute difference between NODDI fin and MEAN fin (|fin(NODDI) − fin(MEANS)|) acquired from a patient with focal cortical dysplasia. Structural images demonstrate subtle cortical dysplasia in the right cerebral hemisphere as evidenced by poor gray-white matter differentiation and T2 hyper-intense signal. The cortical dysplasia lesion region is outlined with red curve in Fig. 3 (b). Consistent with previous NODDI work [18], the lesion shows reduced NODDI fin compared with the contralateral side. Fig. 3 (e) suggests that MEANS fin has slightly better contrast than NODDI fin in this patient. The whole brain mean |fin(NODDI) − fin(MEANS)| is 0.026 ± 0.002 over three patients and the difference is more significant in gray matter than in white matter.
Figure 3.
Images of T1 weighted (a), T2 weighted (b), T2 FLAIR (c), NODDI fin (d), MEANS fin (e) and the absolute difference between NODDI fin and MEANS fin (f) acquired from a representative patient with focal cortical dysplasia. The red curve in (b) outlines the cortical dysplasia lesion.
Discussion
The current study compared NODDI and MEANS for measuring intra-neurite volume fraction on the same human brain data. Both methods are based on multi-compartment tissue modeling. NODDI fits the measured diffusion weighted signals to the analytical model individually. Instead, MEANS focuses on the spherical mean signal and derives intra-neurite volume fraction from spherical mean signal directly. The comparison results suggest that NODDI fin and MEANS fin are approximately equivalent. It should be noted that the current study is not aimed to assess which method is more accurate. Without the information of ground truth, it is unable to assess their accuracy. However, the comparison of different methods may help investigate their similarities and differences.
Compared with NODDI, MEANS is based on a more solid theoretical foundation. NODDI assumes a simple fiber orientation distribution and models the extra-neurite water diffusion in specific form. MEANS is not affected by these assumptions. NODDI and MEANS share two common assumptions: 1) the intra-neurite perpendicular diffusivity is 0 and 2) the intra-neurite intrinsic diffusivity D is constant over the whole brain. Due to the small restricting size [42] and long diffusion time on human scanner [9,43], the first assumption is usually agreed to be reasonable. Recent power law scaling studies demonstrated that the spherical mean signal decay behavior is consistent with Eq. (2) in white matter [37,44]. However, the spherical mean signal decay in gray matter is substantially faster, which makes the first assumption inappropriate for microstructural modeling in gray matter [37,44]. Water permeability [37] and fiber curvedness [45] have been proposed to explain the gray/white matter difference. Thus, the accuracy of fin quantification in gray matter is compromised by the first assumption. Though the measured NODDI fin or MEANS fin is not accurate in gray matter, it may still be able to provide some useful information about structural changes [46,47]. As for the second assumption, it is evident from Eq. (2) that the calculated MEANS fin will be biased when the assumption is violated. A recent study [48] found that the intra-neurite intrinsic diffusivity D is about 2.25 μm2/ms rather than 1.7 μm2/ms as assumed by NODDI. MEANS fin would increase by 15% when the intrinsic diffusivity D changes from 1.7 μm2/ms to 2.25 μm2/ms. And NODDI fin is expected to be affected similarly. Several studies have proposed to estimate fin and D simultaneously using multi-shell diffusion data [26,28–30,32] or novel diffusion sequences [27,49], but the results have not been validated yet. An accurate measurement of D would assist in better understanding the strengths and limitations of NODDI and MEANS.
The main difference between NODDI and MEANS is that NODDI requires at least two b shells. Besides fin, NODDI provides two more valuable indices: fiber orientation dispersion index ODI and isotropic water volume fraction fiso [11]. As mentioned in the original NODDI work [11], ODI can be estimated with just a single shell. Hence, the single shell data used in MEANS can also be used to estimate ODI through the standard NODDI fitting. Alternatively, the single shell data can be used to obtain the full fiber orientation distribution [50], which in turn can be used to calculate the orientation dispersion entropy [26]. The simultaneous fitting of fin and fiso indeed requires at least two shells. But previous studies observed an overestimated NODDI fiso in white matter [11,27]. A simple biexponential-T2 approach may estimate fiso more accurately [51].
NODDI protocol has been optimized and investigated through simulations extensively [11,21,22,52]. It was shown that the maximal b-value could be reduced to 2 ms/μm2 without significant effect on the estimated parameters [11]. As for MEANS, a high b-value (~3 ms/μm2) [9] or even higher [37,44] is required to fully suppress the extra-neurite water contribution. The effect of b-value and number of gradient directions on the accuracy of MEANS fin has been investigated recently [30,37,53,54]. The measured signal at b = 3 ms/μm2 is highly correlated with that at b = 5 ms/μm2 for human brain white matter, which suggests that b = 3 ms/μm2 is sufficient for spherical mean signal based studies in human brain white matter [53]. And it was recommended to use 10 × b / b1 (b1 = 1 ms/μm2) uniformly distributed gradient directions for typical human diffusion studies with signal-to-noise ratio ~ 20 [54].
Compared with NODDI, MEANS fin shows greater potential for clinical applications. First, -based data analysis is independent of fiber orientation distribution [8,25], MEANS fin is likely to be more accurate than NODDI fin. Second, MEANS estimates only one parameter. Diffusion-based microstructural modeling is generally associated with low signal-to-noise ratio and low image resolution [46,55]. Multi-shell diffusion data are needed to estimate extra parameters [11,26,29]. High resolution MEANS fin map is achievable by increasing the number of gradient directions at a single b-value. And the consequential signal-to-noise ratio is expected to be proportional to the square root of the number of gradient directions [49,54]. Third, the contrast is the same with the arithmetic mean signal, which may be generated on the scanner and read by radiologists soon after the scan is complete.
Conclusion
NODDI and MEANS were compared for measuring intra-neurite volume fraction in human brain. The voxel-by-voxel correlation suggests that NODDI fin and MEANS fin are approximately equivalent to each other. Without the need of sophisticated fitting, MEANS may have potential to serve a fast and simple approach to estimate fin in clinics.
Acknowledgements
This work was supported by the National Institutes of Health (grant number R21DC015853). Data were provided by the Human Connectome Project, MGH-USC Consortium (Principal Investigators: Bruce R. Rosen, Arthur W. Toga and Van Wedeen; U01MH093765) funded by the NIH Blueprint Initiative for Neuroscience Research grant; the National Institute of Health grant P41EB015896; and the Instrumentation Grants S10RR023043, 1S10RR023401, 1S10RR019307.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J 1994;66:259–67. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Leow AD, Zhan L, Zhu S, Hageman N, Chiang MC, Barysheva M, et al. White matter integrity measured by fractional anisotropy correlates poorly with actual individual fiber anisotropy. Proc. - 2009 IEEE Int. Symp. Biomed. Imaging From Nano to Macro, ISBI 2009, 2009, p. 622–5. doi: 10.1109/ISBI.2009.5193124. [DOI] [Google Scholar]
- [3].Schilling K, Gao Y, Janve V, Stepniewska I, Landman BA, Anderson AW. Can increased spatial resolution solve the crossing fiber problem for diffusion MRI? NMR Biomed 2017;30. doi: 10.1002/nbm.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the\ndo’s and don’ts of diffusion MRI. Neuroimage 2013;73:239–54. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- [5].Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Van Wedeen J. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med 2002;48:577–82. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- [6].Behrens TEJ, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, et al. Characterization and Propagation of Uncertainty in Diffusion-Weighted MR Imaging. Magn Reson Med 2003;50:1077–88. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- [7].Tournier JD, Calamante F, Gadian DG, Connelly A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. Neuroimage 2004;23:1176–85. doi: 10.1016/j.neuroimage.2004.07.037. [DOI] [PubMed] [Google Scholar]
- [8].Anderson AW. Measurement of fiber orientation distributions using high angular resolution diffusion imaging. Magn Reson Med 2005;54:1194–206. doi: 10.1002/mrm.20667. [DOI] [PubMed] [Google Scholar]
- [9].Raffelt D, Tournier JD, Rose S, Ridgway GR, Henderson R, Crozier S, et al. Apparent Fibre Density: A novel measure for the analysis of diffusion-weighted magnetic resonance images. Neuroimage 2012;59:3976–94. doi: 10.1016/j.neuroimage.2011.10.045. [DOI] [PubMed] [Google Scholar]
- [10].Fieremans E, Jensen JH, Helpern JA. White matter characterization with diffusional kurtosis imaging. Neuroimage 2011;58:177–88. doi: 10.1016/j.neuroimage.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012;61:1000–16. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- [12].Jespersen SN, Kroenke CD, Ostergaard L, Ackerman JJH, Yablonskiy DA. Modeling dendrite density from magnetic resonance diffusion measurements. Neuroimage 2007;34:1473–86. doi: 10.1016/j.neuroimage.2006.10.037. [DOI] [PubMed] [Google Scholar]
- [13].Jespersen SN, Bjarkam CR, Nyengaard JR, Chakravarty MM, Hansen B, Vosegaard T, et al. Neurite density from magnetic resonance diffusion measurements at ultrahigh field: Comparison with light microscopy and electron microscopy. Neuroimage 2010;49:205–16. doi: 10.1016/j.neuroimage.2009.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Adluru G, Gur Y, Anderson JS, Richards LG, Adluru N, DiBella EVR. Assessment of white matter microstructure in stroke patients using NODDI. Conf Proc. Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Annu Conf 2014;2014:742–5. doi: 10.1109/EMBC.2014.6943697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Churchill NW, Caverzasi E, Graham SJ, Hutchison MG, Schweizer TA. White matter microstructure in athletes with a history of concussion: Comparing diffusion tensor imaging (DTI) and neurite orientation dispersion and density imaging (NODDI). Hum Brain Mapp 2017;38:4201–11. doi: 10.1002/hbm.23658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schneider T, Brownlee W, Zhang H, Ciccarelli O, Miller DH, Wheeler-Kingshott CG. Sensitivity of multi-shell NODDI to multiple sclerosis white matter changes: A pilot study. Funct Neurol 2017;32:97–101. doi: 10.11138/FNeur/2017.32.2.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Slattery CF, Zhang J, Paterson RW, Foulkes AJM, Carton A, Macpherson K, et al. ApoE influences regional white-matter axonal density loss in Alzheimer’s disease. Neurobiol Aging 2017;57:8–17. doi: 10.1016/j.neurobiolaging.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Winston GP, Micallef C, Symms MR, Alexander DC, Duncan JS, Zhang H. Advanced diffusion imaging sequences could aid assessing patients with focal cortical dysplasia and epilepsy. Epilepsy Res 2014;108:336–9. doi: 10.1016/j.eplepsyres.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].By S, Xu J, Box BA, Bagnato FR, Smith SA. Application and evaluation of NODDI in the cervical spinal cord of multiple sclerosis patients. NeuroImage Clin 2017;15:333–42. doi: 10.1016/j.nicl.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kunz N, Zhang H, Vasung L, O’Brien KR, Assaf Y, Lazeyras F, et al. Assessing white matter microstructure of the newborn with multi-shell diffusion MRI and biophysical compartment models. Neuroimage 2014;96:288–99. doi: 10.1016/j.neuroimage.2014.03.057. [DOI] [PubMed] [Google Scholar]
- [21].Kodiweera C, Alexander AL, Harezlak J, McAllister TW, Wu YC. Age effects and sex differences in human brain white matter of young to middle-aged adults: A DTI, NODDI, and q-space study. Neuroimage 2016;128:180–92. doi: 10.1016/j.neuroimage.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jelescu IO, Veraart J, Adisetiyo V, Milla SS, Novikov DS, Fieremans E. One diffusion acquisition and different white matter models: How does microstructure change in human early development based on WMTI and NODDI? Neuroimage 2015;107:242–56. doi: 10.1016/j.neuroimage.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tariq M, Schneider T, Alexander DC, Gandini Wheeler-Kingshott CA, Zhang H. Bingham-NODDI: Mapping anisotropic orientation dispersion of neurites using diffusion MRI. Neuroimage 2016;133:207–23. doi: 10.1016/j.neuroimage.2016.01.046. [DOI] [PubMed] [Google Scholar]
- [24].Jespersen SN, Lundell H, Sønderby CK, Dyrby TB. Orientationally invariant metrics of apparent compartment eccentricity from double pulsed field gradient diffusion experiments. NMR Biomed 2013;26:1647–62. doi: 10.1002/nbm.2999. [DOI] [PubMed] [Google Scholar]
- [25].Kaden E, Kruggel F, Alexander DC. Quantitative mapping of the per-axon diffusion coefficients in brain white matter. Magn Reson Med 2016;75:1752–63.doi: 10.1002/mrm.25734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kaden E, Kelm ND, Carson RP, Does MD, Alexander DC. Multi-compartment microscopic diffusion imaging. Neuroimage 2016;139:346–59. doi: 10.1016/j.neuroimage.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lampinen B, Szczepankiewicz F, Mårtensson J, van Westen D, Sundgren PC, Nilsson M. Neurite density imaging versus imaging of microscopic anisotropy in diffusion MRI: A model comparison using spherical tensor encoding. Neuroimage 2017;147:517–31. doi: 10.1016/j.neuroimage.2016.11.053. [DOI] [PubMed] [Google Scholar]
- [28].Reisert M, Kellner E, Dhital B, Hennig J, Kiselev VG. Disentangling micro from mesostructure by diffusion MRI: A Bayesian approach. Neuroimage 2017;147:964–75. doi: 10.1016/j.neuroimage.2016.09.058. [DOI] [PubMed] [Google Scholar]
- [29].Novikov DS, Veraart J, Jelescu IO, Fieremans E. Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion MRI. Neuroimage 2018. doi: 10.1016/J.NEUROIMAGE.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McKinnon ET, Helpern JA, Jensen JH. Modeling white matter microstructure with fiber ball imaging. Neuroimage 2018. doi: 10.1016/j.neuroimage.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zucchelli M, Descoteaux M, Menegaz G. NODDI-SH: a computational efficient NODDI extension for fODF estimation in diffusion MRI. arXiv Prepr. arXiv1708.08999, 2017. [Google Scholar]
- [32].Veraart J, Novikov DS, Fieremans E. TE dependent Diffusion Imaging (TEdDI) distinguishes between compartmental T2relaxation times. Neuroimage 2017. doi: 10.1016/j.neuroimage.2017.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jensen JH, Russell Glenn G, Helpern JA. Fiber ball imaging. Neuroimage 2016;124:824–33. doi: 10.1016/j.neuroimage.2015.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fan Q, Witzel T, Nummenmaa A, Van Dijk KRA, Van Horn JD, Drews MK, et al. MGHUSC Human Connectome Project datasets with ultra-high b-value diffusion MRI. Neuroimage 2016;124:1108–14. doi: 10.1016/j.neuroimage.2015.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- [36].Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage, vol. 23, 2004. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- [37].McKinnon ET, Jensen JH, Glenn GR, Helpern JA. Dependence on b-value of the direction-averaged diffusion-weighted imaging signal in brain. Magn Reson Imaging 2017;36:121–7. doi: 10.1016/j.mri.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Novikov DS, Kiselev VG, Jespersen SN. On modeling. Magn Reson Med 2018;79:3172–93. doi: 10.1002/mrm.27101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gudbjartsson H, Patz S. The rician distribution of noisy mri data. Magn Reson Med 1995;34:910–4. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Andersson JLR, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 2016;125:1063–78. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jenkinson M, Bannister P, Brady M, Smith S. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. Neuroimage 2002;17:825–841. doi: 10.1016/S1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- [42].Liewald D, Miller R, Logothetis N, Wagner HJ, Schüz A. Distribution of axon diameters in cortical white matter: an electron-microscopic study on three human brains and a macaque. Biol Cybern 2014;108:541–57. doi: 10.1007/s00422-014-0626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med 2016;75:1076–85. doi: 10.1002/mrm.25684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Veraart J, Fieremans E, Novikov DS. Universal power-law scaling of water diffusion in human brain defines what we see with MRI. arXiv Prepr. arXiv1609.09145, 2016. [Google Scholar]
- [45].Özarslan E, Yolcu C, Herberthson M, Knutsson H, Westin C-F. Influence of the Size and Curvedness of Neural Projections on the Orientationally Averaged Diffusion MR Signal. Front Phys 2018;6. doi: 10.3389/fphy.2018.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rostampour M, Hashemi H, Najibi SM, Oghabian MA. Detection of structural abnormalities of cortical and subcortical gray matter in patients with MRI-negative refractory epilepsy using neurite orientation dispersion and density imaging. Phys Medica 2018;48:47–54. doi: 10.1016/j.ejmp.2018.03.005. [DOI] [PubMed] [Google Scholar]
- [47].Genç E, Fraenz C, Schlüter C, Friedrich P, Hossiep R, Voelkle MC, et al. Diffusion markers of dendritic density and arborization in gray matter predict differences in intelligence. Nat Commun 2018;9. doi: 10.1038/s41467-018-04268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dhital B, Reisert M, Kellner E, Kiselev VG. Intra-axonal diffusivity in brain white matter. arXiv Prepr. arXiv1712.04565, 2017. [DOI] [PubMed] [Google Scholar]
- [49].Jensen JH, Helpern JA. Characterizing intra-axonal water diffusion with direction-averaged triple diffusion encoding MRI. NMR Biomed 2018;31. doi: 10.1002/nbm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: Non-negativity constrained super-resolved spherical deconvolution. Neuroimage 2007;35:1459–72. doi: 10.1016/j.neuroimage.2007.02.016. [DOI] [PubMed] [Google Scholar]
- [51].Ferizi U, Scherrer B, Schneider T, Alipoor M, Eufracio O, Fick RHJ, et al. Diffusion MRI microstructure models with in vivo human brain Connectome data: results from a multi-group comparison. NMR Biomed 2017;30. doi: 10.1002/nbm.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kamiya K, Hori M, Irie R, Miyajima M, Nakajima M, Kamagata K, et al. Diffusion imaging of reversible and irreversible microstructural changes within the corticospinal tract in idiopathic normal pressure hydrocephalus. NeuroImage Clin 2017;14:663–71. doi: 10.1016/j.nicl.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li H, Chow HM, Chugani DC, Chugani HT. Linking spherical mean diffusion weighted signal with intra-axonal volume fraction. Magn Reson Imaging 2018. doi: 10.1016/j.mri.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li H, Chow HM, Chugani DC, Chugani HT. Minimal number of gradient directions for robust measurement of spherical mean diffusion weighted signal. Magn Reson Imaging 2018;54:148–52. doi: 10.1016/j.mri.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chougar L, Hagiwara A, Maekawa T, Hori M, Andica C, Iimura Y, et al. Limitation of neurite orientation dispersion and density imaging for the detection of focal cortical dysplasia with a “transmantle sign.” Phys Medica Eur J Med Phys 2018. [DOI] [PubMed] [Google Scholar]