Abstract
Amyotrophic Lateral Sclerosis (ALS) is the third most common adult onset neurodegenerative disorder worldwide. It is generally characterized by progressive paralysis starting at the limbs ultimately leading to death caused by respiratory failure. There is no cure and current treatments fail to slow the progression of the disease. As such, new treatment options are desperately needed. Epigenetic targets are an attractive possibility because they are reversible. Epigenetics refers to heritable changes in gene expression unrelated to changes in DNA sequence. Three main epigenetic mechanisms include the methylation of DNA, micro-RNAs and the post-translational modification of histone proteins. Histone modifications occur in many amino acid residues and include phosphorylation, acetylation, methylation as well as other chemical moieties. Recent evidence points to a possible role for epigenetic mechanisms in the etiology of ALS. Here we review recent advances linking ALS and epigenetics, with a strong focus on histone modifications. Both local and global changes in histone modification profiles are associated with ALS drawing attention to potential targets for future diagnostic and treatment approaches.
I. Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the selective loss of both upper and lower motor neurons. (1) As motor neurons in the spinal cord, brain, and brainstem weaken, skeletal muscular atrophy spreads through the patient.(2) Depending on the type of motor neurons affected, ALS can present itself clinically as either limb onset-manifesting as muscle weakness- or bulbar onset manifesting as speech impediments. (1) Prognosis is poor and quality of life is significantly reduced as most patients succumb to the disease within three years of diagnosis.(3) There is no cure for ALS and the only two FDA approved drugs, Rilutek® (riluzole) and Radicava® (edaravone), fail to stop progression of the disease.(4-6)
ALS is classified into two categories: familial and sporadic. Familial ALS represents about 10% of all cases, where the disease can be attributed to a particular gene mutation running in families.(7) Sporadic cases, on the other hand, represent the remaining 90% for which there is no family history of the disease. Numerous genes have been associated to ALS.(8) Some of these include superoxide dismutase 1 (SOD1), chromosome 9 open reading frame 72 (C9orf72), Fused in Sarcoma (FUS), and TAR DNA binding protein 43 (TDP-43).(9) Both familial and sporadic cases share similar pathology; the overwhelming majority of cases present neuronal TDP-43 protein aggregates.(10) Interestingly, not only neuronal cells are affected by these mutations. Glial cells harboring SOD1 mutations have been shown to drive motor neuron degeneration in experimental models.(11-14) More specifically, microglia bearing a SOD1 mutation caused motor neuron degeneration in otherwise healthy mice,(15) and human derived motor neurons co-cultured with these microglia showed signs of motor degeneration as well.(16) Nevertheless, the true origin of ALS remains obscure. Therefore, there is an urgent need to understand the pathogenesis of ALS in order to develop new treatment options. Aberrant epigenetic mechanisms linked to ALS are beginning to be discovered. These are attractive therapeutic targets as they are pharmaceutically accessible and largely reversible.(17)
Epigenetics refers to heritable alterations in gene expression without modification to the genome.(18) Three main epigenetic mechanisms include DNA methylation, micro RNAs (miRNA), and histone post-translational modifications (PTMs). DNA methylation takes place on the 5’ carbon on cytosine bases and is involved in transcription regulation.(19) DNA methylation at enhancers is key in determining cell line fate.(20) Hypermethylation at gene promoters is generally found to silence gene expression, while methylation in the coding region can increase transcription. DNA methylation is a crucial rate-controlling step that occurs in genomic regulatory regions during transcription, making it detectable and possibly even reversible for treatment purposes.(21) miRNAs are key regulators of gene expression.(22) miRNAs are non-coding short pieces of RNA, containing approximately 22 nucleotides, that generally bind to the 3’-untranslated region (3’-UTR) of mRNA, causing degradation of the mRNA and translational silencing.
Histones are proteins which package and organize cellular DNA.(23) 146 bases of DNA coil around two H2A/H2B dimers and one H3/H4 tetramer. The histone N-terminal tails are heavily modified with a multitude of chemical moieties, including mono-, di- and tri-methylation, acetylation, phosphorylation, SUMOylation and ubiquitination occurring in a variety of residues.(24) Histone post-translational modifications (PTMs) play an important role in gene regulation by controlling the accessibility of DNA to the transcriptional machinery. Initially, histone PTMs were thought to only impact gene transcription by controlling how tightly DNA wraps around the histones. For example, histone acetylation on lysine residues lowers the positive charge on histones and the strength of interaction with the negatively charged DNA backbone favoring gene transcription. Conversely, hypoacetylation is associated with transcriptional silencing.(25-28) More recently, it has been established that these modifications actually comprise a “histone code” that other proteins can ‘write,’ ‘erase’ and ‘read.’(29, 30) Histone PTMs can also affect one another through ‘cross talk.’(31) For example, the phosphorylation of Serine 10 on Histone H3 (H3S10ph) promotes acetylation on Lysine 16 on Histone H4 (H4K16ac) and also inhibits tri-methylation on Lysine 9 on Histone H3 (H3K9me3).(32, 33)
While a thorough understanding of the underlying mechanisms resulting in ALS have yet to be reached, recent evidence shows that alterations in the epigenetic landscape might contribute to disease pathology. Here, we review recent evidence showing a link between ALS and epigenetics. While we incorporate exciting findings featuring DNA methylation and miRNA mechanisms, we mostly focus on recent advances linking histone PTMs to ALS.
II. DNA methylation in ALS
DNA methylation on cytosine bases is one the most studied epigenetic factors in neurodegenerative diseases and ALS.(17) In post-mortem spinal tissue from sporadic ALS patients, there are global alterations in DNA methylation and hydroxymethylation.(34) Not surprisingly, many of the genes that were hypo- or hypermethylated showed corresponding changes in decreasing or increasing gene expression. Interestingly, these genes were largely involved with the immune and inflammatory responses. Other reports link Dnmt3a, a de novo DNA methyltransferase, to ALS phenotypes.(20, 35-37) Mice lacking Dnmt3a are hypoactive, underperformed on tests of neuromuscular function and motor coordination and had decreased numbers of motor neurons akin to SOD1 mouse models of ALS.(35) Interestingly, Dnmt3a is required for the development of motor neurons in vitro.(20) Moreover, apoptosis observed in NSC34 cells treated with camptothecin and in wild-type mice with unilateral nerve avulsion was driven by increased activity of Dnmt3a, Dnmt1 – a maintenance DNA mehtyltranserfase – and increased levels of 5-methyltransferase.(36) Remarkably, Dnmt inhibition with RG108, a DNA methyltransferase inhibitor, blocked apoptosis in both cultured neurons and mice. In agreement, levels of Dnmt1, Dnmt3a and 5-methylcytosine are upregulated in the brain and spinal cords of ALS patients. (36) Furthermore, Dnmt3a was found in the mitochondria of the CNS and skeletal muscle of adult wild-type mice, but was significantly reduced in the mitochondria of ALS SOD1 mice.(37) Counter intuitively, there was also an increase in mitochondrial DNA methylation in the SOD1 mice, including at the 16S ribosomal RNA gene, which encodes for the mitochondrial 16 S rRNA subunit. The SOD1 mice also displayed increased mitophagy- the degradation of the mitochondria by way of autophagy- which contributed to neuronal degeneration.
Recently, genome-wide association studies discovered differentially methylated genes involved in pathways important to ALS and frontotemporal dementia(FTD)-ALS.(38) In the FTD-ALS subgroup, pathway analysis revealed three gene sets displaying enriched DNA methylation; among these is the Meissner Brain HCP with H3K4me3 and H3K27me3 gene set. This gene set contains genes with high-CpG-density promoters (HCP) bearing the bivalent histone H3 trimethylation mark at K4 and K27 (H3K4me3 and H3K27me3),(39) suggesting that both DNA methylation and histone PTMs are important in the progression of FTD-ALS. Interestingly, H3K4me3 and H3K27me3 are associated with neurological function.(40) For an in-depth examination of the links between DNA methylation and ALS, we direct the reader to a thorough review by Martin & Wong.(41)
III. miRNA and ALS
miRNAs are an epigenetic mechanism that reduce gene expression by binding Argonaute 2 and forming the RNA-induced silencing complex (RISC).(42, 43) The complex then binds the 3’-UTR of a specific mRNA, and degrades it if there is an extensive match between the mRNA and miRNA or, if there is a less extensive match, just binds the mRNA repressing expression of the bound transcript.(44-46) miRNAs are associated with neurodegenerative diseases such as Alzheimer’s Disease, (47) Parkinson’s Disease,(48) and Huntington’s Disease. (49) Several lines of evidence link ALS to miRNA dysregulation.(17, 50) For instance, there is a global decrease in miRNA levels in the spinal cords of ALS patients.(51) The levels of miRNAs were compared to healthy controls and 90 miRNAs were found to be dysregulated. These miRNAs had mRNA targets involved in cell death, immune response and brain development that may possibly contribute to ALS etiology. Some of these included upregulation of miR-155 and miR-142. These miRNA target ubiquilin 2 (UBQLN2), the RNA binding protein Fox-1 (RBFOX1) and reelin (RELN), a protein secreted by certain neurons to guide neuronal migration.(52) All of these genes have been associated with neurodegeneration.(51) Interestingly, genes involved with immune response were also found to be dysregulated in sALS patients with aberrant DNA methylation patterns.(34) This suggests a role for epigenetics contributing to cellular demise caused by inflammation in ALS. Furthermore, TDP-43 overexpression in AinV15 mouse embryonic stem cells inhibited the endogenous activity of miRNAs regulating the genes EIF2C4/AGO4.(51) These genes are members of the RNA-induced silencing complex- the ribonucleoprotein complex that is responsible for translational silencing by cleaving miRNA and complementary mRNAs- and have been found to be dysregulated in ALS.(53, 54) Moreover, a large number of miRNAs were differentially expressed in the spinal cord motor neurons of sporadic ALS patients.(55) Functional analysis revealed the some of the upregulated miRNAs could bind to the 3’-UTR of low molecular weight neurofilament (NFL),(55) an important cytoskeleton protein involved in mitochondrial transport in motor neurons,(56) leading to a decrease in NFL mRNA levels. Interestingly, protein levels of NFL are found to lowered in ALS.(57-59) Additionally, FUS disrupts the circuitry between miR-409 and miR-495 in motor neurons derived from embryonic mouse cells with a FUS knock-in mutation.(60) miR-409 and miR-495 regulate the expression of Gria2, which encodes for a subunit of α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor. The upregulation of these miRNAs led to a decrease in Gria2 mRNA. The authors validated that miR-409 and miR-495 bind the 3’-UTR of Gria2. Furthermore, FUS stabilizes the interactions between the miRNA and their target, leading to decreased Gria2 expression.(60) Gria2 alteration has been previously implicated in motor neuron degeneration via alterations in Ca2+ homeostasis leading to excitotoxicity.(61-63)
In light of all these alterations, miRNA profiling has been proposed as a potential biomarker panel for neurodegeneration and ALS (Table 1).(50) For example, microarray analysis of leukocytes from sporadic ALS patients compared with healthy controls revealed a eight miRNAs that were dysregulated.(64) Of these miRNA, only miR-338-3p was upregulated, and interestingly it has been previously found to be dysregulated in the brains of ALS patients.(65) A study of skeletal tissue from SOD1 G93A mice revealed that miR-206 was upregulated, and confirmed to also be upregulated in the plasma of ALS patients.(66) Similarly, comparison of circulating miRNAs in plasma form sporadic ALS patients and healthy controls revealed nine dysregulated miRNAs, three of which were upregulated.(67) Two of these miRNAs, has-miR-46469-5p and has-miR4299, were confirmed to be respectively up- and down-regulated in a larger ALS patient cohort. Interestingly, both of these miRNAs target EPHA4, a gene that is has been associated with ALS in animal models.(68) EPHA4 contributes to brain development and neuronal migration, much like RELN discussed above.(51) These results suggest epigenetic mechanisms may be contributing to ALS pathology through alterations in development. Furthermore, a number of miRNAs have been found to be dysregulated in the leukocytes of ALS patients.(69) Specifically, AAKT, which encodes for miR-338, is up-regulated, while DNM2, which encodes for miR-638, is down-regulated. Both of these miRNAs have been found to be involved in other neurodegenerative diseases, such as FTD and hereditary spastic paraplegia. (70, 71) Overall, miRNAs appear to play an important role in the pathology of ALS and show promise as robust diagnostic biomarkers.
Table 1.
List of potential miRNAs biomarkers for Amyotrophic Lateral Sclerosis.
| miRNA | Dysregulation | Tissue | Reference |
|---|---|---|---|
| miR-338-3p | Upregulated | Leukocytes of sporadic ALS patients | 45 |
| miR-206 | Upregulated | Plasma of sporadic ALS patients | 47 |
| has-miR-46469-5p | Upregulated | Plasma of sporadic ALS patients | 48 |
| has-miR-4299 | Downregulated | Plasmas of sporadic ALS patients | 48 |
| miR-338 | Upregulated | Leukocytes of ALS patients | 50 |
| miR-638 | Downregulated | Leukocytes of ALS patients | 50 |
IV. Histone Modifications and Chromatin Remodeling Enzymes in ALS
Compared to other diseases, the study of epigenetic mechanisms involved in ALS is just in its beginning stages and relatively few studies examining the association between epigenetics and the disease have been carried out. While most investigations have focused in DNA methylation, recent evidence has linked aberrant changes in histone PTMs levels to neurodegenerative disease and ALS.(7, 17, 72) Remarkably, many enzymes responsible for installing or removing histone modifications have also been associated with ALS (Table 2). For instance, histone acetyltransferases (HATs) and histone deacetylases (HDACs) are responsible for the addition and removal of acetyl groups on histones, respectively. There are two classes of HDACs that are Zn2+ dependent including HDACs 1-11. A third class, Sirtuins (SIRT) 1-7, are NAD+ dependent.(73) HDACs are drug targets for the treatment of many diseases, including cancer, inflammatory disease and pulmonary disease.(74-76) Correspondingly, there are a variety of kinases responsible for phosphorylating histones, including Aurora B kinase and AMP kinase.(77) Lastly, many histone methyltransferases are responsible for the deposition and removal of methyl groups on lysine and arginine residues on histones.(78) These enzymes can be specific to mono-, di- and tri-methylation, as well as symmetric and asymmetric methylation.(79)
Table 2.
List of Epigenetic Enzymes Involved in Amyotrophic Lateral Sclerosis.
| Class | Enzyme | Activity in ALS | Reference |
|---|---|---|---|
| Histone Acetyltransferases | CBP/p300 | Reduced activity when interacting with FUS | 61 |
| ELP3 | Required for neuronal communication and survival in Drosophila | 63 | |
| Histone Deacetylases | HDAC1 | Mislocalizes to cytoplasm in FUS ALS model | 73 |
| Accumulation in nucleus is neuroprotective | 74 | ||
| HDAC2 | Levels decrease in spinal tissue of ALS patients | 72 | |
| HDAC6 | Promotes SOD1 aggregation | 76 | |
| Overexpression is neuroprotective in mouse model | 77 | ||
| HDAC11 | Levels increase in the spinal tissue of ALS patients | 72 | |
| SIRT1 | Levels increase in the muscles of a mutated SOD1 mouse model | 75 | |
| SIRT2 | Levels decrease in motor neurons of a mutated SOD1 mouse model | 75 | |
| NAD+ * | Depletion leads to motor neuron degeneration | 79 | |
| Histone Methyltransferases | PRMT1 | Overexpression rescues neurite growth in FUS model | 93 |
| Reduced methylation of H4R3, ultimately leading to gene silencing | 94 | ||
| PRMT5 | Genomic instability and R-loop formation | 110 | |
| Chromatin Modifying | CHD2/Chd1 | Inhibited by TDP-43, inhibited stress response required for neuronal survival | 108 |
| POLR2A | Genomic instability and R-loop formation | 110 |
Sirtuin cofactor involved in ALS pathology.
IV.A. Histone Acetylation, HATs and HDACs
Histone acetylation has been repeatedly associated with ALS. In a FUS overexpression model in HeLa cells, FUS binds to and inhibits CBP/p300 HAT at a specific gene promoter and causes hypoacetylation near the CCND1 gene.(80) This causes reduced expression of CCND1, which encodes for cyclin D1, a protein required for progression through the cell cycle.(81) HeLa cell models have been repeatedly used in ALS research.(82-84) Evidencing another link between histone acetylation and ALS, ELP3 – a protein with HAT activity- was linked to motor neuron degeneration in ALS in a microsatellite-based genetic association study.(85) In zebra fish embryos, ELP3 knock down with anti-sense RNA leads to motor neuron degeneration in a dose dependent manner.(85) ELP3 acetylates Lysine 14 on Histone H3 (H3K14) and Lysine 8 on Histone H4 (H4K8).(86) Interestingly, mutagenesis screening in Drosophila revealed that ELP3 is critical for neuronal communication and survival.(85) These results support a role for histone hypoacetylation in ALS pathology. In fact, ELP3 directly regulates heat shock protein 70 (HSP70) expression by acetylation of histones H3 and H4 in yeast;(87) hence, ELP3 defects could result in motor neuron degeneration through a decrease in the transcription of HSP70.(81)
Recently, we characterized global histone acetylation levels in yeast ALS proteinopathy models.(72) Many cellular pathways are conserved from yeast to humans.(88, 89) Furthermore, yeast recapitulate the cellular toxicity and cytoplasmic foci of TDP-43 and FUS seen in human pathology.(90, 91) In fact, yeast models have enabled identification of common ALS genetic risk factors.(92) Yeast overexpressing human FUS showed significantly decreased levels of acetylation on Lysine 14 and Lysine 56 on Histone H3 (H3K14 and H3K56).(72) These histone PTM are conserved in humans. H3K14ac and H3K56ac are both involved in the DNA damage checkpoint activation.(93-95) Furthermore, H3K14ac is also found at the promoter of actively transcribed genes.(96) Interestingly, yeast overexpressing FUS had overall decreased RNA levels, suggesting that histone hypoacetylation leads to reduced transcription.(72) Surprisingly, an analogous TDP-43 yeast proteinopathy model did not display the same decreases in histone acetylation, but instead revealed hyperacetylation on Lysines 12 and 16 on Histone 4 (H4K12 and H4K16). H4K12ac is a modification localized on gene promoters and is associated with gene activation.(97) H4K16ac is a particularly interesting modification because it is associated with both gene expression and repression.(98, 99) Remarkably, each proteinopathy displayed its own distinct histone modification landscape. Puzzlingly, while both of these models share protein aggregation and cytotoxicity features, FUS and TDP-43 overexpression resulted in histone hypo- and hyperacetylation, respectively. This suggests that each proteinopathy and corresponding histone PTM profile may be leading to ALS in unique ways.
In addition to changes on histone acetylation, HDACs have been thoroughly implicated in ALS.(100) For instance, deletion of Set3- a member of a histone deacetylase complex, homolog to the human protein ASH1- was found to suppress the toxicity of TDP-43 inclusions in a yeast model.(101) Moreover, post-mortem analysis of brains and spinal cord tissue derived from ALS patients showed a decrease in HDAC 11 mRNA and an increase of HDAC 2 mRNA.(102) Furthermore, HDAC 1 has been found to mislocalize to the cytoplasm in a FUS knock-in mouse model.(103) Curiously, post-translational modification on the histone modifiers themselves appears to be important. Phosphorylation of serine residues in HDAC1 controls its subcellular localization, and HDAC1 accumulation in the nucleus was neuroprotective in a mouse model.(104) Further evidence for HDAC perturbations in ALS comes from SOD1 mouse models. SIRT1 levels are decreased in neurons and increased in muscles of mice expressing mutant SOD1, while SIRT2 mRNA levels decrease in motor neurons in the same model.(105) In NSC34 and HEK293 cells as well as mice expressing wild type and mutant SOD1G93A, HDAC6 knock-down induced SOD1 aggregation resulting in increased motor neuron loss.(106) In the same mice model, HDAC6 levels drop as ALS symptoms appear and then plummet as the disease progresses.(107) Interestingly, in this mouse model HDAC6 overexpression prolongs life span and delays motor neuron decay by inducing fusion of autophagosomes to lysosomes and promoting autophagy.(107)
HDAC cofactors also appear to play a role in disease.(108) For example, loss of NAD+, a SIRT cofactor, contributes to neurodegeneration.(109) Furthermore, the loss of intracellular nicotinamide phosphoribosyltranferase- the rate limiting enzyme in NAD+ synthesis- leads to motor neuron degeneration in mice where the gene was knocked out.(110) Increased activity of PARP1 has been observed to deplete NAD+ pools, and contributes neurotoxicity.(111) Interestingly, silencing of Parp1 is neuroprotective in a mouse model of cerebellar ataxia,(111) and it is a possible therapeutic target for neurodegeneration.
HDAC inhibition has arisen as a promising therapeutic in various ALS models. Treatment with trichostatin A, an HDAC inhibitor, ameliorated motor neuron degeneration in a SOD1 mouse model.(112) In motor neurons derived from ALS patients, pharmacological inhibition and genetic silencing of HDAC6 reverses axonal transport defects caused by mutant FUS.(113) Furthermore, HDAC inhibition with 4-PB increased motor function and neuroprotection in transgenic ALS mice.(114) Interestingly, treatment with sodium phenylbutryate, a general HDAC inhibitor, extended survival in SOD1G93A ALS mouse model,(115) and its effect is increased when combined with an antioxidant or Riluzole.(116, 117) Phenylbutyrate has shown to be safe and increase histone acetylation levels in ALS patients.(118) Perplexingly, loss of HDAC function and is related to disease in some models,(112-115) while HDAC inhibition is neuroprotective in others.(103, 105-107) Future studies will potentially reconcile these seemingly opposite landscapes and elucidate their contribution to ALS pathways.
IV.B. Histone Methylation and Methyltransferases
Lysine and arginine methylation have varying effects on gene expression. Generally, mono-methylation is associated with transcription activation, while tri-methylation is associated with transcription repression.(119) Recent evidence has linked tri-methylation of lysine residues on H3 and H4 with loss-of-function toxicity in ALS patients with dipeptide repeat expansions (DRE) in C9orf72.(120) There is an increase in H3K9me3, H3K27me3 and H4K20me3 levels around the DREs sequence in brain tissue from ALS patients compared to healthy controls. These histone PTMs are strongly associated with gene silencing.(119) Decreased levels of C9orf72 mRNA in patients support a loss-of-function toxicity model.(120) Interestingly, treating fibroblasts derived from patients bearing C9orf72 mutations with a histone demethylating agent reduced tri-methylation levels near DREs and restored C9orf72 mRNA levels.(120)
In yeast models of ALS, we observed distinct histone methylation profiles for TDP-43 and FUS proteinopathies.(72) Specifically, FUS overexpression is associated with decreased levels of asymmetric di-methylation on Arginine 3 on Histone H4 (H4R3me2asym), while TDP-43 overexpression is associated with decreased levels of tri-methylation on Lysine 36 on Histone H3 (H3K36me3). H34Rme2asym is associated with increased gene expression,(121) whereas H3K36me3 is linked to transcriptional repression by serving as a docking site for HDACs and promoting deacetylation.(122, 123) As for acetylation, these results highlight that different proteinopathies can have unique effects on epigenetic mechanisms. H4Rme2 is linked to H3K14ac,(124) which also decreased in this FUS model, underscoring cross talk between these two modifications.
Protein arginine N-methyltranserfase 1(PRMT1) is a methyltransferase that has been positively implicated in ALS. PRMT1 is responsible H4R3me2asym, which promotes histone acetylation and gene transcription.(125) In a FUSR521C mouse model of ALS, overexpression of PRMT1 was found to rescue neurite growth after oxidative stress.(126) The same study showed that PRMT1 activity is inhibited by interaction with FUS as a stable complex of FUS/PRMT1/Nd1-L mRNA is formed. Nd1-L is an actin stabilizing protein and is under-expressed in this model. Furthermore, in agreement with our FUS yeast model results,(72) loss of PRMT1 function -due to FUS mislocalization-led to the reduction of asymmetric di-methylation of Arginine 3 on Histone H4 (H4R3me2asym) causing a drop in the acetylation of Histone H3 on Lysine 9 and 14, ultimately leading to transcriptional silencing.(127)
IV.C. Histone Phosphorylation and Ubiquitylation
Phosphorylation on serine, threonine and tyrosine histone residues plays an important role in gene expression, transition through the cell cycle and DNA damage repair.(77) Histone phosphorylation and many enzymes responsible for it have been implicated in ALS and other neurodegenerative diseases.(128) For example, loss of FUS through RNAi leads to decreased cell proliferation and an increase in H3 phosphorylation in NSC-34 and HEK-293T cells.(129) This coincides with altered expression of genes involved in cell cycle regulation, cytoskeletal organization, oxidative stress and energy homeostasis, and may point to a loss-of-function mechanism for FUS mutants related to ALS.
In a FUS overexpression model in yeast, we discovered a reduction in phosphorylation levels on Threonine 129 on Histone H2B (H2BT129ph).(72) In yeast, reduced H2BT129ph is associated with decreased gene expression.(130) In the same model, we identified a profound decrease in the levels of phosphorylation on Serine 10 on Histone H3 (H3S10ph). H3S10ph is associated with active gene expression.(77) Furthermore, H3S10ph promotes acetylation of Histone H3 on Lysine 14 (H3K14ac) in yeast,(131) suggesting histone cross-talk may play an important role in disease processes. Interestingly, Ilp1, the yeast homologue of human Aurora B kinase, can phosphorylate both H3S10 and H2BT129. These results suggest Aurora B kinase might be involved in ALS pathology.
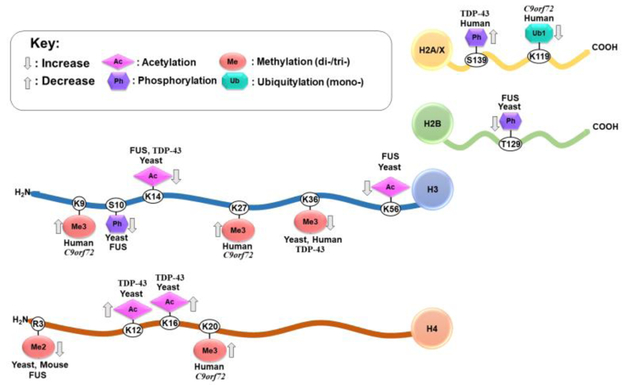
Increased R-Loops (DNA-RNA triple helixes) and DNA double strand breaks were observed in vitro and in the spinal tissue of ALS patients with mutations in C9orf72.(132) This damage was associated with dipeptide repeats in C9orf72 causing an accumulation of P62 and impaired H2A ubiquitylation at lysine 119 (H2AK119ub1). P62 is an autophagy protein related to ALS, and its accumulation inhibits RNF168, the enzyme responsible for ubiquitylation on H2A.(133) Reduced ubiquitylation impaired ATM signaling, an important pathway involved in DNA damage repair,(134) and perturbed DNA double strand break repair.(132) Figure 1 presents all histone modifications linked to ALS to date.
Figure 1.
Histone Post-Translational Modifications Linked to Different Proteinopathies and Genes in Amyotropic Lateral Sclerosis. Arrows denote an increase or a decrease for a given modification in the context of ALS.
One issue that arises whenever discussing histone modification is causality. (135) Are histone modifications a consequence of neurodegenerative proteinopathies or do changes in modifications lead to disease? Further research into how ALS-associated proteins interact with histone ‘writers’, ‘erasers’ and ‘readers’, and perhaps even the histone PTMs themselves is necessary to definitively answer this question. This may still be an emerging field with a number of questions yet to be answered, but as discussed above for HDACs inhibitors, drugs targeting histone modifiers are effective treatments in ALS models.(112-118) Thus, causality might ultimately be inconsequential, as pharmaceutical intervention targeting epigenetic features of ALS would alleviate disease outcomes. A number of histone modifications are associated with ALS, and each enzyme responsible for adding, removing or reading these modifications is a potential drugable target. Aside from HATs and HDACs, further research into chemical modulation of histone kinases, phosphorylases, methyltransferases and demethylases is needed. For example, inhibitors of protein phosphatase 1 and 2, responsible for removal of H3S10ph, (136, 137) as well as inhibitors of JMJD6, a Jumonji-domain-containing protein reported to demethylate H4R3me2(138), may be useful therapeutic targets for ALS.
IV.D. Chromatin Remodeling Enzymes
Eukaryotic DNA is packed into chromatin. DNA wraps around histones to form nucleosomes, the basic units of chromatin. Chromatin structure affects gene expression by controlling the accessibility to DNA.(139) Transcriptionally active chromatin is termed euchromatin and while transcriptionally silent chromatin is referred to as heterochromatin.(24, 140) Chromatin plays a key role in nearly all eukaryotic DNA-templated processes such as mitosis, DNA repair, and transcription. Some histone PTMs directly contribute to chromatin structure. For instance, H4K16ac inhibits the formation of heterochromatin,(141) while H3K9me3 binds Heterochromatin Protein 1 promoting chromatin compaction.(33) Additionally, enzymes that target histone PTMs can also play a role in chromatin compaction. For example, the HDAC SIRT1, which deacetylates H4K16, also has the ability to dephosphorylate the active site of the methyltransferase suppressor of variegation 3-9 homologue 1 (SUV39H1), increasing its activity and consequently increasing H3K27me3 levels and promoting chromatin compaction.(142) Aside from enzymes that directly affect histone PTMs, there a number of ATP-dependent chromatin remodeling enzymes that move, eject or otherwise restructure histones to control chromatin structure.(143, 144)Some of these have been associated to ALS and neurodegenerative disease. For example, in a TDP-43 overexpression Drosophila model, the chromatin-helicase-DNA binding protein Chd1 was inhibited by TDP-43, which in turn inhibited the stress response needed for neuronal survival.(145) Chd1 alters gene accessibility by modifying chromatin structure. (146) This loss of Chd1 function led to decreased expression of protective genes, such as the proteins involved in the REST and Hsp70 pathway. Correspondingly, up-regulation of Chd1 was found to be neuroprotective. The human orthologue of Chd1, CHD2, physically interacted with TDP-43 and was significantly reduced in the temporal cortex of patients with ALS.(145) Therefore, at least some of toxicity associated with TDP-43 seems to be caused by the reduced expression of protective genes caused by chromatin rearrangement.
POLR2A, a subunit of the carboxy-terminal domain of RNA Polymerase II, reads modifications specifying the recruitment of factors that regulate transcription, mRNA processing and chromatin remodeling.(147) Arginine 1810 of POLR2A is symmetrically di-methylated by protein arginine methyltransferase 5 (PRMT5) and this modification recruits the Tudor domain of survival of motor neuron (SMN). SMN interacts with senataxin, which is sometimes mutated in ALS.(148) Perturbations in this pathway lead to the formation of R-loops, genomic instability and pre-mature transcription termination causing neurodegeneration.(147)
Furthermore, RNA Polymerase II, as well as TDP-43 and FUS, colocalize with γH2AX (a histone H2A isoform phosphorylated on serine 139).(149) γH2AX is a sign of DNA double strand breaks.(150) γH2AX co-localization with TDP-43 and FUS suggests these proteins may have a role in double strand break repair. DNA double strand breaks may be harder to resolve when FUS or TDP-43 mislocalize from the nucleus, promoting neurodegeneration and suggesting a loss-of-function toxicity model for these two ALS-associated proteins. A summary of the histone and chromatin modifying enzymes implicated in ALS is presented in Table 2.
V. Conclusions
Mounting evidence highlights the role of epigenetic mechanisms in ALS pathology. DNA methylation, miRNAs and histone PTMs have distinct contributions to neurotoxicity. The evidence reviewed here reveals distinct DNA methylation patterns and dysregulation of miRNAs are associated with ALS. Although understudied, perturbations in the levels of histones modifications lead to alterations of gene expression and can contribute to the neurodegeneration. Remarkably, these alterations seem to be distinctly connected to different proteinopathies. Chromatin remodeling enzymes, which do not specifically impact histone modifications, also contribute to ALS pathology by reducing expression of survival genes and impairing DNA break repair. Though exact knowledge of how the different epigenetic mechanisms, in particular histone PTMs, affect ALS development and progression is still lacking, we are beginning to characterize them and realizing their potential importance in understanding ALS. Although much work remains to elucidate causal relationships and mechanisms linking histone PTMs and neurodegeneration, these findings highlight the critical need for the inclusion of epigenetics in neurodegenerative disease research. Remarkably, some of these altered epigenetic profiles can be detected in blood, and may become useful biomarkers for diagnosis and assessment of disease or treatment progress. We expect discoveries to come in the next decade will take us beyond establishing the links between ALS and epigenetics and move towards elucidating the precise epigenetic mechanisms associated with disease processes and specific symptoms. Pinpointing how epigenetic changes relate to disease can potentially lead to novel diagnostic and therapeutic tools for ALS and other neurodegenerative diseases.
VI.
Funding
Brooklyn College and a NIH NINDS Advanced Postdoctoral Career Transition Award (K22NS09131401) supported M.P.T. The Graduate Center of the City University of New York supported S.N.C.
List of Abbreviations:
- ALS
Amyotrophic Lateral Sclerosis
- FTD
Frontotemporal Dementia
- PTM
Post-Translational Modification
- HAT
Histone Acetyltransferase
- HDAC
Histone Deacetylase
- SIRT
Sirtuin
- PRMT
Protein Arginine N-methyltransferase
- SOD1
Superoxide Dismutase 1
- C9orf72
Chromosome 9 open reading frame 72
- FUS
Fused In Sarcoma
- TDP-43
TAR DNA Binding protein 43
- DRE
Dipeptide Repeat Expansion
- miRNA
micro RNA
- 3’-UTR
3’-Untranslated Region
- NFL
Low Molecular Weight Neurofilament
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole Propionic Acid
- SMN
Survival of Motor Neuron
- HCP
High-CpG-density promoter
- RISC
RNA-induced silencing complex
- SUV39H
Suppressor of variegation 3-9 homologue 1
Footnotes
Conflicts of Interests
The authors have no conflicts of interest to disclose. All authors have read the journal's policy on disclosure of potential conflicts of interest. All named authors have read the journal's authorship agreement and have reviewed and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
VII. References
- 1.Pang SY-Y, Hsu JS, Teo K-C, et al. Burden of rare variants in ALS genes influences survival in familial and sporadic ALS. Neurobiology of Aging. 2017;58:238.e9–.e15. [DOI] [PubMed] [Google Scholar]
- 2.Zarei S, Carr K, Reiley L, et al. A comprehensive review of amyotrophic lateral sclerosis. Surgical Neurology International. 2015;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Scottish Motor Neuron Disease Register: a prospective study of adult onset motor neuron disease in Scotland. Methodology, demography and clinical features of incident cases in 1989. Journal of Neurology, Neurosurgery & Psychiatry. 1992;55(7):536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrari R, Kapogiannis D, Huey ED, et al. FTD and ALS: a tale of two diseases. Current Alzheimer research. 2011;8(3):273–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debove C, Zeisser P, Salzman PM, et al. The Rilutek ® (riluzole) Global Early Access Programme: An open-label safety evaluation in the treatment of amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis and Other Motor Neuron Disorders. 2001;2(3):153–8. [DOI] [PubMed] [Google Scholar]
- 6.Brooks BR, Jorgenson JA, Newhouse BJ, et al. Edaravone in the treatment of amyotrophic lateral sclerosis: efficacy and access to therapy - a roundtable discussion 2018. S175–S86\p. [PubMed] [Google Scholar]
- 7.Belzil VV, Katzman RB, Petrucelli L. ALS and FTD: an epigenetic perspective. Acta Neuropathologica. 2016;132(4):487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nature Reviews Neurology. 2013;9:617. [DOI] [PubMed] [Google Scholar]
- 9.Ling S-C. Synaptic Paths to Neurodegeneration: The Emerging Role of TDP-43 and FUS in Synaptic Functions. Neural Plasticity. 2018;2018:8413496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and Biophysical Research Communications. 2006;351(3):602–11. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez DM, Gregory J, Brennand KJ. The Importance of Non-neuronal Cell Types in hiPSC-Based Disease Modeling and Drug Screening. Frontiers in Cell and Developmental Biology. 2017;5:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clement AM, Nguyen MD, Roberts EA, et al. Wild-Type Nonneuronal Cells Extend Survival of SOD1 Mutant Motor Neurons in ALS Mice. Science. 2003;302(5642): 113. [DOI] [PubMed] [Google Scholar]
- 13.Yamanaka K, Chun SJ, Boillée S, et al. Astrocytes as determinants of disease progression in inherited ALS. Nature neuroscience. 2008;11(3):251–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boillée S, Yamanaka K, Lobsiger CS, et al. Onset and Progression in Inherited ALS Determined by Motor Neurons and Microglia. Science. 2006;312(5778): 1389. [DOI] [PubMed] [Google Scholar]
- 15.Di Giorgio FP, Carrasco MA, Siao MC, et al. Non–cell autonomous effect of glia on motor neurons in an embryonic stem cell–based ALS model. Nature Neuroscience. 2007;10:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Giorgio FP, Boulting GL, Bobrowicz S, et al. Human Embryonic Stem Cell-Derived Motor Neurons Are Sensitive to the Toxic Effect of Glial Cells Carrying an ALS-Causing Mutation. Cell Stem Cell. 2008;3(6):637–48. [DOI] [PubMed] [Google Scholar]
- 17.Paez-Colasante X, Figueroa-Romero C, Sakowski SA, et al. Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat Rev Neurol. 2015;11(5):266–79. [DOI] [PubMed] [Google Scholar]
- 18.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nature Reviews Genetics. 2016;17:487. [DOI] [PubMed] [Google Scholar]
- 19.Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16(1):6–21. [DOI] [PubMed] [Google Scholar]
- 20.Ziller MJ, Ortega JA, Quinlan KA, et al. Dissecting the Functional Consequences of De Novo DNA Methylation Dynamics in Human Motor Neuron Differentiation and Physiology. Cell Stem Cell. 2018;22(4):559–74.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu H, Liu X, Deng Y, et al. DNA methylation, a hand behind neurodegenerative diseases. Frontiers in Aging Neuroscience. 2013;5:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannell Ian G, Kong Yi W, Bushell M. How do microRNAs regulate gene expression? Biochemical Society Transactions. 2008;36(6):1224. [DOI] [PubMed] [Google Scholar]
- 23.Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251. [DOI] [PubMed] [Google Scholar]
- 24.Mazzio EA, Soliman KFA. Basic concepts of epigenetics. Epigenetics. 2012;7(2):119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norton VG, Marvin KW, Yau P, et al. Nucleosome linking number change controlled by acetylation of histones H3 and H4. Journal of Biological Chemistry. 1990;265(32): 19848–52. [PubMed] [Google Scholar]
- 26.Lee DY, Hayes JJ, Pruss D, et al. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72(1):73–84. [DOI] [PubMed] [Google Scholar]
- 27.Hong L, Schroth GP, Matthews HR, et al. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. Journal of Biological Chemistry. 1993;268(1):305–14. [PubMed] [Google Scholar]
- 28.Norton VG, Imai BS, Yau P, et al. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989;57(3):449–57. [DOI] [PubMed] [Google Scholar]
- 29.Garcia BA, Shabanowitz J, Hunt DF. Characterization of histones and their post-translational modifications by mass spectrometry. Current Opinion in Chemical Biology. 2007;11(1):66–73. [DOI] [PubMed] [Google Scholar]
- 30.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41. [DOI] [PubMed] [Google Scholar]
- 31.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14(11):1017–24. [DOI] [PubMed] [Google Scholar]
- 32.Zippo A, Serafini R, Rocchigiani M, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6): 1122–36. [DOI] [PubMed] [Google Scholar]
- 33.Fischle W, Tseng BS, Dormann HL, et al. Regulation of HP1–chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116. [DOI] [PubMed] [Google Scholar]
- 34.Figueroa-Romero C, Hur J, Bender DE, et al. Identification of Epigenetically Altered Genes in Sporadic Amyotrophic Lateral Sclerosis. PLOS ONE. 2012;7(12):e52672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen S, Meletis K, Fu D, et al. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Developmental Dynamics. 2007;236(6):1663–76. [DOI] [PubMed] [Google Scholar]
- 36.Chestnut BA, Chang Q, Price A, et al. Epigenetic Regulation of Motor Neuron Cell Death through DNA Methylation. The Journal of Neuroscience. 2011;31(46):16619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong M, Gertz B, Chestnut B, et al. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Frontiers in Cellular Neuroscience. 2013;7:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taskesen E, Mishra A, van der Sluis S, et al. Susceptible genes and disease mechanisms identified in frontotemporal dementia and frontotemporal dementia with Amyotrophic Lateral Sclerosis by DNA-methylation and GWAS. Scientific Reports. 2017;7(1):8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The N, Pathway Analysis Subgroup of the Psychiatric Genomics C. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nature Neuroscience. 2015;18:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J Martin L, Wong M. Aberrant Regulation of DNA Methylation in Amyotrophic Lateral Sclerosis: A New Target of Disease Mechanisms 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tétreault N, De Guire V. miRNAs: Their discovery, biogenesis and mechanism of action. Clinical Biochemistry. 2013;46(10):842–5. [DOI] [PubMed] [Google Scholar]
- 43.Tolia NH, Joshua-Tor L. Slicer and the Argonautes. Nature Chemical Biology. 2006;3:36. [DOI] [PubMed] [Google Scholar]
- 44.Brennecke J, Stark A, Russell RB, et al. Principles of MicroRNA–Target Recognition. PLOS Biology. 2005;3(3):e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes & Development. 2005;19(5):517–29. [DOI] [PubMed] [Google Scholar]
- 46.Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Development. 2006;20(5):515–24. [DOI] [PubMed] [Google Scholar]
- 47.Basavaraju M, de Lencastre A. Alzheimer's disease: presence and role of microRNAs. Biomolecular concepts. 2016;7(4):241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A, Sen D. MicroRNAs in Parkinson’s disease. Experimental Brain Research. 2017;235(8):2359–74. [DOI] [PubMed] [Google Scholar]
- 49.Reed ER, Latourelle JC, Bockholt JH, et al. MicroRNAs in CSF as prodromal biomarkers for Huntington disease in the PREDICT-HD study. Neurology. 2018;90(4):e264–e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah P, Cho SK, Thulstrup PW, et al. MicroRNA Biomarkers in Neurodegenerative Diseases and Emerging NanoSensors Technology. Journal of Movement Disorders. 2017;10(1): 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Figueroa-Romero C, Hur J, Lunn JS, et al. Expression of microRNAs in human post-mortem amyotrophic lateral sclerosis spinal cords provides insight into disease mechanisms. Mol Cell Neurosci. 2016;71:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tissir F, Goffinet AM. Reelin and brain development. Nature Reviews Neuroscience. 2003;4:496. [DOI] [PubMed] [Google Scholar]
- 53.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biology. 2011;8(6):1149–57. [DOI] [PubMed] [Google Scholar]
- 54.Raman R, Allen Scott P, Goodall Emily F, et al. Gene expression signatures in motor neurone disease fibroblasts reveal dysregulation of metabolism, hypoxia-response and RNA processing functions. Neuropathology and Applied Neurobiology. 2014;41(2):201–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campos-Melo D, Droppelmann CA, He Z, et al. Altered microRNA expression profile in amyotrophic lateral sclerosis: a role in the regulation of NFL mRNA levels. Molecular Brain. 2013;6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gentil BJ, Minotti S, Beange M, et al. Normal role of the low-molecular-weight neurofilament protein in mitochondrial dynamics and disruption in Charcot-Marie-Tooth disease. The FASEB Journal. 2011;26(3):1194–203. [DOI] [PubMed] [Google Scholar]
- 57.Bergeron C, Beric-Maskarel K, Muntasser S, et al. Neurofilament Light and Polyadenylated mRNA Levels Are Decreased in Amyotrophic Lateral Sclerosis Motor Neurons. Journal of Neuropathology & Experimental Neurology. 1994;53(3):221–30. [DOI] [PubMed] [Google Scholar]
- 58.Menzies Fiona M, Grierson Andrew J, Cookson Mark R, et al. Selective loss of neurofilament expression in Cu/Zn superoxide dismutase (SOD1) linked amyotrophic lateral sclerosis. Journal of Neurochemistry. 2004;82(5):1118–28. [DOI] [PubMed] [Google Scholar]
- 59.Wong NKY, He BP, Strong MJ. Characterization of Neuronal Intermediate Filament Protein Expression in Cervical Spinal Motor Neurons in Sporadic Amyotrophic Lateral Sclerosis (ALS). Journal of Neuropathology & Experimental Neurology. 2000;59(11):972–82. [DOI] [PubMed] [Google Scholar]
- 60.Capauto D, Colantoni A, Lu L, et al. A Regulatory Circuitry Between Gria2, miR-409, and miR-495 Is Affected by ALS FUS Mutation in ESC-Derived Motor Neurons. Molecular Neurobiology. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takuma H, Kwak S, Yoshizawa T, et al. Reduction of GluR2 RNA editing, a molecular change that increases calcium influx through AMPA receptors, selective in the spinal ventral gray of patients with amyotrophic lateral sclerosis. Annals of Neurology. 2001;46(6):806–15. [DOI] [PubMed] [Google Scholar]
- 62.Kawahara Y, Ito K, Sun H, et al. RNA editing and death of motor neurons. Nature. 2004;427:801. [DOI] [PubMed] [Google Scholar]
- 63.Hideyama T, Yamashita T, Aizawa H, et al. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiology of Disease. 2012;45(3):1121–8. [DOI] [PubMed] [Google Scholar]
- 64.De Felice B, Guida M, Guida M, et al. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene. 2012;508(1):35–40. [DOI] [PubMed] [Google Scholar]
- 65.Shioya M, Obayashi S, Tabunoki H, et al. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathology and Applied Neurobiology. 2010;36(4):320–30. [DOI] [PubMed] [Google Scholar]
- 66.Toivonen JM, Manzano R, Oliván S, et al. MicroRNA-206: A Potential Circulating Biomarker Candidate for Amyotrophic Lateral Sclerosis. PLOS ONE. 2014;9(2):e89065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi I, Hama Y, Matsushima M, et al. Identification of plasma microRNAs as a biomarker of sporadic Amyotrophic Lateral Sclerosis. Molecular Brain. 2015;8(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Hoecke A, Schoonaert L, Lemmens R, et al. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nature Medicine. 2012;18:1418. [DOI] [PubMed] [Google Scholar]
- 69.Vrabec K, Boštjančič E, Koritnik B, et al. Differential Expression of Several miRNAs and the Host Genes AATK and DNM2 in Leukocytes of Sporadic ALS Patients. Frontiers in Molecular Neuroscience. 2018;11:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferrari R, Grassi M, Salvi E, et al. A genome-wide screening and SNPs-to-genes approach to identify novel genetic risk factors associated with frontotemporal dementia. Neurobiology of Aging. 2015;36(10):2904.e13–.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sambuughin N, Goldfarb LG, Sivtseva TM, et al. Adult-onset autosomal dominant spastic paraplegia linked to a GTPase-effector domain mutation of dynamin 2. BMC Neurology. 2015;15:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen K, Bennett SA, Rana N, et al. Neurodegenerative Disease Proteinopathies Are Connected to Distinct Histone Post-translational Modification Landscapes. ACS Chem Neurosci. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruijter AJMd, Gennip AHv, Caron HN, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochemical Journal. 2003;370(3):737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang L. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases. Journal of Cellular Physiology. 2006;209(3):611–6. [DOI] [PubMed] [Google Scholar]
- 75.Ito K, Ito M, Elliott WM, et al. Decreased Histone Deacetylase Activity in Chronic Obstructive Pulmonary Disease. New England Journal of Medicine. 2005;352(19):1967. [DOI] [PubMed] [Google Scholar]
- 76.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. The Journal of Clinical Investigation. 2014;124(1):30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sawicka A, Seiser C. Histone H3 phosphorylation – A versatile chromatin modification for different occasions. Biochimie. 2012;94(11):2193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson JR, Jing C, Walker PA, et al. Crystal Structure and Functional Analysis of the Histone Methyltransferase SET7/9. Cell. 2002;111(1): 105–15. [DOI] [PubMed] [Google Scholar]
- 79.Hyun K, Jeon J, Park K, et al. Writing, erasing and reading histone lysine methylations. Experimental &Amp; Molecular Medicine. 2017;49:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X, Arai S, Song X, et al. Induced ncRNAs Allosterically Modify RNA Binding Proteins in cis to Inhibit Transcription. Nature. 2008;454(7200): 126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Molecular Cancer. 2007;6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moller A, Bauer CS, Cohen RN, et al. Amyotrophic lateral sclerosis-associated mutant SOD1 inhibits anterograde axonal transport of mitochondria by reducing Miro1 levels. Human Molecular Genetics. 2017;26(23):4668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim SH, Shi Y, Hanson KA, et al. Potentiation of Amyotrophic Lateral Sclerosis (ALS)-associated TDP-43 Aggregation by the Proteasome-targeting Factor, Ubiquilin 1. Journal of Biological Chemistry. 2009;284(12):8083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim SH, Shanware NP, Bowler MJ, et al. Amyotrophic Lateral Sclerosis-associated Proteins TDP-43 and FUS/TLS Function in a Common Biochemical Complex to Co-regulate HDAC6 mRNA. Journal of Biological Chemistry. 2010;285(44):34097–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simpson CL, Lemmens R, Miskiewicz K, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Human Molecular Genetics. 2009;18(3):472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Winkler GS, Kristjuhan A, Erdjument-Bromage H, et al. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proceedings of the National Academy of Sciences. 2002;99(6):3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han Q, Lu J, Duan J, et al. Gcn5- and Elp3-induced histone H3 acetylation regulates Hsp70 gene transcription in yeast. Biochemical Journal. 2008;409(3):779. [DOI] [PubMed] [Google Scholar]
- 88.Jovicic A, Mertens J, Boeynaems S, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18(9):1226–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248(4959):1112. [DOI] [PubMed] [Google Scholar]
- 90.Johnson BS, Snead D, Lee JJ, et al. TDP-43 Is Intrinsically Aggregation-prone, and Amyotrophic Lateral Sclerosis-linked Mutations Accelerate Aggregation and Increase Toxicity. Journal of Biological Chemistry. 2009;284(30):20329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun Z, Diaz Z, Fang X, et al. Molecular Determinants and Genetic Modifiers of Aggregation and Toxicity for the ALS Disease Protein FUS/TLS. PLOS Biology. 2011;9(4):e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elden AC, Kim H-J, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duan M-R, Smerdon MJ. Histone H3 Lysine 14 (H3K14) Acetylation Facilitates DNA Repair in a Positioned Nucleosome by Stabilizing the Binding of the Chromatin Remodeler RSC (Remodels Structure of Chromatin). Journal of Biological Chemistry. 2014;289(12):8353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y, Kallgren SP, Reddy BD, et al. Histone H3 Lysine 14 Acetylation Is Required for Activation of a DNA Damage Checkpoint in Fission Yeast. Journal of Biological Chemistry. 2012;287(6):4386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen C-C, Carson JJ, Feser J, et al. Acetylated Lysine 56 on Histone H3 Drives Chromatin Assembly after Repair and Signals for the Completion of Repair. Cell. 2008;134(2):231–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karmodiya K, Krebs AR, Oulad-Abdelghani M, et al. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13(1):424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40(7):897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blosser TR, Yang JG, Stone MD, et al. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou Y, Grummt I. The PHD Finger/Bromodomain of NoRC Interacts with Acetylated Histone H4K16 and Is Sufficient for rDNA Silencing. Current Biology. 2005;15(15):1434–8. [DOI] [PubMed] [Google Scholar]
- 100.Lazo-Gomez R, Ramirez-Jarquin UN, Tovar YRLB, et al. Histone deacetylases and their role in motor neuron degeneration. Front Cell Neurosci. 2013;7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Armakola M, Higgins MJ, Figley MD, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nature Genetics. 2012;44:1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Janssen C, Schmalbach S, Boeselt S, et al. Differential Histone Deacetylase mRNA Expression Patterns in Amyotrophic Lateral Sclerosis. Journal of Neuropathology & Experimental Neurology. 2010;69(6):573–81. [DOI] [PubMed] [Google Scholar]
- 103.Scekic-Zahirovic J, Sendscheid O, El Oussini H, et al. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. The EMBO Journal. 2016;35(10):1077–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu Y, Vidaurre OG, Adula KP, et al. Subcellular Distribution of HDAC1 in Neurotoxic Conditions Is Dependent on Serine Phosphorylation. J Neurosci. 2017;37(31):7547–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Valle C, Salvatori I, Gerbino V, et al. Tissue-specific deregulation of selected HDACs characterizes ALS progression in mouse models: pharmacological characterization of SIRT1 and SIRT2 pathways. Cell Death Dis. 2014;5:e1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gal J, Chen J, Barnett KR, et al. HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J Biol Chem. 2013;288(21): 15035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Du Z-W, Chen H, Liu H, et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nature Communications. 2015;6:6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hoogeveen RM, Nahrendorf M, Riksen NP, et al. Monocyte and haematopoietic progenitor reprogramming as common mechanism underlying chronic inflammatory and cardiovascular diseases. Eur Heart J. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dobbin MM, Madabhushi R, Pan L, et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nature Neuroscience. 2013;16:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X, Zhang Q, Bao R, et al. Deletion of Nampt in Projection Neurons of Adult Mice Leads to Motor Dysfunction, Neurodegeneration, and Death. Cell Reports. 2017;20(9):2184–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoch NC, Hanzlikova H, Rulten SL, et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature. 2016;541:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yoo Y-E, Ko C-P. Treatment with trichostatin A initiated after disease onset delays disease progression and increases survival in a mouse model of amyotrophic lateral sclerosis. Experimental Neurology. 2011;231(1):147–59. [DOI] [PubMed] [Google Scholar]
- 113.Guo W, Naujock M, Fumagalli L, et al. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun. 2017;8(1): 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Corcoran LJ, Mitchison TJ, Liu Q. A Novel Action of Histone Deacetylase Inhibitors in a Protein Aggresome Disease Model. Current Biology. 2004;14(6):488–92. [DOI] [PubMed] [Google Scholar]
- 115.Ryu H, Smith K, Camelo SI, et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93(5): 1087–98. [DOI] [PubMed] [Google Scholar]
- 116.Petri S, Kiaei M, Kipiani K, et al. Additive neuroprotective effects of a histone deacetylase inhibitor and a catalytic antioxidant in a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiology of Disease. 2006;22(1):40–9. [DOI] [PubMed] [Google Scholar]
- 117.Del Signore SJ, Amante DJ, Kim J, et al. Combined riluzole and sodium phenylbutyrate therapy in transgenic amyotrophic lateral sclerosis mice. Amyotrophic Lateral Sclerosis. 2009;10(2):85–94. [DOI] [PubMed] [Google Scholar]
- 118.Cudkowicz ME, Andres PL, Macdonald SA, et al. Phase 2 study of sodium phenylbutyrate in ALS. Amyotrophic Lateral Sclerosis. 2009;10(2):99–106. [DOI] [PubMed] [Google Scholar]
- 119.Lachner M, Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. Journal of Cell Science. 2003;116(11):2117. [DOI] [PubMed] [Google Scholar]
- 120.Belzil VV, Bauer PO, Prudencio M, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathologica. 2013;126(6):895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gary JD, Lin W-J, Yang mC, et al. The Predominant Protein-arginine Methyltransferase from Saccharomyces cerevisiae. Journal of Biological Chemistry. 1996;271(21):12585–94. [DOI] [PubMed] [Google Scholar]
- 122.Carrozza MJ, Li B, Florens L, et al. Histone H3 Methylation by Set2 Directs Deacetylation of Coding Regions by Rpd3S to Suppress Spurious Intragenic Transcription. Cell. 2005;123(4):581–92. [DOI] [PubMed] [Google Scholar]
- 123.Joshi AA, Struhl K. Eaf3 Chromodomain Interaction with Methylated H3-K36 Links Histone Deacetylation to Pol II Elongation. Molecular Cell. 2005;20(6):971–8. [DOI] [PubMed] [Google Scholar]
- 124.Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes & Development. 2005;19(16):1885–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Litt M, Qiu Y, Huang S. Histone arginine methylations: their roles in chromatin dynamics and transcriptional regulation. Bioscience reports. 2009;29(2):131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jun MH, Ryu HH, Jun YW, et al. Sequestration of PRMT1 and Nd1-L mRNA into ALS-linked FUS mutant R521C-positive aggregates contributes to neurite degeneration upon oxidative stress. Sci Rep. 2017;7:40474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tibshirani M, Tradewell ML, Mattina KR, et al. Cytoplasmic sequestration of FUS/TLS associated with ALS alters histone marks through loss of nuclear protein arginine methyltransferase 1. Hum Mol Genet. 2015;24(3):773–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kwon MJ, Kim S, Han MH, et al. Epigenetic Changes in Neurodegenerative Diseases. Molecules and Cells. 2016;39(11):783–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ward CL, Boggio KJ, Johnson BN, et al. A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death &Amp; Disease. 2014;5:e1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kyriss MNM, Jin Y, Gallegos IJ, et al. Novel Functional Residues in the Core Domain of Histone H2B Regulate Yeast Gene Expression and Silencing and Affect the Response to DNA Damage. Molecular and Cellular Biology. 2010;30(14):3503–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lo W-S, Duggan L, Tolga NC, et al. Snf1--a Histone Kinase That Works in Concert with the Histone Acetyltransferase Gcn5 to Regulate Transcription. Science. 2001;293(5532):1142. [DOI] [PubMed] [Google Scholar]
- 132.Walker C, Herranz-Martin S, Karyka E, et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nature Neuroscience. 2017;20:1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Y, Zhang N, Zhang L, et al. Autophagy Regulates Chromatin Ubiquitination in DNA Damage Response through Elimination of SQSTM1/p62. Molecular Cell. 2016;63(1):34–48. [DOI] [PubMed] [Google Scholar]
- 134.Maréchal A, Zou L. DNA Damage Sensing by the ATM and ATR Kinases. Cold Spring Harbor Perspectives in Biology. 2013;5(9):a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends in Genetics. 2011;27(10):389–96. [DOI] [PubMed] [Google Scholar]
- 136.Nowak SJ, Pai C-Y, Corces VG. Protein Phosphatase 2A Activity Affects Histone H3 Phosphorylation and Transcription in Drosophila melanogaster. Molecular and Cellular Biology. 2003;23(17):6129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Murnion ME, Adams RR, Callister DM, et al. Chromatin-associated Protein Phosphatase 1 Regulates Aurora-B and Histone H3 Phosphorylation. Journal of Biological Chemistry. 2001;276(28):26656–65. [DOI] [PubMed] [Google Scholar]
- 138.Chang B, Chen Y, Zhao Y, et al. JMJD6 Is a Histone Arginine Demethylase. Science. 2007;318(5849):444–7. [DOI] [PubMed] [Google Scholar]
- 139.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 140.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349. [DOI] [PubMed] [Google Scholar]
- 141.Shogren-Knaak M, Ishii H, Sun J-M, et al. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science. 2006;311(5762):844. [DOI] [PubMed] [Google Scholar]
- 142.Vaquero A, Scher M, Erdjument-Bromage H, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440. [DOI] [PubMed] [Google Scholar]
- 143.You Jueng S, Jones Peter A. Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell. 2012;22(1):9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Teif VB, Rippe K. Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Research. 2009;37(17):5641–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Berson A, Sartoris A, Nativio R, et al. TDP-43 Promotes Neurodegeneration by Impairing Chromatin Remodeling. Curr Biol. 2017;27(23):3579–90 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hansen JC. Conformational Dynamics of the Chromatin Fiber in Solution: Determinants, Mechanisms, and Functions. Annual Review of Biophysics and Biomolecular Structure. 2002;31(1):361–92. [DOI] [PubMed] [Google Scholar]
- 147.Yanling Zhao D, Gish G, Braunschweig U, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2015;529:48. [DOI] [PubMed] [Google Scholar]
- 148.Hirano M, Quinzii CM, Mitsumoto H, et al. Senataxin mutations and amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis. 2011;12(3):223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hill SJ, Mordes DA, Cameron LA, et al. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proceedings of the National Academy of Sciences. 2016;113(48):E7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kuo LJ, Yang L-X. γ-H2AX - A Novel Biomarker for DNA Double-strand Breaks. In Vivo. 2008;22(3):305–9. [PubMed] [Google Scholar]